Abstract
Clinical trials frequently include multiple end points that mature at different times. The initial report, typically based on the primary end point, may be published when key planned co‐primary or secondary analyses are not yet available. Clinical Trial Updates provide an opportunity to disseminate additional results from studies, published in JCO or elsewhere, for which the primary end point has already been reported.
Acquired genomic alterations (Acq-GAs), specifically RAS, BRAF, and EGFR-ectodomain mutations and ERBB2 and MET amplifications, are recognized as major mechanisms of resistance to later-line anti–EGFR-antibody therapy in metastatic colorectal cancer (mCRC). However, data regarding emergence of these Acq-GAs under the selective pressure of first-line anti–EGFR-chemotherapy are lacking. We performed next-generation sequencing (Guardant360) on circulating tumor DNA obtained from paired plasma samples (pretreatment and postprogression) from the CALGB/SWOG-80405 trial, which randomly assigned patients with mCRC between first-line chemotherapy with cetuximab (anti–EGFR-chemotherapy) or bevacizumab (anti–VEGF-chemotherapy). The primary objective was to determine the prevalence of Acq-GAs on anti–EGFR-chemotherapy and compare this to the prevalence with anti–VEGF-chemotherapy on trial and pooled estimates (N = 292) seen with later-line anti–EGFR-antibody therapy as reported in the literature. Among the 61 patients on anti–EGFR-chemotherapy, only four (6.6%) developed ≥ 1 Acq-GAs of interest compared with 10.1% (7) on anti–VEGF-chemotherapy (odds ratio, 0.62; 95% CI, 0.20 to 2.11) and 62.0% on anti–EGFR-antibody therapy in later lines (odds ratio, 0.09; 95% CI, 0.03 to 0.23). Acq-GAs, classically associated with anti–EGFR-antibody resistance in later lines (RAS, BRAF, and EGFR-ectodomain mutations; ERBB2 and MET amplifications), were rare with up-front use of anti–EGFR-chemotherapy indicating divergent resistance mechanisms. These findings have critical translational relevance to timing and value of circulating tumor DNA–guided anti-EGFR rechallenge in patients with mCRC, especially those treated with anti-EGFR therapy upfront.
INTRODUCTION
Antiepidermal growth factor receptor antibodies (anti–EGFR-Abs), panitumumab and cetuximab, are highly effective against RAS/BRAF wild-type (WT) metastatic colorectal cancer (mCRC).1,2 Barring patients who have intrinsic or primary resistance to EGFR inhibition, a substantial proportion will derive initial benefit, but eventually progress.1-4 This acquired or secondary resistance remains a major limitation in treating mCRC. Acquisition of genomic alterations under selective pressure exerted by EGFR inhibition, specifically RAS, BRAF, or EGFR-ectodomain mutations and ERBB2 (HER2) or MET amplifications, have been commonly (30%-50%) implicated as key resistance mechanisms to anti–EGFR-Ab therapy in later lines in mCRC (Data Supplement, online only).4-9 Although traditionally anti-EGFR agents have been used in later lines, the current evidence supports use in frontline therapy combined with cytotoxic chemotherapy (FOLFOX or FOLFIRI), especially for left-sidedRAS/BRAF-WT mCRC.10,11 However, data regarding emergence of these acquired alterations associated with anti–EGFR-Ab resistance under the treatment stress of anti–EGFR-Abs in first line combined with cytotoxic chemotherapy, which is now considered the standard of care, are lacking.
To understand the dynamics of tumor evolution and resistance mechanisms to frontline therapy in mCRC, we performed this prospective-retrospective circulating tumor DNA (ctDNA) analyses on serial blood specimens collected from the randomized Cancer and Leukemia B and Southwest Oncology Group (CALGB/SWOG) 80405 trial (cetuximab-chemotherapy v bevacizumab-chemotherapy; ClinicalTrials.gov identifier: NCT00265850).1
METHODS
The CALGB/SWOG-80405 (September 2005-March 2012) was a randomized phase III trial designed to evaluate efficacy of chemotherapy with either cetuximab or bevacizumab (or both) as first-line treatment for (KRAS-WT) mCRC.1 Trial eligibility, design, treatment, and primary analyses have been reported previously.1 The primary end point was overall survival. Among eligible patients, the study found no significant difference in overall survival (median: 30.0 v 29.0 months) between treatment arms.1
This biomarker substudy was a post hoc analysis and included patients treated with either cetuximab or bevacizumab on CALGB/SWOG-80405, who progressed on study treatment at discontinuation and had paired plasma samples (pretreatment and postprogression) available for ctDNA testing.1 Figure 1 depicts study schema, patient selection, and flow for the current study cohort. Sequencing of ctDNA was performed by next-generation sequencing (Guardant360) assay optimized for detecting alterations (mutations and amplifications) in 73 genes (Data Supplement).12 Patient RAS/BRAF status (WT or mutant) was defined by clonal mutations (relative maximum-allele frequency ≥ 25%) in ctDNA. A predefined cutoff of 0.1% for mutant allele frequency was used to determine the presence of mutations. Patients with baseline clonal RAS/BRAF mutations by ctDNA were excluded. Samples without any detectable alterations were also excluded to minimize false negatives (Fig 1).
