Abstract
The interest for machine learning (ML) has grown tremendously in recent years, partly due to the performance leap that occurred with new techniques of deep learning, convolutional neural networks for images, increased computational power, and wider availability of large datasets. Most fields of medicine follow that popular trend and, notably, radiation oncology is one of those that are at the forefront, with already a long tradition in using digital images and fully computerized workflows. ML models are driven by data, and in contrast with many statistical or physical models, they can be very large and complex, with countless generic parameters. This inevitably raises two questions, namely, the tight dependence between the models and the datasets that feed them, and the interpretability of the models, which scales with its complexity. Any problems in the data used to train the model will be later reflected in their performance. This, together with the low interpretability of ML models, makes their implementation into the clinical workflow particularly difficult. Building tools for risk assessment and quality assurance of ML models must involve then two main points: interpretability and data-model dependency. After a joint introduction of both radiation oncology and ML, this paper reviews the main risks and current solutions when applying the latter to workflows in the former. Risks associated with data and models, as well as their interaction, are detailed. Next, the core concepts of interpretability, explainability, and data-model dependency are formally defined and illustrated with examples. Afterwards, a broad discussion goes through key applications of ML in workflows of radiation oncology as well as vendors’ perspectives for the clinical implementation of ML.
1. Introduction
Radiation oncology is a medical field that heavily relies on information technology and computational methods. Even though the goal of radiation therapy can be stated as simply as irradiating the tumor while minimizing the dose to the healthy tissue, numerous and complex calculations are needed to achieve such a goal. From the image reconstruction and analysis steps to locate the tumor and organs, down to the plan optimization process to find the machine parameters that deliver the desired dose, image and data processing algorithms are at the backbone of radiotherapy treatments.
This tight entanglement between software and clinical practice is doing nothing but growing with time and, needless to say, the recent rise of artificial intelligence (AI) tools, specifically machine learning (ML) and deep learning (DL), is disruptively transforming the field of radiation oncology (Thompson et al 2018, Feng et al 2018, Sahiner et al 2019, Boldrini et al 2019). One can find examples of applications of AI/ML/DL algorithms in every block of the radiotherapy workflow, including image segmentation (Seo et al 2020), treatment planning (Wang et al 2020a), quality assurance (Chan et al 2020), and outcome prediction (Isaksson et al 2020), among others (Shan et al 2020, Jarrett et al 2019).
ML/DL has the potential to automate and speed up the whole radiotherapy treatment workflow (Unkelbach et al 2020, Wang et al 2020a, Cardenas et al 2019), freeing time in the physicians schedules to focus on more relevant patient care instead of repetitive and mechanical tasks. More importantly, though, ML/DL can also help standardize and improve the current clinical practice (Sher et al 2021, Thor et al 2021, van der Veen et al 2019), by mitigating variability and suboptimality related to human factors, as well as by transferring the knowledge from more to less experienced centers (e.g., planning of new or emerging treatments, transferring expertise to developing countries, etc). The ESTRO-HERO (Health Economics Radiation Oncology) group has claimed for years a problem of underprovision of radiation therapy (Lievens et al 2020, Korreman et al 2021, Lievens et al 2014), meaning that the optimal utilization benchmark is not met in many countries. With the aging population and the associated increased incidence of cancer, this underprovision will only grow larger. ML/DL can thus play an important role in solving this problem (Korreman et al 2021), but only if we can ensure safe and efficient clinical implementation of this technology.
After a few years of research, the feasibility and potential to use ML models in the clinic has been well demonstrated, and we are now progressively shifting to the implementation phase of either in-house or commercial ML software (Brouwer et al 2020). In 2019 alone, 77 AI/ML-based medical devices were approved by the FDA in the US and 100 were CE-marked in Europe, while back in 2015 the approved devices barely exceeded 10 (Muehlematter et al 2021). Nevertheless, some clinicians may still be reluctant to adopt this technology in the clinical routine. One of the reasons is that they might feel unfamiliar with the technology and its mathematical principles, especially for recent DL models. To overcome this, multiple review articles have been published recently, introducing the main technological pillars of AI/ML/DL to clinicians (Barragán-Montero et al 2021a, Wang et al 2020a, Shen et al 2020b, Cui et al 2020). In parallel, the medical physicists community is working towards a change in the curriculum of the clinicians, to include basic education about AI/ML/DL techniques (Zanca et al 2021, Xing et al 2021). However, the main reason motivating the cautious adoption of ML/DL models in the clinical environment is their sometimes hazardous reliability. Can we guarantee that all outputs provided by the ML model are correct? How can we detect the cases for which the ML prediction has failed? Why or how did the ML model yield that specific result or conclusion? Answering these questions is very often not straightforward for current ML-based applications. This, together with their intrinsic black-box nature, increases the skepticism around ML/DL models and hinders their wide adoption in clinical practice. In the popular acception, a black box is a system whose inner workings are unknown or highly complex. When algorithms are difficult to understand, unveiling their reasoning and their risks of failure becomes very complicated.
The literature is scarce about how to ensure safe clinical implementation of these black-box systems in radiation oncology. But recently, some groups have started gathering recommendations towards that end (Vandewinckele et al 2020, Brouwer et al 2020, He et al 2019, Liu et al 2020, Rivera et al 2020). Developing ML models that guarantee consistently good performance under all circumstances is utopical. However, one can find strategies to increase their transparency and assess the reliability of their answers for each specific situation. Matters of safety and quality standards are addressed by quality assurance (QA) in the broad sense. When processes involve ML/DL, we identify two key concepts that must integrate QA: model interpretability/explainability and data-model dependency.
First, interpretability and explanations of ML models allows the end-user to better understand, debug, and even improve these models (Huff et al 2021, Jia et al 2020, Reyes et al 2020). Often, the terms interpretability and explanability are used interchangeably. However, it is important to distinguish between the property of models to be understandable (i.e. interpretability) and the means that are used to explain non-interpretable models (i.e. explanations). Second, the data-driven nature of ML/DL forces QA to extend beyond the model itself, by investigating the data that feeds it and makes it task-specific, as well as how the model performance depends on it, namely, data-model dependency. On the one hand, the data distribution needs to be carefully analyzed to ensure that it is a faithful representation of the considered problem (Diaz et al 2021, Willemink et al 2020). On the other hand, one can explore how the model performs, for instance, under perturbation of the input data to learn about its robustness (e.g., generalization to similar domains) and precision (e.g., quantification of model uncertainty (Ghoshal et al 2021, Begoli et al 2019)).
In this review, we describe in detail key aspects of interpretability, explainability and data-model dependency in ML/DL, and discuss how they can be applied to increase the reliability and safety of ML/DL applications in the field of radiation oncology. Section 2 starts by reviewing all the possible risks associated with ML/DL models, and provides illustrative examples in the medical field. Section 3 introduces general considerations and technical foundations about interpretability, explainability and data-model dependency in ML. These topics have been studied for years in fundamental ML research, but they only start to integrate the vocabulary of clinical research and practitioners. We believe it is essential to bring this knowledge closer to the clinical environment, in order to provide the radiation oncology community with a well-structured background to develop reliable and safe ML models. Section 4 walks the reader through the radiation oncology workflow and digs into key applications of ML, specifically discussing issues related to interpretability, explainability and data-model dependency. Section 5 wraps-up this manuscript with final conclusions.
2. Risks associated with the use of ML for medical applications
The first step towards a safe clinical implementation of ML models is to become aware of the different risk factors associated with this technology, which is the goal of this section. As ML techniques are essentially data-driven, the main risks associated with their use can then stem from the data itself or the model. Data issues appear when the data used to train our ML algorithm does not reflect the ground truth of the problem at hand, whereas model issues are due to incorrect performance of the model itself. In the following, we identify the main issues in these two categories and provide illustrative examples in the medical field.
2.1. Data
In computer science, the acronym GIGO stands for ‘Garbage In, Garbage Out’, and it refers to the fact that when a system is fed with low-quality data, the output will be deficient likewise. In ML specifically, GIGO can have dramatic consequences as it affects the training of the model. In medical applications of ML, GIGO can affect the patient’s outcome and it is one of the main factors to take into account when aiming at their safe clinical implementation. GIGO has two main roots: insufficient data in quantity and inappropriate data in quality (figure 1).
Figure 1.
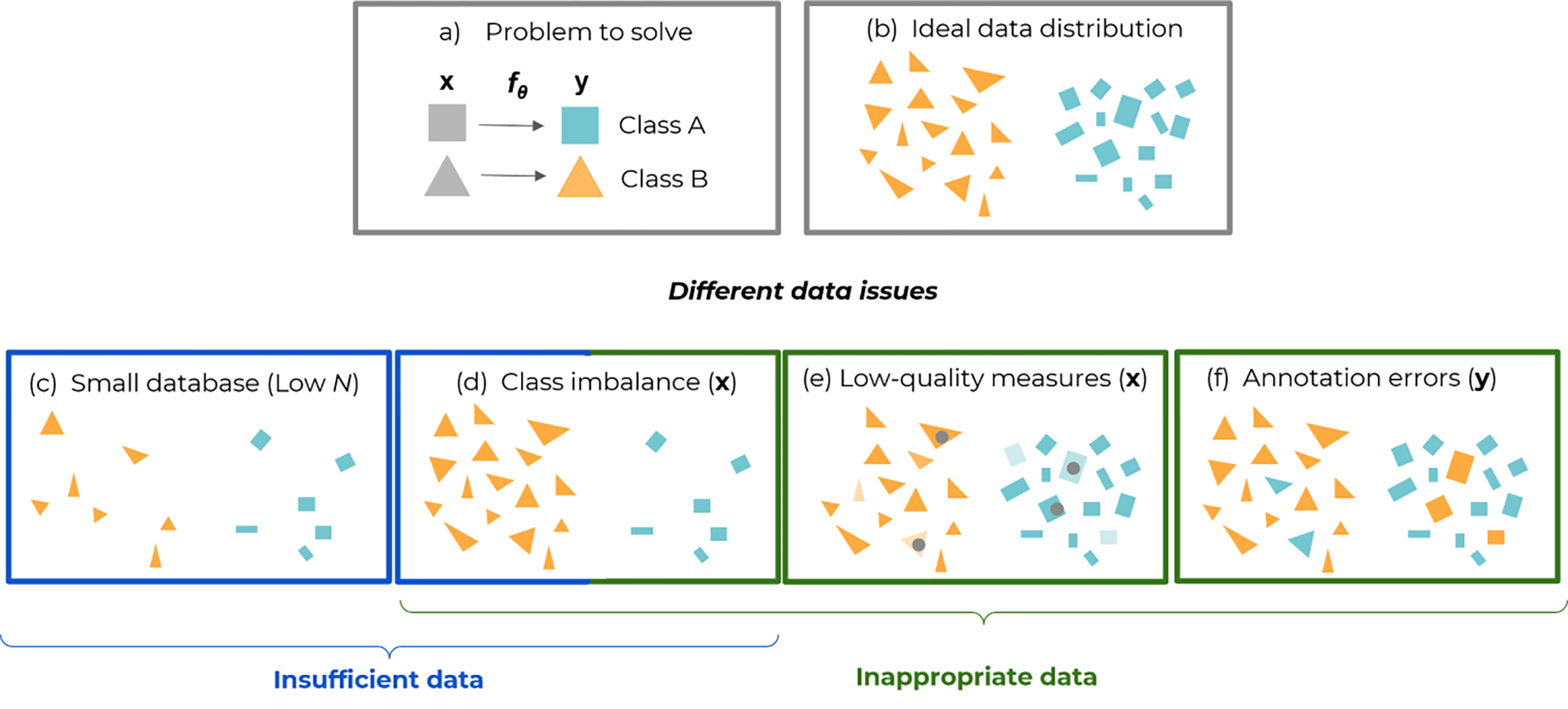
Some data-related pitfalls of supervised learning, exemplified with a binary classification problem. Panel (a) formalizes the problem and how the model maps the inputs (features or images) to the outputs (class labels green and orange). Panel (b) shows an ideal dataset with enough data globally (high N) and in each class. Panel (c) illustrates insufficient data, when the number of total examples N is too low (for all classes). Panels (d) to (f) illustrate cases of inappropriate data: (d) Class imbalance, when class populations are unequal and minor classes might not be given enough importance in the performance figures. (e) Low-quality or corrupted inputs x, e.g., blurred, noisy, or artifacted images, represented by a lighter color and gray dots in the figure. (f) Annotation errors (mistakes in class labels y). To some extent, class imbalance can be seen as a particular case of insufficient data, when one of the classes has a low N with respect to the other(s).
More specifically, most ML applications attempt to learn an unknown phenomenon y = φ(x) in a supervised way, that is, where inputs are mapped to some desired output, with a flexible model y = fθ(x) having parameters θ. A finite dataset of input-output pairs (xi, yi)1≤i≤N is sampled from a population (figure 1.a and 1.b). In this sampling and learning process, insufficient data problems arise when the dataset size N is too low, whereas inappropriate data problems are related to the sampling, measurement, and annotation in the pairs (xi, yi) (figure 1. c–f).
2.1.2. Insufficient data
Insufficient data often result from the difficulty to collect and to annotate data in the medical field, due to cost, ethical issues, or expert availability. A too small dataset is generally unable to reflect all variations that can exist in a (patient) population. The size of the data to be collected typically must grow with the complexity of the task to accomplish. A complicated task usually involves many features or criteria to make a decision. The input dimensionality (e.g., just a few biomarkers, versus images with millions of voxels) and the output dimensionality (e.g., the number of classes or diseases to be distinguished) are typically faithful indicators of complexity. In computer vision, for classification of natural images, rules of thumb state that up to 1000 instances per class can be necessary, and the performance increases logarithmically with the dataset size (Sun et al 2017). In the medical field, the lower availability of data (Willemink et al 2020) is compensated by the greater regularity in images, with simple backgrounds, similar anatomies and orientations in the foreground. For instance, in dose prediction for radiotherapy, models like U-Net are efficient at learning from relatively small datasets (e.g. around 50–100 patients), thanks to a densely connected network architecture (Barragán-Montero et al 2021b, Barragán-Montero et al 2019, Nguyen et al 2019a, Fan et al 2019). Recent applications of U-Net like architectures or yet Generative Adversarial Networks (GANs) for other tasks such as image segmentation (e.g. organ (Nikolov et al 2018) and target volumes (Cardenas et al 2021)), image synthesis (e.g. generation of synthetic CTs from MR images (Maspero et al 2018)), or image registration, have also demonstrated a good performance when trained with databases in the order of one hundred patients or even lower (Sokooti et al 2017). Nevertheless, building a well-curated and up-to-date database of few decens or hundreds (patients) samples still remains a challenge for most medical institutions, and it is often the result of several years of work. For instance, (Grossberg et al 2018) presented the head and neck squamous cell carcinoma collection, comprising data from 215 patients collected during 10 years of treatment (from 2003 and 2013).
2.1.2. Inappropriate data
Inappropriate data covers a wide range of possible problems. In collecting input-output pairs (xi, yi) they can concern the sampling of xi in the population, the measurement of xi, or the annotation yi. Often, medical databases can suffer from several of these issues. Therefore, good data curation algorithms, together with interpretable/explainable ML and the exploration of data-model dependency, can help to properly identify and fix each issue (see Section 3).
Data sampling in the population: domain coverage and class imbalance
To be effective and to generalize to any individual from the population, the collected data must be representative of it, that is, it has to reflect all relevant variations in that population (i.e., domain coverage). In classification tasks, for example, not all variabilities could be represented within a single class or one or several classes might be underrepresented with respect to others in the database used to train the ML model (i.e. minority classes). Often, the technical term used to refer to this situation in ML is “class imbalance” (Johnson and Khoshgoftaar 2019). This results in wrong or reduced accuracy predictions for those underrepresented classes. In fact, the ML model will focus mainly on the majority class during learning, and in extreme cases, may ignore the minority class altogether. Class imbalance can be also seen as a particular case of insufficient data (Section 2.1.1), where the number samples in the minority class(es) (Nm) is much lower than that of the dominating class(es) (Nd), i.e. Nm << Nd (Figure 1). Notice, however, that class imbalance can occur even for models trained with databases containing a large total N, as long as the ratio between classes remains inappropriately balanced. This is the reason why we have decided to include class imbalance in the “inappropriate data” category.
In the medical field, the minority class can be represented by patients groups (e.g. with positive/negative diagnosis, rare diseases, patients under/over certain age, gender, ethnicity, etc…), but also at the pixel level (e.g. 2% of pixels of class A and 98% pixels of class B).
At the patient groups level, a common example of imbalanced datasets are those for skin cancer, which consist predominantly of healthy samples with only a small percentage of malignant ones (Zunair and Ben Hamza 2020, Mikolajczyk and Grochowski 2018, Emara et al 2019). Another example is how gender unbalance between male and female patients in the training database can lead to biased ML models. For instance, a recent study analyzed the effect of gender imbalance when training ML models to diagnose various thoracic diseases (Larrazabal et al 2020). A consistent decrease in performance was observed when using male patients for training and female for testing (and vice versa).
Regarding the pixel level, the most trivial example is the detection or segmentation of small lesions or organs from medical images (Bria et al 2020, Gao et al 2019, 2021). A good illustrative case is the segmentation of organs for head and neck cancer patients, where the ratio between small and big organ volumes can reach a factor 100 (e.g., optic structures versus parotids or oral cavity)(Gao et al 2019). For instance, a difference up to 20% in Dice coefficient for the ML model accuracy can be found between the smallest organs (e.g. optic nerves and chiasm) and the bigger ones (Tong et al 2018)
Data measurement: low quality or corrupted records
As soon as population sampling issues are sorted out, another caveat concerns the quality of the records in that sample. For example, in an application that involves medical images, those can be more or less noisy, blurry, or subject to artifacts (Dodge and Karam 2016). Concepts like image definition, (optical) resolution, contrast, or signal-to-noise ratio are important here and condition even more ML performance than it does for human observers, who can more naturally disregard artifacts and compensate for noise or blur. This is really the classical meaning of ‘garbage in, garbage out’ in signal processing: corrupted data leads to poor performance. Typical examples of noise and artifacts in medical images include CT artifacts due to metal implants (Kalender et al 1987, Barrett and Keat 2004), ring and scatter noise in Cone Beam CT images (Zhu et al 2009), or artifacts due to patient motion (Zaitsev et al 2015). In extreme cases, even slight perturbations can have dramatic effects and can be exploited to defeat or ‘attack’ the model with so-called ‘adversarial examples’ (Finlayson et al 2019, Szegedy et al 2013). For instance, adding adversarial noise to an image of a skin mole, classified by the model as benign, can suddenly make the model change the output to malign (Finlayson et al 2019).
For noise, blur, and low contrast, improving the image acquisition device or tuning its parameters are straightforward recommendations. Data curation to avoid badly corrupted records or the presence of confounding artifacts can also improve performance. Often, this is at the price of lower robustness and generalization capability, since ML models are left totally unaware of these outliers and pathological cases at training time, although they might still show up when the ML model is queried. Some unwanted artifacts in images can also turn into confounders or spurious revealers, like the presence of a plaster cast in radiological images when it comes to spot broken bones, or image tags that correlate with patient, disease, or treatment categories that should be predicted from the image content, not from such side information (Badgeley et al 2019, Zech et al 2018).
Another type of low quality records include the cases for which data is uninformative or not informative enough. The records do not convey all the necessary information to solve the problem at hand. For instance, an image with a small field of view that does not cover (or not entirely) the region of interest for a diagnosis or segmentation model would be considered uninformative. Another example is when the necessary information is spread over several sources and the model has access to only one or few of them. For instance, ML models for segmentation of tumor volumes are often provided with only one image (e.g., CT), while in clinical practice the physician gathers information from several sources to perform the segmentation (e.g., PET, MR, endoscopy images or meta-data like age, patient’s physical condition, other diseases, etc) (Ye et al 2021, Moe et al 2021).
Data annotation: low quality annotation, label noise, or inter-observer variability
In the collected data pairs (xi, yi), yi is responsible for the supervision of the training, that is, to associate the correct output to any input record xi. The quality of this annotation or label is thus of paramount importance (Karimi et al 2020, Frenay and Verleysen 2014).
The most straightforward example of low-quality annotations is the presence of inaccuracies induced by human errors when labeling medical images used for training a ML model. For instance, (Yu et al 2020) recently studied the effect of using inaccurate contours when training an automatic segmentation ML model for the mandible. They showed a decrease in the Dice coefficient between 5% and 15% when the ratio of inaccurate contours increased from 40% to 100%. Another recent study investigated the effect of using erroneous labels when training a ML model for skin cancer classification (Hekler et al 2020), reporting a 10% decrease in accuracy when using the imperfect labels versus the perfect ground truth.
Another major data quality issue in the radiation oncology field is data heterogeneity or variability. Overall, these variabilities can be viewed into two categories: 1) lateral variability and 2) longitudinal variability. Lateral variability describes the difference in data distributions for a given time frame. Some examples include the interobserver variability in radiotherapy treatment planning (Berry et al 2016, Nelms et al 2012), the variability in delineation of tumor and organ volumes across different physicians (Apolle et al 2019, Veen et al 2019, van der Veen et al 2020), or the differences between clinical practices among institutions (Eriguchi et al 2013, Gershkevitsh et al 2014). In contrast, longitudinal variability describes the difference in data distributions over time, such as the evolution of treatment techniques (Shang et al 2015), the introduction of new delineation guidelines (Grégoire et al 2018, Brouwer et al 2015) or fractionation protocols (Parodi 2018, Dearnaley et al 2017).
Lateral and longitudinal variability are often entangled together within retrospective databases containing patients treated with radiotherapy by different physicians, institutions, and at different time points. Although the individual effect of each source of variability is hard to quantify, a recent study has demonstrated that the use of homogeneous data increases the accuracy and the robustness of ML models (Barragán-Montero et al 2021b). The study compared two ML models for radiotherapy dose prediction for esophageal cancer. The first model was trained with a variable database (i.e., retrospective patients, different time frames, planning protocols, treating physicians), while the second was trained with a homogeneous one (i.e., same time frame, same treatment protocol, same physician). The second model was able to reduce the mean absolute error of the predicted dose distribution.
Yet another important issue is the presence of annotation bias. General examples of bias in the medical domain include over-diagnosis of certain diseases (Blumenthal-Barby and Krieger 2015), or bias induced by gender, race or socioeconomic factors (Schulman et al 1999, Bach et al 1999, Forrest et al 2013, Lievens and Grau 2012, Obermeyer et al 2019). For instance, (Bach et al 1999) reported significant racial differences in the treatment of lung cancer. They observed that black patients are less likely to receive surgical treatment than white patients, which entailed a decrease of 8% for the five-year survival rate of this population. Often, one of the most important sources of this kind of bias is the socioeconomic level of the patient, which is also well known to affect the treatment chosen and delivered for cancer patients (Zhou et al 2021, Lievens and Grau 2012, Ou et al 2008, Forrest et al 2013).
Last but not least, variability and biases can somehow co-exist in many scenarios. For instance, in lateral variability, medical experts can disagree persistently about the annotation of some data instances. Across consistent groups of experts, this can be seen as biases, whereas for ML models these discrepancies are seen as a variability around a consensus that might not be agreed upon yet. The framework of supervised learning, with functional models ŷ = f(x) can only produce a single output ŷ for a given input x. If several outputs need nevertheless to be produced, then new explicative inputs must be identified and appended to x. Alternatively, one can also train an individual model for each possible output ŷl, like if several ground truths were possible for a given x. For instance, a recent study about radiotherapy dose prediction for prostate cancer patients illustrated the differences in treatment planning practices between different doctors and institutions, and generated specific ML models for each clinical practice (Kandalan et al 2020).
2.2. Model and learning frameworks
Most current ML methods extend and upscale supervised learning techniques developed by statisticians over the past 100 years (Friedman et al 2001). Supervised learning for ML algorithms do not substantially differ from linear or logistic regression models. In all cases, they find a function y = fθ(x) that models the phenomenon under study y = φ(x). Model fitting amounts to minimizing the discrepancy between the ground truth y, as measured or annotated, and ŷ as yielded by the model. ML tries to identify the relationships that map the features in x to the outputs y. In the following, we present several limitations related to this learning framework, which should be carefully taken into account when implementing ML models in the clinical environment
2.2.2. Non-causal correlations and hidden confounders
When trying to find the relationships that map the features in x to the outputs y, the optimal solution is typically the one that finds strong dependencies between the considered features (e.g., patient’s smoking condition) and outcomes (e.g., probability of lung cancer). However, the weakness of supervised learning, and most ML frameworks in general, is that it cannot infer causality out of the input-output dependencies, which can be either causal and relevant or spurious and confounding in the interpretation of the model. This represents an important risk when it comes to medical applications (Castro et al 2020). For instance, a recent study found that a convolutional neural network (CNN), trained to process X-rays images to predict pneumonia, was using the hospital information to make predictions, often disregarding the areas of the image with radiological findings relevant to the underlying pathology (Zech et al 2018). Specifically, the CNN was trained with databases from multiple hospitals, where the prevalence of pneumonia was very different. The hospital information was retrieved from a hospital-specific token, located in the corner of the image, and other image features indicative of the radiograph’s origin (figure 2). This information was strongly correlated with the prevalence of pneumonia in the considered dataset, without any causality, thus acting as a hidden confounder and leading to the so-called “shortcut learning” (Geirhos et al 2020). One can find many other examples of confounders and spurious correlations in the literature of ML models for medical applications. For instance, another study reported that an artificial neural network, trained to estimate the probability of death from pneumonia in the emergency room, labeled asthmatic patients as having a low risk of death, because in the training data this cohort was seeking care faster than non-asthmatic patients (Cooper et al 2005). Yet another recent study found that colon cancer screening or abnormal breast findings were highly correlated to the risk of having a stroke, with no clinical justification (Mullainathan and Obermeyer 2017).
Figure 2.
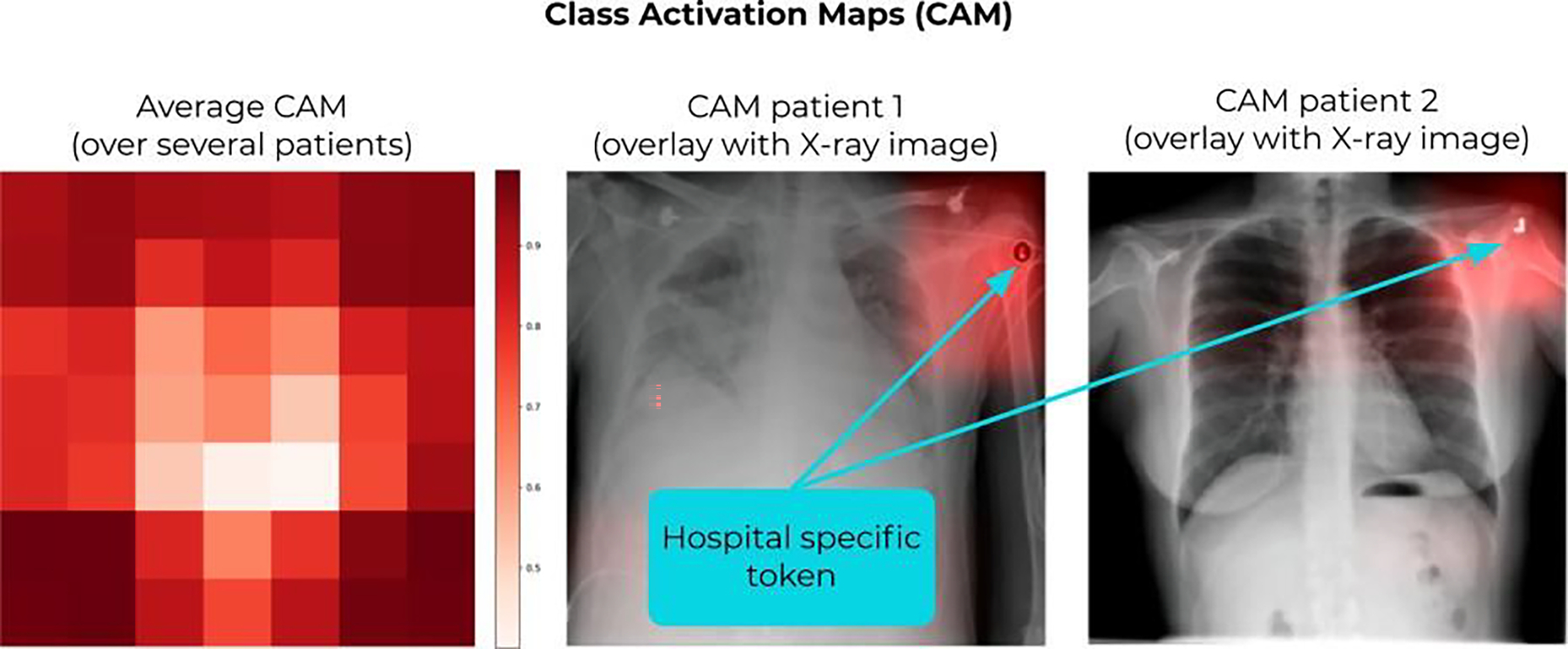
Adapted from (Zech et al 2018). Class activation maps (CAM) showing the relevant regions considered by the CNN to make the prediction. The model in this study was trained to predict pneumonia from X-ray images. By looking at the CAMs, they found out that the model was looking at the corner of the images, and in particular, at the hospital-specific metal token (a hidden confounder) to make the prediction. (Left) CAM averaged over several patients; (middle and right) examples of CAMs for two patients.
2.2.3. Model complexity: size, nonlinearity, and opacity
Beyond the inability to identify relevant causality, the interpretability of ML models can be further impeded by their sheer size and complexity. The advantage of state-of-the-art ML models (i.e. CNNs, GANs, …) over classical linear models is their increased capability to find a function that approximates the problem under study (y = fθ(x)). This is often done by drawing on nonlinear relationships between variables (e.g., patient characteristics) and outcomes (e.g., mortality probability). Finding the final function can be accomplished by either directly estimating the parameters of a nonlinear function of fixed complexity (e.g., an artificial neural network) or estimating the complexity and shape of a nonlinear function (e.g., non-parametric algorithms like gradient boosting) (Friedman et al 2001). In all cases, the consequence of nonlinearity is an increased number of parameters required to build that function fθ(x). A modern ML model can have between a few thousands and several millions of trainable parameters. For instance, Nguyen et al. (2019) compared different ML models for predicting the radiotherapy dose for head and neck cancer patients, reporting between 3 and 40 millions of trainable parameters for the considered models (Nguyen et al 2019a). The bigger the number of parameters, the less tractable the model becomes, thus reducing the interpretability of the provided function and turning it into a black-box. Notice that the same issue happens for big linear models, too. Promoting sparsity, that is, the parsimonious use of the available features and variables, to reduce the number of effective (non-zero) parameters) (Vinga 2021, Oswal 2019, Rish and Grabarnik 2014) can mitigate this issue of size and interpretability of large black-box models. For such models, identifying hidden confounders and non-causal correlations becomes very difficult, which certainly increases the risk when using them for medical applications. This lack of interpretability has been recently highlighted as one of the most important issues to be addressed in the medical domain before ML algorithms can be widely accepted in the clinic (Reyes et al 2020, Luo et al 2019).
2.2.4. Task-specialized learning, static models, and low generalization
Supervised learning is often cast within a simplified framework that ignores time, where all the dataset is supposed to be known at once and engraved in marble for eternity. Any change entails retraining from scratch. In other words, most ML models cannot learn incrementally, interactively, nor in real-time. They are trained with data from past experience and they become fixed and static models as soon as training ends. This represents an important limitation when it comes to their application in the ever-changing medical field: technologies improve (Shang et al 2015), medical protocols evolve (Grégoire et al 2018, Parodi 2018), and the distribution of patient populations change over time (Chai and Jamal 2012). In this fast-moving world, static AI models quickly become irrelevant. Therefore, it is imperative to shift towards models and frameworks that can quickly adapt to new settings or changing distributions over time. The framework of supervised learning is also essentially specific to a task and exclusively driven by performance at that task. This means that a model trained for a particular application offers no real guarantee to be good at other similar tasks, and the learnt skills are hard to reuse and/or generalize. For instance, specific ML models are currently trained to predict the radiotherapy dose for each cancer location (e.g., head and neck (Nguyen et al 2019a), lung (Barragán-Montero et al 2019), breast (Ahn et al 2021), etc.), instead of reusing the learned skills from one location to another. The same issue can be observed for other applications, such as diagnosis or organ segmentation models. In order to be more efficient and increase the generalization capabilities, future ML in the medical field would require stronger models, with an increased capability to reuse the learning skills. This paradigm shift has been coined as the “weak versus strong AI”.
