Abstract
Neuroblastoma (NB) is the second leading extracranial solid tumor of early childhood with about two-thirds of cases presenting before the age of 5, and accounts for roughly 15 percent of all pediatric cancer fatalities in the United States. Treatments against NB are lacking, resulting in a low survival rate in high-risk patients. A repurposing approach using already approved or clinical stage compounds can be used for diseases for which the patient population is small, and the commercial market limited. We have used Bayesian machine learning, in vitro cell assays, and combination analysis to identify molecules with potential use for NB. We demonstrated pyronaridine (SH-SY5Y IC50 1.70 μM, SK-N-AS IC50 3.45 μM), BAY 11-7082 (SH-SY5Y IC50 0.85 μM, SK-N-AS IC50 1.23 μM), niclosamide (SH-SY5Y IC50 0.87 μM, SK-N-AS IC50 2.33 μM) and fingolimod (SH-SY5Y IC50 4.71 μM, SK-N-AS IC50 6.11 μM) showed cytotoxicity against NB. As several of the molecules are approved drugs in the US or elsewhere, they may be repurposed more readily for NB treatment. Pyronaridine was also tested in combinations in SH-SY5Y cells and demonstrated an antagonistic effect with either etoposide or crizotinib. Whereas when crizotinib and etoposide were combined with each other they had a synergistic effect in these cells. We have also described several analogs of pyronaridine to explore the structure-activity relationship against cell lines. We describe multiple molecules demonstrating cytotoxicity against NB and the further evaluation of these molecules and combinations using other NB cells lines and in vivo models will be important in the future to assess translational potential.
Keywords: Drug Discovery, Machine learning, Neuroblastoma, Pyronaridine, Repurposing
BACKGROUND
Neuroblastoma (NB) is a rare cancer of nerve tissue that can arise anywhere in the sympathetic nervous system derived from the embryonic neural crest. NB accounts for 12%–15% of all childhood cancer-related deaths and is the most common and deadly extracranial tumor [1, 2] with a median age at diagnosis of 18 months [1, 3]. Two-thirds of the NB cases present before the age of 5 and 40 percent of patients are diagnosed before one year of age while the number steadily decreases with increasing age [3]. NB develops from immature nerve cells, referred to as neuroblasts. When the neuroblast itself does not mature, a cancer-causing malformation occurs as early as in utero [4]. NB is most common in the adrenal glands however, this cancer can be found in other areas of the body (linked to embryonic development from the neuroblast), including the chest, spinal cord, and abdomen. NB therefore remains a therapeutic challenge and approximately 800 patients are diagnosed each year in the United States [5]. NB is also characterized by a condition known as opsoclonus-myoclonus ataxia syndrome [6]. This syndrome consists of three main symptoms: opsoclonus (conjugate, multidirectional, chaotic eye movements), myoclonus (limb jerking that can also involve the head and face) and truncal ataxia, which can cause gait imbalance, sleep disturbance, cognitive dysfunction, and behavioral changes [3]. Unfortunately, NB mimics the symptoms of several other diseases, and diagnosis often occurs after the cancer has already disseminated throughout the body. Additionally, the prognosis worsens with increased age and advanced stage cancer [7].
Suggested cellular targets to treat NB include the Cannabinoid Receptor 2 (CNR2), Mitogen-Activated Protein Kinase 8 (MAPK8) [8] and Tyrosine kinase ALK/TRK. Multiple inhibitors of ALK/TRK have subsequently been identified for treatment, such as crizotinib, ceritinib, alectinib, lorlatinib and entrectinib [9]. An approach to identify new drugs for diseases that is attracting increasing attention is to use those that are already approved or at the clinical stage and repurpose them. This can be achieved using computational models for the disease or target to virtually screen clinically approved compounds and then test predicted compounds in vitro [10]. An alternative approach is to take a commercial library of FDA-approved drugs or clinical-stage drugs and test them all in vitro [11]. There have been several in vitro drug repurposing studies that have identified FDA-approved drugs that inhibit NB. For example, using a high-content imaging approach, over 300 drugs were tested on 3D spheroids of NB cells in vitro [12]. Two multiple tyrosine kinase inhibitors, ponatinib and axitinib, were proposed as promising candidates from this study as both drugs showed induction of cell-cycle block, apoptosis, and inhibition of colony formation [12]. Ponatinib consistently affected migration and inhibited invasion of NB cells as well as inhibited tumor growth in orthotopic NB mice [12]. The MYC family of transcription factors is a major driver of human cancer and is a potential therapeutic target for NB as MYCN is frequently activated in NB cells [3]. An important pathway regulated by MYC is the CKS1/SKP2/p27Kip1 axis, and the antidepressant drug fluoxetine disrupts CKS1/SKP2/p27Kip1 signaling [13].
Overall, few computational repurposing approaches have been applied to NB. For example, a pathway approach called iPANDA was used to identify the non-FDA approved antimicrobial taurolidine. Taurolidine affects MAPK and IL-10 signaling pathways which are cytotoxic to NB in vitro while in vivo xenograft models showed decreased tumor growth and increased survival [14]. Combining computational, 2D and 3D cell-based approaches [15] as well as patient-derived xenografts [16] may also represent another pathway to identifying new drugs for NB. Recently, analysis of whole-genome, whole-exome and/or transcriptome data from 702 neuroblastoma samples illustrated that forty percent of samples harbor at least one recurrent driver gene alteration, and most aberrations, including MYCN, ATRX, and TERT alterations, differ in frequency by age [17]. COSMIC mutational signature 18, is the most common cause of driver point mutations in NB, including most Anaplastic Lymphoma Kinase (ALK) and Ras-activating variants [17]. Currently, the frontline chemotherapeutic drugs (e.g., etoposide, doxorubicin, and cisplatin) for treating NB are all DNA damaging agents [18]. Several clinical trials for NB have involved ALK inhibitors, but patients with mutations in this protein will likely not respond to treatment and new therapies are therefore still needed. ALK is a receptor tyrosine kinase that falls within the insulin receptor superfamily. ALK expression is often restricted to developing nervous system tissues and expression levels are much lower in mature, adult brains [19]. Mutation and amplification of the ALK gene is associated with both familial (~1/2 familial cases) and somatic (9% primary NB tumors and 14% high risk) NB cases [19]. Single base mutations in regulatory regions of the kinase domain of ALK promote ligand independent signaling of ALK [19]. The three most common single base pair mutations (R1275, F1174, and F1245) represent ~85% of NB ALK mutations [19]. ALK is therefore an ideal target for anti-cancer therapeutics and several tyrosine kinase inhibitors (TKIs) (entrectinib and crizotinib) have shown promise in preclinical studies against NB [20]. However, clinical studies have also shown that TKIs have differential effects on NB cell lines with different ALK mutations [20].
In the absence of other options for potential treatments besides chemotherapy, it is important that additional small molecules are identified for NB. We, therefore, used a combination of machine learning techniques and in vitro cell-based assays with the NB cell lines SH-SY5Y[21], SK-N-AS[22], and fibrosarcoma HT-1080 cell lines for comparison for drug discovery of molecules for NB. We have now identified multiple clinical-stage, approved drugs, and novel molecules with promising in vitro activity against NB that may represent useful starting points for future lead optimization and in vivo testing.
