Abstract
Killing by Entamoeba histolytica requires parasite adherence to host galactose- and N-acetyl-d-galactosamine (Gal/GalNAc)-containing cell surface receptors. A 260-kDa heterodimeric E. histolytica Gal/GalNAc lectin composed of heavy (Hgl) and light (Lgl) subunits has been previously described. Here we present the cloning and characterization of Igl, a 150-kDa intermediate subunit of the Gal/GalNAc lectin. Igl, Hgl, and Lgl colocalized on the surface membrane of trophozoites. Two unlinked copies of genes encoding Igl shared 81% amino acid sequence identity (GenBank accession no. AF337950 and AF337951). They encoded cysteine-rich proteins with amino- and carboxy-terminal hydrophobic signal sequences characteristic of glycosylphosphatidylinositol (GPI)-anchored membrane proteins. The igl genes lacked carbohydrate recognition domains but were members of a large family of amebic genes containing CXXC and CXC motifs. These data indicate that Igl is part of the parasite's multimolecular Gal/GalNAc adhesin required for host interaction.
Carbohydrate-protein interactions initiate the contact-dependent cytotoxicity for which Entamoeba histolytica was named. Parasite recognition of host galactose (Gal) and N-acetyl-d-galactosamine (GalNAc) residues initiates trophozoite adherence to human colonic mucin, colonic epithelium, neutrophils and erythrocytes, certain bacteria, and a variety of cultured cell lines (3–7, 16, 19–22, 27, 36–38). Contact-dependent killing of target cells is >90% inhibited by Gal and GalNAc (34, 37, 41). Additionally, Chinese hamster ovary (CHO) cell glycosylation-deficient mutants lacking terminal Gal/GalNAc residues on N- and O-linked sugars are nearly totally resistant to amebic adherence and cytolytic activity (23, 24, 39).
The E. histolytica 260-kDa Gal/GalNAc lectin is a heterodimer of transmembrane heavy (170 kDa) (Hgl) and GPI-anchored light (35 or 31 kDa) (Lgl) glycoproteins linked by disulfide bonds. It was originally identified by galactose affinity chromatography and with adherence-inhibitory monoclonal antibodies (MAbs) (30, 43). Both Hgl and Lgl are encoded by gene families (28, 35). Antibodies that block or augment parasite Gal/GalNAc binding activity map to the cysteine-rich region (amino acids 356 to 1143) of Hgl (25), and this region (when expressed in Escherichia coli) contains a functional carbohydrate recognition domain (14, 33). The cytoplasmic tail of Hgl has homology to the cytoplasmic domain of β2 and β7 integrins, including regions implicated in binding of the intracellular signaling molecules Shc and Grb2. Overexpression of the cytoplasmic tail results in a dominant negative effect on endogenous lectin activity, with decreased adherence, cytotoxicity, and in vivo virulence (44).
The 150-kDa lectin intermediate subunit (Igl) was originally identified as a trophozoite surface antigen recognized by MAbs which block trophozoite adherence to mammalian cells in vitro (9–11, 42). The EH3015 MAb specific for Igl significantly inhibits adherence of amebae to erythrocytes and CHO cells, erythrophagocytosis by amebae, and amebic cytotoxicity to CHO cells (9). MAb affinity purification of Igl with MAb EH3015 results in copurification of the 260-kDa Hgl-Lgl lectin heterodimer, suggesting that the two proteins are physically associated (10) Igl, separated from the 260-kDa lectin by gel filtration, has galactose-binding activity (10). Immunization with either Igl or the 260-kDa Hgl-Lgl lectin heterodimer provides protection from experimental liver abscess formation in a rodent model (11, 26, 29). Further delineation of the function of Igl requires an understanding of its structure and cellular location.
Colocalization of Igl with Hgl-Lgl.
