Abstract
Background
Non‐small cell lung cancer (NSCLC) is one of the most malignant cancer types, characterized by a poor prognosis. N6‐methyladenosine (m6A) is a prevalent internal modification of mRNA. METTL14, an RNA methyltransferase that mediates m6A modification, is implicated in mRNA biogenesis. However, the biomechanism of METTL14 in NSCLC is not very clear.
Methods
Here, immunohistochemical (IHC) assay was employed to detect METTL14 in NSCLC tissues. The biological functions of METTL14 were demonstrated using cell transfection, cell proliferation assay, cell clone formation assay, cell cycle analysis, cell death analysis, transwell and wound healing assays. Transcriptome and methylated RNA immunoprecipitation (MERIP)‐sequencing were used to explore the pathways and potential mechanism of METTL14 in NSCLC. RNA sequencing, METTL14 rip‐sequencing, and METTL14 merip‐sequencing were conducted to identify the potential targets of METTL14.
Results
METTL14 was significantly correlated with clinical pathological parameters of differentiation and M stage. Additionally, METTL14 promotes cell proliferation, induces cell death, and enhances cell migration and invasion in vitro. Transcriptome and MeRIP‐sequencing reveal oncogenic mechanism of METTL14. RIP‐sequencing highlights CSF1R and AKR1C1 as targets of METTL14. After validation with TCGA dataset, colony stimulating factor 1 receptor (CSF1R) showed significant positive coefficient with METTL14, and was presumed to be one target of METTl14 in lung cancer and verified by the cellular experiments.
Conclusion
In conclusion, our results revealed the clinical significance of m6A RNA modification atlas, the function, and molecular targets CSF1R of METTL14 in NSCLC cell lines. The RNA m6A methyltransferase METTL14 promotes the progression of NSCLC by targeted CSF1R.
Keywords: CSF1R, METTL14, N6‐methyladenosine, non‐small cell lung cancer, profile
METTL14 was significantly highly expressed in lung cancer. By increasing colony stimulating factor 1 receptor (CSF1R) mRNA N6‐methyladenosine, METTL14 elevated CSF1R mRNA expression and contributed to lung cancer progression.
INTRODUCTION
Lung cancer is one of the common malignant tumors and the leading cause of cancer mortality worldwide, and the 5‐year survival rate for patients with lung cancer remains low. 1 Non‐small cell lung cancer (NSCLC) is the main histological type of lung cancer, including adenocarcinoma, squamous cell carcinoma and large cell carcinoma, accounts for approximately 80% of all lung cancers. 2 Despite many advancements have been achieved in NSCLC diagnosis and clinical therapy, the overall survival of NSCLC patients has remained unsatisfactory, with an overall 5‐year survival rate of 10%–15%. 3 Therefore, it is extremely urgent to enhance our understanding of the molecular mechanisms of NSCLC progression and identify a novel biomarker for NSCLC patients.
N6‐methyladenosine (m6A) emerges as one of the most important modifications of mRNAs, and it is distributed at translation start sites, CDS regions, and 3′ UTRs. 4 M6A is a dynamic modification controlled by the cellular m6A machinery, which is the methyl‐transferase complex (METTL3, METTL14 and WTAP) and the demethylase enzymes (FTO and ALKBH5). 5 M6A‐methylated transcripts are recognized by reader proteins that regulate different RNA processing events, such as pre‐mRNA processing, translation, and decay. 6 Additionally, RNA methyltransferase METTL3, 7 , 8 , 9 YTHDF2, 10 ALKBH5 11 were reported to play pivotal role in lung cancer growth, 12 progression 13 and drug resistance. 14 The function of METTL14 has been reported in multiple cancer types, including colorectal cancer, 15 , 16 pancreatic cancer, 17 leukemia, 18 hepatocellular carcinoma 19 and bladder cancer. 20 In lung cancer, there are a limited number of studies of METTL14 in lung cancer. 21 , 22 However, the biological molecular mechanism of METTL14 needs to be further explored in NSCLC.
In this study, we first focused on RNA methyl‐transferase METTL14, and further investigated its clinical significance and function in NSCLC. Finally, we identified the potential targets of METTL14, and revealed the function and molecular targets of METTL14 in NSCLC cell lines.
METHODS
Cell lines and culture
NSCLC cell lines A549 and H1299 were preserved in our laboratory and maintained in RPMI‐1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/ml penicillin, and 100 mg/ml streptomycin. The cells were incubated at 37°C in a humid incubator with 5% CO2. The cells with confluence of 80% were used for the experiments.
METTL14 or CSF1R knockdown and cell transfection
Small interfering RNA (siRNA) against METTL14 (si‐METTL14‐1, si‐METTL14‐2, si‐METTL14‐3) or CSF1R (si‐CSF1R‐1, si‐CSF1R—2, si‐CSF1R‐3) and negative control (si‐NC) were synthesized by Shanghai Gene Pharma Co., Ltd. The siRNA sequences were:
METTLE14‐siRNA1.
Sense: GGAUGAGUUAAUAGCUAAAUC.
Antisense: UUUAGCUAUUAACUCAUCCUU.
METTLE14‐siRNA2.
Sense: GGAUGAUAUUAUGAAGUUAGA.
Antisense: UAACUUCAUAAUAUCAUCCCA.
METTLE14‐siRNA3.
Sense: GACUGGUUGUACAGAAGAAAU.
Antisense: UUCUUCUGUACAACCAGUCAA.
CSF1R—siRNA1.
Sense: GCGUUGAUGUUAACUUUGAUGUCUUdtdt.
Antisense: AAGACAUCAAAGUUAACAUCAACGCdtdt.
CSF1R—siRNA2.
Sense: CUCAACAAUCUGACUUUCAUAAUdtdt.
Antisense: AUUAUGAAAGUCAGAUUGUUGAGdtdt.
CSF1R—siRNA3:
Sense: CCCAGGAGCUACACUGAUAdtdt.
Antisense: UAUCAGUGUAGCUCCUGGGdtdt.
The cells were cultured in 6‐well plates. When the cells reached 75% confluence, SiRNAs were transfected at a final concentration of 50 nM using GeneMut reagent (SignaGen Laboratories).
Lentiviruses from Sangon Biotech (NM_001715.3) were used to overexpress METTL14. Stabling overexpression lentivirus or control lentivirus were transfected into H1299 cells by using lipofectamine 3000 reagent (Invitrogen). Then, H1299 cells were grown in DMEM with 10% FBS added.
RNA extraction and qRT‐PCR
After transfection, cells were extracted using TRIzol reagent. The total cellular RNA was extracted, and reverse transcribed into cDNA using a PrimeScript RT reagent kit. The expression levels of METTL14 were detected by quantitative reverse transcription polymerase chain reaction (qRT‐PCR) and the reaction system used 5 μl of extracted RNA. The glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) gene was used as an internal control, and each sample was performed in triplicate. The METTL14 and GAPHD primer sequences were:
METTL14 forward primer sequence (5′—> 3′): AGTGCCGACAGCATTGGTG; Reverse primerGGAGCAGAGGTATCATAGGAAGC;
GAPDH forward primer sequence (5′—> 3′): GGAGCGAGATCCCTCCAAAAT; Reverse primerGGCTGTTGTCATACTTCTCATGG;
Western blot (WB) analysis
H1299 cells were lysed with a mixed buffer that contained RIPA, NaF, and PMSF (Beyotime) on ice for 30 min. Then, the collected protein was denatured in a 95°C water bath for 10 min and centrifuged at 12 000 rpm at 4°C for 10 min. The upper clear cell lysates were transferred to new tubes. Equal amounts of protein were loaded on gels and separated by SDS‐PAGE. Then, proteins were transferred to PVDF membranes (Immobilon‐P; Millipore) and blocked with 5% milk or bovine serum albumin, followed by incubation with primary and secondary antibodies. 23 Primary antibodies were METTLE14 (1:2000, CST, 51104), CSF1R (1:1000, abcam, ab229188), and GAPDH (1:10000, abcam, ab181602) antibodies.