The low generalization capability of current ML models is widely debated in the literature. In the medical domain, many publications state that, for a successful clinical implementation, ML models should be able to generalize to new data, that is, keep performing well enough on records coming from different hospitals, images from different scanners and vendors, different imaging and treatment protocols, different patient populations, data changes over time, etc. A large number of studies have been published focusing on the question of generalization. For instance, (Liang et al 2020) illustrated the problem of generalization with a ML model trained to convert CBCT into synthetic CT images. The authors trained the model on CBCT images acquired from one vendor’s scanners for head and neck cancer patients, and they quantified the decrease of performance when applying the model to images from another vendor’s scanners and from different locations (e.g., prostate, pancreatic, and cervical cancer). In (Feng et al 2020), the generalization issue was illustrated with a model trained to segment thoracic organs. The model could not generalize to their local dataset because they used an abdominal compression technique, whereas the training set was acquired with free breathing. The subtle shift of thoracic organs due to the abdominal compression caused significantly worse performance on the local dataset. Similarly, (Pan et al 2019) studied the generalization of a ML model to classify abnormal chest radiographs from different institutions. The generalization across different scanners has also been a topic of discussion for models trained to segment MR images (Yan et al 2020, Meyer et al 2021). Other examples include exploring the generalization of ML models for fluence map prediction in radiotherapy treatment planning (Ma et al 2021), generalizability in radiomics modeling (Park et al 2019, Mali et al 2021), or generalization of models for classification of histological images (Lafarge et al 2019). Another well-known example is the study by (Zech et al 2018), already discussed in Section 2.2.1 (Figure 2). The ML model was not able to generalize to radiographs from other hospitals because its learning had been biased by a hidden confounder (i.e., the hospital-specific metallic token).
Generalization is a very abstract term, and the examples above show that poor generalization can be frequent. Recently (D’Amour et al 2020) introduced an umbrella term to cover all the seemingly different failures to generalize in current ML: “underspecification”. It refers to the typical inability of the ML pipeline (training, validation and testing) to ensure that the model has seen and encoded all the relevant variabilities of the underlying system or problem. (Eche et al 2021) discuss how this concept echoes in the medical field, from the perspective of radiologists. They relate underspecification to the aforementioned antagonism of “weak versus strong AI”. They also distinguish narrow and broad generalization. Narrow generalization corresponds to the case that is considered by design in most validation frameworks: test or deployment data are supposed to be independent and identically distributed (i.i.d.) as data in the training and validation sets. Independence guarantees the new data is unseen, while the identity of the underlying distribution ensures consistent predictability. In contrast, broad generalization aims at maintaining predictability if the deployment data are independent but possibly differently distributed. The deployment data distribution can then have other or slightly shifted variabilities than in training and validation. For this reason, broad generalization is also known as (distribution) domain shift or drift. If generalization problems arise, we can refer to our two-fold categories in this section: data and model issues. A model cannot generalize properly if the training data and the actual data at deployment time are not i.i.d., that is, the former is not representative of the latter (see section 2.1), or if the model has not learned correctly, due to hidden confounders, overfitting to (noisy) training data, etc. Broad generalization to non-i.i.d. datasets is a much more ambitious goal and it aims at strong AI, closer to natural intelligence, where general knowledge is acquired and re-used across analogous problems and tasks. Although strongly desirable, broad generalization is controversial. In (Futoma et al 2020) the authors discuss how seeking broad generalisability can be detrimental to the clinical applicability of some ML models, and they provide some illustrative examples. Imagine, for instance, a ML model with an excellent performance for diagnosis of a certain disease in hospital A, properly generalizing to the entire patient population in that hospital. The model might not work with equal performance for hospital B, since the patient population might differ (domain shift and out-of-domain samples). However, trying to change the model to increase the performance for hospital B might be at the cost of lowering the performance for hospital A, in the same way as when individual human experts get replaced with a single all-rounder. For current ML models there is a trade-off between performance and generalization, which must be carefully considered for clinical applications. In this case, building a new (specific) model for hospital B would be more appropriate than using a general model with lower performance. Futoma et al. claim that we should stop demanding broad generalization and focus on understanding how, when, and why a machine learning system works.
3. Interpretability, explainability and data-model dependency
The previous section introduced the different risk factors of ML models for medical applications, clearly distinguishing two categories: data and model issues. However, in practice, data and model issues are often entangled, and identifying the actual risks for a given medical application is not straightforward. In order to properly identify and fix each risk factor, we must implement strategies that enable us to interpret and/or explain the behavior of ML models, as well as to explore the data and how the model performance depends on it. More importantly, this entanglement between data and model issues makes the possible range of solutions a non bijective problem, i.e., a certain technique can be the solution to several of the aforementioned issues in Section 2, and vice-versa, a certain issue can be fixed (or mitigated) by different techniques. For instance, providing explanations about the model behavior may reveal non-causal correlations involving confounders; but they can also be revealed by exploring the performance of the model in different datasets or related tasks. Figure 3 presents a schematic view of the concepts described in this section, in order to guide the reader to understand how these techniques connect and serve as solutions to the risks presented in Section 2, ensuring a safe and efficient clinical implementation of ML. Section 3.1 will cover general concepts and key techniques for interpretability and explainability. These techniques can be used to inspect if a ML model has learnt the underlying problem correctly, thus helping to identify data issues, hidden confounders, etc. Section 3.2 will cover key concepts related to the data and the learning process. On the one hand, targeting directly the data distribution to avoid insufficient and low-quality data will ensure that the ML model is encoding and learning the problem correctly. This includes data curation to detect and fix possible data issues, data augmentation to ensure a sufficient domain coverage, and techniques to efficiently incorporate (expert) prior knowledge about the domain. On the other hand, analyzing how the model reacts to different and external datasets (i.e., test data augmentation or stress testing), and estimating its uncertainty, can serve to further quantify the performance and generalization capacity. Lastly, a full section is dedicated to describe and discuss different learning frameworks proposed in the ML community to achieve robust and efficient learning, becoming one step closer to strong AI models.
Figure 3.
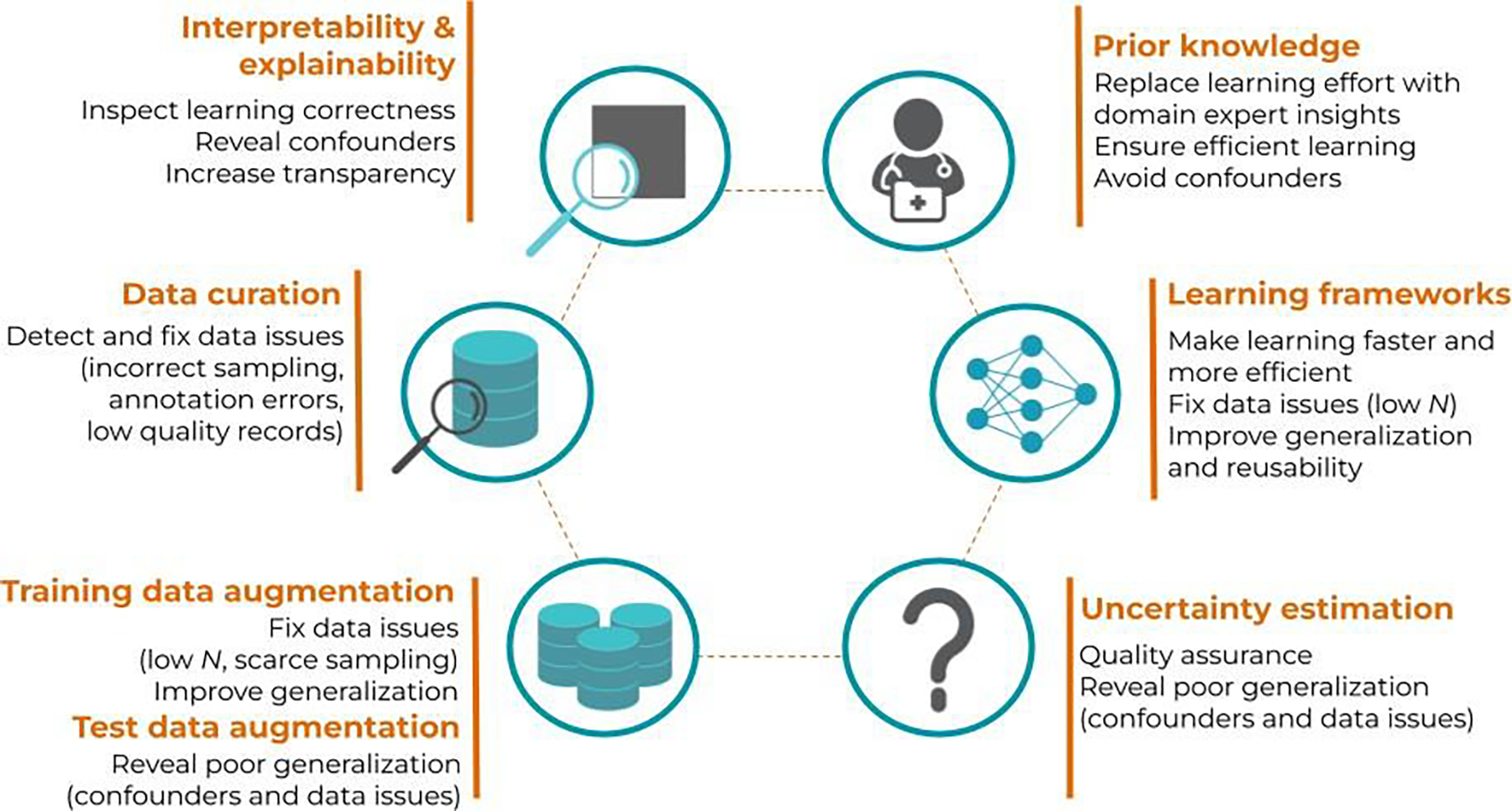
Schematic view of the different concepts presented in this topical review and how they can serve as key solutions to overcome the limitations of current ML models, ultimately ensuring a safe and efficient clinical implementation.
3.1. Interpretability and explainability
Although the terms interpretability and explainability are often used interchangeably (Huff et al 2021, Reyes et al 2020, Luo et al 2019), it is important to stress the difference between the transparency of the model to the end-user (i.e., interpretability), and the techniques used to provide insights about the inner workings of black-box models (i.e., explainability). In this section, we provide basic background knowledge about interpretability and explainability, so that the reader can make a conscious choice when aiming at the clinical implementation of ML methods. Please note that this is not an exhaustive review of all existing methods for interpretable and explainable ML, but rather an introductory section to these topics for the medical community. For extensive technical reviews we refer to (Arrieta et al 2020, Doshi-Velez and Kim 2017).
3.1.2. Interpretability
Interpretability is a property of models (and sometimes decisions) to be understandable by their users (Guidotti et al 2019, Arrieta et al 2020). Although the questions about interpretability have been around for a few decades already (Kodratoff 1994) (Adadi and Berrada 2018), the vocabulary and its conceptualization were not so clear. Until 2015–2016, interpretability was identified in the ML literature by several different terms (interpretability, understandability, comprehensibility, etc.) (Bibal and Frénay 2016). Furthermore, the problems of providing understandable, trustworthy, or justifiable models were confounded. With the growth in use of ML and, in particular, DL, in our society, the ML literature had to focus on interpretability.
In fact, interpretability is a concept that is hard to define because of its subjective nature (Bibal and Frénay 2016). For example, a model can be interpretable for a ML expert, but not for a lay person. In particular, a model that would include and manipulate information that a physician can easily understand can, on the contrary, be difficult to understand by a radiotherapy technician or a dosimetrist. Objectively quantifying interpretability is hard and has mostly been done in the ML literature through the complexity of models, excluding the content of these models. For instance, the bigger a decision tree is (i.e., the more nodes it has), the less interpretable it gets. Similarly, the more non-zero coefficients a linear model has (i.e., the less sparse it is), the less interpretable it is. Some models, specially those with highly nonlinear nature like neural networks (see Section 2.2.2), are assumed to be black boxes in practically all cases, as they always are structurally complex, even if they manipulate understandable information.
Although controversial (Rudin 2019), most researchers rely on the hypothesis that the more complex the model is, the better accuracy it has. For instance, if the underlying relationship between features and outcome is nonlinear, the result will be models with likely better accuracy compared to linear models. Similarly, shallow ML models are often overperformed by deep models (Liang et al 2019a) (Chauhan et al 2019). Hence, what we trade for better accuracy is a higher complexity, and thus worse interpretability of ML models (Valdes et al 2016a, Caruana et al 2015). Those against this hypothesis argue the existence of a set of equally-accurate models for a given problem, with different levels of complexity and interpretability (i.e., Rashomon sets) (Fisher et al 2019, Rudin 2019). Thus, the problem is not the absence of accurate and interpretable models, but the difficulty to find them.
Several authors are actively working in developing accurate and interpretable ML models (Valdes et al 2016a, Caruana et al 2015, Luna et al 2019). For instance, Valdes et al. (2016) developed an improved version of classical decision trees (based on boosting) for a patient stratification tool. The model achieved a high accuracy while being rather transparent, since the subpopulations defined by the leaf nodes of decision trees could easily be interpreted by human experts. Another example is the use of Generalized Additive Models (GAMs), which create nonlinear transformations of individual variables, later combining them into a generalized linear model. The contribution of each variable can be interpreted from the individual graphs representing the nonlinear transformations (Caruana et al 2015). Yet another example is the recent work of Luna et al., who created a further improved decision tree by exploiting the mathematical connection between individual partitions and gradient boosting. The resulting decision trees were smaller and, as such, more accurate (Luna et al 2019). Despite the promising results obtained by these algorithms, whether they can obtain similar performance on more complicated medical problems remains to be seen.
The complexity of the model is only one of the multiple factors that are involved in the concept of interpretability (Guidotti et al 2019). Indeed, this feature does not suffice, as mathematically complex models can be made understandable through their representation. For instance, what makes decision trees interpretable is not the mathematical complexity behind those trees, but the fact that a tree representation is easy to follow by humans. After the complexity of models, the second factor is therefore the possible representations of this model. Third, as previously mentioned, the expertise of the user also plays a major role. The interpretability of decision trees and their useful representation can be low for someone who has never seen any decision tree, while it can be high for a ML expert.
Finally, the time provided to grasp the model is also a factor of interpretability. With an infinite amount of time, all models can be understood. What makes complex models hard to grasp is that they have to be understood in a short period of time. Therefore, the shorter this period of time is, the more difficult it is to interpret the model. This means that in a clinical environment, where the schedules are very tight, for a model to be interpretable, it must largely be less complex than in other contexts with milder time constraints.
Another way to see the aforementioned factors (e.g. complexity, representation, and time) is that if one of them is low, the others have to compensate. For instance, if the period of time to grasp is very short (e.g., in a case of medical emergency), then (1) the intrinsic complexity of the model must be low, and/or (2) the representation of the model must make it easy to grasp, and/or (3) the users (in this example, the emergency caretakers) must be trained to be experts in those models. Note that the concept of explainability (i.e., the ability to explain the inner workings of the model) is also determined by the same factors.
3.1.3. Explainability
When a model is not interpretable (i.e., it is a black box), bu7t its scrutiny is still important or necessary (e.g., by law, to enable a safe clinical implementation or simply to increase trust of the medical practitioners), another property is considered: its explainability (Guidotti et al 2019, Arrieta et al 2020). Explainability is the capacity of a model to be explained, even if not totally interpretable. The question “is the model understandable by itself?” (figure 4) is therefore the first to be answered before unnecessarily using explanation methods if the model is already interpretable. If the answer is negative, there are different approaches to provide explanations, depending on the accessibility of the inner workings of the model (model-specific versus model-agnostic explanations), as well as on the nature of what should be explained (local versus global explanations).
Figure 4.
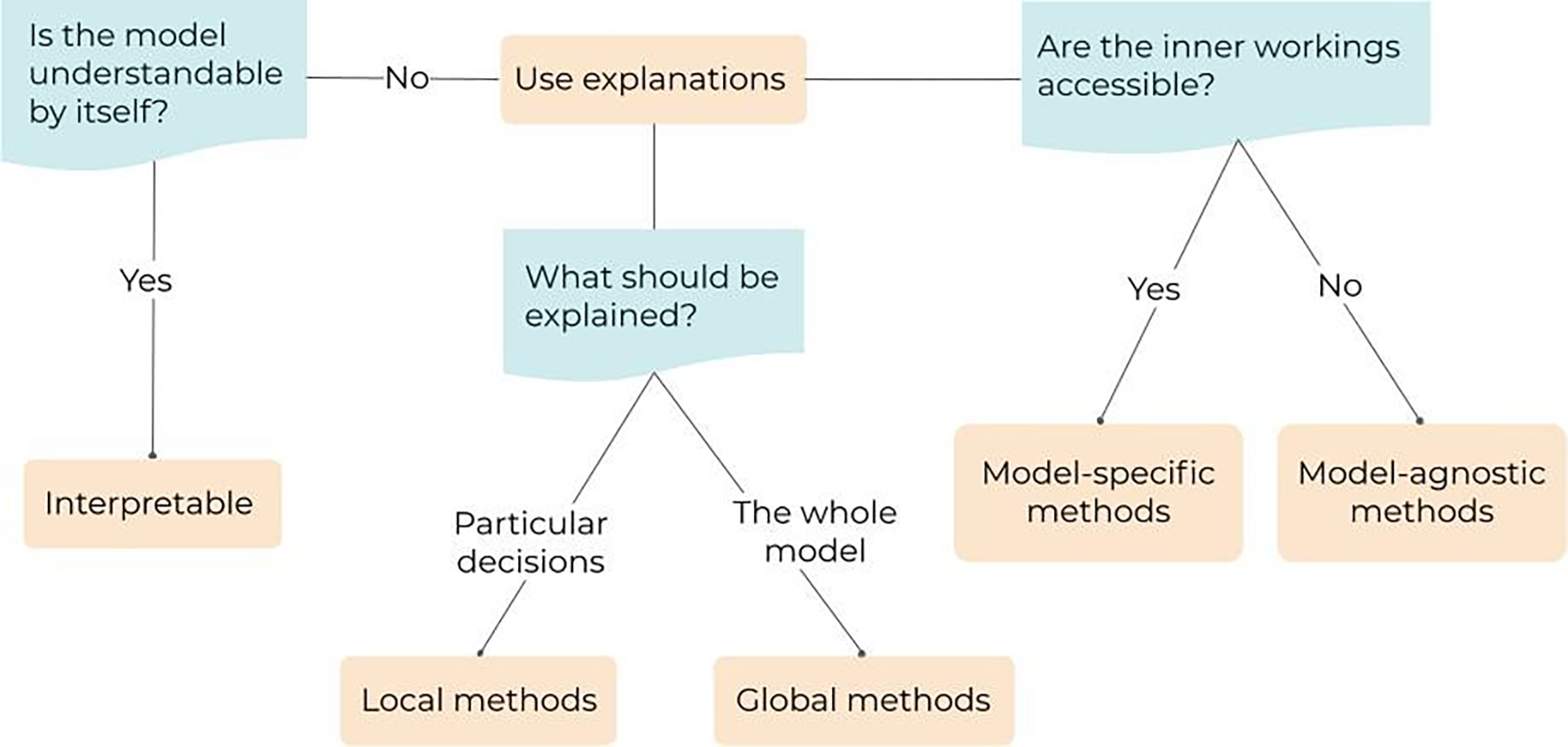
Pipeline describing the typology and the selection of the explanation method. First, if the model is already understandable, it is said to be interpretable. Second, if the model is not understandable, two questions need to be answered to select the right explanation technique: (1) are the inner workings of the model accessible? and (2) what needs to be explained (the whole model or particular decisions)?
Model-specific versus model-agnostic explanations
If the elements of the inner workings of the model are accessible, this information can be used to provide explanations about the model behavior. In these cases, the way the models are built can provide clues about the model decisions. These explanations are model-specific as they cannot be used, as they are, to explain a completely different model. Notice that the difference between the access to these elements of explanation and interpretability is that these elements do not fully explain the model. They are just characteristics of the models that can be exploited to gain insights about its inner workings. These clues may not be enough for gaining the trust of users or, in certain cases, for the law, but it is a first step that makes black boxes a bit more transparent. Two examples detailed just below of model-specific explanations are the feature importance provided by the out-of-bag error in bagging methods like random forests or boosted decision trees, and saliency maps when there is an access to the gradients in artificial or convolutional neural networks (Simonyan et al 2013).
Random forests (Breiman 2001) use different subsets of instances when training the different decision trees in the forest. For each decision tree, the subset of instances that are not used to train the tree (i.e., that are out of the bag) can be used to compute a certain error called the out-of-bag error. The feature importance in the forest is then provided by the effect of perturbing the feature values on the out-of-bag error. If the out-of-bag error changes when perturbing the feature values, this means that the feature is important. For instance, a recent study used the out-of-bag error for highlighting the most important features of a ML model applied to detect lung cancer from CT radiomics and/or semantic features (Bashir et al 2019).
If the gradients of a model are accessible, they can be used to explain the model. For instance, when predicting an image class, convolutional neural networks (CNN) back-propagate the decision on the class to the pixels through the gradients. Looking at the gradients when back-propagating has the effect of providing, for each pixel, the importance of the pixel on the prediction. The resulting image, where pixels are highlighted with respect to their contribution to the prediction, is called a saliency map (Simonyan et al 2013). Other gradient-based explanation techniques have been developed since then, like Grad-CAM (Gradient Class Activation Maps) and all its variants (Selvaraju et al 2017). Gradient-based techniques have been extensively used in medical applications to explain the performance of ML models (Singh et al 2020, Huff et al 2021). A popular example is the study by (Zech et al 2018), already mentioned in Section 2.2.2, where a CNN was trained to predict pneumonia from X-ray images (figure 2). By using class activation maps (CAM) (Zhou et al 2016), they discovered that the CNN was not looking at relevant areas for the disease in the X-ray images. Other examples include the study of (Diamant et al 2019), where a CNN was trained to predict treatment outcome of patients with head and neck cancer, and Grad-CAMs were used to visualize the areas of the CTs that were found to be relevant for the prediction. Yet another example is the study by (Liang et al 2019a), who trained a CNN to predict pneumonitis as a side effect from thoracic radiotherapy, and used Grad-CAM to locate the regions of the dose distribution that were relevant to the prediction.
Another idea is to test whether activations, in a chosen layer, relate to predefined concepts by defining Concept Activation Vectors (CAV) (Kim et al 2018). The idea is similar to saliency maps, except that it is the sensitivity of the activations with regards to predefined concepts that is investigated, instead of a sensitivity with regards to the input (e.g., the pixels). This strategy is sometimes called explanations through semantics (Reyes et al 2020), since it allows us to explain the features learned by the model to the users in terms of human-understandable concepts. Concept Vectors have not yet been used in many medical applications, but a good illustrative example is the study from (M et al 2020). They applied CAV and an extended version of it, Regression Concept Vectors (RCV), to provide explanations for CNNs trained to diagnose breast cancer from histopathological Whole Slide Imaging and retinopathy of prematurity from retinal photographs. They used concepts such as the area or the contrast of the image to describe the visual aspect of the learned features.
In some cases, the black box does not provide any information about its inner workings. This can be, for instance, because the model is property of a company that does not want to provide access to the inside of its black box. In such a case, generic methods for explaining black boxes (also called model-agnostic methods) are used. These agnostic methods work on analyzing the decisions made by the black box when particular inputs are provided.
Agnostic feature importance highlights the input features that seem to be the most important ones when making a decision (Fisher et al 2019). One particularly well-known technique of agnostic feature importance is SHapley Additive exPlanations (SHAP) (Lundberg and Lee 2017). Recently, SHAP has been used to provide explanations of a model trained to predict locoregional relapse for oropharyngeal cancers (Giraud et al 2020), to interpret a model trained to predict 10-year overall survival of breast cancer patients (Jansen et al 2020), or yet to produce heat maps that visualize the areas of melanoma images that are most indicative of the disease (Shorfuzzaman 2021).
Notice that model-agnostic can have two different meanings in the literature. The first one, presented here, considers that the explanation is model-agnostic because no assumption is made about the inner workings of the black box (Guidotti et al 2019, Molnar 2019). The second meaning of “model-agnostic” is that the explanation technique can be applied to a broad range of different models (Arrieta et al 2020, Das and Rad 2020). This distinction makes that saliency maps are not included in the first meaning (because the inner workings are considered through the gradients), but included in the second (because saliency maps can be developed for all differentiable models).
Local versus global explanations
When a local explanation is required, the objective is to provide an explanation that is faithful to the behavior of a black box for a particular decision, and for the decisions on very similar input data. Notice that the categories model-specific/agnostic and global/local are complementary to each other. For instance, the flagship method among model-agnostic local explanation methods is Local Interpretable Model-agnostic Explanations (LIME) (Ribeiro et al 2016). The idea of LIME is to learn an interpretable model (e.g., a linear model) based on instances that are obtained by perturbing the feature values of the instances for which the decision needs to be explained (figure 5). By perturbing the target instance, a neighborhood around this instance is created and the black box is queried for this neighborhood. The interpretable model is then trained to reproduce the decisions of the black box for the instances in this neighborhood, hence the local-aspect of the explanation. Many variants of LIME have been developed, for instance, by making the perturbations in such a way that the neighborhood is realistic (e.g., randomly perturbing pixels of face images will not provide another face image, a smarter perturbation technique would be needed to obtain that (Ivanovs et al 2021)). Applications of LIME in the medical field remain seldom, but an illustrative example is the study by (Palatnik de Sousa et al 2019), who generated explanations on how a CNN detects tumor tissue for lymph nodes metastasis in patches extracted from histology whole slide images. Another example is the study by (Jansen et al 2020), who also used LIME to interpret a model trained to predict 10-year overall survival of breast cancer patients.
Figure 5.
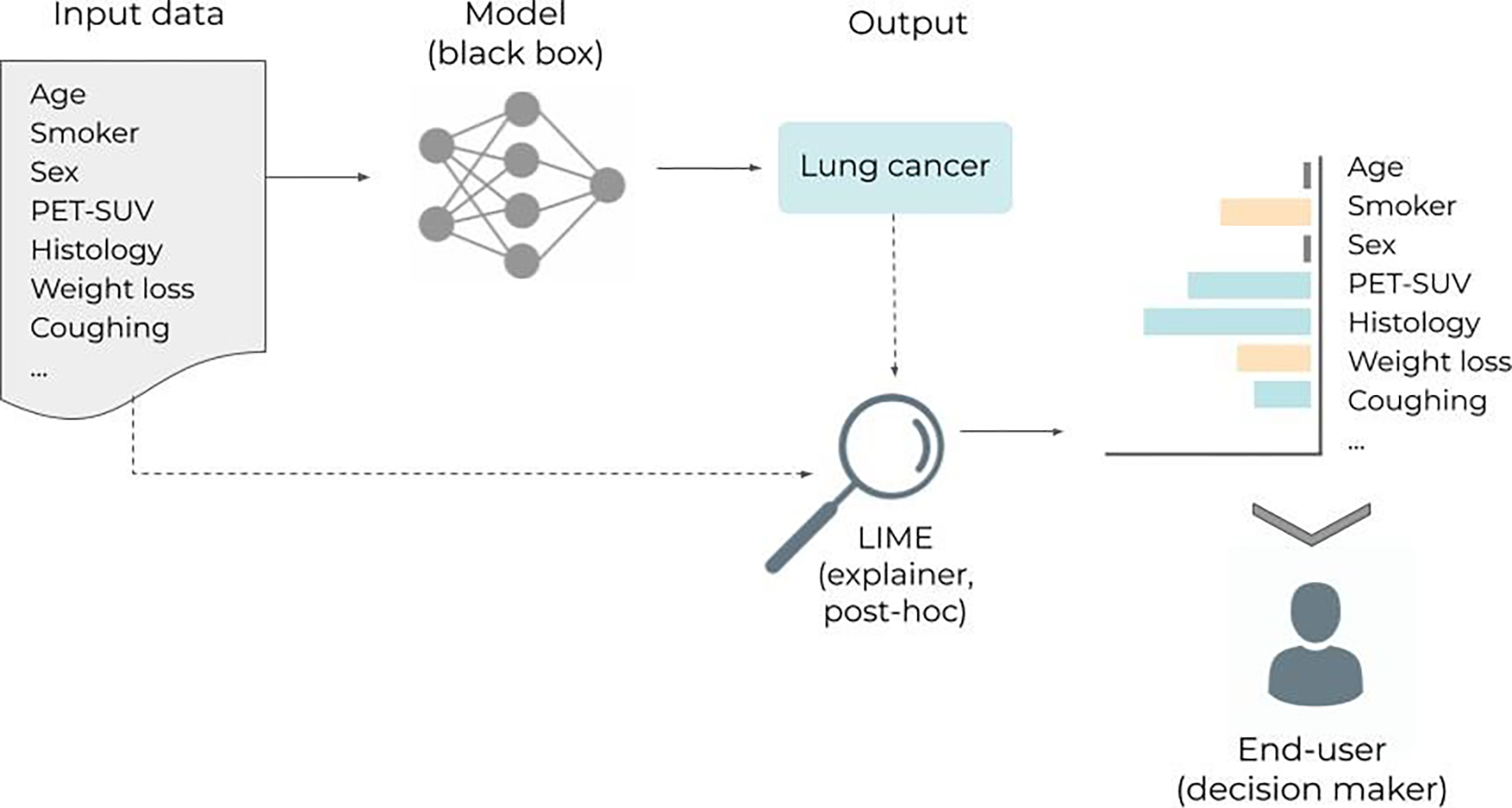
Inspired by (Ribeiro et al 2016). Workflow illustrating the use of LIME (Local Interpretable Model-agnostic Explanations). The idea of LIME is to learn an interpretable model (e.g., a linear model) to explain individual predictions. In the example, a black box model receives a set of variables for a new patient (i.e. age, smoker, …) and classifies the patient as having lung cancer. The LIME model then provides the user with information (i.e. explanations) about the features that most contributed to the prediction. “Age” and “Sex” did not contribute at all, “Smoker” and “Weight-loss” were against it, while “PET-SUV”, “Histology”, and “Coughing” contributed for the positive lung cancer classification.
Regarding model-specific local explanations, attention mechanism is a good example. Attention-based neural networks are models that contain one or several layers designed to focus on the relevant elements of the input for a particular prediction (Bahdanau et al 2014, Vaswani et al 2017). Attention layers have first been developed primarily to increase the performance of models and have afterwards been used as a way to self-explain the model. One particular interest of attention for explanation is that the explanation is learned during the training phase of the model. This means that no post-hoc explanation technique (i.e. after the model is trained), such as LIME, is needed to explain the model in a post-processing phase. Medical applications of attention mechanisms include classification of breast cancer histopathology images (Yang et al 2020a), or segmentation of cardiac substructures on MRI (Sun et al 2020), among others (Chen et al 2020a, Bamba et al 2020, Zhang et al 2017). Notice, however, that the use of attention as an explanation is still debated (Jain and Wallace 2019, Wiegreffe and Pinter 2019).
In the case of a global explanation, like agnostic (Fisher et al 2019, Gevrey et al 2003) or specific (Breiman 2001) global feature importance explanations, the entire inner workings of the black box is approximated. For instance, a neural network can be co-learned with a decision tree to (i) produce a better decision tree thanks to the neural network and (ii) obtain an interpretable representation of the neural network via the decision tree (Nanfack et al n.d.). Another example is the neural decision tree technique proposed in (Yang et al 2018), where any setting of the weights corresponds to a specific decision tree. Notice that a global explanation can be obtained by combining several local explanations that are performed on sufficiently different input instances (Setzu et al 2020). However, the issue is that combining many interpretable models can make the whole combination uninterpretable (e.g., the combination of decision trees in a random forest), which does not solve the problem of explaining the black box.
New trends and limitations
Today, many conferences, workshops and special issues in journals focus on interpretability and explainability. This interest leads to an ever growing literature on the subject. In particular, one hot topic, in addition to the post-hoc methods like LIME, is the subject of disentangled neural networks (Chen et al 2020b, Luo et al 2019). The idea behind neural network disentanglement is to combine the performance of neural networks with the need for interpretability and explanations. In disentangled neural networks, while the network is optimized to solve the problem, the neurons and filters are also constrained to correspond to concepts that are easily identifiable by humans. In the end, when the network is trained and makes a prediction, the activation of the neurons provides important clues on the concepts that have been used to make the decision. Medical applications of disentangled neural networks are rare, since it is a rather new field. But a good example is the work from (Chartsias et al 2019), who explored a factorisation to decompose the input into spatial anatomical and imaging factors. Their model was applied to analyzing cardiovascular MR and CT images. Another example is the study from (Meng et al 2021), who applied disentangled representations to fetal ultrasound images.
Another hot-topic is based on the aforementioned limitation of attention to be an explanation (Jain and Wallace 2019, Wiegreffe and Pinter 2019). While the debate converges towards the idea that attention may not be an explanation, solutions have been developed to address the issue. In particular, effective attention has been found to be the part of attention that can be considered as an explanation (Brunner et al 2019). The idea would therefore be to decompose attention weights into two parts and to use the effective attention part to explain the model.
In general, an important point for discussion is the accuracy of the explanations. For the cases where the approximation of the black box by the explanation is correct, the explanation gives truthful information about how different variables interact to result in a prediction. However, for those cases where the approximation is not correct, algorithms designed to provide explanations about the original black-box model are not a faithful representation of the original model (Jacovi and Goldberg 2020). As such, they provide a false and possibly dangerous sense of confidence. Unfortunately, it is not possible to know beforehand whether the approximation made by the explanation is accurate.
Some authors are also critical of the kind of explanation that is under study. Most, if not all, explanation techniques suppose that an explanation should only be faithful to the model (i.e., accurately reflecting its reasoning) (Jacovi and Goldberg 2020). However, another important aspect of explanations is their plausibility (i.e., how convincing it is to humans) (Riedl 2019). Indeed, one could accept to lose a reasonable amount of faithfulness to make the explanation plausible and, thus, useful, for the user.
