Abstract
Background and Objectives
We previously found that the APOE genotype affects the rate of cognitive decline in mild-to-moderate Alzheimer disease (AD) dementia independently of its effects on AD neuropathologic changes (ADNC) and copathologies. In this study, we tested the hypothesis that the APOE alleles differentially affect the rate of cognitive decline at the normal aging-early AD continuum and that this association is independent of their effects on classical ADNC and copathologies.
Methods
We analyzed APOE associations with the cognitive trajectories (Clinical Dementia Rating scale Sum of Boxes [CDR-SOB] and Mini-Mental State Examination [MMSE]) of more than 1,000 individuals from a national clinicopathologic sample who had either no, mild (sparse neuritic plaques and the Braak neurofibrillary tangle [NFT] stage I/II), or intermediate (moderate neuritic plaques and the Braak NFT stage III/IV) ADNC levels at autopsy via 2 latent classes reverse-time longitudinal modeling.
Results
Carrying the APOEε4 allele was associated with a faster rate of cognitive decline by both CDR-SOB and MMSE relative to APOEε3 homozygotes. This association remained statistically significant after adjusting for ADNC severity, comorbid pathologies, and the effects of ADNC on the slope of cognitive decline. Our modeling strategy identified 2 latent classes in which APOEε4 carriers declined faster than APOEε3 homozygotes, with latent class 1 members representing slow decliners (CDR-SOB: 76.7% of individuals, 0.195 vs 0.146 points/y in APOEε4 vs APOEε3/ε3; MMSE: 88.6% of individuals, −0.303 vs −0.153 points/y in APOEε4 vs APOEε3/ε3), whereas latent class 2 members were fast decliners (CDR-SOB: 23.3% of participants, 1.536 vs 1.487 points/y in APOEε4 vs APOEε3/ε3; MMSE: 11.4% of participants, −2.538 vs −2.387 points/y in APOEε4 vs APOEε3/ε3). Compared with slow decliners, fast decliners were more likely to carry the APOEε4 allele, younger at initial visit and death, more impaired at initial and last visits, and more likely to have intermediate (vs none or mild) ADNC levels, as well as concurrent Lewy bodies and hippocampal sclerosis at autopsy.
Discussion
In a large national sample selected to represent the normal aging-early AD continuum, the APOEε4 allele is associated with a modest but statistically significant acceleration of the cognitive decline rate even after controlling for its effects on ADNC and comorbid pathologies.
Alzheimer disease (AD) is increasingly recognized as a heterogeneous clinicopathologic construct with frequently disparate syndromic presentations, rates of clinical progression, and/or severity and mixture of underlying pathologic findings. This heterogeneity is a major challenge for the success of clinical trials pursuing a disease-modifying effect and therefore represents a research area of the highest priority.1 Clinical and pathologic heterogeneity are linked because individuals with 1 or more comorbid pathologies in the brain have a faster progression of their cognitive decline.2 While the driver(s) of this heterogeneity remains largely unknown, both acquired environmental and genetic factors could conceivably cooperate and account for it. Among the latter, the APOE genotype is a primary suspect given its known effects on clinical and pathologic phenotypes. Regarding APOE clinical correlates, besides a differential impact on age at symptom onset, we and others3,4 have found that, in individuals with dementia due to AD, the APOEε4 allele accelerates and the APOEε2 allele slows down the rate of cognitive decline relative to the APOEε3 reference allele and that they do so independently of their effects on the presence and severity of AD neuropathologic changes (ADNC) and comorbid pathologies. Moreover, the APOEε4 allele has been associated with an amnestic presentation of AD,5-7 whereas the attention/dysexecutive,6-8 the primary progressive aphasia,9 and the posterior cortical atrophy10 presentations of AD appear to have a weaker association with the APOEε4 allele, suggesting an APOEε4-mediated selective vulnerability of temporolimbic networks. Regarding APOE pathologic correlates, it is well-established that the APOEε4 allele is associated with more severe Aβ plaque deposition and cerebral amyloid angiopathy (CAA), and there is growing evidence of similar APOE allele–specific independent effects on the extent of neurofibrillary tangles (NFTs)11 and the co-occurrence and severity of transactive response DNA-binding protein of 43 kDa (TDP-43) proteinopathy,12,13 Lewy body pathology,14-16 and cerebrovascular diseases.17,18
In this study, we tested the hypothesis that the APOE alleles differentially affect the rate of decline of both global cognition and specific cognitive domains (representing distinct neural networks) independently of their effects on ADNC and concurrent pathologies at the normal aging-early AD transition. To this end, we built reverse-time longitudinal models to analyze the cognitive trajectories of a national clinicopathologic sample with either no, mild, or intermediate ADNC levels at autopsy.
Methods
Standard Protocol Approvals, Registrations, and Participant Consents
As determined by the University of Washington Human Subjects Division, the National Alzheimer Coordinating Center (NACC) database itself is exempt from Institutional Review Board review and approval because it does not involve human subjects, as defined by federal and state regulations. However, all contributing AD Centers are required to obtain informed consent from their participants and maintain their own separate Institutional Review Board review and approval from their institution prior to submitting data to NACC.
Eligibility Criteria and Data Collection
The NACC study is a National Institute on Aging-funded longitudinal cohort study of cognitive aging conducted by more than 30 Alzheimer Disease Centers across the United States. Participants undergo baseline and annual follow-up visits in which a Uniform Data Set (UDS)19 is collected that includes demographics, medical history, medication list, and standardized motor, behavioral, functional, and neuropsychological questionnaires and tests.20,21 Participants are also offered to join fluid (plasma/serum and CSF) and imaging biomarkers (MRI and PET) studies and to donate their brain for diagnostic and research purposes following a standardized neuropathologic evaluation protocol.22 The NACC data set was interrogated between September 2005 and March 2021 data freezes. Inclusion criteria included the following: (1) autopsy available; (2) death within 2 years from the last visit; (3) age at death at 50 years or older; and (4) APOE genotype available. Exclusion criteria were as follows: (1) primary neuropathologic diagnosis other than ADNC; (2) APOE genotype not available or ε2/ε4 (due to a possible cancellation of their presumed opposing effects); (3) cognitive impairment due to medical illness, medication adverse side effects, or alcohol; and (4) CERAD neuritic plaque (NP) score frequent or the Braak NFT stage V/VI. The clinical constructs of “normal cognition,” “impaired not mild cognitive impairment,” “mild cognitive impairment,” and “dementia” were deliberately ignored as eligibility criteria to better understand the correlations between APOE genotype, semiquantitative measures of neuropathology, and cognitive trajectories. Data collected included age at first and last visit and death, sex, education (number of years), race and ethnicity, APOE genotype, scores from longitudinal neuropsychological evaluations, and autopsy neuropathologic variables.
Cognitive Outcome Measures
Cognitive outcome measures included the Clinical Dementia Rating scale Sum Of Boxes (CDR-SOB) score of the CDR Dementia Staging Instrument, the Mini-Mental State Examination (MMSE) score, and cognitive domain–specific z scores for memory, attention, executive functions, and language. The latter were derived from the individual neuropsychological tests of the UDS neuropsychological battery following the factor structure reported by Hayden et al.,14 as explained elsewhere.2 In brief, z scores for these tests were obtained for the mean and SD of the group of participants with intact cognition (i.e., CDR-SOB = 0) at first visit. The logical memory immediate and delayed recall were combined to obtain a memory z score; the digits forward and backward and their length to obtain an attention z score; the Trail Making Tests A and B and the Digit Symbol Test to obtain an executive z score; and the semantic verbal fluency (number of animals and vegetables in 1 minute) and the Boston Naming Test to obtain a language z score.