E. histolytica trophozoites of strain HM-1:IMSS were grown at 37°C in TYI-S33 medium (18, 30, 32) with penicillin (100 U/ml) and streptomycin sulfate (100 μg/ml) (Pfizer, Inc., New York, N.Y.) in sealed plastic tissue culture flasks (18, 30, 32). For immunofluorescence staining, amebae were chilled and resuspended in medium M199 (GIBCO BRL, Gaithersburg, Md.) supplemented with 25 mM HEPES (pH 6.8), 5 mM l-cysteine, and 0.5% bovine serum albumin (BSA). Approximately 2 × 105 amebae were transferred to acetone-washed coverslips (Fisher) in 24-well plastic plates. Amebae were allowed to adhere to the coverslips at 37°C for 15 min. Amebae were then fixed in 3.7% paraformaldehyde for 30 min at 37°C, permeabilized in 0.2% Triton X-100 for 1 min, and washed once in phosphate-buffered saline (PBS) and once in 50 mM ammonium chloride. Amebae were incubated in blocking agent (5% bovine serum albumin with 20% goat serum [catalog no. G-6767; Sigma] in PBS) for 1 h at room temperature. The amebae were then incubated with primary antibody, either rabbit anti-260-kDa Hgl-Lgl antiserum (5 μg/ml) or anti-Igl EH3015 (50 μg/ml) in blocking agent for 1 h at room temperature. Amebae were washed three times with PBS and incubated with the appropriate secondary antibody: goat anti-mouse immunoglobulin G-fluorescein isothiocyanate (IgG-FITC) at a 1:64 dilution (catalog no. F-2012; Sigma) or donkey anti-rabbit IgG-Cy3 at a 1:100 dilution (catalog no. 711-165-152; Jackson Immunoresearch Laboratories) in blocking agent for 30 min in the dark. Amebae were washed three times with PBS and once with H2O and were mounted on glass slides using Biomedia Gelmount. Amebae were visualized using a Zeiss LSM 410 laser scanning confocal microscope equipped with an argon-krypton laser. To compile final images, four averages at 8 s each were compiled via a Zeiss 63×, plan-apochromat (numerical aperture, 1:40) objective, with laser excitation at 488 nm for FITC or 568 nm for Cy3. For experiments in which the 260-kDa subunit was capped, approximately 1.25 × 106 amebae were incubated with rabbit polyclonal anti-260-kDa antibodies (15 μg) at 37°C for 15 min prior to fixation and staining for Igl.
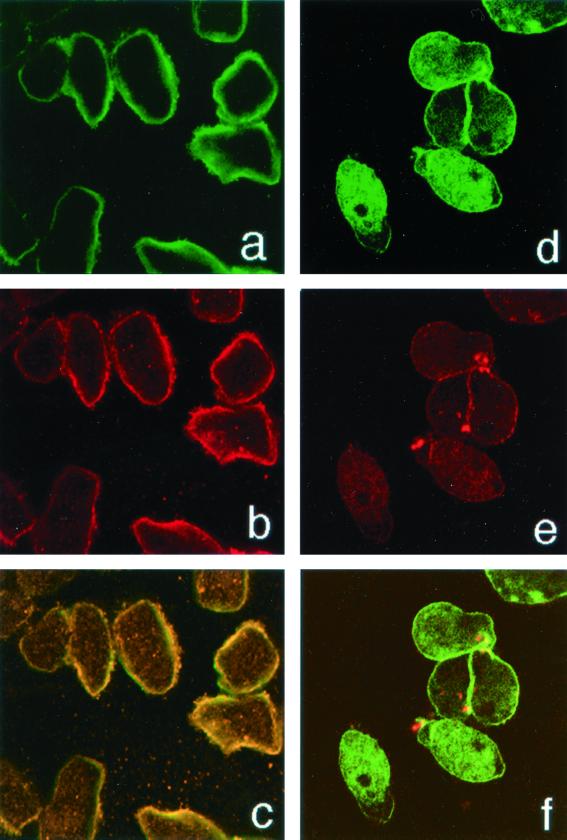
In nonstimulated amebae Igl, Hgl, and Lgl shared the same diffuse surface membrane location, as visualized by immunoflourescence and confocal microscopy (Fig. 1a to c). We also tested for colocalization after first capping Hgl-Lgl at 37°C on the plasma membrane of trophozoites with rabbit antibodies. The amebae were then fixed (but not permeabilized) and reacted with the anti-Igl MAb EII3015. As can be seen from the micrograph (Fig. 1d to f), both proteins were colocalized to membrane caps (yellow). We interpret these data as being consistent with an interaction between the two molecules in the plasma membrane.