Finally, besides the degree of faithfulness and plausibility, the explanation may not be lawful enough (Bibal et al 2021). Indeed, the strength and the type of the explanation can also be constrained by the law. For instance, a feature importance method can have a reasonable level of faithfulness and plausibility, but can fail as an explanation with respect to the law.
3.2. Data-model dependency
As a consequence to the intrinsic data-driven nature of ML algorithms, many of the risks associated with their use are related to the data itself and how it is processed inside the model (see Section 2). Thus, in addition to understanding the behavior of ML models (Section 3.1), acting on the data and analyzing how the model performance depends on it is key to enable a safe and efficient clinical implementation. In the following, we present several lines of action that can help to identify and reduce the risks of failure for ML models in the medical context, as well as to ensure an efficient implementation and use.
3.2.2. Data curation and data augmentation
The most straightforward techniques to ensure sufficient quality and quantity for the data, before training the ML model, are data curation and data augmentation. First, data curation can help detect any errors in the labels or identify missing and incomplete records, among other issues. Second, data augmentation can increase the variability in the training set, thus helping better represent the patient population under study (see Section 2).
Although most of the data curation process is currently done with very simple methods (e.g. scripts for data visualization, dictionaries for correct labeling (Schuler et al 2019, Mayo et al 2016), etc.), some groups have recently started to explore the use of ML models to be used for data curation and label cleaning specifically. For instance, Yang et al. (Yang et al 2020b) used a 3D Non-local Network with Voting (3DNNV) to standardize anatomical nomenclature in radiotherapy treatments. Another interesting approach is the “label cleaning network” or CleanNet, introduced by Lee et al. (Lee et al 2018), although the latter has only been applied to natural images. Another interesting approach is the one presented by (Dakka et al 2021), who trained multiple ML architectures on the data to be cleansed, with several cross-validation sets. The ML models are applied back to the same training (uncleansed) dataset to infer the labels, and those that cannot be consistently classified correctly are considered as poor-quality data. They called the method “untrainable data cleansing”, and illustrated their successful performance in several medical classification problems. Other groups have concentrated efforts in developing crowd-powered algorithms for large-scale medical image annotation (Heim et al 2018). In addition to the data cleaning, pre-processing methods can be used to increase the consistency of the data. For instance, for medical images, it is important to pay attention to things such as the voxel size, the image size, range of the image voxel values, registration between multimodal images, etc. Typical pre-processing techniques are image resampling, cropping and (histogram) normalization. For a comprehensive review of data curation tools and open-access platforms we refer elsewhere (Diaz et al 2021, Willemink et al 2020).
Regarding data augmentation, it works particularly well when dealing with images as input data. Two types of image data augmentation techniques exist: basic image manipulations and deep learning approaches (Shorten and Khoshgoftaar 2019). Basic image manipulation techniques consist of geometric image transformations such as image flipping, translations, random cropping and rotations and photometric image transformation like the addition of noise, mixing images and random erasing. Beyond those more basic approaches, adversarial training (Moosavi-Dezfooli et al 2015, Bowles et al 2018) and neural style transfer (Gatys et al 2015, Jackson et al 2018) are ML-based strategies that can be used for data augmentation. These techniques use neural networks to add transformations to the original data. In the case of adversarial training, two networks compete against each other: the first network (generator) generates synthetic images (the augmented data), while the second network (discriminator) tries to discriminate between real and synthetic images. Thus, the final transformations to generate the augmented data are those that are able to fool the discriminator network, leading to synthetic images that look truly real and have the same characteristics as the original set. In neural style transfer, the transformations are predefined (e.g., night to day) and a single network is used to turn the original data into the new style (Gawlikowski et al 2021, Ma et al 2019). For a complete review of data augmentation techniques we refer to the survey in (Shorten and Khoshgoftaar 2019). Data augmentation is nowadays used in most medical imaging applications to increase the number of training samples and improve generalization (Nalepa et al 2019, Chlap et al 2021). For instance, (Meyer et al 2021) used a data augmentation approach based on Gaussian Mixture Models to increase the variability of a given dataset of MR images in terms of intensities and contrast. This helped to increase the generalization of ML models trained for segmentation of MR images from different scanners. In a similar study, the authors used adversarial training (generative adversarial networks, GANs) to generate synthetic data to overcome generalization issues to different MR manufacturers (Yan et al 2020). Another example is the study by (Zhang et al 2020c), who applied a series of stacked transformations to each image when training the ML model. The idea was to simulate the expected domain shift for a specific medical imaging modality with extensive data augmentation on the source domain, thus improving the generalization to the shifted domains. They applied their model to segment different organs in MR and ultrasound images, showing promising results.
Although data augmentation is typically used to increase the training dataset, the same techniques can also be applied during the testing phase, in order to inspect the robustness and generalization of the ML model to a well-varied data distribution. This is known as test-time data augmentation (Nalepa et al 2019, Wang et al 2019b, Moshkov et al 2020). For instance, (Wang et al 2019b) investigated how test-time augmentation can improve the performance of a ML model for brain tumor segmentation. They augmented the image by 3D rotation, flipping, scaling, and adding random noise. After using test-time augmentation, their results appeared to be more spatially consistent. Recently, (D’Amour et al 2020) proposed a well-controlled framework to analyze the generalization capacity of ML models with the so-called “stress-testing”. The idea is to apply customized tests designed to reproduce the challenges that the model will encounter when deployed in the actual (clinical) world. In particular, two of the proposed tests (i.e., shifted performance and contrastive evaluation) aim to test the model with instances from a shifted domain. This can easily be done with test-time data augmentation, by changing the resolution, contrast, or noise level of the images. Although the concept of stress testing is rather new, the medical community is being encouraged to apply before clinically implementing ML models (Eche et al 2021). For instance, (Young et al 2021) applied stress-testing for ML models trained to diagnose skin lesions. They found inconsistent predictions on images captured repeatedly in the same setting or subjected to simple transformations (e.g., rotation).
In addition, test-time data augmentation can be used as a means to quantify the uncertainty associated with the prediction (see Section 3.2.3) of the ML model (Gawlikowski et al 2021, Wang et al 2019a, Ayhan and Berens 2018). For instance, in the previous example, (Wang et al 2019b) used test-time data augmentation to generate uncertainty maps for the segmented brain volumes.
3.2.3. Prior and domain-specific knowledge
The learning capability of ML models critically depends on the information conveyed by the data used to train them. Beyond this obvious statement that has been discussed in Section 2.1, we can possibly provide and/or guide our ML models with the even more relevant information for improved learning efficiency. Incorporating prior- and domain-specific knowledge into ML models can help achieve this goal and yield more robust models. There are several ways to incorporate this knowledge into an ML model (Xie et al 2021, Deng et al 2020, Muralidhar et al 2018a, Dash et al 2022) and here we present three common approaches: input data, loss function and hand-crafted features.
Input data
Sometimes, we attempt to train the model with incomplete information. For instance, medical images are typically associated with additional information than what is depicted. Certain anatomical features might result from specific diseases or medical procedures (e.g., surgical removal of the tumor), while remaining too stealthy cues. Similarly, a given radiotherapy dose distribution is the result of physician and patient choices regarding secondary effects, treatment protocols, and so on, while having directly this information side channel would ease learning. Training the ML model with the bare images, without including this prior and domain-specific information will result in poor performance. A common strategy to include this prior- and/or domain-knowledge is to modify the input itself. This includes changing the size and/or the format of the input: adding more input channels for CNN models, mixing images and text data as input, etc. When adding more input channels but keeping the same data type (e.g., stacking extra images such as MR or PET on top of CT), no significant changes need to be done in the architecture of the model. However, when using heterogeneous data types (e.g., images, text, scalars, …) several options are possible as to where to merge these sources in the network data path. We refer here to the early fusion, joint fusion and late fusion strategies (Figure 6). In the first, the different input modalities are joined before being fed into a single model. This fusion is done through concatenation and/or pooling, among other strategies. The joint or intermediate fusion consists in joining the features learned from the first layers of the network with other input modalities, before feeding this joint data into a final model. Finally, the late fusion strategy refers to the process of using a combination of outputs coming from multiple models to make a decision (Huang et al. 2020).
Figure 6.
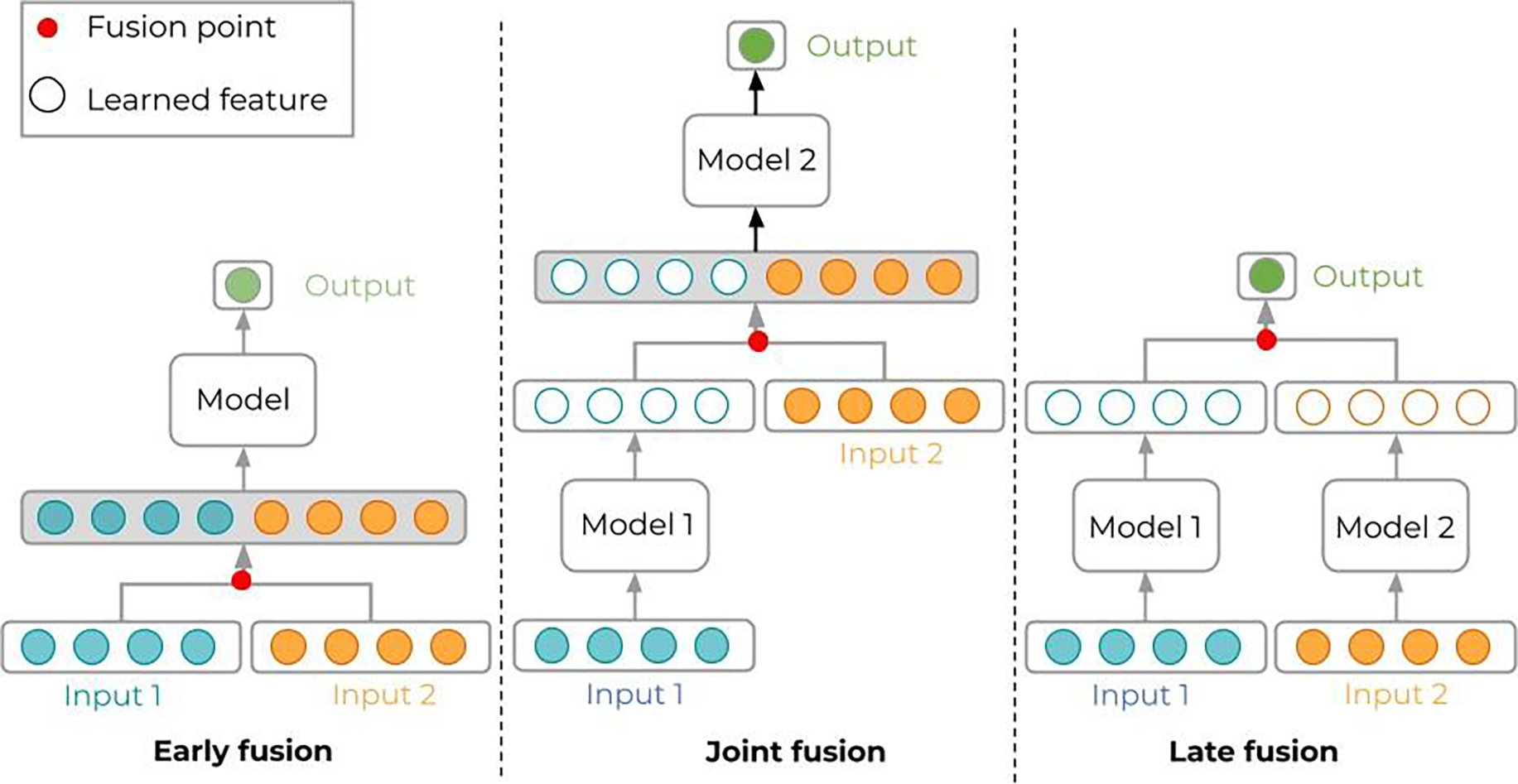
Multi-modal data can be processed in different ways in ML models, depending on how the various modalities are merged. Early fusion (left) is possible if the data types or modalities are not too different. A typical example is given by stacking multiple registered image modalities like CT, MR, PET, which are processed in convolutional layers. Joint fusion (middle) is typical of image data accompanied by simple indicators in vectors or text; then, convolutional layers (model 1) process the images to transform into feature vectors that are then merged (concatenated) with the other indicators to form a longer feature vector to be processed by the final model. Late fusion (right) pushes joint fusion even further: the output is a very simple combination of data coming from separate models dedicated to each data type; to some extent, late fusion bears some similarity with ensemble learning.
Examples of incorporating domain-knowledge into the input data are many. For instance, a study looking into volumetric dose calculation using DL investigated the use of 3D voxel-based distance from source, central beamline distance, radiological depth, and volume density, as entire volumetric inputs (Kontaxis et al 2020). Other photon and proton dose calculation studies investigated having a first-order prior of the dose calculation as input into the model (Xing et al 2020, Wu et al 2020). Similarly, recent studies about dose prediction for radiotherapy have explored the use of auxiliary information (e.g., non-modulated beam doses) to improve the robustness of the ML model (Barragán-Montero et al 2019, Hu et al 2021b). Yet another study about automatic three-dimensional segmentation of organs from CT images improved the performance of the ML model by using as input a two-dimensional contour of the considered organ (Trimpl et al 2021). Examples of mixing different data types include the addition of electronic health records and clinical data, like text and laboratory results, to the image data (Zhen et al 2020, Shehata et al 2020, Huang et al 2020).
These studies demonstrate that, by including these additional domain knowledge-focused inputs, the models outperform those using only more basic input data.
Loss function
In supervised learning, for some input xi, the loss function L(yi, ŷi) measures the mismatch or error between the desired output yi and the actual output ŷi = fθ(xi) for the model with its current parametrization θ. Optimal parameters are found by minimizing the loss for all (xi, yi).
Incorporating domain knowledge in the loss function aims at steering the model to prioritize error minimization for the most relevant data instances (patients), areas (in images), or metrics. Typically, it is done by adding to the loss function penalty terms that encourage outputs with properties imposed by the domain knowledge (output regularization). Commonly used losses in ML, like the mean squared error (MSE) and cross-entropy (CE), are general, domain-agnostic losses that can be applied to many regression and classification problems, respectively. When paired with the proper activation functions in the output layer, their gradients can be well behaved to make the optimization process converge efficiently. However, these generic losses are unable to minimize errors in any targeted manner. In contrast, domain-adapted losses achieve substantially superior performance for ML applications (Muralidhar et al 2018b). This was found to be especially impactful in situations where data is limited and of poor quality, a scenario that is often encountered in the medical field. However, due to the well-behaved gradients of most domain-agnostic losses, it is still preferred to use a combination of the two losses. Highly specific domain-adapted losses will likely have a poorly behaved gradient, and, thus, a well-behaved general loss will be a large driver at the beginning of the optimization. The domain-adapted loss can then fine tune the model further once it gets close to the minima.
Early works of including domain knowledge into the loss function date from the mid nineties (Fu 1995, Dash et al 2022). The penalty terms were based on regularizing embeddings, which are low-dimensional representations of the input variables. The complexity of the embeddings was penalized with first-order logic (Rocktäschel et al 2014). In traditional machine learning models, prior knowledge can also be integrated into the loss function to guide the feature selection process. For instance, (Guan et al 2020) developed a know-guided random forest to incorporate prior knowledge from multiple domains in biomarker discovery. The authors added a penalty coefficient to the Gini index. In nowadays deep learning models, integrating domain knowledge in the loss function is an active field of research. For instance, a recent study investigated the use of both human and learned domain-adapted losses in dose prediction for radiation therapy of prostate cancer with convolutional neural networks (Nguyen et al 2020). They included a differentiable approximation of the dose volume histogram (DVH) into the loss function, which improved the prediction accuracy, particularly for dose-volume metrics. Furthermore, they investigated the inclusion of a learned domain-adapted loss in the form of an adversarial (ADV) loss. Also for a dose prediction task with CNNs, in this case for breast cancer patients, (Bai et al 2021) proposed a dynamically scaled variant of the classical MSE loss, with a scaling factor that decreases in low-dose regions. This “sharp-loss”, as they coined it, aimed at solving the data imbalance issue of dose prediction problems where the region of clinical concern accounts for only a small part of the whole image. Another interesting approach is the focal loss proposed by (Lin et al 2017), which enables the DL model to automatically focus itself onto the most important examples for the training by relying on a defined prior probability for the relevant classes, which helps to overcome data imbalance issues. Recently, (Bird et al 2021) developed a DL model to generate synthetic CT for MR-only radiotherapy, and they used a focal loss function to enhance performance in the hard to predict bone region. Similar to the focal loss concept, (He et al 2020) designed a domain-adapted loss for renal artery segmentation, which sampled the loss region dynamically according to the segmentation quality intra-image, so that the hard-to-segment regions, such as edges, surfaces, ends, etc., will be focused and their segmentation quality will be enhanced. Instead of focusing on specific regions, other studies have explored the incorporation of anatomical priors as output regularization terms in the loss function. For instance, a star shape prior was encoded as a new loss term to improve the segmentation of skin lesions from their surrounding healthy skin (Mirikharaji and Hamarneh 2018). The model penalized the non-star shape segments and guaranteed a global structure in the final segmentation, thus achieving superior results in the ISBI 2017 challenge for skin segmentation. Similar approaches of incorporating anatomical priors as output regularization terms in the loss function can be found for the segmentation of other structures such as liver (Zheng et al 2019), kidney (Ravishankar et al 2017) or cardiac structures (Oktay et al 2018, Zotti et al 2019, Yue et al 2019).
Another interesting approach is to constrain the loss function to fit observed data or to yield predictions that approximately satisfy a given set of physical rules. This has been coined as physics-informed machine learning, and it is becoming increasingly popular. Although still not widely applied to the medical domain, there are some groups that explore this approach. For instance, (Kissas et al 2020) applied physics-informed neural networks to predict arterial blood pressure from non-invasive 4D flow MRI data. They used insights from computational fluid dynamics to ensure that the ML model yields physically consistent predictions. In addition to improved and more efficient learning, physics informed ML models have been claimed to have increased interpretability (Rudin et al 2021).
Handcrafted features (a.k.a. feature engineering)
Beyond the loss function, another way to better guide and interpret the model correctly through the learning process is to include the domain-specific knowledge into the feature selection process. Classical (shallow) ML models rely on humans to define specific features to extract from the data in order to guide the learning process (i.e., handcrafted features or feature engineering). In contrast, modern (deep) ML models (i.e., DL) rely on learning generic, parameterized features, turning feature engineering into an entirely automatic learning process for the model. This has been one of the reasons for the success of DL, since training a model can be done end to end without any human intervention. Moreover, the performance of classical ML models was limited to the adequacy of manually picked features, whereas DL models are assumed to have an improved performance thanks to the many degrees of freedom provided by generic trainable features. However, the automatic feature extraction of modern DL models can sometimes be a double-edged sword. Indeed, a DL model can easily extract thousands of features and, unlike handcrafted ones, these features are very hard to interpret by humans and to relate to relevant concepts in medical applications. Another pitfall of blind feature learning is that, due to the low control on many generic features, there is an increased risk of getting confounding features that are efficient but spurious, irrelevant, or poorly interpretable (see Section 2.2). Thus, incorporating prior- and domain-knowledge into the feature selection process can help improve the performance of ML models and also their interpretability. Although using handcrafted features might seem a step back in the evolution of ML, there are a few studies that start to follow this trend for medical applications (Luo et al 2019, Welch et al 2020).
For instance, radiomics (Lambin et al 2012) is a typical use of ML in medical imaging and oncology relying on handcrafted features. Radiomics assumes that images convey useful but not necessarily visible information for medical tasks like prognosis or therapeutic response prediction (Walls et al 2022, Guiot et al 2022). Feature extraction and selection are then supposed to reveal this information, sometimes called a radiomic signature, gathering a limited number of task-relevant features, while also allowing for automation. After segmentation of the volume of interest, typically a tumor, several types of features can be extracted from it. Geometric features include size measurements (diameters, volumetry, etc.) and shape descriptors (sphericity, compactness, etc.). Image intensity is characterized by histogram features, like energy, entropy, mean, variance, kurtosis, and other similar statistics, which are sometimes specific to imaging modalities like SUVs in PET (Orlhac et al 2021, Leijenaar et al 2015, Jiménez Londoño et al 2022). These first-order intensity features are complemented by second-order features that characterize textures in the images, i.e., the local relationships between nearby image voxels. Those features originate from tools like Haralick’s gray-level co-occurrence matrix (GLCM) (Haralick et al 1973), the gray-level run-length matrix (GLRLM) (Tustison and Gee 2011, Tang 1998), the gray-level size zone matrix (GLSZM) (Thibault et al 2013), the gray-level dependence matrix (GLDM) (Sun and Wee 1982), and the neighborhood gray tone difference matrix (NGTDM) (Amadasun and King 1989). Yet other, higher-order texture characterizations can come from image decompositions in Fourier/Gabor or wavelet/fractal spaces. All these image-related radiomic features can obviously be combined with features of various other origins, like genomics (Lu et al 2021), histology, clinical scores or indicators, etc.
Being slightly anterior to the popularization of DL in medicine, radiomics has historically relied on a classical ML pipeline, starting with handcrafted image preprocessing and feature extraction, followed by optional feature selection and traditional models for classification or regression. However, the field might evolve towards more end-to-end DL models (Lao et al 2017, Diamant et al 2019)(Afshar et al 2019), with trainable features, instead of engineered ones, less sensitivity to the preliminary segmentation of the tumor, at the expense of a higher complexity, lower interpretability, and higher needs in data. Recent publications show how the combination of DL and radiomic handcrafted features improve the results with respect to the classical pipeline. For instance, several studies have investigated the fusion of DL and handcrafted radiomics features to improve the classification performance for benign and malignant ground glass pulmonary nodules (Hu et al 2021c, Xia et al 2020, Cho et al 2021).
Other examples of the combination of DL and handcrafted features include a study where the authors constructed a six-deep-feature signature from MR images by using (sparse) LASSO Cox regression and combined them with clinical risk factors to predict the overall survival of patients with glioblastoma multiforme (Lao et al 2017). Other groups have explored more sophisticated approaches by combining both classical ML and DL models and using latent variables (Cui et al 2019). For instance, (Cui et al 2019) developed a joint architecture with a deep variational autoencoder and a multilayer perceptron (VAE-MLP). The latent variables from the VAE-MLP were used to complement handcrafted features for the prediction of radiation pneumonitis, improving the performance of the model.
Recently, some groups have started to develop strategies to efficiently extract domain-knowledge from a panel of experts, and incorporate it into the ML model for smart feature selection. For instance, (Welch et al 2020) designed different pipelines with varied levels of human interaction to combine clinical knowledge with ML features for prediction of locoregional failure in head and neck cancer (H&N). Another study developed what they called Expert Augmented Machine Learning (EAML), a methodology to automatically acquire problem-specific priors and incorporate them into the ML model (Gennatas et al 2020). These approaches demonstrated to learn more efficiently, increase the interpretability of the ML model by using concepts that medical experts are familiar with, improve the generalization of the model (including out-of-sample distributions), and facilitate the detection of hidden confounders (Gennatas et al 2020).
3.2.4. Uncertainty quantification
Another key aspect to ensure a safe clinical implementation of ML models is to be able to quantify their risk of failure. This can be done by estimating the uncertainty associated with the prediction that the ML model yields for a given input sample (Abdar et al 2021, Gawlikowski et al 2021, Gal 2016, Kendall and Gal 2017). A prediction with a high uncertainty is then a way for the ML model to tell us “I am not confident about the answer“ or even in extreme cases, “I don’t know the answer”. Uncertainty quantification tools can thus alert clinicians when the confidence of the ML model on the output is too low and let them take over to complete the task. Implementing such QA tools is crucial to gain clinicians’ trust in ML technology, since it helps identify the limitations of ML models and avoid the risks associated with uncertain predictions (Begoli et al 2019, Kompa et al 2021).
Several reasons can make a ML prediction uncertain, but given the data-driven nature of ML, many of them are related to the quantity and quality of the data used for training, as well as to the characteristics of the new input sample. In this context, uncertainty is typically categorized in two types: aleatoric and epistemic uncertainty (Hüllermeier and Waegeman 2021, Anon 2009). Aleatoric uncertainty measures the uncertainty inherent to the data (e.g., noisy, inaccurate, or low-quality records and labels, see Section 2.1). It cannot be reduced even if more data is collected. However, increasing the quality of inputs (both training data and new unseen samples) would lead to a reduction. Epistemic uncertainty, on the other hand, represents the lack of knowledge of the model itself and is often referred to as model uncertainty. Epistemic uncertainty can stem from data sampling problems (e.g., the training data does not represent well the population under study, or the new input sample is out of the intended population distribution); or from issues related to the model structure (e.g., the model does not interpolate/extrapolate well enough). Thus, epistemic uncertainty can be reduced by either collecting more data to better sample the problem or by using more appropriate architectures with improved learning abilities (Tanno et al 2021, Gal 2016). Although the two uncertainty types are often combined into the so-called predictive uncertainty (Gal and Ghahramani 2015), distinguishing between them can help us to improve the ML model by tracking and fixing each issue independently (Tanno et al 2021)(Senge et al 2014, Hüllermeier and Waegeman 2021, Depeweg et al 2018).
For simple models, such as linear regression, the standard error of parameter estimates is directly available and it can be used to compute a confidence interval (typically 95%), which is a classical way to estimate the predictive uncertainty. Unfortunately, for more complex models, with a large number of parameters and nonlinear relationships, such as modern deep neural networks, estimating the predictive uncertainty is not straightforward.
Uncertainty quantification for ML/DL is a very active research field, and many different strategies have been proposed in recent years (Gawlikowski et al 2021). One of the traditional approaches is to model uncertainty in a probabilistic way, within a Bayesian framework. Instead of having models that process single point estimates, the idea is to replace them with probability distributions that indicate which values are more likely to happen (Beck and Katafygiotis 1998). In addition to Bayesian methods, another popular and rather simple approach for uncertainty quantification is the use of ensemble methods. We provide a general description of these methods, together with illustrative examples of their application in the medical field. For a detailed description and a full overview of the current state of the art in uncertainty quantification methods we refer to (Gawlikowski et al 2021)
Bayesian methods
Inspired by Bayesian theory, Bayesian DL aims to change conventional DL architectures to have a prior distribution on the weights of the model parameters, instead of a single value (figure 7).
Figure 7.
The fundamental difference between a regular neural network (a) and its Bayesian extension (b), holds in the replacement of scalar values (pointwise estimates) with full-fletched probability distributions with an expected value (equivalent to the pointwise estimate) and a standard deviation, indicative of its associated uncertainty. Dealing with probability distributions is much more demanding computationally and several approximations or surrogates to Bayesian networks exist, like neuron dropout in figure 8 and figure 10.
In this way, the model can easily generate an estimation of the uncertainty, since it will produce a (posterior) probability distribution over the output for a given input sample. The challenge in Bayesian DL architectures is that the inference of the model posterior distribution becomes intractable, due to the high computational complexity required to estimate the weight distributions. This is especially true for complex models with a large number of parameters, such as modern deep neural networks. This is the reason why the research community has focused on developing approximated versions of the full Bayesian framework. One of the most popular approaches is to use Monte Carlo Dropout (MCDO) as Bayesian approximation (Gal and Ghahramani 2016). Dropout is a mechanism initially designed to avoid overfitting during training (Srivastava et al 2014), and it consists in switching off (i.e. dropping) a random fraction of neurons in the network (figure 8). When a neuron is turned off, it is hidden from the network and its output is zero. In MCDO, the neurons that are dropped are sampled from a Bernouilli distribution. Typically, dropout is applied during training, but when using MCDO as Bayesian inference approximation, dropout is also used at testing time. As a consequence, when several (T) predictions are obtained with active MCDO, all T predictions will differ from each other, since they stem from slightly different models, with different sets of neurons that are turned on or off. By performing a sufficient number (T) of predictions, one can have a sort of approximation for the (posterior) probability distribution of the output. This sample of T predictions is used to compute the mean and standard deviation, the former being equivalent to a pointwise prediction and the latter being a surrogate for the predictive uncertainty. In addition to the sample standard deviation, mutual information and predictive entropy are other metrics that can be extracted from the T predictions and are commonly used as a surrogate of the predictive uncertainty (Gawlikowski et al 2021).
Figure 8.
(a) Standard Neural network where all weights are set. (b) A random fraction of the weights are switched off.
The advantage of MCDO is that, as soon as dropout layers are included in the architecture of the network, the implementation and computational efforts to obtain the uncertainty are minimal. On the one hand, the architecture for conventional DL models does not need to be modified to apply MCDO at inference time. On the other hand, despite having to perform T predictions, with current DL models inferring within a few seconds, the uncertainty estimation is rather quick (figure 10(a)).
Figure 10.
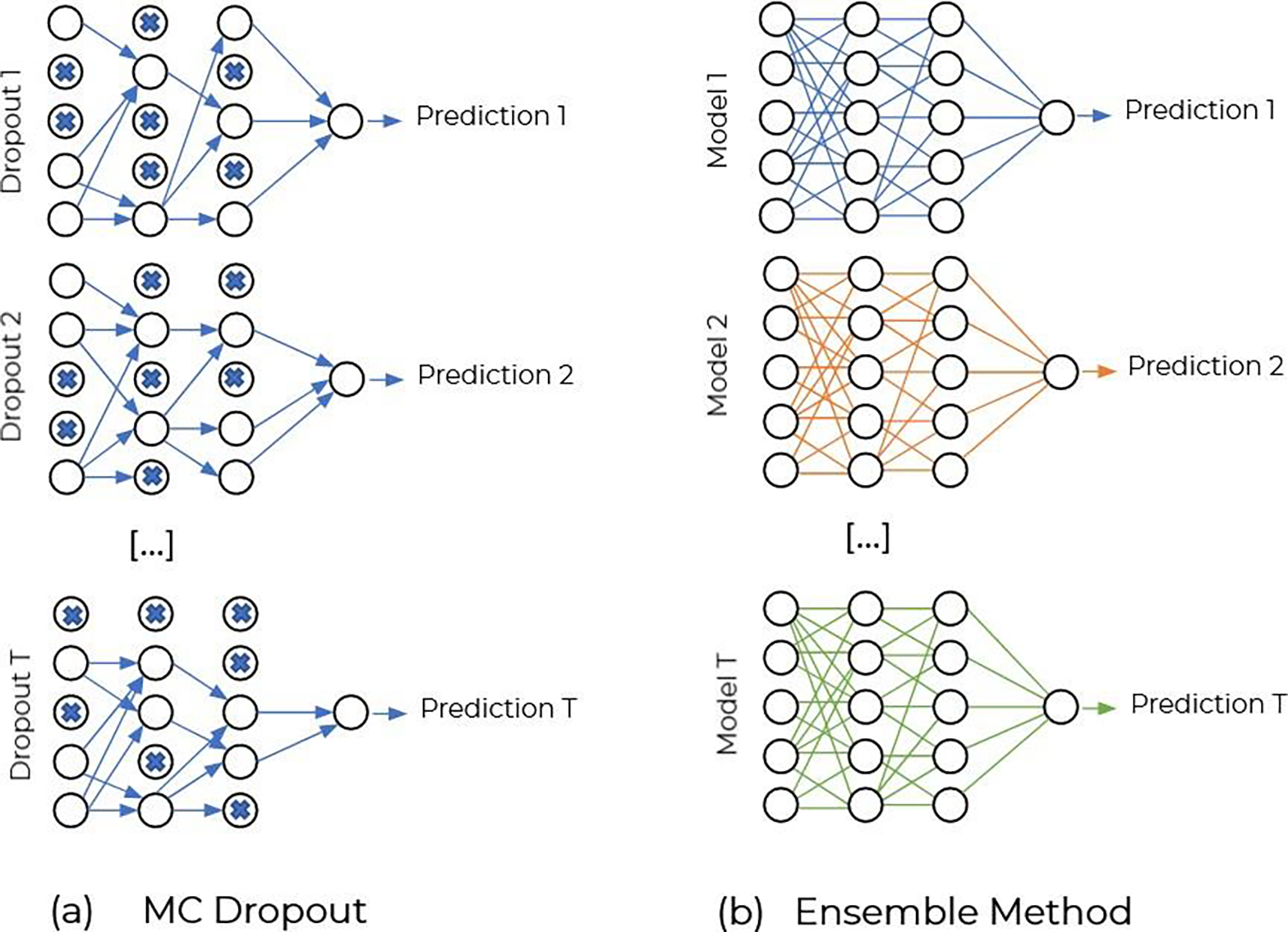
(a) MC Dropout at inference time. The T predictions are obtained from Dropout of different weights. (b) Ensemble method, different models have been previously trained and the T predictions are obtained from each network on the same sample.
