Abstract
The Wnt family of secreted glycolipo-proteins signals through multiple signal transduction pathways and is essential for embryonic development and organ development and homeostasis. The Wnt-pathways are conserved and critical in all metazoans. Wnt signaling pathways comprise the canonical Wnt/β-catenin pathway and several non-canonical signaling branches, of which Wnt-Planar Cell Polarity (PCP) signaling and the Wnt/Calcium pathway have received the most attention and are best understood. nterestingly, all Wnt-pathways have a nuclear signaling branch and also can affect many cellular processes independent of its nuclear transcriptional regulation. Canonical Wnt/β-catenin signaling is the most critical for a nuclear transcriptional response, in both development and disease, yet the mechanism(s) on how the “business end” of the pathway, β-catenin, translocates to the nucleus to act as co-activator to the TCF/Lef transcription factor family still remains obscure. Here we discuss and compare the very different strategies on how the respective Wnt signaling pathways activate a nuclear transcriptional response. We also highlight some recent new insights into how β-catenin is translocated to the nucleus via an IFT-A, Kinesin-2, and microtubule dependent mechanism and how this aspect of canonical Wnt-signaling uses ciliary proteins in a cilium independent manner, conserved between Drosophila and mammalian cells.
1. Overview of Wnt-signaling pathways
The founding member of the Wnt gene family was first discovered in Drosophila, with its original mutant allele lacking wings, and hence it was named wingless (wg) (Sharma & Chopra, 1976). Subsequently, it was shown to be an essential gene and required throughout development including embryogenesis (Nusslein-Volhard & Wieschaus, 1980). Conserved sequences were identified in mice via gene disruption strategies using viral insertions as mutagens in a cancer screening context looking for malignant mammary tissue transformations (Nusse, van Ooyen, Cox, Fung, & Varmus, 1984). As this original insertion sequence in the mouse genome was named int1, and the associated mouse gene was found to be highly homologous to the Drosophila gene, the associated gene family was named Wnt as a combination of Drosophila Wingless and mouse “int.” The family of Wnt/Wg genes is conserved in all metazoans, with multiple members in most species, and encodes proteins of a secreted glycolipoprotein class. Wnt genes are not only essential and critical for animal development, organogenesis, and homeostasis of all metazoans, but are also critically linked to regeneration and stem cell maintenance, as well as being dysregulated in many diseases (Cadigan & Waterman, 2012; Clevers, 2006; Logan & Nusse, 2004; MacDonald, Tamai, & He, 2009).
Strikingly, over the years, it was shown that Wnt protein family associated pathways comprise a fairly large set of distinct signaling branches, which share the Wnt ligands and Frizzled (Fz, or Fzd, for short) receptor family and the downstream effector Dishevelled (Dsh, a.k.a. Dvl in mammals) (see below for review references). Among these, the canonical Wnt/β-catenin signaling pathway stands out as the most studied and overall best understood (Cadigan & Waterman, 2012; Clevers, 2006; Eisenmann, 2005; Logan & Nusse, 2004; MacDonald et al., 2009). For an updated comprehensive pathway overview see also the Wnt homepage (www.stanford.edu/~rnusse/wntwindow.html). In addition to the canonical pathway, there exist several so-called non-canonical Wnt signaling pathways that have emerged and received significant attention. Among these non-canonical pathways, the following received the most attention: Wnt-Planar Cell Polarity (PCP) signaling (reviewed in (Adler, 2002; Goodrich & Strutt, 2011; Harrison, Shao, Strutt, & Strutt, 2020; Humphries & Mlodzik, 2018; Peng & Axelrod, 2012) and the Wnt/Calcium pathway (see reviews Kohn & Moon, 2005; Kuhl, Sheldahl, Park, Miller, & Moon, 2000). These are the most studied and (at least in part) understood and will be discussed further here in the context of nuclear signaling and transcriptional activation. For a complete listing of additional non-canonical Wnt-pathways see (Semenov, Habas, Macdonald, & He, 2007).
Each of the above mentioned three Wnt-pathways have both a nuclear signaling branch and can also affect several cellular processes independently of their effects on nuclear transcriptional regulation. Canonical Wnt/β-catenin signaling is the most critical for a nuclear transcriptional response, in both development and disease (reviewed in (Clevers, 2006; Clevers & Nusse, 2012; MacDonald et al., 2009)). Although quite a bit is understood about the nuclear complexes that activate or repress transcription of target genes in the nucleus with co-repressors and co-activators of the TCF/Lef transcription factor family, the key mechanism(s) of how nuclear translocation of β-catenin is achieved still remains obscure. As β-catenin is the absolutely essential co-activator, around which a transcription activation complex is assembled, this missing link is critical not just for better understanding of the pathway cell biology, but also for potentially better focused therapeutic approaches. In contrast, the two non-canonical Wnt-signaling branches (see below) are better understood at the nuclear signaling level, although there are significant gaps in their understanding mostly at the level of membrane proximal pathway relay mechanisms.
In this chapter, we discuss and compare the very different strategies on how the respective Wnt signaling pathways generate an active nuclear response.
1.1. The canonical Wnt/β-catenin signaling pathway
Canonical Wnt/Wingless signaling is a highly conserved pathway with critical roles in the regulation of a vast catalogue of developmental processes, including but not restricted to tissue patterning as a morphogen across the animal kingdom, cell fate specification, cell proliferation, cell survival, and migration (Cadigan & Waterman, 2012; Clevers, 2006; Logan & Nusse, 2004; MacDonald et al., 2009). Dysregulated pathway components and or misexpression of pathway proteins are in most cases associated with diseases, including several cancer types (e.g. Clevers, 2006; Clevers & Nusse, 2012; Coombs, Covey, & Virshup, 2008; Prestwich & Macdougald, 2007).
At the center of active canonical Wnt signaling is the stabilization of β-catenin (Armadillo/Arm in Drosophila) (Cadigan & Waterman, 2012; MacDonald et al., 2009; Valenta, Hausmann, & Basler, 2012) (see Fig. 1 for simplified schematic). β-catenin/Arm is a multi-functional protein that is critical as both a component of adherens junctions (AJs) and their link to cytoskeletal elements (reviewed in (Bienz, 2005; McEwen, Escobar, & Gottardi, 2012; Tian et al., 2011) and as the key component in Wnt/Wg signaling. In the canonical Wnt pathway, β-catenin/Arm is the essential transcriptional co-activator for Wnt-target gene expression (reviewed in (Cadigan & Waterman, 2012; MacDonald et al., 2009; Valenta et al., 2012). Wnt/Wg proteins bind to Frizzled (Fz/Fzd in mammals) receptors and LRP5/6 (Arrow in Drosophila) co-receptors, resulting in the disassembly of the so-called “destruction complex,” composed of Axin, APC (Adenomatous Polyposis Coli), and the kinases GSK3β and Casein kinase I (CKI), which phosphorylate the cytoplasmic pool of β-catenin/Arm and thus target it for degradation (reviewed in (Cadigan & Waterman, 2012; MacDonald et al., 2009; Valenta et al., 2012)). Note that there are two pools of β-catenin/Arm, one being stably associated with Cadherins in AJ complexes and the other, the cytoplasmic, free pool that is targeted for degradation in the absence of Wnt-signaling. Wnt pathway activation results in the break-up of the destruction complex and re-localization of Axin to the plasma membrane, where it associates with Dsh/Dvl and the Fz/Fzd receptors and LRP5/6 co-receptors, forming large complexes of Axin-Dsh-Fz-LRP5/6 aggregates in endocytosed vesicles, generally referred to as signalosomes (Cadigan & Waterman, 2012; Cliffe, Hamada, & Bienz, 2003; MacDonald et al., 2009). Generally, the Dsh/Dvl and Axin proteins behave like protein scaffolds that hold together either the destruction complex (Axin) or the signalosome (DshDvl) (Cadigan & Waterman, 2012; MacDonald et al., 2009; Valenta et al., 2012). Axin removal from the destruction complex leads not only to complex disassembly, but importantly frees up and thus stabilizes cytoplasmic β-catenin/Arm, which can then translocate to the nucleus to act as a co-activator of the TCF/Lef transcription factors (Cadigan & Waterman, 2012; MacDonald et al., 2009). Again, in the absence of Wnt/Wg presence, β-catenin/Arm is phosphorylated by the “destruction complex” and targeted to the proteasome (Clevers & Nusse, 2012; Niehrs, 2012). While these membrane proximal and cytoplasmic aspects of the pathway are quite well defined, a big mystery remains in how β-catenin/Arm gets into the nucleus. Its primary sequence does not contain a bona fide nuclear localization signal (NLS), and thus while once in the nucleus its molecular function and interaction partners are again well known, the steps in between remain largely unclear. New insight into the potential mechanism(s) of how β-catenin/Arm is translocated in the nucleus have recently emerged (Balmer et al., 2015; Vuong et al., 2018) and these are discussed in a dedicated section below.
Fig. 1.
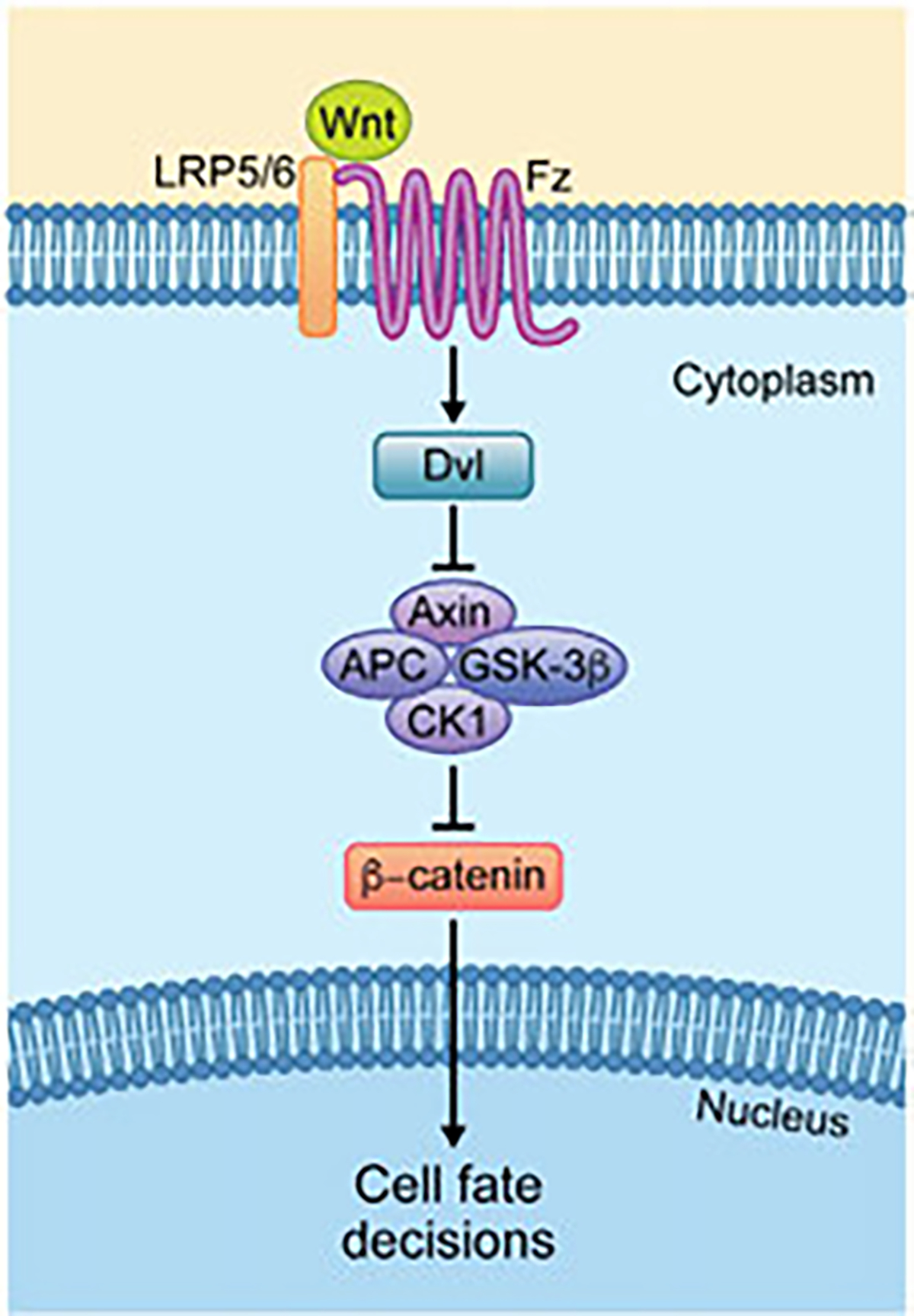
Simplified overview of the canonical Wnt (β-catenin) pathway. Upon pathway activation, Wnt binding causes the dimerization of the Fz (Fzd in vertebrates) receptor and the LRP5/6 co-receptor. The receptor complex then recruit a Dsh/Dvl-Axin multimer, which leads to the disassembly of the “destruction complex,” which—in the inactive state—includes Axin, APC, GSK-3β and CK1 among other proteins, and β-catenin. Thus upon the disassembly of the DC, β-catenin is freed and relieved from proteasome-dependent degradation and accumulates in the cytoplasm. Subsequently, β-catenin translocates to the nucleus, where its associates with the TCF/Lef transcription factors and several co-activators to turn on Wnt target gene transcription, which regulates cell fate decisions, proliferation, and cell survival. See main text for details.
