Abstract
Cocaine self-administration is a complexly determined trait, with a substantial proportion of individual differences being determined by genetic variation. However, the relevant genetic variants that drive heritable differences in cocaine use remain undiscovered. Cocaine intravenous self-administration (IVSA) procedures in laboratory animals provide opportunities to prospectively investigate neurogenetic influences on the acquisition of voluntary cocaine use. Here, we provide information on cocaine (or saline—as a control) IVSA in 84 members of the hybrid mouse diversity panel (HMDP), an array of genetically distinct classical or recombinant inbred strains. We found cocaine IVSA to be substantially heritable in this population, with strain-level intake ranging for near 0 to >25 mg/kg/session. Though saline IVSA was also found to be heritable, a modest genetic correlation between cocaine and saline IVSA indicates that operant responding for the cocaine reinforcer was influenced, at least in part, by unique genetic variants. Genome-wide association studies (GWAS) of infusions earned in cocaine and saline groups revealed significant quantitative trait loci (QTL) on Chromosomes 3 and 14 for cocaine, but not saline, IVSA. Positional candidates were further prioritized through use of bulk RNA sequencing data that revealed genes with cis-eQTL and genetic correlation to number of infusions. Additionally, these data identify reference strains with extreme cocaine IVSA phenotypes, revealing them as polygenic models of risk and resilience to cocaine reinforcement. This work is part of an ongoing effort to characterize genetic variation that moderates cocaine IVSA that may, in turn, provide a more comprehensive understanding of cocaine risk genetics and neurobiology.
Keywords: addiction, cocaine, gene-expression, genetics, GWAS, self-administration
1 |. INTRODUCTION
Cocaine use disorder is a psychiatric condition with a complex aetiology, and genetic factors account for a substantial proportion of associated risk.1,2 Identification of variants and risk genes that ultimately determine individual vulnerability will likely advance our understanding of the neurogenetic mechanisms underlying problematic cocaine use.
Genetic studies in humans have had some success to date.3–5 However, a large proportion of the genetic variants that contribute to heritable influence on cocaine use disorder remain undiscovered. Approaches in laboratory animal populations offer complementary methods with several key advantages. Advanced mouse populations allow for quantitative traits to be characterized prospectively, under controlled conditions. To date, genetic strategies have identified quantitative trait loci (QTL) and subsequent candidate genes for several cocaine-related behaviours. Some of these efforts have focused on cocaine-induced psychomotor activation. Multiple QTL have been associated with cocaine-induced locomotion, with confirmation of some QTL by secondary mapping approaches.6–10
Measures of cocaine reward and reinforcement have also been utilized to map QTL and identify genetic correlations among behavioural and physiological measures.11–18 The prioritization of candidate genes that regulate behavioural responses to cocaine can be further aided by transcriptome profiling of tissue from brain regions implicated in addiction neurobiology, such as the nucleus accumbens (NAc) and medial frontal cortex (mFC; emcompassing the infra- and pre-limbic cortices).12,15,19
Cocaine intravenous self-administration (IVSA) is an assay of voluntary cocaine use and is thought to provide a model of cocaine use with predictive, face and construct validity.20,21 Advances in surgical techniques and devices have allowed for improved catheter patency rates that make characterization of large numbers of genetically diverse mice for cocaine IVSA feasible.13–15,17,18 Applying this assay to advanced mouse populations may enhance our understanding of genetic factors unique to voluntary cocaine use. The BXD-recombinant inbred strain panel was utilized to map cocaine IVSA QTL and genetic correlations,15 and a similar study is underway using collaborative cross and diversity outbred mice (CC/DO).18 Application of cocaine IVSA to additional mouse populations may offer further opportunities to generate replicable and/or reproducible information on the genetics of cocaine use.
The hybrid mouse diversity panel (HMDP) is a large collection of both classic inbred strains and recombinant inbred strains that provides a population with high genetic diversity.22,23 The classical inbred strains provide many meiotic breakpoints that allow for relatively high-resolution QTL mapping. The recombinant inbred strains provide additional breakpoints and also enhance power by adding replicates of alleles.22,23 The stability of the HMDP allows for continued, cumulative study of traits, including characterization of transcript expression that may greatly enhance genetic studies in current and future research. These key characteristics may be utilized to investigate the genetics of cocaine IVSA and related traits.
Here, we describe use of the HMDP to characterize cocaine IVSA, as well as to generate strain-level transcriptome profiles by RNA sequencing for tissue from the NAc and mFC. This is the first use of this population for large-scale study of cocaine IVSA. This approach indicates the heritability of cocaine IVSA and provides a dataset that can be utilized for genome-wide association studies (GWAS) of behavioural outcomes and complimentary use of gene expression to prioritize candidate genes that regulate cocaine self-administration. A saline IVSA control group is included in every strain tested; this control may allow for enhanced precision in discriminating heritable cocaine reinforcement from other heritable factors that may influence behaviour in an IVSA procedure independent of cocaine.
2 |. MATERIALS AND METHODS
2.1 |. Subjects
A total of 956 mice from 84 strains of the HMDP (mean age 11.3 ± 2.2 [SD] weeks) were selected for assessment of cocaine or saline IVSA (see Table 1). A within strain sample size target of six mice/infusate group was selected based on previous power analysis simulations that indicated a sample size of five yields 80% power to detect a QTL of 10% effect size in 100 strains of the HMDP.22 All animals received surgical implantation of chronic indwelling catheters by JAX Surgical Services (The Jackson Laboratory, Bar Harbor ME). Following at least 1 week of recovery, the animals were shipped to Binghamton University. When not being tested, all mice were housed in the home cage and maintained on ad libitum mouse chow (5L0D, Purina Lab Diet) and water. Mice were individually housed in polycarbonate cages (30 × 8 cm) with wood-chip bedding (SANI-CHIPS), a paper nestlet and a red polycarbonate hut. All procedures were approved by the Binghamton University Institutional Animal Care and Use Committee and conducted in accordance with the National Institute of Health “Guide for Care and Use of Laboratory Animals, Eighth Edition” (National Research Council [US] Committee for the Update of the Guide for the Care and Use of Laboratory Animals, 2011).
TABLE 1.
Sample size by strain and infusate group
| STRAIN | Cocaine | Saline |
|---|---|---|
| 129S1/SvlmJ (002448) | 6 | 6 |
| 129X1/SvJ (000691) | 6 | 6 |
| A/J (000649) | 6 | 6 |
| AKR/J (000648) | 6 | 6 |
| BALB/cByJ (001026) | 6 | 6 |
| BALB/cJ (000651) | 7 | 7 |
| BPL/1J (003006) | 6 | 6 |
| BTBRT+Itpr3tf/J (002282) | 7 | 6 |
| C3H/HeJ (000659) | 6 | 6 |
| C3HeB/FeJ (000658) | 6 | 6 |
| C57BL/10J (000665) | 6 | 7 |
| C57BL/6J (000664) | 7 | 6 |
| C57BLKS/J (000662) | 6 | 6 |
| C57BR/cdJ (000667) | 6 | 6 |
| C57L/J (000668) | 6 | 8 |
| C58/J (000668) | 6 | 6 |
| CBA/J (000656) | 6 | 7 |
| DBA/1J (000670) | 6 | 6 |
| DBA/2J (000671) | 6 | 6 |
| FVB/NJ (001800) | 6 | 6 |
| I/LnJ (000674) | 3 | 4 |
| KK/HiJ (02106) | 6 | 6 |
| LP/J (000676) | 6 | 6 |
| MA/MyJ (000677) | 6 | 6 |
| MRL/MpJ (000486) | 7 | 6 |
| NOD/ShiLtJ (001976) | 6 | 7 |
| NZB/BINJ (000684) | 7 | 5 |
| NZO/HlLtJ (02105) | 5 | 7 |
| NZW/LacJ (001058) | 6 | 6 |
| PL/J (000680) | 6 | 6 |
| SJL/J (000686) | 7 | 6 |
| SM/J (000687) | 7 | 6 |
| BXD1/TyJ (000036) | 6 | 7 |
| BXD2/TyJ (000075) | 6 | 5 |
| BXD6/TyJ (000007) | 4 | 3 |
| BXD9/TyJ (000105) | 6 | 6 |
| BXD11/TyJ (000012) | 6 | 5 |
| BXD13/TyJ (000040) | 6 | 5 |
| BXD14/TyJ (000329) | 3 | 5 |
| BXD15/TyJ (000095) | 5 | 6 |
| BXD16/TyJ (000013) | 5 | 6 |
| BXD18/TyJ (000015) | 4 | 5 |
| BXD19/TyJ (000010) | 6 | 6 |
| BXD21/TyJ (000077) | 6 | 4 |
| BXD27/TyJ (000041) | 5 | 5 |
| BXD28/TyJ (000047) | 5 | 6 |
| BXD29/TyJ (010981) | 5 | 4 |
| BXD31/TyJ (000083) | 7 | 6 |
| BXD32/TyJ (000078) | 6 | 6 |
| BXD33/TyJ (003222) | 4 | 5 |
| BXD34/TyJ (003223) | 7 | 7 |
| BXD38/TyJ (003227) | 6 | 6 |
| BXD39/TyJ (003228) | 6 | 6 |
| BXD40/TyJ (003229) | 5 | 6 |
| BXD42/TyJ (03230) | 6 | 6 |
| BXD43/RwwJ (07093) | 6 | 6 |
| BXD48a/RwwJ (007139) | 6 | 6 |
| BXD49/RwwJ (007098) | 5 | 3 |
| BXD50/RwwJ (007099) | 6 | 6 |
| BXD51/RwwJ (007100) | 4 | 5 |
| BXD55/RwwJ (007103) | 5 | 6 |
| BXD56/RwwJ (007104) | 5 | 5 |
| BXD60/RwwJ (007105) | 6 | 7 |
| BXD61/RwwJ (007106) | 6 | 6 |
| BXD62/RwwJ (007107) | 6 | 6 |
| BXD63/RwwJ (007108) | 5 | 6 |
| BXD65/RwwJ (007110) | 6 | 6 |
| BXD68/RwwJ (007113) | 6 | 6 |
| BXD69/RwwJ (007114) | 6 | 4 |
| BXD70/RwwJ (007115) | 6 | 6 |
| BXD71/RwwJ (007116) | 6 | 5 |
| BXD73a/RwwJ (007124) | 6 | 6 |
| BXD75/RwwJ (007119) | 5 | 5 |
| BXD77/RwwJ (007121) | 6 | 6 |
| BXD83/RwwJ (007126) | 6 | 4 |
| BXD84/RwwJ (007127) | 6 | 6 |
| BXD85/RwwJ (007128) | 6 | 5 |
| BXD86/RwwJ (007129) | 6 | 6 |
| BXD89/RwwJ (007132) | 4 | 4 |
| BXD90/RwwJ (007133) | 6 | 5 |
| BXD98/RwwJ (007141) | 6 | 6 |
| BXD99/RwwJ (007142) | 4 | 4 |
| BXD100/RwwJ (007143) | 6 | 6 |
| BXD102/RwwJ (007145) | 2 | 3 |
2.2 |. Surgery
Animals were anaesthetized with tribromoethanol (Avertin, 400 mg/kg) and implanted a chronic indwelling jugular catheter (catalog # CNC-2/3S-082109E/12, Access Technology, IL USA) and access button port (1-VAB62SMBS/25, Instech, PA USA). Carprofen (5 mg/kg) was administered by subcutaneous injection pre- and post-surgically, and bupivacaine (0.1%: 2 mg/kg) was applied topically before incision closure to manage any surgical pain.