MCDO has started to be a popular tool to quantify the predictive uncertainty of ML models for medical applications. For instance, (Mobiny et al 2019) used MCDO to build a risk-aware ML model to detect skin lesions. The model asked for clinician input when the uncertainty of the prediction was too high, and thus, the clinician–ML workflow reached a much higher accuracy than the (non risk-aware) ML model alone. The same group recently published another study (Mobiny et al 2021) where they used a generalized version of Dropout, DropConnect (Wan et al 2013), to quantify the uncertainty in a CNN trained to segment different organs in abdominal 3D CT scans. They used the mutual information to estimate the epistemic uncertainty, since they were interested in knowing the regions of the data space where the model was uncertain. Also for a segmentation task, in prostate cancer patients, (Balagopal et al 2021) used MCDO to estimate and visualize the 95% upper and lower confidence bounds for each prediction, which informed the physicians of areas that might require correction. MCDO has also been used for regression tasks, such as to generate an uncertainty map when predicting the dose for radiotherapy in prostate (Nguyen et al 2021) or head and neck patients (Vanginderdeuren et al 2021) (figure 9). Yet a last example, (Nair et al 2020) provided an interesting comparison of different uncertainty measures derived from MCDO (predictive variance, MC sample variance, predictive entropy, and mutual information) for segmenting lesions in brain MR images. They illustrate how the different metrics do not highlight the same regions.
Figure 9.
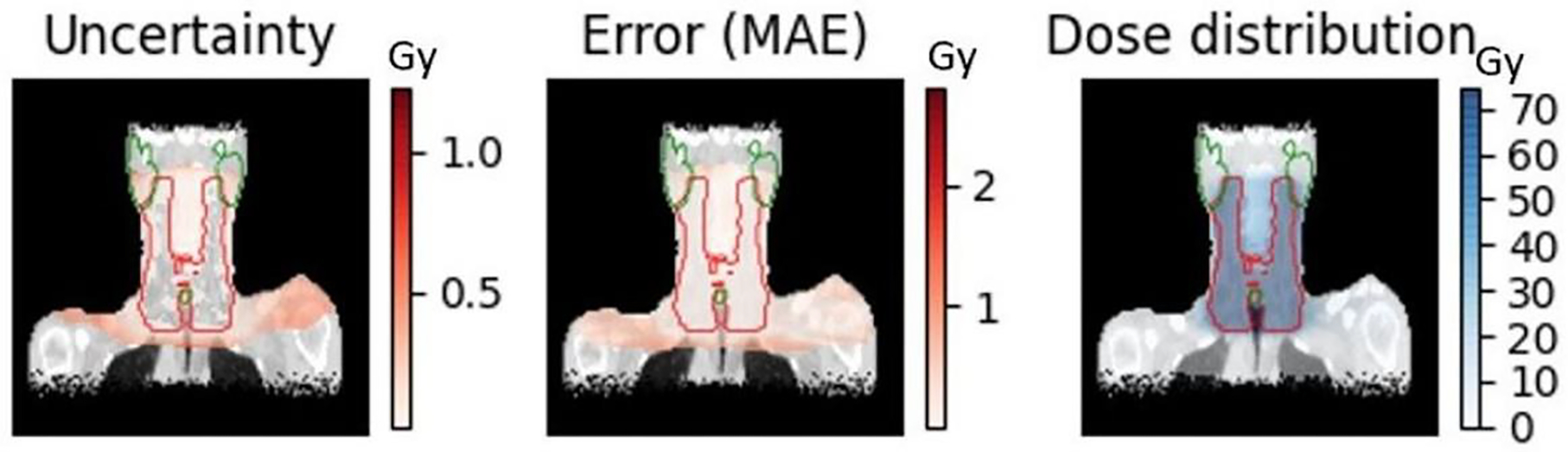
Prediction uncertainty, prediction error and predicted dose distribution for the same slice of a patient with head and neck cancer (Vanginderdeuren et al 2021).
Note that recently, several groups have started to go beyond the MDCO approximation and use an approach closer to the full Bayesian framework. In (LaBonte et al 2020) and (McClure et al 2019), the authors compared MCDO to a CNN where the weights were sampled from a distribution (Blundell et al 2015). In this case, the models learn the parameters of the distributions instead of the weights values. They showed that such models produce better results and more interpretable uncertainty maps as we can decompose aleatoric and epistemic uncertainties (Depeweg et al 2018), as presented also for an ischemic stroke lesion segmentation model (Kwon et al 2020).
Ensemble methods
Ensemble methods deploy concurrent models that solve the same problem and compute a prediction based on the individual predictions of the ensemble members (e.g., average, majority voting, etc) (figure 10(b)). Initially, they were developed to improve the performance of ML models, with stronger generalization and stability. They rely on the hypothesis that a group of decision makers tend to provide better decisions than a single one, since they complement each other’s weaknesses (Sagi and Rokach 2018, Schapire 1989). Having multiple predictions for the same problem allows ensemble methods to represent the model uncertainty on a prediction in a rather simple way: by evaluating the variation among the individual predictions (e.g., with the standard deviation). Ensemble learning was used successfully in (Wickstrom et al 2021) to detect myocardial infarction in echocardiograms by identifying relevant time steps. The drawback of ensemble methods is that they have a higher upfront cost, since multiple models need to be trained individually. However, uncertainty generation at inference time can be as fast as MCDO. To some extent, MCDO is an ensemble method where all models are subnetworks of a complete neural network.
A popular ensemble learning algorithm is bagging (Bootstrap AGGregatING). Bagging uses random subsets of training data (allowing replacement) to build multiple models and averages out their results. Apart from the computational cost, ensemble methods have no technical complexity, and that has motivated their use in different medical applications, often in comparison with Bayesian methods. For instance, the aforementioned examples for dose prediction in radiation therapy (Vanginderdeuren et al 2021, Nguyen et al 2021) compared bagging against MCDO.
3.2.5. Beyond conventional supervised learning
This manuscript has been entirely focused on supervised learning, which is the most used learning framework so far in medical applications. As previously introduced, supervised learning relies on the availability of a dataset that contains input-output (x, y) pairs, where y is in charge of supervising the model training. In other words, supervised learning requires a set of examples x for which the desired answers y, also called labels or annotations, are known. This entails a strong dependency of the model performance on the quantity and quality of the labels y (see Section 2.1). This section presents different learning frameworks that can help reduce this dependency, allowing the model to perform well even with few or low-quality labeled data samples. In addition, we also discuss how some of these learning frameworks can help to overcome the static and task-specific nature of current ML models, improving their generalization capacity (see Section 2.2).
Unsupervised learning
A way to reduce the model performance dependency on the availability of large sets of high quality labeled data is to shift towards learning frameworks with less supervision. Unsupervised learning deals with data x without output values y and it aims at exploring the features and patterns in the distribution of data in x, such as clusters, modes, and outliers (Bengio et al 2013). It is sometimes known as self-organization, since the learning process is blind and cannot rely on unambiguous supervision. Some techniques of unsupervised learning can help reduce the problems of insufficient data due to the cost of manual annotations, as well as those of inappropriate data due to the quality of the annotations. For instance, cluster labels obtained with unsupervised learning can be adopted as class labels in further supervised learning (Peikari et al 2018). The use of unsupervised learning is still less extended than supervised learning, but many groups are starting to explore fully unsupervised or semi-supervised techniques (i.e., when only a part of training data contains known outputs) in the medical domain (Raza and Singh 2021). Examples of unsupervised and semi-supervised learning include clustering to identify patterns across patients suffering from Alzheimer’s disease (Alashwal et al 2019), or medical image analysis like in (Gu et al 2020), where the authors incorporate local structure of unlabeled data into their random forest algorithm. Examples specific to the radiotherapy domain includes the use of unsupervised learning to correct cone-beam CT scans for artifacts (Dong et al 2021), or to learn radiomic features that predict treatment response and overall survival of lung cancer patients (Li et al 2018), among others (Raza and Singh 2021).
Recently, a new variant of unsupervised learning, namely self-supervised learning, has been gaining attention (Lan et al 2019, Jing and Tian 2021, Taleb et al 2020, Hatamizadeh et al 2021). This framework uses unlabelled data but exploits labels that come almost for free, which are intrinsically present in the data and can be extracted from its structure to solve pretext tasks. An example of a pretext task could be rearranging image patches such as parts in a jigsaw (figure 11). Self-supervision works in two steps, the first aiming at obtaining the supervisory outputs (y) by solving a pretext task, whereas the second uses them to solve the actual task of interest.
Figure 11.
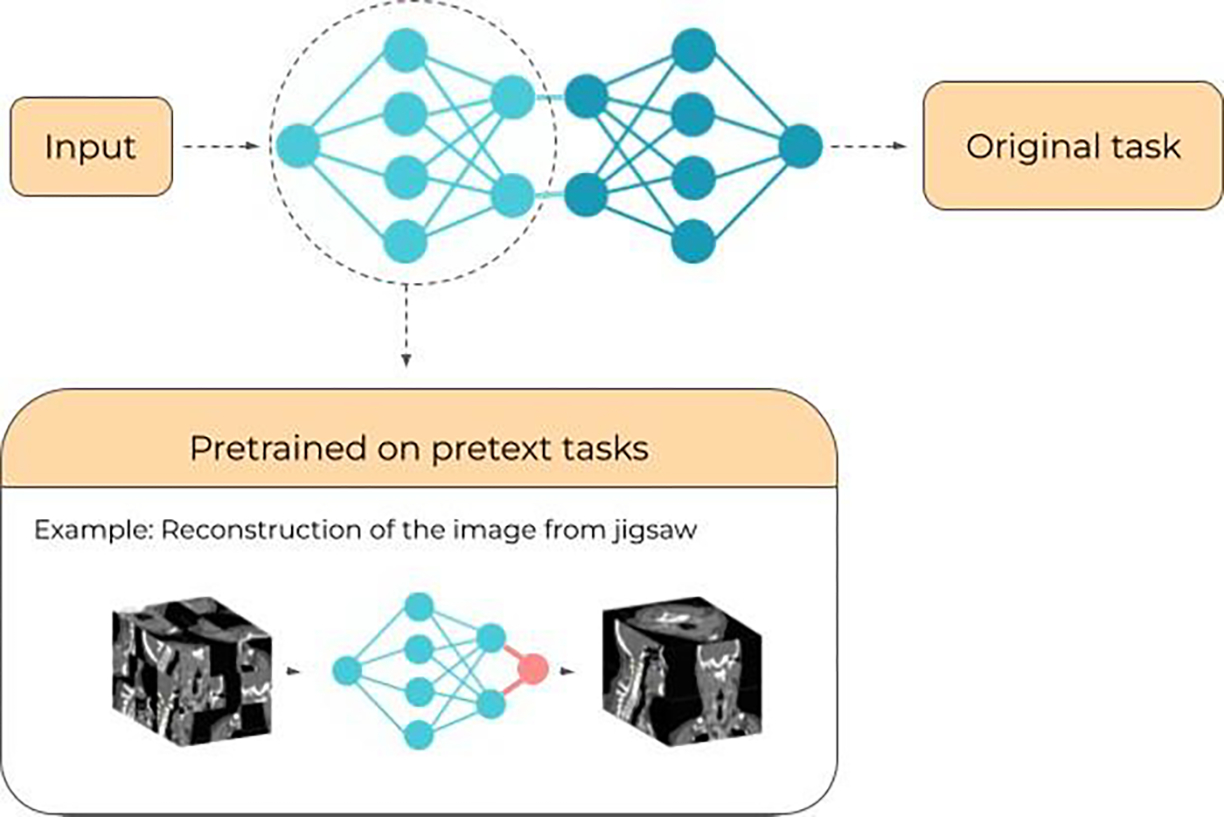
Self-supervised learning workflow with an example of a pretext task where the input is the image from which patches have been mixed up. The aim of this pretext task is to reconstruct the initial image hoping the encoder extracts useful features from the data (Inspired from (Taleb et al 2020). The knowledge acquired by the trained network on the pretext task is later used to carry out the main, original task.
Self-supervised algorithms start only to be used in medical applications, but good illustrative example of their potential is the work of (Chen et al 2019), who used self-supervision for image classification of 2D fetal ultrasound images, organ localization on abdominal CT images, and segmentation on brain MR images (downstream tasks). Their strategy consisted in modifying the spatial distribution of the images, and training a network to restore the original version in order to learn the contextual information (pretext task).
Reinforcement Learning
Together with supervised and unsupervised learning, reinforcement learning is often considered as the third main learning paradigm. In reinforcement learning, the algorithm simulates an agent that interacts with its environment to perform a certain task over time. During training, the agent takes successive actions to change state and eventually reach a final one, like victory or defeat in a game. After each action towards a new state, the environment can either reward or punish the agent who has then to best predict the longer-term consequences of future actions in a trial and error fashion. The difficulty of policy making in reinforcement learning is that immediate rewards are not necessarily correlated with ulterior gains. Hence, feedback partly guides the agent who learns to act based on either past experiences (exploitation) or new choices (exploration).
Reinforcement learning has been used in medical imaging to devise and generate specific treatment plans for cancer patients treated with radiation therapy (Zhang et al 2020b, Shen et al 2019, 2020a) as well as for other diseases (Watts et al 2020). For instance, the study by (Zhang et al 2020b) describes a planning bot based on reinforcement learning to systematically address complex dose tradeoffs and achieve high plan quality for stereotactic body radiation of pancreas cancer patients. The authors defined planning actions to represent steps that human planners would commonly implement to address different planning needs, and they derived a reward function based on the physician-assigned constraints, as one would do in clinical practice. In addition, the authors claimed that the training phase of the bot was tractable and reproducible and that the acquired knowledge was considered to be interpretable by humans. This example shows that, in order to define the environment and actions in reinforcement learning algorithms, significant prior and domain-specific knowledge is needed. In exchange, the advantages of reinforcement learning is that it can help humans to explore new actions (e.g., new planning strategies, new treatments) that have not been previously investigated in clinical practice. It is the case of the study by (Moreau et al 2021), who explored new radiotherapy dose fractionation based on a tumor growth model. Other applications include image segmentation (Zhang et al 2020a, Li and Xia 2020, Winkel et al 2020) or reconstruction (Shen et al 2018).
Active learning
Beyond shifting towards strategies requiring less supervision, another approach to reduce the label workload is active learning (Budd et al 2021, Abdar et al 2021). This learning framework builds upon supervised learning, but starts with a small set of labeled data and later selects the best data to be annotated next for optimal model performance (figure 12). The selection is based on the estimation of the informativeness of each unlabeled data sample. The chosen candidates are labeled by an expert and subsequently added to the training set. Then, the model can be retrained from scratch or fine tuned by using the new labeled data. In short, active learning is a type of iterative supervised learning where the model demands the most relevant data for an optimal performance. As informativeness is not a metric in itself, multiple methods exist to select the samples to be labeled. Most of them are based on uncertainty quantification strategies (Section 3.2.3) and sometimes combined with other quantities such as representativeness (Huang et al 2014) (Du et al 2017). Representativeness is used to select instances that are the most emblematic of the unlabeled dataset and thus contribute to better coverage of the (patient) data distribution domain under study. Using only uncertainty as the selection metric can lead to situations where out-of-data distribution instances are selected because of their high uncertainty, and thus they will instead worsen the model performance once they are included in the training. In their medical image segmentation framework MedAL, detailed in (Smailagic et al 2018), authors use as metric a combination of uncertainty measure and distance between feature descriptors. In (Sourati et al 2018), the Fisher information is used to ensure diversity among queried samples.
Figure 12.
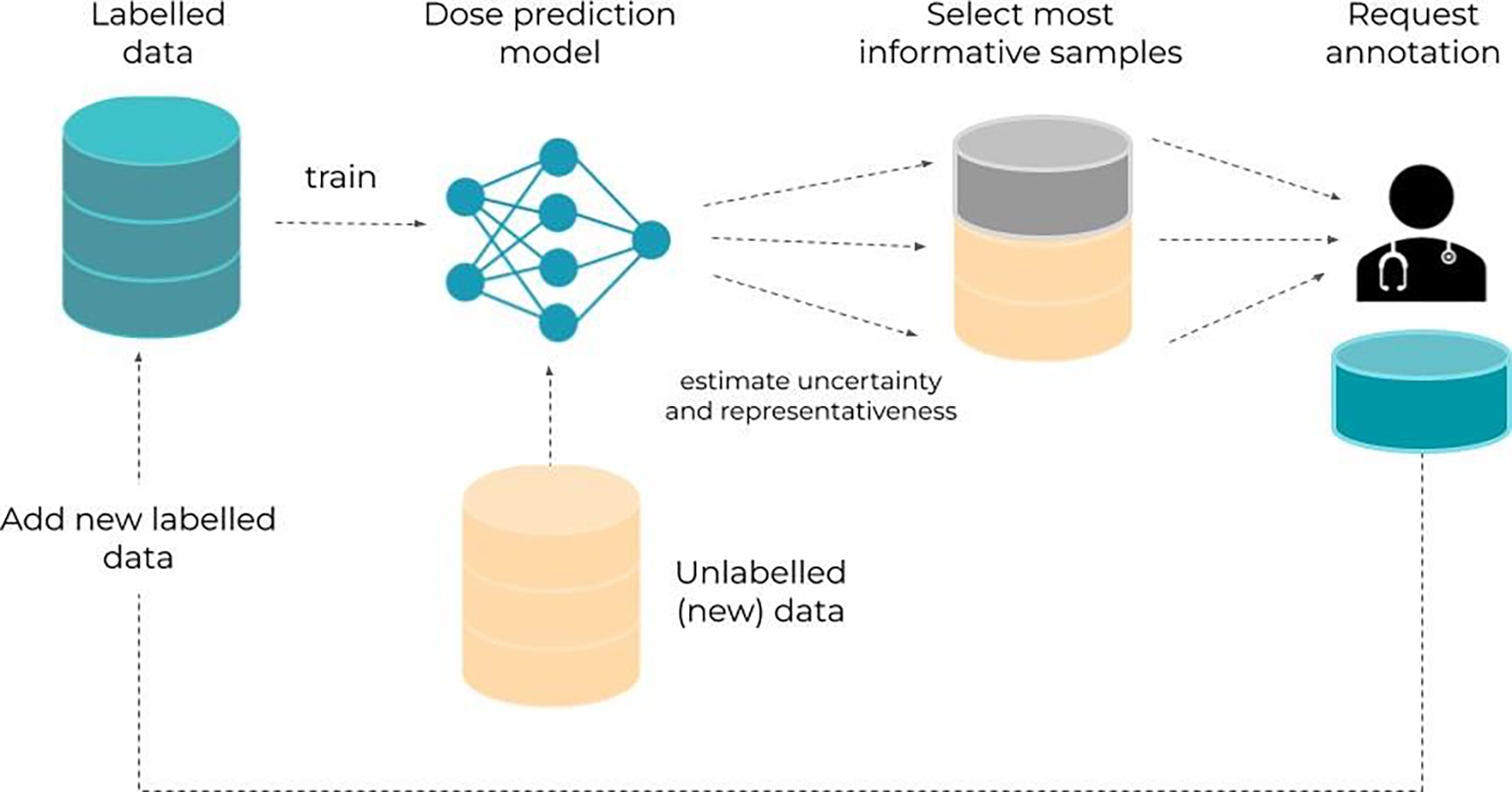
Active learning workflow: unlabelled data gets selected for expert annotation according to the chosen informativeness metric and then added to the training set of the network.
Once the metric is chosen, unlabeled data can be ranked accordingly. First active learning algorithms selected the most informative sample or subset to submit them to human experts for labeling. In (Kirsch et al 2019), authors argue that performing the labeling of a batch is more efficient as it reduces the frequency of expert intervention. Other methods such as CEAL (Cost-Effective Active Learning) (Wang et al 2017) consider that while keeping the human labeling for informative data, samples for which the network is most certain about should be labeled automatically by the model itself.
Transfer learning
Transfer learning (Pan and Yang 2010) reuses part of the architecture and parameters values in a model trained with a given data for a certain task (source domain and task), and tune the model to be applied to a different data or task (target domain and task). Notice that transfer learning is a high-level, abstract framework that can be applied to any model, regardless of the learning paradigm (i.e., supervised, unsupervised or reinforcement learning). The advantages of transfer learning are twofold. On the one hand, one can solve the target task with very little data (figure 13). On the other hand, learning from little data enables the quick generation of new models that work for different tasks, as well as to efficiently update models that were no longer valid due to a change of the data distribution over time. As a consequence, transfer learning is an excellent technique to overcome to some extent the static and task-specific nature of current ML models, improving the generalization to the same domain (i.e., i.i.d. data) or different domains (i.e., shifted distributions) (Section 2.2). The particular use of transfer learning techniques to adapt models to different domains is also known as “domain adaptation” (Wang and Deng 2018, Guan and Liu 2022). Often, the term multi-task learning is also used, when the goal is to learn multiple tasks (Caruana 1998, Zhang and Yang 2021).
Figure 13.
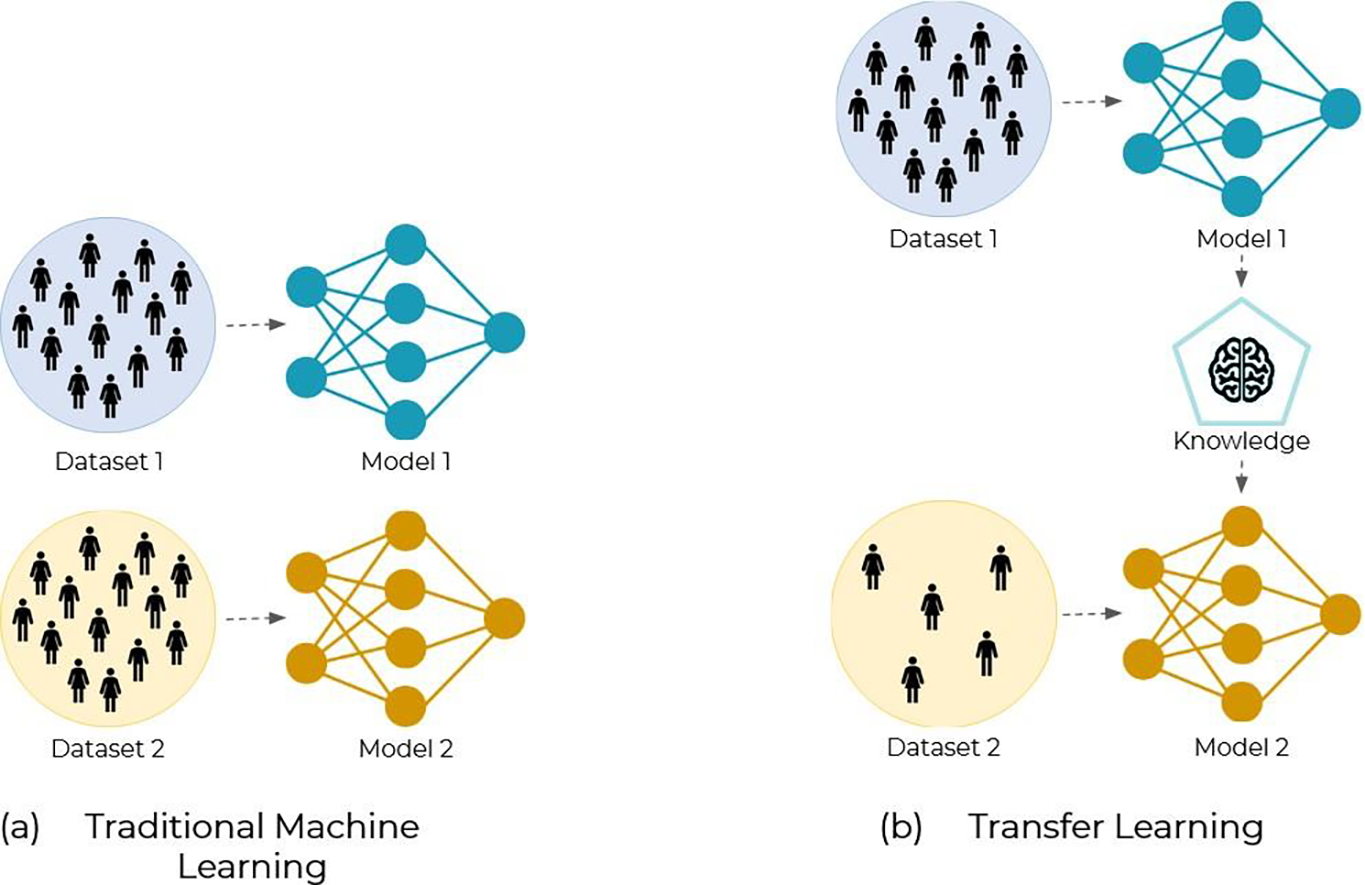
(a) Traditional Machine Learning where to train a model for a new task, we need a large dataset for the training. (b) Transfer Learning, where knowledge is transferred from another network performing a similar task. The advantage is that the required size of Dataset 2 can be reduced significantly.
Examples of the use of transfer learning in the radiotherapy field are many. For instance, a radiotherapy dose prediction study reported several planning styles for prostate cancer patients treated with VMAT and demonstrated that, through the usage of transfer learning, the models were capable of adapting from one planning style to a new target style. Transfer learning significantly reduced errors for clinical dose metrics on target datasets with limited training data size for the target domain, as low as 16 patients (Kandalan et al 2020). Another study, already discussed in section 2.2, focused on CBCT-to-CT image conversion for prostate, pancreatic, and cervical cancer patients. They found that the models were not generalizable across different image scanners, due to different characteristics and parameters in the scanners themselves. Significant improvement in the model performance was observed when using transfer learning to adapt to the target data distribution from a different machine (Liang et al 2020). Yet another example is the recent work from (Mashayekhi et al 2021), who developed, through the use of transfer learning, a site-agnostic radiotherapy dose distribution prediction ML model. The model can leverage data from any treatment site (e.g., prostate, head and neck) and it only requires a brief fine-tuning with a small dataset to be applied to a new site.
The examples above used labeled data from the target domain. When the labels are not present in the target domain dataset, the problem then becomes unsupervised transfer learning, most known as unsupervised domain adaptation (Wilson and Cook 2020, Kouw and Loog 2021). For instance, (Perone et al 2019) explored unsupervised domain adaptation for segmentation of MR images. Similarly, (Kamnitsas et al 2017) used unsupervised domain adaptation for brain lesion segmentation. Another good example is the study by (Brion et al 2021), where the model used unsupervised domain adaptation to leverage a large database of annotated pelvic CTs (source domain) to segment CBCT images (target domain). The target domain database contained CBCT scans that were not annotated. This is extremely useful for the actual clinical practice in radiotherapy, where the manual segmentation is done in CT images while CBCT scans are typically left un-labelled, since they are used chiefly for repositioning or for visual inspection of the anatomy.
Other trends
With the fast evolution of ML, more and more higher level learning frameworks, like transfer learning or active learning, get formalized and investigated. They sometimes combine existing learning paradigms (e.g., supervised learning), frameworks (e.g., active learning) and strategies (e.g., prior knowledge incorporation) to solve a specific problem. A good example is the popular few-shot learning regime (Wang et al 2021, Snell et al 2017, Ravi and Larochelle 2016), somehow the opposite of the big data regime, which tries to address the issue of learning from a very limited number of samples with specific learning techniques. Humans are very good at recognizing new classes (e.g., a book), even when only one or a few examples of that class have been shown to us. Sometimes, we can even distinguish objects from classes that we have never seen, based on our prior knowledge and the (dis)similarity to other known classes (i.e., zero-shot learning (Lampert et al 2009)). Few-shot learning and its extreme variant one-shot learning (Fei-Fei et al 2006, Vinyals et al 2016, Koch et al 2015), try to mimic this human learning feature by integrating prior knowledge into ML models. Few-shot learning is often referred to as a type of meta-learning, a concept that defines algorithms that “learn to learn”, i.e., algorithms that are able to learn from multiple tasks and extrapolate the acquired knowledge to carry out new tasks (Seita n.d., Finn et al 06-11 Aug 2017). Existing few-shot learning studies are essentially supervised learning problems (Wang et al 2021), although one can find some examples of few-shot reinforcement learning (Duan et al 2017, Al-Shedivat et al 2017, Bruce et al 2017), where the goal is to find a solution given only a few state-action pairs. Several strategies have been proposed to efficiently include prior knowledge into ML models; some of them have been already described in detail in Section 3.2.2. For a complete review of all possible strategies to incorporate prior knowledge in the context of few-shot learning we refer to a recent survey by (Wang et al 2021), who identified three main categories: 1) data, using prior knowledge to augment the data from few to many samples (e.g., data augmentation or transfer learning); 2) model, using prior knowledge to reduce the size of the optimization space search; and 3) algorithm, using prior knowledge to alter the search strategy to learn efficiently from few samples. Examples of recents applications of few-shot learning in the medical domain include the study by (Medela et al 2019), who reduced the need of labeled data in diagnosis of histopathological images. They used a popular few-shot learning model, namely, Siamese networks (Koch et al 2015), which distinguished the different classes by ranking the similarity between input images. Other examples include the use of few-shot learning for deformable image registration and motion tracking in 4DCTs (Chi et al 2022, Fechter and Baltas 2020, Zhang et al 2021).
In addition to few-shot and one-shot learning, zero-shot learning studies are also becoming popular (Wang et al 2019c, Palatucci et al 2009, Changpinyo et al 2016, Socher et al 2013), where the aim is to build a ML model that is able to generalize to totally unseen domains. Zero-shot learning can be considered as an extreme subfield of transfer learning. Techniques to solve zero-shot learning problems include simple techniques such as data augmentation (Xu et al 2016) or more sophisticated techniques (Wang et al 2019c). Although the concept of zero-shot learning is not much investigated yet in the medical domain, a recent example of its application is the study by (Paul et al 2021), which presented a zero-shot learning algorithm to diagnose chest X-ray images.
Beside few- to zero-shot learning regimes, other recent or trendy concepts are worth mentioning. For instance, continuous learning (Parisi et al 2019, Pianykh et al 2020, Lee and Lee 2020), where the goal is to build ML models that are not static, meaning that they can adapt to a slowly changing data distribution over time and to their ever-changing environments. Continuous learning can serve to prevent catastrophic forgetting, which is when ML models forget the previous data seen during training, leading to overall reduced performance (Kirkpatrick et al 2017a, Hofmanninger et al 2020). Multiple methods have been proposed which can include, for example, context-dependent gating (Masse et al 2018a) and elastic weight consolidation (Masse et al 2018b, Kirkpatrick et al 2017b). Another example is the Self-Net described in (Mandivarapu et al 2020), that uses an autoencoder to learn a set of low-dimensional representations of the weights learned for different tasks. An example of continuous learning in the medical domain is the study by (Kiyasseh et al 2021), whose model learned to deal with cardiac signals across diseases, time, modalities, and institutions. As the models become more and more used in the clinical setting, developing stable continuous learning methods will become essential for the long-term viability of the models. Notice that continuous learning can also be considered as a meta-learning framework where the ML model learns to learn over time and environmental changes.
Other emerging learning frameworks include automatic machine learning (AutoML), or federated learning. AutoML tries to build ML methods that automatically configure themselves, including data preprocessing, network architecture selection, training, and post-processing for any new task (Hutter et al 2019). The idea behind AutoML is to automate the trial-and-error process that data scientists and practitioners typically carry out manually to find the optimal pre-processing steps and hyperparameters of the ML architecture. A recent example of autoML in the medical field is the increasingly popular nnU-Net, an autoML model for segmenting organs from any medical images (Isensee et al 2021).
Federated learning, also known as distributed learning (Boyd 2010), allows ML models to be trained with data sets of several origins (e.g. hospitals or clinics) without pooling them. As it can maintain patient data confidentiality, federated learning therefore raises much interest in the medical domain (Sheller et al 2019, Chang et al 2018). Instead of bringing all data to a central repository to train an ML model, distributed learning brings the model to the data. This approach facilitates cooperation through coalitions in which each member retains control and responsibility over its own data, including accountability for privacy and consent of the data owners (i.e., patients). Federated learning can also help ML models to better generalize, since they are exposed to training data from different hospitals, better encoding the variability of the problem.
To conclude this section, Figure 14 summarizes some of the issues that have been discussed above, as well as some of the possible solutions (strategies, tools, and frameworks) to mitigate them. As it can be seen, there is no one-to-one mapping between issues and solutions, and practitioners often need some experience to identify the best associations.
Figure 14.
