Abstract
Host factors involved in Chlamydia trachomatis pathogenesis were investigated by random chemical mutagenesis of Chinese hamster ovary (CHO-K1) cells followed by selection for clones resistant to chlamydial infection. A clonal mutant cell line, D4.1–3, refractory to infection by the C. trachomatis L2 serovar was isolated. The D4.1–3 cell line appears to be lacking in a previously undescribed temperature-dependent and heparin-resistant binding step that occurs subsequent to engagement of cell surface heparan sulfate by L2 elementary bodies. This novel binding step differentiates the lymphogranuloma venereum (LGV) serovar from other serovars and may contribute the different pathologies associated with LGV and non-LGV strains.
Strains of Chlamydia trachomatis are the etiologic agents of the most common form of sexually transmitted disease in the United States and of preventable blindness worldwide (21). Chlamydiae are obligate intracellular parasites that possess a unique biphasic life cycle. Infection of a host cell is initiated by the elementary body (EB), which is a metabolically inactive form, while multiplication is achieved by the noninfectious but metabolically active reticulate body. Intracellular survival of chlamydiae occurs within a specialized parasitophorous vacuole, termed an inclusion, that is neither acidified nor fusogenic with lysosomes (18). After infection, an EB very quickly dissociates itself from the endosomal/lysosomal pathway and translocates to the peri-Golgi region, where it begins to intercept exocytic vesicle containing sphingomyelin. The processes of dissociation and translocation depend on chlamydial de novo transcription and synthesis of proteins that are thought to modify the inclusion membrane leading to restricted fusogenicity with the lysosomes (11, 12, 22).
Because of the obligate intracellular nature of chlamydiae, the ability to attach to and enter susceptible host cells is an unconditional requirement. Presently, relatively little is known about the chlamydial entry process. Their obligate intracellular lifestyle, coupled with a lack of a genetic system, poses a formidable barrier in identifying the virulence factors involved in the pathogenesis of this organism. One of the principal virulence factors that would indeed be of great importance in understanding chlamydial pathogenesis, as well as offering a potential target for prophylactic intervention, is the bacterial adhesin. Presently, a number of distinct surface molecules have been proposed to function as adhesins in the attachment of chlamydiae to host cells. They include the major outer membrane protein (23, 24), heat shock protein 70 (Hsp70) (20), OmcB (Omp2) (26), and heparan sulfate (HS)-like glycosaminoglycans (GAGs) (29). Definitive identification of the virulence factors continues to be a challenge.
A number of laboratories have attempted to complement this approach by examining the chlamydial infection process from the perspective of the host cell. Like many viral and bacterial pathogens, some chlamydiae have been shown to require cell surface HS proteoglycans for attachment to host cells (6, 7, 8, 23). Inhibition of infection by exogenous heparin as well as the use of cell lines defective in the synthesis of HS has shown that, at least for some serovars of chlamydia, HS is a requirement for successful infection. However, steps in the infection process subsequent to cell surface attachment that may be unique to chlamydia have not been identified. We addressed this question by conducting random chemical mutagenesis of CHO-K1 cells using ethyl methane sulfonate (EMS) as the mutagen and by isolating clones that had become resistant to chlamydial infection. As a selective agent, we took advantage of the rapid and lytic nature of the C. trachomatis serovar L2 developmental cycle. We have isolated a clonal cell line, D4.1–3, that displays resistance to L2 infection. In this paper, we present a detailed characterization of the resistant phenotype of this novel mutant cell line and demonstrate the existence of a previously undescribed step in chlamydial attachment.