2.3 |. Catheter care and patency
Catheters were maintained with a flush of ~0.05 ml of sterile saline and by then filling the catheter with heparin lock solution (~0.01 ml volume, 500 units/ml concentration, SAI Infusion Technologies, IL) at least once every 3 days during acclimation periods and daily during IVSA testing.
Catheter patency was confirmed by infusions of propofol (~0.02 ml volume, 10 mg/ml concentration, Zoetis, NJ). Immediate but rapidly reversed loss of muscle tone indicated that the catheter was patent. This testing occurred once before the start of IVSA testing (3–4 days prior to testing) and again after the mouse completed the 10th testing session. Any mouse that failed the test was excluded from the experiment. The patency rate after the 10th day of IVSA was 95.1%, reflecting a very high success rate. The overall attrition rate (patency failure, exclusion due to health issues or spontaneous death) was 9.9%. See Table 1 for the numbers of mice by strain and group that successfully completed IVSA.
2.4 |. Cocaine IVSA
All mice were assigned to one of two treatment groups (saline vs. cocaine IVSA), with half of the animals in each strain/sex randomly allocated to each. All subjects were tested for acquisition of self-administration in 10 consecutive daily sessions (Fixed-ratio-1 schedule of reinforcement, 0 or 0.5 mg/kg body weight of cocaine per infusion) that ran until 65 infusions were earned or 2 h passed, whichever came first. Cocaine hydrochloride (Sigma Aldrich; St Louis MO) was dissolved in sterile saline at a concentration of 0.84 mg/ml to produce a freebase dose of 0.5 mg/kg/infusion. Infusion volume was 0.67 ml/kg/infusion for saline and cocaine groups. Testing occurred at the same time each day, during the light phase of a 12/12 h cycle. The animals were tested in Med Associates mouse self-administration chambers (55.69 × 38.1 × 35.56 cm, MED-307W-CT-D1, Med Associates, VT) that were fitted with two retractable ultrasensitive levers and that were housed within sound-attenuating cubicles. Assignment of the active infusion lever (right or left side of the box) was counterbalanced across strains/sex. Assignment of testing chamber was explicitly not random to minimize testing multiple mice from a given strain in the same chamber.
Test sessions began with the activation of the white noise and the illumination of five stimulus lights on the back wall of the chamber. No priming infusion(s) were delivered. When a subject actuated the active lever, an infusion was delivered, the house light flashed, and the aperture lights turned off for 20 s. During this time-out period, contacts on the active lever were recorded but had no programmed consequence. Actuation of the inactive lever had no programmed consequence.
2.5 |. RNA sequencing
NAc and mFC tissue from both infusate groups (saline vs. cocaine) and both sexes of each HMDP strain were dissected 24 h after the final IVSA test session. After decapitation, the brain was removed and placed into saline chilled on ice. After 1 minute, the brain was transferred to a chilled brain matrix (ASI, RBM-2000C). A blade (0.009”, single edge) was placed 0.5 mm anterior of the optic chiasm, and a second blade was placed 1 mm anterior of the first. This slice contained the mFC and was manually cut with a blade to retrieve the tissue medial of the visualized corpus callosum. A third blade was placed 0.5 mm posterior to the first blade. In this slice, the anterior commissure was visualized in both hemispheres, and a 1.5 mm punch was centred on the commissure to dissect the NAc from both hemispheres. Samples were frozen on dry ice and stored at −80°C until they were homogenized in 1 ml of QIAzol reagent (Qiagen). RNA was extracted using the RNeasy Mini Kit (Qiagen) and quantified using a NanoDrop instrument (ThermoFisher). Following quantification, samples were pooled by sex (three samples per pool for each infusate/strain condition), except for four strains (A/J, LP/J, AKR/J and NODShiLtJ40) where individual mice were analysed (data were subsequently collapsed to strain mean).
RNA seq was performed as described, with minor variations.24 Sequencing employed 25 μl of each RNA sample at 40 ng/μl concentration. Libraries were prepared using TruSeq Stranded RNA kits, and 100 ng of RNA following standard protocols provided by Illumina. A total of 392 samples from 41 HMDP strains were analysed. Samples were barcoded and sequenced using 75 bp paired-ends on an Illumina HiSeq3200 machine. Ten samples per lane were used, with each sample distributed round robin style over multiple lanes to minimize variation. Samples were demultiplexed and the fastq.gz files obtained.
An average of 36.6 million (M) and 11.2 M reads were obtained from each of the pooled and non-pooled samples, respectively, giving 72.6 M reads per strain for each infusate group. QC was done on each read (forward and reverse) using FastQC software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).25 The samples had excellent quality, with a median quality score of 40. No trimming or filtering of the sequenced reads was necessary prior to mapping. Mouse genome sequence build GRCm38 was downloaded from the Ensembl website (http://uswest.ensembl.org/Mus_musculus/Info/Index).26 Reference genome indices were built and samples mapped to GRCm38 using STAR aligner version 2.7.9a (https://github.com/alexdobin/STAR/releases/tag/2.7.9a).27 Mapping was performed using the default mismatch value of 10, allowing one multiple mapping per read. HTseq-count was used to obtain expression values for each sample using the sorted BAM files from the alignment output of STAR.28 The RNA sequencing data generated in this study are publicly available from NCBI GEO (https://www.ncbi.nlm.nih.gov/geo) under accession number GSE186981 and from the NCBI BioProject database under SRA accession number PRJNA755328.
2.6 |. Data analyses
Between-subjects factors of strain, infusate group, sex and repeated-measures of IVSA session were assessed by analyses of variance (ANOVAs). Data were transformed by a rank normal inversion transformation to better achieve normality. Heritability was estimated by partial eta squared for the main effect of strain. Where appropriate, post hoc comparisons were made, and Sidak p value correction was utilized to correct for multiple testing. Infusions earned and lever discrimination (active versus inactive lever pressing) were the dependent variables of analyses. A p value less than 0.05 was considered significant.
GWAS was performed with faST-LMM (Python package)29 in order to detect QTL that associate with cocaine or saline self-administration. Genotypes were acquired from the CGD-MDA1 dataset obtained from phenome.jax.org. A minimum minor allele frequency of 5% was utilized. A genome-wide corrected p value of less than 0.05 (4.1 × 10−6) was considered significant.22
RNA seq data were utilized to prioritize positional candidate genes. All genes within the behavioural QTL intervals (2.6 megabase [Mb] interval centred on the SNP with the peak association statistic)22 were assessed for cis-expression QTL (cis-eQTL). False discovery rate (FDR) (Benjamini–Hochberg, q = 0.1) was utilized to correct for multiple comparisons. All genes that demonstrated a cis-eQTL were assessed for potential genetic correlations between expression level and the behavioural trait using Pearson’s correlation and strain means. Expression data from both regions and infusate groups (cocaine and saline) were utilized. Additionally, a strain value difference score between expression in cocaine and saline groups was calculated as a measure of differential expression and utilized for correlations to infusion data. FDR was utilized to correct for multiple comparisons within tissue region and infusate group. Furthermore, only results that displayed correlations to expression in both brain regions or to expression datasets from more than one infusate group or to the expression difference score were included.
3 |. RESULTS
3.1 |. Acquisition of IVSA
Mice from all 84 strains underwent 10 days cocaine or saline IVSA, and the number of infusions earned per session (Figures 1A and 2) was assessed by mixed ANOVA with strain, sex, infusate group and session as factors. Infusions varied over sessions (main effect of session [F{6.6, 4106.5} = 3.4, p = 0.001), with ANOVA also revealing two higher level interactions, including a session-strain–infusate group-sex interaction (F[542.3, 4106.5] = 1.1, p = 0.024]) and a session-strain interaction (F[548.9, 4106.5] = 1.8, p < 0.001).
FIGURE 1.
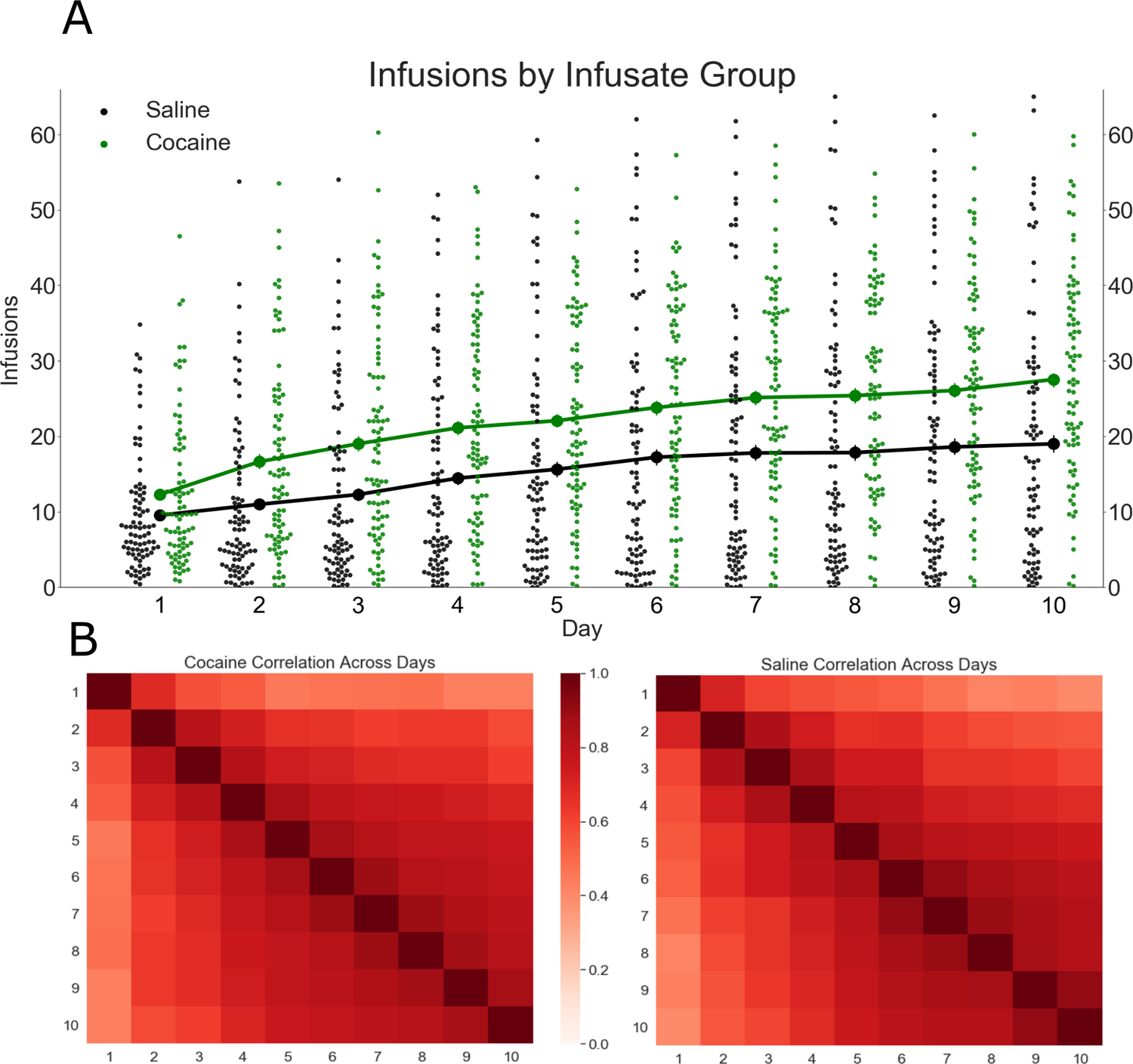
Cocaine infusions over 10 sessions, in 84 strains of the hybrid mouse diversity panel (HMDP). (A) Infusion means by infusate group, over 10 daily sessions (error bars = SEM, scatter points = strain means). Cocaine taking exceeded saline taking overall; however, intravenous self-administration (IVSA) of either infusate was highly strain-dependent. (B) Correlation heatmap for correlation of infusions across sessions, for cocaine and saline groups. The magnitude of session–session correlation increased in later sessions, indicating stabilization of IVSA
FIGURE 2.
