Abstract
Medulloblastoma (MB) is the most common malignant brain tumour in children. High-risk MB patients harbouring MYC amplification or overexpression exhibit a very poor prognosis. Aberrant activation of MYC markedly reprograms cell metabolism to sustain tumorigenesis, yet how metabolism is dysregulated in MYC-driven MB is not well understood. Growing evidence unveiled the potential of BET-bromodomain inhibitors (BETis) as next generation agents for treating MYC-driven MB, but whether and how BETis may affect tumour cell metabolism to exert their anticancer activities remains unknown. In this study, we explore the metabolic features characterising MYC-driven MB and examine how these are altered by BET-bromodomain inhibition. To this end, we employed an NMR-based metabolomics approach applied to the MYC-driven MB D283 and D458 cell lines before and after the treatment with the BETi OTX-015. We found that OTX-015 triggers a metabolic shift in both cell lines resulting in increased levels of myo-inositol, glycerophosphocholine, UDP-N-acetylglucosamine, glycine, serine, pantothenate and phosphocholine. Moreover, we show that OTX-015 alters ascorbate and aldarate metabolism, inositol phosphate metabolism, phosphatidylinositol signalling system, glycerophospholipid metabolism, ether lipid metabolism, aminoacyl-tRNA biosynthesis, and glycine, serine and threonine metabolism pathways in both cell lines. These insights provide a metabolic characterisation of MYC-driven childhood MB cell lines, which could pave the way for the discovery of novel druggable pathways. Importantly, these findings will also contribute to understand the downstream effects of BETis on MYC-driven MB, potentially aiding the development of new therapeutic strategies to combat medulloblastoma.
Subject terms: Biophysics, Cancer, Cell biology, Diseases, Medical research, Oncology
Introduction
Medulloblastoma (MB) is the most common malignant tumour of the central nervous system (CNS) in children, arising from embryonal lesions generated by diverse progenitor cell populations during early brain development1,2. It was first described in 1926 as one subset of glioma3. Since then, advances in molecular genetics ameliorated our understanding of MB, culminating in a consensus definition of four distinct groups: wingless (WNT), sonic hedgehog (SHH), Group 3 and Group 4, which differ in both molecular and clinical characteristics4,5. Unlike the SHH and WNT groups, no common signalling pathway driving the disease has been identified in Group 3 and 4 tumours. The latter ones exhibit the worst prognosis of the four subgroups and recurrent MYC amplification or overexpression has been identified as one of the major biomarkers for this high-risk MB patient group with poor clinical outcome4. The modest understanding of the molecular mechanisms underpinning tumorigenesis in Group 3 and Group 4 MBs has limited the development of targeted therapies. For moderate and high risk-patients trials includes novel chemotherapeutics (pemetrexed and gemcitabine) after standard chemotherapy and risk-adapted radiotherapy. For recurrent and refractory MB patients two trials combine the use of chemotherapeutics with targeted agents such as prexasertib (NCT04023669), a targeted CHK1/2 inhibitor, and ribociclib (NCT01878617), a cyclin-dependent kinase inhibitor6. Current treatments include tumour resection, radiotherapy and chemotherapy. Despite this aggressive multimodal therapy, approximatively 30% of the patients die of disease, while survivors suffer long-term side effects due to the harsh treatments on the developing brain7,8.
To date, several preclinical and clinical studies indicated that BET-bromodomain (BRD) inhibitors (BETis) might be considered as next generation agents for treating MYC-driven MB4,6. These are small molecules that specifically inhibit proteins in the BRD family which contain two bromodomains capable of recognising acetylated lysine residues in histone tails and recruiting transcriptional factors to promote targeted gene transcription9. Their inhibitors prevent the interaction between the bromodomain and the acetyl group, causing the downregulation of certain genes, including c-MYC10. JQ1 was the first BETi to be developed. This is a cell permeable small molecule that promotes differentiation and halts proliferation in cancer cell lines and in diverse murine tumour models10–14. Despite its efficacy, JQ1 has short-lasting effects due to its half-life of around 1 h, greatly limiting the possibilities to translate preclinical findings into clinical benefit11,14,15. OTX-015 (MK-8628) is a BETi developed in the past decade that targets BRD 2/3/416. OTX-015 has been synthesised from ( +)-JQ1 by substituting the tert-butyl ester with a para-hydroxyacetamide. This structural change resulted in an improved pharmacokinetic profile17 (with an half-life of 6 h)16,18,19 which led to its use in several clinical trials including glioblastoma multiforme (GBM), NUT midline carcinoma, triple-negative breast cancer (TNBC), castration-resistant prostate cancer, pancreatic ductal carcinoma and haematological malignancies20. OTX-015 causes an early, strong and long lasting reduction of MYC, and active doses were not toxic in vivo16. Furthermore, in GBM xenograft models this compound has been shown to pass through the blood brain barrier and preferentially bind to cancerous tissue, providing a strong pharmacologic basis for OTX-015 use in brain tumour therapy21. Moreover, OTX-015 prodrugs with significantly increased antitumor activity and reduced toxicity have recently been developed22.
The c-MYC (MYC) transcription factor belongs to the MYC-proto onco-protein family, which also includes MYCN and MYCL23. Heterodimerization with MAX is an essential prerequisite for MYC-driven transcription and cellular transformation. The MYC/MAX dimer binds Enhancer box (E-box) sequences regulating the expression levels of genes involved in growth regulatory networks and oncogenic signalling pathways, including energy production and metabolism23,24. Specifically, MYC activates genes modulating metabolic processes in order to balance nutrient supply and demand and drive cell fate decisions25. This plays a crucial role in tumour development, as cancer cells reprogram their metabolism to fulfil their increased bioenergetic needs, to maintain a high rate of macromolecular biosynthesis, essential for cell growth/division, and to prevent toxic Reactive Oxygen Species (ROS) levels26.