FIG. 1.
The Igl, Hgl, and Lgl lectin subunits associate on the surface of amebic trophozoites. (a to c) Amebic trophozoites adhered to laminin-coated coverslips were fixed with paraformaldehyde and stained for Igl with MAb EH3015 and a secondary FITC-conjugated anti-mouse IgG antibody (green) (a) or polyclonal rabbit anti-Hgl-Lgl subunit antibodies and a secondary Cy3-conjugated anti-rabbit IgG antibody (red) (b). The merged image (yellow) is shown in panel c. (d to f) The 260-kDa subunit was first capped on the surface of trophozoites in suspension culture at 37°C with rabbit anti-260-kDa subunit antibodies. The amebae were then fixed and reacted with the anti-Igl MAb EH3015 and stained as in panels a to c.
Sequence of Igl.
Five milligrams of anti-Igl MAb EH3015 was immobilized on 1 to 2 ml of Affi-Gel 10 (Bio-Rad) according to the manufacturer's instructions. Solubilized amebae (prepared by the method of Petri and Schnaar [31]) were circulated through the MAb column with a peristaltic pump for 48 h at 4°C. The column was then extensively washed (50 to 100 ml) with solubilization buffer and then with PBS. The bound protein was eluted with 4 M MgCl2–10 mM Tris (pH 7.2) (31).
The amino terminus of the immunoaffinity-purified Igl was determined by Edman degradation, and peptides released from sodium dodecyl sulfate (SDS)-polyacrylamide gels by trypsinization were microsequenced by tandem mass spectrometry by the W. M. Keck Biomedical Mass Spectrometry Laboratory at the University of Virginia. Two distinct genes (igl1 and igl2, named for intermediate subunit galactose lectin) (GenBank accession no. AF337950 and AF337951) were identified. The 5′ 62-bp pair sequence of igl1 was obtained by PCR amplification from a cDNA library using degenerate primers based on the amino-terminal sequence. The PCR fragment was used as a probe to screen a lambda gt11 cDNA library of E. histolytica strain HM1:IMSS. A positive clone containing the longest insert was subcloned into pUC19 and then sequenced. To extend the sequence to the 5′ end, rapid amplification of the cDNA end was performed with the 5′-Full RACE Core Set (Takara). The amplified product was cloned into a pCR2.1 vector (Invitrogen) and then sequenced. The initial 5′ 400 bp of igl2 were obtained by sequencing of cDNA amplified using HotStarTaq (Qiagen) and oligonucleotides derived from the N-terminal and tryptic peptide amino acid sequences. The remainder of the igl2 sequence (2.8 kb) was obtained by the sequencing of a DNA fragment amplified from genomic HM1:IMSS DNA using the Expand High Fidelity PCR System (Boehringer Mannheim), an igl2-specific oligonucleotide, and a 3′ igl reverse primer. In all cases, amplified DNA was initially cloned using the TOPO TA Cloning System (Invitrogen) before being used as a sequencing template. Authenticity of PCR products was checked by examining the amplified sequence for the next few amino acids predicted by the peptide sequence but not incorporated into the PCR primer.
igl1 and igl2 shared 81% identity and 84% similarity in amino acid sequence and accounted for the vast majority (>85%) of peptides sequenced from the purified protein (Fig. 2). Of the 1,075 amino acids predicted to be present in the mature protein, 48% (512 of 1,075) were identified in the sequences of the amino terminus and tryptic peptides of the purified protein. Tryptic peptides from the 260-kDa lectin were also present in the digests of the affinity-purified Igl. A search of the International Entamoeba Genome Sequencing Project database identified genomic fragments with near 100% identity to each gene (for igl1, ENTHK43 and ENTEO11; for igl2, ENTJW42TF, ENTCM15, ENTEE14TR, and ENTKW17TR).