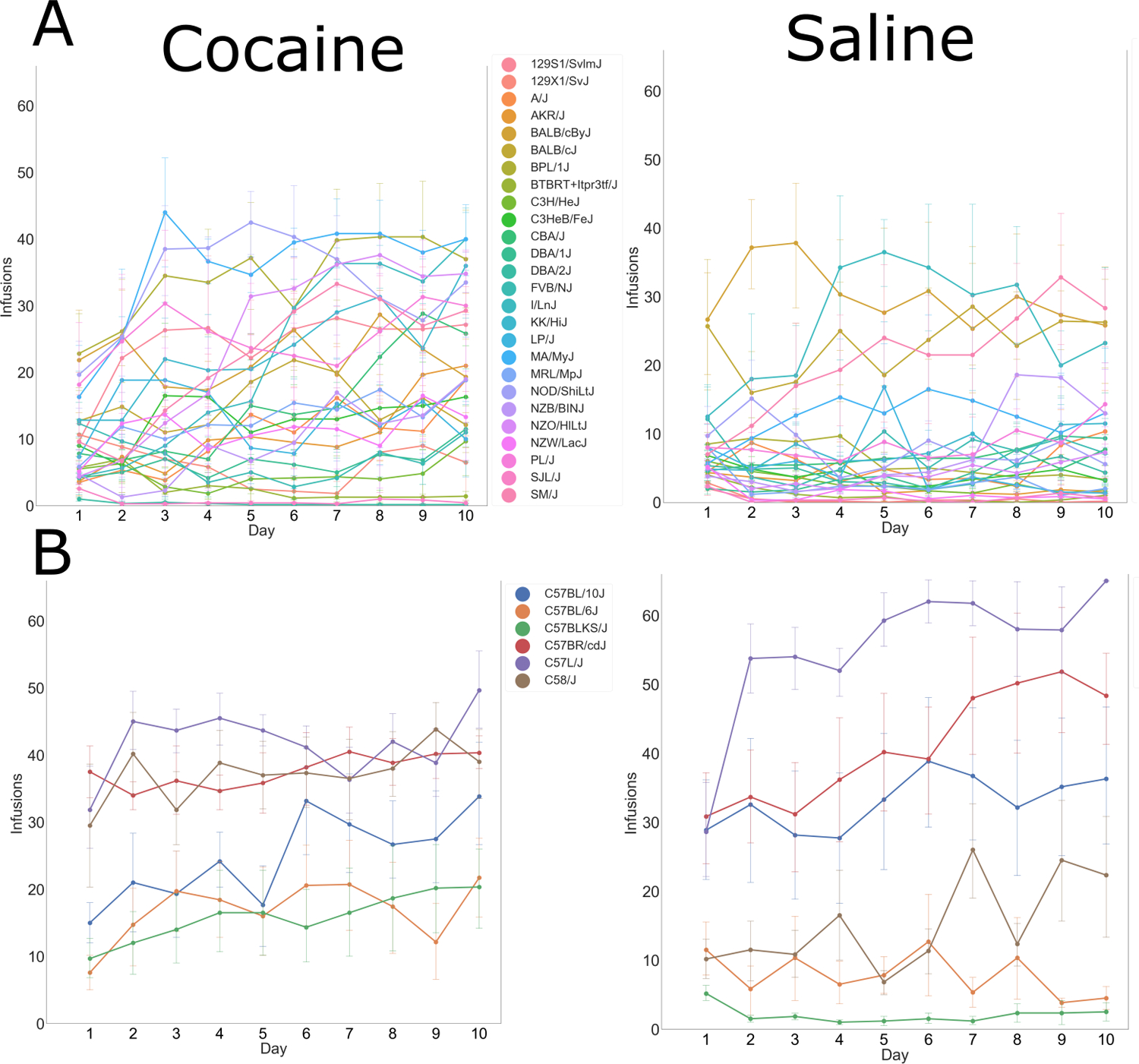
Infusions earned over 10 sessions in individual classic inbred strains (error bars = SEM). Strains are grouped into subcategories, based on relatedness, and graphed separately in order to improve clarity of the graphs. (A) Classic inbred strains and (B) strains of the C57 family
As expected, the cocaine group elicited more infusions overall (main effect of infusate group [F{1, 621} = 12.4, p < 0.001]); however, differences between the two infusate groups were also impacted by strain (strain–infusate group interaction [F{83, 621} = 1.7, p < 0.001]).Separate analyses within each infusate group revealed that variation in intake across sessions was impacted by strain in both groups (session-strain interaction for the cocaine group [F{526.3, 1972.2} = 1.4, p < 0.001] and for the saline group [F{552, 2061.6} = 1.5, p < 0.001], main effect of session in saline group [F{6.7, 2061.6} = 3.5, p = 0.001]); the direction and magnitude of change differed substantially by strain in both infusate groups.
Analysis within infusate groups revealed significant broad sense heritability for cocaine IVSA (main effect strain [F{83, 311} = 4.3, p < 0.001], H2 = 0.53) and saline IVSA (main effect strain [F{83, 310} = 6.1, p < 0.001], H2 = 0.62). Sex interacted with strain to affect the number of infusions earned within the cocaine group (F[82, 311] = 1.5, p = 0.012), but not the saline group, suggesting strain-dependent sex effects impact IVSA when cocaine is the reinforcer.
3.2 |. Post-acquisition IVSA
Correlations between cocaine and saline infusions earned across all 10 test sessions revealed that day-to-day variation in infusions earned diminished as the number of sessions progressed. The correlation between Days 1 and 3 was r = 0.57, p < 0.001 for cocaine and r = 0.60, p < 0.001 for saline; for Days 8 and 10, it was r = 0.82, p < 0.001 for cocaine and r = 0.83, p < 0.001 for saline (Figure 1B; test for difference between correlation r value from Sessions 1–3 to 8–10 by Fisher’s r to z transformation; z = 3.2, p < 0.001 and z = 3.2, p < 0.001 for cocaine and saline, respectively). Similar results were found for strain-level correlations, indicating stabilization of strain-level variance (Days 1 and 3: r = 0.56, p < 0.001 for cocaine; r = 0.54, p < 0.001 for saline; Days 8 and 10: r = 0.94, p < 0.001 for cocaine; r = 0.85, p < 0.001 for saline; the increase in r was significant for cocaine [z = 7.0, p < 0.001] and saline [z = 4.1, p < 0.001]). For this reason, infusions earned in Sessions 8–10 were collapsed for each mouse and utilized as a measure of post-acquisition cocaine or saline IVSA.
Analyses of Sessions 8–10 phenotypes indicated that the groups differed in infusions earned (main effect of infusate group [F{1, 622} = 6.2, p = 0.013], with cocaine intake exceeding saline intake; Figures 2A and 3A). These differences were impacted by strain (strain–infusate group interaction [F{83, 622} = 1.8, p < 0.001]).Analysis within each infusate group revealed significant broad-sense heritability for cocaine taking (main effect strain [F{83, 479} = 3.5, p < 0.001], H2 = 0.49) and saline taking (main effect strain [F{83, 479} = 4.8, p < 0.001], H2 = 0.56).
FIGURE 3.
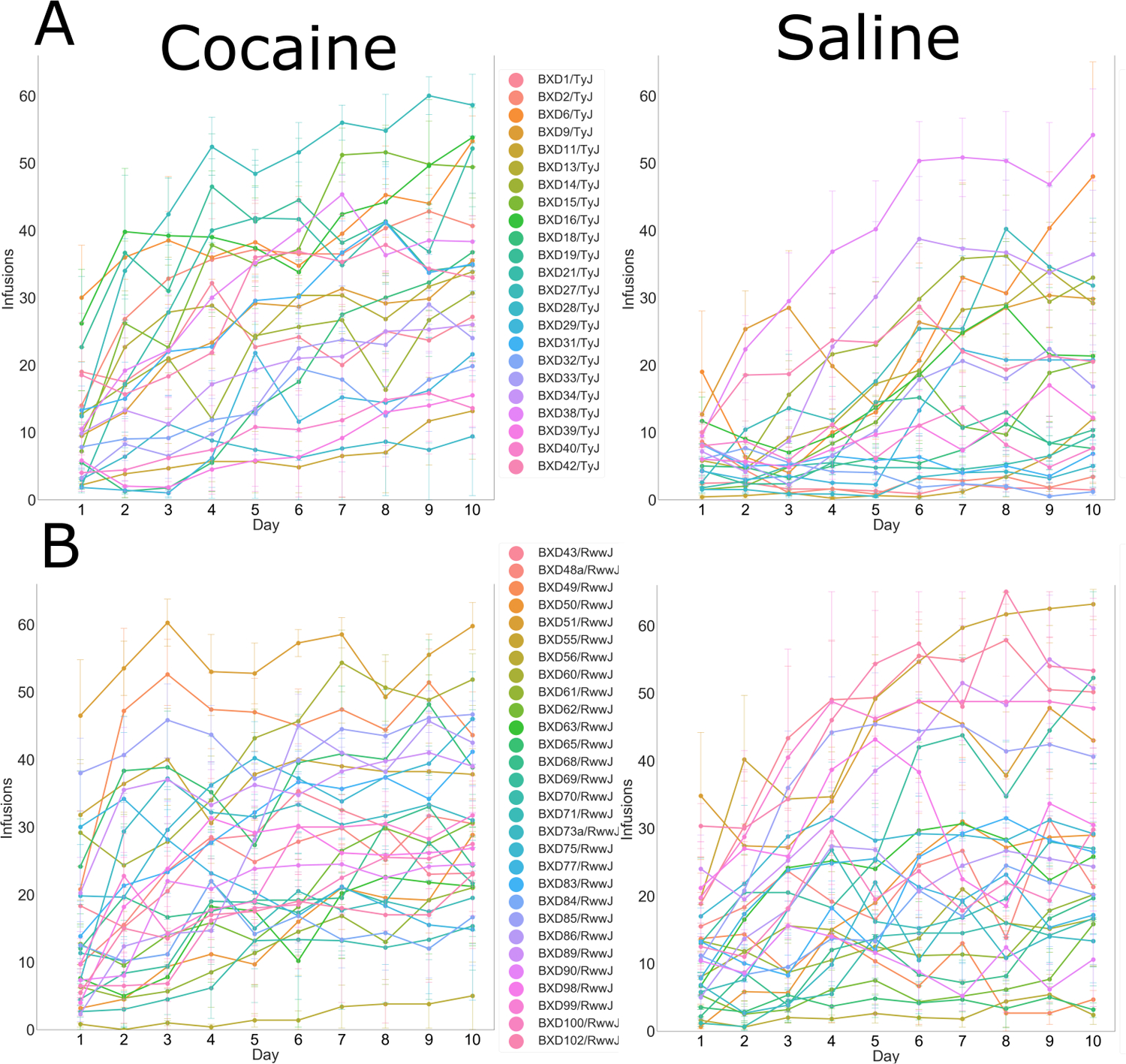
Infusions earned over 10 sessions in individual BXD strains (error bars = SEM). Strains are grouped into subcategories, based on relatedness, and graphed separately in order to improve clarity of the graphs. (A) Taylor BXD strains and (B) Williams BXD strains
Strains demonstrated a broad and continuous range of cocaine taking, with near zero intake at the low end and ~50 infusions (~25 mg/kg) per session at the high end (Figure 4A). The level of cocaine or saline self-administration averaged across Sessions 8–10 were assessed by one sample t tests that compared each strain’s level of cocaine and saline taking to 0. Six of the 84 strains in the cocaine group did not show a number of infusions significantly different from 0 (Figures 4A and 5A). Similarly, five strains in the saline group did not significantly differ from zero infusions (Figures 4A and 5B).