Statistical Analyses
To ascertain the cross-sectional neuropathologic correlates of APOE genotype in this autopsy cohort, we applied logistic regression models with the neuropathologic dependent variable categorized into 2 levels (the CERAD NP score moderate vs none/sparse; the Braak NFT stage III/IV vs 0/I/II; Lewy bodies present vs absent; hippocampal sclerosis [HScl] present vs absent), APOE genotype as an independent variable (i.e., the presence of the APOEε4 or APOEε2 alleles with APOEε3 homozygotes as reference group), and age, sex, and education as covariates. For each neuropathologic variable with multiple ordered categories (CAA mild vs none, moderate vs mild, and severe vs moderate; and arteriolosclerosis mild vs none, moderate vs mild, and severe vs moderate), we applied adjacent categories logit models with APOE genotype as an independent variable and the same covariates as in the logistic regression models, allowing potentially different effects for different adjacent categories.23 Models for the Braak NFT stage and CAA severity were also controlled for the CERAD NP score.
Next, to test whether the APOEε4 (APOEε3/ε4 and APOEε4/ε4) and APOEε2 (APOEε2/ε2 and APOEε2/ε3) participants significantly differed from the APOEε3 (APOEε3/ε3) reference group regarding the rate of cognitive decline (CDR-SOB, MMSE, and cognitive domain–specific composite measures) and whether these effects are independent of APOE allele associations with ADNC and comorbid pathologies, we applied a reverse-time longitudinal modeling strategy as described elsewhere.2,15 In brief, to link the cognitive trajectories to the neuropathologic autopsy findings, the last clinic visit within 2 years from death was treated as the first visit and cognitive score trajectories were modeled in the reverse time toward the first clinic visit, so that the neuropathologic autopsy variables could be appropriately treated as baseline covariates. This approach makes the assumptions that, in a given individual’s brain, NPs increase from none/sparse to moderate, whereas NFTs spread from the entorhinal cortex (the Braak NFT stage I/II) to limbic regions (the Braak NFT stage III/IV) in a sequential manner along the follow-up period; indeed, PET imaging studies support these assumptions.24,25 We did not include the Thal amyloid phases in these models because these do not affect antemortem cognition.26 Moreover, to adjust for the right truncation of cognitive trajectory by time to death, we built a joint latent class model27 for the longitudinal cognitive outcome (mixed-effects submodel) and the time-to-event analyses (i.e., last visit to first visit in reverse time, Cox proportional hazards submodel), with a logistic submodel for latent class membership. We used the Bayes Information Criterion to determine the optimal number of latent classes best supported by the data. Because different cognitive outcomes could potentially have different unobserved participant characteristics associated with them and with the APOE genotype, we allowed the latent classes to vary across longitudinal models, although we found a large membership overlap between latent classes from the CDR-SOB and MMSE models (733/866 [84.6%] after excluding individuals with missing values in covariates). We also implemented a right truncation adjustment for time to last visit by time to death to avoid potential bias due to the oversampling of shorter times to death. We evaluated 3 models in a stepwise fashion: model 1 was adjusted by sex, age, and education; model 2 was further adjusted by the CERAD NP score and Braak NFT stage; and model 3 was further adjusted by the effects of the CERAD NP score and Braak NFT stage on the slope of the cognitive outcome and by comorbid pathologies. The selection of comorbid pathologies for model 3 was based on univariate models in which 1 comorbid pathology variable was added at a time. Because the CERAD NP score frequent and Braak NFT stage V/VI were exclusion criteria, no ceiling or floor effects were detected in either CDR-SOB or MMSE scores or in any of the cognitive domain–specific z scores (not shown), therefore no change point was implemented.2
Data Availability
The NACC database is a publicly available resource available to researchers, and data requests can be submitted online at the following NACC website: nacc.redcap.rit.uw.edu/surveys/?s=KHNPKLJW8TKAD4DA.
Results
Sample Description
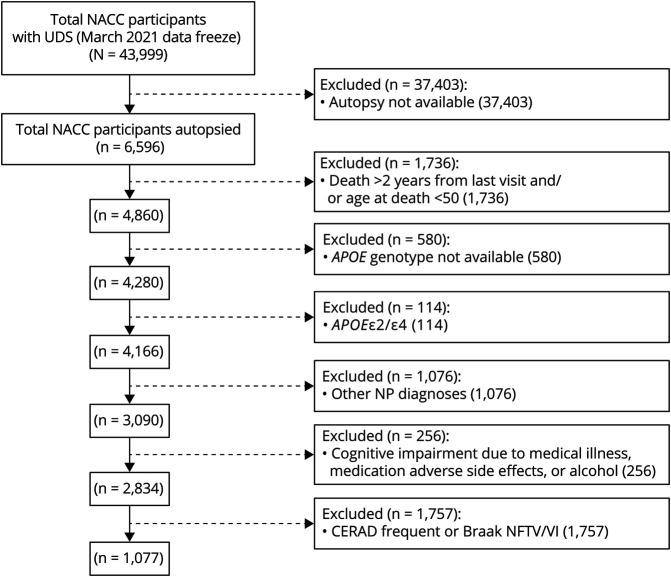
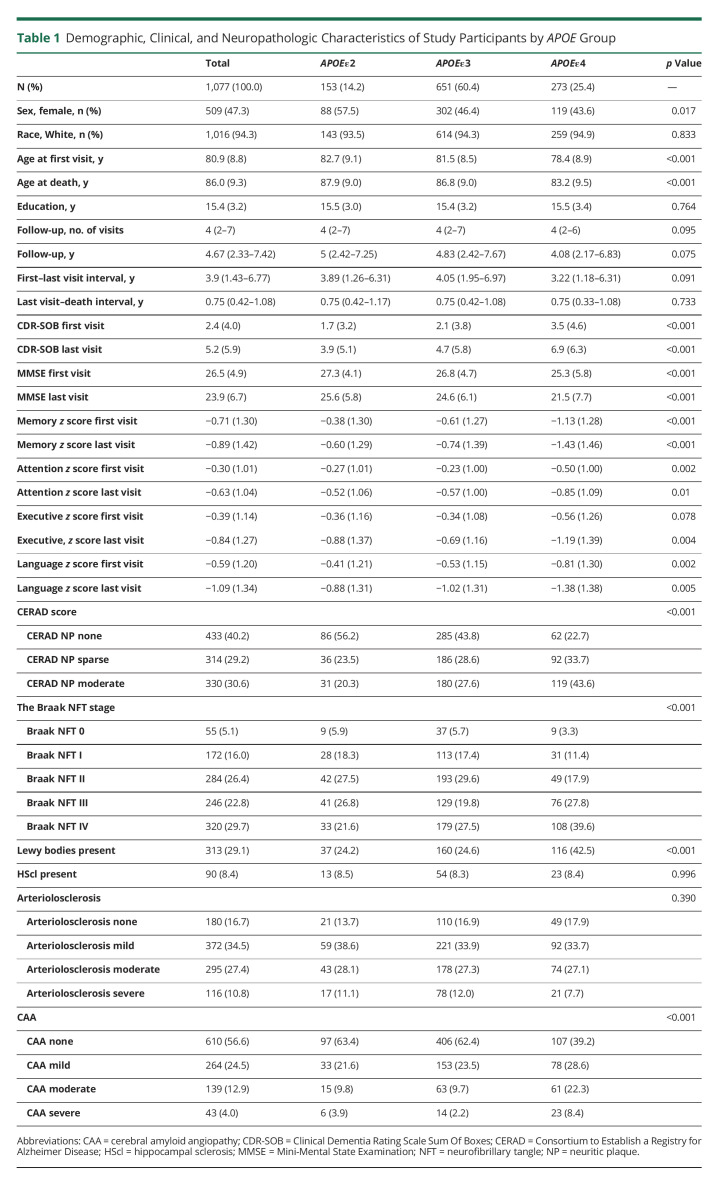
A total of 1,077 participants met all our eligibility inclusion criteria and none of the exclusion criteria (Figure 1), of whom 153 (14.2%) were APOEε2 carriers, 651 (60.4%) were APOEε3 homozygotes, and 273 (25.4%) were APOEε4 carriers. Demographic, clinical, and neuropathologic characteristics of study subjects split by APOE genotype are summarized in Table 1. APOEε4 carriers were more likely to be male, were younger, and had higher CDR-SOB and lower MMSE scores as well as lower memory-, attention-, executive-, and language-specific z scores than APOEε3 homozygotes at first and last visits, whereas opposite trends were observed for APOEε2 carriers. There were no statistically significant differences in education, follow-up duration, or lapse between last visit and death/autopsy. All downstream statistical analyses were adjusted by age at death, sex, and education.