Cell counting kit‐8 (CCK8) assay
SiRNAs transfected A549 and H1299 cells were placed in a 96‐well plate and the serum‐free medium was replaced after being cultured for 24, 48 and 72 h. Then, 10 μl of cell counting kit‐8 (CCK8) reagent was added to each well. The plates were incubated for 2 h, and the optical density (OD) value was measured at 450 nm. Each measurement was repeated in triplicate.
Colony formation assay
After trypsin digestion of cells in logarithmic growth stage, complete medium (basic medium +10% fetal bovine serum) was resuspended into cell suspension and counted. We inoculated 400–1000 cells / well in each experimental group in 6‐well culture plates (generally 700 cells/well according to cell growth). These were cultured until 14 days or the number of cells in most single clones was greater than 50, and the solution was changed every 3 days and the cell state observed. After cloning, the cells were photographed under a microscope, and then washed with PBS once. Then, 1 ml of 4% paraformaldehyde was added to each well, they were fixed for 30–60 min, and washed with PBS once. Next, we added 1 ml of crystal violet dye solution to each well and dyed the cells for 10–20 min. Finally, PBS was used to wash the cells several times, they were air dried, and photographed with a digital camera (taking images of the whole six well plate and each hole separately).
Cell cycle analysis
To determine the function of METTL14 in the cell cycle, H1299 cells were transfected with METTL14 siRNAs. This was achieved by starving the cells in serum‐free DMEM for 24 h. The cells were then fixed in 70% ice‐cold ethanol overnight and subsequently treated with DNase‐free ribonuclease (Takara Bio), stained with propidium iodide (Sigma‐Aldrich), and subjected to a FACSAria flow cytometer (BD Biosciences USA). The data were analyzed using ModFit LT software (Topsham).
Cell death analysis
Then, annexin V fluorescein isothiocyanate (FITC)/propidium iodide (PI) detection kit (Invitrogen) was added to each sample for 15 min in the dark, according to the manufacturer's protocol. The fluorescent signals reflecting cell apoptosis were evaluated by flow cytometry within 1 h. Annexin V + cells were apoptotic cells.
Transwell assay
A transwell assay were performed to assess cell invasion and migration. For the invasion, Matrigel was used to coat the porous membrane and cells were seeded into the upper chamber. The 3 × 105 (without Matrigel) or 5 × 105 cells (with Matrigel) in serum‐free medium were seeded into the upper chamber. The lower chamber was filled with medium supplemented with 10% FBS as a chemoattractant. After 48 h of incubation, the cells on the upper surface of the filter were removed with a cotton swab, and cells that invaded through the filter or Matrigel stuck to the lower surface of the filter were fixed and stained with 0.5% crystal violet, and counted under a light microscope.
Wound healing assay
The confluent cell monolayer was manually damaged by scraping the cells with a 200 μl pipette tip. Photographs were taken using an optical microscope (Olympus) at 0, 24, and 48 h, respectively. The distances were measured by Image‐Pro Plus 6.0 software.
Tissue microarray and immunohistochemistry (IHC)
Tissue microarray was obtained from Outdo Biotech Co., Ltd. Immunohistochemistry was performed to assess the expression of proteins METTL14 in NSCLC samples of tissue microarray. Following deparaffinization and rehydration, sections were blocked for endogenous peroxidase by incubating in 3% H2O2 and then washed in PBS containing 0.05 mol/l EDTA followed by 4% paraformaldehyde. Tissues were incubated in 4% dry milk and 0.3% goat serum in PBS solution for 20 min to block nonspecific binding. Then, 4‐μm sections were incubated overnight at room temperature with anti‐Ki67 (Santa Cruz) antibody in the presence of 10% rabbit serum. After washing, sections were then incubated for another 2 h with horseradish peroxidase (HRP) goat anti‐rabbit IgG secondary antibody. Slides were counterstained with hematoxylin to stain cell nuclei and examined under a light microscope (Olympus).
A PANNORAMIC panoramic slice scanner was used to put the tissue slices on the machine and the slices were gradually move under the lens of the scanner. While moving and imaging, all the tissue information on the tissue slices was scanned and imaged to form a folder containing all tissue information on the tissue section. After the folder is opened with CaseViewer2.4 software, it can be magnified at any multiple of 1–400 times for observation. All tissue points on the tissue chip are scored with the same standard for positive comprehensive score, which is the product of the staining intensity of the target cell and the percentage of positive cells. Positive cells need to be distinguished from background or nonspecific staining. The staining intensity is scored based on the staining characteristics of the target cells: 0 points for no staining (no negative in this experiment), 1 point for light yellow (bottom 1), 2 points for brown (bottom 2), and 3 points for dark brown (bottom 3). Scoring is based on the positive rate of cells: 0 to 5%, 1 to 6%–25%, 2 to 26%–50%, 3 to 51%–75%, and 4 to >75%. Each tissue point on the tissue chip is scored for staining intensity and percentage of positive cells. The positive comprehensive score is the product of the staining intensity and the percentage of positive cells. We defined the expression level of METTL14 according to the median of mean value of METTL14 gene expression in IHC staining scores (=8). We divided the samples with IHC staining scores > = 8 into the high expression group and <8 into the low expression group.
RNA sequencing
Total RNA was isolated from METTL14 silencing and control H1299 cells. The quantity and quality of RNA were assessed by the NanoDrop ND‐1000 spectrophotometer (Thermo Fisher Scientific). RNA integrity was determined by RNA integrity number with the 2200 TapeStation analyzer (Agilent Technologies), and samples that had an RNA integrity number above 7.0 were considered acceptable. The isolated RNAs were used for preparing cDNA libraries by NEBNext Ultra RNA Library Prep Kit for Illumina (ENB). Products were purified with Agencourt AMPure XP beads (Beckman) and diluted to 10 pM for cluster generation in situ on a HiSeq 3000 paired‐end flow cell. The clustering of the index‐coded samples was carried out using a HiSeq Rapid PE Cluster Kit V2 (Illumina). After cluster generation, the libraries were sequenced (2 × 150 bp) on the HiSeq 3000 platform.
RNA immunoprecipitation sequencing (RIP‐Seq)
RIP was performed using the Magna RIP RNA‐binding protein immunoprecipitation kit (Millipore), according to the manufacturer's protocol. Briefly, H1299 cells were washed in PBS and lysed in 10 mM Hepes 7.4, 150 mM KCl, 3 mM MgCl2, 2 mM DTT, 0.5% NP‐40, 1 mM PMSF, and 10% glycerol with protease and RNase inhibitors (Roche). Immunoprecipitation were performed overnight at 4°C with 500–1000 μg of lysate and 5 μg of the following antibodies: Rabbit or mouse total IgG and METTL14 antibody (Santa Cruz Biotechnology). For each IP, 25 μl of protein A/G magnetic beads (Thermo‐Fisher) were added to each sample and incubated for an additional 2 h. Beads were washed 5× with the lysis buffer specified above, and split for RNA and protein analysis. Sequencing reads were aligned to the human genome GRCh37/hg19 by HISAT2. 16 The resulting p‐values were adjusted using Benjamin and Hochberg's approach for controlling the false discovery rate (FDR). The DESeq R package was used to enrich genes with the criterion of Log2[fold change (FC)] > 1 and FDR <0.05.
