SUMMARY
Adaptations to infectious and dietary pressures shape mammalian physiology and disease risk. How such adaptations affect sex-biased diseases remains insufficiently studied. In this study, we show that sex-dependent hepatic gene programs confer a robust (~300%) survival advantage for male mice during lethal bacterial infection. The transcription factor B cell lymphoma 6 (BCL6), which masculinizes hepatic gene expression at puberty, is essential for this advantage. However, protection by BCL6 protein comes at a cost during conditions of dietary excess, which result in overt fatty liver and glucose intolerance in males. Deleting hepatic BCL6 reverses these phenotypes but markedly lowers male survival during infection, thus establishing a sex-dependent trade-off between host defense and metabolic systems. Our findings offer strong evidence that some current sex-biased diseases are rooted in ancient evolutionary trade-offs between immunity and metabolism.
Infections are one of the strongest evolutionary pressures shaping human physiology and disease. As such, the immune system and host defense responses are often prioritized at the expense of other physiological systems (1, 2). As a result, genetic variants that are associated with noninfectious diseases may be maintained in the population if they simultaneously improve survival during infection. For example, variants in human HBB and APOL1 increase the risk of sickle cell anemia and chronic kidney disease (3) but exert a strong protective effect against malaria and trypanosome infections, respectively. These studies highlight the notion of an “evolutionary trade-off” whereby natural selection fails to optimize two traits simultaneously, which causes increased adaptation for one trait at the expense of another and ultimately may elevate disease risk.
Shifting environments also magnify the disease risk associated with trade-offs, resulting in so-called mismatch diseases (4). Thus, the initial benefit of a trait becomes detrimental in a new environment. For example, a mismatch between our genetic legacy and the modern diet that is high in calories, fat, and refined sugars is proposed to account for the high prevalence of chronic metabolic diseases, such as type 2 diabetes (T2D), heart disease, and fatty liver (5). However, although the evolutionary mismatch theory can explain chronic diseases that affect immunity and metabolism, their marked sex bias in the human population is poorly understood. Notably, men carry a much higher disease burden for common metabolic disorders compared with premenopausal women (6–8). Similarly, some infectious diseases exhibit a strong sex bias with poorer outcomes observed in either males or females depending on the pathogen (9). Together, inherent sex differences in physiological systems dictate disease progression in males and females. In this study, we examined the relationship between biological sex during a dietary excess challenge and infection in mice.