RESULTS
Machine learning model and in vitro testing
We built a Bayesian machine learning model using SH-SY5Y inhibition data that is publicly available on CHEMBL which resulted in a five-fold receiver operator characteristic (ROC) value of 0.94 at a threshold of 1.68 μM (Figure 1). All model statistics were >0.73 suggesting it would be a useful model for external validation. We, therefore, used this model to score an in-house library of 260 compounds and prioritize compounds for testing (Table S1). From these, we identified fingolimod that was active (IC50 of 4.71 μM, Figure 2, Table 1). This compound was also tested in SK-N-AS cells and showed an IC50 6.11 μM (Figure 2, Table 1).
Figure 1.
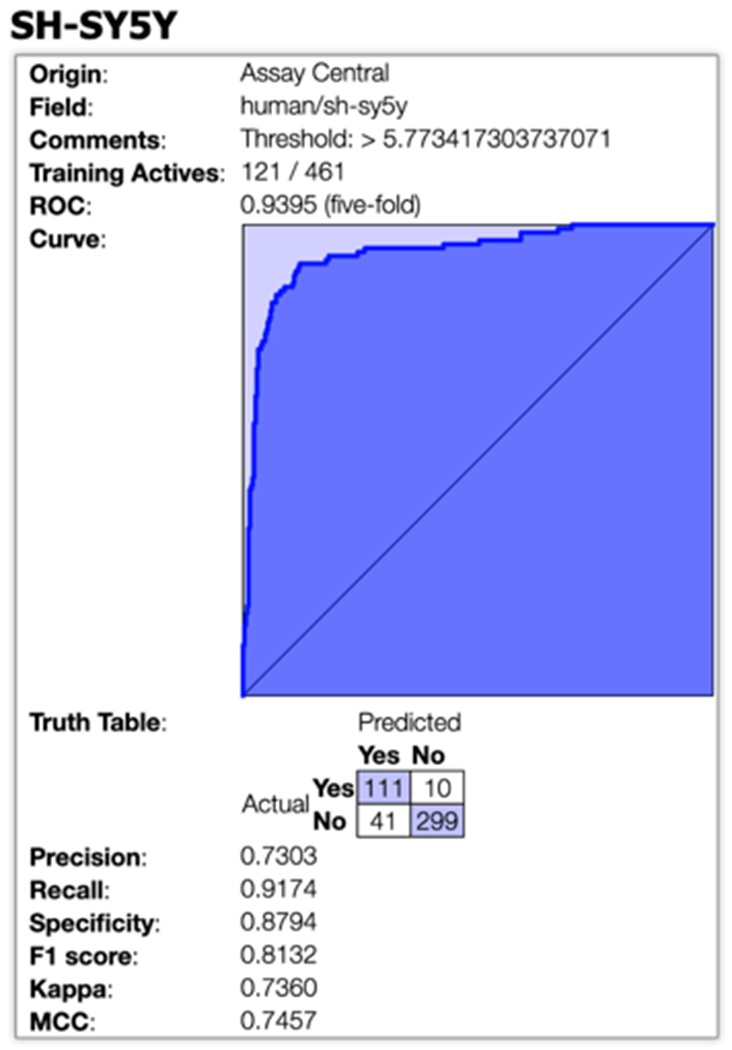
Five-fold ROC plot for the Bayesian neuroblastoma activity model. Abbreviations: Kappa = Cohen’s Kappa, MCC = Matthews Correlation Coefficient.
Figure 2.
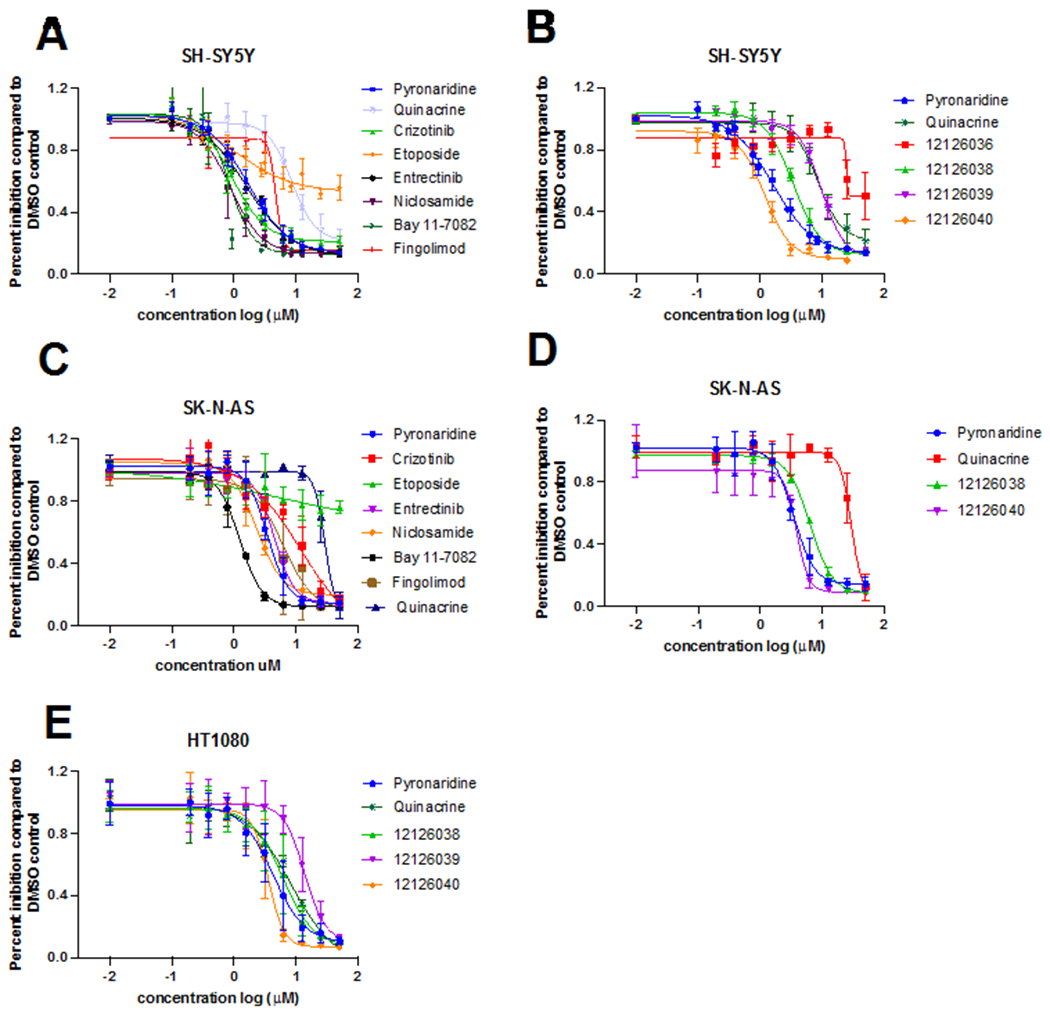
Dose-response curves of compounds tested in SH-SY5Y (A, B), SK-N-AS (C, D) and HT-1080 (E). Control compounds crizotinib IC50 0.92 μM in SH-SY5Y and IC50 12.12 μM in SK-N-AS and entrectinib IC50 1.60 in SH-SY5Y and IC50 4.47 μM in SK-N-AS.
Table 1.