M6A RNA methylation sequencing (MeRIP‐Seq)
MeRIP and library preparation were conducted according to a previously published protocol with minor revisions. 24 Briefly, poly‐A‐purified RNA was fragmented and incubated with an anti‐m6A antibody. The mixture was immunoprecipitated via incubation with protein A beads (Thermo Fisher Scientific). The captured RNA was washed and purified with the RNA clean and concentrator kit (Zymo Research). Total mRNA and 200 ng of immunoprecipitated RNA from each sample were sequenced and used for library construction using the Illumina Hiseq 2000 platform, as per the manufacturer's instructions The completed libraries, qualified by an Agilent 2100 bioanalyzer, were denatured, and diluted to the loading volume of a loading concentration. Clustered libraries were loaded onto a reagent cartridge and forwarded for sequencing runs on an Illumina Hiseq 4000 system by Novogene Biotech Co., Ltd. Sequencing peaks were annotated to the Ensembl database. Sequence motifs were identified using MEME‐ChIP analysis. 25
Statistical analysis
Data were analyzed by Graphpad version 7.0 software and are presented as the mean ± SD. Data comparison was performed by Student's t‐test for two groups and two‐way ANOVA with Bonferroni post‐test for multiple groups. A p‐value < 0.05 was considered to indicate a statistically significant difference.
RESULTS
Clinical significance of RNA methyl‐transferase METTL14 in NSCLC
To explore the clinical significance of METTL14 in NSCLC, we conducted IHC staining of METTL14 protein in 63 paired lung cancer samples of tissue microarray (TMA, Figure S1). The staining score of METTL14 was compared in normal and tumor groups. As a result, METLL14 showed significantly higher expression in the tumor than in the normal group (Figure 1a). Then we analyzed the staining score of METLL14 protein with clinical pathological parameters. As shown in Table 1 and Figure 1b,c, METTL14 was significantly correlated with gender (p = 0.0354) and differentiation (p = 0.0488).
FIGURE 1.
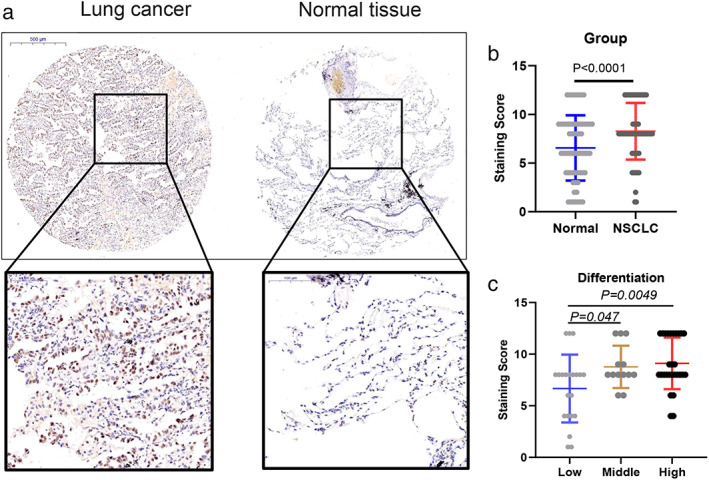
Clinical significance of RNA methyl‐transferase METTL14 in non‐small cell lung cancer (NSCLC). (a) Representative immunohistochemical (IHC) staining data of METTL14 in NSCLC and adjacent normal tissues, respectively. (b) METTL14 staining score in NSCLC tissue and normal samples. (c) METTL14 staining score in low, middle, and high differentiated NSCLC tissue
TABLE 1.
Association between METTL14 expression and clinicopathological characteristics in lung cancer
| Parameter | METTL14 | p‐value | |
|---|---|---|---|
| Age | High | Low | 0.3675 |
| <60 | 24 | 5 | |
| >=60 | 23 | 10 | |
| Gender | 0.0354 | ||
| Female | 18 | 11 | |
| Male | 29 | 4 | |
| Differentiation | 0.0488 | ||
| H | 24 | 4 | |
| L | 12 | 9 | |
| M | 11 | 2 | |
| Location | 0.5777 | ||
| All | 3 | 1 | |
| Left | 18 | 8 | |
| Right | 26 | 6 | |
| Tumor size | 0.7130 | ||
| <5 | 37 | 13 | |
| >=5 | 10 | 2 | |
| Lymph nodes | 0.377816 | ||
| No | 27 | 6 | |
| Yes | 20 | 9 | |
| N stage | 0.719942 | ||
| N0 | 27 | 7 | |
| N1 | 12 | 5 | |
| N2 | 8 | 3 | |
| T stage | 0.519969 | ||
| T1a‐b | 17 | 8 | |
| T2a‐b | 27 | 6 | |
| T4 | 3 | 1 | |
| TNM stage | 0.586341 | ||
| I | 25 | 6 | |
| II | 13 | 5 | |
| III | 9 | 4 | |
Abbreviations: H, high differentiation; L, low differentiation; M, middle differentiation.
METTL14 promotes NSCLC promotion and metastasis in vitro
To investigate the function of METTL14 in NSCLC, three small interference RNAs (siRNAs) targeting METTL14 (METTL14 siRNA1‐3) were designed, and their interference efficiency was analyzed with qRT‐PCR and WB. As shown in Figure 2a,b, siRNA1 showed the best METTL14 silencing efficiency in H1299 cells, and was used for subsequent functional analysis. To explore the function of METTL14 on NSCLC cell proliferation, CCK8 assay was conducted on two cell lines A549 and H1299, respectively. For both cell lines, METTL14 knockdown significantly inhibited the number of cells, especially in H1299 cell line (Figure 2c). Furthermore, we verified that the si‐METTL14 remarkably inhibited the cell proliferation through colony formation assay (Figure 2d), which was consistent with CCK‐8 results. Additionally, cell cycle and cell apoptosis rate were analyzed by FACS. The cell cycle analysis results revealed that METTL14 knockdown significantly induced G0/G1 phase arrest (METTL14 KD 68.35% vs. control 55.14%), and reduced the proportion of G2 phase cells (METTL14 KD 11.03% compared with control 15.02%) (Figure 2e). Furthermore, METTL14 knockdown induced the H1299 cell death rate (METTL14 KD 20.46% vs. control 9.90%, Figure 2f). Then, the function of METTL14 on cell invasion and migration was investigated using transwell and wound healing assays, respectively. As shown in Figure 2g,h, METTL14 knockdown significantly reduced cell invasion and migration. Taken together, the above results suggested that METTL14 knockdown inhibited NSCLC cell proliferation, invasion, and migration in vitro.
FIGURE 2.