FIG 1.
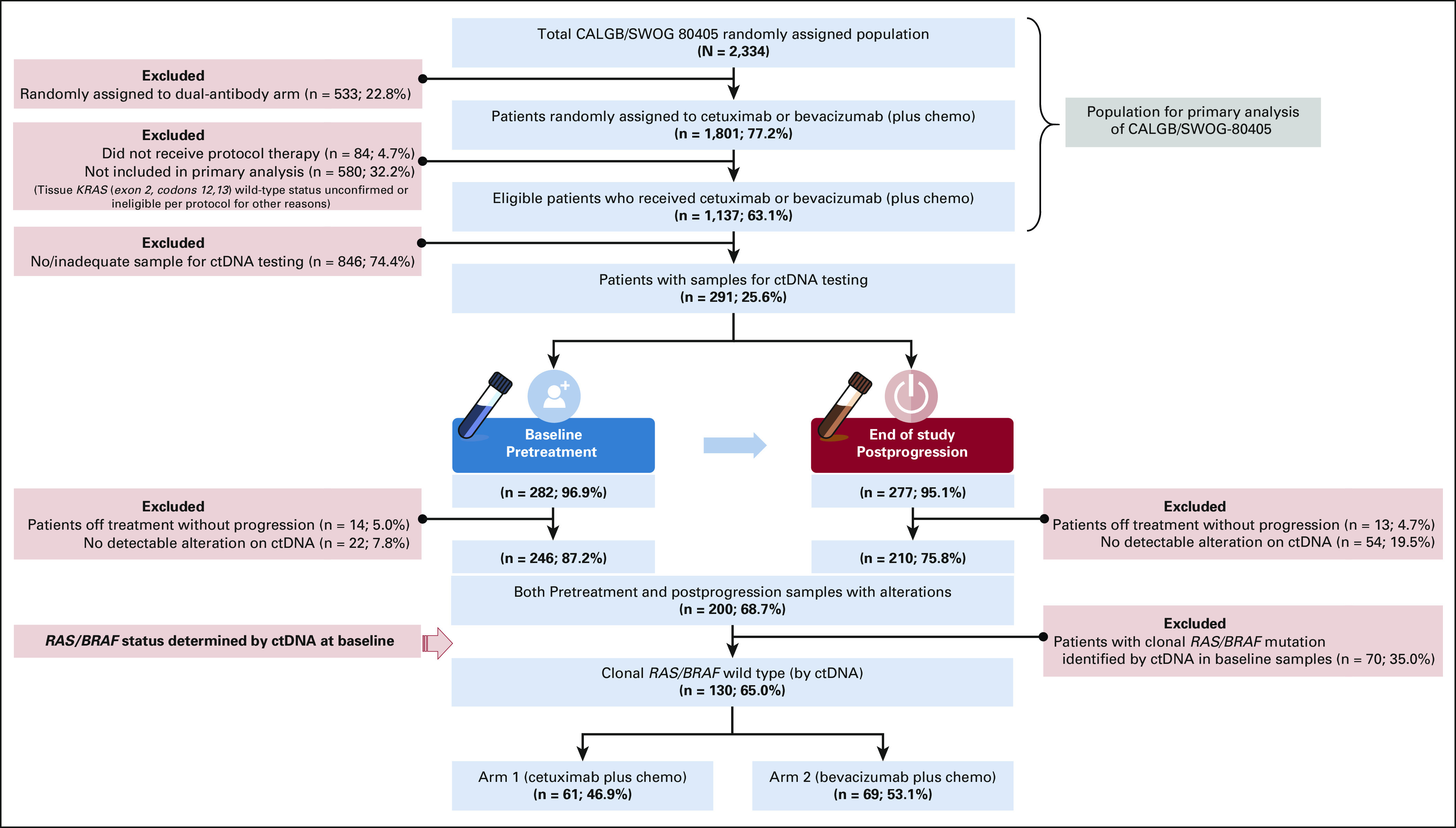
Schematic flow of patients and samples for ctDNA analysis of the CALGB/SWOG-80405 trial. Eligible patients for the current biomarker study included those who were randomly assigned, treated, and subsequently progressed on cetuximab or bevacizumab; had plasma samples (both baseline and postprogression end-of-protocol treatment) available for ctDNA testing; and had at least one detectable genomic alteration. RAS/BRAF status (mutant or wild type) was determined by the presence of clonal mutations (relative mutant allele frequency of ≥ 25% in the sample) in ctDNA, and only RAS/BRAF wild-type patients identified by ctDNA were included in current study. chemo, chemotherapy; ctDNA, circulating tumor DNA.