Monolayers of CHO-K1 cells growing in T-150 flasks were mutagenized for 24 h with 400 μg of EMS/ml in RPMI 1640 medium without fetal bovine serum. After the mutagenesis, high levels of cytotoxicity were observed in all flasks, but a number of surviving CHO cells remained. The mutagenizing medium was removed and replaced with RPMI 1640 medium supplemented with 10% fetal bovine serum, and the cells were allowed to recover and repopulate the flasks for 5 days, at which time there were approximately 5 × 107 cells in each flask. Infection by C. trachomatis L2 serovar was initiated at a multiplicity of infection (MOI) of 3 to ensure infection of all cells in the flasks. At 36 to 40 h postinfection, dead cells were removed from the flasks by rinsing three times with Hanks balanced salt solution (HBSS). The remaining cells, with some harboring inclusions, were reinfected with approximately 5 × 107 L2 EBs. Because the number of cells remaining was much lower than that of the starting population, the effective MOI was very high. A total of three rounds of selection were performed. Cells were allowed to recover to subconfluence between the rounds of selection. Cells without inclusions were observed after the third round of selection and appeared as individual foci. The surviving cells were trypsinized and replated onto 100-mm-diameter tissue culture plates and allowed to grow into foci. Each focus of cells was isolated using a cloning cylinder and plated onto a well in a 24-well plate and allowed to grow for 72 h. A second round of cloning was performed using the method of limiting dilution. A total of 96 clones were isolated, and their resistant phenotype was verified by infection with L2 EBs; 38 clonal lines retained their resistance. One clone, designated D4.1–3, showed the most dramatic resistant phenotype and was characterized further. PCR analysis of this clone using dnaE as a target failed to detect chlamydial DNA. In addition, serial passages (>160 times) of this cell line failed to induce appearance of inclusions; thus, the cell line appeared to be different from the persistently infected cells described by Moulder and colleagues that were also refractory to superinfection (19).
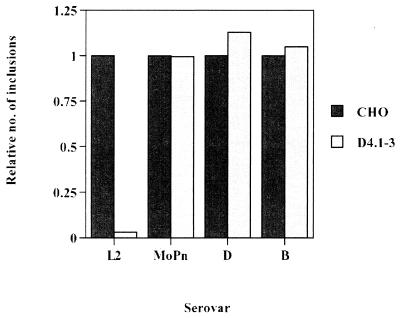
Elementary bodies of C. trachomatis serovars L2, B, D, and MoPn were used to infect wild-type and D4.1–3 mutant cells. The cells were preincubated for 20 min with 42 μg of DEAE-dextran/ml, which increases the efficiency of infection by serovars B, D, and MoPn. Figure 1 shows the number of inclusions, expressed relative to values obtained for wild-type CHO-K1 cells, to allow for direct comparisons between cell lines among all serovars. The mutant cell line remained sensitive to infection by serovars B, D, and MoPn, while retaining its phenotype resistant to L2 infection. Also, the resistance of D4.1–3 to L2 infection was not affected significantly by DEAE-dextran preincubation (data not shown). Resistance to infection appeared to be specific to the lymphogranuloma venereum (LGV) biovar. Similar to that by serovar L2, infection by the L1 and L3 serovars was approximately 20-fold less efficient than that by wild-type CHO-K1 cells (data not shown). Thus, it appears that the D4.1–3 mutation confers resistance to infection by other members of the LGV biovar, indicative of a common mechanism of infection.
FIG. 1.
Susceptibilities of the D4.1–3 mutant cell line to infection by serovars L2, MoPn, B, and D. Cells were preincubated in 42 μg of DEAE-dextran/ml in minimal essential medium for 20 min at 37°C, inoculated with Renografin density gradient-purified (5) L2, MoPn, B, and D EBs (MOI = 0.5) at 37°C for 1 h, washed three times with heparin to remove cell surface-associated EBs, and incubated at 37°C for 24 h. The inclusion-forming assay was performed essentially as described by Furness et al. (10). At 24 h postinfection, infected cells were methanol fixed and processed for immunofluorescence microscopy, using a rabbit polyclonal anti-C. trachomatis L2 EB antibody and a fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin (Zymed, South San Francisco, Calif.). Data are from triplicates and represented as mean ± standard deviation.