FIGURE 4.
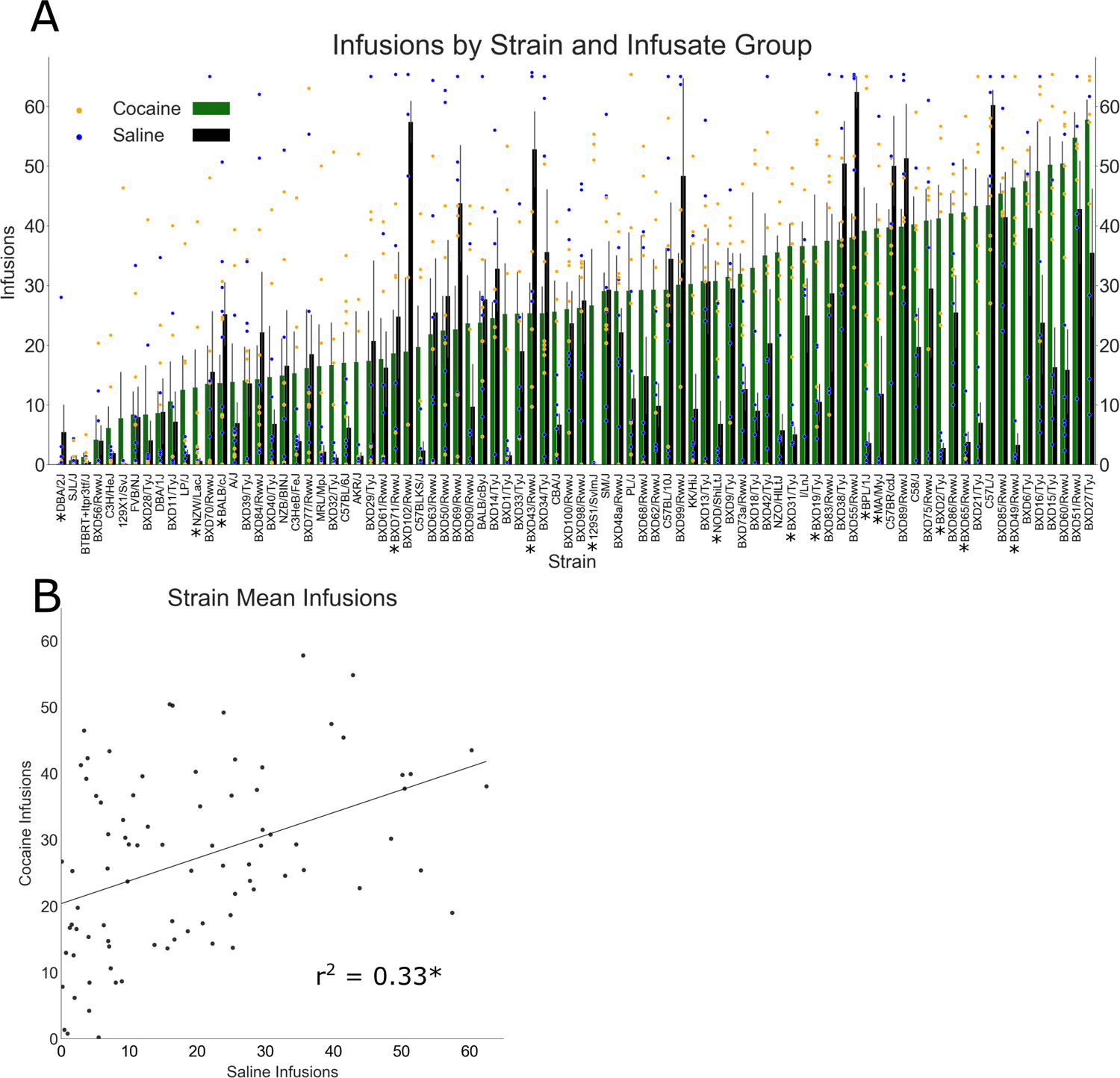
Cocaine and saline intravenous self-administration (IVSA), averaged in the last three sessions. (A) Strain means for cocaine and saline IVSA in the last three sessions. Both cocaine and saline demonstrated heritable variation, with a broad range of intake across strains (error bars = SEM, scatter points = individual mouse values). An * below the strain name on the X-axis indicates that intake of cocaine or saline was significantly different for that strain. (B) Scatterplot of strain means for cocaine and saline IVSA in the last three sessions. There was a modest genetic correlation between saline and cocaine taking
FIGURE 5.
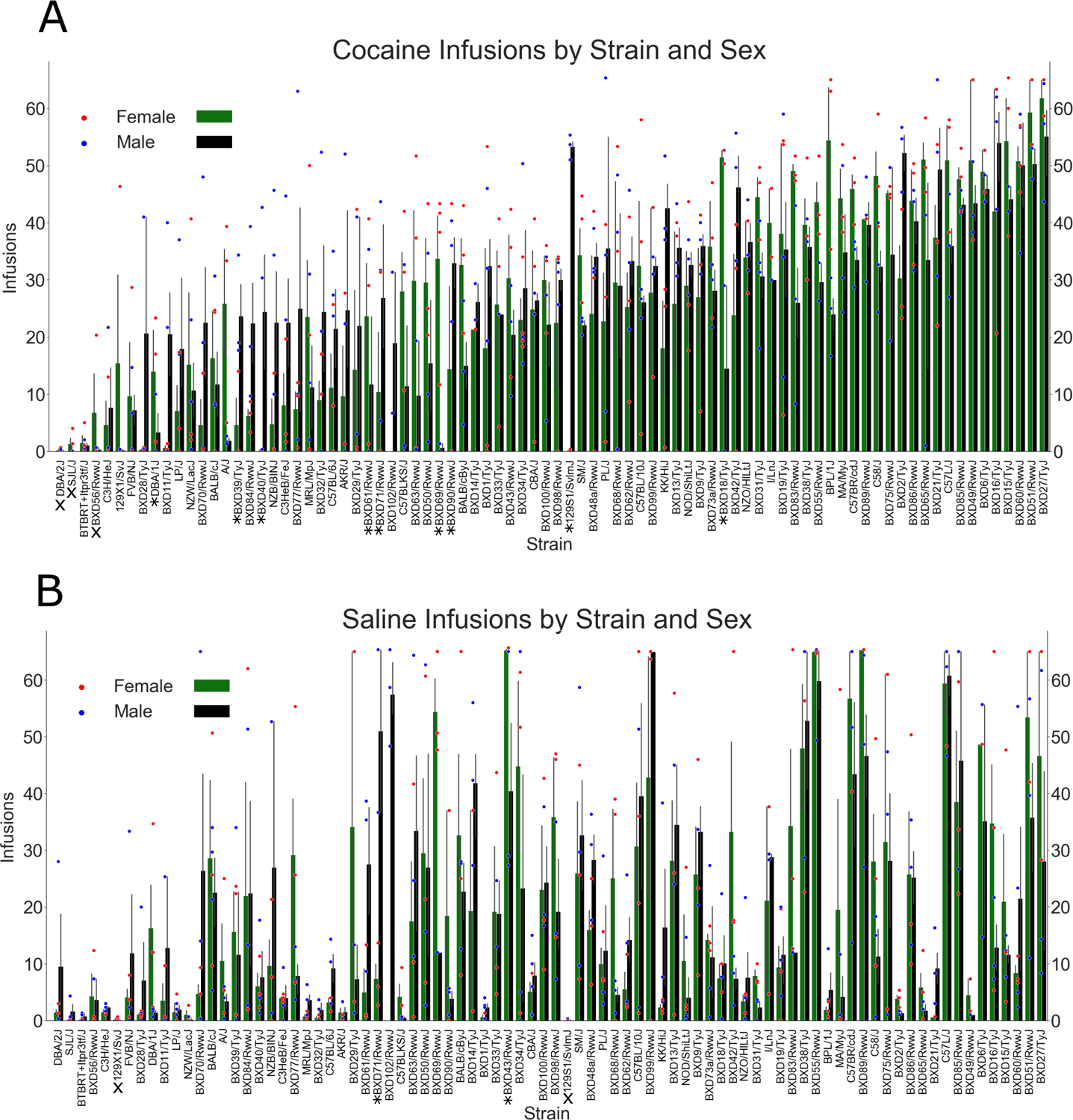
Cocaine and saline taking by strain and sex. (A) Cocaine taking by strain and sex (error bars = SEM, scatter points = individual mouse values). Sex interacted with strain, indicating sex effects are moderated by genetic background. An * below the strain name indicates a significant sex difference within that strain. (B) Saline taking by strain, ranked by levels of cocaine taking, and sex (error bars = SEM, scatter points = individual mouse values). No sex effects were discovered within the saline group. An X below the strain name, in Graphs A and B, indicates that there was no significant difference between the number of infusions and zero for that strain
Although strains demonstrated a broad range of saline taking, post hoc within strain comparisons of the two infusate groups revealed that 14 strains showed significantly different levels of cocaine and saline intake, with a majority of these strains (10 out of 14) having a greater cocaine intake (Figure 4A).
Analysis within each infusate group revealed a strain-sex interaction in the cocaine group (Figure 5A) (F[82, 312] = 1.4, p = 0.037} but not the saline group (F[82, 310] = 0.8, p = 0.940_ (Figure 5B). Post hoc analyses for sex differences, within strain, in the cocaine group revealed that nine strains demonstrated a significant sex difference, with males exhibiting greater cocaine intake in five out of nine strains (see Table S1).
3.3 |. Genetic correlation between cocaine and saline IVSA infusions
To assess the genetic correlation between cocaine and saline IVSA, a strain-level Pearson’s correlation was performed on strain means, examining the average number of infusions earned in Sessions 8–10 on cocaine and saline groups. There was a significant, positive association between IVSA of cocaine and saline (r = 0.58, p < 0.001), revealing that a modest ~33% of the variance was shared between the two infusate group conditions (Figure 4B).
3.4 |. Lever preference
Preference for the active lever (active presses/total presses) was also assessed. The active lever triggers infusion of cocaine or saline and the active lever preference variable reveals whether lever pressing is random (≤50%) or reinforcer-directed (>50%). Because this variable often cannot be calculated early in training (when there are sessions with no presses on either lever), we focused on the later testing sessions (8–10).
The preference ratio was significantly greater in the cocaine group (main effect of infusate group [F{1, 567} = 102.3, p < 0.001]), and lever preference differed by strain (main effect of strain [F{83, 567} = 1.3, p = 0.033]) (Figure 6A).