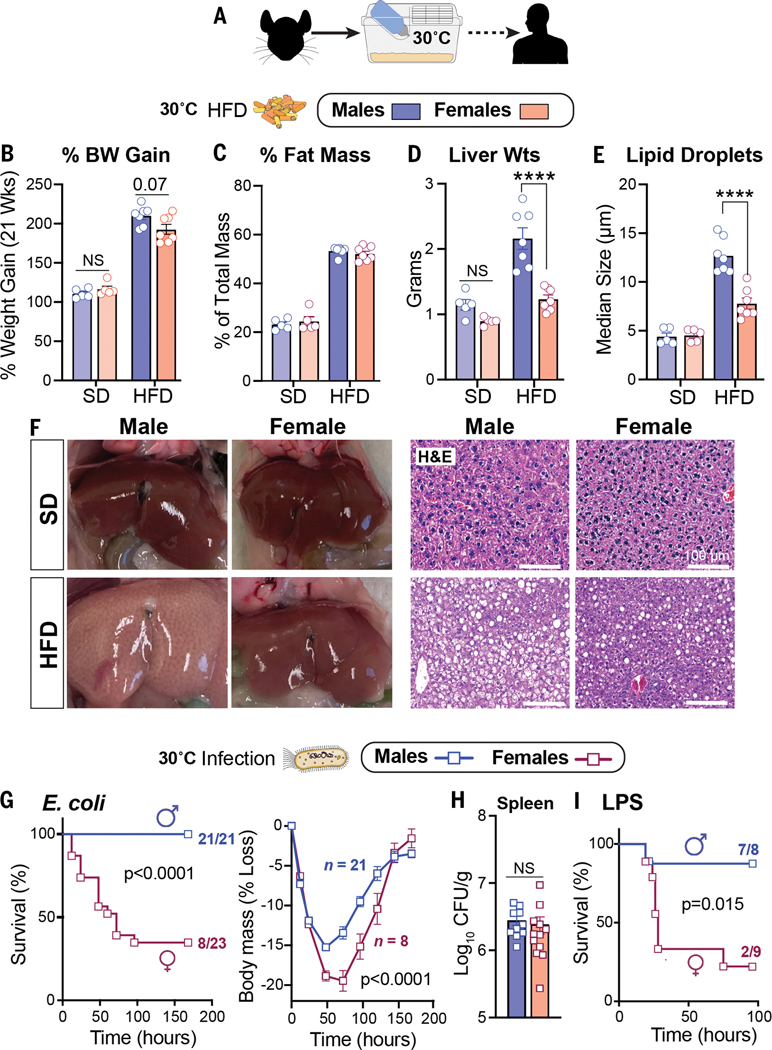
Prior studies found that mice housed at thermoneutral temperature (30°C) are susceptible to the metabolic consequences of chronic dietary excess (10, 11) and infection (12). We therefore used thermoneutral conditions to examine the potential trade-offs between metabolism and host defense mechanisms in C57BL/6J males and females (Fig. 1A). Despite an equivalent increase in body weight and fat mass when fed a high-fat diet (HFD) (Fig. 1, B and C), only male mice developed severe fatty liver and overt macrosteatosis (Fig. 1, D to F, and fig. S1A). Using thermoneutral conditions, we then examined how sex affects host survival during infection with a sublethal dose of Escherichia coli (strain O111:B4) in mice that were fed standard chow. Males were far less susceptible to infection and showed a markedly higher survival rate and body mass preservation at thermoneutrality than females (Fig. 1G and fig. S1, B and C). Spleen bacterial counts were equivalent in both sexes (Fig. 1H), suggesting that males limit their immunopathology and that pathogen clearance fails to account for the sex differences in survival. Greater survival in males was also observed after activation of host immunity by the endotoxin lipopolysaccharide (LPS) (Fig. 1I). Collectively, our results expose a stark relationship, specifically in males, between hepatic fat accumulation after dietary excess and host survival after bacterial infection.
Figure 1.
HFD and infection elicit strong sex-dependent phenotypes in mice.(A) Schematic of housing conditions. (B to F) Body weight gain (B), fat percentage(C), liver weights (Wts) (D), hepatic lipid droplet size (E), and whole livers with corresponding hematoxylin and eosin (H&E) staining (F) after 21 weeks of SD or HFD. (G) Survival curves and body weights of C57BL/6J mice that were infected with E. coli [1 × 108 colony-forming units (CFU)]. Weight curves were analyzed by two-way analysis of variance (ANOVA). (H) Bacterial CFUs of mice that were infected with E. coli (1 × 107 CFU). (I) Survival curves of mice treated with LPS (2 mg/kg). All mice were housed at 30°C. Data are presented as mean ± SEM; NS, not significant; ****p < 0.0001. Scale bars, 100 mm.
