Abstract
Most proteins implicated in neurodegenerative diseases bind metal ions, notably copper and zinc. Metal ion binding may be part of the protein’s function or, alternatively, may promote a deleterious gain of function. With regard to Cu2+ ions, electron paramagnetic resonance techniques have proven to be instrumental in determining the biophysical characteristics of the copper binding sites, as well as structural features of the coordinating protein and how they are impacted by metal binding. Here, the most useful methods are described as they apply to the prion protein, which serves as a model for the broader spectrum of neurodegenerative proteins.
1. Introduction
A wide range of neurodegenerative diseases associated with aging involve the aggregation of specific proteins of the central nervous system. Examples include the Aβ peptide and tau protein in Alzheimer’s disease (Selkoe & Hardy, 2016), α-synuclein in Parkinson’s disease (Proukakis et al., 2013; Spillantini et al., 1997) and the cellular prion protein (PrPC) in the Transmissible Spongiform Encephalopathies (TSEs) (Prusiner, 1982), otherwise known as prion diseases. Definitive functions for most of these proteins are still being worked out. Curiously, however, many of them are metal binding proteins and coordinate copper ions in the form of Cu2+ (Aronoff-Spencer et al., 2000; Burns et al., 2002, 2003; Bush, 2003; Dudzik, Walter, Abrams, Jurica, & Millhauser, 2013; Jobling et al., 2001; Millhauser, 2004, 2007; Proukakis et al., 2013). Considerable effort has been expended on defining the precise details of the copper sites in these proteins with the expectation that such structural information will provide insight into both protein function and the processes involved in aggregation and subsequent neurodegeneration.
Because Cu2+ has a paramagnetic d9 electron configuration, electron paramagnetic resonance (EPR) plays a key role in determining coordination, geometry and reactivity at copper centers. While each neurodegenerative protein exhibits its own unique coordination features and aggregation properties, PrPC is unique insofar that it possesses both structured and unstructured protein domains (Riek, Hornemann, Wider, Glockshuber, & Wuthrich, 1997). This methods chapter will therefore focus on PrPC as a model system for studying both prion and related neurodegerative peptides and proteins in their respective monomeric states. In addition, Zn2+ uptake by PrPC also features in the protein’s putative function. Although Zn2+ is diamagnetic, and therefore EPR silent, EPR methodologies have nevertheless proven useful in identifying its influence on global PrPC structure (Spevacek et al., 2013).
We consider two complementary strategies for defining Cu2+ coordination modes, which, by analogy to modern proteomics (Han, Jin, Breuker, & McLafferty, 2006), we refer to as “bottom-up” and “top-down.” With the bottom-up approach, short segments of a complicated protein are produced by either expression or Solid Phase Peptide Synthesis (SPPS) and examined by EPR in isolation from the full protein. Alternatively, the top-down strategy focuses on EPR of the full protein and select mutants. There are also intermediate strategies where one investigates intact protein domains. As we describe relevant EPR methodologies, the relative benefits of bottom-up and top-down will be elucidated.
The human prion protein is 209 residues in length, after removal of the N-terminal signal peptide, and is decorated by a C-terminal glycophosphatidylinositol (GPI)-anchor, two Asn-linked carbohydrate moieties (Hornemann, Schorn, & Wuthrich, 2004). There are small variations in length, depending on the specific mammalian species. As shown in Fig. 1, residues 23–125 are unstructured random coil (in the absence of metal ions), while the C-terminal domain, residues 126–231, forms a unique disulfide-stabilized structure composed of three α-helices and a short anti-parallel β-sheet (Riek et al., 1997). Also shown are familial mutations that give rise to inherited prion diseases (Prusiner, 1997; Spevacek et al., 2013).
Fig. 1.
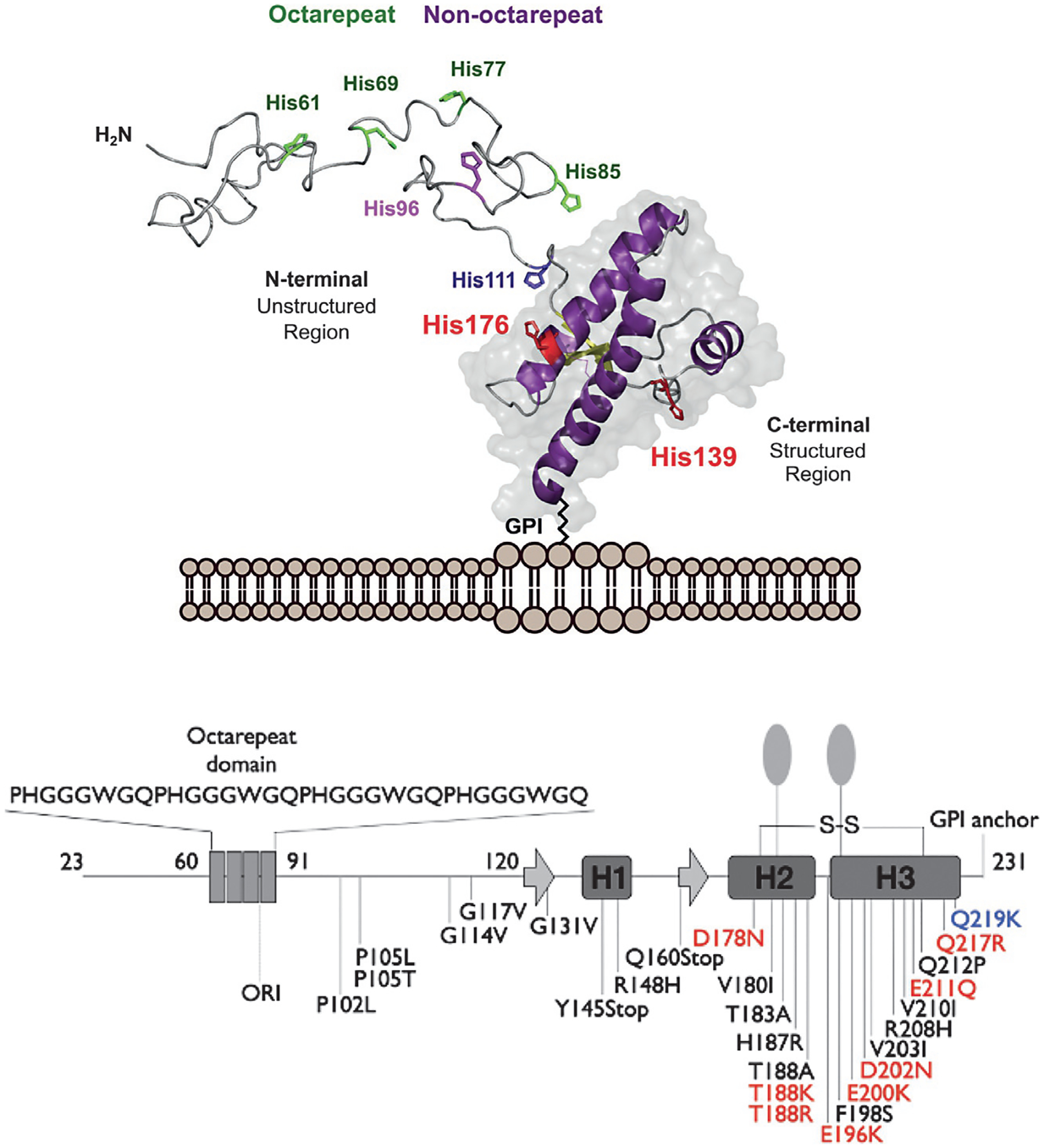
Top, three-dimensional rendering of the cellular prion protein, PrPC, noting the intrinsically disordered N-terminal domain and the helical C-terminal domain. Metal ion binding takes place primarily within the N-terminal domain involving octarepeat His residues (green) and non-octarepeat residues (purple). Bottom, PrPC sequence features revealing locations of the octarepeat domain, secondary structure elements in the C-terminal domain, GPI-anchor, glycan sites, and mutations that confer familial prion disease. Mutations labeled red shift side chain charge to a more positive state. The mutation Q219K (blue) is protective against disease. Figure adapted from Posadas, Y., Lopez-Guerrero, V., Segovia, J., Perez-Cruz, C., & Quintanar, L. (2022) Dissecting the copper bioinorganic chemistry of the functional and pathological roles of the prion protein: Relevance in Alzheimer’s disease and cancer. Current Opinion in Chemical Biology 66, 102098.