To date, despite a plethora of evidence reporting that MYC tunes metabolic networks to foster oncogenesis in diverse tumour types27, data remain scarce and limited, and likewise, the metabolic response of BETi treatment of MYC-driven MB remains unexplored. In this work, we investigate changes in metabolism of MYC-driven MB cell lines prior and after treatment with OTX-015. We chose OTX-015 over other inhibitors, e.g. JQ1, because of its higher promise in clinical trials. This study contributes to the phenotypical characterisation of MYC-driven MB models and reveals metabolic changes caused by OTX-015, providing new clues of the anticancer mechanisms of action for these compounds.
Results
OTX-015 inhibits cell viability and reduces MYC expression in medulloblastoma cell lines
We selected human D283 and D458 MB cell lines as in vitro models of MYC-driven MB. The D283 cell line has been classified as either Group 3 or Group 4 and overexpresses MYC and the transcription factor orthodenticle homeobox 2 (OTX2)28. The latter plays a key role in the pathogenesis of anaplastic MB and treatment with all trans-retinoic acid causes OTX2 downregulation and apoptosis29. D458 cells belong to Group 3 and are characterised by MYC amplification. They derive from the metastasis of a parental tumour and harbour wild type p5328 with R72P polymorphism, a common single nucleotide polymorphism (SNP) that results in either an arginine or proline at position 72 of the protein30,31.
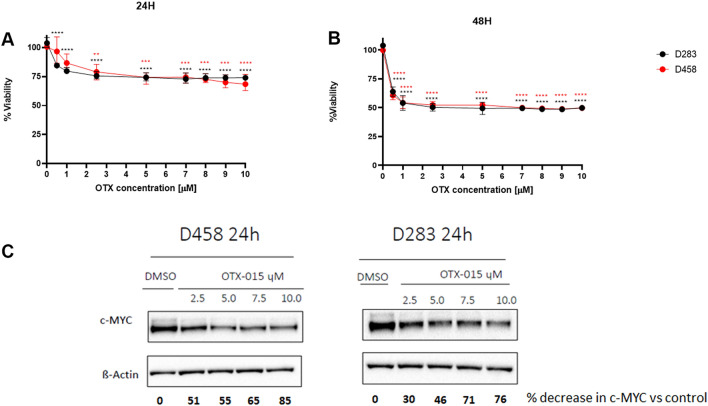
The D283 and D458 cells were treated with a range of OTX-015 doses spanning from 0 to 10 µM for 24 and 48 h, to evaluate the optimal time point to study their metabolism. Western Blot analysis was performed to evaluate MYC protein expression (Fig. 1A–C and Supplementary Fig. S1). Using the MTT assay we demonstrated that OTX-015 decreases cell viability by 50% after 48 h, while Western Blot showed that c-MYC protein levels were already reduced after 24 h of incubation with OTX-015. Specifically, relative protein quantification showed that 7.5 µM of OTX-015 caused a 70% decrease in c-MYC protein levels versus untreated cells. We thus choose to investigate the early effects of BET inhibition and MYC downregulation on cancer metabolism after 24 h of incubation with 7.5 µM of OTX-015.
Figure 1.
OTX-015 inhibits the viability of D458 and D283 MB cell lines. D458 (red) and D283 (black) were treated with different concentrations of OTX-015 spanning from 0 to 10 µM as indicated for (A) 24 and (B) 48 h. Statistical significance of cell viability inhibition was performed using one-way ANOVA with **; *** and **** indicating p < 0.05, < 0.005 and < 0.0001, respectively. (C) Representative Western blot showing MYC downregulation upon treatment with OTX-015 (2.5, 5.0, 7.5, and 10.0 µM) for 24 h. Bold numbers indicate percentage reduction of normalised c-MYC (c-MYC/β-Actin) versus control (DMSO). Original Western blots are presented in Supplementary Fig. 1.
OTX-015 treatment causes a statistically significant metabolic shift in MYC-driven MB cell lines
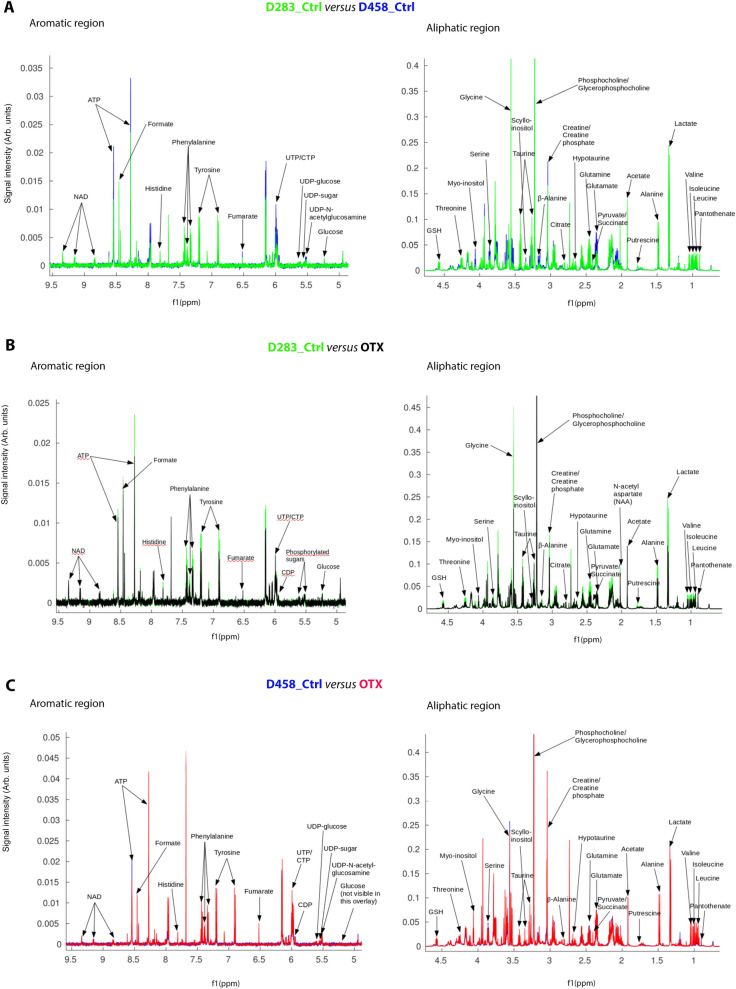
Next, we analysed the metabolism of D283 and D458 cells upon OTX-015 treatment by Nuclear Magnetic Resonance (NMR) spectroscopy. 