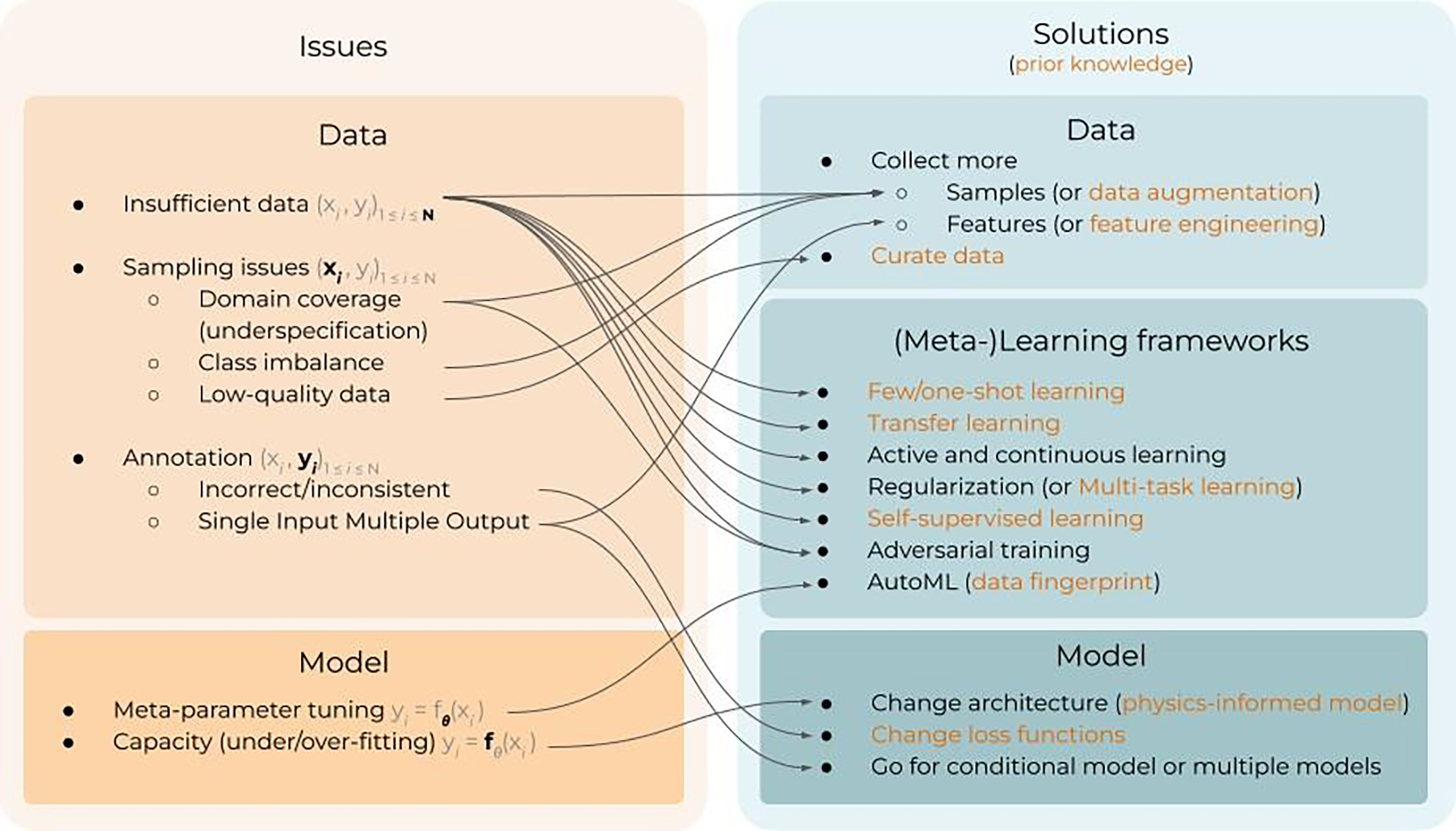
Tentative mapping between common issues that are encountered in the implementation and deployment of machine learning and some possible solutions to overcome them. Since machine learning relies on data-driven models, both the issues and solutions can be seen from the angles of data, modeling, or learning.
4. Discussion: Clinical implementation of ML in radiation oncology — the big picture
The previous sections have provided the reader with a general background about the risks and limitations associated with the use of ML in the clinical environment (Section 2), and the different techniques that are being investigated by the research community to better identify and overcome those issues (Section 3). This section discusses the specific application of ML techniques into the radiation oncology workflow and the implications this has for the clinical practice of this field. First, we start by walking the reader through the radiotherapy workflow, and discuss in detail key tasks that are undergoing a paradigm shift with the introduction of ML. Second, as clinical software is most of the time provided by industrial companies or vendors and implemented in close collaboration with them, we discuss the vendors’ approach and point of view regarding the clinical use of ML.
4. 1. Considerations on the radiation therapy workflow
The typical workflow of radiation oncology can be summarized in a sequence of tasks presented in Figure 15. The inclusion of ML in the workflow aims at reducing human intervention, automating the tasks, standardizing clinical practice, and improving the overall treatment quality. As previously introduced, the gap between expertise and resources between institutions is sometimes quite big, representing one of the greatest inequality and challenges in health-care (Lievens et al 2020). Incorporating ML in the radiotherapy workflow can help to homogenize and improve clinical practice.
Figure 15.
Typical workflow of radiotherapy treatment planning and delivery (top panels, orange boxes) and current applications of ML in the workflow (bottom, blue boxes).
Historically, classical ML and image processing techniques (active contours, watersheds, multi-atlas registration, …) have been long used in an attempt to automate tedious, manual, and time-consuming tasks in the radiotherapy workflow. However, they often still required manual intervention and lacked some form of intelligence and memory. The disruptive change occurred with the advent of modern DL models, i.e, convolutional neural networks and image-to-image architectures like U-Net (Ronneberger et al 2015). Although much less interpretable than the aforementioned classical methods, DL models are now the state of the art.
Cancer diagnosis and treatment choice is the first step in the presented workflow, and involves the analysis of different types of data: medical records, patient’s symptoms, raw images, histopathological data, genomic data, etc. Processing these large amounts of heterogeneous data is becoming a challenge for humans and, thus, the inclusion of intelligent systems for decision support might be of big help. Diagnosis is one of the earliest applications of ML in oncology, and the first studies date from the mid 1990s and early 2000s (Bertsimas and Wiberg 2020), where traditional ML models were used to analyze gene expression profiles and detect cancer biomarkers or to analyze images to detect features indicating the presence of cancer (Wolberg et al 1995). Two of the first cancer locations in which the research community started to focus on were skin and breast cancer. Today, though, a wide range of cancer types and locations benefit from the use of ML as a decision support tool (Kleppe et al 2021, Bertsimas and Wiberg 2020, Iqbal et al 2021). In addition to diagnosis, numerous studies focus on predicting radiation toxicity and possible side effects, in order to aid the physician to select the best treatment protocol (Tran et al 2021, Isaksson et al 2020). While the earliest applications for diagnosis and treatment choice focused either on one type of data (e.g., genomics or images), current ML have the potential to process several types of data simultaneously by fusion of the information at different parts of the model architecture (see Section 3.2.2), therefore making a better informed diagnosis. Progressively, ML models for diagnosis and treatment choice start to be applied in clinical routine (Savage 2020, Benjamens et al 2020); some claim that the ML model rivals with or even outperforms human experts (Esteva et al 2017). However, the truth is that there are still very few ML applications developed in research environments that have made it to the clinic, due to poor generalization or the inability to guarantee the correctness of the answer. To overcome those issues, several solutions have been proposed in this manuscript, which are in line with the recent literature in ML applied to diagnosis. For instance, (Kleppe et al 2021) advocate the evaluation of the ML model in external cohorts, which could be also achieved with extensive data augmentation techniques when external cohorts are not available, as presented in Section 3.2.1. Uncertainty quantification (see Section 3.2.3) is another of the keys advised for diagnosis and decision-making ML models (Begoli et al 2019), which can be combined with techniques for explainability (see Section 3.1) to ensure that there is no learning bias when building the models. Lastly, reinforcement learning algorithms can help to explore new and personalized treatment in a well-controlled framework.
After the treatment protocol has been selected, the second step is to image the patient with a CT scanner, in a controlled setting, simulating the treatment position and with proper immobilization devices to avoid motion. Eventually, other images needed for the treatment can also be acquired (e.g., MR, PET.,...), if they were not already taken in the diagnosis step. Given the excellent performance of modern deep convolutional neural networks to analyze and deal with images, many applications have been developed related to this imaging step (Shan et al 2020). For instance, ML is used in image reconstruction (Ahishakiye et al 2021), to increase the quality of the image by removing artifacts (Xie et al 2018, Dong et al 2021), or to register the different acquired images (Fu et al 2020, Haskins et al 2020). Particularly for image registration, the interest has rapidly increased in the last years, with numerous publications investigating some of the most advanced ML techniques, such as one-shot learning (Zhang et al 2021), unsupervised learning (Balakrishnan et al 2018), or reinforcement learning (Hu et al 2021a), among others. In short, ML models for image registration try either to learn feature maps for the input moving images and fixed images, or to learn new image representations for the original fixed images and moving images (e.g., transform the original images to be better suited for registration) (Fu et al 2020). The use of unsupervised techniques is very helpful for ML registration models, because it suppresses the need for ground-truth deformation fields, which are costly to generate. Another direction to improve future ML models for registration includes boosting their performance by incorporating prior knowledge (see Section 3.2.2). For instance, prior information related to the expected type of deformation, spatial relationship between anatomical structures, and the topology and morphology of anatomical structures, could be added to allow the ML model to perform better (Fu et al 2020).
Another popular task related both to the imaging step and to the treatment delivery, is the conversion or generation of synthetic images. Since the attenuation coefficients in the CT image are needed to perform treatment planning and dose calculation, techniques relying on other images, such as adaptive therapy based on CBCTs or MR-only radiotherapy, largely benefit from the use of ML to generate a synthetic CT. Image synthesis is thus considered the third most popular clinical application (Brouwer et al 2020). Numerous examples of image synthesis have been given throughout the manuscript, such as the use of generative adversarial networks (GANs) to convert MR to CT (Maspero et al 2018, Kazemifar et al 2020, 2019) or CBCT to CT (Liang et al 2019b). A common concern in this field is the generalization of the ML model to different scanners and acquisition protocol (see Section 2.2), and much effort has been put into addressing this issue with different techniques, such as transfer learning (Liang et al 2020) or data augmentation, among others. Beside generalization, a future research line could be developing techniques for interpretability and explainability for image synthesis. However, this is not straightforward, since in contrast to classification and segmentation tasks, the network will not focus on specific parts of the images but rather on the full image to be converted. In this case, Concept Activation Vectors (CAV) could be of help, in order to provide the user with the more relevant concepts for the transformation for verification (Lucieri et al 2020)(Kim et al 2018). In contrast, the risk of failure could be easily assessed with uncertainty quantification tools as described in Section 3.2.3.
Once the images are acquired, the following step is to contour or segment the relevant volumes needed for treatment planning. In particular, the segmentation of most organs from CT images is considered nowadays a pretty much solved problem, with the latest works reporting an accuracy similar to human experts’ performance. For instance, (Nikolov et al 2018) achieved a Dice coefficient over 90% for most organs in the head and neck region. Motivated by these results, many research groups and clinical teams have already attempted a clinical implementation of ML based automatic segmentation, using either in-house or commercial solutions (Vandewinckele et al 2020, Brouwer et al 2020, Cha et al 2021, van der Veen et al 2019). In fact, automatic contouring is today the most used ML application in the clinic and, therefore, we will discuss it in detail in the following paragraphs.
In a survey from 2020, Brouwer et al. reported that 26% of the responders were already using ML-based contouring in their clinics (most of them with commercial software, 76%), and nearly 20% were preparing for its implementation (Brouwer et al 2020). However, despite this large adoption in the clinic, the current ML methods for automatic segmentation still lack QA tools to assess their interpretability and risk of failure. This is today compensated in quite a rudimentary way: the QA of the ML-contours is performed by visual inspection of a medical expert, who edits the contour in the regions where the ML model has failed. Although the time and magnitude of the editions are much shorter than fully manual segmentation (for instance, about 33% shorter for head and neck contours) (van der Veen et al 2019), the process still requires the systematic presence of a medical doctor for QA. When generating the contours offline, before the treatment starts, this can be manageable. But in adaptive radiotherapy workflows, where new contours have to be generated while the patient is on the couch, requiring the presence of a physician for every treatment fraction is truly a big limitation. Hence, it is imperative that clinical ML models start to integrate QA tools similar to those presented in Section 3, in order to ensure their efficient and safe usage. Applying interpretability and explainability techniques during the training and validation phase, in particular those for visualization of the relevant regions contributing to the prediction (e.g., CAM, gradCAM and variants), can help debug the model faster and ensure it works correctly. In contrast, during routine clinical use, especially in adaptive settings, interpretability and explainability techniques might not be the best QA tools due to the tight time constraints. As introduced in Section 3.1, interpretability is a complex concept, involving many factors, and both time and user-expertise play important roles. Unless very intuitive explanations can be provided for a fast evaluation by the medical staff, when time is crucial (i.e., adaptive treatment procedures), uncertainty quantification tools might be a better option. For instance, one could implement a flagging system based on the level of uncertainty associated with the prediction. When uncertainty is low, the treatment can be performed right away with the ML contours, without the need of edits by the medical doctors. When uncertainty is high, the doctors are asked to verify (and edit) the ML contours offline and, eventually, they are provided with explanations that support the ML answer. Such a workflow can save much time for the medical staff and, most importantly, it relieves users of constant QA. Moreover, the manual offline editions can later serve to improve the ML model if an active learning framework is deployed (Section 3.2.4).
It is important to stress that, as in any classification task, the vector of class (organ) probabilities that the model yields for each voxel (i.e., the softmax output) is not a measure of uncertainty, but just a pointwise estimate of a class probability (Gal 2016). Indeed, this probability is often misinterpreted as an uncertainty, which can be misleading and risky. Voxels classified with a high probability can still carry a high uncertainty, especially for cases that are far from the training set (Gal 2016). Instead, techniques such as MCDO or other Bayesian approaches, as well as ensemble methods can be used to estimate the uncertainty and an associated confidence interval (Figure 16).
Figure 16.
For the different slices of four patients, the red line corresponds to the clinical contour, the blue to the prediction and the yellow area the 95% confidence band. The latter can be used as a visual indicator of the model uncertainty. For instance the model is more uncertain for patient four (Balagopal et al 2021).
Concerning the segmentation models for target volumes, there is still much room for improvement to have robust and accurate models. In contrast to the segmentation of organs, which can work rather well by just using the anatomical information in the images, the segmentation of target volumes involves many other variables. For instance, information from several imaging modalities is often used by the physicians to draw the clinical target volumes (e.g. MR, PET, endoscopy, …), together with indicators or reports from clinical examinations. In order to reach human level performance, ML models for target segmentation need to integrate this information and domain-knowledge, using the techniques presented in Section 3.2.3. In addition, interpretability and explainability tools can be of much more importance here than in the case of organ segmentation, since QA cannot be done with a simple visual check due to the large number of variables involved. Apart from visual explanations like CAM and variants, text-based explanations relying on Concept Activation Vectors (Section 3.1.2) could be a QA for the provided contours.
Another strategy that can be of help to have efficient segmentation ML models that brings a real added value to the clinic is the use of techniques requiring less supervision (see Section 3.2.4). This is especially important for image modalities used in adaptive settings (e.g. Cone Beam CT or MR), since retrospective databases of contours on these images are typically unavailable (i.e., the contours are done on the CT but not on the CBCT or MR). In this case, one could apply techniques such as unsupervised domain adaptation (UDA) (Ganin and Lempitsky 2015, Kamnitsas et al 2017, Brion et al 2021). UDA is a sort of unsupervised transfer learning strategy, where the modality for which the labels are available is considered the source domain (e.g., CT), and the modality without labels is the target domain (e.g., CBCT or MR). In all cases, if done properly, the introduction of ML segmentation in the clinic will definitely bring an improvement and standardization of the practice. Instead of having paper guidelines (Apolle et al 2019, Grégoire et al 2014) that are hard to reproduce and are subject to inter-observer variability (Apolle et al 2019), ML models can capture the experts’ knowledge and easily transfer it from one center to another, reducing the inter-observer variability (Veen et al 2019, van der Veen et al 2020, 2019).
After volume segmentation, the next labor-intensive step in the workflow is treatment planning and the optimization of dose distribution. Although current TPSs heavily rely on inverse problem solving and iterative computerized optimization, the definition of the objectives to attain and the constraints to fulfill is often difficult and requires trade-offs that are not easy to formalize mathematically. Once again, ML can memorize from past examples of such tradeoffs and generalize to new patient cases.
One of the first approaches for automatic treatment planning with ML used Random Forest algorithms in combination with multiple atlases (Contextual Atlas Regression Forests) to predict the dose distribution for a new patient based on the information in the atlas (McIntosh and Purdie 2016, McIntosh et al 2017). Almost in parallel, several groups started to explore DL image-to-image networks (like U-Net or GANs), to predict the dose for a new patient anatomy using the CT and organs as input (Nguyen et al 2019b, Fan et al 2019, Kearney et al 2020). Needless to say, none of these approaches is very interpretable, but the one based on multi-altas Random Forests can implicitly report atlas distances representing the most-similar patients from the training set. Recently, this approach has been implemented clinically and analyzed prospectively (McIntosh et al 2021). They reported that these distance metrics could indeed be used to flag lower-quality generated dose distributions and the potential need for human verification, increasing the interpretability and usability of the method. For dose prediction methods based on U-Net or GANs architectures, there is no intrinsic attribute that could provide similar information. However, recent studies have explored the use of MCDO and ensemble methods (Section 3.2.3) to quantify the uncertainty associated with the predicted dose (Vanginderdeuren et al 2021, Nguyen et al 2021). As previously introduced, this uncertainty estimation can be used in a similar way to flag the poor performance of the model, as well as in active learning workflows to further improve the model. These studies reported the correlation coefficient between the estimated uncertainty (using the standard deviation, see Section 3.2.3) and the actual prediction error (difference between ground truth and predicted dose). However, there is still room for improvement in order to achieve accurate metrics for uncertainty quantification in dose prediction, since the reported correlation coefficients were sometimes very low (Vanginderdeuren et al 2021, Nguyen et al 2021).
In addition to risk assessment tools, two other lines of research are worth mentioning in the race for efficient and clinically meaningful ML models for dose prediction. The first one is incorporating domain-knowledge into the ML model, for which several examples have been provided in Section 3.2.2, including the use of domain-specific loss functions (Nguyen et al 2020) and comprehensive input data (Kontaxis et al 2020, Barragán-Montero et al 2019). The second one is the use of transfer learning models to be able, for instance, to generalize to different treatment locations (Mashayekhi et al 2021) and clinical practices (Kandalan et al 2020).
Similar to ML models for segmentation, when properly implemented, ML-based dose prediction can bring significant improvement for clinical practice. For instance, since ML models can infer in a few seconds, one can predict dose distributions for different treatment modalities (e.g., proton therapy versus conventional radiotherapy), in order to refer the patient to the most optimal treatment (Guerreiro et al 2021). This allows for huge time savings and efficient resource usage.
To exploit the predicted dose distribution and to generate the final treatment plan, several options are possible. The most popular one is to use the predicted dose as a voxel-wise objective in the TPS, alone or in combination with dose-volume metrics. This optimization process is often called dose-mimicking, and translates the synthetic predicted dose into a physically deliverable dose (McIntosh et al 2017, Babier et al 2020). The process is the same as in regular treatment planning, using algorithms similar to gradient descent for optimization and analytical or Monte Carlo methods for dose calculation. However, some groups have pushed the use of DL even further, trying to predict the treatment plan (i.e., the machine parameters or fluence maps) from the predicted dose distribution (Wang et al 2020b) (Lee et al 2019). Although these research studies are excellent to explore the potential of DL models in the radiotherapy workflow, we should be extremely cautious when it comes to clinical implementation. Indeed, a distinction should be made between soft computing (e.g., ML models) and scientific computing (e.g., physics-based models, analytical models), with the former not providing any strong guarantees of consistently good performance, whereas the latter does. DL models are excellent methods to be applied when we want to be fast, automatic, and free of any human intervention, like in segmentation or treatment planning. However, when fast and automatic scientific computing models already exist for a given task (e.g., optimization or dose calculation) and soft-computing does not bring any significant improvement in performance, scientific computing and physical models should be encouraged. Recently, another approach attempts to find the sweet spot between these two options, which could be physics-informed ML models (Raissi et al 2019) (section 3.2.2). These ML models have the particularity of being constrained with physical rules, and could help to extract the best from soft- and scientific-computing methods, while also increasing their interpretability (Rudin et al 2021).
Once the treatment is ready for delivery, the next step is to perform quality assurance tests (QA) to ensure that the treatment is delivered as planned. ML applications in radiotherapy QA started to become popular around 2016, and many relevant studies have been published since then (Chan et al 2020, Kalet et al 2020). Some examples include the study by (Li and Chan 2017), who developed a model to predict the performance of a Linac over time; the study by (Osman et al 2020), who trained a model with log files to predict the multi-leaf collimation (MLC) leaf positional deviations; or the study by (Valdes et al 2016b), who designed a ML model to predict passing rates for IMRT QA. Although the use of ML in radiotherapy QA might be very beneficial for the medical physicists team, further automating and improving the QA process, the models developed so far have several limitations. (Kalet et al 2020) claim that data quality and model generalization are among the main limitations. As discussed in Section 2, low-quality and insufficient data might lead to biased performance of the ML model. In order to overcome this issue, (Chan et al 2020) advocates for multi-institutional validation of the developed ML models. In this context, federated learning might help to gather data from several institutions while preserving privacy and security.
During the delivery of the treatment, several of the previously discussed tasks come again into play. For instance, in adaptive or image-guided radiotherapy, we use daily images to monitor the treatment and eventually adapt it to the new anatomy. In this context, ML models for image synthesis or conversion become useful when the monitoring image (e.g. CBCT) needs to be converted into a CT. Similarly, ML models for image registration, automatic segmentation, and treatment planning are useful to generate the adapted plan in a fast and automatic manner. Beside these applications, another task that can benefit from ML and has not been discussed so far is motion management. For instance, (Lin et al 2019) developed a ML model to predict tumor motion by combining features coming from images and Electronic Health Records.
After the treatment has been delivered, the final step is to follow-up the progression of the disease and the possible treatment complications. For this purpose, the patient has regular consultations every few months, where the patient’s condition is analyzed and images are acquired if needed. Treatment outcome prediction can be of help at two time frames: 1) at the beginning of the treatment, to aid the treatment choice; and 2) at the end of the treatment, to predict the locoregional control and survival probabilities for a given patient. For instance, recent studies used ML to predict the treatment response for bladder cancer (Cha et al 2017), lung cancer (Xu et al 2019), and pancreatic cancer(Chen et al 2017), among many others (El Naqa et al 2018, Isaksson et al 2020). In their topical review, (Isaksson et al 2020) claim that, since they play an important role for treatment choice, critical efforts are required to improve the transparency of ML for outcome prediction, making them accessible to the clinical staff, who have little or no specific background on ML. Interpretability and explainability techniques such as the ones presented in section 3.1 could definitely help to reach this goal. Recently, (Luo et al 2019) has published a review about popular applications is outcome prediction, discussing in detail the balance between interpretability and accuracy, and providing techniques to find the optimal settings for their safe clinical implementation To finish, we would like to bring up our point-of-view on how the clinical workflow will change with the introduction of ML. Although the implementation of techniques for interpretability and risk assessment presented here (i.e., data curation, uncertainty quantification, domain-knowledge, …), will reduce the human QA, it will still continue to be very important. Thus, the work of physicians, medical physicists and dosimetrists, will evolve from performing manual tasks to supervising ML models (Korreman et al 2021). Moreover, the medical staff will play a crucial role in data collection and curation to build ML models. The already multidisciplinary nature of this field will become even more important, since that will be the key to achieve comprehensive ML models that efficiently incorporate relevant domain-knowledge. +9*/Note that the need for interpretable and safe ML models start to be also discussed in legal environments and regulatory institutions both in Europe and America (Bibal et al 2021, Anon 2021). A famous example is the General Data Protection Regulation (GDPR), which specifically constrains the use of black box models in certain cases.
4.2. The vendors’ perspective
As previously introduced, a large majority of clinically implemented ML software comes from industrial companies. Thus, the vendors play a crucial role in an efficient and safe deployment, since they are responsible for the released models. In the following, we go through the different phases of the clinical implementation of ML models from the vendor’s perspective.
Model development and commissioning
Developing an ML model includes many steps: data collection and curation, model training, model configuration, and validation (figure 17). As vendors are responsible for the released ML models for their entire life-cycle, all these steps need to be managed and documented by them, not least for the regulatory processes.
Figure 17.
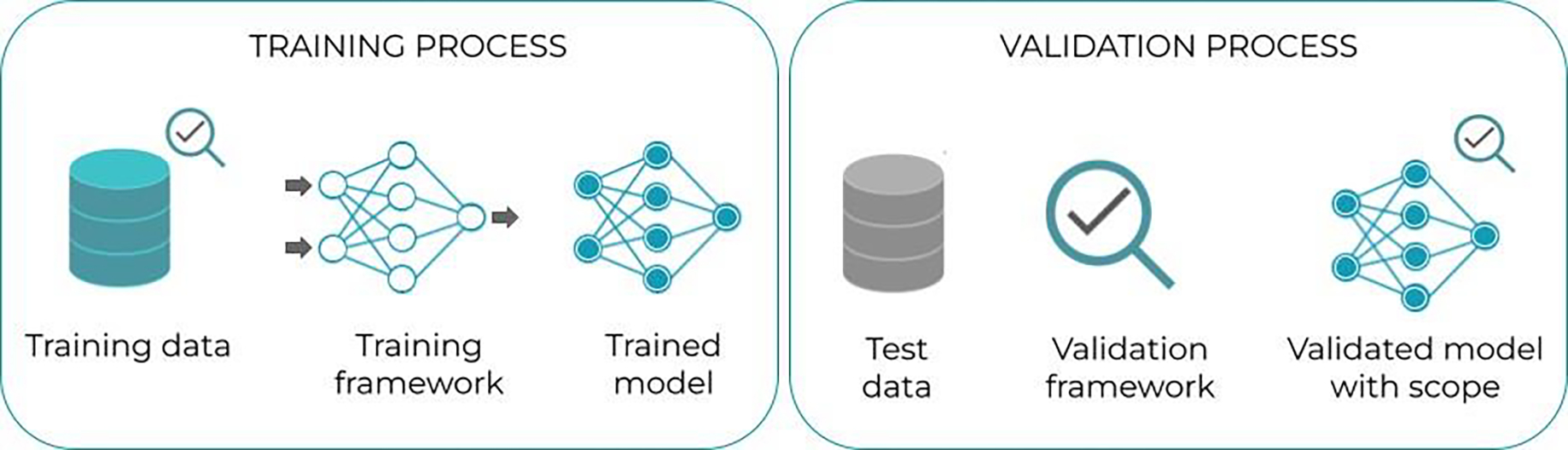
Model development process.
In particular, the data included in model development needs to be accessible to the vendor for future support, model upgrades and regression testing. Often, the data collected, either from public sources or from clinics or both, need to be curated to align with the selected guidelines and protocols and to fit the purpose of model development (Section 3.2.1). Ideally, vendors and clinics should agree and align on interpretation of guidelines and protocols as part of the data curation. Meta data for the datasets also need to be documented, such as versioning, data sources, data creator, protocol, and more. Vendor’s should strive to use datasets from multiple sources in model development to increase model robustness, for instance by using data from multiple continents.
Moreover, it is critical to keep track of the training, validation, and test datasets, as well as data augmentation tools (Section 3.2.1) and hyperparameters, in order to be able to re-train or further develop the model. The training, including infrastructure and computational resources, should be handled by the vendor.
After the training process, the model needs to be properly validated (figure 17) on independent, representative, and diversified data to make sure the model is fit for purpose, and identify the use cases and the limitations of the model (model scope). The resulting validation report can include a model data sheet specifying the training and validation details, as well as the intended use and limitations of the model (figure 18). Such data sheets should always accompany the released ML model when distributed to clinics, which will allow the clinical users to apply the model to relevant cases and reduce the risk of misuse.
Figure 18.

Example of data sheet for a released ML model for automatic planning of radiotherapy treatments for prostate cancer patients. The data sheet contains relevant information about the ML model, including the general overview and scope, as well as information about the training and validation phases. This data sheet should be provided by the vendor together with the model.
When a clinic goes live with a released ML model, they need to commission the model on their local data and use case. For instance, the commissioning of a validated DL segmentation model involves evaluating the model output on image sets and structure sets from the clinic, taking the intended use of the model into account. The commissioning resembles the validation process, and it may involve configuration of settings affecting the postprocessing of the model output to align the commissioned model with the clinical use case, scope, and specific treatment protocol (Figure 19). Notice that the released model itself, e.g., the optimized neural network parameters, is not affected by such a model configuration. The vendor should support the clinic with the commissioning process. After that, the model is locked and no settings affecting the output of the model can be changed. Although active and continuous learning workflows (Section 3.2.4) are very attractive, their feasibility after the commissioning is done is rather complex, due to the risks associated with model changes (Liu et al 2020, Vokinger et al 2021). Thus, they are better suited to be applied during training, when changes in the model are still possible. In case the model becomes not valid anymore because of changes in the data distribution over time (Section 2.1), re-training or re-model configuration could be performed, which would trigger a new commissioning. The commissioning results, which are specific to the clinic, must be stored for future reference and should ideally be shared with the vendor.
Figure 19.
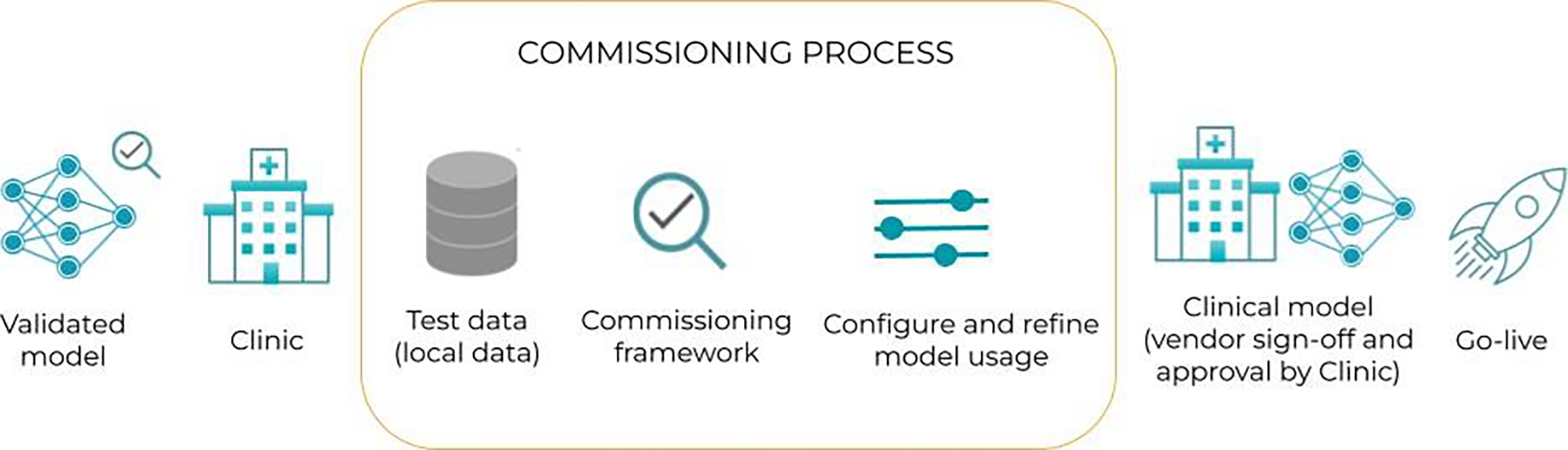
Commissioning and go-live for a released ML model.
Using AI in clinical practice: implementation, model life-cycle and sharing
When going live with an AI model, it is important to monitor the performance of the model in terms of usage, results, adjustments, post-processing and approval times, and problematic cases. The vendor should develop tools for automating such monitoring and QA, to enable a safe and transparent clinical implementation. For instance, the ML generated segmentations can be stored separately and compared to the approved segmentations, allowing for monitoring of the models over time in terms of the manual adjustments needed.
ML models are suitable for sharing as they can be designed not to contain any personal data. We believe clinics will be open to share their knowledge with other clinics through ML models that have been trained and validated on their data, and vendors can provide tools to do that. For clinical purposes, an ML model can be shared if it has been validated and there is a model data sheet specifying its intended use and limitations (Figure 18). Model sharing should be centrally organized rather than bilateral to ensure quality, transparency, model distribution monitoring, and version handling. Also, if a clinically deployed model is deficient, the traceability is important so all affected clinics can be notified. Such centralization of models combined with centralization of outcome data and other relevant input may lead to consensus in how certain treatments should be conducted.
5. Conclusion
Thanks to impressive results in tasks that were previously reserved for human intelligence, like visual object recognition in natural images, machine learning has become very fashionable and has raised much interest in all sorts of applications. Medicine has not escaped that ubiquitous trend and, in particular, specializations that heavily rely on medical imaging, like radiation oncology, try to fully exploit the possibilities of ML models. The sharp turn in that direction leads to a road full of promises but also paved with many pitfalls and poor visibility ahead. In order to address this issue, a twofold approach has been proposed in this review. On the one hand, interpretability and explainability is meant to make ML more trustworthy and its users more confident. On the other hand, exploring the tight relationship between data and model performance can help us to achieve more efficient learning, as well as to develop tools for risk assessment and QA. This review has explored some of the most recent developments in interpretable and explainable machine learning, presented different concepts around the data-model dependency issue, and investigated in the literature how they start being applied in medicine and radiation oncology in particular. In the short term, interpretability is expected to be a topic of growing interest in interdisciplinary conferences and workshops, like “UNSURE” (Uncertainty for Safe Utilization of Machine Learning in Medical Imaging) (Sudre et al 2021) or the “iMIMIC” (Interpretability of Machine Intelligence in Medical Image Computing) (Reyes et al 2021) in MICCAI. In the mid term, interpretability and explainability of ML and artificial intelligence in general are likely to be developed on their legal side by law- and policy-makers, as well as regulatory institutions. For example, Europe has already formed expert groups to discuss and emit recommendations on “responsible artificial intelligence”. These initiatives could follow a similar path as the general data protection regulation (GDPR) or be integrated in it. Finally, in the longer term, more futuristic developments of machine learning and artificial intelligence are aimed at streamlining the interface between human intelligence and its artificial counterpart, most probably by using natural language and other familiar means of communication.