PrPC is localized to extracellular membrane surfaces through insertion of the GPI-anchor into the outer membrane leaflet (Stahl, Borchelt, Hsiao, & Prusiner, 1987). While PrPC is localized predominantly in the central nervous system, it is also found distributed in peripheral tissues (Zomosa-Signoret, Arnaud, Fontes, Alvarez-Martinez, & Liautard, 2008). A notable feature of the unstructured segment of PrPC is the octarepeat (OR) domain, so named because it is composed of tandem, eight-residue PHGGGWGQ repeats (Aronoff-Spencer et al., 2000; Hornshaw, McDermott, & Candy, 1995). Initial work demonstrating copper binding focused in the OR segment, given that it contains four His residues. However, more recent work finds that there are important copper-binding segments outside of this domain (Burns et al., 2003; Chaves et al., 2014; Giachin et al., 2015; Grande-Aztatzi, Rivillas-Acevedo, Quintanar, & Vela, 2013; Qin, Yang, Mastrangelo, & Westaway, 2002; Rivillas-Acevedo et al., 2011) and, moreover, that copper stabilizes global PrPC structure through simultaneous coordination between OR and C-terminal His residues (Evans & Millhauser, 2017; Evans, Pushie, Markham, Lee, & Millhauser, 2016; McDonald et al., 2019; Schilling et al., 2020). Recent reviews describe the links among prion protein copper binding, physiological function and global structure (Evans & Millhauser, 2017; Posadas, Lopez-Guerrero, Segovia, Perez-Cruz, & Quintanar, 2022; Salzano, Giachin, & Legname, 2019).
This chapter will describe the EPR methods listed in Table 1, as they apply to characterizing Cu2+, and to a lesser extent Zn2+, binding to PrPC. As we work through the approaches, we will note differential benefits to bottom-up vs top-down strategies.
Table 1.
EPR methods and their applicability in Bottom up vs Top down investigations.
| EPR method | Bottom up (peptides) | Top down (proteins) |
|---|---|---|
| Continuous Wave (CW) | ✓✓ | ✓ |
| ESEEM | ✓✓ | ✓✓ |
| Isotope edited ESEEM and HYSCORE | ✓✓ | * |
| DEER Nitroxide-Nitroxide | ✓ | ✓✓ |
| DEER Copper-Nitroxide | ✓ | ✓✓ |
| Pulsed ENDOR | ✓ | ✓✓ |
✓✓: ideally suited; ✓: suitable but with limitations; *: requires specialized sample design.
2. Sample preparation
EPR samples typically contain Cu2+ concentrations from 10μM to nearly a 1.0mM. The sample concentration must be adapted to the specific technique. Continuous Wave (CW) EPR is the most sensitive and easily operates at the low end of the feasible concentration range. Buffers are selected to avoid those that directly bind copper and may be degassed before use to avoid line broadening from dissolved oxygen. We find that morpholine-based buffers, such as N-ethylmorpholine (NEM) or 2-(N-morpholino)ethanesulfonic acid (MES), work well for stabilizing solutions in the pH range below 7.0; while other noncomplexing tertiary amine buffers have been reported (Yu, Kandegedara, Xu, & Rorabacher, 1997). Most EPR experiments are performed in frozen solution and, therefore, require a cryoprotectant such as glycerol, which helps maintain a spatially uniform distribution of the proteins. We note that free copper at sample pH values above roughly 6.0 forms diamagnetically-coupled hydroxides and, thus, are EPR silent (Aronoff-Spencer et al., 2000). In other words, if one adds an excess of copper to a protein solution at pH above 6.0, any copper remaining in solution and not bound to the protein, will not give an EPR signal. On the other hand, if working below pH6.0, signals from free Cu2+ may subtracted from acquired spectra. Finally, certain pulsed EPR methods benefit from sample preparation strategies that give long T2 or T1 relaxation times. In such cases, it is often beneficial to use deuterated solvents.
A typical EPR sample solution will have the following:
25mMN-ethylmorpholine (NEM); pH adjusted with HCl or KOH to 6.0 or above (20 to 100mM NaCl may be required for stability of certain proteins)
20–30% by volume glycerol
protein or peptide solution (100μM and above), degassed (if necessary) with nitrogen, helium, or by freeze–pump–thaw cycles
paramagnetic metal ion salt in final concentration 100μM and above, matched to appropriate protein stoichiometry
3. Continuous wave (CW) EPR
CW EPR, typically performed at X-band frequencies (~9.0GHz) is a primary technique for determining EPR spectra. The great benefit of CW EPR for Cu2+ bound to neurodegenerative proteins is that one may quickly extract spectral parameters that report on the coordination environment. Correlations originally developed by Peisach and Blumberg (1974), applied to frozen aqueous solutions, distinguish the degree of nitrogen, oxygen, sulfur bound to the copper center. Moreover, if there are multiple coordination species, these are often represented as superimposed spectra. CW EPR is especially useful for bottom-up strategies since short peptide segments give sharp, well-resolved Cu2+ spectra.
An early experiment demonstrating the benefit of CW EPR applied to PrPC is shown in Fig. 2 (Aronoff-Spencer et al., 2000). With the hypothesis that Cu2+ would coordinate to the OR domain (within residues 57–91), this experiment examined spectra obtained from a series of progressively shorter, OR-derived peptides (Aronoff-Spencer et al., 2000). By simple inspection, one can see that peptides down to the segment HGGGW yield spectra approximately equivalent to the full OR-domain. However, peptides shorter than this (HGGG and HGG) give spectra clearly distinguishable from the OR spectrum. In addition, analysis of the copper hyperfine splittings (I = 3/2), as indicated by the grid markers, suggest that coordination arises from one oxygen and three nitrogen atoms (1O, 3N). These findings were eventually borne out by X-ray crystallography, thus demonstrating the power of this very straightforward technique.
Fig. 2.