In the context of Wnt/Wg-signaling, Drosophila and in particular wing development in the fly serves as an excellent model system and paradigm for pathway dissection and functional analyses. Importantly, once the wing primordia are established and a larval wing discs has formed, Wg is expressed as a two-cell stripe directly at the dorso-ventral (D/V) boundary of the future wing pouch. Emanating from there it forms an extracellular concentration gradient and acts as a morphogen, activating targets in a concentration dependent manner (e.g. (Baena-Lopez, Nojima, & Vincent, 2012; Danielson et al., 1995; Strigini & Cohen, 2000) and thus patterning the wing and specifying future wing structures, including the blade and margin associated cell fates (Neumann & Cohen, 1997; Zecca, Basler, & Struhl, 1996). In this context, high threshold transcriptional targets of Wg, include senseless (sens), a gene responsible for the formation of margin sensory bristles of adult wings, in cells directly adjacent to D/V boundary, while lower threshold target genes expressed in the wing blade include Distalless (Dll), required for wing growth (Neumann & Cohen, 1997; Zecca et al., 1996). Dll is expressed in a graded fashion, decreasing towards the edges of the wing pouch. Similar to most of the genetic dissection of Wg/Wnt signaling having relied on the use of the Drosophila wing, the discovery of a novel and unexpected mechanism for the nuclear translocation of β-catenin/Arm also resulted from genetic screens in this model, using the above mentioned target genes as functional read-out for β-catenin/Arm nuclear function (Balmer et al., 2015; Vuong et al., 2018; Vuong, Mukhopadhyay, & Choi, 2014).
1.2. Wnt/Fz-PCP signaling
Like the canonical Wnt/Wg-signaling pathway, Wnt/Planar Cell Polarity (PCP) signaling was first discovered in insects and is best studied and characterized in Drosophila (e.g. (Adler, 2002; Seifert & Mlodzik, 2007). In flies, PCP is evident in all adult cuticular structures and the compound eye and thus the simple easy of observation has been instrumental in identifying PCP pathway components and assembling these into a Wnt/PCP signaling pathway (e.g. Axelrod, Miller, Shulman, Moon, & Perrimon, 1998; Boutros, Paricio, Strutt, & Mlodzik, 1998; Das, Jenny, Klein, Eaton, & Mlodzik, 2004; Jenny, Reynolds-Kenneally, Das, Burnett, & Mlodzik, 2005; Tree et al., 2002; Tree, Ma, & Axelrod, 2002; Usui et al., 1999; Vinson & Adler, 1987; Vinson, Conover, & Adler, 1989; Wolff & Rubin, 1998) (see Fig. 2 for simplified schematic overview). Cellular patterning in tissues often requires directional information, either at the level of distinct cell fate specification or cellular orientation among equivalent cells. Both, mesenchymal cells and most epithelia are polarized within the planar axis, which is referred to as PCP. Such polarity-type arrangements provide cells and tissues with positional information allowing them to (i) generate polarized structures that are oriented with respect to tissue axes, (ii) embed specialized cells (e.g. sensory cells) with a specific orientation in any given organ, (iii) regulate cellular movement in a directed fashion, or (iv) instruct directed asymmetric cellular differentiation to generate asymmetric clusters of cells with specialized functions. All these PCP aspects are conserved across species and critical for development and homeostasis, and often linked to human disease (see recent reviews (Aw & Devenport, 2017; Butler & Wallingford, 2017; Davey & Moens, 2017; Goodrich & Strutt, 2011; Humphries & Mlodzik, 2018; Peng & Axelrod, 2012; Simons & Mlodzik, 2008)).The feature of directed, asymmetrically oriented differential cell fate specification, involves the nuclear branch of Wnt/Fz-PCP signaling (see dedicated Section 2.2).
Fig. 2.
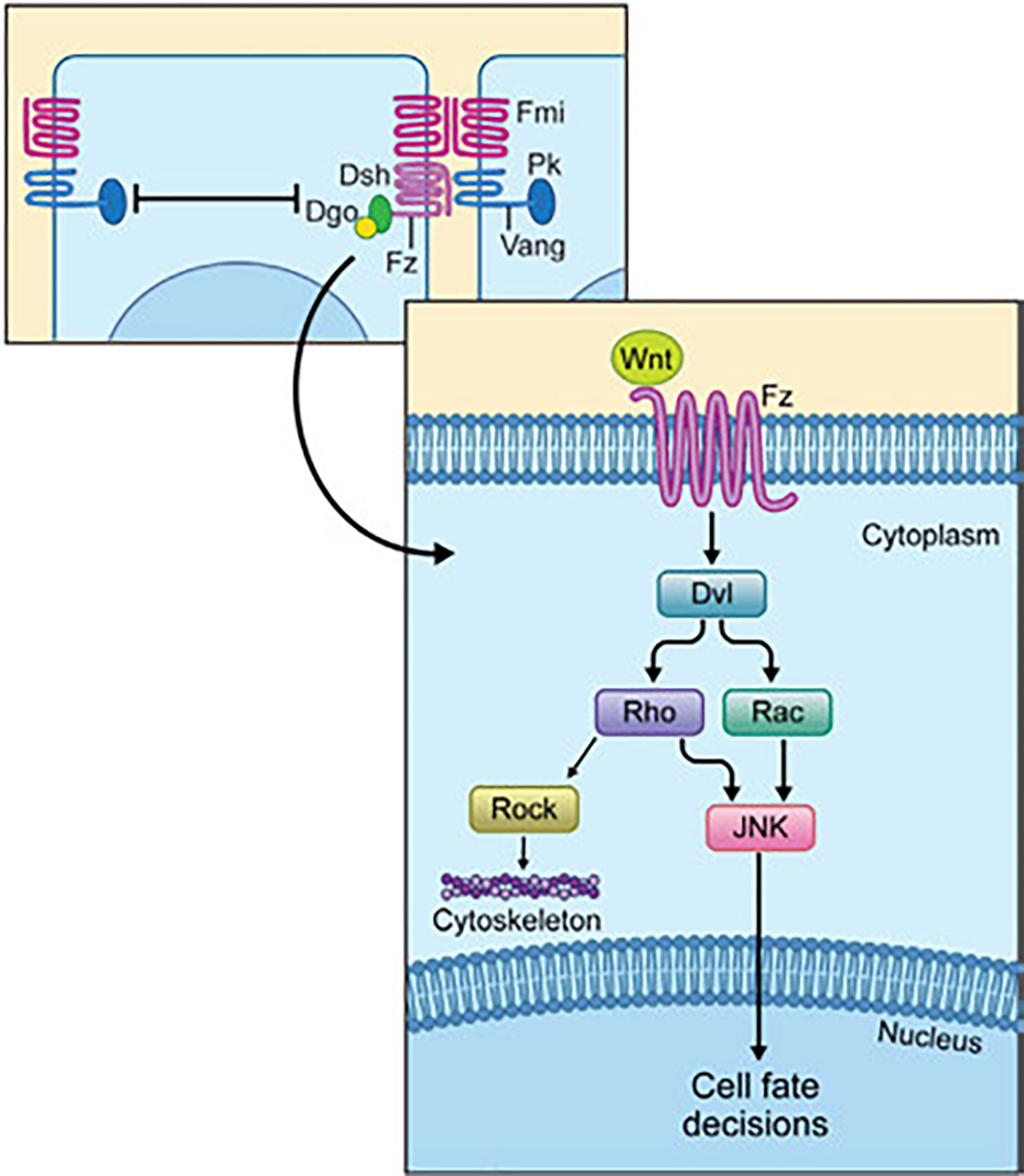
Overview of the non-canonical Wnt-Fz/PCP pathway. Upper part: a simplified diagram of the interactions among the PCP core components, which results in the stable resolution and establishment of two complexes, the Fz-Fmi-Dsh-Dgo complex (presumed to be on the side of the Wnt ligand) and the Vang-Fmi-Pk complex. Lower part: Simplified schematic view of the Wnt/PCP signaling pathway downstream of the Fz-Dsh complex. The PCP pathway is activated when a Wnt ligand binds to a Frizzled (Fz/Fzd) receptor and induces a clustering of the core PCP protein to either the Vang/Vangl-Pk complex or Fz-Dsh-Dgo complex (upper part). Downstream activation of the PCP pathway is associated with Dsh/Dvl phosphorylation and its interactions with downstream effectors Rho and Rac, possibly also cdc42 (not shown), with for example RhoA then acting through either Rock or the JNK-type MAPK cascade, with the respective kinases regulating either the cytoskeleton assembly (Rock) or triggering the expression of the target genes in the nucleus (JNK) regulating cell fate decisions. See main text for details.
For general PCP studies and pathway dissection, two tissues in Drosophila have served as exceptional model systems to unravel key features in Wnt/PCP signaling: these are the wings and eyes. Drosophila wing development, arguably the best understood PCP system, serves as an excellent system to dissect PCP mediated regulation of cytoskeletal orientation and assembly and the establishment and maintenance of the asymmetric distribution of core PCP factors from late larval to pupal stages of development. Here, all wing cells form a single actin-based cellular hair that is oriented in the proximo-distal axis, pointing distally, and this uniform orientation pattern, when disrupted in mutants of PCP components is easily appreciated as such. Similarly, in eyes, the very regular arrangement of ommatidia (or unit eyes, a.k.a. facets) with respect to the anterior-posterior (AP) and dorsal-ventral (DV) axes is altered in PCP mutants (Adler, 2002; Goodrich & Strutt, 2011; Peng & Axelrod, 2012; Seifert & Mlodzik, 2007). Based on the associated phenotypes, a core group of evolutionarily conserved Wnt-PCP genes have been identified, which are generally referred to as the “core Frizzled (Fz)/PCP factors.” The core Fz/PCP molecules include three transmembrane proteins: Fz, Flamingo (Fmi, a.k. a. Starry night/Stan; Celsr in vertebrates), and Van Gogh (Vang; a.k.a. Strabismus/Stbm; Vangl1/2 in vertebrates), and three cytoplasmic factors Dishevelled (Dsh, Dvl in mammals), Prickle (Pk), and Diego (Dgo, Inversin/Diversin in vertebrates) (reviewed in (Aw & Devenport, 2017; Butler & Wallingford, 2017; Davey & Moens, 2017; Goodrich & Strutt, 2011; Humphries & Mlodzik, 2018; Peng & Axelrod, 2012). These core PCP factors interact with each other, resolving into two stable complexes: one composed of Fmi-Fz-Dsh-Dgo and the other of Fmi-Vang-Pk. These complexes are stabilizing each other between cells across cell membranes, with a Fmi-Fz and Fmi-Vang intercellular adhesive interaction, and they antagonize each other within a given cell, via negative interactions between Dsh-Dgo and Pk. As a result these complexes become stably and asymmetrically localized along an axis (Aw & Devenport, 2017; Aw, Heck, Joyce, & Devenport, 2016; Butler & Wallingford, 2017; Davey & Moens, 2017; Goodrich & Strutt, 2011; Humphries & Mlodzik, 2018; Peng & Axelrod, 2012; Yang & Mlodzik, 2015). Generally, it has been demonstrated that the polarization axes are generated by a localized expression of Wnt, with cells orienting towards the Wnt source (for example Carvajal-Gonzalez, Mulero-Navarro, & Mlodzik, 2016; Gao et al., 2011; Minegishi et al., 2017; Wu, Roman, Carvajal-Gonzalez, & Mlodzik, 2013).