The primary objective was to determine the prevalence of key acquired genomic alterations (Acq-GAs) of interest, prespecified as those implicated in anti–EGFR-Ab resistance (Data Supplement), on cetuximab-chemotherapy in the first-line setting. The secondary objective was to compare this prevalence to the prevalence seen with bevacizumab-chemotherapy on current trial and to the pooled prevalence estimates derived from all relevant published studies of anti–EGFR-Ab in later lines of therapy in mCRC (Data Supplement). Descriptive statistics and Fisher's exact test were used. For proportions, 95% CI were calculated using the modified Wald method. All P values are for exploratory purposes and not powered for statistical hypothesis testing. Participating site's institutional review board approval and written informed consent for all patients were obtained for CALGB/SWOG-80405.1 Detailed methodology is provided as Supplementary Methods (Data Supplement).
RESULTS
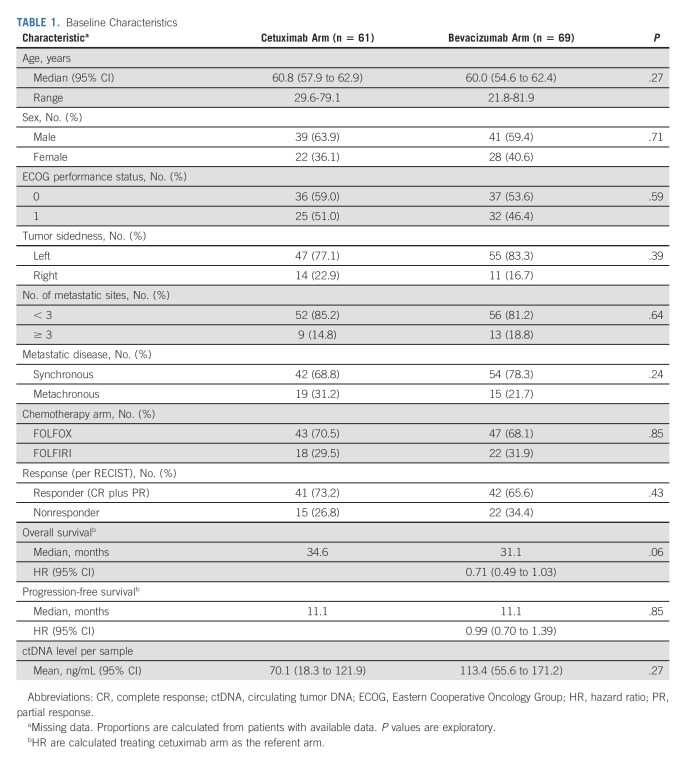
Baseline characteristics of patients who underwent ctDNA testing were similar to the entire CALGB/SWOG-80405 population (Data Supplement). Furthermore, the clinical outcomes with respect to RAS/BRAF status detected by ctDNA at baseline, type of targeted therapy, and tumor sidedness in this cohort were consistent with those observed in the primary 80405 analysis (Data Supplement). Of them, 130 patients met eligibility criteria for current biomarker substudy and were equally distributed between cetuximab (n = 61) and bevacizumab (n = 69) arms. Baseline characteristics of these evaluable patients were comparable between treatment arms (Table 1). Among those treated with cetuximab-chemotherapy, only four (6.6%) of 61 patients developed at least one Acq-GA of interest at progression. This prevalence was similar to that seen with bevacizumab-chemotherapy (7 of 69 [10.1%]; odds ratio [OR] 0.62 [95% CI, 0.20 to 2.11]). No meaningful difference was seen in prevalence of key alterations between treatment arms (cetuximab v bevacizumab): mutations (all: 6.6% v 10.1%) in RAS (4.9% v 5.8%), KRAS (0% v 4.4%), NRAS (4.9% v 1.5%), BRAF (0% v 1.5%), and EGFR-ectodomain (1.6% v 1.5%) and amplifications (all: 1.6% v 4.3%) in ERBB2 (1.6% v 2.9%) and MET (0% v 2.9%; Fig 2A). No differences in key clinical characteristics were seen between patients who had Acq-GAs and those who did not (Fig 2B). The observed prevalence of all Acq-GAs on first-line cetuximab-chemotherapy was also considerably lower than the pooled prevalence (N = 292) on prior studies with anti–EGFR-Ab–based regimen in later lines of therapy (6.6% v 62.0%, OR 0.09 [95% CI, 0.03 to 0.23]), including key alterations such as acquired KRAS mutations (0% v 44%, OR 0.00 [95% CI, 0.00 to 0.08]; Fig 2A and Data Supplement).