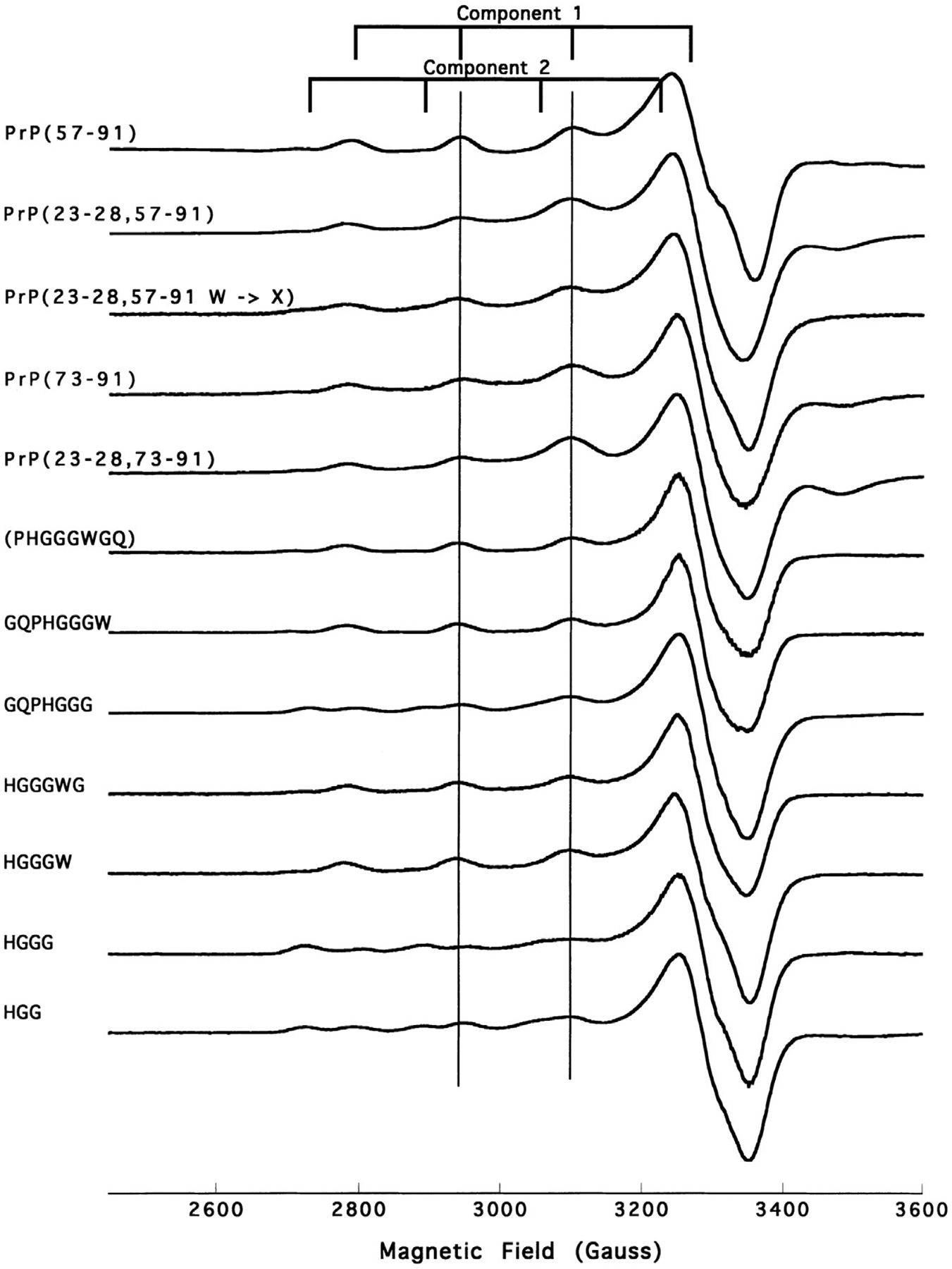
X-band CW EPR spectra, displayed in the first-derivative mode, of OR-derived peptides obtained at pH7.45 and 77K. The grid markers identify the four hyperfine lines from the copper I = 3/2 nucleus. These spectra suggest the existence of two distinct species, initially referred to as Component 1 and Component 2.
Beyond the octarepeat region, PrPC displays two additional Cu2+ binding sites associated to His96 and His111 that have been termed as non-octarepeat sites (Burns et al., 2003). These sites have also been modeled by peptide fragments, such as 92–96 (GGGTH) and 106–115 (KTNMKHMAGA). EPR studies of these peptides and several variants helped elucidate that metal binding to these sites is pH dependent and it involves deprotonated amides that precede the His residue in the sequence (Fig. 3) (Grande-Aztatzi et al., 2013; Rivillas-Acevedo et al., 2011). In both cases, drastic changes in the EPR spectra of the Cu2+ complexes reflect a change in the equatorial coordination mode from a 3N1O to a 4N shell. The fourth nitrogen-based ligand arises from the deprotonation of a backbone amide (pKa ~7.5–7.8) that enters the metal coordination sphere. Sometimes, a highly covalent nitrogen-rich environment may yield strong hyperfine nitrogen couplings that become evident in the EPR spectrum, as is the case of the Cu2+ complex with GGGTH at pH8.5 (Fig. 3). Pulsed EPR techniques can be used to accurately measure nitrogen couplings (vide infra).
Fig. 3.
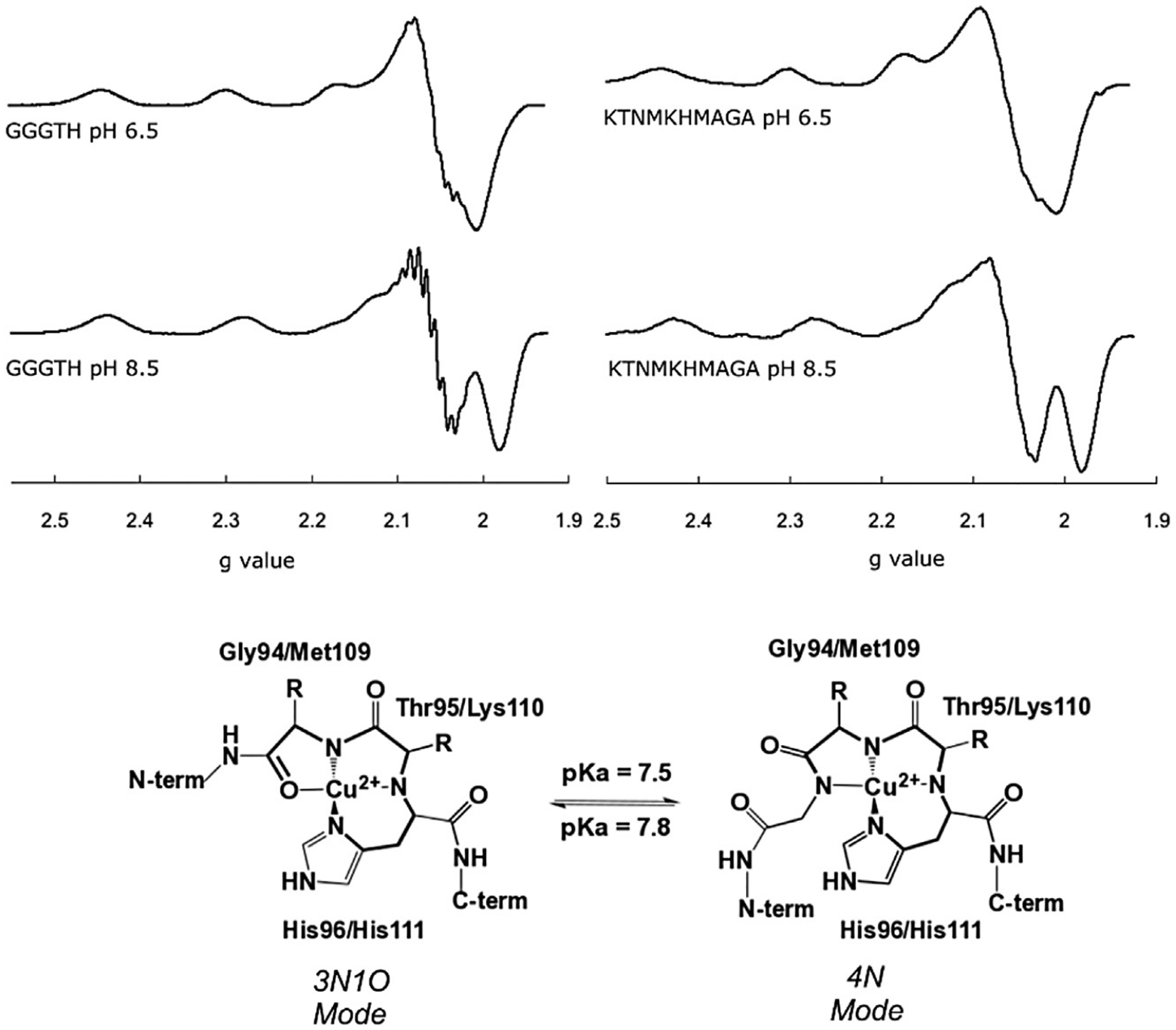
X-band CW EPR spectra of Cu2+ complexes at the His96 (GGGTH) and His111 (KTNMKHMAGA) sites, at pH6.5 and 8.5. Spectral changes upon pH are associated to the conversion of a 3N1O equatorial coordination mode to a 4N, as drawn. The pKa associated to this conversion is 7.8 for the His96 site and 7.5 for the His111 site.