Once a polarization axis is established, largely by the interactions between the core PCP components, the asymmetric localization of the core complexes then regulates downstream effectors to elicit tissue-specific responses affecting both cytoskeletal orientation (including the alignment of the mitotic spindle and localization centrioles and cilia) and nuclear signaling (to induce cell fate) (reviewed in Aw et al., 2016; Butler & Wallingford, 2017; Carvajal-Gonzalez et al., 2016; Goodrich & Strutt, 2011; Humphries & Mlodzik, 2018; Peng & Axelrod, 2012; Segalen & Bellaiche, 2009; Wallingford, 2010). Importantly, not only the core Wnt-Fz/PCP factors are conserved across the animal kingdom, but also the set of downstream cellular responses and read-outs that, like the molecular PCP (see references above). Two of the most obvious and common read-outs (i) polarization of acto-myosin cytoskeletal elements and (ii) positioning of the mitotic spindle/centriole/cilia are discussed in several reviews (see references above). Here, we focus on the nuclear signaling mechanisms of the Wnt/Fz-PCP pathway and compare it to other nuclear Wnt-signaling mechanisms. This read-out, although it shares the membrane proximal regulatory input, can largely be separated from the others downstream of the action of the Dsh/Dvl protein family.
1.3. Wnt/calcium signaling
There are several proposed additional non-canonical Wnt-signaling pathways beyond PCP signaling (see Snapshot: Semenov et al., 2007), which include specific functions of Wnt-signaling in neuronal pathfinding (also linked to PCP) or synaptic functions and bouton formation or stability (reviewed for example in McLeod et al., 2020; McLeod & Salinas, 2018; Teo & Salinas, 2021; Zou, 2020). Of these additional pathways, the Wnt/calcium signaling pathway has had significant attention and is molecularly quite well defined (De, 2011; Kohn & Moon, 2005; Kuhl, Sheldahl, Park, et al., 2000).
The Wnt/calcium pathway leads to the activation of calcium/calmodulin-dependent kinase II (CamKII) and PKC (see Fig. 3 for schematic overview). In this pathway, PKC activates actomyosin cytoskeletal elements via cdc42 and its downstream effectors. The nuclear branch of the Wnt/calcium pathway acts via CamKII and Calcineurin (CNA) on rather general nuclear factors like ATF, NF-AT or NFκB. Wnt/calcium signaling has been suggested to be activated by Wnt5a, in particular, and its analyses have mostly been performed in non-mammalian vertebrate model systems, namely zebrafish and frog/Xenopus (Kohn & Moon, 2005; Kuhl, Sheldahl, Park, et al., 2000). For example, Wnt5 has been linked to intracellular calcium release. Such intracellular calcium signaling is required for body plan specification and shown to affect dorsalization of embryos, and accordingly, zebrafish embryos deficient for Wnt5 display reduced fluxes of calcium and dorsalization defects. Interestingly, such Wnt5 mutant zebrafish embryos (loss-of-function), which display a hyperdorsalization, can in part be rescued by the expression of a constitutively active CamKII, a key proposed effector of the Wnt/calcium pathway (Westfall et al., 2003). Consistent with the studies in zebrafish, expression of dominant negative isoforms of either Wnt11 or CamKII lead to embryonic dorsalization in Xenopus (Kuhl, Sheldahl, Malbon, & Moon, 2000). Other developmental contexts where the Wnt/calcium pathway has been suggested to function include mammalian T-cell differentiation. The NF-AT transcription factor is a nuclear effector regulated by the calcium/calmodulin-dependent phosphatase called Calcineurin/CNA. Co-expression of NF-AT with Wnt5a in Xenopus induces nuclear translocation of NF-AT during embryonic dorsalization (reviewed in (Rao, Luo, & Hogan, 1997). Similarly, treatment of T-cells with Wnt5a results in nuclear translocation and accumulation of NF-AT (Murphy & Hughes, 2002). Taken together, these data are consistent with the notion that a Wnt5/calcium pathway uses CNA to regulate nuclear accumulation of NF-AT as a pathway read out.
Fig. 3.
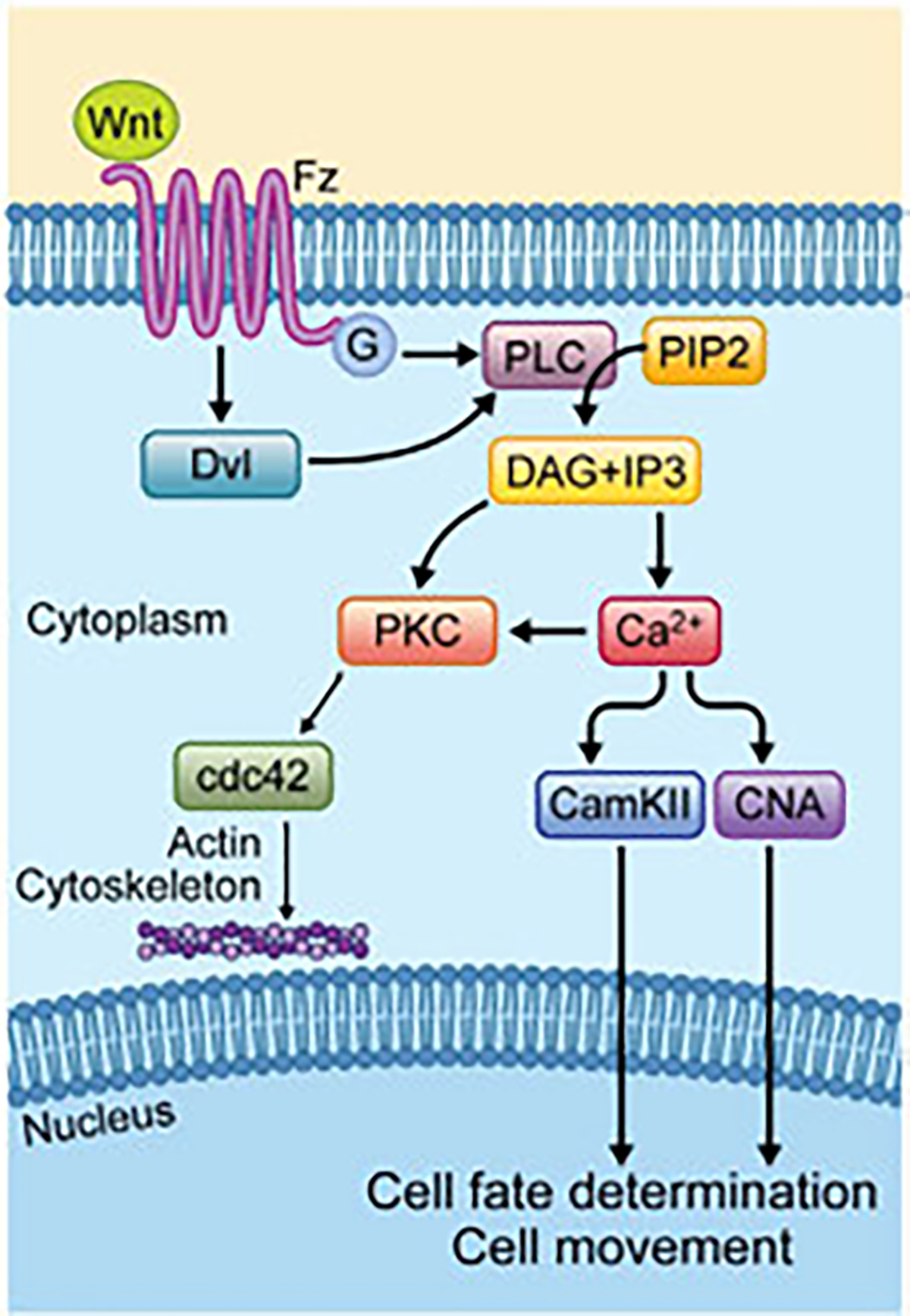
Overview of the Wnt-Ca2+ pathway. Wnt ligand-Fz binding is mediating Dvl activation with possibly trimeric G proteins-coupled to the process. Dvl also can activate PLC to decompose PIP2 into DAG and IP3. DAG is activated by released calcium from ER, which activates PKC, which then activates Cdc42 to regulate actomyosin cytoskeletal elements. In parallel, IP3 leads to intracellular calcium release via ER, which in turn results in phosphorylation and activation of CamKII and CNA. These proteins activate NFkB and NFAT which translocate to the nucleus to regulate cell movement and cell fate decisions.
It has also been suggested that trimeric G-proteins are part of the Wnt/calcium pathway (summarized in (Semenov et al., 2007). Potential evidence for that comes from a study in Drosophila: here a trimeric G-protein complex, Gαo, has been linked to Wnt/Wg signaling in the context of wing development (Katanaev, Ponzielli, Semeriva, & Tomlinson, 2005). However, a link to calcium signaling has not been made in the Drosophila study, and so the proposed Wnt/calcium pathway based on experiments in fish and frogs has not been reconstructed in Drosophila or mice. Whether the potentially associated trimeric G-proteins, as suggested from a study in Drosophila (Katanaev et al., 2005), contribute to these effects or act independently of or parallel to these kinases is not resolved, nor is a formal link established of this trimeric G-protein unit to the Wnt/calcium-pathway.
The direct molecular information flow is less well defined in this Wnt pathway, as compared to canonical Wnt or Wnt/PCP signaling. Interestingly, some of the proposed components of this pathway have been linked to it through derivative analyses in heterologous systems and basically the pathway takes advantage of other existing signaling modules, which were defined in signaling contexts outside to Wnt signaling (reviewed in Kohn & Moon, 2005; Kuhl, Sheldahl, Malbon, & Moon, 2000). Thus, in general, information flow here has been largely derived from the common knowledge base, as again it uses components of other signaling mechanisms (see reviews Kohn & Moon, 2005; Kuhl, Sheldahl, Malbon, & Moon, 2000; Semenov et al., 2007). As such, downstream of Dsh/Dvl, the PLC enzyme gets activated which converts PIP2 into the second messengers di-acylglycerol (DAG) and inositol-3-phosphate (IP3). These either activate PKC directly (DAG) or lead to an increase in intracellular calcium/Ca2+, which in turn activates CamKII and CNA, for example. In summary, Wnt/calcium signaling redeploys these existing, more general, signaling modules, for example a PLC-DAG-PKC activation sequence or PLC-IP3-Ca2+-CAMKII signaling flow, as downstream effector modules of a Wnt-Frizzled-Dsh/Dvl interaction. Similarly, the resulting effectors that would act in the context of cytoskeletal regulation, e.g. PKC-cdc42 etc, or in nuclear contexts, resulting in the activation of NF-AT, ATF2 or NFκB for example, are rather generic cellular or nuclear effectors used downstream of several other pathways.