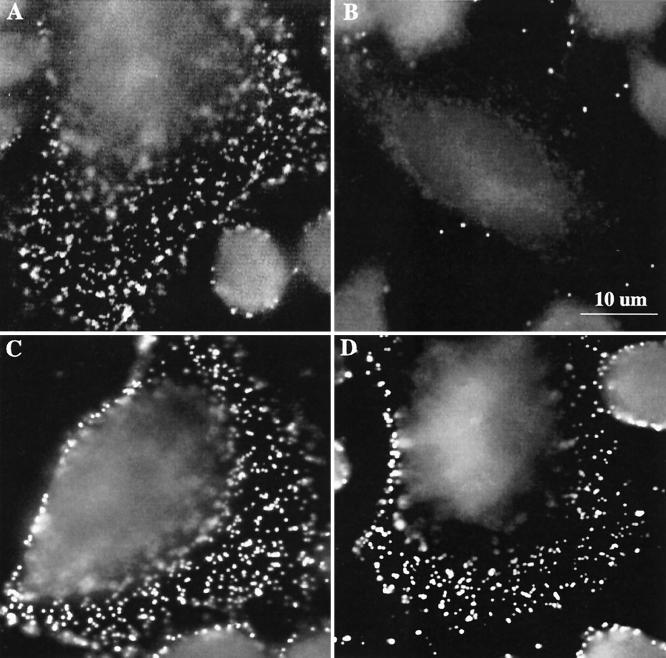
To identify the step in the infection process, i.e., attachment, entry, or intracellular growth, which was disrupted in the D4.1–3 mutant cell line, events at very early time points (10 min postinfection) were investigated using immunofluorescence microscopy. Wild-type CHO-K1 and D4.1–3 mutant cells were inoculated with L2 or D EBs (MOI = 100) at 4°C for 1 h to allow for attachment. Uptake was induced by incubation at 37°C for 10 min, after which the cells were rinsed with cold heparin solution (2 μg/ml) three times to remove surface-associated EBs (see below). The samples were then incubated with a polyclonal antibody that recognizes both L2 and D EBs. Figure 2 demonstrates the markedly reduced number of L2 EBs observed on D4.1–3 cells, while the numbers of serovar D EBs associated with wild-type and mutant cells were comparable.
FIG. 2.
Immunofluorescence images of parental CHO-K1 and D4.1–3 cell lines infected with serovar L2 or serovar D EBs. Cells were inoculated with serovar L2 or D EBs (MOI = 500) at 4°C for 1 h. The cells were washed with cold 1× HBSS three times and were immediately incubated at 37°C for 10 min. The cells were rinsed three times with heparin and fixed with methanol for immunofluorescence microscopy. (A) CHO infected with L2; (B) D4.1–3 infected with L2; (C) CHO infected with D; and (D) D4.1–3 infected with D.