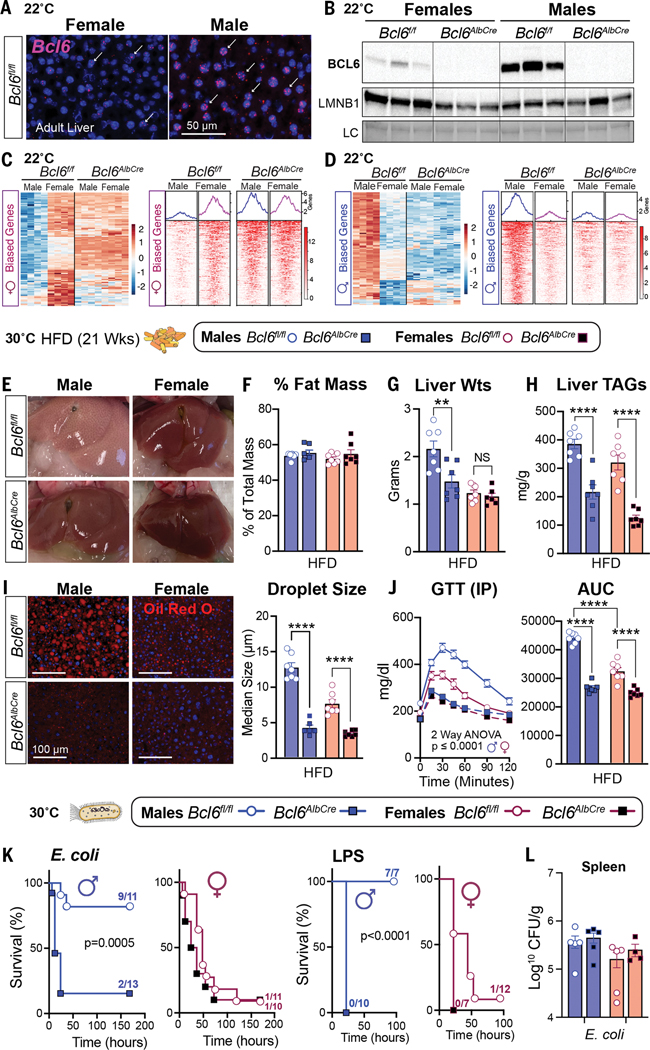
In searching prior literature for a sex-dependent hepatic factor that might mediate these divergent outcomes between the sexes, the transcriptional repressor B cell lymphoma 6 protein (BCL6) emerged as a top candidate given its role in hepatic lipid handling (13, 14) and its enrichment in the male liver as previously shown (15, 16). We found that BCL6 is male biased at both 22°C and 30°C, with Bcl6 transcripts and protein highly expressed in male hepatocytes and livers (Fig. 2, A and B, and fig. S2A). Conditional deletion of Bcl6 in the liver (Bcl6AlbCre) (Fig. 2B) feminizes the adult male liver and eliminates its male-biased gene signature (Fig. 2, C and D; data S1; and fig. S2, B to D). Profiling active enhancers and promoters for acetylated histone 3 lysine 27 (H3K27ac) by chromatin immunoprecipitation sequencing (ChIP-seq) also revealed an essential role of BCL6 in maintaining sex-dependent hepatic chromatin acetylation and male-biased H3K27ac peaks (Fig.2,C and, and fig.S2, E and F).
Figure 2.
BCL6 maintains hepatic maleness and survival to infection but impairs metabolism. (A) In situ hybridization for Bcl6 (magenta, white arrows) in livers of mice at 22°C. Scale bar, 50 mm. (B) Immunoblot for BCL6 and LMNB1 in liver nuclear extracts of 8-week-old Bcl6f/f and Bcl6AlbCre mice at 22°C. (C and D) Heatmaps for top hepatic 100 (C) female- and (D) male-biased genes (filtered by fold change) with corresponding female- or male-biased H3K27ac peaks (adjusted p value for both <0.05) in mice that were housed at 22°C. Scale bars, Z-scores. (E to H) Livers (E), fat percentage (F), liver weights (G), and liver TAGs (H) from mice that were fed a HFD at 30°C. (I) Hepatic Oil Red O staining (ORO) and quantification of lipid droplet (red) size from mice that were fed a HFD for 21 weeks at 30°C. Nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bars, 100 mm. (J) Glucose concentrations and area under the curve (AUC) after an intraperitoneal (IP) glucose tolerance test (GTT) in mice that were fed a HFD for 8 weeks at 30°C. (K and L) Survival curves of mice that were fed a SD and infected with E. coli (1 × 108 CFU) or treated with LPS (1.75 mg/kg) at 30°C (K) and spleen bacterial counts at 30°C (L). Data for control Bcl6f/f mice in (F), (G), and (I) are regraphed from Fig. 1, C to E (HFD). Data are presented as mean ± SEM. LC, loading control (total protein). **p < 0.01; ****p < 0.0001.