2. Induction of nuclear responses downstream of Wnt-signaling
2.1. Mechanisms of canonical Wnt/β-catenin signaling in the nucleus
2.1.1. Wnt/β-catenin mediated transcriptional activation
Upon activation of Wnt signaling, the inhibition of β-catenin destruction leads to increased levels of β-catenin in the cytoplasm, and subsequent β-catenin translocation into the nucleus (Cadigan & Waterman, 2012; MacDonald et al., 2009). Once in the nucleus, β-catenin associates with TCF/Lef through its central region containing the Arm-repeats to activate transcription of Wnt/β-catenin specific target genes (Fig. 4) (Behrens et al., 1996; Cadigan & Waterman, 2012; MacDonald et al., 2009). In the absence of β-catenin, TCF recruits a transcriptional repressor complex with TLE (Groucho in Drosophila), which promotes histone deacetylation and chromatin compaction (Cavallo et al., 1998; Daniels & Weis, 2005). In Drosophila wing imaginal discs, the high threshold Wg transcriptional target gene senseless (sens) directly controls the formation of adult wing margin sensory bristles, while Distalless (Dll) and vestigial (vg) act as medium and low threshold Wg target genes, respectively, and are required for adult wing growth and cell survival (Neumann & Cohen, 1997; Zecca et al., 1996). β-catenin/Arm acts as a transcriptional co-activator and, besides the TCF/Lef transcription factor, also has other nuclear binding partners that play critical roles in the activation of Wnt target genes. For example, through its C-terminal transactivation (TA) domain it interacts with CREB binding protein (CBP)/p300 (Sustmann, Flach, Ebert, Eastman, & Grosschedl, 2008) and SET domain-containing protein 1 (SET-1) (Sierra, Yoshida, Joazeiro, & Jones, 2006) among others (Fig. 4). β-catenin/Arm also interacts directly with BCL9, legless (lgs) in Drosophila (Hoffmans & Basler, 2007). Unlike most other co-activators binding β-catenin/Arm, BCL9/Lgs interacts with the first (most N-terminal) Arm repeat of β-catenin, which is specifically required for β-catenin-dependent transcription and it acts to bridge the complex of TCF-β-catenin/Arm-coactivators to activate transcription of Wnt target genes (Fig. 4) (Mosimann, Hausmann, & Basler, 2009). In Drosophila, the BCL9/Lgs protein is essential for Wg signaling and it acts by recruiting an additional protein, Pygopus, which is believed to provide a link to transcriptional initiation of Wnt signaling target genes. Lack of either BCL9/Lgs or Pygopus phenocopies Arm/β-catenin null mutants (Kramps et al., 2002; Thompson, Townsley, Rosin-Arbesfeld, Musisi, & Bienz, 2002).
Fig. 4.
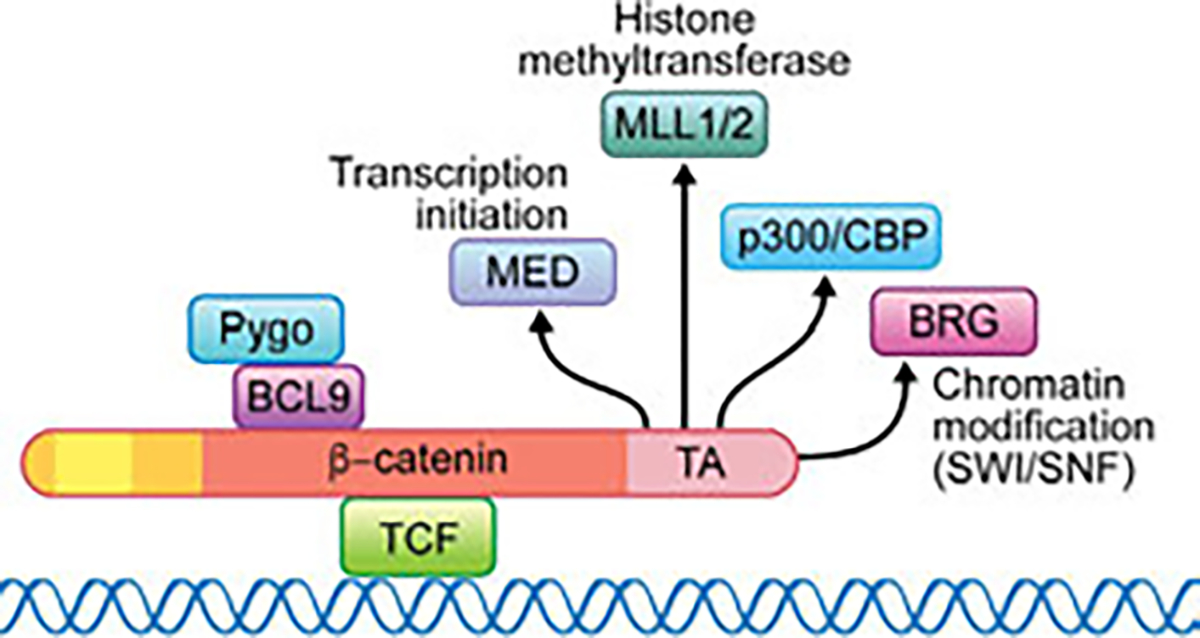
TCF and β-catenin and its co-activator complexes in the nucleus. Upon Wnt signaling activation, β-catenin translocates to the nucleus and binds to TCF. The TCF/β-catenin complex recruits many co-activators required for the initiation and maintenance of active gene expression. These include for example, but are not restricted to, BCL9 (Legless/Lgs) and Pygopus (Pygo), interacting within the Arm repeat region of β-catenin (red region of β-catenin in schematic), and Mediator (for transcription initiation), p300/CBP, MLL1/2 histone methyltransferase and BRG (for chromatin modification (SWI/SNF) in the C-terminal transactivation (TA) region of β-catenin. Note that while the central Arm repeats region of β-catenin (red) associates with TCF, and the more N-terminal Arm-repeats bind to BCL9, most of other co-activator complexes interact with β-catenin in its C-terminal TA domain, creating a dazzling interplay between β-catenin and the transcriptional apparatus and the chromatin. Note that no nuclear interactors have thus far been identified for the most N-terminal region of β-catenin (N-terminal to the Arm repeats, orange and yellow in schematic).
Without question β-catenin is the main regulator of Wnt signaling in the nucleus. However, it is poorly understood how β-catenin is retained inside the nucleus. It has been proposed that the maintenance of β-catenin level in the nucleus is regulated by TCF and BCL9/Lgs via multiple mechanisms. TCF/Lef can enrich β-catenin in the nucleus (Behrens et al., 1996; Huber et al., 1996), and BCL9 and its nuclear interactor Pygopus can also strongly bind to β-catenin and maintain its levels high in that cell compartment (Kramps et al., 2002; Townsley, Cliffe, & Bienz, 2004). This function is tightly regulated through phosphorylation of β-catenin at Tyrosine 142, which not only diminishes β-catenin affinity to α-catenin in the E-cadherin junctional complex but also enhances β-catenin binding to the transcriptional coactivator BCL9/Lgs (Brembeck et al., 2004). This leads, in conjunction with TCF/Lef, to increased transcriptional activity. Another protein that has been proposed to affect β-catenin nuclear localization is the protein kinase AKT, which can phosphorylate β-catenin at Serine 552 and thus promote its nuclear localization (Fang et al., 2007). However, the mechanism by which these factors regulate and keep β-catenin localized in the nucleus, when Wnt is activated, has not been elucidated.
2.1.2. Wnt/β-catenin mediated transcriptional repression
It has been shown that activation of the Wnt/β-catenin pathway not only stimulates transcription but can also repress certain genes. In Drosophila, nearly 20% of Wnt/Wg responsive genes are predicted to be repressed by Wnt/Wg signaling (Franz, Shlyueva, Brunner, Stark, & Basler, 2017). Wnt-repressed genes have TCT/Lef binding sites, but they are distinct from the classical Wnt response Elements (WRE). For these genes, TCF promotes transcription via recruitment of coactivators in the absence of β-catenin; and, in the presence of β-catenin, the formation of a TCF/β-catenin complex represses transcription via recruitment of corepressors (Archbold, Broussard, Chang, & Cadigan, 2014). For example, the Drosophila stripe gene, whose TCF binding site overlaps with a binding site for Cubitus interruptus (Ci), a Hedgehog pathway dependent transcription factor that is essential for stripe transcription. Binding of the TCF/β-catenin complex most likely displaces Ci and thus represses transcription (Piepenburg, Vorbruggen, & Jackle, 2000). It will be interesting to further dissect how the different TCF motifs and β-catenin/Arm regulatory factors bring about activation or repression in a context dependent manner.
2.2. Nuclear branch of Wnt/planar cell polarity (PCP) signaling
During Drosophila eye development, Fz/PCP signaling regulates two patterning process. It establishes specific cell fates in the 5-cell photoreceptor precluster, which governs the general ommatidial patterning via nuclear signaling activating transcription of specific target genes (reviewed in ( Jenny, 2010; Mlodzik, 1999), the mechanism of which will be discussed here, and subsequently it regulates a directed movement, ommatidial rotation, of the photoreceptor cluster, or ommatidium (reviewed in for example (Davey & Moens, 2017; Seifert & Mlodzik, 2007)). Wnt-PCP nuclear signaling has been primarily dissected in the Drosophila eye, and hence we focus on this tissue for its description. However, importantly, it is conserved in other species and contexts (Veeman, Slusarski, Kaykas, Louie, & Moon, 2003).
The Drosophila eye consists of ~800 regularly arranged ommatidia, each consisting of 8 photoreceptor neurons arranged into an invariant trapezoidal pattern and 12 accessory (cone, pigment, and bristle) cells. During larval stages, the eye develops from an epithelial imaginal disc, which is initially composed of identical pluripotent precursor cells. Development and differentiation is initiated when a wave of cell proliferation and differentiation (called morphogenetic furrow, MF) moves across the eye imaginal disc from posterior to anterior; the furrow leaves in its wake evenly spaced preclusters, which comprise initially a 5-cell precluster consisting of the R8, R2/R5, and R3/R4 photoreceptor neurons, of differentiating groups of cells that will mature into ommatidia, consisting of all 8 R-cell neurons and accessory cells (Heberlein, Wolff, & Rubin, 1993; Roignant & Treisman, 2009; Tomlinson & Ready, 1987; Wolff & Ready, 1991). The critical PCP signaling aspect involves, posterior to the furrow, an inductive signaling event specifying the R3 fate within the R3/R4 cell pair, and the associated transcriptional upregulation of R3 specific genes (including upregulation of the Notch ligand Dl to induce the neighboring cell as R4; reviewed in ( Jenny, 2010; Mlodzik, 1999)). In addition, and unrelated to the nuclear branch, Wnt/PCP signaling also coordinates the direction and degree of ommatidial rotation via input to cytoskeletal and cell adhesion regulation. The coordinated induction of R3 in every ommatidial precluster, and the resulting asymmetric, directed signaling to R4, leads to a very regular ommatidial arrangement, with identical chiral forms in each ommatidium in the respective half of the eye, and opposite chiral forms in the ventral and dorsal halves. This type of mirror-symmetric arrangement of ommatidia is thought to be coordinated by Wnt/PCP signaling with Wg and Wnt4, both being expressed on the dorsal and ventral pole of the eye field, providing positional information (reviewed in Jenny, 2010; Mlodzik, 1999; Roignant & Treisman, 2009; Wu et al., 2013).
As discussed above, due to the coordinated inductive nuclear signaling requirement from R3 to R4, loss-of-function (LOF) mutations in core PCP genes generally randomize cell fate specification within the R3/R4 pair (and thus result in random chiral ommatidial arrangements) or even complete loss of chirality, giving rise to R3-R3 or R4-R4 symmetrical ommatidia (reviewed in Jenny, 2010; Mlodzik, 1999), see also (Boutros et al., 1998; Das et al., 1998; Jenny et al., 2005; Rawls & Wolff, 2003; Wolff & Rubin, 1998; Zheng, Zhang, & Carthew, 1995). Similarly, interference with PCP signaling via overexpression of the core components causes phenotypes similar to LOF defects, affecting again R3/R4 cell fate specification and subsequently rotation of ommatidia (Das, Reynolds-Kenneally, & Mlodzik, 2002; Fanto & Mlodzik, 1999; Jenny et al., 2005; Rawls & Wolff, 2003).
The Drosophila eye imaginal disc giving rise to the precisely patterned retina has provided the first and still the most precise insight on how Wnt-Fz/PCP signaling employs a nuclear response. Following an initial bias (presumably provided from the Wnt source at the poles of the eye field) and due to an upregulation of core PCP factors in precursor cells of the R3/R4 pair (Djiane, Yogev, & Mlodzik, 2005), the Fz-Dsh-Dgo complex is enriched on the R3 side of the R3/R4 border, whereas the Vang-Pk complex (inhibiting Fz-Dsh) is enriched on the R4 side (Fig. 2). As such Fz-Dsh signaling is enhanced in R3 and repressed in R4 leading to obvious differences in Fz-Dsh/PCP signaling activity between the two initially equivalent R3/R4 precursors (Djiane et al., 2005). Thus, the cell with the higher Fz-Dsh/PCP activity always becomes specified as R3 via a nuclear signaling branch and then induces its neighbor within the R3/R4 pair to adopt the R4 fate (via Notch signaling activation in R4, (Cooper & Bray, 1999; del Alamo & Mlodzik, 2006; Fanto & Mlodzik, 1999; Tomlinson & Struhl, 1999)). Importantly, specification of R3 and R4 depend on transcriptional responses in the R3 precursor generated by Fz-Dsh/PCP signaling, directly (R3) and indirectly (R4), respectively.