1H NMR profiling of the aqueous cellular extracts allowed the identification of 41 intracellular polar metabolites (Fig. 2, Supplementary Figs. S2–S7, and Supplementary Table S1), assigned by comparing peak chemical shifts, J-couplings and multiplet intensity ratios to NMR spectral resonance patterns available in the Human Metabolome Database (HMDB)32 and the biological magnetic resonance data bank (BMRB)33. Metabolite assignment was confirmed by a 2D NMR approach, performing [1H–1H] TOCSY and [1H–13C] HSQC experiments (Supplementary Figs. S8–S10). Aligned and normalised 1D 1H-NMR spectra overlays of the aliphatic and aromatic region of D283 versus D458, OTX-015-treated D283 (D283_OTX) versus D283 control (D283_Ctrl) and OTX-015-treated D458 (D458_OTX) versus D458 control (D458_Ctrl) (Fig. 2, and Supplementary Figs. S2–S7) show intensity differences for many metabolite peaks, the chemical shift values of which are reported in Supplementary Table S1. Amongst these we detected: glycolysis and tricarboxylic acid (TCA) cycle derivatives including lactate, acetate, alanine, succinate, citric acid, pyruvate, glucose and fumarate; glutamine and glutamine derivatives comprising glutamate N-acetylaspartate and proline; other amino acids such as leucine, isoleucine, valine, phenylalanine, threonine, tyrosine, glycine, serine and histidine; amino acid derivatives including reduced glutathione GSH, creatine, taurine, hypotaurine and formate; phospholipid derivatives including phosphocholine and glycerophosphocholine; nucleotides as ATP, UDP, CTP and NAD; the carbohydrate myo-inositol and its stereoisomer scyllo-inositol; the polyamine putresceine and the vitamin B5 pantothenate. Furthermore, two phosphorylated sugars were observed to change upon OTX-015 treatment. One of these was identified based on the chemical shift of the anomeric proton as UDP-N-acetylglucosamine, whilst the other was dubbed as a generic ‘UDP-sugar’, as the unambiguous assignment of its structure was not possible from our NMR data alone.
Figure 2.
Aligned and normalised overlays of representative one-dimensional 600 MHz 1H NMR spectra of cellular extracts from control and OTX-015 treated cells, focussing on aliphatic and aromatic regions. (A) D283 (D283_Ctrl, green) versus D458 (D458_Ctrl, blue); (B) D283 control (D283_Ctrl, green) versus OTX-015 treated (D283_OTX, black) and (C) D458 control (D458_Ctrl, blue) versus OTX-015 treated (D458_OTX, red) cell lines. Selected peaks of identified metabolites are labelled.
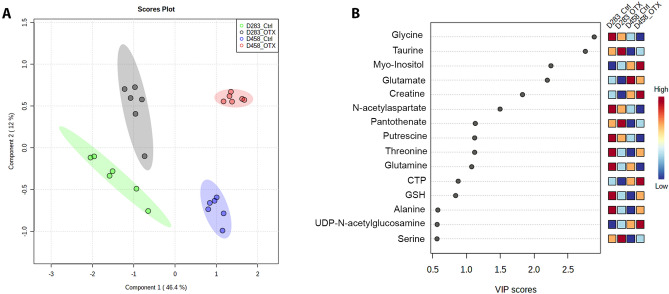
1D 1H NMR spectra were used to perform a Partial least squares discriminant analysis (PLS-DA), a supervised multivariate statistical analysis which is a powerful tool for dealing with large-scale metabolomics studies34. The PLS-DA score scatter plot shows that the metabolome of D283 and D458 cell lines is statistically different, with component 1 and component 2, with a variance of 46.4% and 12% respectively, responsible for group separation (Fig. 3A). Notably, a clear change of the metabolome is detectable in both cell lines upon the treatment with OTX-015, as D283_OTX and D458_OTX are shifted along component 2 compared to their respective controls (D283_Ctrl and D458_Ctrl). Next, to identify the metabolites that contribute to the observed PLS-DA separation, variable importance in projection (VIP) scores were determined for the NMR data set (Fig. 3B). VIP scores ˃1 suggest that dissimilar levels of glycine, taurine, myo-inositol, glutamate, creatine, and N-acetyl aspartate are strongly associated with the separation of D283 and D458 cells, whereas increased myo-inositol and taurine, and reduced glycine, glutamate and N-acetyl aspartate determine PLS-DA separation between treatment and controls.
Figure 3.
(A) Score scatter plot showing PLS-DA analysis of the full spectra of 1H NMR data, where peak intensities of metabolites represent the variables. The spatial distribution of the samples is based on the metabolite pattern. Six biological replicates were used for this analysis. (B) Variable importance of projection (VIP) score was plotted for the metabolites that significantly contributed (VIP ˃ 1) to separation by PLS-DA.