Having established the masculinizing role of BCL6 in hepatic gene signatures, we assessed whether BCL6 is essential for maintaining the distinct sex-specific outcomes of HFD and infection. Indeed, deleting hepatic Bcl6 abolished all morphological hallmarks of fatty liver in males without changing their total fat mass or percent body weight gain (Fig. 2, E and F, and fig. S3, A and B). Liver weights, hepatic triglycerides (TAGs), lipid accumulation, and droplet size were all reduced in Bcl6AlbCre male mice (Fig. 2, G to I), which is consistent with a prior study that found that BCL6 blocks the breakdown of fat by lowering fatty acid oxidation (14). Hepatic TAGs also fell in Bcl6AlbCre mice that were fed a standard diet (SD) (fig. S3C). Eliminating the low amounts of BCL6 present in female livers also attenuated hepatic TAGs, but more subtly (Fig. 2, G to I). The loss of hepatic BCL6 markedly improved glucose homeostasis in mutant male cohorts that were fed either a HFD or a SD and abolished any notable sex differences in this metabolic parameter (Fig. 2J and fig. S3D). In stark contrast to the improved metabolic state in Bcl6AlbCre males, their survival dropped precipitously after E. coli infection or LPS treatment, plummeting to the levels exhibited by control females (Fig. 2K and fig. S3E). Patho- gen clearance in the spleen was unaffected in Bcl6AlbCre mice (Fig. 2L). Thus, high hepatic BCL6 in males is essential for optimizing host survival during infection but drives fatty liver and glucose intolerance during dietary excess, suggesting a strong association between hepatic lipid handling and host defense responses.
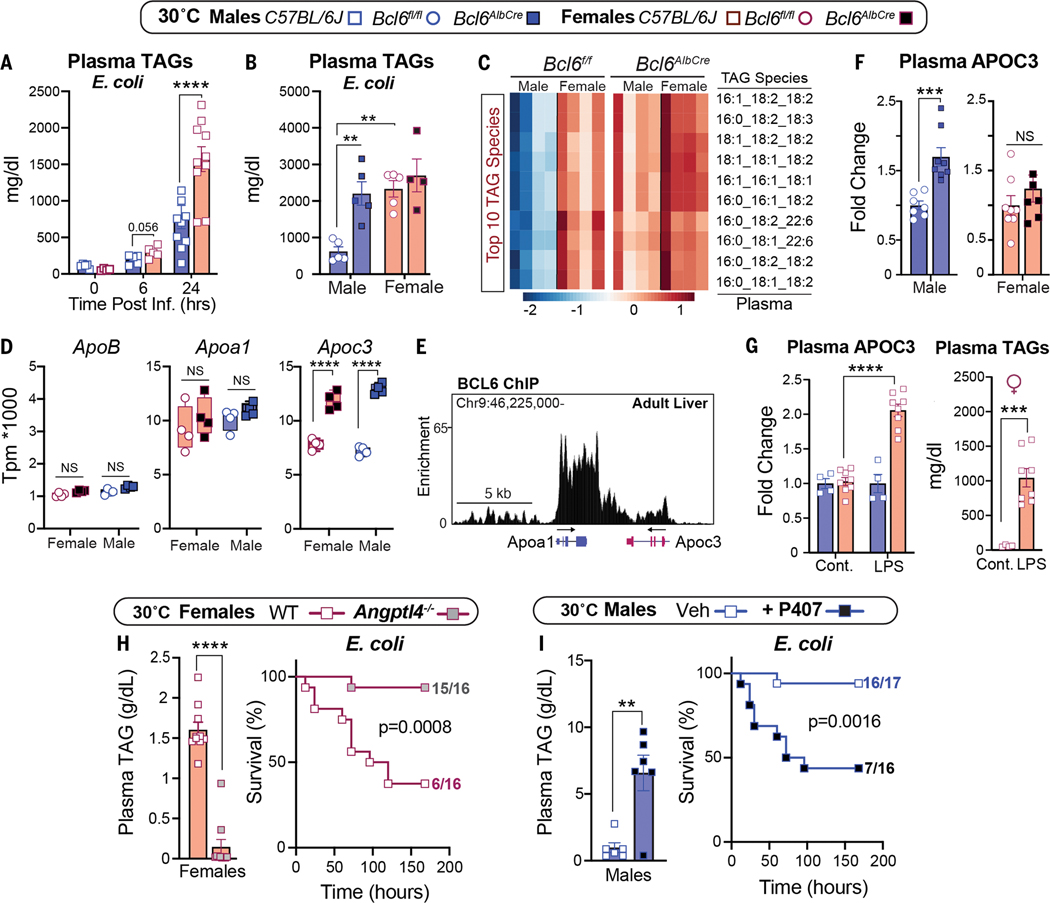
Low survival in females that were fed a SD at 30°C is closely correlated with extremely high plasma TAGs, a condition that is observed in septic humans (17) and rats (18). Infection-induced hyperlipidemia is only observed at thermoneutrality (Fig. 3A and fig. S4A). Likewise, compromised survival in infected Bcl6AlbCre males was linked with a substantial rise in the concentrations of circulating TAG species, similar to those of infected control females (Fig. 3, B and C, and fig. S4B). Increased plasma TAGs in infected Bcl6AlbCre mutant male mice prompted us to investigate whether genes that are crucial in the packaging and clearance of very low-density lipoproteins (VLDLs) and their TAG cargo are regulated by BCL6. Of the three candidate genes examined, hepatic Apoc3, whose gene product inhibits TAG clearance, increased sharply in Bcl6AlbCre mice, whereas ApoB and ApoA1 were unchanged (Fig. 3D and fig. S4C). Reanalysis of the hepatic BCL6 ChIP-seq dataset