Figure 1. Flowchart of Study Participants.
CDR-SOB = Clinical Dementia Rating scale Sum Of Boxes; CERAD = Consortium to Establish a Registry for Alzheimer Disease; MMSE = Mini-Mental State Examination; NFT = neurofibrillary tangle; NP = neuritic plaque.
Table 1.
Demographic, Clinical, and Neuropathologic Characteristics of Study Participants by APOE Group
Neuropathologic Correlates of the APOE Genotype in This Autopsy Sample
eTable 1, links.lww.com/NXG/A574, summarizes the associations between ADNC and comorbid pathologies and APOE genotype in this sample. Relative to APOEε3 homozygotes, carrying the APOEε4 allele was associated with a higher CERAD NP score (moderate vs none/sparse) and Braak NFT stage (III/IV vs 0/I/II), a higher severity of CAA, and the presence of Lewy bodies, but not with the presence of HScl or the severity of arteriolosclerosis. A dose-dependent effect was observed whereby APOEε4 homozygotes were more likely to have a higher CERAD NP score, Braak NFT stage, and CAA severity than APOEε3 homozygotes compared with APOEε3/ε4 participants; however, only APOEε3/ε4 carriers were more likely to have Lewy bodies than APOEε3 homozygotes.
APOEε4 Is Independently Associated With a Faster Cognitive Decline in the Normal Aging-Early AD Continuum
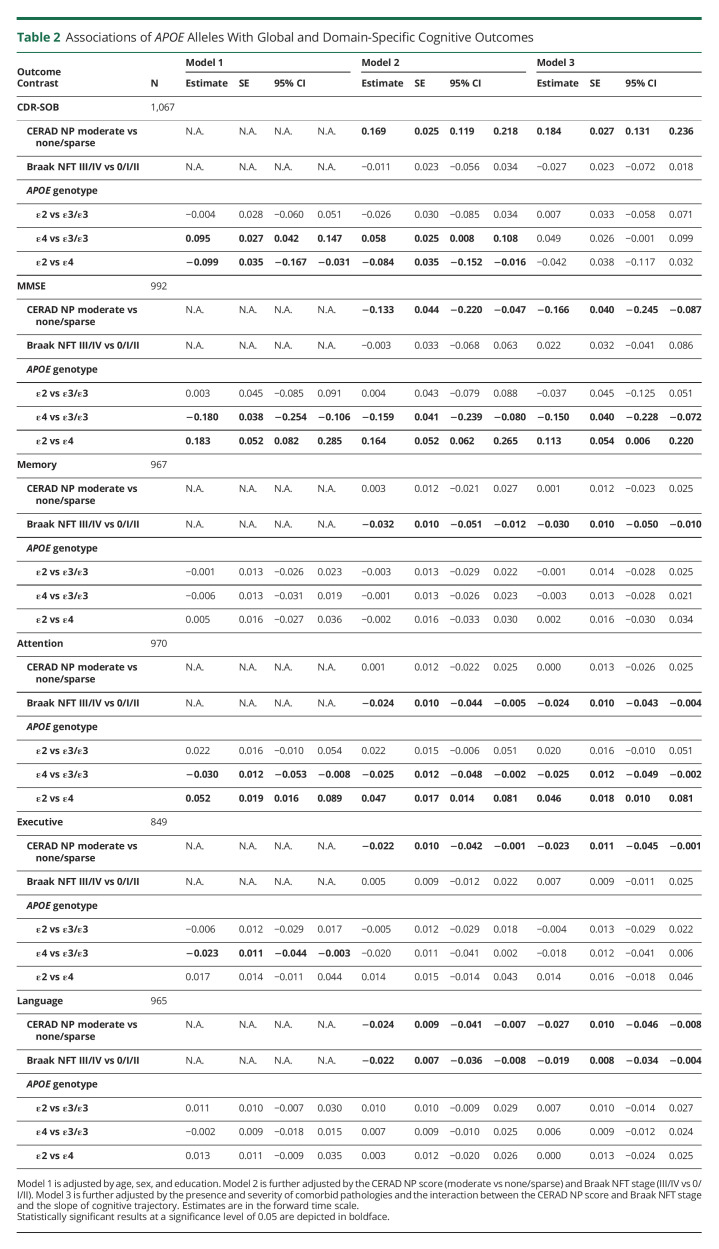
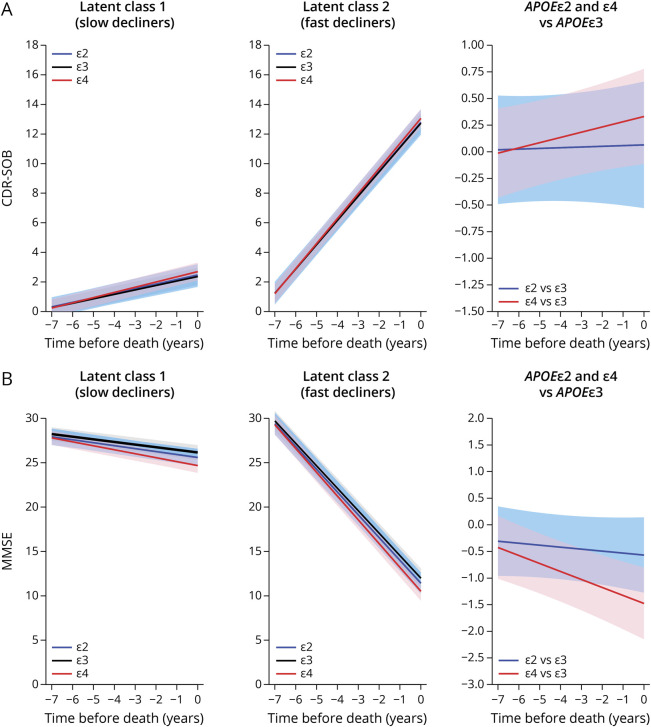
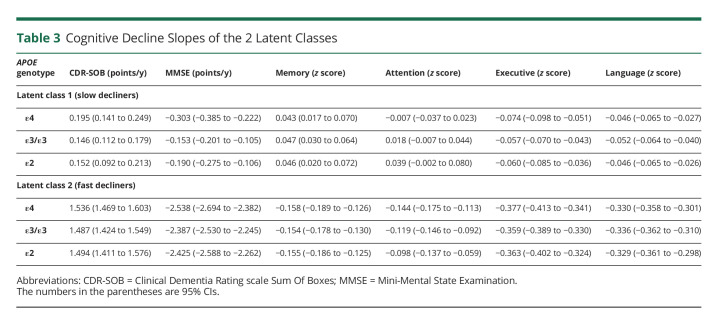
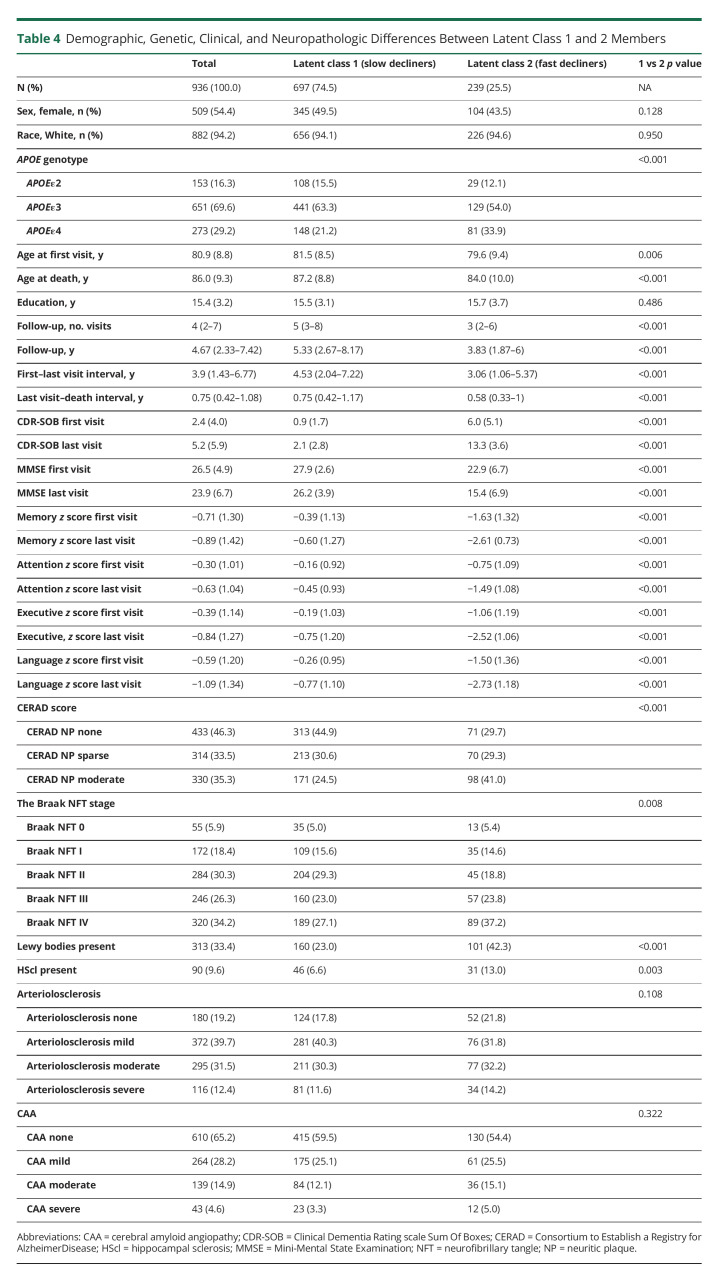
To determine any independent association between the APOE alleles and the rate of cognitive decline, we applied reverse-time longitudinal models, which link the individuals' autopsy findings with their CDR-SOB and MMSE trajectories in reverse time scale by considering the last visit proximate (≤2 years) to death/autopsy as baseline. These models revealed that carrying the APOEε4 allele is associated with a faster rate of cognitive decline by both CDR-SOB and MMSE relative to APOEε3 homozygotes. This association remained statistically significant whether adjusting only for age, sex, and education (model 1); further adjusting for the severity of ADNC (i.e., CERAD NP score and Braak NFT