TABLE 1.
Baseline Characteristics
FIG 2.
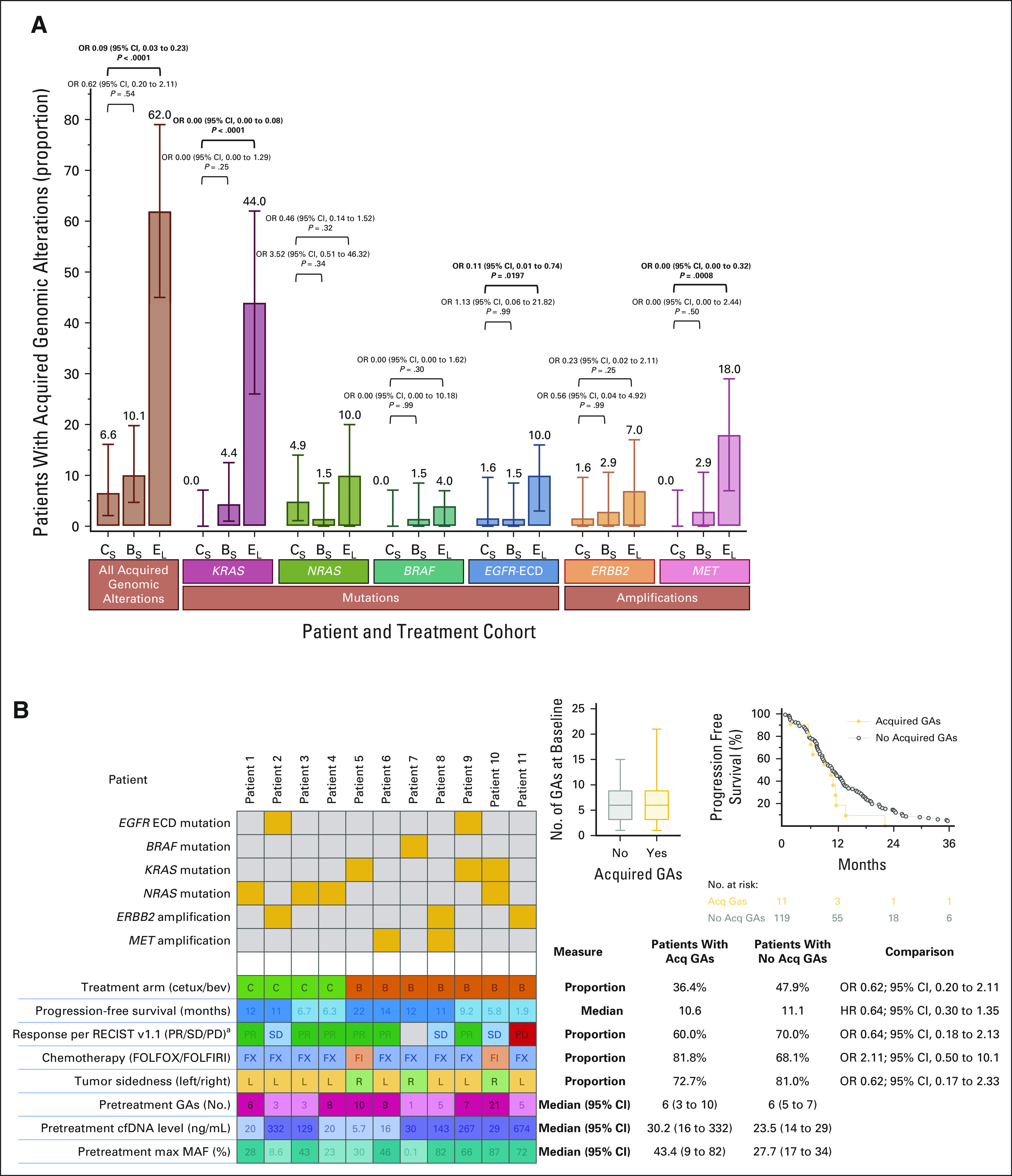
Acquired genomic alterations (mutations and amplifications) in patients with mCRC on systemic therapy. (A) Comparison of acquired genomic alterations between cetuximab-chemotherapy (CS) and bevacizumab-chemotherapy (BS) treatment arms on CALGB/SWOG-80405 study and acquired genomic alterations on anti–EGFR-Ab–based therapy in later lines in mCRC as reported in the literature (EL). The error bars on each represent 95% CIs of the proportion. (B) Key clinical characteristics of patients who acquired genomic alterations and comparisons of these characteristics with those patients who did not develop acquired genomic alterations. All P values reported are for exploratory purposes only and not powered for statistical hypothesis testing. aResponse per RECIST v1.1 (PR/SD/PD). Acq, acquired; anti–EGFR-Abs, antiepidermal growth factor receptor antibodies; bev, bevacizumab; cetux, cetuximab; chemo, chemotherapy; CR, complete response; GAs, genomic alterations; HR, hazard ratio; max MAF, maximum mutant allele frequency; mCRC, metastatic colorectal cancer; OR, odds ratio; PD, progressive disease; PR, partial response; SD, stable disease.