FIGURE 6.
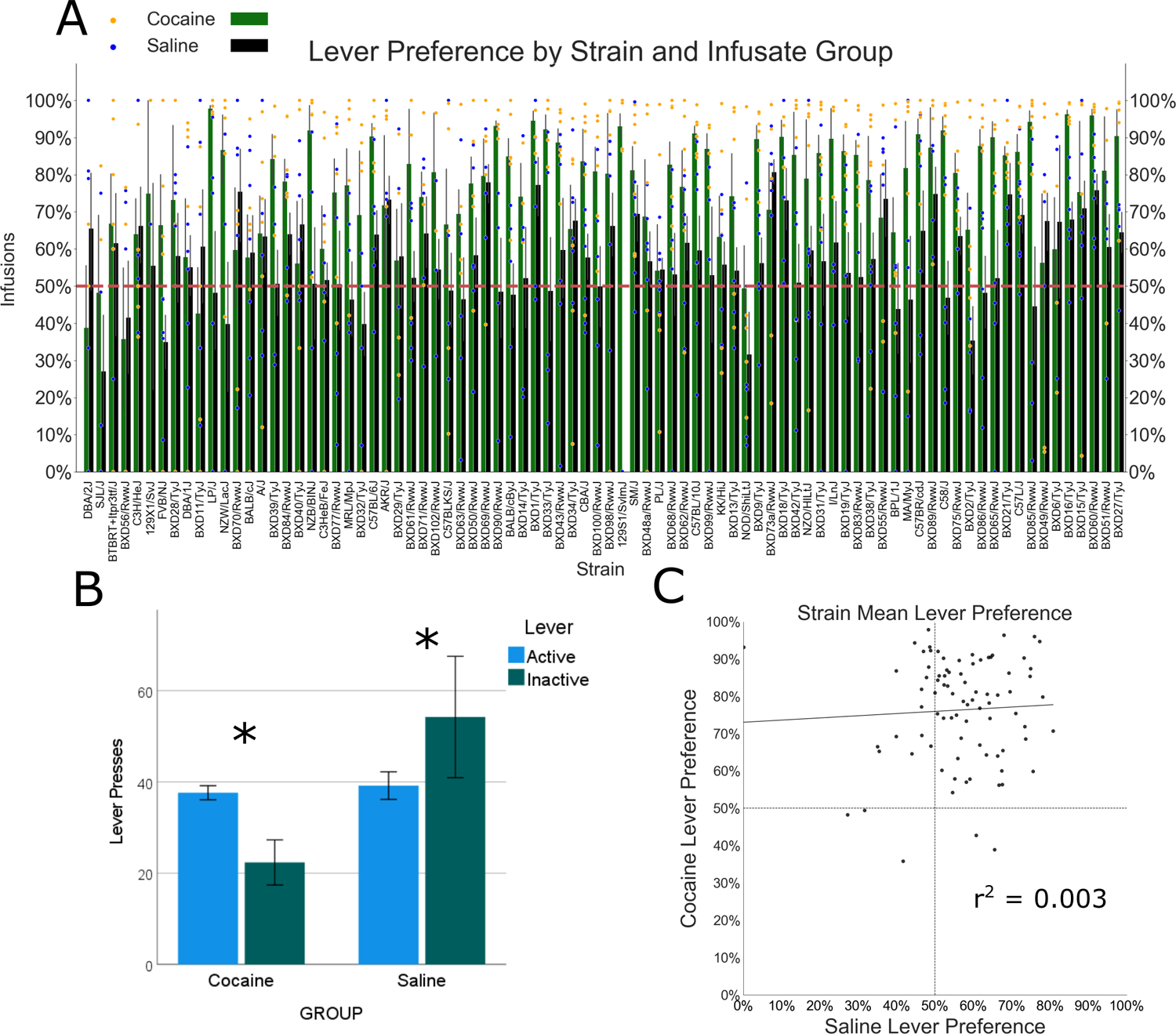
Lever discrimination in the last three sessions of intravenous self-administration (IVSA). (A) Active lever preference by strain and infusate group (error bars = SEM, scatter points = individual mouse values). Lever preference was greater in the cocaine group. (B) Pressing on the active and inactive lever within the cocaine and saline groups. The cocaine group demonstrated greater active lever presses; the saline group showed greater inactive presses. *p < 0.05 for difference between active and inactive lever pressing. (C) Strain mean lever preference. No correlation between saline and cocaine lever preference was discovered
A strain-level correlation was performed to assess genetic relationships between lever preference between cocaine and saline IVSA groups. No significant correlation was found (r = 0.052, p = 0.636) (Figure 6C).
Differences in lever pressing between the active and inactive lever averaged across Sessions 8–10 were assessed by mixed ANOVA with lever as a within subject variable and strain, sex, and infusate group as between-subject factors. A main effect of lever (F[1, 622] = 4.3, p = 0.04) indicated that lever pressing differed between active and inactive levers; however, a lever–infusate group interaction (F[1, 622] = 4.1, p < 0.001) suggested lever discrimination varied between the two infusate groups (Figure 6B). Within group analysis revealed more presses on the active than inactive lever in the cocaine group (F[1, 312] = 41.9, p < 0.001). A strain-sex interaction (F[82, 312] = 1.5, p = 0.012) and a main effect of strain (F[83, 312] = 3.6, p < 0.001) were also found within the cocaine group. In contrast to cocaine, the inactive lever was pressed more than the active in the saline group (F[1, 310] = 13.8, p < 0.001). A main effect of strain (F[83, 310] = 1.7, p < 0.001) indicated that overall lever pressing, irrespective of infusate group, was impacted by strain.
3.5 |. Genome-wide association studies
Cocaine and saline infusion data were binned into 2-day intervals for all 10 days of IVSA (five bins) and subject to GWAS with faST-LMM. A significant QTL on chromosome (chr) 14 (55.088089–57.688089 Mb) was detected for the number of cocaine infusions earned during Days 5 and 6 and 7 and 8 and on chr 3 (50.406218–53.006218 Mb) for Days 5 and 6, 7 and 8 and 9 and 10 (Figure 7A). A suggestive QTL was detected on chr 3 (35.474653–38.074653 Mb) for Days 5 and 6 (Figure 7A). No significant QTL were detected for saline (Figure 7B), suggesting that the cocaine QTL may specifically moderate self-administration of the drug.
FIGURE 7.
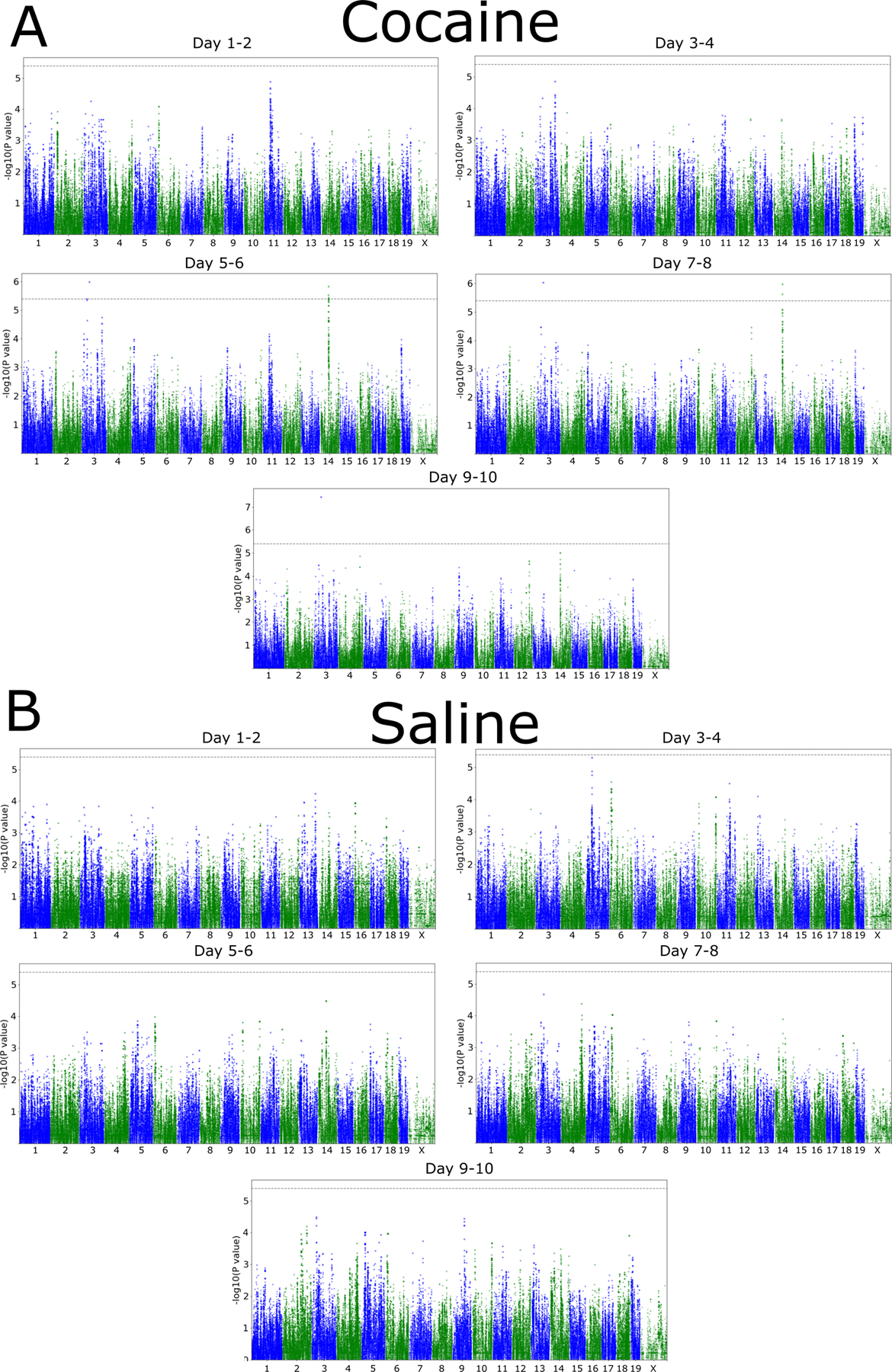
Manhattan plots for genome-wide association studies (GWAS) of infusions earned in 2-day bins over 10 days of IVSA (dotted line = genome-wide p < 0.05). (A) GWAS for cocaine infusions earned. A significant QTL on chromosome (chr) 14 was found for Days 5 and 6 and 7 and 8 (55 088 089-57 688 089 Mb) and on chr 3 for Days 5 and 6, 7 and 8 and 9 and 10 (50 406 218-53 006 218 Mb). A highly suggestive quantitative trait loci (QTL) on chr 3 was found for Days 5 and 6 (50 406 218-53 006 218 Mb). (B) GWAS for saline infusions earned. No significant QTL were detected
3.6 |. Candidate gene selection using RNA sequencing data
Whole transcriptome RNA sequencing data were collected from NAc and mFC tissue of both infusate groups in a subset of the strains (n = 41). We reasoned that strain-level variation in expression of one or more cis-regulated genes expressed from the chr 14 or 3 QTL would predict strain-level variation in cocaine-taking behaviour, supporting its prioritization as a candidate gene. The chr 14 QTL contained 93 positional candidates, and 24 of those genes exhibited significant cis-eQTL (Table 2). These genes were assessed for correlation of strain mean expression to the cocaine infusion traits (Days 5 and 6 and 7 and 8). Four genes (Gjb6, Mphosph8, Nop9 and Zmym5) had expression levels in NAc or mFC that were significantly correlated to the strain mean for cocaine infusions earned (Table 3). All but Nop9 also demonstrated significant correlations to saline infusions.