Compounds tested in NB SH-SY5Y and SK-N-AS, and fibrosarcoma HT-1080. Blank: not tested NA: Ambiguous fit.
| Structure | Compound | Average IC50 SH-SY5Y |
Average IC50 SK-N-AS |
Average IC50 HT-1080 |
|---|---|---|---|---|
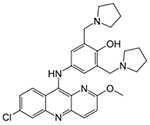
|
Pyronaridine | 1.70 μM | 3.45 μM | 4.23 μM |
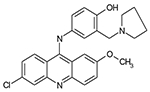
|
12126040 | 1.20 μM | 3.86 μM | 3.6 μM |
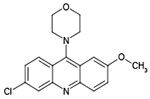
|
12126036 | NA | NA | NA |
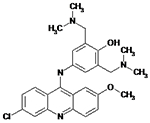
|
12126038 | 3.41 μM | 5.40 μM | 6.09 μM |
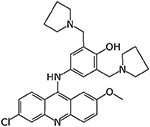
|
12126039 | 9.70 μM | NA | 13.77 μM |
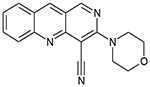
|
12126035 | Drug did not inhibit at 5 μM | NA | NA |
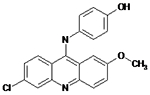
|
12126037 | Drug did not inhibit at 5 μM | NA | NA |
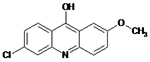
|
12126072 | Drug did not inhibit at 5 μM | NA | NA |
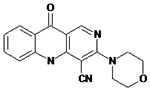
|
10326099 | Drug did not inhibit at 5 μM | NA | NA |
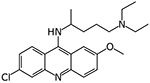
|
Quinacrine | 8.57 μM | NA | 29.1 μM |
|
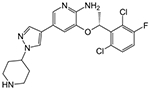
Crizotinib | 0.92 μM | 12.12 μM | NA |
Cell assays
We also performed cell-based screening of compounds (nilotinib, chlorpropamide, GW2961115, lumefantrine, mirtazapine, arbidol, dipyridamole, mefexamide hydrochloride, nimorazole, quinacrine, pyronaridine, BAY 11-7082, niclosamide and pyronaridine) and identified BAY 11-7082 (IC50 0.85 μM), niclosamide (IC50 0.87 μM) and pyronaridine (IC50 1.70 μM) as active against the SH-SY5Y cell line (Figure 2, Table 1). These molecules were also scored with our SH-SY5Y inhibition machine learning model for comparison (Table S1). These compounds were further tested in SK-N-AS cells and had similar levels of inhibition: pyronaridine IC50 3.45 μM, BAY 11-7082 IC50 1.23 μM and niclosamide IC50 2.33 μM (Figure 2, Table 1).
We explored the structure-activity relationship and synthesized analogs of pyronaridine, which were then evaluated in the SH-SY5Y and SK-N-AS NB cell lines as well as in HT1080 fibrosarcoma cell lines. The pyronaridine analogs 12126038 and 12126040 showed potency comparable to pyronaridine, with IC50’s of 3.41 μM and 1.20 μM, respectively in SH-SY5Y (Figure 2B), and in SK-N-AS with IC50’s of 5.40 μM and 3.89 μM (Figure 2D). 12126038 and 12126040 also showed IC50’s of 6.09 μM and 3.6 μM respectively for fibrosarcoma HT1080 cells (Figure 2E). The structures of these and other pyronaridine analogs and IC50 values in different cell lines are described in Table 1.
Combination Analysis
BRAID analysis [23] (Figure S1) of pyronaridine with crizotinib as well as pyronaridine with etoposide in vitro inhibition data from the checkerboard assay indicates an antagonistic effect of these molecules in SH-SY5Y neuroblastoma cells as both have negative κ values. Interestingly, a synergistic affect was identified using BRAID analysis with the combination of crizotinib and etoposide with a κ of 3.46 (Table 2).
Table 2: Combination therapies and BRAID analysis.
The variable κ represents a quantitative synergy value where κ < 0 implies antagonism, κ = 0 implies additivity, and κ > 0 implies synergy. As an additional reference, “strong synergy” corresponds to κ = 2.5, “mild synergy” corresponds to κ = 1, “mild antagonism” corresponds to κ = −0.66, and “strong antagonism” corresponds to κ = −1.
| Compound 1 | Compound 2 | Κ (average of 4 experiments) |
|---|---|---|
| Pyronaridine | Crizotinib | −0.640 |
| Pyronaridine | Etoposide | −0.746 |
| Crizotinib | Etoposide | 3.46 |
Cathepsin D
We evaluated if pyronaridine could inhibit Cathepsin D using a fluorescent enzymatic assay and found pyronaridine inhibited Cathepsin > 50% at the highest concentration tested (200 μM, Figure S2). This might however be physiologically relevant because pyronaridine is a lysosomotropic compound accumulating in the lysosome [24].
DISCUSSION
The efficacy of ALK inhibitors in patients with ALK-mutant neuroblastoma is limited, and there is therefore a need to improve their effectiveness in these patients. ALK F1174L and F1174C mutations were previously identified as acquired resistance mutations in oncogenic ALK fusion proteins during crizotinib and ceritinib treatments, respectively [25, 26]. In the clinical studies of crizotinib and ceritinib in pediatric patients with ALK-driven neuroblastoma, most tumors harboring R1275Q-mutated ALK did not respond to the treatments even though cells with the R1275Q mutation are sensitive to both drugs [27]. This suggests there is still a need for discovery of new molecules for NB. We used SH-SY5Y and SK-N-AS NB cell lines for our drug discovery studies. SH-SY5Y has non-amplified MYCN, it has the ALK F1174 L mutation, and it is TP53 WT [28], while SK-N-AS has a single copy of the MYCN gene (thus showing no overexpression of this gene [29]), the cells express WT ALK at low levels [30] and a truncated p53 (β isoform) [31]. Therefore, in SK-N-AS, the p53β isoform is unable to induce endogenous p21 expression [31]. A previous study has shown that in NB SH-SY5Y cells bearing the wild-type p53, the p53 accumulated in response to cisplatin and can promote apoptosis, whereas SK-N-AS cells, which have a truncated form of p53 did not undergo apoptosis [32].