DISCUSSION
Enhanced understanding of evolving clonal architecture under treatment is crucial to optimizing care and developing effective therapies. This report is the first broad characterization of Acq-GAs to anti–EGFR-Abs in a randomized prospective first-line setting with doublet chemotherapy and demonstrates a distinctly different profile of acquired ctDNA compared with that seen with anti–EGFR-Ab therapy in later lines. Acq-GAs, classically associated with EGFR resistance in later lines, were rare with up-front use of anti–EGFR-Ab combined with highly active chemotherapy and comparable with non–anti-EGFR regimen, suggesting divergent mechanisms of acquired resistance subject to line of therapy and concomitant cytotoxic exposure. This study is exploratory in nature and hypothesis generating, subject to limitations inherent to post hoc analysis and limited sample size, but the results reported represent a randomized cohort. Validation is necessary in future prospective efforts.
Alternate mechanisms of resistance to anti–EGFR-Abs beyond acquisition of resistance conferring genomic modifications (adaptive mutability) exist that can explain this phenomenon. Epigenetic or transcriptional reprogramming and ensuing therapy-induced senescence or EMT (epithelial-to-mesenchymal transition) may enable mCRC to survive the brunt of combined targeted chemotherapy.13,14 EMT gene expression signatures have been associated with both a generic resistance to cytotoxic chemotherapy and EGFR inhibition.15-17 An activated EMT program enables chemotherapy-induced cancer cell plasticity and may confer cross-resistance to cytotoxic and targeted components of frontline therapy in mCRC.18,19 Early evidence shows that acquisition of resistance mechanisms in mCRC can vary with line of therapy and with use of non–anti-EGFR biologics.6,20
Although traditionally used in later lines, there has been a recent trend toward use of anti–EGFR-Abs as frontline therapy, especially for left-sided RAS/BRAF-WT mCRC.1,11 Despite this shift in treatment landscape of mCRC and limited understanding of resistance in this setting, large research efforts and resources are being invested to target previously identified mechanisms, which may not play a sizable role in real world. Furthermore, ctDNA is being increasingly used as a tool to rechallenge patients with anti-EGFR therapy because of clinical benefit seen with application of ctDNA in small prospective investigations.21,22 However, it is unclear whether patients who acquire resistance to first-line cetuximab-chemotherapy without acquisition of any genomic alterations are distinct from those that acquire and loose these genomic alterations with later lines of therapy or the duration for which the phenotypic plasticity induced will confer resistance to subsequent anti-EGFR inhibition. Consequently, urgent efforts are needed to delineate characteristics of this therapeutic resistance in mCRC in the first-line setting and advancing therapeutics directed against these mechanisms. Our findings have critical translational relevance to the timing and value of ctDNA-guided anti-EGFR rechallenge in these patients with mCRC.
ACKNOWLEDGMENT
We would like to thank our patients and their caregivers for participating in this trial. We greatly appreciate the support and guidance of the large group of investigators across community and academic centers throughout the National Clinical Trials Network (NCTN) in the United States and Canada.