Using peptides to model Cu2+ binding sites in PrPC has been a successful approach, given the fact that the N-terminal region is intrinsically disordered. When using peptide fragments to model metal binding sites, it is important to acetylate the N-terminus and amidate the C-terminus to cap the free NH2 and COO− groups of the peptide and prevent them from coordinating to the metal ion. Indeed, having a free NH2 group drastically impacts Cu2+ coordination to a peptide, as shown for the HMAGA fragment, which models how the His111 site in PrPC is impacted upon α-cleavage (Sanchez-Lopez, Fernandez, & Quintanar, 2018). The electron pair of a free-NH2 group acts as a strong anchoring moiety for Cu2+. Indeed, a titration of the NH2-HMAGA peptide by Cu2+ reveals the presence of species where the free-NH2 group dominates metal coordination (Fig. 4).
Fig. 4.
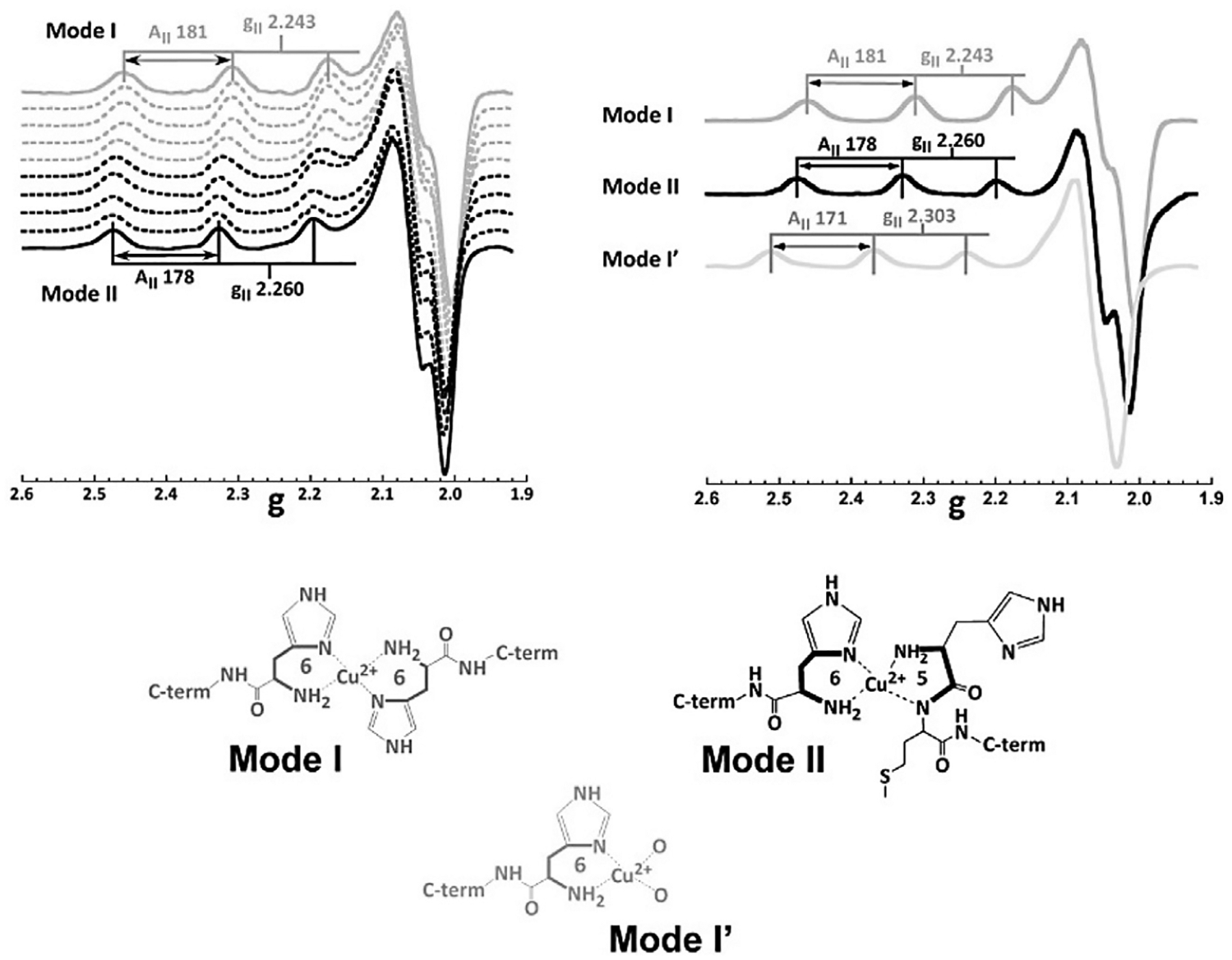
X-band CW EPR spectra of Cu2+ complexes with the NH2-HMAGA fragment (without acetylation), modeling metal binding to His111 in α-cleaved PrPC. A titration of the NH2-HMAGA peptide yields EPR spectra that reveals two species: Mode I favored up to 0.5 equiv. of Cu2+, and Mode II that forms at a 1:1 metal: peptide ratio. At low equiv. of metal ion (0.5 equiv) and low pH (5.0) a third species is favored, termed Mode I′. Proposed coordination models for these species are shown. Figure adapted from Sanchez-Lopez C, Fernandez CO, & Quintanar L (2018) Neuroprotective alpha-cleavage of the human prion protein significantly impacts Cu(II) coordination at its His111 site. Dalton Transactions, 47(28), 9274–9282.
4. Electron spin echo envelope modulation (ESEEM)
Histidine residues, with their imidazole sidechains, invariably play critical roles in both copper and zinc coordination in proteins. ESEEM is a pulsed EPR technique uniquely diagnostic for imidazole coordination to copper centers (Mims & Peisach, 1978). The measurement entails recording the intensity of a spin echo as a function of evolving delays between specific microwave pulses. There are several schemes, the benefits of which are thoroughly reviewed elsewhere (Kevan & Bowman, 1990; Sahu, McCarrick, & Lorigan, 2013). Regardless of specific pulse scheme, the Fourier transform of the time-dependent signal reveals frequencies arising from 14N (I = 1) nuclear quadrupole transitions. With Cu2+ X-band EPR, the observed transitions are intense, arising from the remote nitrogen of the coordinated imidazole ring.
As shown in Fig. 5A, there are three characteristic transitions below 2.0MHz arising from the individual quadrupole resonances among the three 14N spin states. A fourth so-called “double quantum transition” occurs at approximately 4.0MHz. Given the uniqueness of this spectrum, ESEEM is ideally diagnostic for His coordination.
Fig. 5.