Acknowledgements
Ana Barragán and Margerie Huet is funded by the Walloon region in Belgium (PROTHERWAL/CHARP, grant 7289). Gilmer Valdés was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number K08EB026500. Dan Nguyen is supported by the National Institutes of Health (NIH) R01CA237269 and the Cancer Prevention & Research Institute of Texas (CPRIT) IIRA RP150485. Liesbeth Vandewinckele is supported by a Ph.D. fellowship of the research foundation - Flanders (FWO), mandate 1SA6121N. Kevin Souris is funded by the Walloon region (MECATECH / BIOWIN, grant 8090). John A. Lee is a Senior Research Associate with the F.R.S.-FNRS.
References
- Abdar M, Pourpanah F, Hussain S, Rezazadegan D, Liu L, Ghavamzadeh M, Fieguth P, Cao X, Khosravi A, Rajendra Acharya U, Makarenkov V and Nahavandi S 2021. A review of uncertainty quantification in deep learning: Techniques, applications and challenges Information Fusion 76 243–97 Online: 10.1016/j.inffus.2021.05.008 [DOI] [Google Scholar]
- Adadi A and Berrada M 2018. Peeking Inside the Black-Box: A Survey on Explainable Artificial Intelligence (XAI) IEEE Access 6 52138–60 Online: 10.1109/access.2018.2870052 [DOI] [Google Scholar]
- Afshar P, Mohammadi A, Plataniotis KN, Oikonomou A and Benali H 2019. From Handcrafted to Deep-Learning-Based Cancer Radiomics: Challenges and Opportunities IEEE Signal Processing Magazine 36 132–60 Online: 10.1109/msp.2019.2900993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahishakiye E, Van Gijzen M B, Tumwiine J, Wario R and Obungoloch J 2021. A survey on deep learning in medical image reconstruction Intelligent Medicine 1 118–27 Online: 10.1016/j.imed.2021.03.003 [DOI] [Google Scholar]
- Ahn SH, Kim E, Kim C, Cheon W, Kim M, Lee SB, Lim YK, Kim H, Shin D, Kim DY and Jeong JH 2021. Deep learning method for prediction of patient-specific dose distribution in breast cancer Radiat. Oncol. 16 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alashwal H, El Halaby M, Crouse JJ, Abdalla A and Moustafa AA 2019. The Application of Unsupervised Clustering Methods to Alzheimer’s Disease Front. Comput. Neurosci. 13 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shedivat M, Bansal T, Burda Y, Sutskever I, Mordatch I and Abbeel P 2017. Continuous Adaptation via Meta-Learning in Nonstationary and Competitive Environments arXiv [cs.LG] Online: http://arxiv.org/abs/1710.03641 [Google Scholar]
- Amadasun M and King R 1989. Textural features corresponding to textural properties IEEE Transactions on Systems, Man, and Cybernetics 19 1264–74 Online: 10.1109/21.44046 [DOI] [Google Scholar]
- Anon 2009. Aleatory or epistemic? Does it matter? Struct. Saf. 31 105–12 [Google Scholar]
- Anon 2021. The EU medical device regulation: Implications for artificial intelligence-based medical device software in medical physics Phys. Med. 83 1–8 [DOI] [PubMed] [Google Scholar]
- Apolle R, Appold S, Bijl HP, Blanchard P, Bussink J, Faivre-Finn C, Khalifa J, Laprie A, Lievens Y, Madani I, Ruffier A, de Ruysscher D, van Elmpt W and Troost EGC 2019. Inter-observer variability in target delineation increases during adaptive treatment of head-and-neck and lung cancer Acta Oncol. 58 1378–85 [DOI] [PubMed] [Google Scholar]
- Arrieta AB, Díaz-Rodríguez N, Del Ser J, Bennetot A, Tabik S, Barbado A, Garcia S, Gil-Lopez S, Molina D, Benjamins R, Chatila R and Herrera F 2020. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI Information Fusion 58 82–115 Online: 10.1016/j.inffus.2019.12.012 [DOI] [Google Scholar]
- Ayhan MS and Berens P 2018. Test-time Data Augmentation for Estimation of Heteroscedastic Aleatoric Uncertainty in Deep Neural Networks Online: https://openreview.net/pdf?id=rJZz-knjz
- Babier A, Mahmood R, McNiven AL, Diamant A and Chan TCY 2020. The importance of evaluating the complete automated knowledge-based planning pipeline Phys. Med. 72 73–9 [DOI] [PubMed] [Google Scholar]
- Bach PB, Cramer LD, Warren JL and Begg CB 1999. Racial differences in the treatment of early-stage lung cancer N. Engl. J. Med. 341 1198–205 [DOI] [PubMed] [Google Scholar]
- Badgeley MA, Zech JR, Oakden-Rayner L, Glicksberg BS, Liu M, Gale W, McConnell MV, Percha B, Snyder TM and Dudley JT 2019. Deep learning predicts hip fracture using confounding patient and healthcare variables NPJ Digit Med 2 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahdanau D, Cho K and Bengio Y 2014. Neural Machine Translation by Jointly Learning to Align and Translate arXiv [cs.CL] Online: http://arxiv.org/abs/1409.0473 [Google Scholar]
- Bai X, Zhang J, Wang B, Wang S, Xiang Y and Hou Q 2021. Sharp loss: a new loss function for radiotherapy dose prediction based on fully convolutional networks BioMedical Engineering OnLine 20 Online: 10.1186/s12938-021-00937-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal A, Nguyen D, Morgan H, Weng Y, Dohopolski M, Lin M-H, Barkousaraie AS, Gonzalez Y, Garant A, Desai N, Hannan R and Jiang S 2021. A deep learning-based framework for segmenting invisible clinical target volumes with estimated uncertainties for post-operative prostate cancer radiotherapy Med. Image Anal. 72 102101. [DOI] [PubMed] [Google Scholar]
- Balakrishnan G, Zhao A, Sabuncu MR, Dalca AV and Guttag J 2018. An Unsupervised Learning Model for Deformable Medical Image Registration 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition Online: 10.1109/cvpr.2018.00964 [DOI] [Google Scholar]
- Bamba U, Pandey D and Lakshminarayanan V 2020. Classification of brain lesions from MRI images using a novel neural network Multimodal Biomedical Imaging XV Online: 10.1117/12.2543960 [DOI] [Google Scholar]
- Barragán-Montero A, Javaid U, Valdés G, Nguyen D, Desbordes P, Macq B, Willems S, Vandewinckele L, Holmström M, Löfman F, Michiels S, Souris K, Sterpin E and Lee JA 2021a. Artificial intelligence and machine learning for medical imaging: A technology review Phys. Med. 83 242–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragán-Montero AM, Nguyen D, Lu W, Lin M-H, Norouzi-Kandalan R, Geets X, Sterpin E and Jiang S 2019. Three-dimensional dose prediction for lung IMRT patients with deep neural networks: robust learning from heterogeneous beam configurations Medical Physics 46 3679–91 Online: 10.1002/mp.13597 [DOI] [PubMed] [Google Scholar]
- Barragán-Montero AM, Thomas M, Defraene G, Michiels S, Haustermans K, Lee JA and Sterpin E 2021b. Deep learning dose prediction for IMRT of esophageal cancer: The effect of data quality and quantity on model performance Phys. Med. 83 52–63 [DOI] [PubMed] [Google Scholar]
- Barrett JF and Keat N 2004. Artifacts in CT: recognition and avoidance Radiographics 24 1679–91 [DOI] [PubMed] [Google Scholar]
- Bashir U, Kawa B, Siddique M, Mak SM, Nair A, Mclean E, Bille A, Goh V and Cook G 2019. Non-invasive classification of non-small cell lung cancer: a comparison between random forest models utilising radiomic and semantic features Br. J. Radiol. 92 20190159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JL and Katafygiotis LS 1998. Updating Models and Their Uncertainties. I: Bayesian Statistical Framework Journal of Engineering Mechanics 124 455–61 Online: 10.1061/(asce)0733-9399(1998)124:4(455) [DOI] [Google Scholar]
- Begoli E, Bhattacharya T and Kusnezov D 2019. The need for uncertainty quantification in machine-assisted medical decision making Nature Machine Intelligence 1 20–3 Online: 10.1038/s42256-018-0004-1 [DOI] [Google Scholar]
- Bengio Y, Courville A and Vincent P 2013. Representation learning: a review and new perspectives IEEE Trans. Pattern Anal. Mach. Intell. 35 1798–828 [DOI] [PubMed] [Google Scholar]
- Benjamens S, Dhunnoo P and Meskó B 2020. The state of artificial intelligence-based FDA-approved medical devices and algorithms: an online database NPJ Digit Med 3 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry SL, Boczkowski A, Ma R, Mechalakos J and Hunt M 2016. Interobserver variability in radiation therapy plan output: Results of a single-institution study Pract. Radiat. Oncol. 6 442–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsimas D and Wiberg H 2020. Machine Learning in Oncology: Methods, Applications, and Challenges JCO Clin Cancer Inform 4 885–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibal A and Frénay B 2016. Interpretability of machine learning models and representations: an introduction ESANN Online: https://www.researchgate.net/profile/Adrien-Bibal-2/publication/326839249_Interpretability_of_Machine_Learning_Models_and_Representations_an_Introduction/links/5b6861c aa6fdcc87df6d58e4/Interpretability-of-Machine-Learning-Models-and-Representations-an-Introduction.pdf [Google Scholar]
- Bibal A, Lognoul M, de Streel A and Frénay B 2021. Legal requirements on explainability in machine learning Artificial Intelligence and Law 29 149–69 Online: 10.1007/s10506-020-09270-4 [DOI] [Google Scholar]
- Bird D, Nix MG, McCallum H, Teo M, Gilbert A, Casanova N, Cooper R, Buckley DL, Sebag-Montefiore D, Speight R, Al-Qaisieh B and Henry AM 2021. Multicentre, deep learning, synthetic-CT generation for ano-rectal MR-only radiotherapy treatment planning Radiother. Oncol. 156 23–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal-Barby JS and Krieger H 2015. Cognitive biases and heuristics in medical decision making: a critical review using a systematic search strategy Med. Decis. Making 35 539–57 [DOI] [PubMed] [Google Scholar]
- Blundell C, Cornebise J, Kavukcuoglu K and Wierstra D 2015. Weight Uncertainty in Neural Networks 32nd International Conference on Machine Learning, ICML 2015 Online: http://arxiv.org/abs/1505.05424 [Google Scholar]
- Boldrini L, Bibault J-E, Masciocchi C, Shen Y and Bittner M-I 2019. Deep Learning: A Review for the Radiation Oncologist Front. Oncol. 9 977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles C, Chen L, Guerrero R, Bentley P, Gunn R, Hammers A, Dickie DA, Hernández MV, Wardlaw J and Rueckert D 2018. GAN Augmentation: Augmenting Training Data using Generative Adversarial Networks Online: http://arxiv.org/abs/1810.10863
- Boyd S 2010. Distributed Optimization and Statistical Learning via the Alternating Direction Method of Multipliers Online: 10.1561/9781601984616 [DOI]
- Breiman L 2001. Random Forests Mach. Learn. 45 5–32 [Google Scholar]
- Bria A, Marrocco C and Tortorella F 2020. Addressing class imbalance in deep learning for small lesion detection on medical images Comput. Biol. Med. 120 103735. [DOI] [PubMed] [Google Scholar]
- Brion E, Léger J, Barragán-Montero AM, Meert N, Lee JA and Macq B 2021. Domain adversarial networks and intensity-based data augmentation for male pelvic organ segmentation in cone beam CT Comput. Biol. Med. 131 104269. [DOI] [PubMed] [Google Scholar]
- Brouwer CL, Dinkla AM, Vandewinckele L, Crijns W, Claessens M, Verellen D and van Elmpt W 2020. Machine learning applications in radiation oncology: Current use and needs to support clinical implementation Phys Imaging Radiat Oncol 16 144–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer CL, Steenbakkers RJHM, Bourhis J, Budach W, Grau C, Grégoire V, van Herk M, Lee A, Maingon P, Nutting C, O’Sullivan B, Porceddu SV, Rosenthal DI, Sijtsema NM and Langendijk JA 2015. CT-based delineation of organs at risk in the head and neck region: DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and TROG consensus guidelines Radiother. Oncol. 117 83–90 [DOI] [PubMed] [Google Scholar]
- Bruce J, Suenderhauf N, Mirowski P, Hadsell R and Milford M 2017. One-Shot Reinforcement Learning for Robot Navigation with Interactive Replay arXiv [cs.AI] Online: http://arxiv.org/abs/1711.10137 [Google Scholar]
- Brunner G, Liu Y, Pascual D, Richter O, Ciaramita M and Wattenhofer R 2019. On Identifiability in Transformers arXiv [cs.CL] Online: http://arxiv.org/abs/1908.04211 [Google Scholar]
- Budd S, Robinson EC and Kainz B 2021. A survey on active learning and human-in-the-loop deep learning for medical image analysis Med. Image Anal. 71 102062. [DOI] [PubMed] [Google Scholar]
- Cardenas CE, Beadle BM, Garden AS, Skinner HD, Yang J, Rhee DJ, McCarroll RE, Netherton TJ, Gay SS, Zhang L and Court LE 2021. Generating High-Quality Lymph Node Clinical Target Volumes for Head and Neck Cancer Radiation Therapy Using a Fully Automated Deep Learning-Based Approach Int. J. Radiat. Oncol. Biol. Phys. 109 801–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas CE, Yang J, Anderson BM, Court LE and Brock KB 2019. Advances in Auto-Segmentation Semin. Radiat. Oncol. 29 185–97 [DOI] [PubMed] [Google Scholar]
- Caruana R 1998. Multitask Learning Learning to Learn 95–133 Online: 10.1007/978-1-4615-5529-2_5 [DOI] [Google Scholar]
- Caruana R, Lou Y, Gehrke J, Koch P, Sturm M and Elhadad N 2015. Intelligible Models for HealthCare Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining - KDD ‘15 Online: 10.1145/2783258.2788613 [DOI] [Google Scholar]
- Castro DC, Walker I and Glocker B 2020. Causality matters in medical imaging Nat. Commun. 11 3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha E, Elguindi S, Onochie I, Gorovets D, Deasy JO, Zelefsky M and Gillespie EF 2021. Clinical implementation of deep learning contour autosegmentation for prostate radiotherapy Radiother. Oncol. 159 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J and Jamal MM 2012. Esophageal malignancy: a growing concern World J. Gastroenterol. 18 6521–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha KH, Hadjiiski L, Chan H-P, Weizer AZ, Alva A, Cohan RH, Caoili EM, Paramagul C and Samala RK 2017. Bladder Cancer Treatment Response Assessment in CT using Radiomics with Deep-Learning Sci. Rep. 7 8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K, Balachandar N, Lam C, Yi D, Brown J, Beers A, Rosen B, Rubin DL and Kalpathy-Cramer J 2018. Distributed deep learning networks among institutions for medical imaging J. Am. Med. Inform. Assoc. 25 945–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changpinyo S, Chao WL and Gong B 2016. Synthesized classifiers for zero-shot learning Proc. IEEE Online: https://www.cv-foundation.org/openaccess/content_cvpr_2016/html/Changpinyo_Synthesized_Classifiers_for_CVPR_2016_paper.html [Google Scholar]
- Chan MF, Witztum A and Valdes G 2020. Integration of AI and Machine Learning in Radiotherapy QA Front Artif Intell 3 577620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartsias A, Joyce T, Papanastasiou G, Semple S, Williams M, Newby DE, Dharmakumar R and Tsaftaris SA 2019. Disentangled representation learning in cardiac image analysis Med. Image Anal. 58 101535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Vig L, De Filippo De Grazia M, Corbetta M, Ahmad S and Zorzi M 2019. A Comparison of Shallow and Deep Learning Methods for Predicting Cognitive Performance of Stroke Patients From MRI Lesion Images Front. Neuroinform. 0 Online: 10.3389/fninf.2019.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Bentley P, Mori K, Misawa K, Fujiwara M and Rueckert D 2019. Self-supervised learning for medical image analysis using image context restoration Med. Image Anal. 58 101539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Dong W, Wang J, Lu X, Kaymak U and Huang Z 2020a. Interpretable clinical prediction via attention-based neural network BMC Med. Inform. Decis. Mak. 20 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Oshima K, Schott D, Wu H, Hall W, Song Y, Tao Y, Li D, Zheng C, Knechtges P, Erickson B and Li XA 2017. Assessment of treatment response during chemoradiation therapy for pancreatic cancer based on quantitative radiomic analysis of daily CTs: An exploratory study PLoS One 12 e0178961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Bei Y and Rudin C 2020b. Concept whitening for interpretable image recognition Nature Machine Intelligence 2 772–82 Online: 10.1038/s42256-020-00265-z [DOI] [Google Scholar]
- Chi W, Xiang Z and Guo F 2022. Few-shot learning for deformable image registration in 4DCT images Br. J. Radiol. 95 20210819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlap P, Min H, Vandenberg N, Dowling J, Holloway L and Haworth A 2021. A review of medical image data augmentation techniques for deep learning applications Journal of Medical Imaging and Radiation Oncology 65 545–63 Online: 10.1111/1754-9485.13261 [DOI] [PubMed] [Google Scholar]
- Cho H-H, Lee HY, Kim E, Lee G, Kim J, Kwon J and Park H 2021. Radiomics-guided deep neural networks stratify lung adenocarcinoma prognosis from CT scans Commun Biol 4 1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GF, Abraham V, Aliferis CF, Aronis JM, Buchanan BG, Caruana R, Fine MJ, Janosky JE, Livingston G, Mitchell T, Monti S and Spirtes P 2005. Predicting dire outcomes of patients with community acquired pneumonia J. Biomed. Inform. 38 347–66 [DOI] [PubMed] [Google Scholar]
- Cui S, Luo Y, Tseng H-H, Ten Haken R K and El Naqa I 2019. Combining handcrafted features with latent variables in machine learning for prediction of radiation-induced lung damage Med. Phys. 46 2497–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Tseng H-H, Pakela J, Ten Haken R K and El Naqa I 2020. Introduction to machine and deep learning for medical physicists Med. Phys. 47 e127–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakka MA, Nguyen TV, Hall JMM, Diakiw SM, VerMilyea M, Linke R, Perugini M and Perugini D 2021. Automated detection of poor-quality data: case studies in healthcare Sci. Rep. 11 18005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amour A, Heller K, Moldovan D, Adlam B, Alipanahi B, Beutel A, Chen C, Deaton J, Eisenstein J, Hoffman MD, Hormozdiari F, Houlsby N, Hou S, Jerfel G, Karthikesalingam A, Lucic M, Ma Y, McLean C, Mincu D, Mitani A, Montanari A, Nado Z, Natarajan V, Nielson C, Osborne TF, Raman R, Ramasamy K, Sayres R, Schrouff J, Seneviratne M, Sequeira S, Suresh H, Veitch V, Vladymyrov M, Wang X, Webster K, Yadlowsky S, Yun T, Zhai X and Sculley D 2020. Underspecification Presents Challenges for Credibility in Modern Machine Learning arXiv [cs.LG] Online: http://arxiv.org/abs/2011.03395 [Google Scholar]
- Das A and Rad P 2020. Opportunities and Challenges in Explainable Artificial Intelligence (XAI): A Survey arXiv [cs.CV] Online: http://arxiv.org/abs/2006.11371 [Google Scholar]
- Dash T, Chitlangia S, Ahuja A and Srinivasan A 2022. A review of some techniques for inclusion of domain-knowledge into deep neural networks Sci. Rep. 12 1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearnaley D, Syndikus I, Gulliford S and Hall E 2017. Hypofractionation for Prostate Cancer: Time to Change Clinical Oncology 29 3–5 Online: 10.1016/j.clon.2016.09.020 [DOI] [PubMed] [Google Scholar]
- Deng C, Ji X, Rainey C, Zhang J and Lu W 2020. Integrating Machine Learning with Human Knowledge iScience 23 101656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depeweg S, Hernandez-Lobato J-M, Doshi-Velez F and Udluft S 2018. Decomposition of Uncertainty in Bayesian Deep Learning for Efficient and Risk-sensitive Learning International Conference on Machine Learning International Conference on Machine Learning (PMLR) pp 1184–93 [Google Scholar]
- Diamant A, Chatterjee A, Vallières M, Shenouda G and Seuntjens J 2019. Deep learning in head & neck cancer outcome prediction Sci. Rep. 9 2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz O, Kushibar K, Osuala R, Linardos A, Garrucho L, Igual L, Radeva P, Prior F, Gkontra P and Lekadir K 2021. Data preparation for artificial intelligence in medical imaging: A comprehensive guide to open-access platforms and tools Phys. Med. 83 25–37 [DOI] [PubMed] [Google Scholar]
- Dodge S and Karam L 2016. Understanding how image quality affects deep neural networks 2016 Eighth International Conference on Quality of Multimedia Experience (QoMEX) Online: 10.1109/qomex.2016.7498955 [DOI] [Google Scholar]
- Dong G, Zhang C, Liang X, Deng L, Zhu Y, Zhu X, Zhou X, Song L, Zhao X and Xie Y 2021. A Deep Unsupervised Learning Model for Artifact Correction of Pelvis Cone-Beam CT Frontiers in Oncology 11 Online: 10.3389/fonc.2021.686875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi-Velez F and Kim B 2017. Towards A Rigorous Science of Interpretable Machine Learning arXiv [stat.ML] Online: http://arxiv.org/abs/1702.08608 [Google Scholar]
- Duan Y, Andrychowicz M, Stadie B, Jonathan Ho O, Schneider J, Sutskever I, Abbeel P and Zaremba W 2017. One-shot imitation learning Adv. Neural Inf. Process. Syst. 30 Online: https://proceedings.neurips.cc/paper/2017/hash/ba3866600c3540f67c1e9575e213be0a-Abstract.html [Google Scholar]
- Du B, Wang Z, Zhang L, Zhang L, Liu W, Shen J and Tao D 2017. Exploring Representativeness and Informativeness for Active Learning IEEE Trans Cybern 47 14–26 [DOI] [PubMed] [Google Scholar]
- Eche T, Schwartz LH, Mokrane F-Z and Dercle L 2021. Toward Generalizability in the Deployment of Artificial Intelligence in Radiology: Role of Computation Stress Testing to Overcome Underspecification Radiology: Artificial Intelligence 3 Online: 10.1148/ryai.2021210097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Naqa I, Pandey G, Aerts H, Chien J-T, Andreassen CN, Niemierko A and Ten Haken R K 2018. Radiation Therapy Outcomes Models in the Era of Radiomics and Radiogenomics: Uncertainties and Validation Int. J. Radiat. Oncol. Biol. Phys. 102 1070–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara T, Afify HM, Ismail FH and Hassanien AE 2019. A Modified Inception-v4 for Imbalanced Skin Cancer Classification Dataset 2019 14th International Conference on Computer Engineering and Systems (ICCES) Online: 10.1109/icces48960.2019.9068110 [DOI] [Google Scholar]
- Eriguchi T, Takeda A, Oku Y, Ishikura S, Kimura T, Ozawa S, Nakashima T, Matsuo Y, Nakamura M, Matsumoto Y, Yamazaki S, Sanuki N and Ito Y 2013. Multi-institutional comparison of treatment planning using stereotactic ablative body radiotherapy for hepatocellular carcinoma - benchmark for a prospective multi-institutional study Radiat. Oncol. 8 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM and Thrun S 2017. Dermatologist-level classification of skin cancer with deep neural networks Nature 542 115–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Wang J, Chen Z, Hu C, Zhang Z and Hu W 2019. Automatic treatment planning based on three-dimensional dose distribution predicted from deep learning technique Med. Phys. 46 370–81 [DOI] [PubMed] [Google Scholar]
- Fechter T and Baltas D 2020. One-Shot Learning for Deformable Medical Image Registration and Periodic Motion Tracking IEEE Trans. Med. Imaging 39 2506–17 [DOI] [PubMed] [Google Scholar]
- Fei-Fei L, Fergus R and Perona P 2006. One-shot learning of object categories IEEE Trans. Pattern Anal. Mach. Intell. 28 594–611 [DOI] [PubMed] [Google Scholar]
- Feng M, Valdes G, Dixit N and Solberg TD 2018. Machine Learning in Radiation Oncology: Opportunities, Requirements, and Needs Front. Oncol. 8 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Bernard ME, Hunter T and Chen Q 2020. Improving accuracy and robustness of deep convolutional neural network based thoracic OAR segmentation Physics in Medicine & Biology 65 07NT01 Online: 10.1088/1361-6560/ab7877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson SG, Bowers JD, Ito J, Zittrain JL, Beam AL and Kohane IS 2019. Adversarial attacks on medical machine learning Science 363 1287–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn C, Abbeel P and Levine S 06−11 Aug 2017. Model-Agnostic Meta-Learning for Fast Adaptation of Deep Networks Proceedings of the 34th International Conference on Machine Learning Proceedings of Machine Learning Research vol 70, ed Precup D and The YW (PMLR) pp 1126–35 [Google Scholar]
- Fisher A, Rudin C and Dominici F 2019. All Models are Wrong, but Many are Useful: Learning a Variable’s Importance by Studying an Entire Class of Prediction Models Simultaneously J. Mach. Learn. Res. 20 Online: https://www.ncbi.nlm.nih.gov/pubmed/34335110 [PMC free article] [PubMed] [Google Scholar]
- Forrest LF, Adams J, Wareham H, Rubin G and White M 2013. Socioeconomic Inequalities in Lung Cancer Treatment: Systematic Review and Meta-Analysis PLoS Med. 10 e1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenay B and Verleysen M 2014. Classification in the Presence of Label Noise: A Survey IEEE Transactions on Neural Networks and Learning Systems 25 845–69 Online: 10.1109/tnnls.2013.2292894 [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T and Tibshirani R 2001. The elements of statistical learning vol 1 (Springer series in statistics; New York: ) [Google Scholar]
- Fu L 1995. Introduction to knowledge-based neural networks Knowledge-Based Systems 8 299–300 Online: 10.1016/0950-7051(96)81914-9 [DOI] [Google Scholar]
- Futoma J, Simons M, Panch T, Doshi-Velez F and Celi LA 2020. The myth of generalisability in clinical research and machine learning in health care Lancet Digit Health 2 e489–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Lei Y, Wang T, Curran WJ, Liu T and Yang X 2020. Deep learning in medical image registration: a review Phys. Med. Biol. 65 20TR01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal Y. Uncertainty in deep learning. 2016.