by Waxman’s group (16) revealed that BCL6 binds directly to the Apoc3 locus to dampen its expression (Fig. 3E). Thus, as predicted, increased plasma APOC3 occurs after eliminating high hepatic BCL6 in uninfected males. This relationship is not as clear-cut in Bcl6AlbCre females, who fail to exhibit high APOC3 levels despite an increase in Apoc3 transcripts. These results suggest that posttranscriptional factors are at play during the packaging of female hepatic APOC3 into lipoproteins (Fig. 3F). Nevertheless, wild-type females that are treated with LPS exhibit a notable increase in both circulating APOC3 and TAGs (Fig. 3G), which is consistent with the essential role of APOC3 in maintaining plasma TAGs (19). Our findings support the postulate by Scholl and colleagues (18) that decreased clearance of plasma TAGs by lipoprotein lipase (LPL) contributes to sepsis-induced hyperlipidemia.
Figure 3.
Sex-dependent hyperlipidemia is linked to host defense responses. (A and B) Plasma TAGs of (A) wild-type mice over time and (B) Bcl6f/f and Bcl6AlbCre mice that were infected with E. coli (1 × 107 CFU) at 30°C. (C) Top 10 most abundant TAG species measured by lipidomics in infected Bcl6f/f and Bcl6AlbCre mice. Scale bar, Z-scores. (D) Transcript abundances of hepatic ApoB, Apoa1, and Apoc3 in Bcl6f/f and Bcl6AlbCre mice (RNA-seq) at 30°C. TPM, transcripts per million. (E) Genomic binding of BCL6 in Apoc3/Apoa1 locus in the male liver (ChIP-seq) at 22°C. (F) Plasma APOC3 in Bcl6f/f and Bcl6AlbCre mice that were housed at 30°C. (G) Plasma APOC3 and TAGs in control and LPS-treated (0.5 mg/kg) C57BL/6J female mice at 30°C. Cont., control. (H) Plasma TAGs (1 × 107 CFU) and survival curves (1 × 108 CFU) of female mice that were infected with E. coli at 30°C. (I) Plasma TAGs (1 × 107 CFU) and survival curves (1 × 108 CFU) of male mice that were infected with E. coli at 30°C. Data are presented as mean ± SEM. **p < 0.01; ***p < 0.001; ****p < 0.0001.
To determine whether high plasma TAGs contribute directly to poor survival in females, we used ANGPTL4 knockout (KO) mice (20) that clear out TAGs because of increased LPL activity (Fig. 3H). Normalizing TAGs in infected Angptl4−/− females restored both survival and body weights (Fig. 3H and fig. S4D). Angptl4−/− males also showed a drop in TAGs and remained resistant to infection (fig. S4, E and F). Conversely, increasing plasma TAGs by using poloxamer 407 (P407), a synthetic inhibitor of LPL, worsened the survival of males after infection (Fig. 3I). Our results establish that the marked sex differences in infection outcomes are tightly linked with the amounts of circulating TAGs.