How is a Fz-Dsh/PCP signal relayed to the nucleus? First indications that such a pathway branch exists came from the realization that Fz-Dsh/PCP signaling acts through Rho family GTPases and can activate the JNK-type MAPK cascade (Boutros et al., 1998; Strutt, Weber, & Mlodzik, 1997). Strikingly, these studies revealed that the Dsh isoforms/truncations containing domains that are required in PCP signaling also activate the JNK MAPK-cascade in both cell culture assays and in vivo in Xenopus (Boutros et al., 1998; Shulman, Perrimon, & Axelrod, 1998). These insights came initially from gain of function assays, but they were quickly confirmed in genetic LOF interactions in Drosophila (Boutros et al., 1998; Paricio, Feiguin, Boutros, Eaton, & Mlodzik, 1999). Subsequently, a key requirement for Rho-family GTPases, as downstream effectors of Fz and Dsh in nuclear PCP signaling, was established, which confirmed the existence of such a pathway and provided new insights into it (Boutros et al., 1998; Fanto & Mlodzik, 1999; Paricio et al., 1999; Strutt et al., 1997), see below for more detail. The combination of these studies and related experiments in vertebrates established not only that the canonical Wnt-pathway and Wnt-PCP signaling diverge at the level of Dsh, downstream of the Wnt-Frizzled interactions, but also firmly linked Dsh to the activation of a JNK-type MAPK cascade via Rho-family GTPases (reviewed in Boutros & Mlodzik, 1999; Wallingford & Habas, 2005).
The above insights led the way to further dissecting the requirements of the Rho GTPases as Dsh effectors. The initial identification of RhoA (a.k.a. Rho1 in Drosophila) as a mediator of PCP signaling and characterization of its mutant alleles (Strutt et al., 1997) already suggested that RhoA is likely to act on both the cytoskeletal branch and a nuclear branch of the Fz-Dsh-RhoA PCP pathway. This expectation of a pathway split at the level of RhoA was subsequently confirmed and refined by demonstrating that one of the RhoA effectors, Rho-kinase (Rock; dROK in Drosophila), only affects cytoskeletal signaling in both the Drosophila eye and wing (Winter et al., 2001), while RhoA itself also affects nuclear signaling (Boutros et al., 1998; Strutt et al., 1997). A detailed dissection of RhoA signaling mutants, which allow to distinguish between cytoplasmic and nuclear signaling branches, further confirmed this notion, and defined RhoA mutant isoforms that acted predominantly via nuclear effectors and these affected the transcription dependent cell fate outcomes within the R3/R4 pair (Fanto, Weber, Strutt, & Mlodzik, 2000). Subsequent work analyzing the potential function of the Rac GPTases confirmed this and also pointed to a nuclear effect of this in R3/R4 cell fate induction (Munoz-Descalzo, Gomez-Cabrero, Mlodzik, & Paricio, 2007). In both cases, the GTPases activate the JNK-cascade, which translocates into the nucleus to act as an activator of the AP-1 transcription factor, consisting of Jun and Fos (reviewed for example in (Kockel, Homsy, & Bohmann, 2001)).
Jun and Fos, the dimerization partners in the AP-1 transcription factor (reviewed in (Kockel et al., 2001; Shaulian & Karin, 2001), are required to both specify R3 (Weber, Paricio, & Mlodzik, 2000) and also to upregulate transcription of Dl and neuralized (neur) in R3 to induce the R4 fate in the neighboring cell (Cooper & Bray, 1999; del Alamo & Mlodzik, 2006; Fanto & Mlodzik, 1999; Weber et al., 2000). In this nuclear signaling branch, the Wnt-PCP pathway takes advantage of an existing nuclear signaling module, activating a MAPK (JNK)-kinase cascade with ultimately JNK, the MAPK, entering the nucleus to phosphorylate and activate the AP-1 members Jun and Fos (kayak/kay in Drosophila). This in turn allows the Jun-Fos homo- or hetero-dimers to recruit coactivators and turn on transcription of tissue and cell type specific target genes in the R3 cell (Weber et al., 2000; Weber, Pataki, Mihaly, & Mlodzik, 2008) and other contexts (Veeman et al., 2003). Thus, the Wnt/PCP pathway uses a rather generic module/set of factors for its nuclear response, one that is also used downstream of several other signaling pathways. There is slight a variation of the theme though, as the common link of the MAPK cascade to AP-1 (Jun/Fos) requires factors from both families, Jun and Fos (for example in RTK/Ras/MAPK signaling), downstream of nuclear Wnt/PCP signaling Jun appears at least in part dispensable, as displayed by redundancy with Fos (Weber et al., 2000). As in Drosophila there is only one family member for each factor, Jun and Fos (kay in Drosophila), this is real redundancy and cannot be explained by another Jun family member taking over. Moreover, there is no such redundancy in other signaling context in Drosophila, notably dorsal closure and thorax closure for example, where both Jun and Fos are essential (Kockel, Zeitlinger, Staszewski, Mlodzik, & Bohmann, 1997; Zeitlinger et al., 1997). As such this variation of the JNK-module signaling to Jun/Fos is unique in the Wnt/PCP context.
Similar to the MAPK-Jun/Fos signaling module downstream of other pathways, the Wnt-PCP-JNK branch signaling through Jun/Fos cooperates with other nuclear signaling pathways in its response. For example, in the context of the R3 and R4 cell fate specification, nuclear Wnt/PCP signaling acts in concert of Egfr (RTK)/Ras/MAPK signaling and its nuclear effectors of the ETS transcription factor family (Weber et al., 2008), and its effects on Notch signaling in R4 are amplified through feedback loops that also affect R3. It is worth noting that a similar link between Wnt/PCP signaling and asymmetric Notch pathway activation has been documented in the fish lateral line neuromasts—the hearing and balance organ of fish—which is also comprised of a multicellular sensory unit system comparable to the ommatidia. Here also the Wnt/PCP pathway sets up the direction of Notch signaling (Dale, Sisson, & Topczewski, 2009; Mirkovic, Pylawka, & Hudspeth, 2012; Navajas Acedo et al., 2019). Possibly, Wnt/PCP signaling and the Notch pathway also link up to govern the asymmetric formation of the feather buds in birds (Crowe, Henrique, Ish-Horowicz, & Niswander, 1998; Widelitz et al., 1999).
In summary, the core Fz/PCP nuclear signaling branch uses a well-known (and not pathway specific) set of effectors, both cytoplasmic kinases and nuclear factors, and it also cooperates with several other signaling pathways during R3/R4 specification, and likely in general. Thus the analysis of nuclear Wnt/PCP signaling also underscores how multiple signaling pathways and local cellular interactions are linked in achieving precision in a highly regulated cell fate induction process.
3. How does β-catenin get transported into the nucleus
3.1. New insights into β-catenin nuclear translocation
β-catenin is a multifunctional protein associated with adherens junctions, linking these to the actomyosin cytoskeleton, and canonical Wnt signaling, where it acts as a nuclear co-factor (see above). As such its subcellular localization needs to be very tightly regulated. The entry of β-catenin into the nucleus is a key signaling step in the Wnt/β-catenin pathway. Although the high concentration of free cytoplasmic β-catenin appears to be a prerequisite for its subsequent nuclear translocation, it does not explain how β-catenin gets into the nucleus to promote transcription (Henderson & Fagotto, 2002; Stadeli, Hoffmans, & Basler, 2006). In the classic nuclear transport model, nuclear localization sequence (NLS) containing proteins bind to Importin-α, which is brought to the nuclear pore complex (NPC) and transported through the nuclear pore via the interaction with Importin-β. With the assistance of Ran GTPase, the cargo is then moved through the nuclear envelope and released inside the nucleus (Fagotto, Gluck, & Gumbiner, 1998; Yokoya, Imamoto, Tachibana, & Yoneda, 1999). However, β-catenin has no identifiable NLS (Suh & Gumbiner, 2003). It has been shown that β-catenin can enter the nucleus in a Ran-independent manner (Yokoya et al., 1999). Since β-catenin and Importin share similar Armadillo repeat domains (Kutay, Bischoff, Kostka, Kraft, & Gorlich, 1997), β-catenin might directly interact with nuclear core components, bypassing the β-importin/karyopherin proteins (Fagotto et al., 1998; Yokoya et al., 1999). Another proposed model for the nuclear translocation of β-catenin is its transport into the nucleus in a complex with other proteins, such as FOXM1 or BCL9 (Townsley et al., 2004). Once in the nucleus, interaction with TCF, BCL9 and Pygopus may function as a bridge to β-catenin (Krieghoff, Behrens, & Mayr, 2006; Tolwinski & Wieschaus, 2001). Furthermore, a role for Rac1 GTPase and Jun N-terminal kinase 2 (JNK2) has been suggested in the nuclear localization of β-catenin upon Wnt pathway activation, although it remains unclear how Rac1, normally linked to Wnt/Fz-PCP signaling (see above), is involved in canonical Wnt signaling (Wu et al., 2008).
Recently, a genetic screen to test for potential roles of ciliary proteins in non-ciliated epithelial cells in Drosophila identified Intraflagellar Transport complex A (IFT-A) proteins to modulate canonical Wnt/Wg signaling, independently of the ciliary role of IFTs (Balmer et al., 2015). Among the five conserved core IFT-A proteins in Drosophila, mutants for four of the corresponding genes, IFT122, IFT121, IFT140 and IFT143, displayed growth defects and wing notches or missing wing margin bristles in adult wings, indicating that they play a role in Drosophila wing development and canonical Wg signaling. It remained unclear whether the cytoplasmic IFT-A proteins associate with microtubular structures and whether such association is required for their function in Wnt/Wg signaling. Interestingly, we have shown separately that Klp64D, the Drosophila motor protein Kinesin 2 subunit, also plays a role in Wg/Wnt signaling and Arm/β-catenin trafficking (Vuong et al., 2014). Klp64D recruits Arm/β-catenin and its interacting partner Dishevelled (Dsh) for Wingless signal transduction. In the absence of Kinesin 2 function, Arm/β-catenin abnormally accumulated in the cytoplasm, indicating that Klp64D might be involved in trafficking of Arm/β-catenin for Wg/Wnt signaling pathway.
Subsequent studies demonstrated that IFT-A proteins, and IFT140 in particular, associate with Kinesin 2 and promote nuclear translocation of β-catenin upon Wnt/Wg pathway activation (Vuong et al., 2018). This requirement, acting downstream of β-catenin stabilization, has been also shown to be conserved from Drosophila to mammalian cells, e.g. mouse embryonic fibroblasts (Vuong et al., 2018). Loss of function mutants of either Kinesin 2 or IFT-A proteins in Drosophila wing tissues displayed indistinguishable effects on Wnt target gene expression and wing development. Kinesin 2 and IFT140 interact genetically and physically with each other. Kap 3, a non-motor component of Kinesin 2, directly interacts with IFT140, and acts as the cargo domain to transport IFT140 along microtubules. Both single mutant clones and double mutant clones for kinesin 2 and ift140 or kap 3 fail to activate Wg/Wnt signaling targets in Drosophila. Moreover, double mutant clones for either kinesin 2 or IFT140 with axin in Drosophila displayed high levels of stabilized cytoplasmic Arm/β-catenin in both wing imaginal disc cells and salivary glands, but its nuclear translocation was markedly reduced as nicely visualized in the large salivary gland cells (Vuong et al., 2018). Similarly, ift140−/− mutant mouse embryonic fibroblast (MEFs) showed a striking reduction of nuclear β-catenin upon Wnt3a mediated pathway activation, which importantly was not affected in control ift88 mutant MEFs (IFT88 is a component of IFT-B), confirming the non-ciliary role of IFT-A in Wnt-signaling in mammalian contexts. Taken together, these data suggest a conserved non-ciliary role for the Kinesin 2/IFT-A complex in transporting Arm/β-catenin to the nucleus (Vuong et al., 2018).