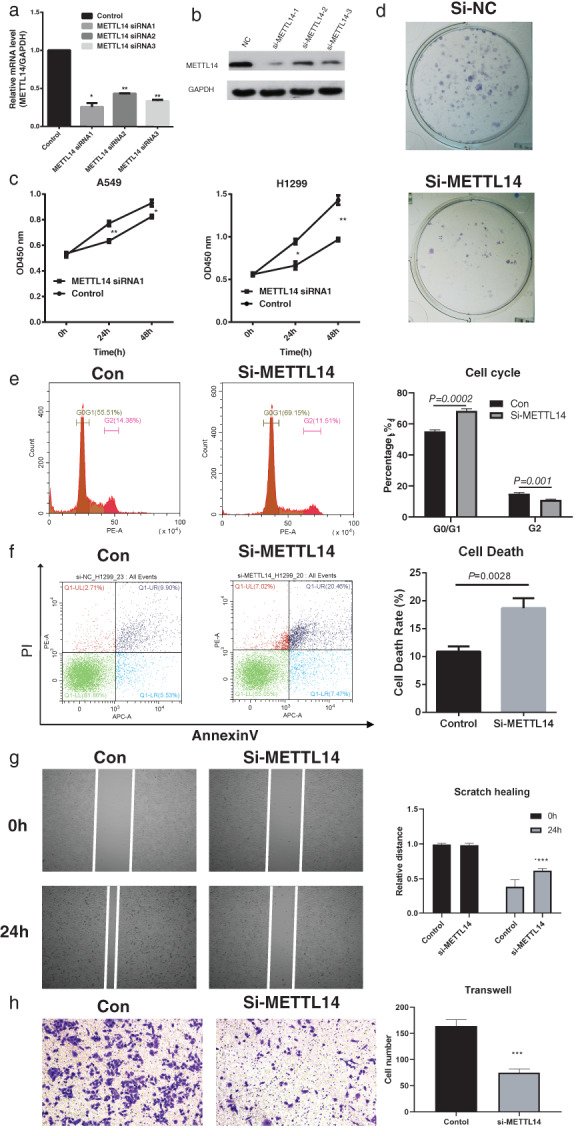
Functional study of METTL14 silencing on non‐small cell lung cancer (NSCLC) cell lines in vitro. (a) The silencing efficiency of 3 METTL14 targeting siRNAs was checked with qRT‐PCR. (b) The silencing efficiency of 3 METTL14 targeting siRNAs was checked with WB. (c) Cell counting kit‐8 was conducted to examine the proliferating phenotype of METTL14 on A549 and H1299 cells. (d) The colony formation assay showed the proliferating phenotype of METTL14 on H1299 cells. (e) In H1299 cells, cell cycle distribution was evaluated with FACS. (f) In H1299 cells, cell apoptosis death rate was examined with annexin V and PI staining. (g) In H1299 cells, the migration capability of METTL14 silenced cells were studied with scratch healing assay. (h) In H1299 cells, the invasion capability of METTL14 silenced cells were studied with transwell assay. Data are presented as means ± SEM, n = 3. *p < 0.05; **p < 0.01 versus control group
Transcriptome and MeRIP‐sequencing reveals oncogenic mechanism of METTL14
To uncover the pathways and potential mechanism of METTL14 in NSCLC, we conducted RNA sequencing and methylated RNA immunoprecipitation (MeRIP) sequencing in H1299 cells. As shown in Figure 3a and Table S1, 574 differential expression genes were explored (fold change > = 2 or < =0.5 and p‐adj < =0.05) in H1299 cells, including 269 protein coding genes, 187 lncRNAs and 118 other type RNAs (processed pseudogenes, snoRNAs and misc_RNAs), suggesting a pervasive role of METTL14 on RNA types. Of all 574 differentially expressed genes (DEGs), 455 were upregulated and 119 were downregulated. Then, we analyzed the function of the coding genes and found that well‐known cancer related pathways such as the Rap1 signaling pathway 26 (genes like CSF1R and MRAS) and TGF‐beta signaling pathway 27 , 28 (genes like ID2 and THBS1) were significantly enriched (Figure 3b). Interestingly, we also observed several deregulated lncRNAs with oncogenic or tumor suppressing function in lung cancer, such as KCNQ1OT1, 29 DHRS4‐AS1 30 and NEAT1. 31 , 32
FIGURE 3.
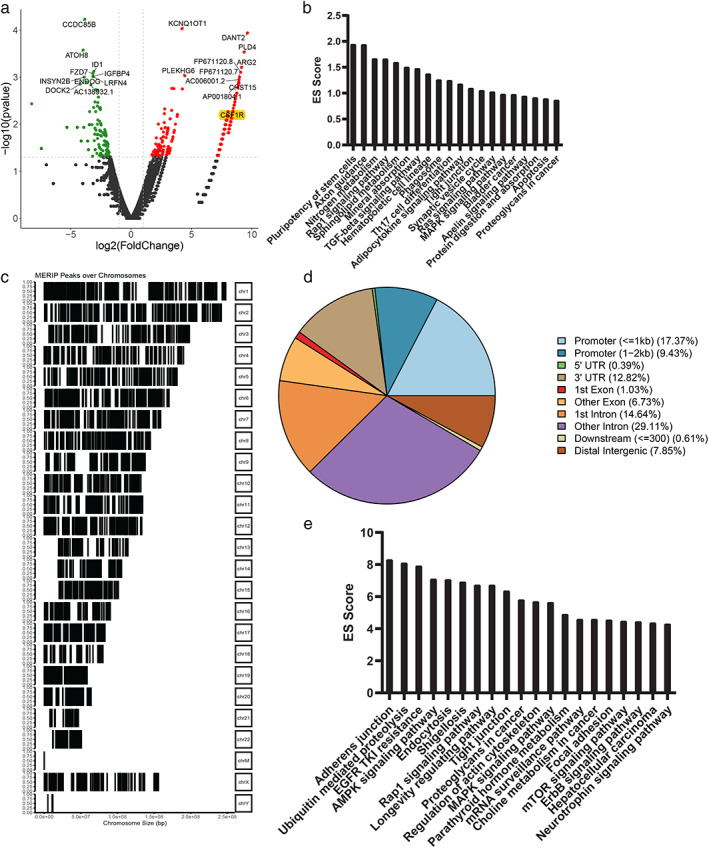
Transcriptome and MeRIP‐sequencing reveals oncogenic mechanism of METTL14. (a) Heatmap showing the deregulated genes in METTL14 knockdown H1299 cells. (b) Functional enriched pathways of deregulated genes in METTL14 knockdown H1299 cells; ES score, enrichment score = −log10 p‐adj. (c) Representative map of M6A peaks in chromosomes, in METTL14 knockdown H1299 cells. (d) Peak locations of DMGs in gene body locations. (e) Functional enriched pathways of M6A peak genes in METTL14 knockdown H1299 cells; ES score, enrichment score = −log10 p‐adj
For MeRIP sequencing result, differential analysis enriched 7158 peaks of 4480 genes in H1299 cells (Figure 3c and Table S2). Gene body analysis of these peaks showed that these peaks locate mainly in introns, 3'UTR and promoter regions of protein coding and noncoding genes (Figure 3d and Table S2). Functionally, protein‐coding genes were mainly involved in well‐known cancer related pathways like Rap1 signaling pathway (genes like ITGB1, CTNND1 and IGF1R), Proteoglycans in cancer (genes like RAC1, PRKCG and WNT5B) and MAPK signaling pathway (genes like FLT3, HSPB1and ARRB2) (Figure 3e). Similar to RNA sequencing results, oncogenic or tumor suppressing lncRNAs were also identified, and contained SNHG3, 33 MALAT1 34 , 35 and TUG1. 36 Finally, we explored the 65 common genes in both differential expression and peak, including 46 protein‐coding genes and 14 lncRNAs (Table S3).
RIP‐sequencing highlights CSF1R and AKR1C1 as targets of METTL14
Furthermore, to identify RNAs bound by METTL14, RNA immunoprecipitation (RIP) sequencing of METTL14 was conducted in H1299 cells. As a result, 2100 peaks of 1024 genes were identified, including 906 protein‐coding mRNAs and 28 lncRNAs (Figure 4a and Table S4). As expected, the protein‐coding mRNAs were involved in RNA metabolism pathways like spliceosome, mRNA surveillance pathway and RNA transport. Additionally, genes in the Notch signaling pathway (NCOR2, CREBBP, KAT2A, EP300 and DVL3) were one of the top10 enriched items (Figure 4b). For lncRNAs, METTL14 bound oncogenic or tumor suppressing lncRNAs were also identified, exemplified by HOXB‐AS3 37 and PVT1. 38 Next, we compared the METTL14 binding mRNAs with 65 common genes by RNA sequencing and MeRIP sequencing. Five genes were commonly identified, including four protein coding genes (PPIEL, AKR1C1, SARS2 and CSF1R) and one lncRNA ASAP1‐IT2 (Figure 4c). Then, the m6A methylation level of 5 genes were analyzed in the above mentioned 7 paired NSCLC tissue samples, and only CSF1R showed differential methylation level (Figures 4d–f and S2). Therefore, CSF1R was presumed to be the target of METTl14 in lung cancer. In cancer cells, aldo‐keto reductase family 1 member C1 (AKR1C1) showed robust role in tumor genesis, 39 , 40 , 41 including lung cancer. 42 , 43 , 44 Colony stimulating factor 1 receptor (CSF1R) is a cytokine which controls the production, differentiation, and function of macrophages. Notably, CSF1R also showed oncogenic function and performed as prognostic marker in lung cancer, which is consistent with the function and clinical significance of METTL14. Finally, to validate CSF1R as the target of METTL14, METTL14/CSF1R coexpression was calculated in TCGA lung cancer datasets. Both genes showed significant positive coefficient in lung adenocarcinoma (R = 0.35, p = 2.4 e−15) and lung squamous carcinoma (R = 0.35, p = 5.3 e−15) samples (Figure 4g). Altogether, we uncovered an m6A RNA modification atlas, revealed the function and clinical significance of METTL14 in lung cancer.