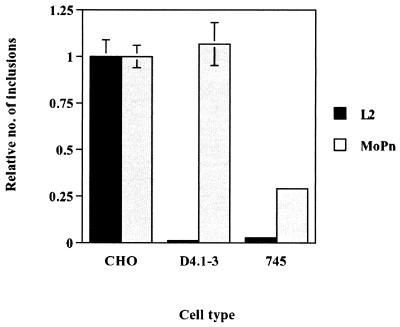
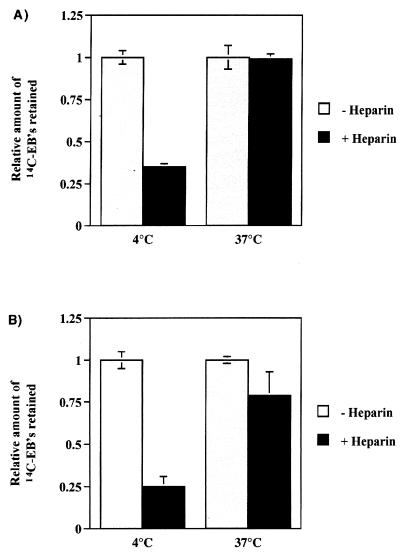
Previous studies have shown a requirement for HS-like GAGs in the attachment of C. trachomatis L2 and MoPn, as well as Chlamydia psittaci serovar GPIC (6, 7, 8, 23, 26). Lack of cell surface HS in the D4.1–3 cells would result in its resistance to infection by serovar L2. We therefore compared the phenotypes of wild-type CHO-K1 and D4.1–3 with that of pgsA-745 cells, a cell line derived from CHO-K1 that is unable to synthesize GAGs (9) and displays reduced attachment of C. trachomatis (23). We performed an inclusion-forming assay using both L2 and MoPn EBs on wild-type CHO cells and the two mutant cell lines. Figure 3 shows the resistance of pgsA-745 cells to infection by both L2 and MoPn. In contrast, the D4.1–3 mutant exhibited resistance only to L2 infection. Therefore, the D4.1–3 mutant appears to be different from the GAG-deficient pgsA-745 cell line. The susceptibility of D4.1–3 cells to serovar MoPn, along with the significant resistance of the GAG-deficient pgsA-745 cell line to MoPn infection, indicates that the mutation conferring the resistant phenotype of D4.1–3 to L2 infection is not associated with a defect in the synthesis of GAG. To distinguish the processes of chlamydial attachment and internalization, we employed paraformaldehyde-fixed cells, prepared by a modification of the protocol described by Hatch et al. (13), to inhibit the process of internalization without disruption to the attachment step. Briefly, cells were fixed with freshly prepared 4% paraformaldehyde for 40 min at room temperature. The cells were rinsed with phosphate-buffered saline three times, and the remaining paraformaldehyde was neutralized by incubation with 10 mM NH4Cl for 30 min at 4°C, followed by an incubation with 3% bovine serum albumin for 30 min at 4°C. The inhibition of chlamydial internalization was confirmed by electron microscopy (data not shown). Viable and paraformaldehyde-fixed CHO cells were incubated with 14C-labeled EBs at an MOI of 500 and were incubated at 4 or 37°C for 30 min. Figure 4 demonstrates the efficacy of heparin in removing surface-associated EBs from both live cells and cells incubated at 4°C. Following incubation at 37°C, however, heparin is ineffective in removing EBs associated with either live or fixed cells. In live cells, this observation can be attributed to the concomitant internalization of bound EBs. In contrast, a high level of 14C-EBs remains associated with paraformaldehyde-fixed cells even after the heparin wash. Taken together, the data indicate that a heparin-resistant association of EBs to host cells occurs at 37°C and that washing with exogenous heparin distinguishes an initial, reversible stage followed by an irreversible step that is temperature dependent.
FIG. 3.
Differential susceptibility of D4.1–3 mutant cells and GAG-deficient pgsA-745 cells to infection by C. trachomatis serovar MoPn EBs. Cells were inoculated with L2 or MoPn EBs (MOI = 0.5) for 1 h at 37°C. The cells were rinsed three times with HBSS and were incubated at 37°C for 24 h. Inclusions were visualized by immunofluorescence microscopy and counted. Data are from triplicates and are represented as mean ± standard deviation.
FIG. 4.
Effects of incubation temperature and heparin treatment on the attachment of 14C-labeled EBs to live (A) or paraformaldehyde-fixed (B) CHO-K1 cells. Cells were infected with 14C-labeled EBs (MOI = 500) for 30 min at either 4 or 37°C. Quality of binding was assessed by release with HBSS or heparin washes. Retained radioactivity was counted by scintillation counting. Data are from triplicates and are represented as mean ± standard deviation.