stage) (model 2); or additionally adjusting for comorbid pathologies and the effects of ADNC on the slope (model 3) (Table 2 and Figure 2). By contrast, no significant difference was observed between APOEε2 carriers and APOEε3 homozygotes in any of the models. Noteworthy, our modeling strategy identified 2 latent classes in which the differences between APOEε4 carriers and APOEε3 homozygotes were statistically significant. The slope of these 2 latent classes differed markedly, with latent class 1 members representing slow decliners (CDR-SOB: 76.7% of individuals, 0.195 vs 0.146 points/y in APOEε4 vs APOEε3/ε3; MMSE: 88.6% of individuals, −0.303 vs −0.153 points/y in APOEε4 vs APOEε3/ε3), whereas latent class 2 members were fast decliners (CDR-SOB: 23.3% of individuals, 1.536 vs 1.487 points/y in APOEε4 vs APOEε3/ε3; MMSE: 11.4% of individuals, −2.538 vs −2.387 points/y in APOEε4 vs APOEε3/ε3) (Table 3). Compared with slow decliners, fast decliners were not only more likely to carry the APOEε4 allele but also younger and more impaired at initial and last visits and more likely to have intermediate (vs none or mild) ADNC levels as well as concurrent Lewy bodies and HScl at autopsy (Table 4).
Table 2.
Associations of APOE Alleles With Global and Domain-Specific Cognitive Outcomes
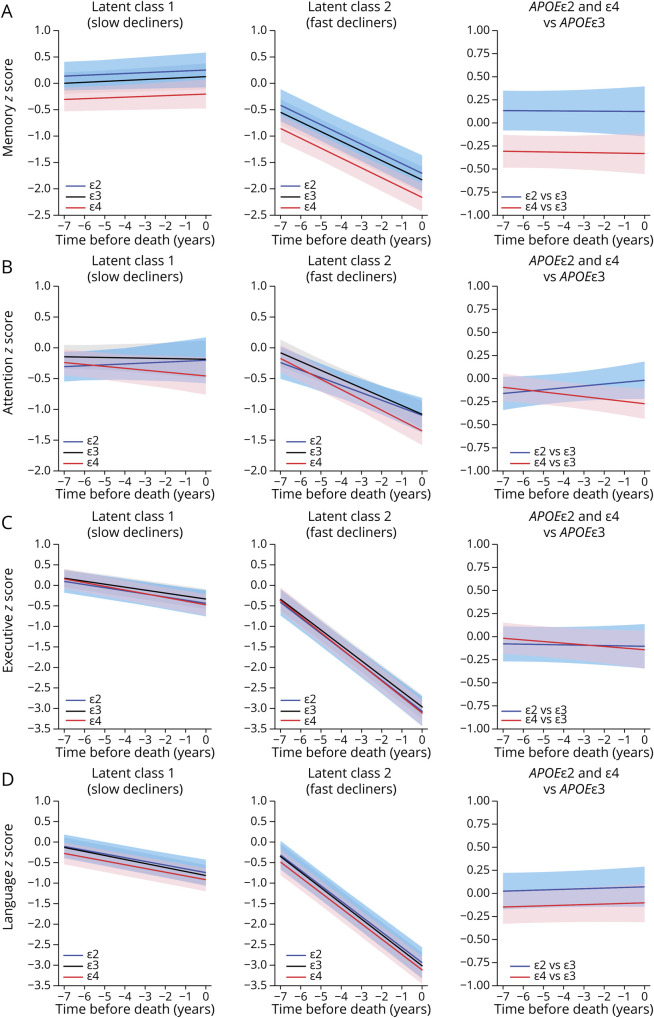
Figure 2. Effects of APOE Genotype on CDR-SOB and MMSE Trajectories.
Model 3–based trajectories for CDR-SOB (A) and MMSE (B) scores with the intercept at the time of death calculated for an 86-year-old woman with 15 years of education and autopsy findings of CERAD NP score moderate and Braak NFT stage III/IV. Graphs depict the 3 APOE groups in latent class 1 (slow decliners) and latent class 2 (fast decliners) as well as the differences APOEε4 vs APOEε3 and APOEε2 vs APOEε3. Shaded areas correspond to the 95% CIs. CDR-SOB = Clinical Dementia Rating scale Sum Of Boxes; CERAD = Consortium to Establish a Registry for Alzheimer Disease; MMSE = Mini-Mental State Examination; NFT = neurofibrillary tangle; NP = neuritic plaque.
Table 3.
Cognitive Decline Slopes of the 2 Latent Classes
Table 4.