FIG. 2.
Deduced amino acid sequences of the two genes (igl1 and igl2) encoding Igl. Residues in black type on a gray background are identical in the two genes, and residues in white on a gray background represent conservative substitutions.
The sequence of Igl1 and Igl2 revealed proteins with hydrophobic amino- and carboxy-terminal signal sequences consistent with a GPI-anchored plasma membrane protein. Igl1 and Igl2 had calculated molecular masses of 119,512 and 120,386 Da and predicted isoelectric points (pI) of 5.52 and 5.17, respectively. In contrast, the estimated molecular mass and pI of the native protein were 150 kDa and 6.9 (10), suggesting the existence of posttranslational modifications in the native protein. The most abundant amino acid residues were cysteines (12.3%), lysines (9.5%), and threonines and serines (both 8.9%). The amino acid sequences predicted 12 potential N-glycosylation sites and three O-glycosylation sites.
The Igl proteins lacked a carbohydrate recognition motif but had limited sequence identity with the variant surface glycoproteins of Giardia lamblia (for example, BLAST e value of 2e-42; 22% identity of amino acids 32 to 1036 with amino acids 51 to 1126 of pir T42017) (1, 8, 15). The sequence identity included some of the CXXC motifs of the variant surface glycoproteins implicated in protein-protein interactions. They also had limited sequence identity with the furin-like protease 2 precursor of Drosophila melanogaster (dFurin2) (BLAST value of 4e-11; 23% identity of amino acids 486 to 985 with dFurin2 amino acids 994 to 1452). The region of sequence identity of Igl with dFurin2 encompassed the furin cysteine-rich domain, which is dispensable for furin endoproteinase activity in vitro (13, 17).
Southern blots demonstrate two unlinked copies of igl.
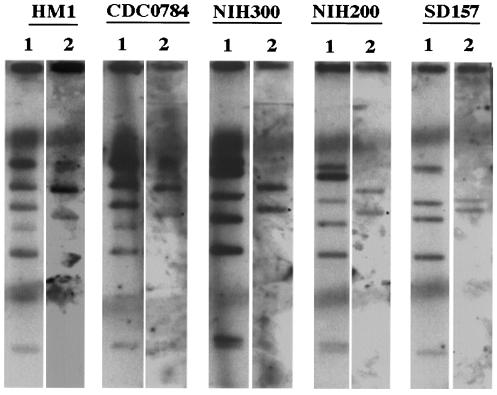
Clamped homogenous electric field (CHEF) gels of genomic DNA digested with HindIII were electrophoresed in a Bio-Rad CHEF DRIII apparatus as described previously (35). CHEF gels were dried down and used directly in hybridization. The gel was denatured in 0.5 M NaOH–0.15 M NaCl and neutralized in 0.5 M Tris-HCl (pH 7.2)–0.15 M NaCl before hybridizing overnight with random-primed (Boehringer-Mannheim) 32P-labeled fragments of the hgl1 and igl1 genes. The hgl1 probe corresponded to nucleotides 1492 to 3560 of the hgl gene, and the igl1 probe corresponded to igl nucleotides 1017 to 1237. This igl region is 97% identical at the nucleotide level between igl1 and igl2. Hybridization was in 6× SSC (1× SSC is 0.15 M Nacl plus 0.015 M sodium citrate), 5× Denhardt's with 0.1% SDS, and 100-μg/ml denatured salmon sperm DNA (Sigma) at 56°C. Gels were washed in 2× SSC–0.1% SDS and then in 0.1×SSC–0.1% SDS at 56°C before being exposed to autoradiography film. Genomic DNA from several different isolates of E. histolytica was digested with HindIII and electrophoresed in a Bio-Rad CHEF DRIII apparatus. The CHEF gel was dried down, and duplicate lanes were directly probed with the igl and hgl probes. Both the igl and hgl probes lack a HindIII site, so only one band would be expected to be seen on Southern blots for a single gene. In fact, all isolates of E. histolytica demonstrated two igl bands, consistent with a minimum of two unlinked igl genes (Fig. 3). The hgl probe hybridized with five to seven bands, depending on the strain, as has been previously reported (35).