Key metabolites regulated by OTX-015 are shared in both MB cell lines
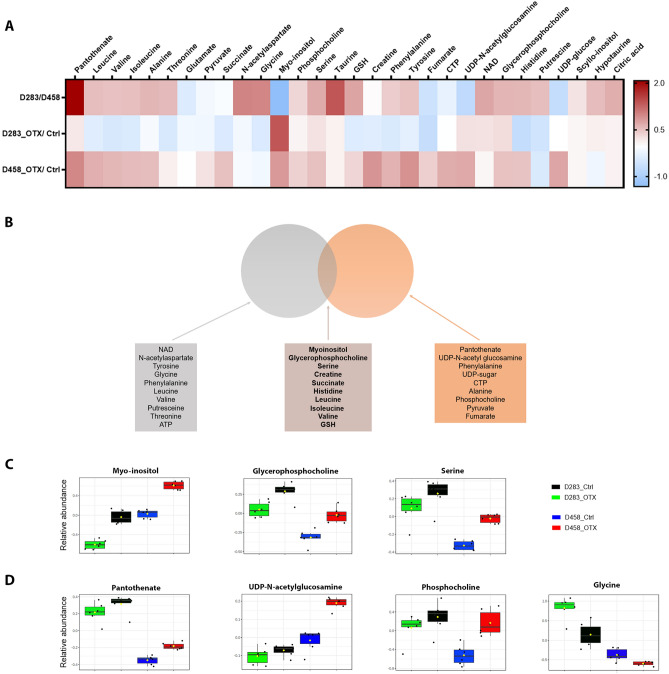
A heat map representing a quantitative targeted analysis confirmed key metabolic differences between the groups identified by PLS-DA (Fig. 4A). One-way analysis of variance (ANOVA) for this metabolite quantification is presented in Table 1. Fisher's Least Significant Difference (LSD) indicates that D283 and D458 are characterised by different levels of amino acids and derivatives (N-acetylaspartate, taurine, hypotaurine, tyrosine, glycine, phenylalanine, serine, leucine, isoleucine, valine, alanine, GSH, glutamate, and threonine); carbohydrates (myo-inositol, scyllo-inositol, UDP-N-acetylglucosamine, and UDP-sugar); phospholipid derivatives (phosphocholine and glycerophosphocholine); nucleotides (NAD and CTP); the polyamine putresceine, and the vitamin B5 pantothenate.
Figure 4.
(A) Heatmap showing metabolite quantification (Log2 fold change values) in D283 versus D458 cell lines, D283_OTX versus D283_Ctrl and D458_OTX versus D458_Ctrl. Metabolites were quantified with MATLAB, according to intensity of metabolite signal in the 1H NMR spectrum and calculated using an internal standard (n = 6). (B,C) Boxes depicting metabolites commonly deregulated in D283_OTX versus D283_Ctrl and D458_OTX versus D458_Ctrl and statistically significant in both the comparisons (B) or at least in one of them (C). Statistical analysis has been performed using one way-ANOVA (see Supplementary Table 1).
Table 1.
Features identified by one-way ANOVA and post hoc analysis using Metaboanalyst 5.0.
| Metabolite | f.value | p.value | −log10(p) | FDR | Fishers LSD |
|---|---|---|---|---|---|
| Myo-inositol | 142.29 | 1.17E−13 | 12.932 | 4.79E–12 | D283_OTX–D283_Ctrl; D458_Ctrl–D283_Ctrl; D458_OTX–D283_Ctrl; D458_OTX–D283_OTX; D458_OTX–D458_Ctrl |
| NAD | 89.231 | 9.44E−12 | 11.025 | 1.94E–10 | D283_OTX–D283_Ctrl; D283_Ctrl–D458_Ctrl; D283_Ctrl–D458_OTX; D283_OTX–D458_Ctrl; D283_OTX–D458_OTX |
| Pantothenate | 80.609 | 2.41E−11 | 10.617 | 3.22E–10 | D283_Ctrl–D458_Ctrl; D283_Ctrl–D458_OTX; D283_OTX–D458_Ctrl; D283_OTX–D458_OTX; D458_OTX–D458_Ctrl |
| N-acetylaspartate | 78.341 | 3.14E−11 | 10.503 | 3.22E–10 | D283_Ctrl–D283_OTX; D283_Ctrl–D458_Ctrl; D283_Ctrl–D458_OTX; D283_OTX–D458_Ctrl; D283_OTX–D458_OTX |
| Taurine | 68.362 | 1.09E−10 | 9.9632 | 8.92E–10 | D283_Ctrl–D458_Ctrl; D283_Ctrl–D458_OTX; D283_OTX–D458_Ctrl; D283_OTX–D458_OTX |
| Creatine | 59.695 | 3.69E−10 | 9.4327 | 2.52E–09 | D283_Ctrl–D283_OTX; D458_OTX–D283_Ctrl; D458_Ctrl–D283_OTX; D458_OTX–D283_OTX; D458_OTX–D458_Ctrl |
| Tyrosine | 56.716 | 5.83E−10 | 9.2343 | 3.26E–09 | D283_Ctrl–D283_OTX; D283_Ctrl–D458_Ctrl; D458_OTX–D283_Ctrl; D283_OTX–D458_Ctrl; D458_OTX–D283_OTX; D458_OTX–D458_Ctrl |
| Hypotaurine | 56.159 | 6.37E−10 | 9.1961 | 3.26E–09 | D283_Ctrl–D458_Ctrl; D283_Ctrl–D458_OTX; D283_OTX–D458_Ctrl; D283_OTX–D458_OTX |
| UDP-N-acetylglucosamine | 53.675 | 9.51E−10 | 9.0218 | 4.33E–09 | D458_Ctrl–D283_Ctrl; D458_OTX–D283_Ctrl; D458_Ctrl–D283_OTX; D458_OTX–D283_OTX; D458_OTX–D458_Ctrl |