FIGURE 4.
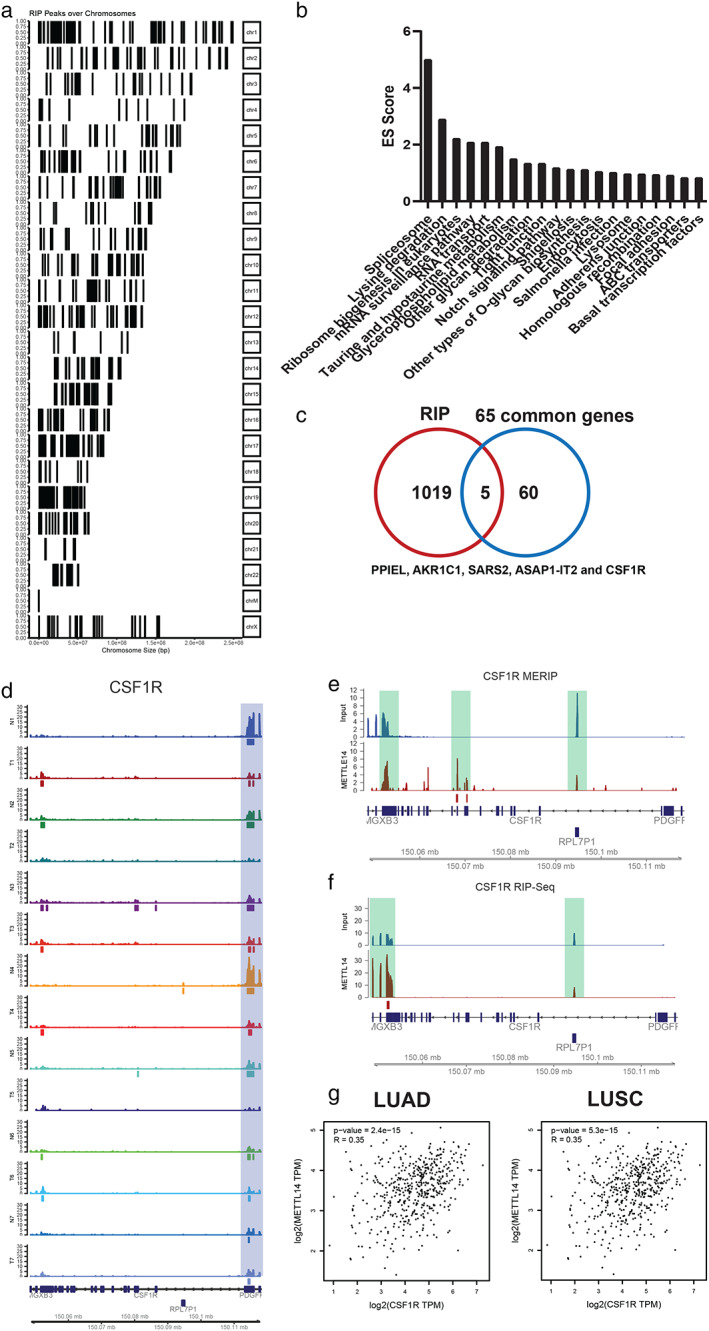
RIP‐sequencing highlights CSFR1 as a target of METTL14. (a) Peak locations of RIP‐sequencing identified peaks in gene body locations. (b) Functional enriched pathways of RIP‐sequencing identified peak genes in METTL14 knockdown H1299 cells; ES score, enrichment score = −log10 p‐adj. (c) Venn plot showed five genes PPIEL, AKR1C1, SARS2, CSF1R and one lncRNA ASAP1‐IT2 as METTL14 targets; (d) M6A peaks of CSF1R in seven paired samples of NSCLC and adjacent samples. (e) MeRIP and RIP peaks of CSF1R in H1299 cell; (f) RIP peaks of CSF1R in H1299 cell. (g) Spearman correlation analysis validation of METTL14 with CSF1R in TCGA LUAD and LUSC samples
METTL14 in the involvement of CSF1R in NSCLC proliferation and metastasis
Finally, we conducted cellular experiments for validation of the function of CSF1R in NSCLC proliferation and metastasis. Three siRNAs targeting CSF1R (si‐CSF1R 1–3) were designed, and their interference efficiency was detected by WB. As shown in Figure 5a, WB analysis showed that si‐CSF1R‐3 significantly decreased the expression of CSF1R, and si‐METTL14‐1 also remarkably suppressed its expression, thus the si‐CSF1R‐3 and si‐METTL14‐1 were selected for further experiments. The CCK‐8 analysis showed that H1299 cells treated with si‐CSF1R and si‐METTL14 at the same time significantly inhibited cell proliferation compared to the control group (Figure 5b). In addition, the transwell and wound healing assays demonstrated that CSF1R knockdown significantly reduced the invasion cell number and migration distance (Figure 5c,d), and METTL14 was involved in this process. In addition, we detected the mRNA level after the overexpression of METTL14. As shown in Figure 6a, overexpression of METTL14 could enhance the CSF1R expression. According to the CCK‐8 analysis, transwell and wound healing assays, CSF1R knockdown can block the proliferation and metastasis effect of METTL14 overexpression in H1299 cell line (p < 0.05, Figure 6b–d). In summary, these results revealed that the siCSF1R significantly inhibited the proliferation, migration, and invasion of HT1299 cells, which involved with the regulation of METTL14.
FIGURE 5.
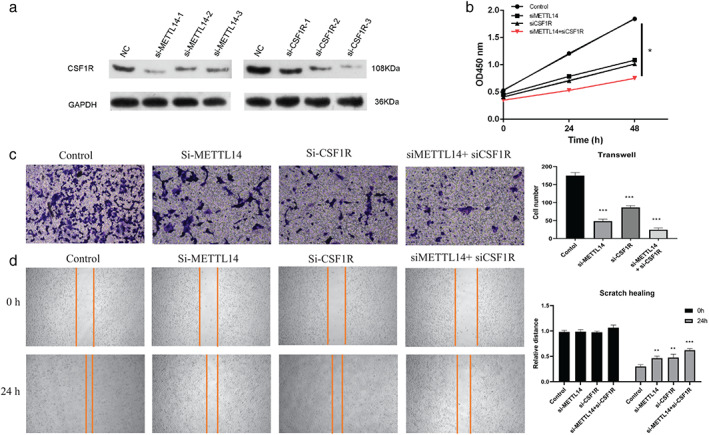
si‐CSF1R suppressed non‐small cell lung cancer (NSCLC) cell proliferation and metastasis. (a) The silencing effect of si‐METTL14 siRNA1‐3 or 3 si‐CSF1R siRNA1‐3 on expression of CSF1R was detected by Western blot. (b) Cell counting kit‐8 assay was applied to detect the proliferation ability of silence of METTL14 and CSF1R in HT1299 cells, p < 0.05. (c) Transwell assay was used to detect the migration capability of METTL14 or CSF1R silenced H1299 cells. (d) Wound healing assay was used to detect the invasion capability of METTL14 or CSF1R silenced H1299 cells. Data are presented as means ± SEM, n = 3. *p < 0.05; **p < 0.01 versus control group
FIGURE 6.