Our SH-SY5Y inhibition Bayesian model generated with publicly available data had excellent 5-fold cross validation statistics (Figure 1) and fingolimod (Figure 2) was identified using this. We have also described a cell-based screening for NB which resulted in the identification of several new hits with promising activity in vitro. In our study, fingolimod showed an IC50 4.71 μM in SH-SY5Y and an IC50 6.11 μM in SK-N-AS (Figure 2, Table 1) and represents an off-patent FDA approved drug that targets sphingosine-1-phosphate as a receptor modulator for lymphocyte circulation and in turn, acts as an immunomodulator. Fingolimod is currently used to treat multiple sclerosis [33]. Interestingly, this molecule has previously been identified as active against NB [34] and the sphingosine-1-phosphate receptor is considered an important target for chemoresistance [35]. We also identified BAY-7082, an NF-κB inhibitor, that is also known to induce apoptosis and S-phase arrest in gastric cancer cells [36]. BAY 11-7082 is an anti-inflammatory agent due to its ability to prevent NF-KB activation [37, 38] by inhibiting protein tyrosine phosphorylation and thereby blocking the function of IKK, effectively maintaining NFKB in an in-active form [37]. Inflammation is associated with cancer phenotypes as it promotes DNA damage, angiogenesis, and cell proliferation (activated NF-KB prevents apoptosis) [39]. BAY 11-7082 showed an improved IC50 in NB cells over crizotinib and entrectinib, demonstrating an IC50 of 0.85 μM in SH-SY5Y and IC50 1.35 μM in SK-N-AS NB cell lines (Figure 2 A, C). BAY 11-7082 (30 μM) induced complete cell death in all 10 multiple myeloma cell lines as well as in the large majority of primary multiple myeloma samples [40]. BAY 11-7082 has previously shown anti-tumor effects on bladder, breast, esophageal, lung, and gastric cancers [41-44]. However, BAY 11-7082 has not previously been in clinical trials against any cancers to our knowledge. The antiparasitic niclosamide was also identified in this study (IC50 0.87 μM in SH-SY5Y and IC50 2.33 μM in SK-N-AS cells, Figure 2A, C) and is a drug used for treating tapeworm, which is part of World Health Organization’s List of Essential Medicines. This drug has been shown to have several other uses [45, 46] although it is poorly orally available. Niclosamide was recently identified for NB using a gene expression-based approach and demonstrated efficacy in a xenograft model [47] SK-N-DZ (MYCN-amplified) and SK-N-AS (MYCN-nonamplified). Notably, treatment of these xenografts with niclosamide led to reduced tumor growth and improved survival with no apparent toxicity [47].
A compound of interest, based on our previous work [48], is the antimalarial pyronaridine, which we have now shown has an IC50 of 1.7 μM in SH-SY5Y (Figure 2A) and IC50 of 3.45 μM in the SK-N-AS cell lines. We have previously demonstrated cytotoxicity for this molecule against Vero (IC50 1.3 μM [49]) HeLa (IC50 3.1-4.1 μM [50]) and most recently tested in A549+ACE2 and other human cell lines (IC50 >10 μM [48]) cells. Artesunate-pyronaridine (Pyramax) is a combination therapy for malaria [51], that has been approved in Europe since 2017. Pyronaridine is an approved drug for malaria despite having a Mannich base group which has been associated with promiscuous compounds. Our group has been testing the activity of this compound extensively among many targets, and we have found that it does not appear to be promiscuous. For example, screening of pyronaridine (tested at 1 μM) against 485 kinases identified only two as having mean percent inhibition greater than 30% including CAMK1 (35%) and MELK (31%) [52]. Pyronaridine has a well-characterized antiviral activity against select viruses e.g. Ebola and COVID-19 in vitro [48, 53] and in vivo [49, 52] but does not appear to target many other viruses as described in these publications. Pyronaridine has previously been shown to have potent cytotoxicity on human breast and hematological cancer cells through induction of apoptosis [54]. Pyronaridine has been screened by the National Cancer Institute developmental therapeutics program against various breast, central nervous system, colon, leukemia, melanoma, non-small cell lung cancer, ovarian, prostate and renal cancer cell lines [55] but not neuroblastoma. A study of pyronaridine as a single oral dose (400 mg) given to a healthy volunteer found a Cmax in plasma of 495.8 ng/ml at a Tmax of 0.5 h [51, 56], which provides a concentration close to 1 μM, however, pyronaridine preferentially associates with blood cells and is highly plasma protein bound [51], which suggests that it can potentially reach the concentration necessary to have an effect on NB cells at a higher dose. Pyronaridine induced apoptosis via mitochondrial depolarization, Caspase 3 activation, inhibition of cell cycle progression and by directly intercalating with cellular DNA at high concentrations [54]. The pro-apoptotic activity of pyronaridine and its capacity to inhibit cancer cell growth have led to the potential use for the treatment of cancer. In addition, we have found inhibition of Cathepsin D at high concentrations (Figure S2). This target is unlikely to have a role in the cytotoxicity observed in NB, since pyronaridine is a lysosomotropic drug [24]. Cathepsin is well known aspartyl protease and is an important regulator of apoptotic pathways in cells [57]. A study performed on the human SH-SY5Y cell line demonstrated the cathepsin D involvement in the mitophagy process [58]. High levels of cathepsin D were also observed in breast, ovarian, colorectal, prostate, bladder cancer and melanoma [57]. Cathepsin D was additionally shown to have pro-angiogenesis, pro-apoptotic, pro-invasive and pro-metastatic properties [57]. In recent research, it has become evident that the lysosome is important in drug resistance phenotypes [59]. As pyronaridine is a lysosomotropic compound [24], we evaluated combination therapies of pyronaridine with etoposide and pyronaridine with crizotinib, to verify if they had any synergistic effect. Because of the weak base characteristics of lysosomotropic compounds, they can accumulate in the lysosomal lumen and elevate pH, inducing lysosomal membrane permeabilization [60]. One possibility for combination therapies to overcome resistance is to explore the use of lysosomotropic adjuvants, that could cause intracellular redistribution of chemotherapic drugs from the lysosomal lumen to the cytosol and subsequently to their sites of action [59]. Interestingly, pyronaridine did not enhance the effect of etoposide, showing a κ −0.746 in the BRAID analysis, showing a mild antagonistic effect if these drugs are used in combination (Table 2 and Fig S1B). Etoposide is a widely used chemotherapy medication for the treatment of several types of cancer including testicular cancer, lung cancer, lymphoma, leukemia, neuroblastoma, and ovarian cancer. Etoposide forms a ternary complex with DNA and the topoisomerase II (Topo II) enzyme. Our combination treatment in vitro showed that etoposide and crizotinib work synergistically (Table 2, Fig S1C) as determined by BRAID analysis with a κ of 3.46. This was expected, since crizotinib and another topoisomerase inhibitor, topotecan, are reported to also act synergistically [61, 62]. Cancer cells rely on Topo II more than healthy cells since they divide more rapidly. Therefore, this causes errors in DNA synthesis and promotes apoptosis of the cancer cell [63]. Interestingly, pyronaridine has been suggested to function as an inhibitor of Topo II in P. falciparum in vitro, with a concentration of 11 μM required for complete inhibition of DNA decatenation by the parasite enzyme [64]. Subsequently, it was found that pyronaridine showed little inhibitory activity against P. falciparum Topo II in situ, with the parasite maintained under continuous culture in vitro and it was concluded that P. falciparum Topo II is not the specific target of pyronaridine in this parasite [64]. To our knowledge, the activity of pyronaridine against human Topo II has not been investigated. Pyronaridine and crizotinib showed a mild antagonist effect, with a κ −0.640 (Table 2, Figure S1A).
Treatment of NB patients diverges extensively between the different risk groups [1] as treatment of high-risk patients involves an intense induction chemotherapy regimen that includes cisplatin, vincristine, carboplatin, etoposide and cyclophosphamide [1]. NB is therefore one of the deadliest malignancies of childhood and needs refinement of the current treatment protocols, including the use of more targeted therapies, (both pharmacologically and cellular) which more closely reflects the molecular landscape of individual tumors. This will require the continued analysis of the influence of cells within the tumor microenvironment in order to search for novel therapy options [1]. We used a combination of machine learning techniques and cell-based assays to identify molecules with potential use for NB treatment. We identified several molecules, with low μM NB activity for this cancer including niclosamide, BAY-7082, pyronaridine and pyronaridine analogs (12126038 and 12126040) in the SH-SY5Y NB cell line.