FIG. 3.
Southern blots demonstrate two unlinked copies of igl in E. histolytica. DNA from five different E. histolytica isolates was digested with HindIII and separated by CHEF gel electrophoresis. Lanes 1 were hybridized with a probe for hgl, and lanes 2 were hybridized with a probe for igl.
A family of proteins containing CXXC motifs is present in E. histolytica.
A BLAST search (2) of the E. histolytica genome database at www.tigr.org/tdb/edb2/enta/htmls/found igl1 and igl2 gene sequences and at least 100 putative open reading frames (ORFs) containing CXXC motifs. Since the E. histolytica genome project is incomplete and not yet assembled, it is difficult to estimate the exact size of this family of proteins or determine if they share any other structural features such as a GPI anchor. Six unique ORFs that had the highest similarity to Igl (BLAST search e values of 7.7e-17 to 4.6e-20) were selected for further analysis. We named these putative proteins Cxp1 to Cxp6. These ORFs ranged from 270 to 519 amino acids in length. Only Cxp6 contained start and stop codons. Using the “Bestfit” program (Wisconsin Package [version 10.1]; Genetics Computer Group [GCG], Madison, Wis.), these proteins, including Igl, were 32 to 46% identical to each other. Figure 4 highlights the presence of the repeated CXXC motif in Igl and the Cxp proteins. Aside from the CXXC motifs there was little similarity between the proteins.
FIG. 4.
CXXC motifs present in Cxp proteins. The amino acid sequences of Igl1 and Igl2 were aligned with six Cxp proteins using the “PileUp” program from the Wisconsin Package (version 10.1) (Genetics Computer Group). The CXXC motifs are highlighted in gray. Numbering refers to the Igl proteins. Only a selected region is shown. Nucleic acid sequences corresponding to the Cxp ORFs can be found in the GSS division of GenBank. Accession numbers for the following ORFs are as indicated: Cxp1, AZ541185; Cxp2, AZ672865; Cxp3, AZ682291; Cxp4, AZ685201; Cxp5, AZ529058; Cxp-6, AZ500184.
Determination of the primary structure of Igl is an important step in the understanding its function. While its novel sequence is currently uninformative, its colocalization with the 260-kDa lectin subunit in the plasma membrane of E. histolytica suggests a cooperative role in host-parasite interaction.
Acknowledgments
We thank Hideo Tsukamoto for N-terminal amino acid sequencing.
This work was supported by National Institutes of Health grants AI 26649 (W.A.P.) and AI 32615 (B.J.M.) and by grants from the Ministry of Education, Science and Culture and the Ministry of Health and Welfare of Japan (H.T.). W.A.P. is a Burroughs Wellcome Scholar in Molecular Parasitology, and C.D.H. is a recipient of a Howard Hughes Postdoctoral Fellowship for Physicians. The Entamoeba sequencing effort is supported the National Institute of Allergy and Infectious Diseases of the National Institutes of Health and the Burroughs Wellcome Fund.