TABLE 2.
Genes in cocaine infusion QTL intervals with cis-eQTL Abbreviation: QTL, quantitative trait loci.
| Gene | SNP | Position (base) | PValue |
|---|---|---|---|
| Chromosome 14 QTL (55088089–57688089 Mb) | |||
| 1700039M10Rik | rs51263087 | 57489083 | 0.00095225 |
| A730061H03Rik | rs13466742 | 55569551 | 0.00063818 |
| Adcy4 | rs31504942 | 55430117 | 5.5884E-06 |
| Atp12a | rs30459253 | 55892786 | 3.5173E-06 |
| Cbln3 | rs30171265 | 56758423 | 1.6125E-05 |
| Cenpj | rs50348120 | 56057592 | 0.005584 |
| Cma1 | rs48616584 | 56378025 | 0.00058376 |
| Ctsg | rs49142937 | 56757039 | 0.00020934 |
| Dhrs4 | rs51685240 | 56281873 | 0.0010645 |
| Eef1akmt1 | rs48220977 | 56380012 | 0.00060539 |
| Fitm1 | rs51815357 | 55236235 | 0.0002076 |
| Gja3 | rs46981153 | 55667604 | 0.00040251 |
| Gjb2 | rs47936586 | 56488999 | 0.00063947 |
| Gjb6 | rs30321242 | 55195266 | 0.00390434 |
| Khnyn | rs30494831 | 55785585 | 0.00063033 |
| Mcpt2 | rs50348120 | 56057592 | 1.4081E-05 |
| Mcpt-ps1 | rs49142937 | 56757039 | 0.00199293 |
| Mphosph8 | rs48814297 | 56745552 | 0.00018757 |
| Nfatc4 | rs49877866 | 55208920 | 0.00093986 |
| Nop9 | rs30158589 | 55602148 | 0.00062249 |
| Nrl | rs51166292 | 56619218 | 0.0001555 |
| Rnf17 | rs30171265 | 56758423 | 1.0878E-05 |
| Tssk4 | rs48814297 | 56745552 | 0.00083562 |
| Zmym5 | rs30158589 | 55602148 | 0.0021784 |
| Chromosome 3 QTL (50406218–53006218 Mb) | |||
| 5830415G21Rik | rs30006707 | 50528633 | 2.0826E-06 |
| Gm20750 | rs51015053 | 52526207 | 0.00060342 |
| Gm2447 | rs30318630 | 52978064 | 0.00216844 |
| Mgst2 | rs51491672 | 51777632 | 4.3354E-05 |
| Ndufc1 | rs6327143 | 50851828 | 0.00250785 |
| Noct | rs48461564 | 51681793 | 0.00452224 |
| Setd7 | rs30061622 | 50484160 | 0.0019422 |
| Chromosome 3 suggestive QTL (35474653–38074653 Mb) | |||
| 1810062G17Rik | rs30628771 | 35988796 | 0.00016971 |
| 4932438A13Rik | rs29940534 | 36549366 | 0.00025951 |
| 8430422M14Rik | rs30682559 | 35989005 | 7.3436E-06 |
| Adad1 | rs30000745 | 36997342 | 3.283E-05 |
| Anxa5 | rs31048427 | 35996039 | 0.00039342 |
| Bbs12 | rs30066305 | 37388360 | 4.098E-05 |
| Bbs7 | rs30134973 | 37967875 | 2.1414E-05 |
| Ccdc144b | rs30940497 | 36078026 | 0.00030446 |
| Ccna2 | rs30115173 | 37791344 | 8.4813E-05 |
| Cetn4 | rs3146895 | 36601849 | 2.3154E-05 |
| D3Ertd254e | rs3158450 | 37532093 | 1.8564E-06 |
| Exosc9 | rs3151914 | 36599358 | 2.2197E-06 |
| Gm11549 | rs29965498 | 36638158 | 0.00049008 |
| Gm5148 | rs3694769 | 36764538 | 4.6464E-07 |
| Il2 | rs30115173 | 37791344 | 0.00010122 |
| Il21 | rs3146895 | 36601849 | 6.0003E-05 |
| Mccc1 | rs36586121 | 35988926 | 0.00022018 |
| Mccc1os | rs36586121 | 35988926 | 6.4348E-11 |
| Mir7009 | rs50498826 | 36202521 | 1.1785E-05 |
| Nudt6 | rs31006974 | 36514604 | 0.00020501 |
| Qrfpr | rs29930642 | 36347389 | 0.0004952 |
| Spata5 | rs3158450 | 37532093 | 0.00048346 |
TABLE 3.
Genes with cis-eQTL and genetic correlation to cocaine infusions
| Gene | Tissue Region | Tissue Group | IVSA Days | Correlation to Cocaine Infusions (r, p) | Correlation to Saline Infusions (r, p) | ePheWAS Results |
|---|---|---|---|---|---|---|
| Chromosome 14 QTL (55088089–57688089 Mb) | ||||||
| Gjb6 | mFC | Cocaine | 7–8 | 0.53, 0.000458* | 0.28, 0.088546 | none |
| 5–6 | 0.56, 0.000184* | 0.35, 0.028524 | ||||
| Difference Score | 7–8 | 0.6, 0.000051* | 0.45, 0.003659* | |||
| 5–6 | 0.55, 0.000288* | 0.53, 0.000549* | ||||
| NAc | Difference Score | 7–8 | 0.54, 0.000385* | 0.17, 0.298484 | ||
| 5–6 | 0.4, 0.012292* | 0.26, 0.108479 | ||||
| Mphosph8 | mFC | Saline | 7–8 | −0.52, 0.000766* | −0.56, 0.000188* | none |
| 5–6 | −0.53, 0.000492* | −0.56, 0.000194* | ||||
| NAc | Difference Score | 7–8 | 0.53, 0.000544* | 0.44, 0.004742* | ||
| 5–6 | 0.53, 0.000543* | 0.54, 0.00035* | ||||
| Saline | 7–8 | −0.52, 0.000687* | −0.53, 0.000473* | |||
| 5–6 | −0.52, 0.000609* | −0.6, 0.000053* | ||||
| Nop9 | mFC | Cocaine | 7–8 | 0.44, 0.005564* | 0.32, 0.046507 | BXD Epoch of production (GN12689) |
| NAc | Difference Score | 7–8 | 0.38, 0.016632* | 0.25, 0.117628 | ||
| Zmym5 | mFC | Difference Score | 7–8 | 0.45, 0.003662* | 0.3, 0.059557 | none |
| 5–6 | 0.43, 0.006051* | 0.33, 0.039581 | ||||
| Saline | 7–8 | −0.55, 0.000282* | −0.2, 0.219552 | |||
| 5–6 | −0.53, 0.000578* | −0.43, 0.006124* | ||||
| NAc | Difference Score | 7–8 | 0.46, 0.002915* | 0.48, 0.00211* | ||
| 5–6 | 0.47, 0.002322* | 0.55, 0.000277* | ||||
| Saline | 7–8 | −0.51, 0.000927* | −0.42, 0.007298* | |||
| 5–6 | −0.49, 0.001555* | −0.53, 0.000545* | ||||
| Chromosome 3 QTL (50406218–53006218 Mb) | ||||||
| Gm20750 | mFC | Cocaine | 5–6 | −0.34, 0.031558* | −0.37, 0.021814 | none |
| 7–8 | −0.34, 0.033746* | −0.37, 0.020983 | ||||
| NAc | Difference Score | 5–6 | 0.34, 0.033408* | 0.42, 0.00828* | ||
| 7–8 | 0.34, 0.033736* | 0.4, 0.012112* | ||||
| Saline | 7–8 | −0.44, 0.005551* | −0.22, 0.171097 | |||
| Ndufc1 | mFC | Cocaine | 9–1 | 0.44, 0.005277* | 0.21, 0.193813 | BXD Epoch of production (GN12689) |
| 5–6 | 0.42, 0.007577* | 0.22, 0.173639 | ||||
| 7–8 | 0.47, 0.00235* | 0.21, 0.194256 | ||||
| Difference Score | 9–1 | 0.55, 0.000257* | 0.27, 0.095173 | |||
| 5–6 | 0.51, 0.000942* | 0.36, 0.023188 | ||||
| 7–8 | 0.58, 0.000126* | 0.31, 0.053681 | ||||
| NAc | Cocaine | 9–1 | 0.51, 0.000831* | 0.23, 0.161079 | ||
| 5–6 | 0.53, 0.000569* | 0.36, 0.022425* | ||||
| 7–8 | 0.58, 0.000108* | 0.3, 0.06051 | ||||
| Difference Score | 9–1 | 0.5, 0.001244* | 0.23, 0.161079 | |||
| 5–6 | 0.44, 0.004634* | 0.33, 0.041757* | ||||
| 7–8 | 0.52, 0.000658* | 0.29, 0.070091 | ||||
| Noct | NAc | Cocaine | 9–1 | 0.35, 0.027096* | −0.1, 0.564003 | Dock1 (D2 medium spiny neuron expression signature)(GN15553), D1 expression (GN15185), Dopamine receptor ratio (Drd1 to Drd2)(GN15554), Tibia gracility index (GN18419), Ebf1 (D1 medium spiny neuron expression signature)(GN15552) |
| 5–6 | 0.41, 0.010415* | 0.34, 0.033079* | ||||
| 7–8 | 0.44, 0.005052* | 0.26, 0.106702 | ||||
| Difference Score | 5–6 | 0.31, 0.054767* | 0.23, 0.165352 | |||
| 7–8 | 0.32, 0.050066* | 0.23, 0.155525 | ||||
| Setd7 | mFC | Cocaine | 5–6 | −0.39, 0.013922* | −0.26, 0.114428 | Activity in closed quadrants using an elevated zero maze(GN12354), BXD Epoch of production (GN12689) |
| 7–8 | −0.37, 0.021821* | −0.15, 0.358531 | ||||
| NAc | Difference Score | 9–1 | 0.42, 0.007962* | 0.45, 0.004333* | ||
| 5–6 | 0.53, 0.000501* | 0.47, 0.002436* | ||||
| 7–8 | 0.54, 0.000369* | 0.35, 0.026742* | ||||
| Chromosome 3 suggestive QTL (35474653–38074653 Mb) | ||||||
| 4932438A13Rik | NAc | Difference Score | 5–6 | 0.41, 0.009087* | 0.58, 0.000094* | none |
| mFC | Saline | 5–6 | −0.39, 0.015236* | −0.51, 0.000796* | ||
| 8430422M14Rik | NAc | Difference Score | 5–6 | 0.4, 0.012174* | 0.17, 0.30793 | none |
| D3Ertd254e | mFC | Cocaine | 5–6 | −0.47, 0.00241* | −0.52, 0.000769* | none |
| Saline | 5–6 | −0.49, 0.001601* | −0.58, 0.000123* | |||
| NAc | Saline | 5–6 | −0.47, 0.002594* | −0.68, 0.000002* | ||
| Exosc9 | mFC | Difference Score | 5–6 | 0.4, 0.012199* | 0.36, 0.025473 | none |
| NAc | 5–6 | 0.46, 0.003264* | 0.38, 0.015552* | |||
| Spata5 | mFC | Difference Score | 5–6 | 0.41, 0.008782* | 0.41, 0.009916 | D1 expression(GN15185), Dopamine receptor ratio (Drd1 to Drd2)(GN15554), Ebf1 (D1 medium spiny neuron expression signature)(GN15552) |
| Saline | 5–6 | −0.48, 0.002111* | 0.27, 0.100151 | |||
| NAc | Difference Score | 5–6 | 0.5, 0.001247* | 0.62, 0.000031* | ||
Abbreviations: IVSA, intravenous self-administration; mFC, medial frontal cortex; NAc, nucleus accumben; QTL, quantitative trait loci.
indicates significance after correction for multiple comparisons.