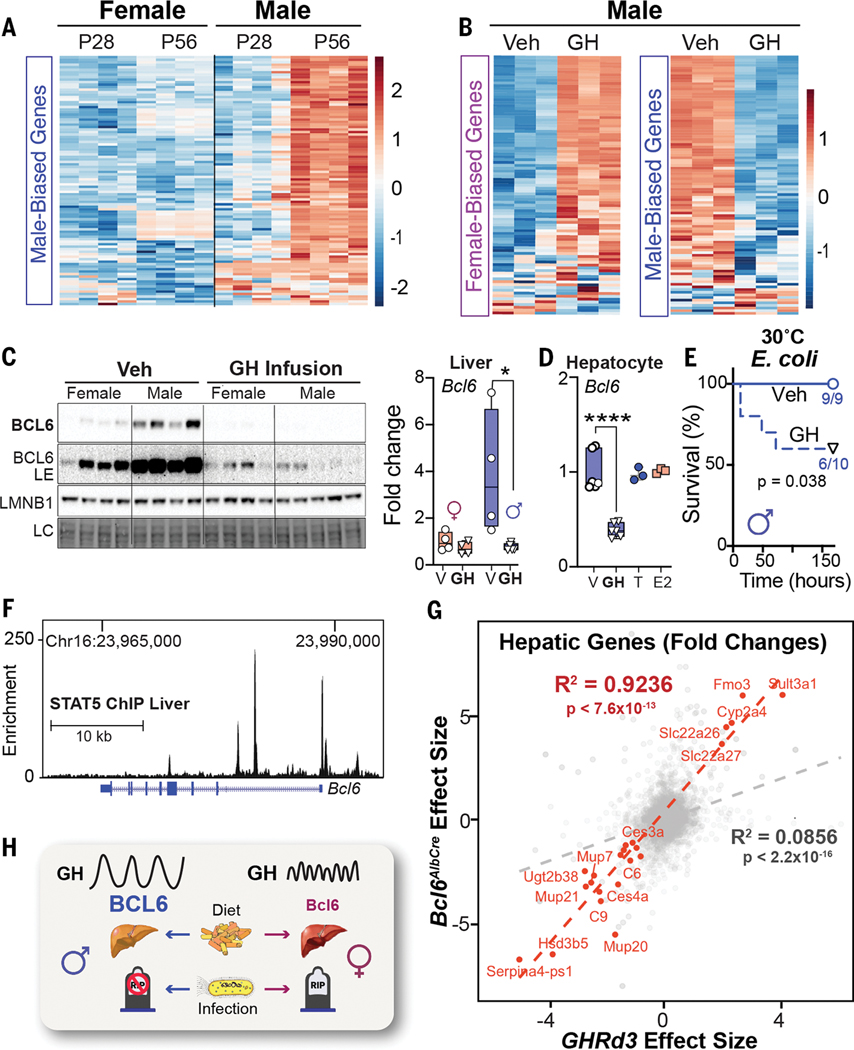
We next investigated what factors enable BCL6 to control the hepatic gene programs in male mice. The appearance of male-biased genes coincides with puberty, which becomes apparent at 8 weeks of age (Fig. 4A and fig. S5A). Surgical castration (GDX) of prepubescent males enhanced female-biased gene expression in the liver (fig. S5B), led to a steep drop in survival that was accompanied by elevated plasma TAGs after E. coli infection (fig. S5, C and D), and diminished hepatic BCL6 levels, which were partially restored by a testosterone (T) treatment (fig. S5E). Pulsatile secretion of growth hormone (GH) from the anterior pituitary is distinctive in males and consists of peaks with prolonged extended dips; this pattern is required for male-biased hepatic gene expression in mice (16, 21). Indeed, after reanalyzing datasets from (22), focusing on our set of 200 sex-biased genes, we confirmed that continuous infusion of GH feminizes male livers (Fig. 4B and fig. S5F). We also found that GH treatment strongly represses hepatic BCL6 protein and transcripts (Fig. 4C). Although saturating levels of GH in primary hepatocytes also suppressed Bcl6, T and estradiol (E) had no effect in this setting, which implies that the T-induced rescue of BCL6 expression in vivo must be indirect (Fig. 4D and fig. S5E). Expectedly, disrupting normal GH pulsatility by continuous GH infusion reduced hepatic BCL6 and diminished host survival in males (Fig. 4E). As shown by Waxman’s group, the major effector of hepatic GH signaling, STAT5, binds to the Bcl6 locus (16), which provides a direct molecular link between GH and BCL6 levels (Fig. 4F).