Demographic, Genetic, Clinical, and Neuropathologic Differences Between Latent Class 1 and 2 Members
APOEε4 Is Independently Associated With a Faster Decline in Attention and Executive Function, but Not Memory or Language
Next, to investigate possible differential associations between APOE alleles and specific cognitive domains corresponding to different brain networks, we applied similar models with memory-, attention-, executive-, and language-specific composites as cognitive outcome measures (Table 2 and Figure 3). Unexpectedly, APOEε4 carriers did not significantly differ from APOEε3 homozygotes in the trajectory of the memory domain in any of the models. Of note, APOEε4 carriers had poorer memory z scores than APOEε2 carriers and APOEε3 homozygotes at both initial and last visits (Table 1 and Figure 2), suggesting a potential floor effect; however, 2 subsequent sensitivity analyses restricting the sample to individuals with a CDR-SOB score of ≤0.5 either at their first visit or at their last visit (within 2 years from death) did not change these results. Because of our prespecified CERAD NP score frequent and Braak NFT stage V/VI exclusion criteria, only 29 of the 273 APOEε4 carriers in this sample were APOEε4 homozygotes, therefore precluding a conclusive APOEε4 allele dose-response analysis.
Figure 3. Effects of APOE Genotype on Cognitive Domain–Specific Trajectories.
Model 3–based trajectories for memory- (A), attention- (B), executive- (C), and language-specific (D) composite z scores with the intercept at the time of death calculated for an 86-year-old woman with 15 years of education and autopsy findings of CERAD NP score moderate and Braak NFT stage III/IV. Graphs depict the 3 APOE groups in latent class 1 (slow decliners) and latent class 2 (fast decliners) as well as the differences APOEε4 vs APOEε3 and APOEε2 vs APOEε3. Shaded areas correspond to the 95% CIs. CERAD = Consortium to Establish a Registry for Alzheimer Disease; NFT = neurofibrillary tangle; NP = neuritic plaque.
Similarly, no differences were found in the language domain in any of the models. By contrast, APOEε4 carriers declined significantly faster than APOEε3 homozygotes in the attention domain, but this difference reached statistical significance only in latent class 2 (fast decliners). Last, the executive domain declined significantly faster in APOEε4 carriers than APOEε3 homozygotes when adjusting for age, sex, and education (model 1, p = 0.028); however, this difference was only marginally significant when further adjusting by the CERAD NP score and Braak NFT stage (model 2, p = 0.077, 95% CI [−0.041 to 0.002]) and lost statistical significance when further adjusting by comorbid pathologies and the effect of ADNC on the slope (model 3, p = 0.135, 95% CI [−0.041 to 0.006]) likely due to insufficient power and the compounding effect of comorbid pathologies on executive function. Indeed, model 3 revealed that Lewy bodies are independently associated with a faster decline in all 4 cognitive domains; moderate and severe (vs none) arteriolosclerosis with faster executive dysfunction; HScl with faster memory decline; and severe (vs none) CAA with faster language impairment. Model 3 also revealed seemingly neuropathology-specific associations between ADNC and cognitive domains, with the Braak NFT stage (III/IV vs 0/I/II) independently correlating with a faster decline in memory and attention, whereas the CERAD NP score (moderate vs none/sparse) was independently correlated with a faster executive dysfunction, and both ADNC were associated with a faster decline in language.
Consistent with the aforementioned results relative to CDR-SOB and MMSE trajectories, examination of the cognitive domain–specific composite outcome measures revealed no significant differences in the trajectory of any of these cognitive domains between APOEε2 carriers and APOEε3 homozygotes in any of these models (Table 2). Last, we performed a sensitivity analysis removing the 29 APOEε4 homozygotes to determine whether they were driving the APOEε4 vs APOEε3 differences described earlier, and the results remained largely unchanged (eTable 2, links.lww.com/NXG/A575); although some of the contrasts lost statistical significance, this was due to the loss of statistical power, as indicated by their 95% CI boundary close to zero.
Discussion
While it is well-established that the APOE genotype dramatically affects the risk of developing AD and the conversion rate from mild cognitive impairment to dementia, these effects are generally attributed to its effects on the accrual of ADNC, especially Aβ plaques. In this study, we investigated the independent contribution of the APOE genotype to the heterogeneity of the rate of clinical progression at the earliest stages of the normal aging-AD continuum in a national clinicopathologic sample with no, mild, or moderate ADNC at autopsy. We found that the APOEε4 allele is associated with an acceleration of the rate of global (CDR-SOB and MMSE) and attention function decline independently of its effects on ADNC and comorbid pathologies.
This longitudinal study extends our similar previous analysis of an NACC sample selected to represent the other end of the AD spectrum, i.e., individuals who end up having moderate or frequent NPs and the Braak NFT stage III/IV or V/VI at autopsy examination.4 By focusing here on the normal aging-early AD continuum (i.e., excluding participants with frequent NPs and Braak NFT stage V/VI), we circumvented the ceiling and floor effects typical of these cognitive measures in advanced AD dementia and, consequently, the need for a change point in our longitudinal models. While both analyses concluded that, relative to APOEε3 homozygosis, carrying the APOEε4 allele is associated with a statistically significant faster cognitive decline independently of ADNC and comorbid pathologies, the differences in this normal aging-early AD sample are quantitatively smaller and clinically less relevant compared with those found in the autopsy sample with moderate/high ADNC4 (0.04 CDR-SOB points/y and −0.15 MMSE points/y here vs 0.68 CDR-SOB points/y and −0.42 MMSE points/y in our prior study). These observations support a nonlinear association of APOE genotype with the rate of cognitive decline along the course of ADNC accrual and the use of nonlinear28 or segmented linear29 models to investigate this relationship. Of note, the 2-latent class modeling enabled us to recognize 2 subgroups of individuals with distinct slopes of clinical progression, with fast decliners being more likely to be APOEε4 carriers, younger at first visit and death, more impaired at first and last visits, and more likely to have intermediate ADNC (the Braak NFT stage III/IV and moderate NPs), Lewy bodies, and HScl at autopsy, but not more severe vascular pathologies (CAA or arteriolosclerosis). We speculate that this variance in the rate of progression may be explained by other factors, both intrinsic (i.e., polygenic risk score,30 microglia immune responses,31 and the interaction between diverse tau species and genetic background32,33) and modifiable (i.e., cognitive reserve, environmental exposures, and vascular) risk factors.34
It has been reported that the APOEε4 allele favors an amnestic over executive/attentional, aphasic, or visuospatial/perceptive presentations of AD, suggesting that APOEε4 confers a selective vulnerability of the temporolimbic network to ADNC.5-10,35,36 Others have shown that the APOEε4 allele has some beneficial effects on a visual working memory task even in the presence of Aβ plaques.37 Although APOEε4 carriers had worse memory z scores at first and last visits than APOEε3 homozygotes and APOEε2 carriers, we did not observe a significant association between the APOEε4 allele and the rate of memory decline in this sample. Our sensitivity analyses argue against the possibility that a floor effect in the memory composite underlies this lack of association. The small number of APOEε4 homozygotes (n = 29) precluded a meaningful allele dose-response analysis. The language trajectory did not differ between APOEε4 carriers and APOEε3 homozygotes, whereas the association between the APOEε4 allele and faster executive dysfunction was not independent of APOEε4 effects on ADNC and comorbid pathologies. Conversely, we did observe a selective association between the APOEε4 allele and a faster rate of decline in the attention domain (as assessed with the digits forward and backward test), which was independent of ADNC and comorbid pathologies. The neuropathology-independent nature of this association raises the question whether the APOEε4 allele may have a deleterious effect on attention already in mid-adulthood. However, prior longitudinal studies investigating APOE allele associations with performance in attentional/executive processes in middle-age adults have yielded conflicting results, with some reporting a disadvantage of APOEε4 carriers38-40 and others, no difference41 or even an advantage42,43 (reviewed in reference 44).