Ryan Sun
Consulting or Advisory Role: Boehringer Ingelheim
Research Funding: Sanofi
Scott Kopetz
Stock and Other Ownership Interests: MolecularMatch, Lutris, Iylon, Frontier Medicines
Consulting or Advisory Role: Genentech, EMD Serono, Merck, Holy Stone Healthcare, Novartis, Lilly, Boehringer Ingelheim, Boston Biomedical, AstraZeneca/MedImmune, Bayer Health, Pierre Fabre, EMD Serono, Redx Pharma, Ipsen, Daiichi Sankyo, Natera, HalioDx, Lutris, Jacobio, Pfizer, Repare Therapeutics, Inivata, GlaxoSmithKline, Jazz Pharmaceuticals, Iylon, Xilis, AbbVie, Amal Therapeutics, Gilead Sciences, Mirati Therapeutics, Flame Biosciences, Servier, Carina Biotech, Bicara Therapeutics, Endeavor BioMedicines, Numab, Johnson & Johnson/Janssen, Genomic Health, Frontier Medicines, Replimune, Taiho Pharmaceutical
Research Funding: Sanofi, Biocartis, Guardant Health, Array BioPharma, Genentech/Roche, EMD Serono, MedImmune, Novartis, Amgen, Lilly, Daiichi Sankyo
Federico Innocenti
Employment: AbbVie
Stock and Other Ownership Interests: AbbVie
Honoraria: Tempus
Consulting or Advisory Role: Symberix, Emerald Lake Safety
Patents, Royalties, Other Intellectual Property: United States Patent: "Flavopiridol drug combinations and methods with reduced side effects", Ratain M.J., Innocenti F., Iyer L. Filed on April 12, 2001, serial number 09/835082, United States Patent: "Optimization of cancer treatment with irinotecan", Ratain M.J., Innocenti F., Karabatsos P., Grimsley C., Di Rienzo A. Filed on February 12, 2003, serial number 60/446942, United States Patent: "Methods of identifying risk of bevacizumab-induced proteinuria and hypertension", Innocenti F., Quintanilha J., Lin D., Owzar K., Wang J. Filed on July 17, 2020, serial number 16/932002, United States Provisional Patent Application: "Plasma levels of angiopoietin-2, VEGF-A, and VCAM-1 as markers of bevacizumab induced hypertension", Innocenti F., Quintanilha J. Filed on April 1, 2021, serial number 63/169301
Travel, Accommodations, Expenses: AbbVie
Heinz-Josef Lenz
Honoraria: Merck Serono, Roche, Bayer, Boehringer Ingelheim, Isofol Medical, GlaxoSmithKline, G1 THerapeutics, Jazz Pharmaceuticals, Oncocyte, Fulgent Genetics
Consulting or Advisory Role: Merck Serono, Roche, Bayer, BMS, GlaxoSmithKline
Travel, Accommodations, Expenses: Merck Serono, Bayer, BMS
Kanwal Raghav
Consulting or Advisory Role: AstraZeneca, Bayer, Eisai, Daiichi Sankyo
Speakers' Bureau: Bayer
Research Funding: Daiichi Sankyo/Lilly (Inst), Bayer (Inst), Roche/Genentech (Inst), Guardant Health (Inst)
Alan P. Venook
Consulting or Advisory Role: Merck Sharp & Dohme, Amgen, GlaxoSmithKline, Exelixis, BridgeBio Pharma, Bayer Health, Gilead Sciences, Exact Sciences, Bristol Myers Squibb Foundation/Janssen
Research Funding: Amgen
Patents, Royalties, Other Intellectual Property: Royalties from Now-UptoDate for authoring and maintaining two chapters
No other potential conflicts of interest were reported.
See accompanying editorial on page 436
DISCLAIMER
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
PRIOR PRESENTATION
Presented at the ASCO Gastrointestinal Cancers Symposium, San Francisco, CA, January 22, 2022.
SUPPORT
Research reported in this publication was supported by the National Cancer Institute (NCI) of the National Institutes of Health (NIH) under award numbers R01CA184843 (S.K.); U10CA180821 and U10CA180882 (Alliance for Clinical Trials in Oncology); U24CA196171 (Biospecimens Repository); UG1CA180830, UG1CA233329, and U10CA180888 (SWOG). Detailed support to Alliance available at https://acknowledgments.alliancefound.org.
CLINICAL TRIAL INFORMATION
DATA SHARING STATEMENT
Data collected for the study, including deidentified individual patient data and a data dictionary, will be made available to others. Please contact Alliance for Clinical Trials in Oncology via email: concepts@alliancenctn.org. Data will be made available as required for specific, approved analyses, and all requests will be reviewed prior to approval. Data will be provided from locked, cleaned, and deidentified study database starting with the date of publication.