Figure 4.
Sex-dependent GH signaling controls BCL6 expression and survival to infection. (A and B) Heatmaps of top 100 (A) male-biased genes and (B) female-/male-biased genes at postnatal day 28 (P28) and P56 or in male mice after GH treatment at 22°C. Scale bars, Z-scores. (C) Immunoblotting and reverse transcription–quantitative polymerase chain reaction (RT-QPCR) for hepatic BCL6 protein and transcript in adult mice infused with vehicle (Veh) or recombinant mouse GH for 15 days at 22°C. (D) Bcl6 mRNA expression in primary mouse hepatocytes treated with vehicle (V), T, estradiol benzoate (E2)(n = 3), or GH (n = 6). (E) Survival curves of male mice infused with Veh or GH for 13 days and then infected with E. coli (1 × 108 CFU) at 30°C. (F) STAT5 binding to Bcl6 locus in the male liver (ChIP-seq) at 22°C. Chr, chromosome. (G) Effect size correlation of all (gray) or differentially expressed (red) transcripts in livers of Bcl6AlbCre and Ghrd3 male mice. (H) Schematic of the GH–BCL6 axis in regulating sex-dependent endpoints when challenged by diet or infection. LE, long exposure. Data are presented as mean ± SEM;*p < 0.05, ****p < 0.0001.
To extend these findings, we leveraged a mouse model that carries the common human variant of growth hormone receptor (GHRd3, deletion of exon 3) that mimics increased GH signaling and confers a marked protective effect in humans (~4-fold) against developing T2D (23). This variant feminizes livers in male mice and is thought to impart an evolutionary advantage during periods of food scarcity in humans (24). The significant (R2 = 0.92, p < 7.6 × 10−13) overlap in hepatic gene changes detected in Ghrd3 and Bcl6AlbCre mutant males (Fig. 4G and fig. S5G) suggests that this common GHR variant attenuates hepatic BCL6 function. In contrast to the notable hepatic gene changes with the onset of puberty or after castration, we failed to find any significant changes in our sex-biased gene signatures in estrogen receptor alpha (Esr1) liver KO mice after reanalyzing datasets by Maggi and colleagues (25) (fig. S5H). The ability of the GHRd3 variant to stave off nutritional stress (24), coupled with our study, might suggest that the GH–BCL6 signaling axis creates a trade-off for females that diminishes survival during infection while enhancing survival in the fasted state. This notion is partially supported by the fact that survival rates for women outpace men during famine (26).