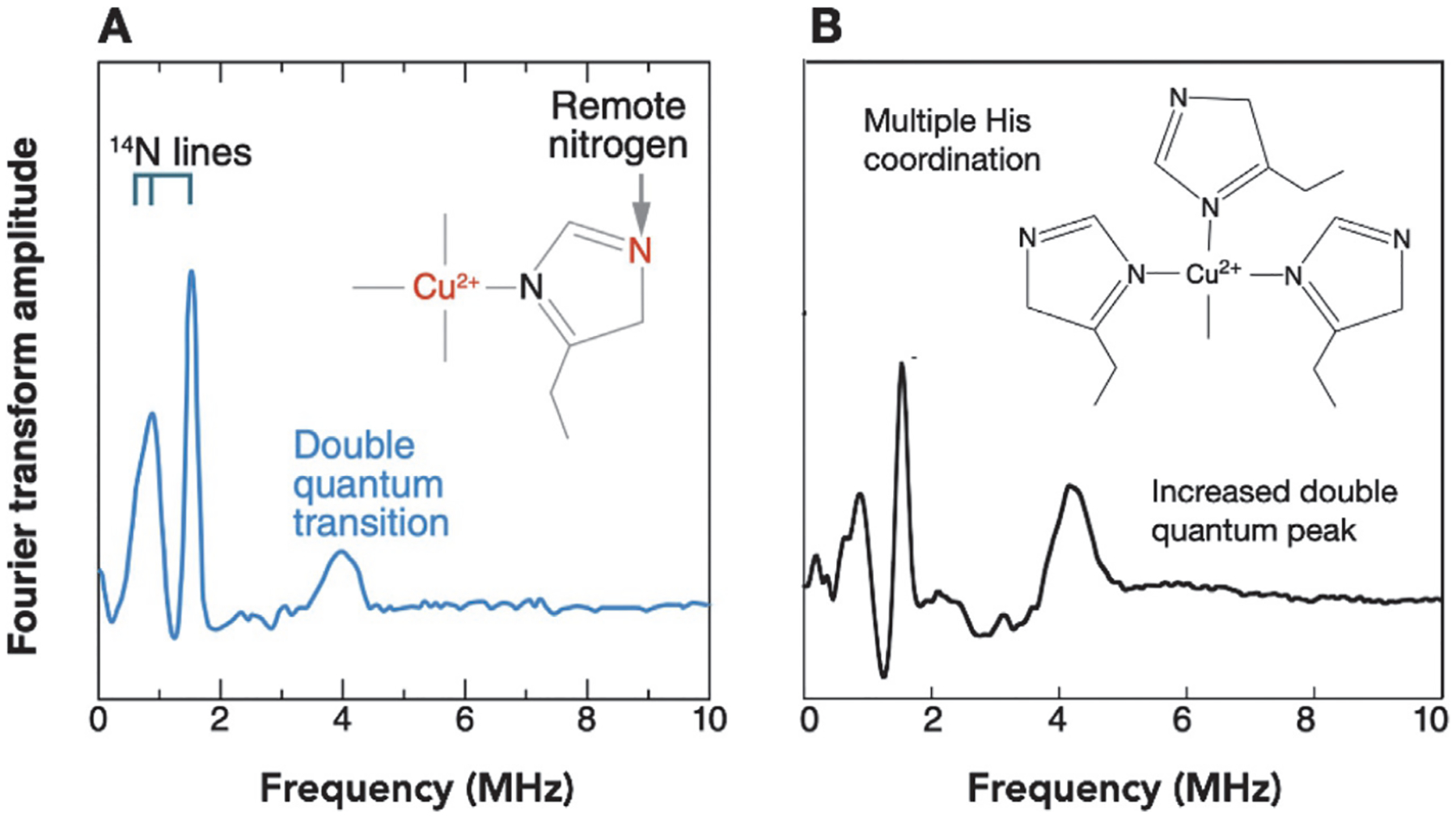
Characteristic ESEEM spectra for (A) single His coordination and (B) multiple His coordination. The three lines below 2.0MHz in each case arise from the remote 14N (I = 1) nuclear quadrupole transitions. The intensity of the transition at ~4MHz is indicative of the number of coordinated His residues.
ESEEM is readily applied in both bottom-up and top-down strategies. Copper proteins at typical EPR concentrations (100μM and above), and temperatures 20K or lower, give quality ESEEM spectra with fully resolvable lines (Chattopadhyay et al., 2005). Moreover, when developing bottom-up peptide models of protein copper centers, ESEEM spectra may be used to compare and validate preservation of the coordination environment. There are important benefits that extend beyond the simple identification of imidazole coordination. For example, Fig. 5B shows a resulting ESEEM spectrum for multiple His coordination with a diagnostic increase in the double quantum transition (Chattopadhyay et al., 2005). With PrPC, this unique spectroscopic signature was combined with CW EPR measurements to demonstrate concentration-dependent changes in His coordination in the OR domain as a function of copper:domain ratio. Specifically, when saturated at high [Cu2+], each His coordinates a single Cu2+ ion, which is referred to as component 1 binding. However, at low [Cu2+], approximately 1:1 copper to protein, each copper ion is coordinated by multiple His residues (referred to as component 3 binding). These observations offer perspective on how to carry out characterization with other neurodegenerative proteins. If there are multiple, proximal His residues in a flexible, disordered polypeptide segment, concentration-dependent measurements combined with ESEEM are essential.
With the bottom-up approach, peptides prepared with selectively placed stable isotopes or unnatural amino acids may to used to directly test proposed coordination models. For example, ESEEM obtained from a peptide corresponding to the OR domain, fully Cu2+ saturated (Component 1), yields the ESEEM spectrum shown in Fig. 6. There are multiple lines beyond those expected from the transitions due to the single His imidazole. Molecular modeling, pH titrations and companion EPR measurements, suggested the structure shown in Fig. 6.
Fig. 6.
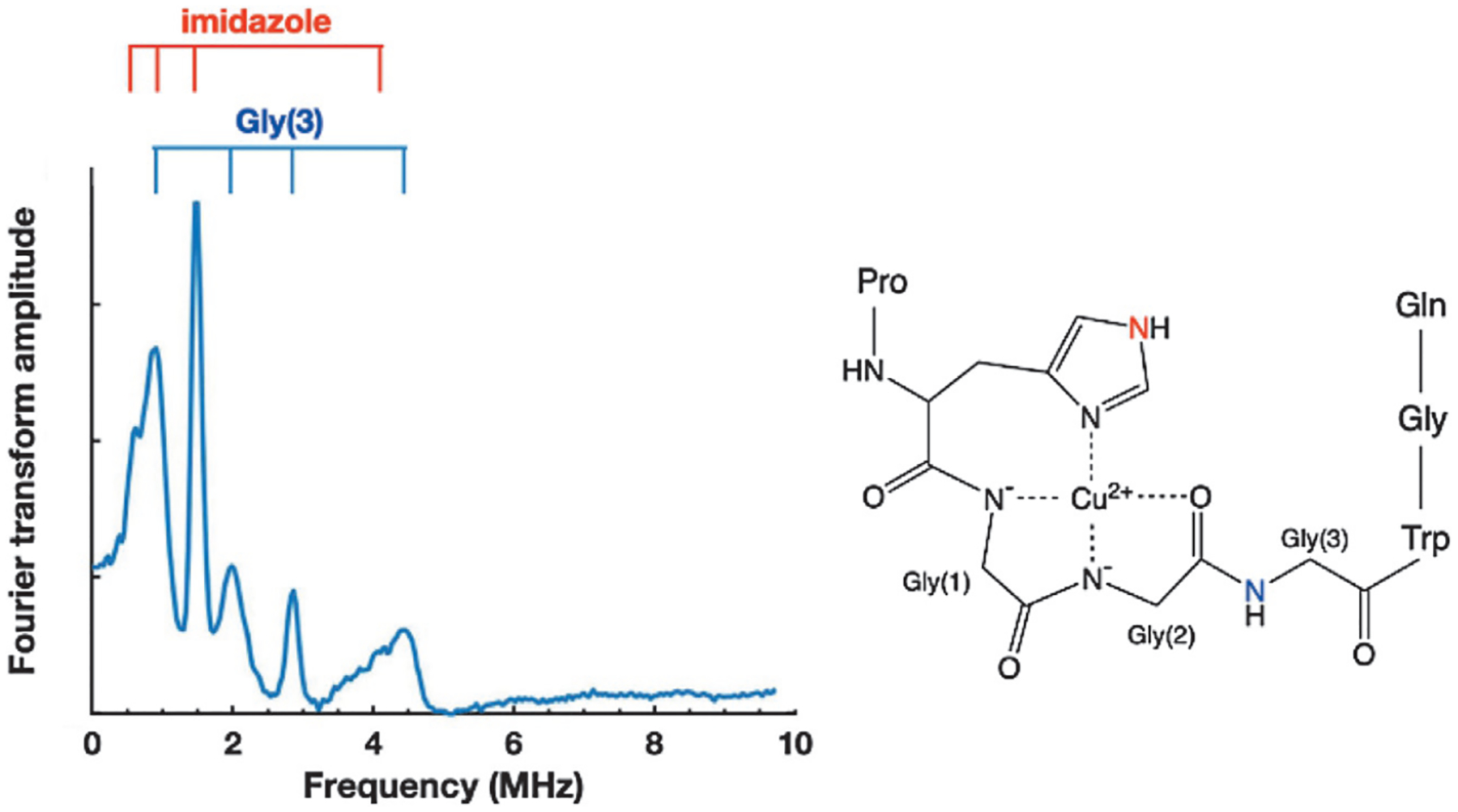
ESEEM spectra obtained from Cu2+ coordinated by a single repeat segment. Transitions beyond those assigned to the remote nitrogen of the single His side chain (red) arise from the backbone nitrogen of Gly(3) (blue). The assignment of Gly(3) was verified by the introduction of a non-coordinating residue at Gly(2).