Unlike our prior analysis of a NACC sample with moderate/high ADNC, in this normal aging-early AD sample, we did not detect a statistically significant protective effect of the APOEε2 allele over the APOEε3 allele for any of the cognitive outcome measures, whereas the APOEε2 vs APOEε4 contrast did yield a protective effect of APOEε2 based on MMSE, CDR-SOB (except in model 3), and attention domain trajectories. Only 7 of the 153 APOEε2 participants in our sample were homozygotes—a rare genotype associated with an extremely low likelihood of developing AD45 and a slower rate of cognitive decline46—precluding any dose-response analysis.
The findings reported in this study have potentially important pathophysiologic implications. Applying the resistance vs resilience framework47 to this study and our prior analyses of the NACC autopsy cohort,4,11,23 it appears that the APOEε4 allele not only promotes ADNC (including CAA) but also lowers the brain ability to cope with them (i.e., resilience), with an effect size augmenting as ADNC severity increases. Conversely, the APOEε2 allele clearly provides resistance against the accumulation of ADNC11,45 but would only confer detectable resilience against cognitive impairment as substantial ADNC (i.e., moderate and frequent NPs and the Braak NFT stage ≥ III) accumulate in the brain.4,11 Furthermore, the demonstration of APOEε4–linked neuropathology-independent effects on cognitive decline raises the question of what the unmeasured mediator(s) of such deleterious effects might be. Microglial and blood-brain barrier dysfunction are primary suspects because recent studies have reported that APOEε4 exacerbates microglial prophagocytic and proinflammatory transcriptional programs48-50 and disrupts the blood-brain barrier integrity and function.18,51 But neuronal and synaptic mechanisms have also been proposed, and a multihit scenario is plausible.52 Additionally, these results have relevant implications for the design and interpretation of preventative and therapeutic clinical trials targeting early AD. Specifically, they are in agreement with simulation trials indicating that a strategy of enrichment in APOEε4 carriers may enhance the power to detect significant effect differences between the treatment and placebo groups within the duration of the trial, provided that the intervention tested has an equal response in APOEε4 carriers and noncarriers53,54 (but see also references 55 and 56). Moreover, they reinforce the goal of developing APOEε4–directed therapies regardless of their downstream effects on ADNC levels.57
Some limitations of our methodology should be acknowledged. First, the ethnic/racial composition of this sample limits the generalizability of our findings to other racial/ethnic groups. The APOEε4 allele is known to have a weaker association with AD risk in Blacks and Hispanics,57 but its impact on rate of cognitive decline needs to be investigated. Second, we could not perform an ε4 allele dose-response analysis because of the low number of APOEε4 homozygotes. Third, we did not include interaction terms such as the interaction between APOE genotype and comorbid pathologies and between these and the slope of cognitive decline to avoid overfitting of the models. Fourth, we could not evaluate the impact of APOE alleles on the visuospatial domain because the appropriate tests for this domain (e.g., Benson figure copy) were only available in the version 3 of the UDS (data freeze March 2014).21 Last, TDP-43 data were not available for a large proportion of subjects because this was incorporated to the NACC neuropathology protocol in 201422; therefore, HScl was used instead as an imperfect proxy of TDP-43 proteinopathy.58
In summary, in a sample selected to represent the normal aging-early AD continuum, the APOEε4 allele is associated with a modest but statistically significant acceleration of the cognitive decline rate even after controlling for its effects on ADNC and comorbid pathologies. Further studies to identify the mechanism(s) of these differential effects of APOE alleles on cognition in the normal aging-AD continuum are warranted.
Acknowledgment
The NACC database is funded by the NIA/NIH Grant U24 AG072122. NACC data are contributed by the NIA-funded ADRCs: P30 AG062429 (PI James Brewer, MD, PhD), P30 AG066468 (PI Oscar Lopez, MD), P30 AG062421 (PI Bradley Hyman, MD, PhD), P30 AG066509 (PI Thomas Grabowski, MD), P30 AG066514 (PI Mary Sano, PhD), P30 AG066530 (PI Helena Chui, MD), P30 AG066507 (PI Marilyn Albert, PhD), P30 AG066444 (PI John Morris, MD), P30 AG066518 (PI Jeffrey Kaye, MD), P30 AG066512 (PI Thomas Wisniewski, MD), P30 AG066462 (PI Scott Small, MD), P30 AG072979 (PI David Wolk, MD), P30 AG072972 (PI Charles DeCarli, MD), P30 AG072976 (PI Andrew Saykin, PsyD), P30 AG072975 (PI David Bennett, MD), P30 AG072978 (PI Neil Kowall, MD), P30 AG072977 (PI Robert Vassar, PhD), P30 AG066519 (PI Frank LaFerla, PhD), P30 AG062677 (PI Ronald Petersen, MD, PhD), P30 AG079280 (PI Eric Reiman, MD), P30 AG062422 (PI Gil Rabinovici, MD), P30 AG066511 (PI Allan Levey, MD, PhD), P30 AG072946 (PI Linda Van Eldik, PhD), P30 AG062715 (PI Sanjay Asthana, MD, FRCP), P30 AG072973 (PI Russell Swerdlow, MD), P30 AG066506 (PI Todd Golde, MD, PhD), P30 AG066508 (PI Stephen Strittmatter, MD, PhD), P30 AG066515 (PI Victor Henderson, MD, MS), P30 AG072947 (PI Suzanne Craft, PhD), P30 AG072931 (PI Henry Paulson, MD, PhD), P30 AG066546 (PI Sudha Seshadri, MD), P20 AG068024 (PI Erik Roberson, MD, PhD), P20 AG068053 (PI Justin Miller, PhD), P20 AG068077 (PI Gary Rosenberg, MD), P20 AG068082 (PI Angela Jefferson, PhD), P30 AG072958 (PI Heather Whitson, MD), and P30 AG072959 (PI James Leverenz, MD).
Glossary
- AD
Alzheimer disease
- ADNC
AD neuropathologic changes
- CAA
cerebral amyloid angiopathy
- CDR-SOB
Clinical Dementia Rating scale Sum Of Boxes
- HScl
hippocampal sclerosis
- MMSE
Mini-Mental State Examination
- NACC
National Alzheimer Coordinating Center
- NFTs
neurofibrillary tangles
- NP
neuritic plaque
- TDP-43
transactive response DNA-binding protein of 43 kDa
- UDS
Uniform Data Set
Appendix. Authors
Study Funding
This study was supported by the NIH (NIA K08AG064039 to A. Serrano-Pozo, NINDS R01NS094610 to J. Qian and R.A. Betensky, the Massachusetts Alzheimer's Disease Research Center NIA P30 AG062421 to A. Serrano-Pozo and B.T. Hyman, and the New York University Alzheimer's Disease Research Center NIA P30 AG066512 to R.A. Betensky) and The Jack Satter Foundation, Inc. (A. Serrano-Pozo).
Disclosure
J. Qian reports no disclosures; Y. Zhang currently works for Sanofi Genzyme; R.A. Betensky serves on the data safety monitoring boards of Biogen, Reata, and PTC Therapeutics, serves as a consultant for Cowen Inc., Pfizer, and Amgen, and has served as an expert witness for Amarin, Actavis, Amazon, Teva, Padagis, and Apotex; B.T. Hyman has a family member who works at Novartis and owns stock in Novartis, serves on the scientific advisory board of Dewpoint and owns stock, serves on a scientific advisory board or is a consultant for Abbvie, Arvinas, Biogen, Novartis, Cell Signaling Technologies, Sangamo, Sanofi, Takeda, US Department of Justice, and Vigil, and his laboratory is supported by sponsored research agreements with Abbvie, F Prime, and Spark as well as research grants from the NIH (PI), the Cure Alzheimer's Fund (PI), the Tau Consortium (PI), The JPB Foundation (PI), the Alzheimer's Association (mentor), and BrightFocus (mentor); A. Serrano-Pozo's laboratory is funded by the NIH/NIA, the Alzheimer's Association, The Jack Satter Foundation Inc., and The Karen Toffler Charitable Trust, and he has a material transfer agreement with Ionis Pharmaceuticals, Inc. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NG.