The chr 3 QTL contains 39 positional candidates, of which seven exhibited a significant cis-eQTL (Table 2). Four of these genes (Gm20750, Ndufc1, Noct and Setd7) had expression levels that genetically correlated to cocaine infusions (Table 3). All but Setd7 also demonstrated significant correlations to saline infusions.
The chr 3 suggestive QTL contained 43 positional candidates and 22 with significant cis-eQTL (Table 2). Five of these genes (4932438A13Rik, 8430422M14Rik, D3Ertd254e, Exosc9 and Spata5) had expression levels that genetically correlated to cocaine infusions (Days 5 and 6) (Table 3). All but 8430422M14Rik also demonstrated significant correlations to saline infusions.
All genes that demonstrated both cis-eQTL and genetic correlation to cocaine infusions were investigated for genetic correlation to all traits in the phenome database of genenetwork.org30 using the ePheWAS analysis tool provided at systems-genetics.org.31 The striatum (GN285) and mFC (GN135) array datasets were utilized. Expression levels of the genes Noct and Spata5, in striatal tissue, significantly associate with measures of dopamine receptor 1 and 2 expressions. Noct expression in the striatum also associates with tibia gracility (GN18419). Setd7 expression in the mFC associates with activity in elevated zero maze (GN12689). Setd7I, Ndufc1 and Nop9 expression in mFC associates with the epoch of BXD production (GN12689). See Table 3 for a summary of these results.
4 |. DISCUSSION
We found that cocaine IVSA is substantially heritable in the mouse genetic reference population of the HMDP, with more than half of all phenotypic variation being explained by strain. Strain-level infusions demonstrated a broad and continuous range of cocaine intake, from near zero to ~25 mg/kg/session. Given that the HMDP is suited for forward genetics analysis,22,23 we conducted GWAS to identify QTL associated with cocaine taking. Furthermore, individual strains can be identified as extreme responders and may serve as models of polygenic risk for high-level cocaine intake (see Table S2 for lowest and highest three strains for each trait). These models may also be utilized to investigate the neurobiological mediators that underlie extreme differences in cocaine IVSA responding.
Mice were tested in 10 daily sessions for cocaine or saline IVSA. We found that IVSA behaviour changed over the 10 sessions in a strain-dependent manner. Many strains in the cocaine group increased their levels of intake over the sessions, consistent with experience-dependent acquisition of operant responding. Some strains did not increase intake to the same degree. or at all, and consequently, the variability in intake between strains increased as the training sessions progressed. Furthermore, correlations between sessions, at the individual mouse-level and strain-level, were assessed, revealing that the magnitude of intersession correlations increased across sessions and that strain-level variance was highly stable. In the final sessions (8–10), 78 of the strains demonstrated cocaine intake that was statistically significantly greater than zero. These results reveal genetically moderated operant responding for an intravenous cocaine reinforcer. A similar pattern of effects was observed in the saline infusate group, including both strain-dependent increases in responding across the sessions and greater intersession stability in the final sessions. Notably, 14 of the strains exhibited significantly different intake of cocaine compared with saline; given the modest number of mice per strain per infusate condition studied here, this is likely an underestimate of the real differences in cocaine and saline taking behaviour. Ten, 2-h sessions are a relatively modest duration of testing that was selected to capture genetic regulation of cocaine-induced differential transcript expression at an early IVSA timepoint (these analyses will be the subject of a future manuscript) and to ensure sufficient catheter patency rates. Additional testing sessions or alternative IVSA paradigms (extinction, reinstatement and choice procedures) may reveal additional genetic information.
The HMDP is a useful population for genetic association studies due to genetic diversity and high recombination that allows for relatively fine resolution mapping of QTL. Here, we identified significant QTL for cocaine infusions on chr 14 and 3 and a suggestive QTL on chr 3. These QTL were not detected in the saline group, suggesting that the effects of these loci are specific to cocaine self-administration. The detection of these QTL in behavioural data gathered in the latter, but not earlier, half of the training sessions suggests that the effects of this QTL may be most prominent after acquisition has progressed. These QTL are a critical step in identifying the genes that moderate cocaine intake.
The chr 14 QTL, chr 3 QTL and chr 3 suggestive QTL intervals contained 93, 39 and 43 positional candidate genes, respectively. To prioritize positional candidates, we identified genes exhibiting cis-eQTL and genetic correlation to cocaine infusions. These efforts narrowed the list of candidates to 13 genes across all behavioural QTL. These genes were further investigated for genetic correlation to relevant phenotypes by ePHeWAS on systems-genetics.org. Two genes, Spata5 and Noct, demonstrate ePheWAS associations to dopamine receptor expression in the striatum and may be considered as priority candidates given the role of striatal dopamine in cocaine IVSA.32 The gene Gjb6 is reported to impact anxiety measures and neuromodulator levels, including dopamine, serotonin and acetylcholine, in a mouse knock out model33 and may be considered as a candidate considering the comorbidity of anxiety disorders and SUDs.34 In addition to cis-eQTL analysis, other approaches may aid in prioritizing candidates, including identification of trans-eQTL hotspots within significant QTLs. Trans-eQTL tend to have smaller effect sizes than cis-eQTL.35 After more strains have been sequenced, we intend to examine this issue. Additionally, cell-type-specific RNA sequencing may reveal richer information. Bulk tissue sequencing was used here to maintain throughput and ensure a sufficient number of strains could be evaluated. However, all remaining brain tissue has been stored so that future studies may elect other approaches in additional brain regions of interest.
Saline and cocaine IVSA are both heritable phenotypes in the HMDP; however, the genetic correlation between cocaine to saline IVSA was modest (~33% shared variance), with many strains demonstrating disparate cocaine and saline intake. Where intake differed within strain, cocaine IVSA was most often found to be greater than saline IVSA. These results suggest that some common genetic factors may influence self-administration of either infusate; however, those are less impactful than the unique genetic factors that influence intake of one infusate, but not the other. These saline data may be utilized to increase precision when measuring genetic influences on cocaine-specific reinforcement. Furthermore, these data suggest that some strains demonstrate equivalent cocaine and saline intake or greater intake of saline; with high levels of intake for both infusate types in some strains (e.g., C57L/J). Previous research found similar effects, with high, strain-dependent lever pressing in the absence of cocaine delivery and poor lever discrimination in the same strain during cocaine IVSA.17 The degree to which high levels of cocaine taking in these strains reflects cocaine-seeking may be unclear, and these outcomes highlight the importance of including saline controls, particularly when investigating strains for the first time.
Lever pressing and infusions earned may be the result of either associative or non-associative mechanisms. We found evidence that infusion-reinforced lever pressing occurred to a greater degree in the cocaine group. In contrast, the inactive lever was pressed more, on average, in the saline group. In both infusate groups, activation of the active lever triggers a potentially perceptible intravenous infusion and a flashing house light; it is possible that either serves as a reinforcer or affects behavioural arousal. Similar discrete light stimuli were sufficient to reinforce instrumental actions in the BXD population, and this trait demonstrated heritability.36,37 However, active lever preference was lower in the saline group in the present study, so reinforcement due to perceptible cues after an active lever press may not be a prominent motivator of responding for saline infusions. Alternatively, lever pressing in the saline infusate group may be the consequence of other phenotypes (e.g., increased exploratory motor activity in the test chamber, increased wakefulness, etc.). Taken together, these results suggest that there are some genetic variants that affect the potential to engage in lever pressing per se, with an additional set of variants affecting responding when cocaine is available. While beyond the scope of this current study, future efforts may further dissect the unique and shared variance between cocaine and saline taking and apply forward genetic analyses in order to further dissect the genetics of these traits.
Sex was found to moderate strain effects on responding for cocaine, but not saline. Sex effects have been previously reported for mouse and rat cocaine IVSA.38–43 Most often, females are found to have greater cocaine intake or faster acquisition. Here, we found that sex effects were highly strain-dependent and, in strains that demonstrate a sex effect, the direction of the effect is also strain-dependent. These results suggest that sex effects interact with genetic background to determine the presence and direction of the effect. The HMDP may be utilized to identify the genetic variants that interact with sex to affect cocaine IVSA. Investigation of gene–sex hormone interactions and sex chromosome complement may be especially informative as both can have a large impact on cocaine taking and account for some of the sex differences in cocaine IVSA observed in rodent populations.42,44–50
Our data demonstrate that cocaine IVSA is a highly heritable trait in the HMDP and that it is mostly genetically distinct from saline IVSA. We have identified a QTL associated with cocaine infusions and utilized our gene expression data to prioritize positional candidate genes. Ongoing efforts will continue to dissect the genetic of cocaine self-administration and validate prospective candidate genes. This research may contribute to a better understanding of cocaine use disorder genetics and lead to more effective treatments.
Supplementary Material
ACKNOWLEDGEMENTS
These studies were supported, in part, by National Institute on Drug Abuse grants U01-DA041602 (DJS, JDJ) and P50-DA039841(JDJ) and National Institute on Alcohol Abuse and Alcoholism T32-AA025606 (JDJ and JRB). We would like to thank Barbara Force, Anthony Corsi and Jesse Filippazzo for their assistance and technical support.
Funding information
National Institute on Alcohol Abuse and Alcoholism, Grant/Award Number: T32-AA025606; National Institute on Drug Abuse, Grant/Award Numbers: P50-DA039841, U01-DA041602
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
DATA AVAILABILITY STATEMENT
The RNA sequencing data generated in this study are publicly available from NCBI GEO (https://www.ncbi.nlm.nih.gov/geo) under accession number GSE186981 and from the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under SRA accession number PRJNA755328.