| Glycine | 47.974 | 2.55E−09 | 8.5934 | 1.05E–08 | D283_Ctrl–D283_OTX; D283_Ctrl–D458_Ctrl; D283_Ctrl–D458_OTX; D283_OTX–D458_Ctrl; D283_OTX–D458_OTX |
| Citric acid | 45.963 | 3.70E−09 | 8.4316 | 1.38E–08 | D283_Ctrl–D458_Ctrl; D283_Ctrl–D458_OTX; D283_OTX–D458_Ctrl; D283_OTX–D458_OTX |
| Succinate | 42.472 | 7.32E−09 | 8.1356 | 2.50E–08 | D283_Ctrl–D283_OTX; D458_OTX–D283_Ctrl; D458_Ctrl–D283_OTX; D458_OTX–D283_OTX; D458_OTX–D458_Ctrl |
| Glycerophosphocholine | 34.849 | 3.90E−08 | 7.4091 | 1.23E–07 | D283_OTX–D283_Ctrl; D283_Ctrl–D458_Ctrl; D283_OTX–D458_Ctrl; D283_OTX–D458_OTX; D458_OTX–D458_Ctrl |
| Phenylalanine | 32.012 | 7.86E−08 | 7.1045 | 2.30E–07 | D283_Ctrl–D283_OTX; D283_Ctrl–D458_Ctrl; D458_OTX–D283_Ctrl; D458_OTX–D283_OTX; D458_OTX–D458_Ctrl |
| UDP-sugar | 29.259 | 1.63E−07 | 6.7869 | 4.46E–07 | D458_Ctrl–D283_Ctrl; D458_OTX–D283_Ctrl; D458_Ctrl–D283_OTX; D458_OTX–D283_OTX; D458_OTX–D458_Ctrl |
| Histidine | 25.497 | 4.88E−07 | 6.3117 | 1.25E–06 | D283_Ctrl–D283_OTX; D283_Ctrl–D458_Ctrl; D458_OTX–D283_OTX; D458_OTX–D458_Ctrl |
| Serine | 25.026 | 5.64E−07 | 6.2484 | 1.36E–06 | D283_OTX–D283_Ctrl; D283_Ctrl–D458_Ctrl; D283_OTX–D458_Ctrl; D283_OTX–D458_OTX; D458_OTX–D458_Ctrl |
| Leucine | 20.657 | 2.44E−06 | 5.6126 | 5.56E–06 | D283_Ctrl–D283_OTX; D283_Ctrl–D458_Ctrl; D458_OTX–D283_OTX; D458_OTX–D458_Ctrl |
| CTP | 19.627 | 3.56E−06 | 5.4482 | 7.69E–06 | D458_Ctrl–D283_Ctrl; D458_OTX–D283_Ctrl; D458_Ctrl–D283_OTX; D458_OTX–D283_OTX; D458_OTX–D458_Ctrl |
| GSH | 17.53 | 8.07E−06 | 5.093 | 1.66E–05 | D283_Ctrl–D283_OTX; D283_Ctrl–D458_Ctrl; D283_Ctrl–D458_OTX; D283_OTX–D458_Ctrl; D458_OTX–D458_Ctrl |
| Isoleucine | 16.708 | 1.13E−05 | 4.9454 | 2.12E–05 | D283_Ctrl–D283_OTX; D283_Ctrl–D458_Ctrl; D458_OTX–D283_OTX; D458_OTX–D458_Ctrl |
| Valine | 16.702 | 1.14E−05 | 4.9443 | 2.12E–05 | D283_Ctrl–D283_OTX; D283_Ctrl–D458_Ctrl; D458_OTX–D283_OTX; D458_OTX–D458_Ctrl |
| Alanine | 16.271 | 1.37E−05 | 4.8648 | 2.43E–05 | D283_Ctrl–D458_Ctrl; D283_OTX–D458_Ctrl; D458_OTX–D458_Ctrl |
| Putrescine | 12.322 | 8.68E−05 | 4.0613 | 0.00014834 | D283_Ctrl–D283_OTX; D283_Ctrl–D458_Ctrl; D283_Ctrl–D458_OTX |
| Glutamate | 11.426 | 0.00013918 | 3.8564 | 0.00022825 | D458_Ctrl–D283_Ctrl; D458_OTX–D283_Ctrl; D458_Ctrl–D283_OTX; D458_OTX–D283_OTX |
| Phosphocholine | 10.27 | 0.00026472 | 3.5772 | 0.00041744 | D283_Ctrl–D458_Ctrl; D283_OTX–D458_Ctrl; D458_OTX–D458_Ctrl |
| Threonine | 9.7105 | 0.00036679 | 3.4356 | 0.00055698 | D283_Ctrl–D283_OTX; D283_Ctrl–D458_Ctrl; D283_Ctrl–D458_OTX |
| Pyruvate | 9.4295 | 0.00043384 | 3.3627 | 0.00063527 | D458_OTX–D283_Ctrl; D458_Ctrl–D283_OTX; D458_OTX–D283_OTX; D458_OTX–D458_Ctrl |
| Scyllo-inositol | 9.1696 | 0.000508 | 3.2941 | 0.00071821 | D283_Ctrl–D458_Ctrl; D283_Ctrl–D458_OTX; D283_OTX–D458_Ctrl; D283_OTX–D458_OTX |
| Fumarate | 5.5076 | 0.0063516 | 2.1971 | 0.0086805 | D458_OTX–D283_Ctrl; D458_OTX–D283_OTX; D458_OTX–D458_Ctrl |
| ATP | 4.3404 | 0.016453 | 1.7837 | 0.021761 | D283_Ctrl–D283_OTX; D458_Ctrl–D283_OTX; D458_OTX–D283_OTX |
Fisher's least significant difference (LSD) shows significant pairwise comparisons of the conditions under study.