In male mice, the hepatic GH–BCL6 axis is essential for mounting protective defenses against infection while promoting substantial hepatic fat accumulation and glucose intolerance during caloric excess (Fig. 4H). Although sex differences in this pathway remain to be documented in humans, it has been noted that patients with hypopituitarism and low GH develop fatty liver, which improves after GH therapy (27). On the basis of the conserved features of metabolic programs across mammals, we speculate that the current prevalence of fatty liver in males might stem from older host defense mechanisms that coevolved from increased exposure to pathogens due to aggressive behaviors required for mating and social status (28). Our study leads us to propose that adaptations to infectious and dietary pressures sculpt sexually dimorphic pathways, contributing to modern sex-biased diseases.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Chawla and Ingraham labs for comments on the manuscript and X. Cui and J. Argiris for assistance with mouse husbandry. We also thank A. Capra and E. Goldberg for intellectual discussions. In addition, we thank S. Koliwad for providing Angptl4−/− mice and J. Hellman for the E. coli strain that was used in experiments. Funding: The authors’ work was supported by grants from NIH (AG062331, DK121657, and GCRLE Senior Scholar Award to H.A.I.), NIH (DK094641 and DK101064 to A.C.), EMBO (ALTF 1185-2017 to J.N.), HFSPO (LT000446/ 2018-L to J.N.), UCSF PBBR (7000/7002124 to J.N.), NIH (DK129763 to J.N.), and NHMRC (GNT1142229 to Y.A.L.). Mass spectrometry equipment was obtained through NIH Shared Instrumentation Grant 1S10OD016232-01 (J.E.C.), 1S10OD018210-01A1 (J.E.C.), and 1S10OD021505-01 (J.E.C.). Author contributions: J.N., Y.A.L., W.C.K., E.J.W., A.C. and H.A.I. conceived and designed the experiments, interpreted the results, and wrote the paper. J.N., Y.A.L., and W.C.K. performed the experiments. D.D. assisted with the analysis of RNA sequencing (RNA-seq) and ChIP-seq datasets, and J.A.M., T.V.R., and J.E.C. performed lipidomics on plasma and liver samples. O.G. performed the analysis comparing Bcl6AlbCre and Ghrd3 liver transcriptomes and provided conceptual input. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The RNA-seq and ChIP-seq datasets generated and analyzed during the study are available in the Gene
Expression Omnibus (GEO) repository (ncbi.nlm.nih.gov/geo/) under the SuperSeries accession number GSE138396. License information: Copyright © 2022 the authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original US government works. https://www.science.org/about/science-licenses-journal-article-reuse
REFERENCES AND NOTES
- 1.Wang A, Luan HH, Medzhitov R, Science 363, eaar3932 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stearns SC, Medzhitov R, Evolutionary Medicine (Sinauer, 2016). [Google Scholar]
- 3.Benton ML et al. , Nat. Rev. Genet 22, 269–283 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Rienzo A, Hudson RR, Trends Genet. 21, 596–601 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Manus MB, Evol. Med. Public Health 2018, 190–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lonardo A. et al. , Hepatology 70, 1457–1469 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosca L, Barrett-Connor E, Kass Wenger N, Circulation 124, 2145–2154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tramunt B. et al. , Diabetologia 63, 453–461 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein SL, Flanagan KL, Nat. Rev. Immunol 16, 626–638 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Fischer AW, Cannon B, Nedergaard J, Mol. Metab 26, 1–3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giles DA et al. , Nat. Med 23, 829–838 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganeshan K. et al. , Cell 177, 399–413.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salisbury DA et al. , Nat. Metab 3, 940–953 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sommars MA et al. , eLife 8, e43922 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer RD, Laz EV, Su T, Waxman DJ, Mol. Endocrinol 23, 1914–1926 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Laz EV, Waxman DJ, Mol. Cell. Biol 32, 880–896 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallin JI, Kaye D, O’Leary WM, Engl N. J. Med 281, 1081–1086 (1969). [DOI] [PubMed] [Google Scholar]
- 18.Scholl RA, Lang CH, Bagby GJ, J. Surg. Res 37, 394–401 (1984). [DOI] [PubMed] [Google Scholar]
- 19.Maeda N. et al. , J. Biol. Chem 269, 23610–23616 (1994). [PubMed] [Google Scholar]
- 20.Kersten S, Curr. Opin. Lipidol 30, 205–211 (2019). [DOI] [PubMed] [Google Scholar]
- 21.MacLeod JN, Pampori NA, Shapiro BH, J. Endocrinol 131, 395–399 (1991). [DOI] [PubMed] [Google Scholar]
- 22.Lau-Corona D, Suvorov A, Waxman DJ, Mol. Cell. Biol 37, e00301–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strawbridge RJ et al. , Growth Horm. IGF Res 17, 392–398 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Saitou M. et al. , Sci. Adv 7, eabi4476 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Della Torre S. et al. , Cell Metab. 28, 256–267.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zarulli V. et al. , Proc. Natl. Acad. Sci. U.S.A 115, E832–E840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang SJ et al. , Endocr. Pract 27, 1149–1155 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Klein SL, Behav. Processes 51, 149–166 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.