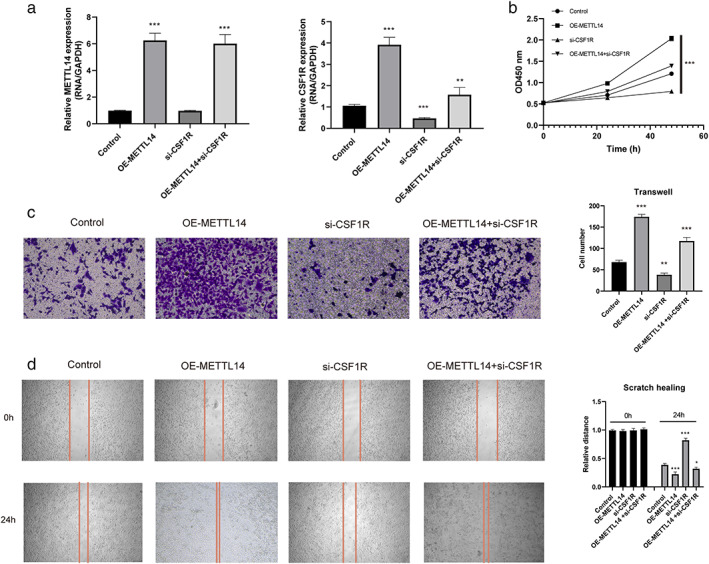
si‐CSF1R can block the proliferation and metastasis effect of METTL14 overexpression. (a) The expression of METTL14 and CSF1R under different conditions were detected by RT‐qPCR. (b) Cell counting kit‐8 assay was applied to detect the proliferation ability in HT1299 cells. (c) Transwell assay was used to detect the migration capability of overexpression of METTL14 or silence of CSF1R in H1299 cells. (d) Wound healing assay was used to detect the invasion capability of overexpression of METTL14 or silence of CSF1R in H1299 cells. Data are presented as means ± SEM, n = 3. *p < 0.05; **p < 0.01 versus control group
DISCUSSION
As one of the most common RNA modifications, N6‐methyladenosine (m6A) modified RNAs (mRNAs, lncRNAs and other RNAs) are widely involved in RNA biogenesis, transport, splicing, stability, translation, binding affinity and other RNA metabolism processes. 45 , 46 The deregulated m6A modifications have been previously reported to disturb physiological signaling transduction and to be involved in tumor initiation, progression, treatment response and prognosis. 45 , 47 , 48 , 49 , 50 , 51 , 52 The RNA expression atlas and altered m6A modification mRNA have been revealed in lung cancer in multiple projects. 53 , 54 Pervasive m6A signals were detected in all samples, and most genes (84.65% and 79.61%) were both m6A modified in lung cancer and adjacent samples, with a minority of tumor and normal specific genes. It is noteworthy that even genes m6A modified in both groups were the same, corresponding transcripts and peak locations are still distinct; for example, cancer stemness maker gene CD44, 55 of which transcript ENST00000279452.10 and ENST00000526669.6 were m6A modified in normal and tumor, respectively. As m6A modification has been shown to regulate RNA splicing, decay and translation, the transcripts and peak location discrepancy may reflect the molecular complexity between m6A modification and RNA metabolism. As for RNA type, in addition to well‐known modifications on mRNAs and lncRNAs, we also detected m6A signals on other types of RNAs, such as small nucleolar RNAs (snoRNAs) and small nuclear RNA (snRNAs), which is consist with the miCLIP method result in 2015. 56 However, the role of m6A modified snoRNAs/snRNAs remains largely unknown in cancer.
METTL14, acting as the central component of N6‐methytransferse complex, has been verified to be dysregulated and involved in the initiation and progression of various malignancies. A previous study suggested that METTL14 is significantly downregulated in CRC and overexpression of MTTL14 inhibits the ability of migration and invasion in the CRC cell line. 15 In this study, METLL14 showed significantly higher expression in the tumor than in the normal group. METTL14 was significantly correlated with gender (p = 0.0354) and differentiation (p = 0.0488). Knockdown of METTL14 significantly reduced cell invasion and migration. Taken together, the above results suggest that knockdown of METTL14 inhibited NSCLC cell proliferation, invasion, and migration in vitro. Their results confirmed that METTL14 function as an oncogenic gene in NSCLC. In colorectal cancer, METTL14 promotes cancer progression by regulating miR‐375, 57 and inhibits metastasis via regulating SOX4. 16 In pancreatic cancer, METTL14 promotes cancer growth and metastasis 58 by m6A modifying PERP mRNA. 59 In bladder cancer, the tumor suppressing function of METTL14 was found to be dependent on m6A modification of Notch1 and tumor initiating cells (TIC) self‐renewal, 20 which was also observed in this study, with genes in the Notch signaling pathway (NCOR2, CREBBP, KAT2A, EP300 and DVL3) as METTL14 binding mRNAs. In leukemia, METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via MYB/MYC mRNA m6A modification. 18 Collectively, METTL14 showed distinct or even converse effect in multiple cancer types, even in the same cancer type. In our opinion, the effect of METTL14 may be attributed to its targets which is complicated regulated in tissue and cancer dependent manner.
To further address the role of METTL14, RNA‐Seq and MeRIP‐Seq to reveal that the mRNAs of PPIEL, AKR1C1, SARS2 and CSF1Rand lncRNA ASAP1‐IT2 were observed as direct or indirect METTL14 targets, implying that noncoding RNA can also serve as METTL14 targets, such as lncRNA XIST 15 in colorectal cancer, LNCAROD in head and neck squamous cell carcinoma, 60 and microRNA‐126 processing. 19 As in lung cancer, METTL14 was involved in IL‐37 conferred antitumor activity. 61
Interestingly, CSF1R also showed differential m6A peaks in tissue sample, which is identified as METTL14 target in lung cancer. Aldo‐keto reductase 1 family member C1 (AKR1C1) is a type of aldosterone reductase and plays a regulatory role in a variety of key metabolic pathways. 62 In cancer, AKR1C1 showed a robust role in cancer genesis, 39 , 40 , 41 including lung cancer. 42 , 43 , 44 Colony stimulating factor 1 receptor (CSF1R) is a cytokine which controls the production, differentiation, and function of macrophages. In lung cancer and other cancer types, the oncogenic role of CSF1R was mostly reported in tumor‐associated macrophages (TAMs). 63 , 64 , 65 , 66 Notably, METTL14 has also been reported to act in TAM mediated CD8 T cell dysfunction and tumor growth, 67 indicating an alternative role of METTL14 modified CSF1R in lung cancer immune microenvironment. Moreover, there are limited studies of PPIEL, SARS2 and lncRNA ASAP1‐IT2 in cancer, and their role and clinical significance should be studied in the future.
In conclusion, we elucidated the key role of METTL14‐mediated m6A modification in human NSCLC progression and regulatory mechanism. We revealed that CSF1R and AKR1C1as targets of METTL14. The RNA m6A methyltransferase METTL14 promotes the procession of non‐small cell lung cancer by targeted CSF1R. The discovery of the METTL14/CSF1R axis and its impact on NSCLC metastasis will aid in further NSCLC study and in exploring efficient therapeutic strategies against NSCLC.
AUTHOR CONTRIBUTIONS
Y.H. and S.R.: study design; Y.X., L.Y., and S.R.: experimental studies; Y.H.: tissue samples collection; Y.H., and S.R.: data analysis; Y.H. and S.R.: manuscript editing. All authors read and approved the final manuscript.
CONFLICT OF INTEREST
The authors declare no competing interests.
Supporting information
Figure S1. IHC staining data of METTL14 in NSCLC and adjacent normal tissues.
Figure S2. M6A peaks of AKR1C1, ASAP1‐IT2, PPIEL, SARS2 in 7 paired samples of NSCLC and adjacent samples.