Upon treatment with OTX-015, a metabolic response was detected in both D283 and D458 cell lines and interestingly a few metabolites appear to be commonly regulated. These include myo-inositol, glycerophosphocholine, serine, pantothenate, UDP-N-acetylglucosamine, phosphocholine and glycine, all of which increased in both D283 and D458 after treatment (Fig. 4B–D). Notably, OTX-015 treatment induced additional changes in the endogenous metabolome of D283 and D458, albeit the specific alterations differ between the two cell lines (Fig. 4A and Supplementary Fig. S11).
Metabolite set enrichment analysis indicates significantly enriched metabolic pathways in MYC-driven MB cells
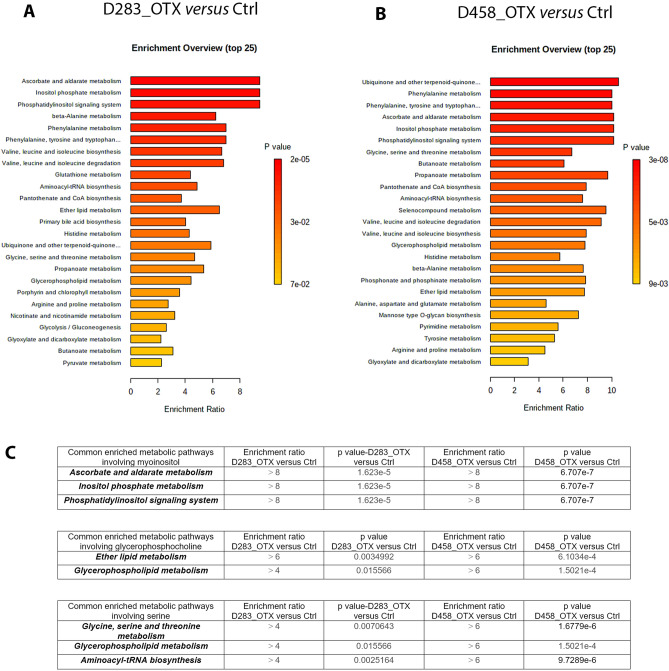
To understand how metabolic rewiring caused by the OTX-015 treatment impact on functional signalling pathways, we performed a metabolite set enrichment analysis (MSEA) on quantified metabolites (described in Table 1), along with their relative concentrations, using the web-based platform MetaboAnalyst35 (Fig. 5). The top 25 pathways significantly enriched in D283_OTX versus untreated control and D458_OTX versus untreated control cells are shown in Fig. 5A,B. We focused on the common enriched pathways in which the metabolites myo-inositol, glycerophosphocholine and serine increased significantly upon OTX-015 treatment in both cell lines (Fig. 5).
Figure 5.
Summary plot of the metabolite set enrichment analysis (MSEA) of the identified metabolites in (A) D283_OTX versus D283_Ctrl and (B) D458_OTX versus D458_Ctrl. Horizontal graph bars indicate the pathways that are enriched in these groups. The colour code corresponds to the calculated p-value. Vertical red lines indicate pathways with a p-value < 0.05. (C) Summary table of the pathways commonly enriched in both the cell lines upon OTX-015 treatment and with a p-value < 0.05 are shown.
These data confirm that OTX-015 triggers a different metabolic reprogramming in D283 and D458 MB cell lines, reflecting their diverse basal metabolism. Nonetheless, in both cell lines OTX-015 affects ascorbate and aldarate metabolism, inositol phosphate metabolism, phosphatidylinositol signalling system, ether lipid metabolism, glycerophospholipid metabolism, glycine serine and threonine metabolism and aminoacyl tRNA biosynthesis (Fig. 5C).
Discussion
MYC-driven MB is an aggressive prevalent paediatric tumour, which is refractory to intensive multimodal therapy4,36. In this scenario, BETis are emerging as promising agents in MYC-dependent MB as well as in other tumours, because of their ability to impair MYC expression and to modulate the epigenetic machinery of cancer cells4,20. In this work, we studied the effect of the BETi OTX-015 on the metabolism of the D283 and D458 MYC-driven childhood MB cell lines. OTX-015 has been developed from the conventional BETi JQ1 to improve its pharmacokinetic profile and thus its potential in clinical use16,17,22.
It is already known that MYC regulates metabolism which in turn controls MYC levels27. MYC promotes cell proliferation and induces metabolic alterations because of increased anabolic demand. On the other hand, MYC is also downstream of multiple control mechanisms regulated by nutrient levels and metabolic stress27. Despite the well-recognised crosstalk between MYC and metabolism in cancer, how metabolism is deregulated in MYC-driven MB is still unknown. Furthermore, although inhibition of BRD proteins is known to cause metabolic reprogramming37,38, whether BETi-dependent metabolic changes can be used to monitor drug response towards BETis is not understood. Here, we investigated the metabolome of MYC-driven MB cell models and interrogated how OTX-015 would modulate their metabolic phenotype using an NMR-based approach. NMR spectroscopy has proven to be of high value as an accurate and quantitative method to investigate metabolic alterations related to disease, through detecting metabolic signatures for early diagnosis and prognosis evaluation, unveiling cellular mechanisms and discovering pharmacological actions and potential therapeutic significance of drug compounds39–41.