References
- 1.Veitch DP, Weiner MW, Aisen PS, et al. Understanding disease progression and improving Alzheimer's disease clinical trials: recent highlights from the Alzheimer's Disease Neuroimaging Initiative. Alzheimers Dement. 2019;15(1):106-152. doi: 10.1016/j.jalz.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Boyle PA, Wang T, Yu L, et al. To what degree is late life cognitive decline driven by age-related neuropathologies? Brain. 2021;144(7):2166-2175. doi: 10.1093/brain/awab092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu L, Boyle P, Schneider JA, et al. APOE ε4, Alzheimer's disease pathology, cerebrovascular disease, and cognitive change over the years prior to death. Psychol Aging. 2013;28(4):1015-1023. doi: 10.1037/a0031642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qian J, Betensky RA, Hyman BT, Serrano-Pozo A. Association of APOE genotype with heterogeneity of cognitive decline rate in Alzheimer disease. Neurology. 2021;96(19):e2414-e2428. doi: 10.1212/wnl.0000000000011883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Flier WM, Schoonenboom SNM, Pijnenburg YaL, Fox NC, Scheltens P. The effect of APOE genotype on clinical phenotype in Alzheimer disease. Neurology. 2006;67(3):526-527. doi: 10.1212/01.wnl.0000228222.17111.2a. [DOI] [PubMed] [Google Scholar]
- 6.Wolk DA, Dickerson BC, Alzheimer's Disease Neuroimaging Initiative. Apolipoprotein E (APOE) genotype has dissociable effects on memory and attentional-executive network function in Alzheimer's disease. Proc Natl Acad Sci USA. 2010;107(22):10256-10261. doi: 10.1073/pnas.1001412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickerson BC, Wolk DA, the Alzheimer's Disease Neuroimaging Initiative. Dysexecutive versus amnesic phenotypes of very mild Alzheimer's disease are associated with distinct clinical, genetic and cortical thinning characteristics. J Neurol Neurosurg Psychiatry. 2011;82(1):45-51. doi: 10.1136/jnnp.2009.199505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mez J, Cosentino S, Brickman AM, Huey ED, Manly JJ, Mayeux R. Dysexecutive versus amnestic Alzheimer disease subgroups: analysis of demographic, genetic, and vascular factors. Alzheimer Dis Assoc Disord. 2013;27(3):218-225. doi: 10.1097/wad.0b013e31826a94bd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weintraub S, Teylan M, Rader B, et al. APOE is a correlate of phenotypic heterogeneity in Alzheimer disease in a national cohort. Neurology. 2020;94(6):e607-e612. doi: 10.1212/wnl.0000000000008666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schott JM, Crutch SJ, Carrasquillo MM, et al. Genetic risk factors for the posterior cortical atrophy variant of Alzheimer's disease. Alzheimers Dement. 2016;12(8):862-871. doi: 10.1016/j.jalz.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serrano-Pozo A, Qian J, Monsell SE, Betensky RA, Hyman BT. APOEε2 is associated with milder clinical and pathological Alzheimer disease. Ann Neurol. 2015;77(6):917-929. doi: 10.1002/ana.24369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wennberg AM, Tosakulwong N, Lesnick TG, et al. Association of apolipoprotein E ε4 with transactive response DNA-binding protein 43. JAMA Neurol. 2018;75(11):1347-1354. doi: 10.1001/jamaneurol.2018.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H-S, Yu L, White CC, et al. Evaluation of TDP-43 proteinopathy and hippocampal sclerosis in relation to APOE ε4 haplotype status: a community-based cohort study. Lancet Neurol. 2018;17(9):773-781. doi: 10.1016/s1474-4422(18)30251-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickson DW, Heckman MG, Murray ME, et al. APOE ε4 is associated with severity of Lewy body pathology independent of Alzheimer pathology. Neurology. 2018;91(12):e1182-e1195. doi: 10.1212/wnl.0000000000006212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis AA, Inman CE, Wargel ZM, et al. APOE genotype regulates pathology and disease progression in synucleinopathy. Sci Transl Med. 2020;12(529):eaay3069. doi: 10.1126/scitranslmed.aay3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Y, Li F, Sonoustoun B, et al. APOE4 exacerbates α-synuclein seeding activity and contributes to neurotoxicity in Alzheimer's disease with Lewy body pathology. Acta Neuropathol. 2022;143(6):641-662. doi: 10.1007/s00401-022-02421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamar M, Yu L, Rubin LH, et al. APOE genotypes as a risk factor for age-dependent accumulation of cerebrovascular disease in older adults. Alzheimers Dement. 2019;15(2):258-266. doi: 10.1016/j.jalz.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montagne A, Nation DA, Sagare AP, et al. APOE4 leads to blood-brain barrier dysfunction predicting cognitive decline. Nature. 2020;581(7806):71-76. doi: 10.1038/s41586-020-2247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beekly DL, Ramos EM, Lee WW, et al. The national Alzheimer's coordinating center (NACC) database: the Uniform data Set. Alzheimer Dis Assoc Disord. 2007;21(3):249-258. doi: 10.1097/wad.0b013e318142774e. [DOI] [PubMed] [Google Scholar]
- 20.Weintraub S, Salmon D, Mercaldo N, et al. The Alzheimer's disease Centers' Uniform data Set (UDS): the neuropsychologic test battery. Alzheimer Dis Assoc Disord. 2009;23(2):91-101. doi: 10.1097/wad.0b013e318191c7dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weintraub S, Besser L, Dodge HH, et al. Version 3 of the Alzheimer Disease Centers' neuropsychological test battery in the uniform data Set (UDS). Alzheimer Dis Assoc Disord. 2018;32(1):10-17. doi: 10.1097/wad.0000000000000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Besser LM, Kukull WA, Teylan MA, et al. The revised national Alzheimer's coordinating center's neuropathology form-available data and new analyses. J Neuropathol Exp Neurol. 2018;77(8):717-726. doi: 10.1093/jnen/nly049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serrano-Pozo A, Qian J, Monsell SE, Frosch MP, Betensky RA, Hyman BT. Examination of the clinicopathologic continuum of Alzheimer disease in the autopsy cohort of the national Alzheimer coordinating center. J Neuropathol Exp Neurol. 2013;72(12):1182-1192. doi: 10.1097/nen.0000000000000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen WJ, Janssen O, Tijms BM, et al. Prevalence estimates of amyloid abnormality across the Alzheimer disease clinical spectrum. JAMA Neurol. 2022;79(3):228-243. doi: 10.1001/jamaneurol.2021.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleisher AS, Pontecorvo MJ, Devous MD, et al. Positron emission tomography imaging with [18F]flortaucipir and postmortem assessment of Alzheimer disease neuropathologic changes. JAMA Neurol. 2020;77(7):829-839. doi: 10.1001/jamaneurol.2020.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serrano-Pozo A, Qian J, Muzikansky A, et al. Thal amyloid stages do not significantly impact the correlation between neuropathological change and cognition in the Alzheimer disease continuum. J Neuropathol Exp Neurol. 2016;75(6):516-526. doi: 10.1093/jnen/nlw026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Proust-Lima C, Séne M, Taylor JMG, Jacqmin-Gadda H. Joint latent class models for longitudinal and time-to-event data: a review. Stat Methods Med Res. 2014;23(1):74-90. doi: 10.1177/0962280212445839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martins CaR, Oulhaj A, de Jager CA, Williams JH. APOE alleles predict the rate of cognitive decline in Alzheimer disease: a nonlinear model. Neurology. 2005;65(12):1888-1893. doi: 10.1212/01.wnl.0000188871.74093.12. [DOI] [PubMed] [Google Scholar]