ACKNOWLEDGMENTS
This work was supported by Natural Science Foundation of Hunan Province, China (2019JJ30038) and the Scientific Research Program of Hunan Provincial Health Commission (B2018‐0541).
Ren S, Xiao Y, Yang L, Hu Y. RNA m6A methyltransferase METTL14 promotes the procession of non‐small cell lung cancer by targeted CSF1R . Thorac Cancer. 2023;14(3):254–266. 10.1111/1759-7714.14741
Funding information Natural Science Foundation of Hunan Province, Grant/Award Number: 2019JJ30038; Scientific Research Program of Hunan Provincial Health Commission, Grant/Award Number: B2018‐0541
DATA AVAILABILITY STATEMENT
The raw data has been stored in the GEO (GSE179659). Additional information and request for the materials and methods detail should be directed and will be fulfilled by the corresponding author.
REFERENCES
- 1. Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, et al. Cancer treatment and survivorship statistics, 2019. CA‐Cancer J Clin. 2019;69(5):363–85. [DOI] [PubMed] [Google Scholar]
- 2. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non‐small cell lung cancer. Nature. 2018;553(7689):446–54. [DOI] [PubMed] [Google Scholar]
- 3. Alexander M, Kim SY, Cheng H. Update 2020: management of non‐small cell lung cancer. Lung. 2020;198(6):897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6‐methyladenosine in cancer progression. Mol Cancer. 2020;19(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang CF, Lin SP, Chiang CP, Wu YH, H'ng WS, Chang CP, et al. Loss of GPNMB causes autosomal‐recessive amyloidosis cutis dyschromica in humans. Am J Hum Genet. 2018;102:219–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dai D, Wang H, Zhu L, Jin H, Wang X. N6‐methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9(2):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu Y, Chang N, Zhang Y, Zhang X, Xu L, Che Y, et al. METTL3‐mediated m(6)A mRNA modification of FBXW7 suppresses lung adenocarcinoma. J Exp Clin Cancer Res. 2021;40(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jin D, Guo J, Wu Y, du J, Yang L, Wang X, et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1‐miR‐1914‐3p‐YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12(1):135. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Cheng C, Wu Y, Xiao T, Xue J, Sun J, Xia H, et al. METTL3‐mediated m(6)A modification of ZBTB4 mRNA is involved in the smoking‐induced EMT in cancer of the lung. Mol Ther Nucleic Acids. 2021;23:487–500. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10. Hou G, Zhao X, Li L, Yang Q, Liu X, Huang C, et al. SUMOylation of YTHDF2 promotes mRNA degradation and cancer progression by increasing its binding affinity with m6A‐modified mRNAs. Nucleic Acids Res. 2021;49(5):2859–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jin D, Guo J, Wu Y, Yang L, Wang X, du J, et al. m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs‐mediated YAP expression and inhibiting miR‐107/LATS2‐mediated YAP activity in NSCLC. Mol Cancer. 2020;19(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yin H, Zhang X, Yang P, Zhang X, Peng Y, Li D, et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat Commun. 2021;12(1):1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang S, Luo S, Gong C, Liang L, Xiao Y, Li M, et al. MTTL3 upregulates microRNA‐1246 to promote occurrence and progression of NSCLC via targeting paternally expressed gene 3. Mol Ther Nucleic Acids. 2021;24:542–53. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Shen C et al. Both p53 codon 72 Arg/Arg and pro/Arg genotypes in glioblastoma multiforme are associated with a better prognosis in bevacizumab treatment. BMC Cancer. 2020;20(1):709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down‐regulating oncogenic long non‐coding RNA XIST. Mol Cancer. 2020;19(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, et al. METTL14‐mediated N6‐methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen L, Hu L, Zhu X, Wang Y, Li Q, Ma J, et al. MALAT1 overexpression attenuates AS by inhibiting ox‐LDL‐stimulated dendritic cell maturation via miR‐155‐5p/NFIA axis. Cell Cycle. 2020;19(19):2472–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22(2):191–205 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) ‐methyladenosine‐dependent primary MicroRNA processing. Hepatology. 2017;65(2):529–43. [DOI] [PubMed] [Google Scholar]
- 20. Gu C, Wang Z, Zhou N, Li G, Kou Y, Luo Y, et al. Mettl14 inhibits bladder TIC self‐renewal and bladder tumorigenesis through N(6)‐methyladenosine of Notch1. Mol Cancer. 2019;18(1):168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shen Y, Li C, Zhou L, Huang JA. G protein‐coupled oestrogen receptor promotes cell growth of non‐small cell lung cancer cells via YAP1/QKI/circNOTCH1/m6A methylated NOTCH1 signalling. J Cell Mol Med. 2021;25(1):284–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gu C, Shi X, Qiu W, Huang Z, Yu Y, Shen F, et al. Comprehensive analysis of the prognostic role and mutational characteristics of m6A‐related genes in lung squamous cell carcinoma. Front Cell Dev Biol. 2021;9:661792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han Z, Liu D, Chen L, He Y, Tian X, Qi L, et al. PNO1 regulates autophagy and apoptosis of hepatocellular carcinoma via the MAPK signaling pathway. Cell Death Dis. 2021;12(6):552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dominissini D, Moshitch‐Moshkovitz S, Salmon‐Divon M, Amariglio N, Rechavi G. Transcriptome‐wide mapping of N(6)‐methyladenosine by m(6)A‐seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8(1):176–89. [DOI] [PubMed] [Google Scholar]
- 25. Machanick P, Bailey TL. MEME‐ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27(12):1696–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khattar E, Maung KZY, Chew CL, Ghosh A, Mok MMH, Lee P, et al. Rap1 regulates hematopoietic stem cell survival and affects oncogenesis and response to chemotherapy. Nat Commun. 2019;10(1):5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elliott RL, Blobe GC. Role of transforming growth factor Beta in human cancer. J Clin Oncol. 2005;23(9):2078–93. [DOI] [PubMed] [Google Scholar]
- 28. Derynck R, Akhurst RJ, Balmain A. TGF‐beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29(2):117–29. [DOI] [PubMed] [Google Scholar]
- 29. He H, Song X, Yang Z, Mao Y, Zhang K, Wang Y, et al. Upregulation of KCNQ1OT1 promotes resistance to stereotactic body radiotherapy in lung adenocarcinoma by inducing ATG5/ATG12‐mediated autophagy via miR‐372‐3p. Cell Death Dis. 2020;11(10):883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yan F, Zhao W, Xu X, Li C, Li X, Liu S, et al. LncRNA DHRS4‐AS1 inhibits the stemness of NSCLC cells by sponging miR‐224‐3p and upregulating TP53 and TET1. Front Cell Dev Biol. 2020;8:585251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kong X, Zhao Y, Li X, Tao Z, Hou M, Ma H. Overexpression of HIF‐2alpha‐dependent NEAT1 promotes the progression of non‐small cell lung cancer through miR‐101‐3p/SOX9/Wnt/beta‐catenin signal pathway. Cell Physiol Biochem. 2019;52(3):368–81. [DOI] [PubMed] [Google Scholar]
- 32. Jiang P, Chen A, Wu X, Zhou M, ul Haq I, Mariyam Z, et al. NEAT1 acts as an inducer of cancer stem cell‐like phenotypes in NSCLC by inhibiting EGCG‐upregulated CTR1. J Cell Physiol. 2018;233(6):4852–63. [DOI] [PubMed] [Google Scholar]