In this work, we discovered that D283 exhibits diverse metabolic characteristics compared to D458 cells, consistent with their different origin and genetic background28. The most significantly different metabolites between the two cell lines were: amino acids and their derivatives such as hypotaurine and GSH; the TCA cycle metabolite citric acid; the phospholipid derivatives phosphocholine and glycerol-phosphocholine; the vitamin B5 pantothenate; NAD and scylloinositol. Upon treatment of D283 and D458 with OTX-015, common features were revealed in addition to cell-specific metabolic changes. In both cell lines, quantitative NMR data showed an increase in myo-inositol, a metabolite well known to control cell osmoregulation, whose role in pathophysiological contexts is tissue-and tumour-dependent42. Notably, myo-inositol targets critical cancer liabilities43–45 by: (i) inhibiting pRB phosphorylation and subsequently blocking the cell cycle; (ii) reducing PI3K, Akt, ERK and NF-kB and (iii) impairing invasiveness43–45. The observed increase in myo-inositol levels therefore may contribute to the anti-proliferative effect observed in D283 and D458 cell lines upon treatment with OTX-015.
Metabolite set enrichment analysis (MSEA) revealed that OTX-015 treatment causes a marked common enrichment in metabolic pathways involving myo-inositol (enrichment ratio ˃8). These include ascorbate and aldarate metabolism, inositol phosphate metabolism and phosphatidylinositol signalling system pathways in both cell lines. To our knowledge, ascorbate and aldarate metabolism has never previously been linked to MB, albeit it has been described as markedly altered in other cancers46. Notably, both ascorbate (AA) and its oxidised form dehydroascorbate (DHA), which together represent the two existing redox states of Vitamin C, are important for neural function and antioxidant/oxidant balance47. Indeed, aberrant doses of DHA induce oxidative effects selectively killing cancer cells48. In agreement with previous work49, our results suggest that, beyond MYC downregulation, OTX-015, and perhaps other BETis, could elicit their antitumour capabilities also by disrupting the metabolic equilibrium between antioxidants and ROS, thereby eventually leading to cancer cell death.
Our data also show that inositol phosphate metabolism and phosphatidylinositol signalling system pathways are associated with OTX-015 treatment. These two pathways are well known to be initiated upon stimulation of phospholipase C, which metabolises phosphatidylinositol 4,5-bisphosphate into the intracellular second messengers 1,2-diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3)50. DAG activates protein kinase C, whilst IP3 regulates cellular calcium, and both can be converted into more polar inositol phosphates (IPs) and diphosphoryl inositol phosphates (PP-IPs). Inositols, of which the most abundant stereoisomer is myo-inositol51, play a crucial role in several biological functions and deregulation of their synthesis has been linked to disease including cancer43,44. IPs and PP-IPs are key in transcription control, mRNA export and DNA repair, and are also important for glucose sensing in β-pancreatic cells52. As mentioned above, inositols can impair oncogenic signalling using several mechanisms including inhibition of the PI3K/Akt pathway43. Notably, this has already been recognised in medulloblastoma as one of the relevant oncogenic pathways contributing to the disease53 and, accordingly, a selective pan-class I PI3K inhibitor namely BKM120 has been revealed as a potential novel therapy for MB54. Thus, in our MYC-driven cell models OTX-015 treatment may upregulate myo-inositol production, counteracting PI3K/Akt/mTOR signalling and eventually triggering tumour cell death.
Our NMR analysis recorded higher concentrations of glycerophosphocholine and phosphocholine upon OTX-015 treatment. Increase in these metabolites has been strongly associated with the well-known accumulation of lipid droplets55,56. Previous work demonstrated that MYC inhibition, achieved either with the c-MYC/Max inhibitor 10058-F4 or by MYC depletion, triggers mitochondrial dysfunction and β-fatty acid oxidation impairment, eventually leading to accumulation of intracellular lipid droplets57,58. Notably, BRD4 inhibition has also been shown to regulate OXPHOS capacity and to rewire metabolism in complex I (CI) mutant cybrid cells37. Consistently with this, our data appear to suggest that one of the putative anticancer mechanisms of OTX-015 may be hindering mitochondrial function, which would lead to inefficient lipid catabolism and increased levels of glycerophosphocholine and phosphocholine. Further experiments are however needed to investigate the intriguing possibility of a crosstalk between BETis, the mitochondrial machinery and lipid metabolism in MYC-driven MB.
In agreement with the increased levels of glycerophosphocholine, our MSEA analysis revealed that OTX-015 also causes a common enrichment of two intertwined metabolic pathways involving this metabolite, specifically ether lipid metabolism and glycerophospholipid metabolism. Although diverse roles of these pathways in cancer have been reported59–62, certain types of ether lipids function as endogenous antioxidants and are involved in cell differentiation63, whilst glycerophospholipids have been found decreased in glioma stem-like cells64. Changes in these two pathways may therefore have an impact on the grade of cell differentiation of MYC-driven MB.
Upon treatment with OTX-015, we also detected a significant concentration rise of serine in both cell lines. This amino acid is synthesised from the glycolytic intermediate 3-phosphoglycerate and can be transformed into glycine, which in turn is used in one carbon metabolism processes to sustain the synthesis of macromolecule building blocks such as purines, and/or for GSH synthesis to counteract ROS effects65. Importantly, serine-glycine conversion is catalysed by the serine hydroxymethyltransferase (SHMT), of which both the cytoplasmic (SHMT1) and mitochondrial (SHMT2) isoforms are known to be upregulated by MYC66. Thus, we could speculate that downregulation of MYC caused by OTX-015 impairs SHMT1/2 activity, resulting in serine accumulation. Consistent with this, we also detected a concomitant reduction in glycine in both cell lines and revealed that a commonly affected pathway following OTX-015 treatment is glycine, serine and threonine metabolism. Importantly, glycine is a precursor of purines and thus a key metabolite for rapidly proliferating cancer cells67. Accordingly, a recent study unveiled that glycine levels are increased in MYC-amplified MB with respect to the normal cerebellum68.