- 29.Chen XR, Shao Y, Sadowski MJ, for the Alzheimer's Disease Neuroimaging Initiative. Segmented linear mixed model analysis reveals association of the APOEɛ4 allele with faster rate of Alzheimer's disease dementia progression. J Alzheimers Dis. 2021;82(3):921-937. doi: 10.3233/jad-210434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sims R, Hill M, Williams J. The multiplex model of the genetics of Alzheimer's disease. Nat Neurosci. 2020;23(3):311-322. doi: 10.1038/s41593-020-0599-5. [DOI] [PubMed] [Google Scholar]
- 31.Boche D, Gordon MN. Diversity of transcriptomic microglial phenotypes in aging and Alzheimer's disease. Alzheimers Dement. 2022;18(2):360-376. doi: 10.1002/alz.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dujardin S, Commins C, Lathuiliere A, et al. Tau molecular diversity contributes to clinical heterogeneity in Alzheimer's disease. Nat Med. 2020;26(8):1256-1263. doi: 10.1038/s41591-020-0938-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dujardin S, Fernandes A, Bannon R, et al. Tau propagation is dependent on the genetic background of mouse strains. Brain Commun. 2022;4(2):fcac048. doi: 10.1093/braincomms/fcac048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serrano-Pozo A, Growdon JH. Is Alzheimer's disease risk modifiable?. J Alzheimers Dis. 2019;67(3):795-819. doi: 10.3233/jad181028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emrani S, Arain HA, DeMarshall C, Nuriel T. APOE4 is associated with cognitive and pathological heterogeneity in patients with Alzheimer's disease: a systematic review. Alzheimers Res Ther. 2020;12(1):141. doi: 10.1186/s13195-020-00712-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frisoni GB, Altomare D, Thal DR, et al. The probabilistic model of Alzheimer disease: the amyloid hypothesis revised. Nat Rev Neurosci. 2022;23(1):53-66. doi: 10.1038/s41583-021-00533-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu K, Nicholas JM, Pertzov Y, et al. Dissociable effects of APOE-ε4 and β-amyloid pathology on visual working memory. Nat Aging. 2021;1(11):1002-1009. doi: 10.1038/s43587-021-00117-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenwood PM, Lambert C, Sunderland T, Parasuraman R. Effects of apolipoprotein E genotype on spatial attention, working memory, and their interaction in healthy, middle-aged adults: results from the National Institute of Mental Health's BIOCARD study. Neuropsychology. 2005;19(2):199-211. doi: 10.1037/0894-4105.19.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lancaster C, Tabet N, Rusted J. The APOE paradox: do attentional control differences in mid-adulthood reflect risk of late-life cognitive decline. Neurobiol Aging. 2016;48:114-121. doi: 10.1016/j.neurobiolaging.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 40.Zimmermann J, Alain C, Butler C. Impaired memory-guided attention in asymptomatic APOE4 carriers. Sci Rep. 2019;9(1):8138. doi: 10.1038/s41598-019-44471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lancaster C, Forster S, Tabet N, Rusted J. Putting attention in the spotlight: the influence of APOE genotype on visual search in mid adulthood. Behav Brain Res. 2017;334:97-104. doi: 10.1016/j.bbr.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 42.Evans S, Dowell NG, Tabet N, Tofts PS, King SL, Rusted JM. Cognitive and neural signatures of the APOE E4 allele in mid-aged adults. Neurobiol Aging. 2014;35(7):1615-1623. doi: 10.1016/j.neurobiolaging.2014.01.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gharbi-Meliani A, Dugravot A, Sabia S, et al. The association of APOE ε4 with cognitive function over the adult life course and incidence of dementia: 20 years follow-up of the Whitehall II study. Alz Res Ther. 2021;13(1):5. doi: 10.1186/s13195-020-00740-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lancaster C, Tabet N, Rusted J. The elusive nature of APOE ε4 in mid-adulthood: understanding the cognitive profile. J Int Neuropsychol Soc. 2017;23(3):239-253. doi: 10.1017/s1355617716000990. [DOI] [PubMed] [Google Scholar]
- 45.Reiman EM, Arboleda-Velasquez JF, Quiroz YT, et al. Exceptionally low likelihood of Alzheimer's dementia in APOE2 homozygotes from a 5, 000-person neuropathological study. Nat Commun. 2020;11(1):667. doi: 10.1038/s41467-019-14279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sweigart B, Andersen SL, Gurinovich A, et al. APOE E2/E2 is associated with slower rate of cognitive decline with age. J Alzheimers Dis. 2021;83(2):853-860. doi: 10.3233/jad-201205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arenaza-Urquijo EM, Vemuri P. Resistance vs resilience to Alzheimer disease: clarifying terminology for preclinical studies. Neurology. 2018;90(15):695-703. doi: 10.1212/wnl.0000000000005303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Y, Yamada K, Liddelow SA, et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature. 2017;549(7673):523-527. doi: 10.1038/nature24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serrano-Pozo A, Li Z, Noori A, et al. Effect of APOE alleles on the glial transcriptome in normal aging and Alzheimer's disease. Nat Aging. 2021;1(10):919-931. doi: 10.1038/s43587-021-00123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kloske CM, Dugan AJ, Weekman EM, et al. Inflammatory pathways are impaired in Alzheimer disease and differentially associated with apolipoprotein E status. J Neuropathol Exp Neurol. 2021;80(10):922-932. doi: 10.1093/jnen/nlab085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson RJ, Meltzer JC, Nguyen H, et al. APOE4 derived from astrocytes leads to blood-brain barrier impairment. Brain. 2021;145(10):3582-3593. doi: 10.1093/brain/awab478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steele OG, Stuart AC, Minkley L, et al. A multi-hit hypothesis for an APOE4-dependent pathophysiological state. Eur J Neurosci. 2022;56(9):5476-5515. doi: 10.1111/ejn.15685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ballard C, Atri A, Boneva N, et al. Enrichment factors for clinical trials in mild-to-moderate Alzheimer's disease. Alzheimers Dement (N Y). 2019;5(1):164-174. doi: 10.1016/j.trci.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jutten RJ, Sikkes SAM, Van der Flier WM, et al. Finding treatment effects in Alzheimer trials in the face of disease progression heterogeneity. Neurology. 2021;96(22):e2673-e2684. doi: 10.1212/wnl.0000000000012022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kennedy RE, Cutter GR, Schneider LS. Effect of APOE genotype status on targeted clinical trials outcomes and efficiency in dementia and mild cognitive impairment resulting from Alzheimer's disease. Alzheimers Dement. 2014;10(3):349-359. doi: 10.1016/j.jalz.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kennedy RE, Cutter GR, Wang G, Schneider LS. Post hoc analyses of ApoE genotype-defined subgroups in clinical trials. J Alzheimers Dis. 2016;50(4):1205-1215. doi: 10.3233/jad-150847. [DOI] [PubMed] [Google Scholar]
- 57.Serrano-Pozo A, Das S, Hyman BT. APOE and Alzheimer's disease: advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 2021;20(1):68-80. doi: 10.1016/s1474-4422(20)30412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nelson PT, Dickson DW, Trojanowski JQ, et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain. 2019;142(6):1503-1527. doi: 10.1093/brain/awz099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The NACC database is a publicly available resource available to researchers, and data requests can be submitted online at the following NACC website: nacc.redcap.rit.uw.edu/surveys/?s=KHNPKLJW8TKAD4DA.