In this context, IFT140 and Arm/β-catenin directly interact through a small domain (53 amino acids) at N-terminal region of Arm/β-catenin, Arm34–87 (Vuong et al., 2018). The deletion of this small region of Arm is a known mutant stable isoform, ArmS10, which is commonly used in Drosophila as a “constitutively active” isoform of Arm/β-catenin (Blauwkamp, Chang, & Cadigan, 2008). Strikingly, ArmS10 can enter the nucleus independently of the Kinesin 2/IFT140 function, which suggests that this small region is critical for the differential behavior relative to wild-type stable Arm/β-catenin in terms of Kinesin 2/IFT140 effects.
Taken together, these findings allowed us to propose a new molecular mechanism for the entry of Arm/β-catenin to the nucleus. When Wg/Wnt signaling is activated, the complex of Kinesin 2/Kap3/IFT-A binds cytoplasmic Arm/β-catenin via IFT140 and this complex might then be transported along the microtubules to the nucleus (Fig. 5) (Vuong et al., 2018). Once near (or in) the nucleus, Arm/β-catenin may be released from the IFT-A complex to bind to TCF/Lef to mediate transcriptional activation of Wnt target genes. Moreover, as binding of Arm/β-catenin to the Kinesin2/IFT-A complex promotes nuclear localization, it is likely that IFT140/IFT-A with other proteins for Arm/β-catenin binding to the specific domain in the N-terminal region of Arm/β-catenin. Such cytoplasmic proteins could trap/sequester Arm/β-catenin in the cytoplasm, and therefore limit its capacity to enter the nucleus.
Fig. 5.
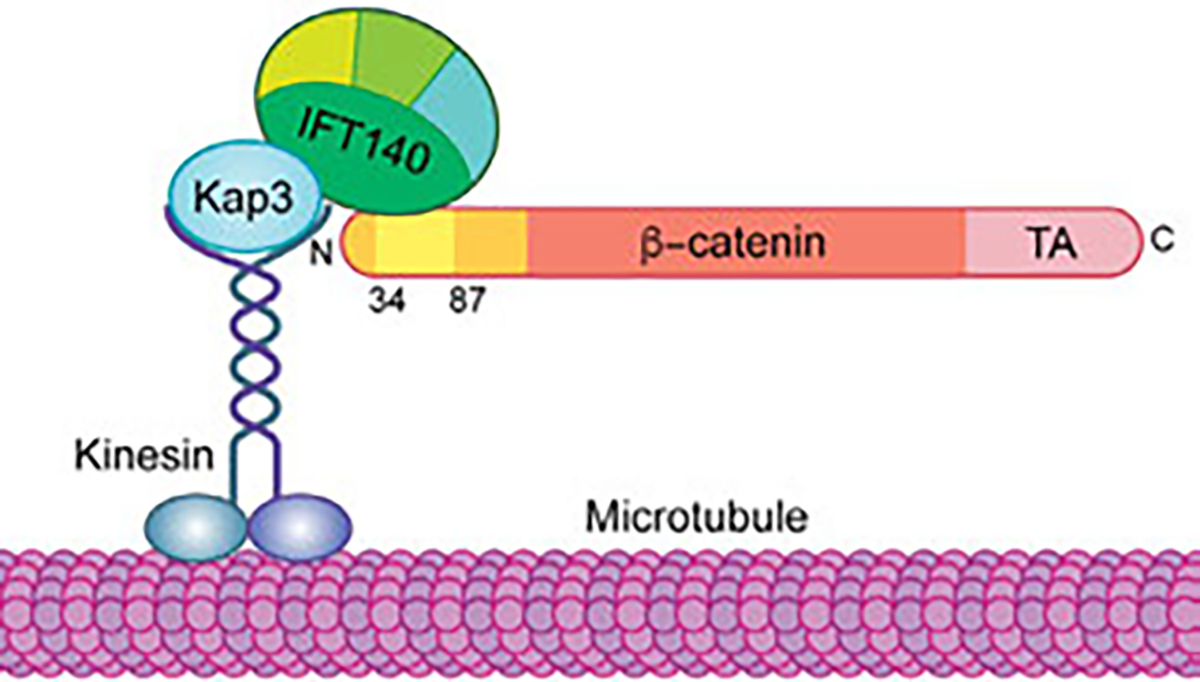
Proposed model for β-catenin associated complex that mediates its translocation to the nucleus. Upon Wnt/β-catenin signaling activation, Kinesin 2 interacts with IFT-A via a direct Kap3-IFT140 interaction. Note that this complex forms on cytoplasmic microtubules. IFT140 also binds free cytoplasmic β-catenin in the N-terminal region of β-catenin (yellow in schematic), thus recruiting β-catenin to the Kinesin 2/IFT-A complex, which could allow its movement along microtubule towards the nucleus. See main text for details.
3.2. Missing information in the context β-catenin nuclear translocation
Our study (Vuong et al., 2018) has identified a novel mechanistic function of the kinesin2/IFT-A complex in Wnt signaling, independent of their role in the cilium. The Kinesin 2/IFT-A complex might promote nuclear translocation of Arm/β-catenin by two unrelated mechanisms: (i) protecting it from binding by a cytoplasmic inhibitor/retention factor, and (ii) moving it along microtubules to the nucleus. As these mechanistic features of the complex are conserved in mammalian cells, these exciting observations are potentially amenable to therapeutic treatment.
Nonetheless, several questions remain and should be the focus of future studies in Drosophila and in mammalian contexts: (i) How does the Kinesin-2/IFT-A complex promote nuclear localization of Arm/β-catenin? While the existing data suggest that the complex moves along microtubules to transport β-catenin to the nucleus (Vuong et al., 2018), this remains a hypothesis and needs to be confirmed by live imaging in Drosophila and mammalian cells. (ii) What is the identity of the antagonistic factors that compete with IFT140 for binding to the β-catenin/Arm N-terminal region, and do they really serve an inhibitory function for β-catenin/Arm nuclear localization as cytoplasmic retention factors? The APC/Axin complex was suggested as a cytoplasmic anchor, keeping β-catenin/Arm out of the nucleus (Tolwinski & Wieschaus, 2001) and our data in the salivary glands assay are consistent with this function (Vuong et al., 2018). However, Axin can be excluded as the inhibitory factor competing with IFT140, because in the absence of Axin, β-catenin/Arm fails to translocate to the nucleus in Klp64D or IFT140 mutant backgrounds. Efforts to identify IFT140 competing factors await further investigation. And (iii), it remains to be tested whether the small N-terminal domain of β-catenin/Arm, required for binding to IFT140 (Arm34–87), plays a critical role in Wg/Wnt signaling. If so, it might serve as an entry point for potential new drug development and/or therapeutic application to inhibit an overactive Wnt/β-catenin signaling scenario.
4. Concluding remarks
β-catenin (Armadillo/Arm in Drosophila) is a multitasking and evolutionary conserved molecule that exerts a crucial role in a multitude of developmental and homeostatic processes in metazoans. β-catenin has been established to be a critical component of the activation of the Wnt signaling pathway. In this canonical Wnt pathway, β-catenin is the key effector responsible for transduction of the signal to the nucleus, as it triggers transcriptional responses of Wnt-specific target genes and is thus responsible for the control of cell fate decisions and growth in many cells and tissues. The second well defined type of Wnt signaling, the Wnt/PCP pathway, is independent of β-catenin function. β-catenin is however not just a component of the Wnt signaling cascade. In the late 1980s, β-catenin was identified together with two other molecules (∝-catenin and γ-catenin/plakoglobin), as critical proteins associated with E-cadherin, the key molecule of Ca2+ - dependent cell adhesion (Ozawa, Baribault, & Kemler, 1989). How can β-catenin as a single molecule affect so many cellular phenomena in animals, and regulate and mediate so many different outputs and read-outs? It is a complex question that will require many more studies to be fully answered.
In this review chapter, we have mainly focused on the role of β-catenin in the canonical Wnt/β-catenin signaling pathway and how it can translocate into the nucleus to activate the target genes of the pathway. As we proposed, upon Wnt signaling activation, the complex of Kinesin-2/Kap3/IFT-A binds to β-catenin and transports it to the nucleus by moving along cytoplasmic microtubules. The binding of β-catenin to microtubules has been first reported during neuronal morphogenesis in mice. In this context, β-catenin is transported to microtubule ends in a complex with APC and N-cadherin. Kap3 is required for localization of β-catenin to the plasma membrane of neuroepithelial cells (Teng et al., 2005). Binding of APC to Kap3 is required for localization of APC to the tip of epithelial extensions ( Jimbo et al., 2002). However, it remains unclear whether these proteins are transported in the same kinesin complex and, importantly, how APC might regulate the transport of β-catenin and N-cadherin. Furthermore, APC has also been reported to play a role in the transport of β-catenin to the nucleus upon activation of Wnt signaling (Tolwinski & Wieschaus, 2001). Therefore, future studies should tackle these questions to investigate the mechanism of how the Kinesin2/IFT-A complex might promote β-catenin translocation to the nucleus and whether there are other proteins involved in this complex. Moreover, since the N-terminal region of β-catenin is required for direct binding to IFT140 (within the IFT-A), it is necessary to establish the role of this domain of β-catenin in the nuclear translocation and the Wnt signaling pathway in general. A better understanding of the β-catenin structure, as this N-terminal region is thus far reported as unstructured, as well as the identification and characterization of new factors involved in nuclear Wnt signaling will not only provide significant new insights into the mechanism(s) of how β-catenin is translocated and retained inside the nucleus, but might also facilitate therapeutic approaches and development of drugs to target this pathway in the context Wnt/β-catenin-driven cancers and other disease in humans.
In contrast to canonical Wnt/β-catenin signaling, the nuclear branches of the non-canonical Wnt pathways discussed here seem rather derived, with the redeployment of fairly standard kinase cascade-transcription factor interactions that were defined in other signaling contexts initially. As such, β-catenin is the only Wnt signaling dedicated nuclear effector. Nonetheless, novel insights will be exciting for either Wnt signaling pathway, and importantly the complexity of interactions, positive and negative, between the distinct Wnt signaling branches promise lots of unexpected discoveries to follow.
Acknowledgments
We are grateful to all Mlodzik lab members for their helpful discussions and support, and Philippe Soriano for thoughtful comments on the manuscript. We wish to thank Jill Gregory at the Academic Medical Illustration Department of the Icahn School of Medicine at Mount Sinai for excellent assistance with figure preparation. Related research in the Mlodzik laboratory is supported by a National Institutes of Health grant from the NIGMS R35 GM127103.