The relapse and chemoresistance in cancers, including NB, is often associated with the inactivation of the p53 tumor suppressor. p53 mutations are unusual in human NB but, when they do occur, are found in post-chemotherapy tumors. We studied the compounds identified for SH-SY5Y in SK-N-AS NB cells. SK-N-AS presents C-terminal homozygous deletion in p53 [32]. Pyronaridine and analogs 12126038 and 12126040 were also tested in SK-N-AS cells, with similar IC50s (Table 1, Fig 2 D) suggesting no impact for p53. Interestingly, pyronaridine, pyronaridine analogs, niclosamide and BAY-7082 showed increased potency when compared to crizotinib in the SK-N-AS NB cell lines (Table 1). Some of these compounds could perhaps be used as a starting point to develop higher affinity inhibitors for NB and further understand their mechanism. Besides NB SH-SY5Y and SK-N-AS cells, these compounds were also active against fibrosarcoma HT-1080 cells which might suggest a use in further cancers (Table 1). Pyronaridine provides a relatively new scaffold to be explored for cancer treatments as the major developments in this field over the past decades have been focused on developing kinase inhibitors. Since it has been shown that pyronaridine is safe to use in humans suffering from malaria from the various clinical trials [51], it could also have potential use as a therapeutic against cancer. The combination of artesunate-pyronaridine is on the WHO list of medicines for malaria. A single-oral dose (400 mg) of pyronaridine given to a healthy volunteer showed a Cmax in plasma of 495.8 ng/mL at a Tmax of 0.5 h, which gives a concentration close to 1 μM [51, 56]. In our study, pyronaridine showed an IC50 1.70 μM in SH-SY5Y which is very close to a Cmax of 1 μM. We also describe BAY 11-7082 as having an improved IC50 against NB than crizotinib and entrectinib in NB SH-SY5Y and SK-N-AS cells, and this compound might also be worthy of further evaluation in other NB cells lines that are refractory to treatment with ALK inhibitors.
In conclusion, this work builds on our earlier efforts demonstrating the application of Bayesian machine learning models for chordoma and their application to scoring compounds prior to in vitro testing [65]. We have demonstrated how both computational and focused in vitro screening offer compound selection and repurposing opportunities for testing against other cancers in vitro.
METHODS
Compounds
Compounds fingolimod, BAY 11-7082, niclosamide, entrectinib and crizotinib were purchased from MedChemExpress (MCE, Monmouth Junction, NJ). Pyronaridine tetraphosphate was purchased from BOC Sciences (Shirley, NY). Etoposide was purchased from Chemspace (Monmouth Junction, NJ). Quinacrine hydrochloride was purchased from Cayman Chemical (Ann Arbor, MI). Compounds were dissolved in DMSO before dilution in cell culture media for cell-based assays. Pyronaridine analogs were synthesized as previously reported [66].
Machine learning
Public data from ChEMBL [67] was obtained for compounds screened against the SH-SY5Y cell line (IC50 data from Target ID 614910) and then used to generate a Bayesian machine learning model with Assay Central software [68] and the extended connectivity fingerprint (ECFP6) descriptor. Assay Central models include the following metrics for internal predictive performance by five-fold cross-validation: Recall, Precision, Specificity, F1-Score, Receiver Operating Characteristic (ROC) curve, Cohen’s Kappa, and the Matthews Correlation Coefficient. Classification models require an activity threshold, and we have applied a threshold selection to optimize performance. This model was then used to score an in-house library (totaling 260 compounds at the time) before the selection of several compounds for testing. Assay Central predictions include a probability-like score (were values above 0.5 considered an active prediction) and an applicability score which assesses the representation of the predicted molecule within the training set [69].
Cell lines and culture
SH-SY5Y NB cells were generously provided by the Catalyst for Rare Diseases (UNC Eshelman School of Pharmacy). SK-N-AS NB and CCD-sk fibroblast cells were purchased from the Tissue Culture Facility at the University of North Carolina at Chapel Hill. HT1080 fibrosarcoma cells were purchased from Sigma Aldrich. All cell lines were cultured with a 1:1 mixture of DMEM and Ham’s F12 Medium with L-glutamine and 15 mM HEPES (Gibco, Life Technologies, NY, USA) supplemented with 10% Fetal Bovine Serum (FBS) (Invitrogen). All cells were cultured in an atmosphere of 37°C and 5% CO2.
Cell viability assay and IC50 determination
NB (SH-SY5Y and SK-N-AS) and fibrosarcoma (HT1080) cells were seeded into clear, flat bottom 96-well plates at a density of 1x104 cells/well in a total volume of 200 μl culture media containing drugs dissolved in DMSO starting 50 μM and subsequently diluted two-fold for an additional 8 dilutions (final 0.195 μM) or DMSO alone. DMSO was present in all wells at a final concentration of 0.5%. Cells containing drug or DMSO alone were incubated for 48 h at 37°C, 5% CO2 after which resazurin (Sigma Aldrich) was added to a final concentration of 0.0125 mg/ml to measure cell viability. Fluorescence was measured after incubating the cells with resazurin for 8 h for NB cells and for 3 h for fibrosarcoma cells by Spectramax ID5 (Molecular Devices). Fluorescence intensity was measured using an excitation filter of 544 nm and an emission filter of 590 nm. Each experiment contained 3 replicates of each drug concentration and all experiments were performed at least twice. Percent inhibition of each drug was calculated relative to the DMSO-only control. Non-linear regression analysis was performed using Prism version (Version 8.4.1, GraphPad Software, San Diego, CA) was used to determine the 50% inhibitory concentration (IC50) from fitted curves (four parameter log agonist versus response variable slope). Error bars of dose-response curves represent the SEM of replicates.
Dual compound synergy assays and BRAID analysis
NB SH-SY5Y cells were seeded into 96-well plates at a density of 1x104 cells/well in a total volume of 200 μl culture media containing either a single drug dissolved in DMSO, two drugs (combination) both of which were dissolved in DMSO, or DMSO alone (0.5%). Drug combinations were set up in a 6 x 6 matrix with 1:3 dilutions of drug starting at 25 μM. Cells were incubated in the presence of a single drug, drugs in combination, or DMSO alone for 48 h 37°C, 5% CO2, after which resazurin (Sigma Aldrich) was added to a final concentration of 0.0125 mg/ml to measure cell viability. Fluorescence was measured after incubating the cells with resazurin for 8 h using an Spectramax ID5 (Molecular Devices) and an excitation filter of 544 nm and an emission filter of 590 nm. Each experiment was repeated in quadruplicate. Determination of synergy was analyzed for synergistic, additive, or antagonistic effects of the drugs in combination using or BRAID software [23] where k represents a quantitative synergy value, k < 0 implies antagonism, k = 0 implies additivity, and k > 0 implies synergy [23].
Cathepsin D
We tested the activity of pyronaridine against Cathepsin D using the Abcam (Cambridge, UK) Cathepsin Inhibitor Screening Kit, which is a fluorescence-based assay that utilizes the preferred cathepsin D substrate sequence GKPILFFRLK(Dnp)-D-R-NH2) labeled with MCA according to manufacturer’s instructions with a few modifications. The assay was performed using 384 well plates in a total volume of 25 μL reaction. Dose-response curves were performed at 37 °C for 2 h, and pyronaridine was pre-incubated with Cathepsin D for 15 min prior to adding the substrate. Cathepsin D cleaves the synthetic substrate to release the quenched fluorescent group MCA, which might then easily be measured employing a fluorometer or fluorescence plate reader at Ex/Em = 320/420 nm. The relative efficacy of test inhibitors is compared to the positive control inhibitor, Pepstatin A (IC50 < 0.1 nM).