A necessary consequence of this structure is that the amide nitrogen of Gly(3) (blue) is rigidly held at a distance compatible with X-band ESEEM transitions. Therefore, a prediction from the structure in Fig. 6 is that disruption of Gly(2) coordination to Cu2+ would eliminate the fixed position of Gly(3). To test the feasibility of this structure, OR peptides prepared with N-methylglycine (sarcosine) at Gly(2) were examined by ESEEM (Chattopadhyay et al., 2005). The resulting spectra were equivalent to that of Fig. 5A, thus confirming assignment of the additional lines to 14N from Gly(3) and, importantly, further validating the proposed component 1 structure in Fig. 6.
Hyperfine Sublevel Correlation (HYSCORE) spectroscopy is a two-dimensional variation of ESEEM that often reveals additional electron-nuclear couplings (Hofer, Grupp, & Mehring, 1986). For example, while ESEEM at X-band is mainly limited to weakly coupled I = 1 nuclei, such as those demonstrated in the examples above, HYSCORE expands this capability to anisotropically broadened transitions observed for dipolar coupled I = ½ nuclei (Burns et al., 2002). This capability was used in a bottom-up refinement of the component 1 structure shown in Fig. 6. Specifically, replacement of Gly(3) with its 15N-labeled analog in the peptide HGGGW revealed pronounced cross peaks with frequencies commensurate of a non-liganded 15N nucleus, and consistent with the distance between the copper center and the nitrogen of the third glycine. The significant advantage in this example is that, as opposed to editing out unassigned peaks by disruption of the coordination structure, as was done with the introduction of sarcosine, or by elimination of peaks with 15N labels, HYSCORE provides positive spectroscopic confirmation with output that one can use to quantitatively assess dipolar-coupled distances.
5. Double electron-electron resonance (DEER) EPR
Intramolecular distance measurements are often crucial for identifying structural features and organization. The canonical methods for assessing monomeric peptide and protein structure are, of course, X-ray crystallography and NMR. However, in the case of proteins with large, disordered segments, or with bound paramagnetic species such as Cu2+, application of these methods may not be feasible. In the case of PrPC, both methods have yielded critical structural details for the ordered C-terminal domain, but neither has succeeded in resolving structure or detailed Cu2+-binding properties of the intrinsically disordered N-terminal domain.
DEER EPR is a two frequency, pump-probe pulsed technique that provides detailed distances and distance distributions in the range from approximately 20Å to 100Å and beyond (Jeschke & Polyhach, 2007). Typically, DEER EPR is applied to distances between pairs of nitroxide spin labels, with each label engineered to a specific site within a protein or protein complex. Measurements are carried out at cryogenic temperatures of 80K or below, depending on sample and instrument set up. The theory and detailed application of DEER EPR, along with benefits and shortcomings, has been described in numerous reviews (Schiemann et al., 2021). Modern instrumentation readily obtains quality distance data from samples of less than 80μL and 30μM doubly-labeled protein or peptide. The focus here will be on the application of DEER EPR to domain organization within PrPC.
DEER EPR has thus far provided essential information for assessing how both Zn2+ (Spevacek et al., 2013) and Cu2+ (Evans et al., 2016) impact PrPC structure, with measurements performed on the full protein, and hence, a top-down strategy. Regarding Zn2+ coordination to PrPC, stoichiometry and chemical protection assays initially demonstrated that the ion coordinates to the four His imidazoles within the OR domain (Walter, Stevens, Visconte, & Millhauser, 2007). However, physiological experiments suggested that Zn2+ might introduce structural organization beyond OR coordination. To address this with DEER EPR, a family of PrPC constructs were developed where each has one spin label in the C-terminal domain and a second in, or near, the OR domain. Nitroxide spin labeling vicinal to the OR domain used the standard approach of introducing a unique Cys residue, followed by reaction with the methane-thiosulfonate spin label (MTSL) reagent. However, because the C-terminal domain possesses two conserved Cys residues that form a critical disulfide bond, spin labeling with MTSL is problematic. Consequently, labeling in this domain utilized an expanded genetic code approach to incorporate para-acetyl phenylalanine, followed by reaction with a hydroxylamine nitroxide reagent, to produce the ketoxime-linked K1 side chain (Fig. 7) (Fleissner et al., 2009).
Fig. 7.
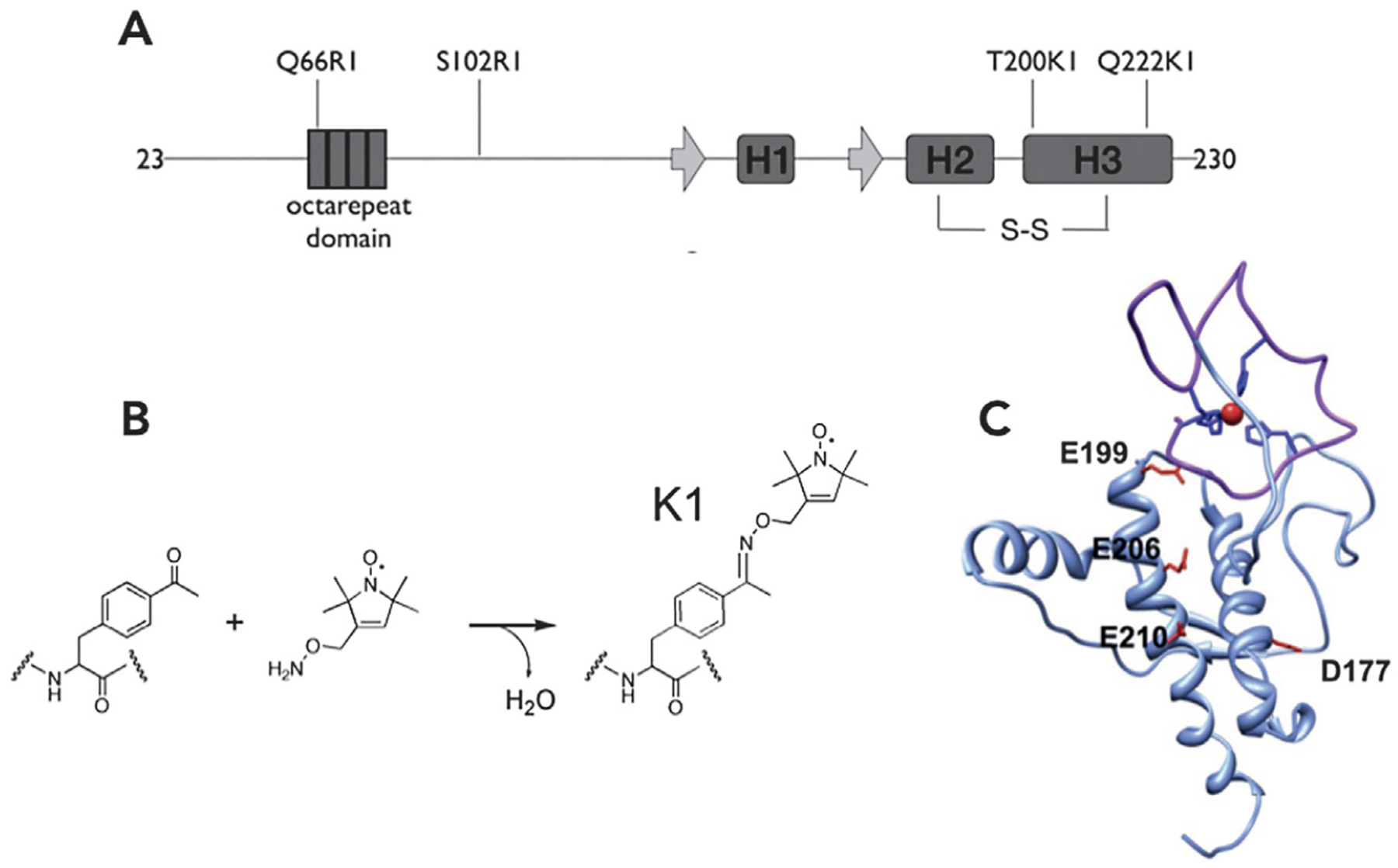
Nitroxide labeling strategy and resulting PrPC conformation resulting from Zn2+ coordination in the OR domain. (A) N-terminal labeling was achieved by introduction of a selectively placed Cys residue, followed by reaction with MTSL, resulting in the R1 side chain. (B) Labeling in the C-terminal domain involved incorporation of the unnatural amino acid para-acetyl phenylalanine, which was then reacted with hydroxylamine nitroxide reagent. (C) Energy minimization using distances derived from the nitroxide-nitroxide DEER EPR results identified a compact structure with the Zn2+-OR forming an interdomain cis interaction with the regulatory C-terminal domain.