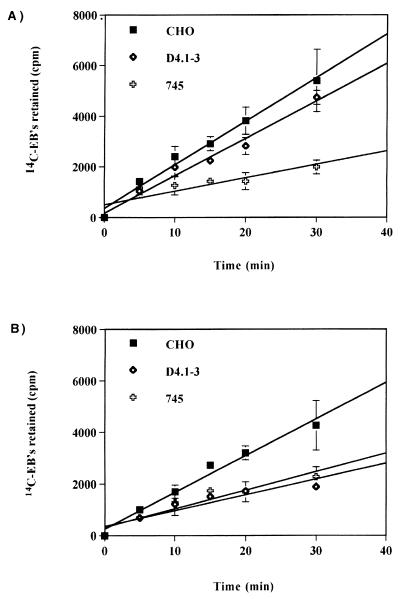
To further investigate this apparent additional step in chlamydial attachment, we characterized the binding of 14C-labeled EBs to paraformaldehyde-fixed CHO, D4.1–3, and pgsA-745 cells. By monitoring remaining EBs associated with the fixed cells, we could detect and quantify the existence of the hypothesized secondary binding after washing with HBSS and heparin-HBSS to distinguish reversible and irreversible attachment, respectively. In Fig. 5A, in which the formation of both GAG-dependent and independent binding was monitored, the slopes of the lines generated by wild-type and D4.1–3 cells were similar (m = 172 versus 148). However, the slope generated by the data set obtained from GAG-deficient cells (m = 53) was significantly lower than that for either wild-type or D4.1–3 cells. These results confirm the HS requirement for L2 EB attachment and support our conclusion that the D4.1–3 cell line is distinct from the GAG-deficient pgsA-745 cell line.
FIG. 5.
Kinetics of binding of 14C-labeled L2 EBs to paraformaldehyde-fixed CHO, D4.1–3, and pgsA-745 cells. 14C-labeled L2 EBs in 200 μl of HBSS (≈2.4 × 104 cpm) were added to each well. The radioactive inoculum was removed at the indicated times after incubation at 37°C, and the cells were rinsed three times, either with HBSS alone (A) or supplemented with 2 μg of heparin/ml (B) (USB, Cleveland, Ohio). The fixed cells were solubilized with 2% sodium dodecyl sulfate in phosphate-buffered saline, and associated radioactivity was monitored by scintillation counting. The data was plotted using Cricket Graph version 1.5.1 Data are from triplicates and are represented as mean ± standard deviation.
We then used a more stringent wash that included 2 μg of heparin/ml in HBSS (Fig. 5B). In contrast to wild-type CHO-K1 cells, D4.1–3 mutant cells appeared to be defective in this secondary binding step. The slope of the line obtained was similar to that of the pgsA-745 mutant cells (m = 61 versus 72). The slope obtained for the wild-type cells was markedly different (m = 142) from those of D4.1–3 and pgsA-745 mutants. Taken together, the binding kinetics demonstrate that chlamydial attachment to host cell surfaces can be dissected into at least two steps: (i) reversible binding to cell surface GAGs and (ii) irreversible attachment to an unknown cell surface component subsequent to GAG binding.
EMS mutagenesis of CHO cells has been used extensively to dissect the genetic components involved in biological processes, such as regulation of cholesterol metabolism (17), resistance to viral pathogens (1, 16, 25), and GAG biosynthesis (9). Here we demonstrate the utility of this technique in identification of host factors that are involved in chlamydial infection and provide a detailed description of a novel CHO cell mutant that is resistant to chlamydial infection. The cellular basis for this resistance is the inability of the host cell to fully support attachment, specifically, at a previously undescribed binding step downstream of GAG binding. This newly discovered step in chlamydial attachment is resistant to dissociation by exogenous heparin, although the formation of this binding may require the prior engagement of EB particles by cell surface HS. The data supports a model of LGV attachment that involves an initial, reversible step likely mediated by electrostatic interactions with HS-like GAGs, followed by an irreversible step involving binding to an as-yet-unknown receptor that mediates the actual entry process. A similar model of attachment has been proposed by Su et al., with MoPn major outer membrane protein being a critical component responsible for at least the initial electrostatic interactions with cell surface HS (23). The present result, however, would suggest that the secondary receptor differs between serovars L2 and MoPn. Such a mechanism is reminiscent of that described for varicella-zoster virus, where GAG mediates an initial attachment to host cell surfaces but where endocytosis is dependent upon binding to the mannose-6-phosphate receptor (30). The identity of the host cell receptors that mediate chlamydial entry remains to be identified.