References
- Adler PN (2002). Planar signaling and morphogenesis in Drosophila. Developmental Cell, 2, 525–535. [DOI] [PubMed] [Google Scholar]
- Archbold HC, Broussard C, Chang MV, & Cadigan KM (2014). Bipartite recognition of DNA by TCF/Pangolin is remarkably flexible and contributes to transcriptional responsiveness and tissue specificity of wingless signaling. PLoS Genetics, 10, e1004591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw WY, & Devenport D (2017). Planar cell polarity: Global inputs establishing cellular asymmetry. Current Opinion in Cell Biology, 44, 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw WY, Heck BW, Joyce B, & Devenport D (2016). Transient tissue-scale deformation coordinates alignment of planar cell polarity junctions in the mammalian skin. Current Biology, 26, 2090–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod JD, Miller JR, Shulman JM, Moon RT, & Perrimon N (1998). Differential recruitment of dishevelled provides signaling specificity in the planar cell polarity and wingless signaling pathways. Genes & Development, 12, 2610–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Lopez LA, Nojima H, & Vincent JP (2012). Integration of morphogen signalling within the growth regulatory network. Current Opinion in Cell Biology, 24, 166–172. [DOI] [PubMed] [Google Scholar]
- Balmer S, Dussert A, Collu GM, Benitez E, Iomini C, & Mlodzik M (2015). Components of intraflagellar transport complex A function independently of the cilium to regulate canonical Wnt signaling in Drosophila. Developmental Cell, 34, 705–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, et al. (1996). Functional interaction of beta-catenin with the transcription factor LEF-1. Nature, 382, 638–642. [DOI] [PubMed] [Google Scholar]
- Bienz M (2005). beta-Catenin: A pivot between cell adhesion and Wnt signalling. Current Biology, 15, R64–R67. [DOI] [PubMed] [Google Scholar]
- Blauwkamp TA, Chang MV, & Cadigan KM (2008). Novel TCF-binding sites specify transcriptional repression by Wnt signalling. The EMBO Journal, 27, 1436–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, & Mlodzik M (1999). Dishevelled: At the crossroads of divergent intracellular signaling pathways. Mechanisms of Development, 83, 27–37. [DOI] [PubMed] [Google Scholar]
- Boutros M, Paricio N, Strutt DI, & Mlodzik M (1998). Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell, 94, 109–118. [DOI] [PubMed] [Google Scholar]
- Brembeck FH, Schwarz-Romond T, Bakkers J, Wilhelm S, Hammerschmidt M, & Birchmeier W (2004). Essential role of BCL9–2 in the switch between beta-catenin’s adhesive and transcriptional functions. Genes & Development, 18, 2225–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MT, & Wallingford JB (2017). Planar cell polarity in development and disease. Nature Reviews. Molecular Cell Biology, 18, 375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, & Waterman ML (2012). TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harbor Perspectives in Biology, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Gonzalez JM, Mulero-Navarro S, & Mlodzik M (2016). Centriole positioning in epithelial cells and its intimate relationship with planar cell polarity. BioEssays, 38, 1234–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, et al. (1998). Drosophila Tcf and Groucho interact to repress wingless signalling activity. Nature, 395, 604–608. [DOI] [PubMed] [Google Scholar]
- Clevers H (2006). Wnt/beta-catenin signaling in development and disease. Cell, 127, 469–480. [DOI] [PubMed] [Google Scholar]
- Clevers H, & Nusse R (2012). Wnt/beta-catenin signaling and disease. Cell, 149, 1192–1205. [DOI] [PubMed] [Google Scholar]
- Cliffe A, Hamada F, & Bienz M (2003). A role of dishevelled in relocating axin to the plasma membrane during wingless signaling. Current Biology, 13, 960–966. [DOI] [PubMed] [Google Scholar]
- Coombs GS, Covey TM, & Virshup DM (2008). Wnt signaling in development, disease and translational medicine. Current Drug Targets, 9, 513–531. [DOI] [PubMed] [Google Scholar]
- Cooper MT, & Bray SJ (1999). Frizzled regulation of notch signalling polarizes cell fate in the Drosophila eye. Nature, 397, 526–530. [DOI] [PubMed] [Google Scholar]
- Crowe R, Henrique D, Ish-Horowicz D, & Niswander L (1998). A new role for Notch and Delta in cell fate decisions: Patterning the feather array. Development, 125, 767–775. [DOI] [PubMed] [Google Scholar]
- Dale RM, Sisson BE, & Topczewski J (2009). The emerging role of Wnt/PCP signaling in organ formation. Zebrafish, 6, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, & Weis WI (2005). Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nature Structural & Molecular Biology, 12, 364–371. [DOI] [PubMed] [Google Scholar]
- Danielson KG, Pillarisetti J, Cohen IR, Sholehvar B, Huebner K, Ng LJ, et al. (1995). Characterization of the complete genomic structure of the human WNT-5A gene, functional analysis of its promoter, chromosomal mapping, and expression in early human embryogenesis. The Journal of Biological Chemistry, 270, 31225–31234. [DOI] [PubMed] [Google Scholar]
- Das G, Jenny A, Klein TJ, Eaton S, & Mlodzik M (2004). Diego interacts with prickle and Strabismus/Van Gogh to localize planar cell polarity complexes. Development, 131, 4467–4476. [DOI] [PubMed] [Google Scholar]
- Das P, Maduzia LL, Wang H, Finelli AL, Cho SH, Smith MM, et al. (1998). The Drosophila gene Medea demonstrates the requirement for different classes of Smads in dpp signaling. Development, 125, 1519–1528. [DOI] [PubMed] [Google Scholar]
- Das G, Reynolds-Kenneally J, & Mlodzik M (2002). The atypical cadherin Flamingo links frizzled and notch signaling in planar polarity establishment in the Drosophila eye. Developmental Cell, 2, 655–666. [DOI] [PubMed] [Google Scholar]
- Davey CF, & Moens CB (2017). Planar cell polarity in moving cells: Think globally, act locally. Development, 144, 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De A (2011). Wnt/Ca2 + signaling pathway: A brief overview. Acta Biochimica et Biophysica Sinica Shanghai, 43, 745–756. [DOI] [PubMed] [Google Scholar]
- del Alamo D, & Mlodzik M (2006). Frizzled/PCP-dependent asymmetric neuralized expression determines R3/R4 fates in the Drosophila eye. Developmental Cell, 11, 887–894. [DOI] [PubMed] [Google Scholar]
- Djiane A, Yogev S, & Mlodzik M (2005). The apical determinants aPKC and dPatj regulate frizzled-dependent planar cell polarity in the Drosophila eye. Cell, 121, 621–631. [DOI] [PubMed] [Google Scholar]
- Eisenmann DM (2005). Wnt signaling (pp. 1–17). WormBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Gluck U, & Gumbiner BM (1998). Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of beta-catenin. Current Biology, 8, 181–190. [DOI] [PubMed] [Google Scholar]
- Fang D, Hawke D, Zheng Y, Xia Y, Meisenhelder J, Nika H, et al. (2007). Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. The Journal of Biological Chemistry, 282, 11221–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanto M, & Mlodzik M (1999). Asymmetric notch activation specifies photoreceptors R3 and R4 and planar polarity in the Drosophila eye. Nature, 397, 523–526. [DOI] [PubMed] [Google Scholar]
- Fanto M, Weber U, Strutt DI, & Mlodzik M (2000). Nuclear signaling by Rac and Rho GTPases is required in the establishment of epithelial planar polarity in the Drosophila eye. Current Biology, 10, 979–988. [DOI] [PubMed] [Google Scholar]
- Franz A, Shlyueva D, Brunner E, Stark A, & Basler K (2017). Probing the canonicity of the Wnt/wingless signaling pathway. PLoS Genetics, 13, e1006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Song H, Bishop K, Elliot G, Garrett L, English MA, et al. (2011). Wnt signaling gradients establish planar cell polarity by inducing Vangl2 phosphorylation through Ror2. Developmental Cell, 20, 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, & Strutt D (2011). Principles of planar polarity in animal development. Development, 138, 1877–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C, Shao H, Strutt H, & Strutt D (2020). Molecular mechanisms mediating asymmetric subcellular localisation of the core planar polarity pathway proteins. Biochemical Society Transactions, 48, 1297–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein U, Wolff T, & Rubin GM (1993). The TGF beta homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell, 75, 913–926. [DOI] [PubMed] [Google Scholar]
- Henderson BR, & Fagotto F (2002). The ins and outs of APC and beta-catenin nuclear transport. EMBO Reports, 3, 834–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmans R, & Basler K (2007). BCL9–2 binds Arm/beta-catenin in a Tyr142-independent manner and requires Pygopus for its function in Wg/Wnt signaling. Mechanisms of Development, 124, 59–67. [DOI] [PubMed] [Google Scholar]
- Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, & Kemler R (1996). Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mechanisms of Development, 59, 3–10. [DOI] [PubMed] [Google Scholar]
- Humphries AC, & Mlodzik M (2018). From instruction to output: Wnt/PCP signaling in development and cancer. Current Opinion in Cell Biology, 51, 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A (2010). Planar cell polarity signaling in the Drosophila eye. Current Topics in Developmental Biology, 93, 189–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Reynolds-Kenneally J, Das G, Burnett M, & Mlodzik M (2005). Diego and Prickle regulate Frizzled planar cell polarity signalling by competing for dishevelled binding. Nature Cell Biology, 7, 691–697. [DOI] [PubMed] [Google Scholar]
- Jimbo T, Kawasaki Y, Koyama R, Sato R, Takada S, Haraguchi K, et al. (2002). Identification of a link between the tumour suppressor APC and the kinesin superfamily. Nature Cell Biology, 4, 323–327. [DOI] [PubMed] [Google Scholar]
- Katanaev VL, Ponzielli R, Semeriva M, & Tomlinson A (2005). Trimeric G protein-dependent frizzled signaling in Drosophila. Cell, 120, 111–122. [DOI] [PubMed] [Google Scholar]
- Kockel L, Homsy JG, & Bohmann D (2001). Drosophila AP-1: Lessons from an invertebrate. Oncogene, 20, 2347–2364. [DOI] [PubMed] [Google Scholar]
- Kockel L, Zeitlinger J, Staszewski LM, Mlodzik M, & Bohmann D (1997). Jun in Drosophila development: Redundant and nonredundant functions and regulation by two MAPK signal transduction pathways. Genes & Development, 11, 1748–1758. [DOI] [PubMed] [Google Scholar]
- Kohn AD, & Moon RT (2005). Wnt and calcium signaling: Beta-catenin-independent pathways. Cell Calcium, 38, 439–446. [DOI] [PubMed] [Google Scholar]
- Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, et al. (2002). Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell, 109, 47–60. [DOI] [PubMed] [Google Scholar]
- Krieghoff E, Behrens J, & Mayr B (2006). Nucleo-cytoplasmic distribution of beta-catenin is regulated by retention. Journal of Cell Science, 119, 1453–1463. [DOI] [PubMed] [Google Scholar]
- Kuhl M, Sheldahl LC, Malbon CC, & Moon RT (2000). Ca(2+)/calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. The Journal of Biological Chemistry, 275, 12701–12711. [DOI] [PubMed] [Google Scholar]
- Kuhl M, Sheldahl LC, Park M, Miller JR, & Moon RT (2000). The Wnt/Ca2+ pathway: A new vertebrate Wnt signaling pathway takes shape. Trends in Genetics, 16, 279–283. [DOI] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, & Gorlich D (1997). Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell, 9, 1061–1071. [DOI] [PubMed] [Google Scholar]
- Logan CY, & Nusse R (2004). The Wnt signaling pathway in development and disease. Annual Review of Cell and Developmental Biology, 20, 781–810. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, & He X (2009). Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Developmental Cell, 17, 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen AE, Escobar DE, & Gottardi CJ (2012). Signaling from the adherens junction. Sub-Cellular Biochemistry, 60, 171–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod F, Boyle K, Marzo A, Martin-Flores N, Moe TZ, Palomer E, et al. (2020). Wnt signaling through nitric oxide synthase promotes the formation of multi-innervated spines. Frontiers in Synaptic Neuroscience, 12, 575863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod F, & Salinas PC (2018). Wnt proteins as modulators of synaptic plasticity. Current Opinion in Neurobiology, 53, 90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi K, Hashimoto M, Ajima R, Takaoka K, Shinohara K, Ikawa Y, et al. (2017). A Wnt5 activity asymmetry and intercellular signaling via PCP proteins polarize node cells for left-right symmetry breaking. Developmental Cell, 40(439–452), e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovic I, Pylawka S, & Hudspeth AJ (2012). Rearrangements between differentiating hair cells coordinate planar polarity and the establishment of mirror symmetry in lateral-line neuromasts. Biology Open, 1, 498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M (1999). Planar polarity in the Drosophila eye: A multifaceted view of signaling specificity and cross-talk. The EMBO Journal, 18, 6873–6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, & Basler K (2009). Beta-catenin hits chromatin: Regulation of Wnt target gene activation. Nature Reviews. Molecular Cell Biology, 10, 276–286. [DOI] [PubMed] [Google Scholar]