AUTHOR CONTRIBUTIONS
Conception and design: Kanwal Raghav, Scott Kopetz
Financial support: Scott Kopetz
Administrative support: Alan P. Venook, Scott Kopetz
Provision of study materials or patients: Kanwal Raghav, Federico Innocenti, Heinz-Josef Lenz, Scott Kopetz
Collection and assembly of data: Kanwal Raghav, Fang-Shu Ou, Alan P. Venook, Federico Innocenti, Heinz-Josef Lenz, Scott Kopetz
Data analysis and interpretation: Kanwal Raghav, Ryan Sun, Heinz-Josef Lenz, Scott Kopetz
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Acquired Genomic Alterations on First-Line Chemotherapy With Cetuximab in Advanced Colorectal Cancer: Circulating Tumor DNA Analysis of the CALGB/SWOG-80405 Trial (Alliance)
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Ryan Sun
Consulting or Advisory Role: Boehringer Ingelheim
Research Funding: Sanofi
Scott Kopetz
Stock and Other Ownership Interests: MolecularMatch, Lutris, Iylon, Frontier Medicines
Consulting or Advisory Role: Genentech, EMD Serono, Merck, Holy Stone Healthcare, Novartis, Lilly, Boehringer Ingelheim, Boston Biomedical, AstraZeneca/MedImmune, Bayer Health, Pierre Fabre, EMD Serono, Redx Pharma, Ipsen, Daiichi Sankyo, Natera, HalioDx, Lutris, Jacobio, Pfizer, Repare Therapeutics, Inivata, GlaxoSmithKline, Jazz Pharmaceuticals, Iylon, Xilis, AbbVie, Amal Therapeutics, Gilead Sciences, Mirati Therapeutics, Flame Biosciences, Servier, Carina Biotech, Bicara Therapeutics, Endeavor BioMedicines, Numab, Johnson & Johnson/Janssen, Genomic Health, Frontier Medicines, Replimune, Taiho Pharmaceutical
Research Funding: Sanofi, Biocartis, Guardant Health, Array BioPharma, Genentech/Roche, EMD Serono, MedImmune, Novartis, Amgen, Lilly, Daiichi Sankyo
Federico Innocenti
Employment: AbbVie
Stock and Other Ownership Interests: AbbVie
Honoraria: Tempus
Consulting or Advisory Role: Symberix, Emerald Lake Safety
Patents, Royalties, Other Intellectual Property: United States Patent: "Flavopiridol drug combinations and methods with reduced side effects", Ratain M.J., Innocenti F., Iyer L. Filed on April 12, 2001, serial number 09/835082, United States Patent: "Optimization of cancer treatment with irinotecan", Ratain M.J., Innocenti F., Karabatsos P., Grimsley C., Di Rienzo A. Filed on February 12, 2003, serial number 60/446942, United States Patent: "Methods of identifying risk of bevacizumab-induced proteinuria and hypertension", Innocenti F., Quintanilha J., Lin D., Owzar K., Wang J. Filed on July 17, 2020, serial number 16/932002, United States Provisional Patent Application: "Plasma levels of angiopoietin-2, VEGF-A, and VCAM-1 as markers of bevacizumab induced hypertension", Innocenti F., Quintanilha J. Filed on April 1, 2021, serial number 63/169301
Travel, Accommodations, Expenses: AbbVie
Heinz-Josef Lenz
Honoraria: Merck Serono, Roche, Bayer, Boehringer Ingelheim, Isofol Medical, GlaxoSmithKline, G1 THerapeutics, Jazz Pharmaceuticals, Oncocyte, Fulgent Genetics
Consulting or Advisory Role: Merck Serono, Roche, Bayer, BMS, GlaxoSmithKline
Travel, Accommodations, Expenses: Merck Serono, Bayer, BMS
Kanwal Raghav
Consulting or Advisory Role: AstraZeneca, Bayer, Eisai, Daiichi Sankyo
Speakers' Bureau: Bayer
Research Funding: Daiichi Sankyo/Lilly (Inst), Bayer (Inst), Roche/Genentech (Inst), Guardant Health (Inst)
Alan P. Venook
Consulting or Advisory Role: Merck Sharp & Dohme, Amgen, GlaxoSmithKline, Exelixis, BridgeBio Pharma, Bayer Health, Gilead Sciences, Exact Sciences, Bristol Myers Squibb Foundation/Janssen
Research Funding: Amgen
Patents, Royalties, Other Intellectual Property: Royalties from Now-UptoDate for authoring and maintaining two chapters
No other potential conflicts of interest were reported.