Human strains of C. trachomatis are placed into two biovars, LGV and trachoma, that differ clinically and biologically. In vitro, the biovars are distinguished by their interactions with cultured cells. Whereas attachment and entry of the trachoma biovar are enhanced by DEAE-dextran or centrifugation of the inoculum onto the monolayer, the LGV biovar is unaffected by these treatments. In addition, polyanions, such as heparin or dextran sulfate, inhibit LGV attachment much more efficiently than they do trachoma biovars (2, 3, 6, 7, 14, 15). It has long been suspected that chlamydiae, as obligate intracellular parasites, may have adapted to exploit multiple means of entry. That LGV strains preferentially utilize receptors different from those used by trachoma strains is further demonstrated by the inability of the different biovars to competitively inhibit binding of each other (14, 28). However, this finding is not consistent between laboratories, with some groups observing sufficient competition between LGV and trachoma biovars to suggest that they interact with common receptors (6, 27). The D4.1–3 mutation specifically reduced attachment by members of the LGV biovar but not by serovars D, B, and MoPn. This difference in susceptibility may be indicative of different receptor utilization consistent with the proposed HS-dependent and independent binding of the distinct C. trachomatis biovars. Different receptor utilization may contribute to the differences in the pathologies associated with infections by the LGV and trachoma biovars. In contrast to members of the trachoma biovar, which are localized to ocular and genital mucosal surfaces, members of the LGV biovar penetrate the surface epithelium and the submucosa to initiate systemic infection and lymphadenopathy. Differential expression of the LGV- or trachoma-specific secondary receptors in vivo would reflect the pathologies associated with each biovar. A similar hypothesis has been proposed by Wyrick and colleagues (28), attributing the colonization and infection of the submucosal layer by the L2 serovar, which is primarily a submucosal pathogen, to the enrichment of HS in the extracellular matrix (4). In addition to restricted cell surface expression of HS, one could also envision the secondary receptor providing an additional level of specificity that further defines the differences in pathology associated with LGV and non-LGV serovars.
Unlike the case for many bacterial pathogens, there are no existing means of stably introducing recombinant DNA into chlamydiae to definitively test chlamydial proteins that are candidates for adhesins. Furthermore, because chlamydiae are obligate intracellular pathogens, generating unconditional mutants that are defective in attachment is not possible. We have mutagenized the host cell in an effort to determine host factors that are involved in chlamydial infection, with the hope of utilizing new information gained from these experiments to identify chlamydial virulence factors. Distinct CHO cell mutants collectively identify two stages in the attachment process. The pgsA-745 mutant cell line (9) is deficient in host cell surface GAG expression and therefore displays diminished attachment of those chlamydial strains that require HS-like GAGs for their initial interaction (6, 7, 8, 23, 24). The D4.1–3 cell line represents a different category of mutants that support the initial electrostatic interaction of LGV strains with the cell surface but do not progress to a secondary irreversible binding. The CHO cell mutagenesis experiments described here generated a number of mutants that demonstrate significant levels of resistance to L2 infection. Mutants affected in additional stages of chlamydial infection, such as entry or intracellular growth, may be represented in our panel of CHO cell mutants, thus making these cell lines potentially invaluable in elucidating chlamydial biology.