Nitroxide-nitroxide X-band DEER EPR was performed on the family of doubly labeled proteins both in the absence and presence of one equivalent of Zn2+ (labeling scheme shown in Fig. 7A). Upon addition of zinc, a clear decrease in interdomain distances was observed, consistent with docking between the Zn2+-occupied OR domain and a well-defined surface patch on the C-terminal domain, as shown in Fig. 7. Refined with energy minimization, DEER EPR distance restraints place the Zn2+-occupied OR over a conserved cluster of negatively charged residues comprised of D177, E199, E206 and E210 (mouse sequence, Fig. 7C).
Interestingly, families who carry charge altering mutation at D177, E199, or E210 are predisposed to inherited prion disease. These findings led to the hypothesis that the Zn2+-promoted interdomain interaction is protective against PrPC-mediated neuronal toxicity. While the initial experiments described here were performed at X-band, it is now well recognized that Q-band (34GHz) EPR is vastly more sensitive for nitroxide-nitroxide DEER EPR measurements.
The physiological implications raised by the studies with Zn2+ motivated parallel investigations with copper, specifically probing whether Cu2+ would drive similar domain-domain contacts. The intrinsic paramagnetism of Cu2+ enables direct DEER EPR distance measurements between this ion and site-specifically attached spin labels. To keep both the DEER nitroxide pump and copper probe frequencies within the resonator bandwidth, this experiment is currently limited to X-band EPR. Detailed measurements from four PrPC constructs, with K1 spin labels in the C-terminal domain, resulted in precise localization of the copper ion above the conserved, negatively charged patch described above (Evans et al., 2016).
Because DEER EPR does not resolve distances below ~15Å, it may be important to consider the possibility that shorter Cu-nitroxide distances exist in the distributions derived from these spin-labeled PrP variants. To address this, a room temperature CW EPR on the same samples, along with careful measurements of nitroxide line intensity reduction that results from dipolar broadening, identifies conformations resulting from copper ion—nitroxide distances within 8–25Å (Voss, Salwinski, Kaback, & Hubbell, 1995). When applied to the Cu2+-PrPC complex, the resulting distances supported the distributions derived from the copper-nitroxide DEER EPR measurements.
An exciting hypothesis from the studies above is that copper and zinc stabilize conformations of PrPC that resist cellular toxicity, as implicated in neurodegenerative disease. Although beyond the scope of this EPR-centered chapter, it is noteworthy that subsequent electrophysiological experiments performed on cultured cells demonstrated that the metal ion-promoted a cis interaction that is indeed protective against PrPC-caused membrane leakage and consequent cell death (Schilling et al., 2020; Wu et al., 2017).
6. Concluding remarks
The methods described in this chapter demonstrate how both basic and advanced EPR methods further the understanding and reveal critical features of partially ordered proteins, relevant to neurodegenerative diseases. Speciation and structural details of copper coordination sites in proteins can be obtained from EPR studies. Beyond those approaches described here, one may also apply pulsed Electron-Nuclear Double Resonance (ENDOR) to elucidate fine structural details of copper coordination sites. While specific metal binding sites in intrinsically disordered proteins may be successfully modeled by peptide fragments (“bottom-up” approach), correlation to studies of the full protein (“top-down” approach) provide important insights into the impact of metal binding in protein structure. What’s truly exciting at this juncture is that EPR methods applied to PrPC revealed unanticipated, metal ion-assisted conformations that regulate the protein’s intrinsic toxicity. Given the remarkable spectrum of proteins involved in neurodegenerative processes, there is little doubt that EPR will uncover similar stories as they apply to other protein-driven dementias.
Acknowledgments
Authors would like to thank Yanahi Posadas and Carolina Sánchez-López for assistance with adapting Figs. 3 and 4. L.Q. thanks funding from Ministry of Education of Mexico (SEP-Cinvestav). G.L.M. gratefully acknowledges funding in support of this work from the National Institutes of Health (R35 GM131781).
References
- Aronoff-Spencer E, Burns CS, Avdievich NI, Gerfen GJ, Peisach J, Antholine WE, et al. (2000). Identification of the Cu2+ binding sites in the N-terminal domain of the prion protein by EPR and CD spectroscopy. Biochemistry, 39, 13760–13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CS, Aronoff-Spencer E, Dunham CM, Lario P, Avdievich NI, Antholine WE, et al. (2002). Molecular features of the copper binding sites in the octarepeat domain of the prion protein. Biochemistry, 41, 3991–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CS, Aronoff-Spencer E, Legname G, Prusiner SB, Antholine WE, Gerfen GJ, et al. (2003). Copper coordination in the full-length, recombinant prion protein. Biochemistry, 42(22), 6794–6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush AI (2003). The metallobiology of Alzheimer’s disease. Trends in Neurosciences, 26(4), 207–214. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay M, Walter ED, Newell DJ, Jackson PJ, Aronoff-Spencer E, Peisach J, et al. (2005). The Octarepeat domain of the prion protein binds cu(II) with three distinct coordination modes at pH 7.4. Journal of the American Chemical Society, 127(36), 12647–12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves JA, Sanchez-Lopez C, Gomes MP, Sisnande T, Macedo B, de Oliveira VE, et al. (2014). Biophysical and morphological studies on the dual interaction of non-octarepeat prion protein peptides with copper and nucleic acids. Journal of Biological Inorganic Chemistry, 19(6), 839–851. [DOI] [PubMed] [Google Scholar]
- Dudzik CG, Walter ED, Abrams BS, Jurica MS, & Millhauser GL (2013). Coordination of copper to the membrane-bound form of alpha-synuclein. Biochemistry, 52(1), 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans EGB, & Millhauser GL (2017). Copper- and zinc-promoted Interdomain structure in the prion protein: A mechanism for autoinhibition of the neurotoxic N-terminus. Progress in Molecular Biology and Translational Science, 150, 35–56. [DOI] [PubMed] [Google Scholar]
- Evans EG, Pushie MJ, Markham KA, Lee HW, & Millhauser GL (2016). Interaction between prion Protein’s copper-bound Octarepeat domain and a charged C-terminal pocket suggests a mechanism for N-terminal regulation. Structure, 24(7), 1057–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner MR, Brustad EM, Kalai T, Altenbach C, Cascio D, Peters FB, et al. (2009). Site-directed spin labeling of a genetically encoded unnatural amino acid. Proceedings of the National Academy of Sciences of the United States of America, 106(51), 21637–21642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachin G, Mai PT, Tran TH, Salzano G, Benetti F, Migliorati V, et al. (2015). The non-octarepeat copper binding site of the prion protein is a key regulator of prion conversion. Scientific Reports, 5, 15253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande-Aztatzi R, Rivillas-Acevedo L, Quintanar L, & Vela A (2013). Structural models for cu(II) bound to the fragment 92–96 of the human prion protein. The Journal of Physical Chemistry. B, 117(3), 789–799. [DOI] [PubMed] [Google Scholar]