REFERENCES
- 1. Venook AP, Niedzwiecki D, Lenz HJ, et al. Effect of first-line chemotherapy combined with cetuximab or bevacizumab on overall survival in patients with KRAS wild-type advanced or metastatic colorectal cancer: A randomized clinical trial. JAMA. 2017;317:2392–2401. doi: 10.1001/jama.2017.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 3. Misale S, Di Nicolantonio F, Sartore-Bianchi A, et al. Resistance to anti-EGFR therapy in colorectal cancer: From heterogeneity to convergent evolution. Cancer Discov. 2014;4:1269–1280. doi: 10.1158/2159-8290.CD-14-0462. [DOI] [PubMed] [Google Scholar]
- 4. Parseghian CM, Napolitano S, Loree JM, et al. Mechanisms of innate and acquired resistance to anti-EGFR therapy: A review of current knowledge with a focus on rechallenge therapies. Clin Cancer Res. 2019;25:6899–6908. doi: 10.1158/1078-0432.CCR-19-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pietrantonio F, Vernieri C, Siravegna G, et al. Heterogeneity of acquired resistance to anti-EGFR monoclonal antibodies in patients with metastatic colorectal cancer. Clin Cancer Res. 2017;23:2414–2422. doi: 10.1158/1078-0432.CCR-16-1863. [DOI] [PubMed] [Google Scholar]
- 6. Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diaz LA, Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arena S, Bellosillo B, Siravegna G, et al. Emergence of multiple EGFR extracellular mutations during cetuximab treatment in colorectal cancer. Clin Cancer Res. 2015;21:2157–2166. doi: 10.1158/1078-0432.CCR-14-2821. [DOI] [PubMed] [Google Scholar]
- 9. Kim TW, Peeters M, Thomas A, et al. Impact of emergent circulating tumor DNA RAS mutation in panitumumab-treated chemoresistant metastatic colorectal cancer. Clin Cancer Res. 2018;24:5602–5609. doi: 10.1158/1078-0432.CCR-17-3377. [DOI] [PubMed] [Google Scholar]
- 10. Abrams TA, Meyer G, Schrag D, et al. Chemotherapy usage patterns in a US-wide cohort of patients with metastatic colorectal cancer. J Natl Cancer Inst. 2014;106:djt371. doi: 10.1093/jnci/djt371. [DOI] [PubMed] [Google Scholar]
- 11. Goldberg RM, Montagut C, Wainberg ZA, et al. Optimising the use of cetuximab in the continuum of care for patients with metastatic colorectal cancer. ESMO Open. 2018;3:e000353. doi: 10.1136/esmoopen-2018-000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res. 2018;24:3539–3549. doi: 10.1158/1078-0432.CCR-17-3831. [DOI] [PubMed] [Google Scholar]
- 13. Russo M, Crisafulli G, Sogari A, et al. Adaptive mutability of colorectal cancers in response to targeted therapies. Science. 2019;366:1473–1480. doi: 10.1126/science.aav4474. [DOI] [PubMed] [Google Scholar]
- 14. Faheem MM, Seligson ND, Ahmad SM, et al. Convergence of therapy-induced senescence (TIS) and EMT in multistep carcinogenesis: Current opinions and emerging perspectives. Cell Death Discov. 2020;6:51. doi: 10.1038/s41420-020-0286-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Basu D, Nguyen TT, Montone KT, et al. Evidence for mesenchymal-like sub-populations within squamous cell carcinomas possessing chemoresistance and phenotypic plasticity. Oncogene. 2010;29:4170–4182. doi: 10.1038/onc.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Byers LA, Diao L, Wang J, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang AD, Fan F, Camp ER, et al. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res. 2006;12:4147–4153. doi: 10.1158/1078-0432.CCR-06-0038. [DOI] [PubMed] [Google Scholar]
- 19. Boumahdi S, de Sauvage FJ. The great escape: Tumour cell plasticity in resistance to targeted therapy. Nat Rev Drug Discov. 2020;19:39–56. doi: 10.1038/s41573-019-0044-1. [DOI] [PubMed] [Google Scholar]
- 20. Bray SM, Lee J, Kim ST, et al. Genomic characterization of intrinsic and acquired resistance to cetuximab in colorectal cancer patients. Sci Rep. 2019;9:15365. doi: 10.1038/s41598-019-51981-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sartore-Bianchi A, Pietrantonio F, Lonardi S, et al. Phase II study of anti-EGFR rechallenge therapy with panitumumab driven by circulating tumor DNA molecular selection in metastatic colorectal cancer: The CHRONOS trial J Clin Oncol 39 3506 3506 2021. 34270348 [Google Scholar]
- 22. Cremolini C, Rossini D, Dell'Aquila E, et al. Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal cancer with acquired resistance to first-line cetuximab and irinotecan: A phase 2 single-arm clinical trial. JAMA Oncol. 2019;5:343–350. doi: 10.1001/jamaoncol.2018.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data collected for the study, including deidentified individual patient data and a data dictionary, will be made available to others. Please contact Alliance for Clinical Trials in Oncology via email: concepts@alliancenctn.org. Data will be made available as required for specific, approved analyses, and all requests will be reviewed prior to approval. Data will be provided from locked, cleaned, and deidentified study database starting with the date of publication.