Supplementary Material
ACKNOWLEDGMENTS
Dr. Thomas Lane is kindly acknowledged for advice on BRAID. Dr. Alex Clark is acknowledged for assistance with Assay Central.
Grant information
SE kindly acknowledges NIH funding R44GM122196-02A1 from NIH NIGMS, and NIEHS for 1R43ES031038-01. “Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number R43ES031038. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.” AJH acknowledges FY2018 UNC Research Opportunities Initiative (ROI) Award. The authors acknowledge the generous support provided by the Eshelman Institute of Innovation and the Bryson Foundation at the UNC Eshelman School of Pharmacy.
ABBREVIATIONS USED
- CNR2
Cannabinoid Receptor 2
- MAPK8
Mitogen-Activated Protein Kinase 8
- NB
Neuroblastoma
- TKIs
tyrosine kinase inhibitors
Footnotes
Competing interests
L.R., A.P.R., K.M.Z., D.H.F., and S.E. are employees of Collaborations Pharmaceuticals, Inc. All other authors do not have competing interests.
DATA AND SOFTWARE AVAILABILITY
Public data from ChEMBL [67] was obtained for compounds screened against the SH-SY5Y cell line (IC50 data from Target ID 614910) and then used to generate a Bayesian machine learning model with our proprietary software Assay Central[68].
REFERENCES
- [1].Johnsen JI, Dyberg C, Wickström M, Neuroblastoma-A Neural Crest Derived Embryonal Malignancy, Front Mol Neurosci, 12 (2019) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Moreno L, Barone G, DuBois SG, Molenaar J, Fischer M, Schulte J, Eggert A, Schleiermacher G, Speleman F, Chesler L, Geoerger B, Hogarty MD, Irwin MS, Bird N, Blanchard GB, Buckland S, Caron H, Davis S, De Wilde B, Deubzer HE, Dolman E, Eilers M, George RE, George S, Jaroslav S, Maris JM, Marshall L, Merchant M, Mortimer P, Owens C, Philpott A, Poon E, Shay JW, Tonelli R, Valteau-Couanet D, Vassal G, Park JR, Pearson ADJ, Accelerating drug development for neuroblastoma: Summary of the Second Neuroblastoma Drug Development Strategy forum from Innovative Therapies for Children with Cancer and International Society of Paediatric Oncology Europe Neuroblastoma, Eur J Cancer, 136 (2020) 52–68. [DOI] [PubMed] [Google Scholar]
- [3].Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, Weiss WA, Neuroblastoma, Nat Rev Dis Primers, 2 (2016) 16078. [DOI] [PubMed] [Google Scholar]
- [4].Voute PA Jr., Wadman SK, van Putten WJ, Congenital neuroblastoma. Symptoms in the mother during pregnancy, Clin Pediatr (Phila), 9 (1970) 206–207. [DOI] [PubMed] [Google Scholar]
- [5].Izbicki T, Mazur J, Izbicka E, Epidemiology and etiology of neuroblastoma: an overview, Anticancer Res, 23 (2003) 755–760. [PubMed] [Google Scholar]
- [6].Meena JP, Seth R, Chakrabarty B, Gulati S, Agrawala S, Naranje P, Neuroblastoma presenting as opsoclonus-myoclonus: A series of six cases and review of literature, J Pediatr Neurosci, 11 (2016) 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Stiller CA, Bunch KJ, Trends in survival for childhood cancer in Britain diagnosed 1971-85, Br J Cancer, 62 (1990) 806–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Almstedt E, Elgendy R, Hekmati N, Rosen E, Warn C, Olsen TK, Dyberg C, Doroszko M, Larsson I, Sundstrom A, Arsenian Henriksson M, Pahlman S, Bexell D, Vanlandewijck M, Kogner P, Jornsten R, Krona C, Nelander S, Integrative discovery of treatments for high-risk neuroblastoma, Nat Commun, 11 (2020) 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pacenta HL, Macy ME, Entrectinib and other ALK/TRK inhibitors for the treatment of neuroblastoma, Drug Des Devel Ther, 12 (2018) 3549–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Urbina F, Puhl AC, Ekins S, Recent advances in drug repurposing using machine learning, Curr Opin Chem Biol, 65 (2021) 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Vignaux PA, Minerali E, Lane TR, Foil DH, Madrid PB, Puhl AC, Ekins S, The Antiviral Drug Tilorone Is a Potent and Selective Inhibitor of Acetylcholinesterase, Chem Res Toxicol, 34 (2021) 1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sidarovich V, De Mariano M, Aveic S, Pancher M, Adami V, Gatto P, Pizzini S, Pasini L, Croce M, Parodi F, Cimmino F, Avitabile M, Emionite L, Cilli M, Ferrini S, Pagano A, Capasso M, Quattrone A, Tonini GP, Longo L, A High-Content Screening of Anticancer Compounds Suggests the Multiple Tyrosine Kinase Inhibitor Ponatinib for Repurposing in Neuroblastoma Therapy, Mol Cancer Ther, 17 (2018) 1405–1415. [DOI] [PubMed] [Google Scholar]
- [13].Bibbo S, Lamolinara A, Capone E, Purgato S, Tsakaneli A, Panella V, Sallese M, Rossi C, Ciufici P, Nieddu V, De Laurenzi V, Iezzi M, Perini G, Sala G, Sala A, Repurposing a psychoactive drug for children with cancer: p27(Kip1)-dependent inhibition of metastatic neuroblastomas by Prozac, Oncogenesis, 9 (2020) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Swift L, Zhang C, Kovalchuk O, Boklan J, Trippett T, Narendran A, Dual functionality of the antimicrobial agent taurolidine which demonstrates effective anti-tumor properties in pediatric neuroblastoma, Invest New Drugs, (2019). [DOI] [PubMed] [Google Scholar]
- [15].Nolan JC, Frawley T, Tighe J, Soh H, Curtin C, Piskareva O, Preclinical models for neuroblastoma: Advances and challenges, Cancer Lett, 474 (2020) 53–62. [DOI] [PubMed] [Google Scholar]
- [16].Kamili A, Gifford AJ, Li N, Mayoh C, Chow SO, Failes TW, Eden GL, Cadiz R, Xie J, Lukeis RE, Norris MD, Haber M, McCowage GB, Arndt GM, Trahair TN, Fletcher JI, Accelerating development of high-risk neuroblastoma patient-derived xenograft models for preclinical testing and personalised therapy, Br J Cancer, 122 (2020) 680–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Brady SW, Liu Y, Ma X, Gout AM, Hagiwara K, Zhou X, Wang J, Macias M, Chen X, Easton J, Mulder HL, Rusch M, Wang L, Nakitandwe J, Lei S, Davis EM, Naranjo A, Cheng C, Maris JM, Downing JR, Cheung NV, Hogarty MD, Dyer MA, Zhang J, Pan-neuroblastoma analysis reveals age- and signature-associated driver alterations, Nat Commun, 11 (2020) 5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hara J, Development of treatment strategies for advanced neuroblastoma, Int J Clin Oncol, 17 (2012) 196–203. [DOI] [PubMed] [Google Scholar]