REFERENCES
- 1.Adam R D, Aggarwal A, Lal A A, de La Cruz V F, McCutchan T, Nash T E. Antigenic variation of a cysteine-rich protein in Giardia lamblia. J Exp Med. 1988;167:109–118. doi: 10.1084/jem.167.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arhets P, Olivo J C, Sansonetti P, Guillen N. Virulence and functions of myosin II are inhibited by overexpression of light meromyosin in Entamoeba histolytica. Mol Biol Cell. 1998;8:1537–1547. doi: 10.1091/mbc.9.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berninghausen O, Leippe M. Necrosis versus apoptosis as the mechanism of target cell death induced by Entamoeba histolytica. Infect Immun. 1997;65:3615–3621. doi: 10.1128/iai.65.9.3615-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bracha R, Mirelman D. Adherence and ingestion of Escherichia coli serotype O55 by trophozoites of Entamoeba histolytica. Infect Immun. 1983;40:882–887. doi: 10.1128/iai.40.3.882-887.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burchard G D, Bilke R. Adherence of pathogenic and nonpathogenic Entamoeba histolytica strains to neutrophils. Parasitol Res. 1992;78:146–153. doi: 10.1007/BF00931657. [DOI] [PubMed] [Google Scholar]
- 7.Chadee K, Petri W A, Jr, Innes D J, Ravdin J I. Rat and human colonic mucins bind to and inhibit the adherence lectin of Entamoeba histolytica. J Clin Investig. 1987;80:1245–1254. doi: 10.1172/JCI113199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen N, Upcroft J A, Upcroft P. A Giardia duodenalis gene encoding a protein with multiple repeats of a toxin homologue. Parasitology. 1995;111:423–431. doi: 10.1017/s0031182000065926. [DOI] [PubMed] [Google Scholar]
- 9.Cheng X J, Kaneda Y, Tachibana H. A monoclonal antibody against the 150 kDa surface antigen of Entamoeba histolytica inhibits adherence and cytotoxicity to mammalian cells. Med Sci Res. 1997;25:159–161. [Google Scholar]
- 10.Cheng X J, Tsukamoto H, Kaneda Y, Tachibana H. Identification of the 150 kDa surface antigen of Entamoeba histolytica as a galactose- and N-acetyl-d-galactosamine-inhibitable lectin. Parasitol Res. 1998;84:632–639. doi: 10.1007/s004360050462. [DOI] [PubMed] [Google Scholar]
- 11.Cheng X J, Tachibana H. Protection of hamsters from amebic liver abscess formation by immunization with the 150- and 170-kDa surface antigens of Entamoeba histolytica. Parasitol Res. 2001;87:126–130. doi: 10.1007/s004360000323. [DOI] [PubMed] [Google Scholar]
- 12.Cho J, Eichinger D. Crithidia fasciculata induces encystation of Entamoeba invadens in a galactose-dependent manner. J Parasitol. 1998;84:705–710. [PubMed] [Google Scholar]
- 13.de Bie I, Savaria D, Roebroek A J M, Day R, Lazure C, van de Ven W J M, Seidah N G. Processing specificity and biosynthesis of the Drosophila melaogaster convertases dfurin1, dfurin1-CRR, dfurin1-X, and dfurin2. J Biol Chem. 1995;270:1020–1028. doi: 10.1074/jbc.270.3.1020. [DOI] [PubMed] [Google Scholar]
- 14.Dodson J M, Lenkowski P W, Jr, Eubanks A C, Jackson T F H G, Napodano J, Lyerly D M, Lockhart L A, Mann B J, Petri W A., Jr Role of the Entamoeba histolytica adhesin carbohydrate recognition domain in infection and immunity. J Infect Dis. 1999;179:460–466. doi: 10.1086/314610. [DOI] [PubMed] [Google Scholar]
- 15.Gillin F D, Hagblom P, Harwood J, Aley S B, Reiner D S, McCaffery M, So M, Guiney D G. Isolation and expression of the gene for a major surface protein of Giardia lamblia. Proc Natl Acad Sci USA. 1990;87:4463–4467. doi: 10.1073/pnas.87.12.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godbold G, Mann B J. Cell killing by the human parasite Entamoeba histolytica is inactivated by the Rho-inactivating enzyme C3 exoenzyme. Mol Biochem Parasitol. 2000;108:147–151. doi: 10.1016/s0166-6851(00)00207-3. [DOI] [PubMed] [Google Scholar]