- Gal Y and Ghahramani Z 2015. Dropout as a Bayesian Approximation: Representing Model Uncertainty in Deep Learning Online: http://dx.doi.org/
- Gal Y and Ghahramani Z 2016. Dropout as a Bayesian Approximation: Representing Model Uncertainty in Deep Learning International Conference on Machine Learning International Conference on Machine Learning (PMLR) pp 1050–9 [Google Scholar]
- Ganin Y and Lempitsky V 2015. Unsupervised Domain Adaptation by Backpropagation Proceedings of the 32nd International Conference on Machine Learning Proceedings of Machine Learning Research vol 37, ed Bach F and Blei D (Lille, France: PMLR; ) pp 1180–9 [Google Scholar]
- Gao Y, Huang R, Chen M, Wang Z, Deng J, Chen Y, Yang Y, Zhang J, Tao C and Li H 2019. FocusNet: Imbalanced Large and Small Organ Segmentation with an End-to-End Deep Neural Network for Head and Neck CT Images Lecture Notes in Computer Science 829–38 Online: 10.1007/978-3-030-32248-9_92 [DOI] [Google Scholar]
- Gao Y, Huang R, Yang Y, Zhang J, Shao K, Tao C, Chen Y, Metaxas DN, Li H and Chen M 2021. FocusNetv2: Imbalanced large and small organ segmentation with adversarial shape constraint for head and neck CT images Medical Image Analysis 67 101831 Online: 10.1016/j.media.2020.101831 [DOI] [PubMed] [Google Scholar]
- Gatys LA, Ecker AS and Bethge M 2015. A Neural Algorithm of Artistic Style Online: http://arxiv.org/abs/1508.06576
- Gawlikowski J, Tassi CRN, Ali M, Lee J, Humt M, Feng J, Kruspe A, Triebel R, Jung P, Roscher R, Shahzad M, Yang W, Bamler R and Zhu XX 2021. A Survey of Uncertainty in Deep Neural Networks arXiv [cs.LG] Online: http://arxiv.org/abs/2107.03342 [Google Scholar]
- Geirhos R, Jacobsen J-H, Michaelis C, Zemel R, Brendel W, Bethge M and Wichmann FA 2020. Shortcut learning in deep neural networks Nature Machine Intelligence 2 665–73 Online: 10.1038/s42256-020-00257-z [DOI] [Google Scholar]
- Gennatas ED, Friedman JH, Ungar LH, Pirracchio R, Eaton E, Reichmann LG, Interian Y, Luna JM, Simone CB, Auerbach A, Delgado E, van der Laan MJ, Solberg TD and Valdes G 2020. Expert-augmented machine learning Proc. Natl. Acad. Sci. U. S. A. 117 4571–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershkevitsh E, Pesznyak C, Petrovic B, Grezdo J, Chelminski K, do Carmo Lopes M, Izewska J and Van Dyk J 2014. Dosimetric inter-institutional comparison in European radiotherapy centres: Results of IAEA supported treatment planning system audit Acta Oncol. 53 628–36 [DOI] [PubMed] [Google Scholar]
- Gevrey M, Dimopoulos I and Lek S 2003. Review and comparison of methods to study the contribution of variables in artificial neural network models Ecol. Modell. 160 249–64 [Google Scholar]
- Ghoshal B, Tucker A, Sanghera B and Wong WL 2021. Estimating uncertainty in deep learning for reporting confidence to clinicians in medical image segmentation and diseases detection Computational Intelligence 37 701–34 Online: 10.1111/coin.12411 [DOI] [Google Scholar]
- Giraud P, Giraud P, Nicolas E, Boisselier P, Alfonsi M, Rives M, Bardet E, Calugaru V, Noel G, Chajon E, Pommier P, Morelle M, Perrier L, Liem X, Burgun A and Bibault JE 2020. Interpretable Machine Learning Model for Locoregional Relapse Prediction in Oropharyngeal Cancers Cancers 13 Online: 10.3390/cancers13010057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grégoire V, Ang K, Budach W, Grau C, Hamoir M, Langendijk JA, Lee A, Le Q-T, Maingon P, Nutting C, O’Sullivan B, Porceddu SV and Lengele B 2014. Delineation of the neck node levels for head and neck tumors: a 2013 update. DAHANCA, EORTC, HKNPCSG, NCIC CTG, NCRI, RTOG, TROG consensus guidelines Radiother. Oncol. 110 172–81 [DOI] [PubMed] [Google Scholar]
- Grégoire V, Evans M, Le Q-T, Bourhis J, Budach V, Chen A, Eisbruch A, Feng M, Giralt J, Gupta T, Hamoir M, Helito JK, Hu C, Hunter K, Johansen J, Kaanders J, Laskar SG, Lee A, Maingon P, Mäkitie A, Micciche’ F, Nicolai P, O’Sullivan B, Poitevin A, Porceddu S, Składowski K, Tribius S, Waldron J, Wee J, Yao M, Yom SS, Zimmermann F and Grau C 2018. Delineation of the primary tumour Clinical Target Volumes (CTV-P) in laryngeal, hypopharyngeal, oropharyngeal and oral cavity squamous cell carcinoma: AIRO, CACA, DAHANCA, EORTC, GEORCC, GORTEC, HKNPCSG, HNCIG, IAG-KHT, LPRHHT, NCIC CTG, NCRI, NRG Oncology, PHNS, SBRT, SOMERA, SRO, SSHNO, TROG consensus guidelines Radiother. Oncol. 126 3–24 [DOI] [PubMed] [Google Scholar]
- Grossberg AJ, Mohamed ASR, Elhalawani H, Bennett WC, Smith KE, Nolan TS, Williams B, Chamchod S, Heukelom J, Kantor ME, Browne T, Hutcheson KA, Gunn GB, Garden AS, Morrison WH, Frank SJ, Rosenthal DI, Freymann JB and Fuller CD 2018. Imaging and clinical data archive for head and neck squamous cell carcinoma patients treated with radiotherapy Sci Data 5 180173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H and Liu M 2022. Domain Adaptation for Medical Image Analysis: A Survey IEEE Trans. Biomed. Eng. 69 1173–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Runger G and Liu L 2020. Dynamic incorporation of prior knowledge from multiple domains in biomarker discovery BMC Bioinformatics 21 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro F, Seravalli E, Janssens GO, Maduro JH, Knopf AC, Langendijk JA, Raaymakers BW and Kontaxis C 2021. Deep learning prediction of proton and photon dose distributions for paediatric abdominal tumours Radiother. Oncol. 156 36–42 [DOI] [PubMed] [Google Scholar]
- Guidotti R, Monreale A, Ruggieri S, Turini F, Giannotti F and Pedreschi D 2019. A Survey of Methods for Explaining Black Box Models ACM Computing Surveys 51 1–42 Online: 10.1145/3236009 [DOI] [Google Scholar]
- Guiot J, Vaidyanathan A, Deprez L, Zerka F, Danthine D, Frix A-N, Lambin P, Bottari F, Tsoutzidis N, Miraglio B, Walsh S, Vos W, Hustinx R, Ferreira M, Lovinfosse P and Leijenaar RTH 2022. A review in radiomics: Making personalized medicine a reality via routine imaging Med. Res. Rev. 42 426–40 [DOI] [PubMed] [Google Scholar]
- Gu L, Zhang X, You S, Zhao S, Liu Z and Harada T 2020. Semi-Supervised Learning in Medical Images Through Graph-Embedded Random Forest Front. Neuroinform. 0 Online: 10.3389/fninf.2020.601829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralick RM, Shanmugam K and I ‘hak Dinstein 1973. Textural Features for Image Classification IEEE Transactions on Systems, Man, and Cybernetics SMC-3 610–21 Online: 10.1109/tsmc.1973.4309314 [DOI] [Google Scholar]
- Haskins G, Kruger U and Yan P 2020. Deep learning in medical image registration: a survey Mach. Vis. Appl. 31 8 [Google Scholar]
- Hatamizadeh A, Yang D, Roth H and Xu D 2021. UNETR: Transformers for 3D Medical Image Segmentation arXiv [eess.IV] Online: http://arxiv.org/abs/2103.10504 [Google Scholar]
- Heim E, Roß T, Seitel A, März K, Stieltjes B, Eisenmann M, Lebert J, Metzger J, Sommer G, Sauter AW, Schwartz FR, Termer A, Wagner F, Kenngott HG and Maier-Hein L 2018. Large-scale medical image annotation with crowd-powered algorithms J Med Imaging (Bellingham) 5 034002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Baxter SL, Xu J, Xu J, Zhou X and Zhang K 2019. The practical implementation of artificial intelligence technologies in medicine Nature Medicine 25 30–6 Online: 10.1038/s41591-018-0307-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekler A, Kather JN, Krieghoff-Henning E, Utikal JS, Meier F, Gellrich FF, Upmeier Zu Belzen J, French L, Schlager JG, Ghoreschi K, Wilhelm T, Kutzner H, Berking C, Heppt MV, Haferkamp S, Sondermann W, Schadendorf D, Schilling B, Izar B, Maron R, Schmitt M, Fröhling S, Lipka DB and Brinker TJ 2020. Effects of Label Noise on Deep Learning-Based Skin Cancer Classification Front. Med. 7 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Yang G, Yang J, Chen Y, Kong Y, Wu J, Tang L, Zhu X, Dillenseger J-L, Shao P, Zhang S, Shu H, Coatrieux J-L and Li S 2020. Dense biased networks with deep priori anatomy and hard region adaptation: Semi-supervised learning for fine renal artery segmentation Med. Image Anal. 63 101722. [DOI] [PubMed] [Google Scholar]
- Hofmanninger J, Perkonigg M, Brink JA, Pianykh O, Herold C and Langs G 2020. Dynamic Memory to Alleviate Catastrophic Forgetting in Continuous Learning Settings Medical Image Computing and Computer Assisted Intervention – MICCAI 2020 359–68 Online: 10.1007/978-3-030-59713-9_35 [DOI] [Google Scholar]
- Huang S-C, Pareek A, Seyyedi S, Banerjee I and Lungren MP 2020. Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines NPJ Digit Med 3 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S-J, Jin R and Zhou Z-H 2014. Active Learning by Querying Informative and Representative Examples IEEE Trans. Pattern Anal. Mach. Intell. 36 1936–49 [DOI] [PubMed] [Google Scholar]
- Huff DT, Weisman AJ and Jeraj R 2021. Interpretation and visualization techniques for deep learning models in medical imaging Phys. Med. Biol. 66 04TR01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Luo Z, Wang X, Sun S, Yin Y, Cao K, Song Q, Lyu S and Wu X 2021a. End-to-end multimodal image registration via reinforcement learning Med. Image Anal. 68 101878. [DOI] [PubMed] [Google Scholar]
- Hu J, Song Y, Wang Q, Bai S and Yi Z 2021b. Incorporating historical sub-optimal deep neural networks for dose prediction in radiotherapy Med. Image Anal. 67 101886. [DOI] [PubMed] [Google Scholar]
- Hüllermeier E and Waegeman W 2021. Aleatoric and epistemic uncertainty in machine learning: an introduction to concepts and methods Mach. Learn. 110 457–506 [Google Scholar]
- Hutter F, Kotthoff L and Vanschoren J 2019. Automated machine learning: methods, systems, challenges (Springer Nature; ) [Google Scholar]
- Hu X, Gong J, Zhou W, Li H, Wang S, Wei M, Peng W and Gu Y 2021c. Computer-aided diagnosis of ground glass pulmonary nodule by fusing deep learning and radiomics features Phys. Med. Biol. 66 065015. [DOI] [PubMed] [Google Scholar]
- Iqbal MJ, Javed Z, Sadia H, Qureshi IA, Irshad A, Ahmed R, Malik K, Raza S, Abbas A, Pezzani R and Sharifi-Rad J 2021. Clinical applications of artificial intelligence and machine learning in cancer diagnosis: looking into the future Cancer Cell International 21 Online: 10.1186/s12935-021-01981-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaksson LJ, Pepa M, Zaffaroni M, Marvaso G, Alterio D, Volpe S, Corrao G, Augugliaro M, Starzyńska A, Leonardi MC, Orecchia R and Jereczek-Fossa BA 2020. Machine Learning-Based Models for Prediction of Toxicity Outcomes in Radiotherapy Front. Oncol. 10 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee F, Jaeger PF, Kohl SAA, Petersen J and Maier-Hein KH 2021. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation Nat. Methods 18 203–11 [DOI] [PubMed] [Google Scholar]
- Ivanovs M, Kadikis R and Ozols K 2021. Perturbation-based methods for explaining deep neural networks: A survey Pattern Recognition Letters 150 228–34 Online: 10.1016/j.patrec.2021.06.030 [DOI] [Google Scholar]
- Jackson PT, Atapour-Abarghouei A, Bonner S, Breckon T and Obara B 2018. Style Augmentation: Data Augmentation via Style Randomization Online: http://arxiv.org/abs/1809.05375 [Google Scholar]
- Jacovi A and Goldberg Y 2020. Towards Faithfully Interpretable NLP Systems: How Should We Define and Evaluate Faithfulness? Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics Online: 10.18653/v1/2020.acl-main.386 [DOI] [Google Scholar]
- Jain S and Wallace BC 2019. Attention is not Explanation arXiv [cs.CL] Online: http://arxiv.org/abs/1902.10186 [Google Scholar]
- Jansen T, Geleijnse G, Van Maaren M, Hendriks MP, Ten Teije A and Moncada-Torres A 2020. Machine Learning Explainability in Breast Cancer Survival Stud. Health Technol. Inform. 270 307–11 [DOI] [PubMed] [Google Scholar]
- Jarrett D, Stride E, Vallis K and Gooding MJ 2019. Applications and limitations of machine learning in radiation oncology Br. J. Radiol. 92 20190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Ren L and Cai J 2020. Clinical implementation of AI technologies will require interpretable AI models Med. Phys. 47 1–4 [DOI] [PubMed] [Google Scholar]
- Jiménez Londoño G A, García Vicente A M, Bosque JJ, Amo-Salas M, Pérez-Beteta J, Honguero-Martinez AF, Pérez-García VM and Soriano Castrejón Á M 2022. SUVmax to tumor perimeter distance: a robust radiomics prognostic biomarker in resectable non-small cell lung cancer patients Eur. Radiol. Online: 10.1007/s00330-021-08523-3 [DOI] [PubMed] [Google Scholar]
- Jing L and Tian Y 2021. Self-Supervised Visual Feature Learning With Deep Neural Networks: A Survey IEEE Trans. Pattern Anal. Mach. Intell. 43 4037–58 [DOI] [PubMed] [Google Scholar]
- Johnson JM and Khoshgoftaar TM 2019. Survey on deep learning with class imbalance Journal of Big Data 6 Online: 10.1186/s40537-019-0192-5 [DOI] [Google Scholar]
- Kalender WA, Hebel R and Ebersberger J 1987. Reduction of CT artifacts caused by metallic implants Radiology 164 576–7 Online: 10.1148/radiology.164.2.3602406 [DOI] [PubMed] [Google Scholar]
- Kalet AM, Luk SMH and Phillips MH 2020. Radiation Therapy Quality Assurance Tasks and Tools: The Many Roles of Machine Learning Med. Phys. 47 e168–77 [DOI] [PubMed] [Google Scholar]
- Kamnitsas K, Baumgartner C, Ledig C, Newcombe V, Simpson J, Kane A, Menon D, Nori A, Criminisi A, Rueckert D and Glocker B 2017. Unsupervised Domain Adaptation in Brain Lesion Segmentation with Adversarial Networks Lecture Notes in Computer Science 597–609 Online: 10.1007/978-3-319-59050-9_47 [DOI] [Google Scholar]
- Kandalan RN, Nguyen D, Rezaeian NH, Barragán-Montero AM, Breedveld S, Namuduri K, Jiang S and Lin M-H 2020. Dose Prediction with Deep Learning for Prostate Cancer Radiation Therapy: Model adaptation to Different Treatment Planning Practices Radiother. Oncol. Online: 10.1016/j.radonc.2020.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi D, Dou H, Warfield SK and Gholipour A 2020. Deep learning with noisy labels: Exploring techniques and remedies in medical image analysis Med. Image Anal. 65 101759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemifar S, Barragán Montero A M, Souris K, Rivas ST, Timmerman R, Park YK, Jiang S, Geets X, Sterpin E and Owrangi A 2020. Dosimetric evaluation of synthetic CT generated with GANs for MRI-only proton therapy treatment planning of brain tumors Journal of Applied Clinical Medical Physics 21 76–86 Online: 10.1002/acm2.12856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemifar S, McGuire S, Timmerman R, Wardak Z, Nguyen D, Park Y, Jiang S and Owrangi A 2019. MRI-only brain radiotherapy: Assessing the dosimetric accuracy of synthetic CT images generated using a deep learning approach Radiotherapy and Oncology 136 56–63 Online: 10.1016/j.radonc.2019.03.026 [DOI] [PubMed] [Google Scholar]
- Kearney V, Chan JW, Wang T, Perry A, Descovich M, Morin O, Yom SS and Solberg TD 2020. DoseGAN: a generative adversarial network for synthetic dose prediction using attention-gated discrimination and generation Sci. Rep. 10 11073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall A and Gal Y 2017. What Uncertainties Do We Need in Bayesian Deep Learning for Computer Vision? arXiv [cs.CV] Online: http://arxiv.org/abs/1703.04977 [Google Scholar]
- Kim B, Wattenberg M, Gilmer J, Cai C, Wexler J, Viegas F and Sayres R 2018. Interpretability Beyond Feature Attribution: Quantitative Testing with Concept Activation Vectors (TCAV) Proceedings of the 35th International Conference on Machine Learning Proceedings of Machine Learning Research vol 80, ed Dy J and Krause A (PMLR; ) pp 2668–77 [Google Scholar]
- Kirkpatrick J, Pascanu R, Rabinowitz N, Veness J, Desjardins G, Rusu AA, Milan K, Quan J, Ramalho T, Grabska-Barwinska A, Hassabis D, Clopath C, Kumaran D and Hadsell R 2017a. Overcoming catastrophic forgetting in neural networks Proc. Natl. Acad. Sci. U. S. A. 114 3521–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick J, Pascanu R, Rabinowitz N, Veness J, Desjardins G, Rusu AA, Milan K, Quan J, Ramalho T, Grabska-Barwinska A, Hassabis D, Clopath C, Kumaran D and Hadsell R 2017b. Overcoming catastrophic forgetting in neural networks Proc. Natl. Acad. Sci. U. S. A. 114 3521–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch A, van Amersfoort J and Gal Y 2019. BatchBALD: Efficient and Diverse Batch Acquisition for Deep Bayesian Active Learning Online: http://dx.doi.org/
- Kissas G, Yang Y, Hwuang E, Witschey WR, Detre JA and Perdikaris P 2020. Machine learning in cardiovascular flows modeling: Predicting arterial blood pressure from non-invasive 4D flow MRI data using physics-informed neural networks Computer Methods in Applied Mechanics and Engineering 358 112623 Online: 10.1016/j.cma.2019.112623 [DOI] [Google Scholar]
- Kiyasseh D, Zhu T and Clifton D 2021. A clinical deep learning framework for continually learning from cardiac signals across diseases, time, modalities, and institutions Nat. Commun. 12 4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppe A, Skrede O-J, De Raedt S, Liestøl K, Kerr DJ and Danielsen HE 2021. Designing deep learning studies in cancer diagnostics Nat. Rev. Cancer 21 199–211 [DOI] [PubMed] [Google Scholar]
- Koch G, Zemel R, Salakhutdinov R and Others 2015. Siamese neural networks for one-shot image recognition ICML deep learning workshop vol 2 (Lille; ) p 0 [Google Scholar]
- Kodratoff Y. The comprehensibility manifesto. KDD Nugget Newsletter. 1994;94 [Google Scholar]
- Kompa B, Snoek J and Beam AL 2021. Second opinion needed: communicating uncertainty in medical machine learning npj Digital Medicine 4 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontaxis C, Bol GH, Lagendijk JJW and Raaymakers BW 2020. DeepDose: Towards a fast dose calculation engine for radiation therapy using deep learning Phys. Med. Biol. 65 075013. [DOI] [PubMed] [Google Scholar]
- Korreman S, Eriksen JG and Grau C 2021. The changing role of radiation oncology professionals in a world of AI - Just jobs lost - Or a solution to the under-provision of radiotherapy? Clin Transl Radiat Oncol 26 104–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouw WM and Loog M 2021. A Review of Domain Adaptation without Target Labels IEEE Trans. Pattern Anal. Mach. Intell. 43 766–85 [DOI] [PubMed] [Google Scholar]
- Kwon Y, Won J-H, Kim BJ and Paik MC 2020. Uncertainty quantification using Bayesian neural networks in classification: Application to biomedical image segmentation Computational Statistics & Data Analysis 142 106816 Online: 10.1016/j.csda.2019.106816 [DOI] [Google Scholar]
- LaBonte TM, Martinez C and Roberts SA 2020. We Know Where We Don’t Know: 3D Bayesian CNNs for Uncertainty Quantification of Binary Segmentations for Material Simulations Online: https://www.osti.gov/servlets/purl/1783605
- Lafarge MW, Pluim JPW, Eppenhof KAJ and Veta M 2019. Learning Domain-Invariant Representations of Histological Images Front. Med. 6 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, Zegers CML, Gillies R, Boellard R, Dekker A and Aerts HJWL 2012. Radiomics: extracting more information from medical images using advanced feature analysis Eur. J. Cancer 48 441–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert CH, Nickisch H and Harmeling S 2009. Learning to detect unseen object classes by between-class attribute transfer 2009 IEEE Conference on Computer Vision and Pattern Recognition pp 951–8 [Google Scholar]
- Lan Z, Chen M, Goodman S, Gimpel K, Sharma P and Soricut R 2019. ALBERT: A Lite BERT for Self-supervised Learning of Language Representations arXiv [cs.CL] Online: http://arxiv.org/abs/1909.11942 [Google Scholar]
- Lao J, Chen Y, Li Z-C, Li Q, Zhang J, Liu J and Zhai G 2017. A Deep Learning-Based Radiomics Model for Prediction of Survival in Glioblastoma Multiforme Sci. Rep. 7 10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrazabal AJ, Nieto N, Peterson V, Milone DH and Ferrante E 2020. Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis Proc. Natl. Acad. Sci. U. S. A. 117 12592–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS and Lee AY 2020. Clinical applications of continual learning machine learning The Lancet Digital Health 2 e279–81 Online: 10.1016/s2589-7500(20)30102-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kim H, Kwak J, Kim YS, Lee SW, Cho S and Cho B 2019. Fluence-map generation for prostate intensity-modulated radiotherapy planning using a deep-neural-network Sci. Rep. 9 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K-H, He X, Zhang L and Yang L 2018. CleanNet: Transfer Learning for Scalable Image Classifier Training with Label Noise 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition Online: 10.1109/cvpr.2018.00571 [DOI] [Google Scholar]
- Leijenaar RTH, Nalbantov G, Carvalho S, van Elmpt WJC, Troost EGC, Boellaard R, Aerts HJW, Gillies RJ and Lambin P 2015. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis Scientific Reports 5 Online: 10.1038/srep11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Tian Y, Chen X, Yan H, Yan L, Zhang T, Zhou Z, Wang L and Dai J 2019a. Prediction of Radiation Pneumonitis With Dose Distribution: A Convolutional Neural Network (CNN) Based Model Front. Oncol. 9 1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Chen L, Nguyen D, Zhou Z, Gu X, Yang M, Wang J and Jiang S 2019b. Generating synthesized computed tomography (CT) from cone-beam computed tomography (CBCT) using CycleGAN for adaptive radiation therapy Phys. Med. Biol. 64 125002. [DOI] [PubMed] [Google Scholar]
- Liang X, Nguyen D and Jiang SB 2020. Generalizability issues with deep learning models in medicine and their potential solutions: illustrated with Cone-Beam Computed Tomography (CBCT) to Computed Tomography (CT) image conversion Machine Learning: Science and Technology Online: 10.1088/2632-2153/abb214 [DOI] [Google Scholar]
- Lievens Y, Borras JM and Grau C 2020. Provision and use of radiotherapy in Europe Mol. Oncol. 14 1461–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens Y, Defourny N, Coffey M, Borras JM, Dunscombe P, Slotman B, Malicki J, Bogusz M, Gasparotto C, Grau C, HERO Consortium, Kokobobo A, Sedlmayer F, Slobina E, Coucke P, Gabrovski R, Vosmik M, Eriksen JG, Jaal J, Dejean C, Polgar C, Johannsson J, Cunningham M, Atkocius V, Back C, Pirotta M, Karadjinovic V, Levernes S, Maciejewski B, Trigo ML, Šegedin B, Palacios A, Pastoors B, Beardmore C, Erridge S, Smyth G and Cleries Soler R 2014. Radiotherapy staffing in the European countries: final results from the ESTRO-HERO survey Radiother. Oncol. 112 178–86 [DOI] [PubMed] [Google Scholar]
- Lievens Y and Grau C 2012. Health economics in radiation oncology: introducing the ESTRO HERO project Radiother. Oncol. 103 109–12 [DOI] [PubMed] [Google Scholar]
- Li H, Galperin-Aizenberg M, Pryma D, Simone CB 2nd and Fan Y 2018. Unsupervised machine learning of radiomic features for predicting treatment response and overall survival of early stage non-small cell lung cancer patients treated with stereotactic body radiation therapy Radiother. Oncol. 129 218–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Zou W, Li T, Feigenberg SJ, Teo B-KK and Dong L 2019. A Super-Learner Model for Tumor Motion Prediction and Management in Radiation Therapy: Development and Feasibility Evaluation Scientific Reports 9 Online: 10.1038/s41598-019-51338-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T-Y, Goyal P, Girshick R, He K and Dollár P 2017. Focal loss for dense object detection Proceedings of the IEEE international conference on computer vision pp 2980–8 [Google Scholar]
- Li Q and Chan MF 2017. Predictive time-series modeling using artificial neural networks for Linac beam symmetry: an empirical study Ann. N. Y. Acad. Sci. 1387 84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, The SPIRIT-AI and CONSORT-AI Working Group, Rivera SC, Moher D, Calvert MJ and Denniston AK 2020. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension Nature Medicine 26 1364–74 Online: 10.1038/s41591-020-1034-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z and Xia Y 2020. Deep Reinforcement Learning for Weakly-Supervised Lymph Node Segmentation in CT Images IEEE J Biomed Health Inform PP Online: 10.1109/JBHI.2020.3008759 [DOI] [PubMed] [Google Scholar]
- Lucieri A, Bajwa MN, Alexander Braun S, Malik MI, Dengel A and Ahmed S 2020. On interpretability of deep learning based skin lesion classifiers using concept activation vectors 2020 International Joint Conference on Neural Networks (IJCNN) 2020 International Joint Conference on Neural Networks (IJCNN) (IEEE) Online: https://ieeexplore.ieee.org/document/9206946/ [Google Scholar]
- Lu C, Shiradkar R and Liu Z 2021. Integrating pathomics with radiomics and genomics for cancer prognosis: A brief review Chin. J. Cancer Res. 33 563–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna JM, Gennatas ED, Ungar LH, Eaton E, Diffenderfer ES, Jensen ST, Simone CB, Friedman JH, Solberg TD and Valdes G 2019. Building more accurate decision trees with the additive tree Proc. Natl. Acad. Sci. U. S. A. 116 19887–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg SM and Lee S-I 2017. A unified approach to interpreting model predictions Proceedings of the 31st international conference on neural information processing systems pp 4768–77 [Google Scholar]
- Luo Y, Tseng H-H, Cui S, Wei L, Ten Haken R K and El Naqa I 2019. Balancing accuracy and interpretability of machine learning approaches for radiation treatment outcomes modeling BJR Open 1 20190021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Ji Z and Gao M 2019. Neural Style Transfer Improves 3D Cardiovascular MR Image Segmentation on Inconsistent Data Lecture Notes in Computer Science 128–36 Online: 10.1007/978-3-030-32245-8_15 [DOI] [Google Scholar]
- Ma L, Chen M, Gu X and Lu W 2021. Generalizability of deep learning based fluence map prediction as an inverse planning approach Online: 10.48550/arXiv.2104.15032 [DOI]
- Mali SA, Ibrahim A, Woodruff HC, Andrearczyk V, Müller H, Primakov S, Salahuddin Z, Chatterjee A and Lambin P 2021. Making Radiomics More Reproducible across Scanner and Imaging Protocol Variations: A Review of Harmonization Methods J Pers Med 11 Online: 10.3390/jpm11090842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandivarapu JK, Camp B and Estrada R 2020. Self-Net: Lifelong Learning via Continual Self-Modeling Front Artif Intell 3 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashayekhi M, Tapia IR, Balagopal A, Zhong X, Barkousaraie AS, McBeth R, Lin M-H, Jiang S and Nguyen D 2021. Site-Agnostic 3D Dose Distribution Prediction with Deep Learning Neural Networks arXiv [cs.LG] Online: http://arxiv.org/abs/2106.07825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maspero M, Savenije MHF, Dinkla AM, Seevinck PR, Intven MPW, Jurgenliemk-Schulz IM, Kerkmeijer LGW and van den Berg CAT 2018. Dose evaluation of fast synthetic-CT generation using a generative adversarial network for general pelvis MR-only radiotherapy Phys. Med. Biol. 63 185001. [DOI] [PubMed] [Google Scholar]
- Masse NY, Grant GD and Freedman DJ 2018a. Alleviating catastrophic forgetting using context-dependent gating and synaptic stabilization Proc. Natl. Acad. Sci. U. S. A. 115 E10467–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse NY, Grant GD and Freedman DJ 2018b. Alleviating catastrophic forgetting using context-dependent gating and synaptic stabilization Proc. Natl. Acad. Sci. U. S. A. 115 E10467–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo CS, Pisansky TM, Petersen IA, Yan ES, Davis BJ, Stafford SL, Garces YI, Miller RC, Martenson JA, Mutter RW, Choo R, Hallemeier CL, Laack NN, Park SS, Ma DJ, Olivier KR, Keole SR, Fatyga M, Foote RL and Haddock MG 2016. Establishment of practice standards in nomenclature and prescription to enable construction of software and databases for knowledge-based practice review Pract. Radiat. Oncol. 6 e117–26 [DOI] [PubMed] [Google Scholar]
- McClure P, Rho N, Lee JA, Kaczmarzyk JR, Zheng CY, Ghosh SS, Nielson DM, Thomas AG, Bandettini P and Pereira F 2019. Knowing What You Know in Brain Segmentation Using Bayesian Deep Neural Networks Frontiers in Neuroinformatics 13 Online: 10.3389/fninf.2019.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh C, Conroy L, Tjong MC, Craig T, Bayley A, Catton C, Gospodarowicz M, Helou J, Isfahanian N, Kong V, Lam T, Raman S, Warde P, Chung P, Berlin A and Purdie TG 2021. Clinical integration of machine learning for curative-intent radiation treatment of patients with prostate cancer Nat. Med. 27 999–1005 [DOI] [PubMed] [Google Scholar]
- McIntosh C and Purdie TG 2016. Contextual Atlas Regression Forests: Multiple-Atlas-Based Automated Dose Prediction in Radiation Therapy IEEE Trans. Med. Imaging 35 1000–12 [DOI] [PubMed] [Google Scholar]
- McIntosh C, Welch M, McNiven A, Jaffray DA and Purdie TG 2017. Fully automated treatment planning for head and neck radiotherapy using a voxel-based dose prediction and dose mimicking method Phys. Med. Biol. 62 5926–44 [DOI] [PubMed] [Google Scholar]
- Medela A, Picon A, Saratxaga CL, Belar O, Cabezón V, Cicchi R, Bilbao R and Glover B 2019. Few Shot Learning in Histopathological Images:Reducing the Need of Labeled Data on Biological Datasets 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) pp 1860–4 [Google Scholar]
- Meng Q, Matthew J, Zimmer VA, Gomez A, Lloyd DFA, Rueckert D and Kainz B 2021. Mutual Information-Based Disentangled Neural Networks for Classifying Unseen Categories in Different Domains: Application to Fetal Ultrasound Imaging IEEE Transactions on Medical Imaging 40 722–34 Online: 10.1109/tmi.2020.3035424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MI, de la Rosa E, Pedrosa de Barros N, Paolella R, Van Leemput K and Sima DM 2021. A Contrast Augmentation Approach to Improve Multi-Scanner Generalization in MRI Front. Neurosci. 15 708196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M G, V A, S M-M and H M 2020. Concept attribution: Explaining CNN decisions to physicians Comput. Biol. Med. 123 103865. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk A and Grochowski M 2018. Data augmentation for improving deep learning in image classification problem 2018 International Interdisciplinary PhD Workshop (IIPhDW) Online: 10.1109/iiphdw.2018.8388338 [DOI] [Google Scholar]
- Mirikharaji Z and Hamarneh G 2018. Star Shape Prior in Fully Convolutional Networks for Skin Lesion Segmentation Medical Image Computing and Computer Assisted Intervention – MICCAI 2018 (Springer International Publishing; ) pp 737–45 [Google Scholar]
- Mobiny A, Singh A and Van Nguyen H 2019. Risk-Aware Machine Learning Classifier for Skin Lesion Diagnosis J. Clin. Med. Res. 8 Online: 10.3390/jcm8081241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobiny A, Yuan P, Moulik SK, Garg N, Wu CC and Van Nguyen H 2021. DropConnect is effective in modeling uncertainty of Bayesian deep networks Scientific Reports 11 Online: 10.1038/s41598-021-84854-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moe YM, Groendahl AR, Tomic O, Dale E, Malinen E and Futsaether CM 2021. Deep learning-based auto-delineation of gross tumour volumes and involved nodes in PET/CT images of head and neck cancer patients Eur. J. Nucl. Med. Mol. Imaging 48 2782–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar C 2019. Interpretable Machine Learning (Lulu.com)
- Moosavi-Dezfooli S-M, Fawzi A and Frossard P 2015. DeepFool: a simple and accurate method to fool deep neural networks Online: http://arxiv.org/abs/1511.04599
- Moreau G, François-Lavet V, Desbordes P and Macq B 2021. Reinforcement Learning for Radiotherapy Dose Fractioning Automation Biomedicines 9 Online: 10.3390/biomedicines9020214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkov N, Mathe B, Kertesz-Farkas A, Hollandi R and Horvath P 2020. Test-time augmentation for deep learning-based cell segmentation on microscopy images Sci. Rep. 10 5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlematter UJ, Daniore P and Vokinger KN 2021. Approval of artificial intelligence and machine learning-based medical devices in the USA and Europe (2015–20): a comparative analysis Lancet Digit Health 3 e195–203 [DOI] [PubMed] [Google Scholar]
- Mullainathan S and Obermeyer Z 2017. Does Machine Learning Automate Moral Hazard and Error? Am. Econ. Rev. 107 476–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidhar N, Islam MR, Marwah M, Karpatne A and Ramakrishnan N 2018a. Incorporating Prior Domain Knowledge into Deep Neural Networks 2018 IEEE International Conference on Big Data (Big Data) pp 36–45 [Google Scholar]
- Muralidhar N, Islam MR, Marwah M, Karpatne A and Ramakrishnan N 2018b. Incorporating Prior Domain Knowledge into Deep Neural Networks 2018 IEEE International Conference on Big Data (Big Data) Online: 10.1109/bigdata.2018.8621955 [DOI] [Google Scholar]
- Nair T, Precup D, Arnold DL and Arbel T 2020. Exploring uncertainty measures in deep networks for Multiple sclerosis lesion detection and segmentation Med. Image Anal. 59 101557. [DOI] [PubMed] [Google Scholar]
- Nalepa J, Marcinkiewicz M and Kawulok M 2019. Data Augmentation for Brain-Tumor Segmentation: A Review Front. Comput. Neurosci. 13 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanfack G, Temple P and Frénay B Global explanations with decision rules: A co-learning approach Online: https://auai.org/uai2021/pdf/uai2021.231.preliminary.pdf
- Nelms BE, Robinson G, Markham J, Velasco K, Boyd S, Narayan S, Wheeler J and Sobczak ML 2012. Variation in external beam treatment plan quality: An inter-institutional study of planners and planning systems Pract. Radiat. Oncol. 2 296–305 [DOI] [PubMed] [Google Scholar]
- Nguyen D, Jia X, Sher D, Lin M-H, Iqbal Z, Liu H and Jiang S 2019a. 3D radiotherapy dose prediction on head and neck cancer patients with a hierarchically densely connected U-net deep learning architecture Phys. Med. Biol. 64 065020. [DOI] [PubMed] [Google Scholar]
- Nguyen D, Long T, Jia X, Lu W, Gu X, Iqbal Z and Jiang S 2019b. A feasibility study for predicting optimal radiation therapy dose distributions of prostate cancer patients from patient anatomy using deep learning Sci. Rep. 9 1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, McBeth R, Sadeghnejad Barkousaraie A, Bohara G, Shen C, Jia X and Jiang S 2020. Incorporating human and learned domain knowledge into training deep neural networks: A differentiable dose-volume histogram and adversarial inspired framework for generating Pareto optimal dose distributions in radiation therapy Med. Phys. 47 837–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Sadeghnejad Barkousaraie A, Bohara G, Balagopal A, McBeth R, Lin M-H and Jiang SB 2021. A comparison of Monte Carlo dropout and bootstrap aggregation on the performance and uncertainty estimation in radiation therapy dose prediction with deep learning neural networks Phys. Med. Biol. Online: 10.1088/1361-6560/abe04f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov S, Blackwell S, Zverovitch A, Mendes R, Livne M, De Fauw J, Patel Y, Meyer C, Askham H, Romera-Paredes B, Kelly C, Karthikesalingam A, Chu C, Carnell D, Boon C, D’Souza D, Moinuddin SA, DeepMind Radiographer Consortium, Montgomery H, Rees G, Suleyman M, Back T, Hughes C, Ledsam JR and Ronneberger O 2018. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy arXiv [cs.CV] Online: http://arxiv.org/abs/1809.04430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermeyer Z, Powers B, Vogeli C and Mullainathan S 2019. Dissecting racial bias in an algorithm used to manage the health of populations Science 366 447–53 [DOI] [PubMed] [Google Scholar]
- Oktay O, Ferrante E, Kamnitsas K, Heinrich M, Bai W, Caballero J, Cook SA, de Marvao A, Dawes T, O’Regan DP, Kainz B, Glocker B and Rueckert D 2018. Anatomically Constrained Neural Networks (ACNNs): Application to Cardiac Image Enhancement and Segmentation IEEE Trans. Med. Imaging 37 384–95 [DOI] [PubMed] [Google Scholar]
- Orlhac F, Nioche C, Klyuzhin I, Rahmim A and Buvat I 2021. Radiomics in PET Imaging PET Clinics 16 597–612 Online: 10.1016/j.cpet.2021.06.007 [DOI] [PubMed] [Google Scholar]
- Osman AFI, Maalej NM and Jayesh K 2020. Prediction of the individual multileaf collimator positional deviations during dynamic IMRT delivery priori with artificial neural network Med. Phys. 47 1421–30 [DOI] [PubMed] [Google Scholar]
- Oswal UK. Leveraging Structured Sparsity for Data-efficient and Interpretable Machine Learning. 2019.