- Munoz-Descalzo S, Gomez-Cabrero A, Mlodzik M, & Paricio N (2007). Analysis of the role of the Rac/Cdc42 GTPases during planar cell polarity generation in Drosophila. The International Journal of Developmental Biology, 51, 379–387. [DOI] [PubMed] [Google Scholar]
- Murphy LL, & Hughes CC (2002). Endothelial cells stimulate T cell NFAT nuclear translocation in the presence of cyclosporin A: Involvement of the wnt/glycogen synthase kinase-3 beta pathway. Journal of Immunology, 169, 3717–3725. [DOI] [PubMed] [Google Scholar]
- Navajas Acedo J, Voas MG, Alexander R, Woolley T, Unruh JR, Li H, et al. (2019). PCP and Wnt pathway components act in parallel during zebrafish mechanosensory hair cell orientation. Nature Communications, 10, 3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann CJ, & Cohen SM (1997). Long-range action of wingless organizes the dorsal-ventral axis of the Drosophila wing. Development, 124, 871–880. [DOI] [PubMed] [Google Scholar]
- Niehrs C (2012). The complex world of WNT receptor signalling. Nature Reviews. Molecular Cell Biology, 13, 767–779. [DOI] [PubMed] [Google Scholar]
- Nusse R, van Ooyen A, Cox D, Fung YK, & Varmus H (1984). Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature, 307, 131–136. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, & Wieschaus E (1980). Mutations affecting segment number and polarity in Drosophila. Nature, 287, 795–801. [DOI] [PubMed] [Google Scholar]
- Ozawa M, Baribault H, & Kemler R (1989). The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. The EMBO Journal, 8, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paricio N, Feiguin F, Boutros M, Eaton S, & Mlodzik M (1999). The Drosophila STE20-like kinase misshapen is required downstream of the Frizzled receptor in planar polarity signaling. The EMBO Journal, 18, 4669–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, & Axelrod JD (2012). Asymmetric protein localization in planar cell polarity: Mechanisms, puzzles, and challenges. Current Topics in Developmental Biology, 101, 33–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O, Vorbruggen G, & Jackle H (2000). Drosophila segment borders result from unilateral repression of hedgehog activity by wingless signaling. Molecular Cell, 6, 203–209. [PubMed] [Google Scholar]
- Prestwich TC, & Macdougald OA (2007). Wnt/beta-catenin signaling in adipogenesis and metabolism. Current Opinion in Cell Biology, 19, 612–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Luo C, & Hogan PG (1997). Transcription factors of the NFAT family: Regulation and function. Annual Review of Immunology, 15, 707–747. [DOI] [PubMed] [Google Scholar]
- Rawls AS, & Wolff T (2003). Strabismus requires Flamingo and Prickle function to regulate tissue polarity in the Drosophila eye. Development, 130, 1877–1887. [DOI] [PubMed] [Google Scholar]
- Roignant JY, & Treisman JE (2009). Pattern formation in the Drosophila eye disc. The International Journal of Developmental Biology, 53, 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segalen M, & Bellaiche Y (2009). Cell division orientation and planar cell polarity pathways. Seminars in Cell & Developmental Biology, 20, 972–977. [DOI] [PubMed] [Google Scholar]
- Seifert JR, & Mlodzik M (2007). Frizzled/PCP signalling: A conserved mechanism regulating cell polarity and directed motility. Nature Reviews, 8, 126–138. [DOI] [PubMed] [Google Scholar]
- Semenov MV, Habas R, Macdonald BT, & He X (2007). SnapShot: Noncanonical Wnt signaling pathways. Cell, 131, 1378. [DOI] [PubMed] [Google Scholar]
- Sharma RP, & Chopra VL (1976). Effect of the wingless (wg1) mutation on wing and haltere development in Drosophila melanogaster. Developmental Biology, 48, 461–465. [DOI] [PubMed] [Google Scholar]
- Shaulian E, & Karin M (2001). AP-1 in cell proliferation and survival. Oncogene, 20, 2390–2400. [DOI] [PubMed] [Google Scholar]
- Shulman JM, Perrimon N, & Axelrod JD (1998). Frizzled signaling and the developmental control of cell polarity. Trends in Genetics, 14, 452–458. [DOI] [PubMed] [Google Scholar]
- Sierra J, Yoshida T, Joazeiro CA, & Jones KA (2006). The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes & Development, 20, 586–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, & Mlodzik M (2008). Planar cell polarity signaling: From fly development to human disease. Annual Review of Genetics, 42, 517–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadeli R, Hoffmans R, & Basler K (2006). Transcription under the control of nuclear Arm/beta-catenin. Current Biology, 16, R378–R385. [DOI] [PubMed] [Google Scholar]
- Strigini M, & Cohen SM (2000). Wingless gradient formation in the Drosophila wing. Current Biology, 10, 293–300. [DOI] [PubMed] [Google Scholar]
- Strutt DI, Weber U, & Mlodzik M (1997). The role of RhoA in tissue polarity and frizzled signalling. Nature, 387, 292–295. [DOI] [PubMed] [Google Scholar]
- Suh EK, & Gumbiner BM (2003). Translocation of beta-catenin into the nucleus independent of interactions with FG-rich nucleoporins. Experimental Cell Research, 290, 447–456. [DOI] [PubMed] [Google Scholar]
- Sustmann C, Flach H, Ebert H, Eastman Q, & Grosschedl R (2008). Cell-type-specific function of BCL9 involves a transcriptional activation domain that synergizes with beta-catenin. Molecular and Cellular Biology, 28, 3526–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng J, Rai T, Tanaka Y, Takei Y, Nakata T, Hirasawa M, et al. (2005). The KIF3 motor transports N-cadherin and organizes the developing neuroepithelium. Nature Cell Biology, 7, 474–482. [DOI] [PubMed] [Google Scholar]
- Teo S, & Salinas PC (2021). Wnt-frizzled signaling regulates activity-mediated synapse formation. Frontiers in Molecular Neuroscience, 14, 683035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B, Townsley F, Rosin-Arbesfeld R, Musisi H, & Bienz M (2002). A new nuclear component of the Wnt signalling pathway. Nature Cell Biology, 4, 367–373. [DOI] [PubMed] [Google Scholar]
- Tian X, Liu Z, Niu B, Zhang J, Tan TK, Lee SR, et al. (2011). E-cadherin/beta-catenin complex and the epithelial barrier. Journal of Biomedicine & Biotechnology, 2011, 567305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolwinski NS, & Wieschaus E (2001). Armadillo nuclear import is regulated by cytoplasmic anchor Axin and nuclear anchor dTCF/Pan. Development, 128, 2107–2117. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, & Ready DF (1987). Neuronal differentiation in Drosophila ommatidium. Developmental Biology, 120, 366–376. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, & Struhl G (1999). Decoding vectorial information from a gradient: Sequential roles of the receptors frizzled and notch in establishing planar polarity in the Drosophila eye. Development, 126, 5725–5738. [DOI] [PubMed] [Google Scholar]
- Townsley FM, Cliffe A, & Bienz M (2004). Pygopus and Legless target Armadillo/beta-catenin to the nucleus to enable its transcriptional co-activator function. Nature Cell Biology, 6, 626–633. [DOI] [PubMed] [Google Scholar]
- Tree DR, Ma D, & Axelrod JD (2002). A three-tiered mechanism for regulation of planar cell polarity. Seminars in Cell & Developmental Biology, 13, 217–224. [DOI] [PubMed] [Google Scholar]
- Tree DR, Shulman JM, Rousset R, Scott MP, Gubb D, & Axelrod JD (2002). Prickle mediates feedback amplification to generate asymmetric planar cell polarity signaling. Cell, 109, 371–381. [DOI] [PubMed] [Google Scholar]
- Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, Schwarz TL, et al. (1999). Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of frizzled. Cell, 98, 585–595. [DOI] [PubMed] [Google Scholar]
- Valenta T, Hausmann G, & Basler K (2012). The many faces and functions of beta-catenin. The EMBO Journal, 31, 2714–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman MT, Slusarski DC, Kaykas A, Louie SH, & Moon RT (2003). Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Current Biology, 13, 680–685. [DOI] [PubMed] [Google Scholar]
- Vinson CR, & Adler PN (1987). Directional non-cell autonomy and the transmission of polarity information by the frizzled gene of Drosophila. Nature, 329, 549–551. [DOI] [PubMed] [Google Scholar]
- Vinson CR, Conover S, & Adler PN (1989). A Drosophila tissue polarity locus encodes a protein containing seven potential transmembrane domains. Nature, 338, 263–264. [DOI] [PubMed] [Google Scholar]
- Vuong LT, Iomini C, Balmer S, Esposito D, Aaronson SA, & Mlodzik M (2018). Kinesin-2 and IFT-A act as a complex promoting nuclear localization of beta-catenin during Wnt signalling. Nature Communications, 9, 5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong LT, Mukhopadhyay B, & Choi KW (2014). Kinesin-II recruits Armadillo and dishevelled for wingless signaling in Drosophila. Development, 141, 3222–3232. [DOI] [PubMed] [Google Scholar]
- Wallingford JB (2010). Planar cell polarity signaling, cilia and polarized ciliary beating. Current Opinion in Cell Biology, 22, 597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB, & Habas R (2005). The developmental biology of dishevelled: An enigmatic protein governing cell fate and cell polarity. Development, 132, 4421–4436. [DOI] [PubMed] [Google Scholar]
- Weber U, Paricio N, & Mlodzik M (2000). Jun mediates frizzled-induced R3/R4 cell fate distinction and planar polarity determination in the Drosophila eye. Development, 127, 3619–3629. [DOI] [PubMed] [Google Scholar]
- Weber U, Pataki C, Mihaly J, & Mlodzik M (2008). Combinatorial signaling by the Frizzled/PCP and Egfr pathways during planar cell polarity establishment in the Drosophila eye. Developmental Biology, 316, 110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall TA, Brimeyer R, Twedt J, Gladon J, Olberding A, Furutani-Seiki M, et al. (2003). Wnt-5/pipetail functions in vertebrate axis formation as a negative regulator of Wnt/beta-catenin activity. The Journal of Cell Biology, 162, 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widelitz RB, Jiang TX, Chen CW, Stott NS, Jung HS, & Chuong CM (1999). Wnt-7a in feather morphogenesis: Involvement of anterior-posterior asymmetry and proximal-distal elongation demonstrated with an in vitro reconstitution model. Development, 126, 2577–2587. [DOI] [PubMed] [Google Scholar]
- Winter CG, Wang B, Ballew A, Royou A, Karess R, Axelrod JD, et al. (2001). Drosophila Rho-associated kinase (Drok) links frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell, 105, 81–91. [DOI] [PubMed] [Google Scholar]
- Wolff T, & Ready DF (1991). Cell death in normal and rough eye mutants of Drosophila. Development, 113, 825–839. [DOI] [PubMed] [Google Scholar]
- Wolff T, & Rubin GM (1998). Strabismus, a novel gene that regulates tissue polarity and cell fate decisions in Drosophila. Development, 125, 1149–1159. [DOI] [PubMed] [Google Scholar]
- Wu J, Roman AC, Carvajal-Gonzalez JM, & Mlodzik M (2013). Wg and Wnt4 provide long-range directional input to planar cell polarity orientation in Drosophila. Nature Cell Biology, 15, 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Tu X, Joeng KS, Hilton MJ, Williams DA, & Long F (2008). Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell, 133, 340–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, & Mlodzik M (2015). Wnt-Frizzled/planar cell polarity signaling: Cellular orientation by facing the wind (Wnt). Annual Review of Cell and Developmental Biology, 31, 623–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoya F, Imamoto N, Tachibana T, & Yoneda Y (1999). beta-Catenin can be transported into the nucleus in a Ran-unassisted manner. Molecular Biology of the Cell, 10, 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M, Basler K, & Struhl G (1996). Direct and long-range action of a wingless morphogen gradient. Cell, 87, 833–844. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Kockel L, Peverali FA, Jackson DB, Mlodzik M, & Bohmann D (1997). Defective dorsal closure and loss of epidermal decapentaplegic expression in Drosophila fos mutants. The EMBO Journal, 16, 7393–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Zhang J, & Carthew RW (1995). Frizzled regulates mirror-symmetric pattern formation in the Drosophila eye. Development, 121, 3045–3055. [DOI] [PubMed] [Google Scholar]
- Zou Y (2020). Breaking symmetry—Cell polarity signaling pathways in growth cone guidance and synapse formation. Current Opinion in Neurobiology, 63, 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]