- 17.Hatsuzawa K, Murakami K, Nakayama K. Molecular and enzymatic proterties of furin, a Kex2-like endoproteinase involved in precursor cleavage at Arg-X-Lys/Arg-Arg sites. J Biochem (Tokyo) 1992;111:296–301. doi: 10.1093/oxfordjournals.jbchem.a123753. [DOI] [PubMed] [Google Scholar]
- 18.Huston C D, Houpt E R, Mann B J, Hahn C S, Petri W A., Jr Caspase 3 dependent killing of human cells by the parasite Entamoeba histolytica. Cell Microbiol. 2000;2:617–625. doi: 10.1046/j.1462-5822.2000.00085.x. [DOI] [PubMed] [Google Scholar]
- 19.Leippe M, Ebel S, Schoenberger O L, Horstmann R D, Muller-Eberhard H J. Pore-forming protein of pathogenic Entamoeba histolytica. Proc Natl Acad Sci USA. 1991;88:7659–7663. doi: 10.1073/pnas.88.17.7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leippe M, Tannich E, Nickel R, van der Goot G, Pattus F, Horstmann R D, Muller-Eberhard H J. Primary and secondary structure of the pore-forming, peptide of pathogenic Entamoeba histolytica. EMBO J. 1992;11:3501–3506. doi: 10.1002/j.1460-2075.1992.tb05432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leroy A, De Bruyne G, Mareel M, Nokkaew C, Bailey G, Nelis H. Contact-dependent transfer of the galactose-specific lectin of Entamoeba histolytica to the lateral surface of enterocytes in culture. Infect Immun. 1995;63:4253–4260. doi: 10.1128/iai.63.11.4253-4260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leroy A, Lauwert T, De Bruyne G, Corenlissen M, Mareel M. Entamoeba histolytica disturbs the tight junction complex in human enteric T84 cell layers. FASEB J. 2000;14:1139–1146. doi: 10.1096/fasebj.14.9.1139. [DOI] [PubMed] [Google Scholar]
- 23.Li E, Becker A, Stanley S L. Use of Chinese hamster ovary cells with altered glycosylation patterns to define the carbohydrate specificity of Entamoeba histolytica adhesion. J Exp Med. 1988;167:1725–1730. doi: 10.1084/jem.167.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li E, Becker A, Stanley S L. Chinese hamster ovary cells deficient in N-acetylglucosaminyltransferase I activity are resistant to Entamoeba histolytica-mediated cytotoxicity. Infect Immun. 1989;57:8–12. doi: 10.1128/iai.57.1.8-12.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mann B J, Chung C Y, Dodson J M, Ashley L S, Braga L L, Snodgrass T L. Neutralizing monoclonal antibody epitopes of the Entamoeba histolytica galactose adhesin map to the cysteine-rich extracellular domain of the 170-kilodalton subunit. Infect Immun. 1993;61:1772–1778. doi: 10.1128/iai.61.5.1772-1778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann B J, Burkholder B V, Lockhart L A. Protection in a gerbil model of amebiasis by oral immunization with Salmonella expressing the galactose/N-acetyl d-galactosamine inhibitable lectin of Entamoeba histolytica. Vaccine. 1997;15:659–663. doi: 10.1016/s0264-410x(96)00236-8. [DOI] [PubMed] [Google Scholar]
- 27.McCoy J J, Mann B J, Vedvick T, Petri W A., Jr Sequence analysis of genes encoding the Entamoeba histolytica galactose-specific adhesin light subunit. Mol Biochem Parasitol. 1993;61:325–328. doi: 10.1016/0166-6851(93)90079-d. [DOI] [PubMed] [Google Scholar]
- 28.McCoy J J, Mann B J, Petri W A., Jr Adherence and cytotoxicity of Entamoeba histolytica, or how lectins let parasites stick around. Infect Immun. 1994;62:3045–3050. doi: 10.1128/iai.62.8.3045-3050.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petri W A, Jr, Ravdin J I. Protection of gerbils from amebic liver abscess by immunization with the galactose-specific adherence lectin of Entamoeba histolytica. Infect Immun. 1991;59:97–101. doi: 10.1128/iai.59.1.97-101.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petri W A, Jr, Smith R D, Schlesinger P H, Murphy C F, Ravdin J I. Isolation of the galactose binding adherence lectin of Entamoeba histolytica. J Clin Investig. 1987;80:1238–1244. doi: 10.1172/JCI113198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petri W A, Jr, Schnaar R L. Purification and characterization of the galactose- and N-acetylgalactosamine-(Gal/GalNAc) specific adherence lectin of Entamoeba histolytica. Methods Enzymol. 1995;253:98–104. doi: 10.1016/s0076-6879(95)53011-8. [DOI] [PubMed] [Google Scholar]