- [19].Trigg RM, Turner SD, ALK in Neuroblastoma: Biological and Therapeutic Implications, Cancers (Basel), 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Umapathy G, Mendoza-Garcia P, Hallberg B, Palmer RH, Targeting anaplastic lymphoma kinase in neuroblastoma, APMIS, 127 (2019) 288–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kovalevich J, Santerre M, Langford D, Considerations for the Use of SH-SY5Y Neuroblastoma Cells in Neurobiology, Methods Mol Biol, 2311 (2021) 9–23. [DOI] [PubMed] [Google Scholar]
- [22].Acosta S, Lavarino C, Paris R, Garcia I, de Torres C, Rodríguez E, Beleta H, Mora J, Comprehensive characterization of neuroblastoma cell line subtypes reveals bilineage potential similar to neural crest stem cells, BMC Dev Biol, 9 (2009) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Twarog NR, Stewart E, Hammill CV, A AS, BRAID: A Unifying Paradigm for the Analysis of Combined Drug Action, Sci Rep, 6 (2016) 25523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lane TR, Dyall J, Mercer L, Goodin C, Foil DH, Zhou H, Postnikova E, Liang JY, Holbrook MR, Madrid PB, Ekins S, Repurposing Pyramax® for the Treatment of Ebola Virus Disease: Additivity of the Lysosomotropic Pyronaridine and Non-Lysosomotropic Artesunate Antiviral Research, 182 (2020) 104908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sasaki T, Okuda K, Zheng W, Butrynski J, Capelletti M, Wang L, Gray NS, Wilner K, Christensen JG, Demetri G, Shapiro GI, Rodig SJ, Eck MJ, Jänne PA, The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers, Cancer Res, 70 (2010) 10038–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Friboulet L, Li N, Katayama R, Lee CC, Gainor JF, Crystal AS, Michellys PY, Awad MM, Yanagitani N, Kim S, Pferdekamper AC, Li J, Kasibhatla S, Sun F, Sun X, Hua S, McNamara P, Mahmood S, Lockerman EL, Fujita N, Nishio M, Harris JL, Shaw AT, Engelman JA, The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer, Cancer Discov, 4 (2014) 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mossé YP, Lim MS, Voss SD, Wilner K, Ruffner K, Laliberte J, Rolland D, Balis FM, Maris JM, Weigel BJ, Ingle AM, Ahern C, Adamson PC, Blaney SM, Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: a Children’s Oncology Group phase 1 consortium study, Lancet Oncol, 14 (2013) 472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang HQ, Halilovic E, Li X, Liang J, Cao Y, Rakiec DP, Ruddy DA, Jeay S, Wuerthner JU, Timple N, Kasibhatla S, Li N, Williams JA, Sellers WR, Huang A, Li F, Combined ALK and MDM2 inhibition increases antitumor activity and overcomes resistance in human ALK mutant neuroblastoma cell lines and xenograft models, Elife, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Suenaga Y, Kaneko Y, Matsumoto D, Hossain MS, Ozaki T, Nakagawara A, Positive auto-regulation of MYCN in human neuroblastoma, Biochem Biophys Res Commun, 390 (2009) 21–26. [DOI] [PubMed] [Google Scholar]
- [30].Ota Y, Yoda H, Inoue T, Watanabe T, Shinozaki Y, Takatori A, Nagase H, Targeting anaplastic lymphoma kinase (ALK) gene alterations in neuroblastoma by using alkylating pyrrole-imidazole polyamides, PLoS One, 16 (2021) e0257718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Goldschneider D, Horvilleur E, Plassa LF, Guillaud-Bataille M, Million K, Wittmer-Dupret E, Danglot G, de The H, Benard J, May E, Douc-Rasy S, Expression of C-terminal deleted p53 isoforms in neuroblastoma, Nucleic Acids Res, 34 (2006) 5603–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nakamura Y, Ozaki T, Niizuma H, Ohira M, Kamijo T, Nakagawara A, Functional characterization of a new p53 mutant generated by homozygous deletion in a neuroblastoma cell line, Biochem Biophys Res Commun, 354 (2007) 892–898. [DOI] [PubMed] [Google Scholar]
- [33].Chiba K, Discovery of fingolimod based on the chemical modification of a natural product from the fungus, Isaria sinclairii, J Antibiot (Tokyo), 73 (2020) 666–678. [DOI] [PubMed] [Google Scholar]
- [34].Li MH, Hla T, Ferrer F, FTY720 inhibits tumor growth and enhances the tumor-suppressive effect of topotecan in neuroblastoma by interfering with the sphingolipid signaling pathway, Pediatr Blood Cancer, 60 (2013) 1418–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lifshitz V, Priceman SJ, Li W, Cherryholmes G, Lee H, Makovski-Silverstein A, Borriello L, DeClerck YA, Yu H, Sphingosine-1-Phosphate Receptor-1 Promotes Environment-Mediated and Acquired Chemoresistance, Mol Cancer Ther, 16 (2017) 2516–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chen L, Ruan Y, Wang X, Min L, Shen Z, Sun Y, Qin X, BAY 11-7082, a nuclear factor-κB inhibitor, induces apoptosis and S phase arrest in gastric cancer cells, J Gastroenterol, 49 (2014) 864–874. [DOI] [PubMed] [Google Scholar]
- [37].Krishnan N, Bencze G, Cohen P, Tonks NK, The anti-inflammatory compound BAY-11-7082 is a potent inhibitor of protein tyrosine phosphatases, FEBS J, 280 (2013) 2830–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lee J, Rhee MH, Kim E, Cho JY, BAY 11-7082 is a broad-spectrum inhibitor with anti-inflammatory activity against multiple targets, Mediators Inflamm, 2012 (2012) 416036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Singh N, Baby D, Rajguru JP, Patil PB, Thakkannavar SS, Pujari VB, Inflammation and cancer, Ann Afr Med, 18 (2019) 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rauert-Wunderlich H, Siegmund D, Maier E, Giner T, Bargou RC, Wajant H, Stühmer T, The IKK inhibitor Bay 11-7082 induces cell death independent from inhibition of activation of NFκB transcription factors, PLoS One, 8 (2013) e59292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang Q, Mao Z, Sun J, NF-κB inhibitor, BAY11-7082, suppresses M2 tumor-associated macrophage induced EMT potential via miR-30a/NF-κB/Snail signaling in bladder cancer cells, Gene, 710 (2019) 91–97. [DOI] [PubMed] [Google Scholar]
- [42].Zhang L, Xie J, Gan R, Wu Z, Luo H, Chen X, Lu Y, Wu L, Zheng D, Synergistic inhibition of lung cancer cells by EGCG and NF-κB inhibitor BAY11-7082, J Cancer, 10 (2019) 6543–6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Viola K, Kopf S, Huttary N, Vonach C, Kretschy N, Teichmann M, Giessrigl B, Raab I, Stary S, Krieger S, Keller T, Bauer S, Hantusch B, Szekeres T, de Martin R, Jäger W, Mikulits W, Dolznig H, Krupitza G, Grusch M, Bay11-7082 inhibits the disintegration of the lymphendothelial barrier triggered by MCF-7 breast cancer spheroids; the role of ICAM-1 and adhesion, Br J Cancer, 108 (2013) 564–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yan Y, Qian H, Cao Y, Zhu T, Nuclear factor-κB inhibitor Bay11-7082 inhibits gastric cancer cell proliferation by inhibiting Gli1 expression, Oncol Lett, 21 (2021) 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chen W, Mook RA Jr., Premont RT, Wang J, Niclosamide: Beyond an antihelminthic drug, Cell Signal, 41 (2018) 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Xu J, Shi PY, Li H, Zhou J, Broad Spectrum Antiviral Agent Niclosamide and Its Therapeutic Potential, ACS Infect Dis, 6 (2020) 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Huang CT, Hsieh CH, Lee WC, Liu YL, Yang TS, Hsu WM, Oyang YJ, Huang HC, Juan HF, Therapeutic Targeting of Non-oncogene Dependencies in High-risk Neuroblastoma, Clin Cancer Res, 25 (2019) 4063–4078. [DOI] [PubMed] [Google Scholar]