Acknowledgments
We thank J. D. Esko for the pgsA cell line and H. Caldwell and B. Whitmire for their gift of purified EBs of serovars MoPn, B, D, L1, and L3. We appreciate the technical assistance of J. Sager and thank E. Fischer for the electron microscopy. We also thank G. McClarty and H. Caldwell and thank members of the Hackstadt laboratory for valuable suggestions and review of the manuscript.
REFERENCES
- 1.Bair C-H, Chung C-S, Vasilevskaya I A, Chang W. Isolation and characterization of a Chinese hamster ovary mutant cell line with altered sensitivity to vaccinia virus killing. J Virol. 1996;70:4655–4666. doi: 10.1128/jvi.70.7.4655-4666.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker Y, Hochberg E, Zakay-Rones Z. Interaction of trachoma elementary bodies with host cells. Isr J Med Sci. 1969;5:121–124. [PubMed] [Google Scholar]
- 3.Bose S K, Paul R G. Purification of Chlamydia trachomatis lymphogranuloma venereum elementary bodies and their interaction with HeLa cells. J Gen Microbiol. 1982;128:1371–1379. doi: 10.1099/00221287-128-6-1371. [DOI] [PubMed] [Google Scholar]
- 4.Boutin E L, Sanderson R D, Bernfield M, Cunha G R. Epithelial-mesenchymal interactions in uterus and vagina alter the expression of the cell surface proteoglycan, syndecan. Dev Biol. 1991;143:63–74. doi: 10.1016/0012-1606(91)90317-v. [DOI] [PubMed] [Google Scholar]
- 5.Caldwell H D, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J C, Stephens R S. Trachoma and LGV biovars of Chlamydia trachomatis share the same glycosaminoglycan-dependent mechanism for infection of eukaryotic cells. Mol Microbiol. 1994;11:501–507. doi: 10.1111/j.1365-2958.1994.tb00331.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen J C, Stephens R S. Chlamydia trachomatis glycosaminoglycan-dependent and independent attachment to eukaryotic cells. Microb Pathog. 1997;22:23–30. doi: 10.1006/mpat.1996.0087. [DOI] [PubMed] [Google Scholar]
- 8.Davis C H, Wyrick P B. Differences in the association of Chlamydia trachomatis serovar E and serovar L2 with epithelial cells in vitro may reflect biological differences in vivo. Infect Immun. 1997;65:2914–2924. doi: 10.1128/iai.65.7.2914-2924.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esko J D, Steward T E, Taylor W H. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc Natl Acad Sci USA. 1985;82:3197–3201. doi: 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furness G, Graham D M, Reeve P. The titration of trachoma and inclusion blennorrhoea viruses in cell cultures. J Gen Microbiol. 1960;23:613–619. doi: 10.1099/00221287-23-3-613. [DOI] [PubMed] [Google Scholar]
- 11.Hackstadt T, Rockey D D, Heinzen R A, Scidmore M A. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- 12.Hackstadt T, Scidmore M A, Rockey D D. Lipid metabolism in Chlamydia trachomatis infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc Natl Acad Sci USA. 1995;92:4877–4881. doi: 10.1073/pnas.92.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatch T P, Vance D W, Jr, Al-Hossainy E. Attachment of Chlamydia psittaci to formaldehyde-fixed and unfixed L cells. J Gen Microbiol. 1981;125:273–283. doi: 10.1099/00221287-125-2-273. [DOI] [PubMed] [Google Scholar]
- 14.Kuo C-C, Grayston J T. Interaction of Chlamydia trachomatis organisms and HeLa 229 cells. Infect Immun. 1976;13:1103–1109. doi: 10.1128/iai.13.4.1103-1109.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo C C, Wang S-P, Grayston J T. Effect of polycations, polyanions, and neuraminidase on the infectivity of trachoma-inclusion conjunctivitis and lymphogranuloma venereum organisms in ZheLa cells: sialic acid residues as possible receptors for trachoma-inclusion conjunctivitis. Infect Immun. 1973;8:74–79. doi: 10.1128/iai.8.1.74-79.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mento S J, Siminovitch L. Isolation and preliminary characterization of Sindbis virus-resistant Chinese hamster ovary cells. Virology. 1981;111:320–330. doi: 10.1016/0042-6822(81)90336-6. [DOI] [PubMed] [Google Scholar]