- 32.Petri W A, Jr, Chapman M D, Snodgrass T, Mann B J, Broman J, Ravdin J I. Subunit structure of the galactose and N-acetyl-d-galactosamine-inhibitable adherence lectin of Entamoeba histolytica. J Biol Chem. 1989;264:3007–3012. [PubMed] [Google Scholar]
- 33.Pillai D R, Wan P S K, Yau Y C W, Ravdin J I, Kain K C. The cysteine-rich region of the Entamoeba histolytica adherence lectin (170-kilodalton subunit) is sufficient for high affinity Gal/GalNAc-specific binding in vitro. Infect Immun. 1999;67:3836–3841. doi: 10.1128/iai.67.8.3836-3841.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ragland B D, Ashley L S, Vaux D L, Petri W A., Jr Entamoeba histolytica: target cells killed by trophozoites undergo apoptosis which is not blocked by bcl-2. Exp Parasitol. 1994;79:460–467. doi: 10.1006/expr.1994.1107. [DOI] [PubMed] [Google Scholar]
- 35.Ramakrishnan G, Ragland B D, Purdy J E, Mann B J. Physical mapping and expression of gene families encoding the N-acetyl d-galactosamine adherence lectin of Entamoeba histolytica. Mol Microbiol. 1996;19:91–100. doi: 10.1046/j.1365-2958.1996.356885.x. [DOI] [PubMed] [Google Scholar]
- 36.Ravdin J I, Guerrant R L. Role of adherence in cytopathogenic mechanisms of Entamoeba histolytica. Study with mammalian tissue culture cells and human erythrocytes. J Clin Investig. 1981;68:1305–1313. doi: 10.1172/JCI110377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravdin J I, Croft B Y, Guerrant R L. Cytopathogenic mechanisms of Entamoeba histolytica. J Exp Med. 1980;152:377–390. doi: 10.1084/jem.152.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ravdin J I, John J E, Johnston L I, Innes D J, Guerrant R L. Adherence of Entamoeba histolytica to rat and human colonic mucosa. Infect Immun. 1985;48:292–297. doi: 10.1128/iai.48.2.292-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ravdin J I, Stanley P, Murphy C F, Petri W A., Jr Characterization of cell surface carbohydrate receptors for Entamoeba histolytica adherence lectin. Infect Immun. 1989;57:2179–2186. doi: 10.1128/iai.57.7.2179-2186.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rini J M. Lectin structure. Annu Rev Biophys Biomol Struct. 1995;24:551–577. doi: 10.1146/annurev.bb.24.060195.003003. [DOI] [PubMed] [Google Scholar]
- 41.Saffer L D, Petri W A., Jr Role of the galactose-specific lectin of Entamoeba histolytica in contact-dependent killing of mammalian cells. Infect Immun. 1991;59:4681–4683. doi: 10.1128/iai.59.12.4681-4683.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tachibana H, Kobayashi S, Cheng X J, Hiwatashi E. Differentiation of Entamoeba histolytica from Entamoeba dispar facilitated by monoclonal antibodies against a 150 kDa surface antigen. Parasitol Res. 1997;83:435–439. doi: 10.1007/s004360050276. [DOI] [PubMed] [Google Scholar]
- 43.Tannich E, Ebert F, Horstmann R D. Primary structure of the 170-kDa surface lectin of pathogenic Entamoeba histolytica. Proc Natl Acad Sci USA. 1991;88:1849–1853. doi: 10.1073/pnas.88.5.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vines R R, Ramakrishnan G, Rogers J, Lockhart L, Mann B J, Petri W A., Jr Regulation of adherence and virulence by the Entamoeba histolytica lectin cytoplasmic domain, which contains an β2 integrin motif. Mol Biol Cell. 1998;9:2069–2079. doi: 10.1091/mbc.9.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]