- Han X, Jin M, Breuker K, & McLafferty FW (2006). Extending top-down mass spec-trometry to proteins with masses greater than 200 kilodaltons. Science, 314(5796), 109–112. [DOI] [PubMed] [Google Scholar]
- Hofer P, Grupp A, & Mehring M (1986). High-resolution time-domain electron-nuclear-sublevel spectroscopy by pulsed coherence transfer. Phys Rev A Gen Phys, 33(5), 3519–3522. [DOI] [PubMed] [Google Scholar]
- Hornemann S, Schorn C, & Wuthrich K (2004). NMR structure of the bovine prion protein isolated from healthy calf brains. EMBO Reports, 5(12), 1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornshaw MP, McDermott JR, & Candy JM (1995). Copper binding to the N-terminal tandem repeat regions of mammalian and avian prion protein. Biochem. Biophys. Res. Comm, 207, 621–629. [DOI] [PubMed] [Google Scholar]
- Jeschke G, & Polyhach Y (2007). Distance measurements on spin-labelled bio-macromolecules by pulsed electron paramagnetic resonance. Physical Chemistry Chemical Physics, 9(16), 1895–1910. [DOI] [PubMed] [Google Scholar]
- Jobling MF, Huang X, Stewart LR, Barnham KJ, Curtain C, Volitakis I, et al. (2001). Copper and zinc binding modulates the aggregation and neurotoxic properties of the prion peptide PrP106–126. Biochemistry, 40, 8073–8084. [DOI] [PubMed] [Google Scholar]
- Kevan L, & Bowman MK (1990). Modern pulsed and continuous-wave electron spin resonance. New York: Wiley. [Google Scholar]
- McDonald AJ, Leon DR, Markham KA, Wu B, Heckendorf CF, Schilling K, et al. (2019). Altered domain structure of the prion protein caused by cu(2+) binding and functionally relevant mutations: Analysis by cross-linking, MS/MS, and NMR. Structure, 27(6), 907–922 e905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millhauser GL (2004). Copper binding in the prion protein. Accounts of Chemical Research, 37(2), 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millhauser GL (2007). Copper and the prion protein: Methods, structures, function, and disease. Annual Review of Physical Chemistry, 58, 299–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mims WB, & Peisach J (1978). The nuclear modulation effect in electron spin echoes for complexes of Cu2+ and imidazole with 14N and 15N. The Journal of Chemical Physics, 69, 4921–4930. [Google Scholar]
- Peisach J, & Blumberg WE (1974). Structural implications derived from the analysis of electron paramagnetic resonance spectra of natural and artificial copper proteins. Archives of Biochemistry and Biophysics, 165, 691–708. [DOI] [PubMed] [Google Scholar]
- Posadas Y, Lopez-Guerrero V, Segovia J, Perez-Cruz C, & Quintanar L (2022). Dissecting the copper bioinorganic chemistry of the functional and pathological roles of the prion protein: Relevance in Alzheimer’s disease and cancer. Current Opinion in Chemical Biology, 66, 102098. [DOI] [PubMed] [Google Scholar]
- Proukakis C, Dudzik CG, Brier T, MacKay DS, Cooper JM, Millhauser GL, et al. (2013). A novel alpha-synuclein missense mutation in Parkinson disease. Neurology, 80(11), 1062–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB (1982). Novel proteinaceous infectious particles cause scrapie. Science, 216, 136–144. [DOI] [PubMed] [Google Scholar]
- Prusiner SB (1997). Prion diseases and the BSE crisis. Science, 278, 245–251. [DOI] [PubMed] [Google Scholar]
- Qin K, Yang Y, Mastrangelo P, & Westaway D (2002). Mapping cu(II) bindings sites in prion proteins by diethylpyrocarbonate modification and MALDI-TOF mass spectro-metric “footprinting”. The Journal of Biological Chemistry, 277, 1981–1990. [DOI] [PubMed] [Google Scholar]
- Riek R, Hornemann S, Wider G, Glockshuber R, & Wuthrich K (1997). NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231). FEBS Letters, 413, 282–288. [DOI] [PubMed] [Google Scholar]
- Rivillas-Acevedo L, Grande-Aztatzi R, Lomeli I, Garcia JE, Barrios E, Teloxa S, et al. (2011). Spectroscopic and electronic structure studies of copper(II) binding to His111 in the human prion protein fragment 106–115: Evaluating the role of protons and methionine residues. Inorganic Chemistry, 50(5), 1956–1972. [DOI] [PubMed] [Google Scholar]
- Sahu ID, McCarrick RM, & Lorigan GA (2013). Use of electron paramagnetic resonance to solve biochemical problems. Biochemistry, 52(35), 5967–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzano G, Giachin G, & Legname G (2019). Structural consequences of copper binding to the prion protein. Cell, 8(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Lopez C, Fernandez CO, & Quintanar L (2018). Neuroprotective alpha-cleavage of the human prion protein significantly impacts Cu(II) coordination at its His111 site. Dalton Transactions, 47(28), 9274–9282. [DOI] [PubMed] [Google Scholar]
- Schiemann O, Heubach CA, Abdullin D, Ackermann K, Azarkh M, Bagryanskaya EG, et al. (2021). Benchmark test and guidelines for DEER/PELDOR experiments on nitroxide-labeled biomolecules. Journal of the American Chemical Society, 143(43), 17875–17890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling KM, Tao L, Wu B, Kiblen JTM, Ubilla-Rodriguez NC, Pushie MJ, et al. (2020). Both N-terminal and C-terminal histidine residues of the prion protein are essential for copper coordination and neuroprotective self-regulation. Journal of Molecular Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ, & Hardy J (2016). The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Molecular Medicine, 8(6), 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spevacek AR, Evans EG, Miller JL, Meyer HC, Pelton JG, & Millhauser GL (2013). Zinc drives a tertiary fold in the prion protein with familial disease mutation sites at the interface. Structure, 21(2), 236–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, & Goedert M (1997). Alpha-synuclein in Lewy bodies. Nature, 388(6645), 839–840. [DOI] [PubMed] [Google Scholar]
- Stahl N, Borchelt DR, Hsiao K, & Prusiner SB (1987). Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell, 51(2), 229–240. [DOI] [PubMed] [Google Scholar]
- Voss J, Salwinski L, Kaback HR, & Hubbell WL (1995). A method for distance determination in proteins using a designed metal ion binding site and site-directed spin labeling: Evaluation with T4 lysozyme. Proceedings of the National Academy of Sciences of the United States of America, 92(26), 12295–12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter ED, Stevens DJ, Visconte MP, & Millhauser GL (2007). The prion protein is a combined zinc and copper binding protein: Zn2+ alters the distribution of Cu2+ coordination modes. Journal of the American Chemical Society, 129(50), 15440–15441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, McDonald AJ, Markham K, Rich CB, McHugh KP, Tatzelt J, et al. (2017). The N-terminus of the prion protein is a toxic effector regulated by the C-terminus. eLife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Kandegedara A, Xu Y, & Rorabacher DB (1997). Avoiding interferences from Good’s buffers: A contiguous series of noncomplexing tertiary amine buffers covering the entire range of pH 3–11. Analytical Biochemistry, 253(1), 50–56. [DOI] [PubMed] [Google Scholar]
- Zomosa-Signoret V, Arnaud JD, Fontes P, Alvarez-Martinez MT, & Liautard JP (2008). Physiological role of the cellular prion protein. Veterinary Research, 39(4), 9. [DOI] [PubMed] [Google Scholar]