- 17.Metherall J E, Goldstein J L, Luskey K L, Brown M S. Loss of transcriptional repression of three sterol-regulated genes in mutant hamster cells. J Biol Chem. 1989;264:15634–15641. [PubMed] [Google Scholar]
- 18.Moulder J W. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moulder J W, Levy N J, Schulman L P. Persistent infection of mouse fibroblasts (L cells) with Chlamydia psittaci: evidence for a cryptic chlamydial form. Infect Immun. 1980;30:874–883. doi: 10.1128/iai.30.3.874-883.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raulston J E, Davis C H, Schmiel D H, Morgan M W, Wyrick P B. Molecular characterization and outer membrane association of a Chlamydia trachomatis protein related to the Hsp70 family of proteins. J Biol Chem. 1993;268:23139–23147. [PubMed] [Google Scholar]
- 21.Schachter J. Infection and disease epidemiology. In: Stephens R S, editor. Chlamydia: intracellular biology, pathogenesis, and immunity. Washington, D.C.: ASM Press; 1999. pp. 139–169. [Google Scholar]
- 22.Scidmore M A, Rockey D D, Fischer E R, Heinzen R A, Hackstadt T. Vesicular interactions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect Immun. 1996;64:5366–5372. doi: 10.1128/iai.64.12.5366-5372.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su H, Raymond L, Rockey D D, Fischer E, Hackstadt T, Caldwell H D. A recombinant Chlamydia trachomatis major outer membrane protein binds to heparan sulfate receptors on epithelial cells. Proc Natl Acad Sci USA. 1996;93:11143–11148. doi: 10.1073/pnas.93.20.11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su H, Watkins N G, Zhang Y X, Caldwell H D. Chlamydia trachomatis-host cell interactions: role of the chlamydial major outer membrane protein as an adhesin. Infect Immun. 1990;58:1017–1025. doi: 10.1128/iai.58.4.1017-1025.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taber R, Alexander V, Wald N., Jr The selection of virus-resistant Chinese hamster ovary cells. Cell. 1976;8:529–533. doi: 10.1016/0092-8674(76)90221-x. [DOI] [PubMed] [Google Scholar]
- 26.Ting L- M, Hsia R-C, Haidaris C G, Bavoil P M. Interaction of outer envelope protein of Chlamydia psittaci GPIC with the HeLa cell surface. Infect Immun. 1995;63:3600–3608. doi: 10.1128/iai.63.9.3600-3608.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vretou E, Goswami P C, Bose S K. Adherence of multiple serovars of Chlamydia trachomatis to a common receptor on HeLa and McCoy cells is mediated by thermolabile protein(s) J Gen Microbiol. 1989;135:3229–3237. doi: 10.1099/00221287-135-12-3229. [DOI] [PubMed] [Google Scholar]
- 28.Wyrick P B, Choong J, Davis C H, Knight S T, Royal M O, Maslow A S, Bagnell C R. Entry of genital Chlamydia trachomatis into polarized human epithelial cells. Infect Immun. 1989;57:2378–2389. doi: 10.1128/iai.57.8.2378-2389.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J P, Stephens R S. Mechanism of C. trachomatis attachment to eukaryotic host cells. Cell. 1992;69:861–869. doi: 10.1016/0092-8674(92)90296-o. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Z, Gershon M D, Ambron R, Gabel C, Gershon A A. Infection of cells by varicella-zoster virus: inhibition of viral entry by mannose-6-phosphate and heparin. Proc Natl Acad Sci USA. 1995;92:3546–3550. doi: 10.1073/pnas.92.8.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]