- 33. Xuan Y, Wang Y. Long non‐coding RNA SNHG3 promotes progression of gastric cancer by regulating neighboring MED18 gene methylation. Cell Death Dis. 2019;10(10):694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jen J, Tang YA, Lu YH, Lin CC, Lai WW, Wang YC. Oct4 transcriptionally regulates the expression of long non‐coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol Cancer. 2017;16(1):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schmidt LH, Spieker T, Koschmieder S, Schäffers S, Humberg J, Jungen D, et al. The long noncoding MALAT‐1 RNA indicates a poor prognosis in non‐small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6(12):1984–92. [DOI] [PubMed] [Google Scholar]
- 36. Niu Y, Ma F, Huang W, Fang S, Li M, Wei T, et al. Long non‐coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer. 2017;16(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang W, Kai J, Li D, Wei Z, Wang Y, Wang W. lncRNA HOXB‐AS3 exacerbates proliferation, migration, and invasion of lung cancer via activating the PI3K‐AKT pathway. J Cell Physiol. 2020;235(10):7194–203. [DOI] [PubMed] [Google Scholar]
- 38. Olivero CE, Martínez‐Terroba E, Zimmer J, Liao C, Tesfaye E, Hooshdaran N, et al. p53 activates the long noncoding RNA Pvt1b to inhibit Myc and suppress tumorigenesis. Mol Cell. 2020;77(4):761–774 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chang WM, Chang YC, Yang YC, Lin SK, Chang PMH, Hsiao M. AKR1C1 controls cisplatin‐resistance in head and neck squamous cell carcinoma through cross‐talk with the STAT1/3 signaling pathway. J Exp Clin Cancer Res. 2019;38(1):245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jiang Y, Li Y, Cheng J, Ma J, Li Q, Pang T. Upregulation of AKR1C1 in mesenchymal stromal cells promotes the survival of acute myeloid leukaemia cells. Br J Haematol. 2020;189(4):694–706. [DOI] [PubMed] [Google Scholar]
- 41. Huebbers CU, Verhees F, Poluschkin L, Olthof NC, Kolligs J, Siefer OG, et al. Upregulation of AKR1C1 and AKR1C3 expression in OPSCC with integrated HPV16 and HPV‐negative tumors is an indicator of poor prognosis. Int J Cancer. 2019;144(10):2465–77. [DOI] [PubMed] [Google Scholar]
- 42. Zhu H, Hu Y, Zeng C, Chang L, Ge F, Wang W, et al. The SIRT2‐mediated deacetylation of AKR1C1 is required for suppressing its pro‐metastasis function in non‐small cell lung cancer. Theranostics. 2020;10(5):2188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhu H et al. AKR1C1 activates STAT3 to promote the metastasis of non‐small cell lung cancer. Theranostics. 2018;8(3):676–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang HW, Lin CP, Chiu JH, Chow KC, Kuo KT, Lin CS, et al. Reversal of inflammation‐associated dihydrodiol dehydrogenases (AKR1C1 and AKR1C2) overexpression and drug resistance in nonsmall cell lung cancer cells by wogonin and chrysin. Int J Cancer. 2007;120(9):2019–27. [DOI] [PubMed] [Google Scholar]
- 45. Hu BB, Wang XY, Gu XY, Zou C, Gao ZJ, Zhang H, et al. N(6)‐methyladenosine (m(6)A) RNA modification in gastrointestinal tract cancers: roles, mechanisms, and applications. Mol Cancer. 2019;18(1):178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roignant JY, Soller M. m(6)A in mRNA: An ancient mechanism for fine‐tuning gene expression. Trends Genet. 2017;33(6):380–90. [DOI] [PubMed] [Google Scholar]
- 47. Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12(1):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pan Y, Ma P, Liu Y, Li W, Shu Y. Multiple functions of m(6)A RNA methylation in cancer. J Hematol Oncol. 2018;11(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Uddin MB, Wang Z, Yang C. The m(6)A RNA methylation regulates oncogenic signaling pathways driving cell malignant transformation and carcinogenesis. Mol Cancer. 2021;20(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou Z, Lv J, Yu H, Han J, Yang X, Feng D, et al. Mechanism of RNA modification N6‐methyladenosine in human cancer. Mol Cancer. 2020;19(1):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. B JWA et al. RNA sequencing (RNA‐Seq) and its application in ovarian cancer. Gynecol Oncol. 2019;152(1):194–201. [DOI] [PubMed] [Google Scholar]
- 53. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chang JT, Lee YM, Huang RS. The impact of the cancer genome atlas on lung cancer. Transl Res. 2015;166(6):568–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zoller M. CD44: can a cancer‐initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11(4):254–67. [DOI] [PubMed] [Google Scholar]
- 56. Linder B, Grozhik AV, Olarerin‐George AO, Meydan C, Mason CE, Jaffrey SR. Single‐nucleotide‐resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12(8):767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen X, Xu M, Xu X, Zeng K, Liu X, Sun L, et al. METTL14 suppresses CRC progression via regulating N6‐methyladenosine‐dependent primary miR‐375 processing. Mol Ther. 2020;28(2):599–612. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58. Chen S, Yang C, Wang ZW, Hu JF, Pan JJ, Liao CY, et al. CLK1/SRSF5 pathway induces aberrant exon skipping of METTL14 and cyclin L2 and promotes growth and metastasis of pancreatic cancer. J Hematol Oncol. 2021;14(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang M, Liu J, Zhao Y, He R, Xu X, Guo X, et al. Upregulation of METTL14 mediates the elevation of PERP mRNA N(6) adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol Cancer. 2020;19(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ban Y, Tan P, Cai J, Li J, Hu M, Zhou Y, et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol Oncol. 2020;14(6):1282–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Iida Y, Yoshikawa R, Murata A, Kotani H, Kazuki Y, Oshimura M, et al. Local injection of CCL19‐expressing mesenchymal stem cells augments the therapeutic efficacy of anti‐PD‐L1 antibody by promoting infiltration of immune cells. J Immunother Cancer. 2020;8(2):e000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wei X, Wei Z, Li Y, Tan Z, Lin C. AKR1C1 contributes to cervical cancer progression via regulating TWIST1 expression. Biochem Genet. 2021;59(2):516–30. [DOI] [PubMed] [Google Scholar]
- 63. Priceman SJ, Sung JL, Shaposhnik Z, Burton JB, Torres‐Collado AX, Moughon DL, et al. Targeting distinct tumor‐infiltrating myeloid cells by inhibiting CSF‐1 receptor: combating tumor evasion of antiangiogenic therapy. Blood. 2010;115(7):1461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shih CT, Shiau CW, Chen YL, Chen LJ, Chao TI, Wang CY, et al. TD‐92, a novel erlotinib derivative, depletes tumor‐associated macrophages in non‐small cell lung cancer via down‐regulation of CSF‐1R and enhances the anti‐tumor effects of anti‐PD‐1. Cancer Lett. 2021;498:142–51. [DOI] [PubMed] [Google Scholar]
- 65. Inamura K, Shigematsu Y, Ninomiya H, Nakashima Y, Kobayashi M, Saito H, et al. CSF1R‐expressing tumor‐associated macrophages, smoking and survival in lung adenocarcinoma: analyses using quantitative phosphor‐integrated dot staining. Cancers. 2018;10(8):252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sedighzadeh SS, Khoshbin AP, Razi S, Keshavarz‐Fathi M, Rezaei N. A narrative review of tumor‐associated macrophages in lung cancer: regulation of macrophage polarization and therapeutic implications. Transl Lung Cancer Res. 2021;10(4):1889–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dong L, Chen C, Zhang Y, Guo P, Wang Z, Li J, et al. The loss of RNA N(6)‐adenosine methyltransferase Mettl14 in tumor‐associated macrophages promotes CD8(+) T cell dysfunction and tumor growth. Cancer Cell. 2021;39:945–957.e10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. IHC staining data of METTL14 in NSCLC and adjacent normal tissues.
Figure S2. M6A peaks of AKR1C1, ASAP1‐IT2, PPIEL, SARS2 in 7 paired samples of NSCLC and adjacent samples.
Data Availability Statement
The raw data has been stored in the GEO (GSE179659). Additional information and request for the materials and methods detail should be directed and will be fulfilled by the corresponding author.