- Ou S-HI, Zell JA, Ziogas A and Anton-Culver H 2008. Low socioeconomic status is a poor prognostic factor for survival in stage I nonsmall cell lung cancer and is independent of surgical treatment, race, and marital status Cancer 112 2011–20 Online: 10.1002/cncr.23397 [DOI] [PubMed] [Google Scholar]
- Palatnik de Sousa I, Maria Bernardes Rebuzzi Vellasco M and Costa da Silva E 2019. Local Interpretable Model-Agnostic Explanations for Classification of Lymph Node Metastases Sensors 19 Online: 10.3390/s19132969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatucci M, Pomerleau D, Hinton GE and Mitchell TM 2009. Zero-shot learning with semantic output codes Adv. Neural Inf. Process. Syst. 22 Online: https://proceedings.neurips.cc/paper/3650-zero-shot-learning-with-semantic-output-codes [Google Scholar]
- Pan I, Agarwal S and Merck D 2019. Generalizable Inter-Institutional Classification of Abnormal Chest Radiographs Using Efficient Convolutional Neural Networks J. Digit. Imaging 32 888–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan SJ and Yang Q 2010. A Survey on Transfer Learning IEEE Trans. Knowl. Data Eng. 22 1345–59 [Google Scholar]
- Parisi GI, Kemker R, Part JL, Kanan C and Wermter S 2019. Continual lifelong learning with neural networks: A review Neural Netw. 113 54–71 [DOI] [PubMed] [Google Scholar]
- Park JE, Park SY, Kim HJ and Kim HS 2019. Reproducibility and Generalizability in Radiomics Modeling: Possible Strategies in Radiologic and Statistical Perspectives Korean J. Radiol. 20 1124–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi K 2018. The biological treatment planning evolution of clinical fractionated radiotherapy using high LET Int. J. Radiat. Biol. 94 752–5 [DOI] [PubMed] [Google Scholar]
- Paul A, Shen TC, Lee S, Balachandar N, Peng Y, Lu Z and Summers RM 2021. Generalized Zero-Shot Chest X-Ray Diagnosis Through Trait-Guided Multi-View Semantic Embedding With Self-Training IEEE Trans. Med. Imaging 40 2642–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peikari M, Salama S, Nofech-Mozes S and Martel AL 2018. A Cluster-then-label Semi-supervised Learning Approach for Pathology Image Classification Sci. Rep. 8 7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perone CS, Ballester P, Barros RC and Cohen-Adad J 2019. Unsupervised domain adaptation for medical imaging segmentation with self-ensembling Neuroimage 194 1–11 [DOI] [PubMed] [Google Scholar]
- Pianykh OS, Langs G, Dewey M, Enzmann DR, Herold CJ, Schoenberg SO and Brink JA 2020. Continuous Learning AI in Radiology: Implementation Principles and Early Applications Radiology 297 6–14 [DOI] [PubMed] [Google Scholar]
- Raissi M, Perdikaris P and Karniadakis GE 2019. Physics-informed neural networks: A deep learning framework for solving forward and inverse problems involving nonlinear partial differential equations J. Comput. Phys. 378 686–707 [Google Scholar]
- Ravishankar H, Venkataramani R, Thiruvenkadam S, Sudhakar P and Vaidya V 2017. Learning and Incorporating Shape Models for Semantic Segmentation Medical Image Computing and Computer Assisted Intervention − MICCAI 2017 203–11 Online: 10.1007/978-3-319-66182-7_24 [DOI] [Google Scholar]
- Ravi S and Larochelle H 2016. Optimization as a Model for Few-Shot Learning Online: https://openreview.net/pdf?id=rJY0-Kcll
- Raza K and Singh NK 2021. A Tour of Unsupervised Deep Learning for Medical Image Analysis Curr. Med. Imaging Rev. 17 1059–77 [DOI] [PubMed] [Google Scholar]
- Reyes M, Abreu PH, Cardoso J, Hajij M, Zamzmi G, Rahul P and Thakur L 2021. Interpretability of Machine Intelligence in Medical Image Computing, and Topological Data Analysis and Its Applications for Medical Data: 4th International Workshop, iMIMIC 2021, and 1st International Workshop, TDA4MedicalData 2021, Held in Conjunction with MICCAI 2021, Strasbourg, France, September 27, 2021, Proceedings (Springer Nature) [Google Scholar]
- Reyes M, Meier R, Pereira S, Silva CA, Dahlweid F-M, von Tengg-Kobligk H, Summers RM and Wiest R 2020. On the Interpretability of Artificial Intelligence in Radiology: Challenges and Opportunities Radiol Artif Intell 2 e190043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro M, Singh S and Guestrin C 2016. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Demonstrations Online: 10.18653/v1/n16-3020 [DOI] [Google Scholar]
- Riedl MO 2019. Human-centered artificial intelligence and machine learning Human Behavior and Emerging Technologies 1 33–6 Online: 10.1002/hbe2.117 [DOI] [Google Scholar]
- Rish I and Grabarnik G 2014. Sparse Modeling: Theory, Algorithms, and Applications (CRC Press; ) [Google Scholar]
- Rivera SC, The SPIRIT-AI and CONSORT-AI Working Group, Liu X, Chan A-W, Denniston AK, Calvert MJ, SPIRIT-AI and CONSORT-AI Steering Group and SPIRIT-AI and CONSORT-AI Consensus Group 2020. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension Nature Medicine 26 1351–63 Online: 10.1038/s41591-020-1037-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocktäschel T, Bošnjak M, Singh S and Riedel S 2014. Low-Dimensional Embeddings of Logic Proceedings of the ACL 2014 Workshop on Semantic Parsing Online: 10.3115/v1/w14-2409 [DOI] [Google Scholar]
- Ronneberger O, Fischer P and Brox T 2015. U-Net: Convolutional Networks for Biomedical Image Segmentation Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 (Springer International Publishing; ) pp 234–41 [Google Scholar]
- Rudin C 2019. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead Nature Machine Intelligence 1 206–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin C, Chen C, Chen Z, Huang H, Semenova L and Zhong C 2021. Interpretable Machine Learning: Fundamental Principles and 10 Grand Challenges arXiv [cs.LG] Online: http://arxiv.org/abs/2103.11251 [Google Scholar]
- Sagi O and Rokach L 2018. Ensemble learning: A survey WIREs Data Mining and Knowledge Discovery 8 Online: 10.1002/widm.1249 [DOI] [Google Scholar]
- Sahiner B, Pezeshk A, Hadjiiski LM, Wang X, Drukker K, Cha KH, Summers RM and Giger ML 2019. Deep learning in medical imaging and radiation therapy Medical Physics 46 e1–36 Online: 10.1002/mp.13264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage N 2020. How AI is improving cancer diagnostics Nature 579 S14–6 Online: 10.1038/d41586-020-00847-2 [DOI] [PubMed] [Google Scholar]
- Schapire RE 1989. The strength of weak learnability 30th Annual Symposium on Foundations of Computer Science Online: 10.1109/sfcs.1989.63451 [DOI] [Google Scholar]
- Schuler T, Kipritidis J, Eade T, Hruby G, Kneebone A, Perez M, Grimberg K, Richardson K, Evill S, Evans B and Gallego B 2019. Big Data Readiness in Radiation Oncology: An Efficient Approach for Relabeling Radiation Therapy Structures With Their TG-263 Standard Name in Real-World Data Sets Adv Radiat Oncol 4 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman KA, Berlin JA, Harless W, Kerner JF, Sistrunk S, Gersh BJ, Dubé R, Taleghani CK, Burke JE, Williams S, Eisenberg JM and Escarce JJ 1999. The effect of race and sex on physicians’ recommendations for cardiac catheterization N. Engl. J. Med. 340 618–26 [DOI] [PubMed] [Google Scholar]
- Seita D Learning to Learn The Berkeley Artificial Intelligence Research Blog Online: http://bair.berkeley.edu/blog/2017/07/18/learning-to-learn/ [Google Scholar]
- Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D and Batra D 2017. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization 2017 IEEE International Conference on Computer Vision (ICCV) Online: 10.1109/iccv.2017.74 [DOI] [Google Scholar]
- Senge R, Bösner S, Dembczyński K, Haasenritter J, Hirsch O, Donner-Banzhoff N and Hüllermeier E 2014. Reliable classification: Learning classifiers that distinguish aleatoric and epistemic uncertainty Information Sciences 255 16–29 Online: 10.1016/j.ins.2013.07.030 [DOI] [Google Scholar]
- Seo H, Badiei Khuzani M, Vasudevan V, Huang C, Ren H, Xiao R, Jia X and Xing L 2020. Machine learning techniques for biomedical image segmentation: An overview of technical aspects and introduction to state-of-art applications Med. Phys. 47 e148–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setzu M, Guidotti R, Monreale A and Turini F 2020. Global Explanations with Local Scoring Machine Learning and Knowledge Discovery in Databases 159–71 Online: 10.1007/978-3-030-43823-4_14 [DOI] [Google Scholar]
- Shang Q, Shen ZL, Ward MC, Joshi NP, Koyfman SA and Xia P 2015. Evolution of treatment planning techniques in external-beam radiation therapy for head and neck cancer Appl Radiat Oncol 4 18–25 [Google Scholar]
- Shan H, Jia X, Yan P, Li Y, Paganetti H and Wang G 2020. Synergizing medical imaging and radiotherapy with deep learning Machine Learning: Science and Technology 1 021001 Online: 10.1088/2632-2153/ab869f [DOI] [Google Scholar]
- Shehata M, Shalaby A, Switala AE, El-Baz M, Ghazal M, Fraiwan L, Khalil A, El-Ghar MA, Badawy M, Bakr AM, Dwyer A, Elmaghraby A, Giridharan G, Keynton R and El-Baz A 2020. A multimodal computer-aided diagnostic system for precise identification of renal allograft rejection: Preliminary results Med. Phys. 47 2427–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheller MJ, Anthony Reina G, Edwards B, Martin J and Bakas S 2019. Multi-institutional Deep Learning Modeling Without Sharing Patient Data: A Feasibility Study on Brain Tumor Segmentation Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries 92–104 Online: 10.1007/978-3-030-11723-8_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Gonzalez Y, Chen L, Jiang SB and Jia X 2018. Intelligent Parameter Tuning in Optimization-Based Iterative CT Reconstruction via Deep Reinforcement Learning IEEE Trans. Med. Imaging 37 1430–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Gonzalez Y, Klages P, Qin N, Jung H, Chen L, Nguyen D, Jiang SB and Jia X 2019. Intelligent inverse treatment planning via deep reinforcement learning, a proof-of-principle study in high dose-rate brachytherapy for cervical cancer Phys. Med. Biol. 64 115013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Nguyen D, Chen L, Gonzalez Y, McBeth R, Qin N, Jiang SB and Jia X 2020a. Operating a treatment planning system using a deep-reinforcement learning-based virtual treatment planner for prostate cancer intensity-modulated radiation therapy treatment planning Med. Phys. 47 2329–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Nguyen D, Zhou Z, Jiang SB, Dong B and Jia X 2020b. An introduction to deep learning in medical physics: advantages, potential, and challenges Phys. Med. Biol. 65 05TR01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher DJ, Godley A, Park Y, Carpenter C, Nash M, Hesami H, Zhong X and Lin M-H 2021. Prospective study of artificial intelligence-based decision support to improve head and neck radiotherapy plan quality Clin Transl Radiat Oncol 29 65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorfuzzaman M 2021. An explainable stacked ensemble of deep learning models for improved melanoma skin cancer detection Multimedia Systems Online: 10.1007/s00530-021-00787-5 [DOI] [Google Scholar]
- Shorten C and Khoshgoftaar TM 2019. A survey on Image Data Augmentation for Deep Learning Journal of Big Data 6 Online: 10.1186/s40537-019-0197-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K, Vedaldi A and Zisserman A 2013. Deep Inside Convolutional Networks: Visualising Image Classification Models and Saliency Maps arXiv [cs.CV] Online: http://arxiv.org/abs/1312.6034 [Google Scholar]
- Singh A, Sengupta S and Lakshminarayanan V 2020. Explainable Deep Learning Models in Medical Image Analysis J. Imaging Sci. Technol. 6 Online: 10.3390/jimaging6060052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smailagic A, Costa P, Noh HY, Walawalkar D, Khandelwal K, Galdran A, Mirshekari M, Fagert J, Xu S, Zhang P and Campilho A 2018. MedAL: Accurate and Robust Deep Active Learning for Medical Image Analysis 2018 17th IEEE International Conference on Machine Learning and Applications (ICMLA) Online: 10.1109/icmla.2018.00078 [DOI] [Google Scholar]
- Snell J, Swersky K and Zemel R 2017. Prototypical networks for few-shot learning Adv. Neural Inf. Process. Syst. 30 Online: https://proceedings.neurips.cc/paper/6996-prototypical-networks-for-few-shot-learning [Google Scholar]
- Socher R, Ganjoo M, Manning CD and Ng A 2013. Zero-shot learning through cross-modal transfer Adv. Neural Inf. Process. Syst. 26 Online: https://proceedings.neurips.cc/paper/2013/hash/2d6cc4b2d139a53512fb8cbb3086ae2e-Abstract.html [Google Scholar]
- Sokooti H, de Vos B, Berendsen F, Lelieveldt BPF, Išgum I and Staring M 2017. Nonrigid Image Registration Using Multi-scale 3D Convolutional Neural Networks Medical Image Computing and Computer Assisted Intervention − MICCAI 2017 (Springer International Publishing; ) pp 232–9 [Google Scholar]
- Sourati J, Gholipour A, Dy JG, Kurugol S and Warfield SK 2018. Active Deep Learning with Fisher Information for Patch-wise Semantic Segmentation Deep Learn Med Image Anal Multimodal Learn Clin Decis Support (2018) 11045 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava N, Hinton G, Krizhevsky A, Sutskever I and Salakhutdinov R 2014. Dropout: A Simple Way to Prevent Neural Networks from Overfitting J. Mach. Learn. Res. 15 1929–58 [Google Scholar]
- Sudre CH, Licandro R, Baumgartner C, Melbourne A, Dalca A, Hutter J, Tanno R, Turk EA, Van Leemput K, Barrena JT, Wells WM and Macgowan C 2021. Uncertainty for Safe Utilization of Machine Learning in Medical Imaging, and Perinatal Imaging, Placental and Preterm Image Analysis: 3rd International Workshop, UNSURE 2021, and 6th International Workshop, PIPPI 2021, Held in Conjunction with MICCAI 2021, Strasbourg, France, October 1, 2021, Proceedings (Springer Nature; ) [Google Scholar]
- Sun C, Shrivastava A, Singh S and Gupta A 2017. Revisiting Unreasonable Effectiveness of Data in Deep Learning Era 2017 IEEE International Conference on Computer Vision (ICCV) Online: 10.1109/iccv.2017.97 [DOI] [Google Scholar]
- Sun C and Wee WG 1982. Neighboring gray level dependence matrix for texture classification Computer Graphics and Image Processing 20 297 Online: 10.1016/0146-664x(82)90093-4 [DOI] [Google Scholar]
- Sun J, Darbehani F, Zaidi M and Wang B 2020. SAUNet: Shape Attentive U-Net for Interpretable Medical Image Segmentation Medical Image Computing and Computer Assisted Intervention – MICCAI 2020 797–806 Online: 10.1007/978-3-030-59719-1_77 [DOI] [Google Scholar]
- Szegedy C, Zaremba W, Sutskever I, Bruna J, Erhan D, Goodfellow I and Fergus R 2013. Intriguing properties of neural networks arXiv [cs.CV] Online: http://arxiv.org/abs/1312.6199 [Google Scholar]
- Taleb A, Loetzsch W, Danz N, Severin J, Gaertner T, Bergner B and Lippert C 2020. 3D Self-Supervised Methods for Medical Imaging arXiv [cs.CV] Online: http://arxiv.org/abs/2006.03829 [Google Scholar]
- Tang X 1998. Texture information in run-length matrices IEEE Transactions on Image Processing 7 1602–9 Online: 10.1109/83.725367 [DOI] [PubMed] [Google Scholar]
- Tanno R, Worrall DE, Kaden E, Ghosh A, Grussu F, Bizzi A, Sotiropoulos SN, Criminisi A and Alexander DC 2021. Uncertainty modelling in deep learning for safer neuroimage enhancement: Demonstration in diffusion MRI Neuroimage 225 117366. [DOI] [PubMed] [Google Scholar]
- Thibault G, Fertil B, Navarro C, Pereira S, Cau P, Levy N, Sequeira J and Mari J-L 2013. SHAPE AND TEXTURE INDEXES APPLICATION TO CELL NUCLEI CLASSIFICATION International Journal of Pattern Recognition and Artificial Intelligence 27 1357002 Online: 10.1142/s0218001413570024 [DOI] [Google Scholar]
- Thompson RF, Valdes G, Fuller CD, Carpenter CM, Morin O, Aneja S, Lindsay WD, Aerts HJWL, Agrimson B, Deville C Jr, Rosenthal SA, Yu JB and Thomas CR Jr 2018. Artificial intelligence in radiation oncology: A specialty-wide disruptive transformation? Radiother. Oncol. 129 421–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor M, Apte A, Haq R, Iyer A, LoCastro E and Deasy JO 2021. Using Auto-Segmentation to Reduce Contouring and Dose Inconsistency in Clinical Trials: The Simulated Impact on RTOG 0617 Int. J. Radiat. Oncol. Biol. Phys. 109 1619–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong N, Gou S, Yang S, Ruan D and Sheng K 2018. Fully automatic multi-organ segmentation for head and neck cancer radiotherapy using shape representation model constrained fully convolutional neural networks Med. Phys. 45 4558–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran KA, Kondrashova O, Bradley A, Williams ED, Pearson JV and Waddell N 2021. Deep learning in cancer diagnosis, prognosis and treatment selection Genome Med. 13 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimpl MJ, Boukerroui D, Stride EPJ, Vallis KA and Gooding MJ 2021. Interactive contouring through contextual deep learning Med. Phys. 48 2951–9 [DOI] [PubMed] [Google Scholar]
- Tustison NJ and Gee J 2011. Run-Length Matrices For Texture Analysis The Insight Journal Online: 10.54294/ex0itu [DOI] [Google Scholar]
- Unkelbach J, Bortfeld T, Cardenas CE, Gregoire V, Hager W, Heijmen B, Jeraj R, Korreman SS, Ludwig R, Pouymayou B, Shusharina N, Söderberg J, Toma-Dasu I, Troost EGC and Vasquez Osorio E 2020. The role of computational methods for automating and improving clinical target volume definition Radiother. Oncol 153 15–25 [DOI] [PubMed] [Google Scholar]
- Valdes G, Luna JM, Eaton E, Simone CB 2nd, Ungar LH and Solberg TD 2016a. MediBoost: a Patient Stratification Tool for Interpretable Decision Making in the Era of Precision Medicine Sci. Rep. 6 37854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes G, Scheuermann R, Hung CY, Olszanski A, Bellerive M and Solberg TD 2016b. A mathematical framework for virtual IMRT QA using machine learning Med. Phys. 43 4323. [DOI] [PubMed] [Google Scholar]
- Vandewinckele L, Claessens M, Dinkla A, Brouwer C, Crijns W, Verellen D and van Elmpt W 2020. Overview of artificial intelligence-based applications in radiotherapy: Recommendations for implementation and quality assurance Radiother. Oncol. 153 55–66 [DOI] [PubMed] [Google Scholar]
- Vanginderdeuren A, Huet-Dastarac M, Barragan AM and Lee J 2021. Estimating uncertainty in radiation oncology dose prediction with dropout and bootstrap in U-Net models ESANN 2021 proceedings Online: 10.14428/esann/2021.es2021-117 [DOI] [Google Scholar]
- Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł and Polosukhin I 2017. Attention is all you need Advances in neural information processing systems pp 5998–6008 [Google Scholar]
- Veen J van der, van der Veen J, Gulyban A and Nuyts S 2019. Interobserver variability in delineation of target volumes in head and neck cancer Radiotherapy and Oncology 137 9–15 Online: 10.1016/j.radonc.2019.04.006 [DOI] [PubMed] [Google Scholar]
- van der Veen J, Willems S, Bollen H, Maes F and Nuyts S 2020. Deep learning for elective neck delineation: More consistent and time efficient Radiother. Oncol. 153 180–8 [DOI] [PubMed] [Google Scholar]
- van der Veen J, Willems S, Deschuymer S, Robben D, Crijns W, Maes F and Nuyts S 2019. Benefits of deep learning for delineation of organs at risk in head and neck cancer Radiother. Oncol. 138 68–74 [DOI] [PubMed] [Google Scholar]
- Vinga S 2021. Structured sparsity regularization for analyzing high-dimensional omics data Brief. Bioinform. 22 77–87 [DOI] [PubMed] [Google Scholar]
- Vinyals O, Blundell C, Lillicrap T, Kavukcuoglu K and Wierstra D 2016. Matching networks for one shot learning Adv. Neural Inf. Process. Syst. 29 Online: https://proceedings.neurips.cc/paper/2016/hash/90e1357833654983612fb05e3ec9148c-Abstract.html [Google Scholar]
- Vokinger KN, Feuerriegel S and Kesselheim AS 2021. Continual learning in medical devices: FDA’s action plan and beyond The Lancet Digital Health 3 e337–8 Online: 10.1016/s2589-7500(21)00076-5 [DOI] [PubMed] [Google Scholar]
- Walls GM, Osman SOS, Brown KH, Butterworth KT, Hanna GG, Hounsell AR, McGarry CK, Leijenaar RTH, Lambin P, Cole AJ and Jain S 2022. Radiomics for Predicting Lung Cancer Outcomes Following Radiotherapy: A Systematic Review Clin. Oncol. 34 e107–22 [DOI] [PubMed] [Google Scholar]
- Wang G, Li W, Aertsen M, Deprest J, Ourselin S and Vercauteren T 2019a. Aleatoric uncertainty estimation with test-time augmentation for medical image segmentation with convolutional neural networks Neurocomputing 335 34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Li W, Ourselin S and Vercauteren T 2019b. Automatic Brain Tumor Segmentation Using Convolutional Neural Networks with Test-Time Augmentation Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries (Springer International Publishing; ) pp 61–72 [Google Scholar]
- Wang K, Zhang D, Li Y, Zhang R and Lin L 2017. Cost-effective active learning for deep image classification IEEE Trans. Circuits Syst. Video Technol. 27 2591–600 [Google Scholar]
- Wang M and Deng W 2018. Deep visual domain adaptation: A survey Neurocomputing 312 135–53 Online: 10.1016/j.neucom.2018.05.083 [DOI] [Google Scholar]
- Wang M, Zhang Q, Lam S, Cai J and Yang R 2020a. A Review on Application of Deep Learning Algorithms in External Beam Radiotherapy Automated Treatment Planning Front. Oncol. 10 580919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Sheng Y, Wang C, Zhang J, Li X, Palta M, Czito B, Willett CG, Wu Q, Ge Y, Yin F-F and Wu QJ 2020b. Fluence Map Prediction Using Deep Learning Models – Direct Plan Generation for Pancreas Stereotactic Body Radiation Therapy Front Artif Intell 3 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zheng VW, Yu H and Miao C 2019c. A Survey of Zero-Shot Learning: Settings, Methods, and Applications ACM Trans. Intell. Syst. Technol. 10 1–37 [Google Scholar]
- Wang Y, Yao Q, Kwok JT and Ni LM 2021. Generalizing from a Few Examples ACM Computing Surveys 53 1–34 Online: 10.1145/3386252 [DOI] [Google Scholar]
- Wan L, Zeiler M, Zhang S, Le Cun Y and Fergus R 2013. Regularization of Neural Networks using DropConnect Proceedings of the 30th International Conference on Machine Learning Proceedings of Machine Learning Research vol 28, ed Dasgupta S and McAllester D (Atlanta, Georgia, USA: PMLR; ) pp 1058–66 [Google Scholar]
- Watts J, Khojandi A, Vasudevan R and Ramdhani R 2020. Optimizing Individualized Treatment Planning for Parkinson’s Disease Using Deep Reinforcement Learning Conf. Proc. IEEE Eng. Med. Biol. Soc. 2020 5406–9 [DOI] [PubMed] [Google Scholar]
- Welch ML, McIntosh C, McNiven A, Huang SH, Zhang B-B, Wee L, Traverso A, O’Sullivan B, Hoebers F, Dekker A and Jaffray DA 2020. User-controlled pipelines for feature integration and head and neck radiation therapy outcome predictions Phys. Med. 70 145–52 [DOI] [PubMed] [Google Scholar]
- Wickstrom K, Mikalsen KO, Kampffmeyer M, Revhaug A and Jenssen R 2021. Uncertainty-Aware Deep Ensembles for Reliable and Explainable Predictions of Clinical Time Series IEEE J Biomed Health Inform 25 2435–44 [DOI] [PubMed] [Google Scholar]
- Wiegreffe S and Pinter Y 2019. Attention is not not Explanation arXiv [cs.CL] Online: http://arxiv.org/abs/1908.04626 [Google Scholar]
- Willemink MJ, Koszek WA, Hardell C, Wu J, Fleischmann D, Harvey H, Folio LR, Summers RM, Rubin DL and Lungren MP 2020. Preparing Medical Imaging Data for Machine Learning Radiology 295 4–15 Online: 10.1148/radiol.2020192224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G and Cook DJ 2020. A Survey of Unsupervised Deep Domain Adaptation ACM Trans Intell Syst Technol 11 1–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel DJ, Weikert TJ, Breit H-C, Chabin G, Gibson E, Heye TJ, Comaniciu D and Boll DT 2020. Validation of a fully automated liver segmentation algorithm using multi-scale deep reinforcement learning and comparison versus manual segmentation Eur. J. Radiol. 126 108918. [DOI] [PubMed] [Google Scholar]
- Wolberg WH, Street WN and Mangasarian OL 1995. Image analysis and machine learning applied to breast cancer diagnosis and prognosis Anal. Quant. Cytol. Histol. 17 77–87 [PubMed] [Google Scholar]
- Wu C, Nguyen D, Xing Y, Barragan A, Schuemann J, Shang H, Pu Y and Jiang SB 2020. Improving Proton Dose Calculation Accuracy by Using Deep Learning Machine Learning: Science and Technology Online: 10.1088/2632-2153/abb6d5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Gong J, Hao W, Yang T, Lin Y, Wang S and Peng W 2020. Comparison and Fusion of Deep Learning and Radiomics Features of Ground-Glass Nodules to Predict the Invasiveness Risk of Stage-I Lung Adenocarcinomas in CT Scan Front. Oncol. 10 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Zheng X, Chen Y, Xie L, Liu J, Zhang Y, Yan J, Zhu H and Hu Y 2018. Artifact Removal using Improved GoogLeNet for Sparse-view CT Reconstruction Sci. Rep. 8 6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Niu J, Liu X, Chen Z, Tang S and Yu S 2021 A survey on incorporating domain knowledge into deep learning for medical image analysis Med. Image Anal. 69 101985. [DOI] [PubMed] [Google Scholar]
- Xing L, Goetsch S and Cai J 2021. Point/Counterpoint. Artificial intelligence should be part of medical physics graduate program curriculum Med. Phys. 48 1457–60 [DOI] [PubMed] [Google Scholar]
- Xing Y, Zhang Y, Nguyen D, Lin M-H, Lu W and Jiang S 2020. Boosting radiotherapy dose calculation accuracy with deep learning J. Appl. Clin. Med. Phys. Online: 10.1002/acm2.12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Hospedales TM and Gong S 2016. Multi-Task Zero-Shot Action Recognition with Prioritised Data Augmentation Computer Vision – ECCV 2016 343–59 Online: 10.1007/978-3-319-46475-6_22 [DOI] [Google Scholar]
- Xu Y, Hosny A, Zeleznik R, Parmar C, Coroller T, Franco I, Mak RH and Aerts HJWL 2019. Deep Learning Predicts Lung Cancer Treatment Response from Serial Medical Imaging Clin. Cancer Res. 25 3266–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Kim J-Y, Kim H and Adhikari SP 2020a. Guided Soft Attention Network for Classification of Breast Cancer Histopathology Images IEEE Trans. Med. Imaging 39 1306–15 [DOI] [PubMed] [Google Scholar]
- Yang Q, Chao H, Nguyen D and Jiang S 2020b. Mining Domain Knowledge: Improved Framework Towards Automatically Standardizing Anatomical Structure Nomenclature in Radiotherapy IEEE Access 8 105286–300 Online: 10.1109/access.2020.2999079 [DOI] [Google Scholar]
- Yang Y, Morillo IG and Hospedales TM 2018. Deep Neural Decision Trees Online: http://dx.doi.org/
- Yan W, Huang L, Xia L, Gu S, Yan F, Wang Y and Tao Q 2020. MRI Manufacturer Shift and Adaptation: Increasing the Generalizability of Deep Learning Segmentation for MR Images Acquired with Different Scanners Radiol Artif Intell 2 e190195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Guo D, Tseng C-K, Ge J, Hung T-M, Pai P-C, Ren Y, Zheng L, Zhu X, Peng L, Chen Y, Chen X, Chou C-Y, Chen D, Yu J, Chen Y, Jiao F, Xin Y, Huang L, Xie G, Xiao J, Lu L, Yan S, Jin D and Ho T-Y 2021. Multi-Institutional Validation of Two-Streamed Deep Learning Method for Automated Delineation of Esophageal Gross Tumor Volume Using Planning CT and FDG-PET/CT Front. Oncol. 11 785788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AT, Fernandez K, Pfau J, Reddy R, Cao NA, von Franque M Y, Johal A, Wu BV, Wu RR, Chen JY, Fadadu RP, Vasquez JA, Tam A, Keiser MJ and Wei ML 2021. Stress testing reveals gaps in clinic readiness of image-based diagnostic artificial intelligence models NPJ Digit Med 4 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Q, Luo X, Ye Q, Xu L and Zhuang X 2019. Cardiac Segmentation from LGE MRI Using Deep Neural Network Incorporating Shape and Spatial Priors Lecture Notes in Computer Science 559–67 Online: 10.1007/978-3-030-32245-8_62 [DOI] [Google Scholar]
- Yu S, Chen M, Zhang E, Wu J, Yu H, Yang Z, Ma L, Gu X and Lu W 2020. Robustness study of noisy annotation in deep learning based medical image segmentation Phys. Med. Biol. 65 175007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitsev M, Maclaren J and Herbst M 2015. Motion artifacts in MRI: A complex problem with many partial solutions J. Magn. Reson. Imaging 42 887–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanca F, Hernandez-Giron I, Avanzo M, Guidi G, Crijns W, Diaz O, Kagadis GC, Rampado O, Lønne PI, Ken S, Colgan N, Zaidi H, Zakaria GA and Kortesniemi M 2021. Expanding the medical physicist curricular and professional programme to include Artificial Intelligence Phys. Med. 83 174–83 [DOI] [PubMed] [Google Scholar]
- Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ and Oermann EK 2018. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study PLoS Med. 15 e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Chen B and Li S 2020a. Sequential conditional reinforcement learning for simultaneous vertebral body detection and segmentation with modeling the spine anatomy Med. Image Anal. 67 101861. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wang C, Sheng Y, Palta M, Czito B, Willett C, Zhang J, Jensen PJ, Yin F-F, Wu Q, Ge Y and Wu QJ 2020b. An interpretable planning bot for pancreas stereotactic body radiation therapy Int. J. Radiat. Oncol. Biol. Phys. Online: 10.1016/j.ijrobp.2020.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang X, Yang D, Sanford T, Harmon S, Turkbey B, Wood BJ, Roth H, Myronenko A, Xu D and Xu Z 2020c. Generalizing Deep Learning for Medical Image Segmentation to Unseen Domains via Deep Stacked Transformation IEEE Trans. Med. Imaging 39 2531–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu X, Gach HM, Li H and Yang D 2021. GroupRegNet: a groupwise one-shot deep learning-based 4D image registration method Phys. Med. Biol. 66 045030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y and Yang Q 2021. A Survey on Multi-Task Learning IEEE Transactions on Knowledge and Data Engineering 1–1 Online: 10.1109/tkde.2021.307020336506788 [DOI] [Google Scholar]
- Zhang Z, Xie Y, Xing F, McGough M and Yang L 2017. MDNet: A Semantically and Visually Interpretable Medical Image Diagnosis Network 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Online: 10.1109/cvpr.2017.378 [DOI] [Google Scholar]
- Zheng H, Lin L, Hu H, Zhang Q, Chen Q, Iwamoto Y, Han X, Chen Y-W, Tong R and Wu J 2019. Semi-supervised Segmentation of Liver Using Adversarial Learning with Deep Atlas Prior Medical Image Computing and Computer Assisted Intervention – MICCAI 2019 (Springer International Publishing; ) pp 148–56 [Google Scholar]
- Zhen S-H, Cheng M, Tao Y-B, Wang Y-F, Juengpanich S, Jiang Z-Y, Jiang Y-K, Yan Y-Y, Lu W, Lue J-M, Qian J-H, Wu Z-Y, Sun J-H, Lin H and Cai X-J 2020. Deep Learning for Accurate Diagnosis of Liver Tumor Based on Magnetic Resonance Imaging and Clinical Data Front. Oncol. 10 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Khosla A, Lapedriza A, Oliva A and Torralba A 2016. Learning Deep Features for Discriminative Localization 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Online: 10.1109/cvpr.2016.319 [DOI] [Google Scholar]
- Zhou K, Shi H, Chen R, Cochuyt JJ, Hodge DO, Manochakian R, Zhao Y, Ailawadhi S and Lou Y 2021. Association of Race, Socioeconomic Factors, and Treatment Characteristics With Overall Survival in Patients With Limited-Stage Small Cell Lung Cancer JAMA Netw Open 4 e2032276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Wang J and Xing L 2009. Noise suppression in scatter correction for cone-beam CT Med. Phys. 36 741–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotti C, Luo Z, Lalande A and Jodoin P-M 2019. Convolutional Neural Network With Shape Prior Applied to Cardiac MRI Segmentation IEEE Journal of Biomedical and Health Informatics 23 1119–28 Online: 10.1109/jbhi.2018.2865450 [DOI] [PubMed] [Google Scholar]
- Zunair H and Ben Hamza A 2020. Melanoma detection using adversarial training and deep transfer learning Physics in Medicine & Biology 65 135005 Online: 10.1088/1361-6560/ab86d3 [DOI] [PubMed] [Google Scholar]