- [48].Puhl AC, Fritch EJ, Lane TR, Tse LV, Yount BL, Sacramento CQ, Fintelman-Rodrigues N, Tavella TA, Maranhão Costa FT, Weston S, Logue J, Frieman M, Premkumar L, Pearce KH, Hurst BL, Andrade CH, Levi JA, Johnson NJ, Kisthardt SC, Scholle F, Souza TML, Moorman NJ, Baric RS, Madrid PB, Ekins S, Repurposing the Ebola and Marburg Virus Inhibitors Tilorone, Quinacrine, and Pyronaridine: In Vitro Activity against SARS-CoV-2 and Potential Mechanisms, ACS Omega, 6 (2021) 7454–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lane TR, Massey C, Comer JE, Anantpadma M, Freundlich JS, Davey RA, Madrid PB, Ekins S, Repurposing the antimalarial pyronaridine tetraphosphate to protect against Ebola virus infection, PLoS Negl Trop Dis, 13 (2019) e0007890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lane TR, Massey C, Comer JE, Freiberg AN, Zhou H, Dyall J, Holbrook MR, Anantpadma M, Davey RA, Madrid PB, Ekins S, Pyronaridine tetraphosphate efficacy against Ebola virus infection in guinea pig, Antiviral Res, 181 (2020) 104863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Croft SL, Duparc S, Arbe-Barnes SJ, Craft JC, Shin CS, Fleckenstein L, Borghini-Fuhrer I, Rim HJ, Review of pyronaridine anti-malarial properties and product characteristics, Malar J, 11 (2012) 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Puhl AC, Gomes GF, Damasceno S, Godoy AS, Noske GD, Nakamura AM, Gawriljuk VO, Fernandes RS, Monakhova N, Riabova O, Lane TR, Makarov V, Veras FP, Batah SS, Fabro AT, Oliva G, Cunha FQ, Alves-Filho JC, Cunha TM, Ekins S, Pyronaridine Protects against SARS-CoV-2 Infection in Mouse, ACS Infect Dis, 8 (2022) 1147–1160. [DOI] [PubMed] [Google Scholar]
- [53].Ekins S, Freundlich JS, Clark AM, Anantpadma M, Davey RA, Madrid P, Machine learning models identify molecules active against the Ebola virus in vitro, F1000Res, 4 (2015) 1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Villanueva PJ, Martinez A, Baca ST, DeJesus RE, Larragoity M, Contreras L, Gutierrez DA, Varela-Ramirez A, Aguilera RJ, Pyronaridine exerts potent cytotoxicity on human breast and hematological cancer cells through induction of apoptosis, PLoS One, 13 (2018) e0206467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].NCI, NCI-60 Human Tumor Cell Lines Screen, 2020.
- [56].Jayaraman SD, Ismail S, Nair NK, Navaratnam V, Determination of pyronaridine in blood plasma by high-performance liquid chromatography for application in clinical pharmacological studies, J Chromatogr B Biomed Sci Appl, 690 (1997) 253–257. [DOI] [PubMed] [Google Scholar]
- [57].Mijanovic O, Petushkova AI, Brankovic A, Turk B, Solovieva AB, Nikitkina AI, Bolevich S, Timashev PS, Parodi A, Zamyatnin AA, Cathepsin D-Managing the Delicate Balance, Pharmaceutics, 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ivankovic D, Chau KY, Schapira AH, Gegg ME, Mitochondrial and lysosomal biogenesis are activated following PINK1/parkin-mediated mitophagy, J Neurochem, 136 (2016) 388–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Geisslinger F, Müller M, Vollmar AM, Bartel K, Targeting Lysosomes in Cancer as Promising Strategy to Overcome Chemoresistance-A Mini Review, Front Oncol, 10 (2020) 1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Aits S, Jäättelä M, Lysosomal cell death at a glance, J Cell Sci, 126 (2013) 1905–1912. [DOI] [PubMed] [Google Scholar]
- [61].Krytska K, Ryles HT, Sano R, Raman P, Infarinato NR, Hansel TD, Makena MR, Song MM, Reynolds CP, Mossé YP, Crizotinib Synergizes with Chemotherapy in Preclinical Models of Neuroblastoma, Clin Cancer Res, 22 (2016) 948–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zhang L, Wu B, Baruchel S, Oral Metronomic Topotecan Sensitizes Crizotinib Antitumor Activity in ALK, Transl Oncol, 10 (2017) 604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hande KR, Etoposide: four decades of development of a topoisomerase II inhibitor, Eur J Cancer, 34 (1998) 1514–1521. [DOI] [PubMed] [Google Scholar]
- [64].Bailly C, Pyronaridine: An update of its pharmacological activities and mechanisms of action, Biopolymers, 112 (2021) e23398. [DOI] [PubMed] [Google Scholar]
- [65].Anderson E, Havener TM, Zorn KM, Foil DH, Lane TR, Capuzzi SJ, Morris D, Hickey AJ, Drewry DH, Ekins S, Synergistic drug combinations and machine learning for drug repurposing in chordoma, Sci Rep, 10 (2020) 12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Puhl AC, Gomes GF, Damasceno S, Godoy AS, Noske GD, Nakamura AM, Gawriljuk VO, Fernandes RS, Monakhova N, Riabova O, Lane TR, Makarov V, Veras FP, Batah SS, Fabro AT, Oliva G, Cunha FQ, Alves-Filho JC, Cunha TM, Ekins S, Pyronaridine Protects Against SARS-CoV-2 in Mouse, bioRxiv, (2021) 2021.2009.2030.462449. [DOI] [PubMed] [Google Scholar]
- [67].Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, Mutowo P, Atkinson F, Bellis LJ, Cibrian-Uhalte E, Davies M, Dedman N, Karlsson A, Magarinos MP, Overington JP, Papadatos G, Smit I, Leach AR, The ChEMBL database in 2017, Nucleic Acids Res, 45 (2017) D945–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Lane TR, Foil DH, Minerali E, Urbina F, Zorn KM, Ekins S, Bioactivity Comparison across Multiple Machine Learning Algorithms Using over 5000 Datasets for Drug Discovery, Mol Pharm, 18 (2021) 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Russo DP, Zorn KM, Clark AM, Zhu H, Ekins S, Comparing Multiple Machine Learning Algorithms and Metrics for Estrogen Receptor Binding Prediction, Mol Pharm, 15 (2018) 4361–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Public data from ChEMBL [67] was obtained for compounds screened against the SH-SY5Y cell line (IC50 data from Target ID 614910) and then used to generate a Bayesian machine learning model with our proprietary software Assay Central[68].