REFERENCES
- 1.Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6(7):521–532. doi: 10.1038/nrg1635 [DOI] [PubMed] [Google Scholar]
- 2.Kendler KS, Karkowski LM, Neale MC, Prescott CA. Illicit psychoactive substance use, heavy use, abuse, and dependence in a US population-based sample of male twins. Arch Gen Psychiatry. 2000;57(3):261–269. doi: 10.1001/archpsyc.57.3.261 [DOI] [PubMed] [Google Scholar]
- 3.Cabana-Domínguez J, Shivalikanjli A, Fernàndez-Castillo N,Cormand B. Genome-wide association meta-analysis of cocaine dependence: Shared genetics with comorbid conditions. ProgNeuroPsychopharmacol Biol Psych. 2019;94:109667. doi: 10.1016/j.pnpbp.2019.109667 [DOI] [PubMed] [Google Scholar]
- 4.Gelernter J, Sherva R, Koesterer R, et al. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Molec Psychiat. 2014;19(6):717–723. doi: 10.1038/mp.2013.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marees AT, Gamazon ER, Gerring Z, et al. Post-GWAS analysis of six substance use traits improves the identification and functional interpretation of genetic risk loci. Drug Alcohol Depend. 2020;206:107703.doi: 10.1016/j.drugalcdep.2019.107703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones BC, Tarantino LM, Rodriguez LA, et al. Quantitative-trait loci analysis of cocaine-related behaviours and neurochemistry. Pharmacogenetics. 1999;9(5):607–617. doi: 10.1097/00008571-199910000-00007 [DOI] [PubMed] [Google Scholar]
- 7.Kumar V, Kim K, Joseph C, et al. C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science. 2013;342(6165):1508–1512. doi: 10.1126/science.1245503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phillips TJ, Huson MG, McKinnon CS. Localization of genes mediating acute and sensitized locomotor responses to cocaine in BXD/Ty recombinant inbred mice. J Neurosci. 1998;18(8):3023–3034. doi: 10.1523/JNEUROSCI.18-08-03023.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tolliver BK, Carney JM. Sensitization to stereotypy in DBA/2J but not C57BL/6J mice with repeated cocaine. Pharmacol Biochem Behav. 1994;48(1):169–173. doi: 10.1016/0091-3057(94)90513-4 [DOI] [PubMed] [Google Scholar]
- 10.Boyle AE, Gill KJ. A verification of previously identified QTLs for cocaine-induced activation using a panel of B6.A chromosome substitution strains (CSS) and A/J × C57Bl/6J F2 mice. Psychopharmacology(Berl). 2009;207(2):325–334. doi: 10.1007/s00213-009-1656-7 [DOI] [PubMed] [Google Scholar]
- 11.Bagley JR, Adams J, Bozadjian RV, Bubalo L, Kippin TE. Strain differences in maternal neuroendocrine and behavioral responses to stress and the relation to offspring cocaine responsiveness. Int J Dev Neurosci. 2019;78:130–138. doi: 10.1016/j.ijdevneu.2019.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bagley JR, Szumlinski KK, Kippin TE. Discovery of early life stress interacting and sex-specific quantitative trait loci impacting cocaine responsiveness. Br J Pharmacol. Published online 2019, 176, 21, 4159, 4172. doi: 10.1111/bph.14661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cervantes MC, Laughlin RE, Jentsch JD. Cocaine self-administration behavior in inbred mouse lines segregating different capacities for inhibitory control. Psychopharmacology (Berl). 2013;229(3):515–525. doi: 10.1007/s00213-013-3135-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickson PE, Ndukum J, Wilcox T, et al. Association of novelty-related behaviors and intravenous cocaine self-administration in diversity outbred mice. Psychopharmacology (Berl). 2014;232(6):1011–1024. doi: 10.1007/s00213-014-3737-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickson PE, Miller MM, Calton MA, et al. Systems genetics of intravenous cocaine self-administration in the BXD recombinant inbred mouse panel. Psychopharmacology (Berl). Published online November 19, 2015;233(4):701–714. doi: 10.1007/s00213-015-4147-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kippin TE, Campbell JC, Ploense K, Knight CP, Bagley J. Prenatal stress and adult drug-seeking behavior: interactions with genes and relation to nondrug-related behavior. Adv Neurobiol. 2015;10:75–100. doi: 10.1007/978-1-4939-1372-5_5 [DOI] [PubMed] [Google Scholar]
- 17.Roberts AJ, Casal L, Huitron-Resendiz S, Thompson T, Tarantino LM. Intravenous cocaine self-administration in a panel of inbred mouse strains differing in acute locomotor sensitivity to cocaine. Psychopharmacology (Berl). 2018;235(4):1179–1189. doi: 10.1007/s00213-018-4834-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saul MC, Bagley JR, Bailey LS, et al. Consideration of genetic and sex effects in mice enhances consilience with human addiction studies. bioRxiv. 2020. doi: 10.1101/2020.02.14.949784 [DOI] [Google Scholar]
- 19.Laughlin RE, Grant TL, Williams RW, Jentsch JD. Genetic dissection of behavioral flexibility: reversal learning in mice. Biol Psychiatry. 2011; 69(11):1109–1116. doi: 10.1016/j.biopsych.2011.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belin-Rauscent A, Fouyssac M, Bonci A, Belin D. How preclinical models evolved to resemble the diagnostic criteria of drug addiction. Biol Psychiatry. 2016;79(1):39–46. doi: 10.1016/j.biopsych.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connor EC, Chapman K, Butler P, Mead AN. The predictive validity of the rat self-administration model for abuse liability. Neurosci Bio behav Rev. 2011;35(3):912–938. doi: 10.1016/j.neubiorev.2010.10.012 [DOI] [PubMed] [Google Scholar]
- 22.Bennett BJ, Farber CR, Orozco L, et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20(2):281–290. doi: 10.1101/gr.099234.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghazalpour A, Rau CD, Farber CR, et al. Hybrid mouse diversity panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mamm Genome. 2012;23(9):680–692. doi: 10.1007/s00335-012-9411-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hasin-Brumshtein Y, Khan AH, Hormozdiari F, et al. Hypothalamic transcriptomes of 99 mouse strains reveal trans eQTL hotspots, splicing QTLs and novel non-coding genes. Elife. 2016;5:e15614. doi: 10.7554/eLife.15614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews S FastQC: a quality control tool for high throughput sequence data. Published online 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc
- 26.Howe KL, Achuthan P, Allen J, et al. Ensembl 2021. Nucleic Acids Res. 2021;49(D1):D884–D891. doi: 10.1093/nar/gkaa942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8(10):833–835. doi: 10.1038/nmeth.1681 [DOI] [PubMed] [Google Scholar]
- 30.Mulligan MK, Mozhui K, Prins P, Williams RW. GeneNetwork: a toolbox for systems genetics. In: Schughart K, Williams RW, eds. Systems Genetics: Methods and Protocols. Springer; 2017:75–120. doi: 10.1007/978-1-4939-6427-7_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Wang X, Rukina D, et al. An integrated systems genetics and omics toolkit to probe gene function. Cell Syst. 2018;6(1):90–102.e4. doi: 10.1016/j.cels.2017.10.016 [DOI] [PubMed] [Google Scholar]
- 32.Wise RA, Robble MA. Dopamine and addiction. Annu Rev Psychol. 2020;71(1):79–106. doi: 10.1146/annurev-psych-010418-103337 [DOI] [PubMed] [Google Scholar]
- 33.Dere E, Souza-Silva MAD, Frisch C, et al. Connexin30-deficient mice show increased emotionality and decreased rearing activity in the open-field along with neurochemical changes. Eur J Neurosci. 2003;18(3):629–638. doi: 10.1046/j.1460-9568.2003.02784.x [DOI] [PubMed] [Google Scholar]
- 34.Regier DA, Rae DS, Narrow WE, Kaelber CT, Schatzberg AF. Prevalence of anxiety disorders and their comorbidity with mood and addictive disorders. Br J Pharmacol. 1998;173(S34):24–28. doi: 10.1192/S0007125000293483 [DOI] [PubMed] [Google Scholar]
- 35.Pierce BL, Tong L, Chen LS, et al. Mediation analysis demonstrates that trans-eQTLs are often explained by cis-mediation: a genome-wide analysis among 1,800 South Asians. PLoS Genet. 2014;10(12):e1004818. doi: 10.1371/journal.pgen.1004818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dickson PE, Mittleman G. Stimulus complexity and mouse strain drive escalation of operant sensation seeking within and across sessions in C57BL/6J and DBA/2J mice. Front Behav Neurosci. 2020;13:286–286. doi: 10.3389/fnbeh.2019.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dickson PE, Roy TA, McNaughton KA, Wilcox TD, Kumar P, Chesler EJ. Systems genetics of sensation seeking. Genes Brain Behav. 2019;18(3):e12519. doi: 10.1111/gbb.12519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carroll ME, Morgan AD, Lynch WJ, Campbell UC, Dess NK. Intravenous cocaine and heroin self-administration in rats selectively bred for differential saccharin intake: phenotype and sex differences. Psychopharmacology (Berl). 2002;161(3):304–313. doi: 10.1007/s00213-002-1030-5 [DOI] [PubMed] [Google Scholar]
- 39.Davis BA, Clinton SM, Akil H, Becker JB. The effects of novelty-seeking phenotypes and sex differences on acquisition of cocaine self-administration in selectively bred high-responder and low-responder rats. Pharmacol Biochem Behav. 2008;90(3):331–338. doi: 10.1016/j.pbb.2008.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jackson LR, Robinson TE, Becker JB. Sex differences and hormonal influences on acquisition of cocaine self-administration in rats. Neuropsychopharmacology. 2005;31(1):129–138. doi: 10.1038/sj.npp.1300778 [DOI] [PubMed] [Google Scholar]
- 41.Kerstetter KA, Aguilar VR, Parrish AB, Kippin TE. Protracted time-dependent increases in cocaine-seeking behavior during cocaine withdrawal in female relative to male rats. Psychopharmacology (Berl). 2008;198(1):63–75. doi: 10.1007/s00213-008-1089-8 [DOI] [PubMed] [Google Scholar]
- 42.Lynch WJ, Arizzi MN, Carroll ME. Effects of sex and the estrous cycle on regulation of intravenously self-administered cocaine in rats. Psychopharmacology (Berl). 2000;152(2):132–139. doi: 10.1007/s002130000488 [DOI] [PubMed] [Google Scholar]
- 43.Roth ME, Carroll ME. Sex differences in the escalation of intravenous cocaine intake following long- or short-access to cocaine self-administration. Pharmacol Biochem Behav. 2004;78(2):199–207. doi: 10.1016/j.pbb.2004.03.018 [DOI] [PubMed] [Google Scholar]
- 44.Bagley JR, Adams J, Bozadjian RV, Bubalo L, Ploense KL, Kippin TE. Estradiol increases choice of cocaine over food in male rats. Physiol Behav. 2019;203:18–24. doi: 10.1016/j.physbeh.2017.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu M, Becker JB. Acquisition of cocaine self-administration in ovariectomized female rats: effect of estradiol dose or chronic estradiol administration. Drug Alcohol Depend. 2008;94(1–3):56–62. doi: 10.1016/j.drugalcdep.2007.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kerstetter KA, Ballis MA, Duffin-Lutgen S, Carr AE, Behrens AM, Kippin TE. Sex differences in selecting between food and cocaine reinforcement are mediated by estrogen. Neuropsychopharmacology. 2012;37(12):2605–2614. doi: 10.1038/npp.2012.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kippin TE, Fuchs RA, Mehta RH, et al. Potentiation of cocaine-primed reinstatement of drug seeking in female rats during estrus. Psychopharmacology (Berl). 2005;182(2):245–252. doi: 10.1007/s00213-005-0071-y [DOI] [PubMed] [Google Scholar]
- 48.Martini M, Irvin JW, Lee CG, Lynch WJ, Rissman EF. Sex chromosome complement influences vulnerability to cocaine in mice. Horm Behav. 2020;125:104821. doi: 10.1016/j.yhbeh.2020.104821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perry AN, Westenbroek C, Becker JB. Impact of pubertal and adult estradiol treatments on cocaine self-administration. Horm Behav. 2013;64(4):573–578. doi: 10.1016/j.yhbeh.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramoa CP, Doyle SE, Naim DW, Lynch WJ. Estradiol as a mechanism for sex differences in the development of an addicted phenotype following extended access cocaine self-administration. Neuropsychopharmacology. 2013;38(9):1698–1705. doi: 10.1038/npp.2013.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data generated in this study are publicly available from NCBI GEO (https://www.ncbi.nlm.nih.gov/geo) under accession number GSE186981 and from the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under SRA accession number PRJNA755328.