Another pathway that is affected in both cell lines in response to OTX-015 treatment is aminoacyltRNA biosynthesis. Notably, supporting our results, aminoacyltRNA biosynthesis has been reported as a pathway signature for group 3γ MB tumours, which is characterised by increased MYC copy number and negative prognosis69. AminoacyltRNA synthases are key enzymes in protein synthesis that during evolution acquired further domains, which provided these enzymes with additional roles beyond translation. Upregulation of seryl-tRNA synthetase induced cellular senescence and inhibited the growth of cervical tumour xenografts in mice by causing the senescence of tumour cells70,71. We found that MYC-driven MB cells treated with OTX-015 have significant higher levels of serine compared to untreated cells, and this is compatible with an upregulation of seryl-tRNA synthetase70. OTX-015 may therefore promote senescence by enhancing seryl-tRNA synthetase biosynthesis, although any link between OTX-015 and seryl-tRNA biosynthesis awaits to be proven.
In summary, in this work we describe the metabolic profile of MYC-driven MB cell models and reveal key metabolic changes triggered by OTX-015 treatment that could be either MYC-dependent or -independent. These novel insights contribute to the phenotypical characterisation of MYC-driven MB and could lay the foundation for uncovering new druggable signalling pathways beyond MYC-inhibition. Notably, our analysis revealed how OTX-015 was able to alter the metabolism of MYC-driven MB cell models. This provides key additional information on possible mechanisms of actions elicited by this compound at the cellular level to inhibit the growth of susceptible cancer cells, whilst also generating potentially useful clues for the use of BETis in the treatment of medulloblastoma. This first metabolic characterisation is likely to promote, guide and pave the ways to further analyses, which will likely reveal additional metabolic features and possible ramifications for the medulloblastoma treatment with OTX-015 or other BETis.
Materials and methods
Cell lines
The D283 and D458 cell lines were kindly provided by Associate Professor JI Johnsen (Karolinska Institutet, Stockholm, Sweden). Cells were maintained in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Life Technologies, Waltham, MA, USA), 10 mM Hepes, 2 mM L-glutamine, 100 IU/ml penicillin G, and 100 μg/ml streptomycin Sigma, St. Louis, MO, USA) at 37 °C in a humidified 5% CO2 atmosphere.
Western blot
Whole-cell lysates were prepared using RIPA buffer (Sigma-Aldrich) supplemented with Halt Protease and Phosphatase Inhibitor Mixture (Thermo Scientific). Samples were resolved on SDS/PAGE and transferred to Polyvinylidene fluoride (PVDF) membranes (BioRad, 1704156). The c-Myc antibody (9E10) (NB600-302, Novus Biologicals Bio-Techne, United Kingdom 1:2000) was used to identify c-MYC, while anti-β-actin (Santa Cruz, SC47778 1:10,000) was used as loading control. HRP-conjugated secondary antibodies were from GE Healthcare. Bands were detected using SuperSignal West Dura chemiluminescent substrate (Thermo Fisher Scientific, 34075) and images were obtained in a ChemiDoc XRS + (BioRad). Image processing was performed with ImageLab (Life Science Research, BioRad). All Western blot data are the result of three independent biological replicates. The quantification was performed by normalising to β-actin and the percentage of decrease in c-MYC in treated versus control cells was calculated.
Cell viability assay
Cell viability was evaluated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction assay (Sigma-Aldrich). Cells were seeded in 96-well plates at a density of 135.000 cells/mL. Twenty-four hours after seeding, either drug (OTX-015, Sigma-Aldrich) or vehicle (DMSO, Sigma-Aldrich) was added to the media. After 48 h, 10 μL of MTT solution 5 mg/mL (Sigma-Aldrich) was added to each well. Two hours later, MTT was dissolved adding SDS 10% 0.01 M HCl and incubated overnight. The next day, absorbance at 570 nm was measured using a microplate reader (SpectraMax i3x Multi-Mode Microplate Reader, Molecular Devices). Experiments were repeated at least three times and viability was normalised to mean of untreated control, set as 100% viability.
NMR experiment and analysis
Cell culture and extraction
Cells were seeded at a density of 5 × 106 cells/mL in 150 mm dishes and incubated the next day with 7.5 µM OTX-015. After 24 h, cells were collected in 50 mL falcon tube, immerged in a dry ice/ethanol bath for 30 s and then centrifuged at + 4 °C at 200 g for 5 min. Media was discarded and the pellet suspend by adding 2.0 mL of -20 °C-cold MeOH. Afterwards, 2 mL of ice cold CH3Cl and MilliQ H2O were added and the solution was mixed by inversion and incubated on ice for 10 min to allow the formation of a stable bilayer. Subsequently, samples were centrifuged at + 4 °C at 300g for 45 min to separate the top aqueous layer from the bottom organic layer, containing metabolites soluble in the polar and apolar fraction, respectively. The polar fraction was dried in an Eppendorf concentrator (Concentrator plus, Eppendorf) and analysed by NMR.
NMR acquisition, processing and statistical analysis
Prior to NMR experiments, samples were resuspended in a 90/10 H2O/D2O buffer, with 100 mM Na2HPO4, 5 mM Trimethylsilylpropanoic acid (TSP) and 4 mM NaN3. NMR spectroscopy was performed on a Bruker Avance NEO 600 MHz NMR spectrometer equipped with a TCI CryoProbe Prodigy (Bruker). Spectra were acquired at 298 K and consisted for each sample of a 1D 1H PURGE spectrum72, which was then phase corrected, baseline corrected. The chemical shifts were referenced to the TSP peak at 0.0 ppm.
Spectra were then exported to MATLAB, using a toolbox written by A. Edison and co-workers73. Spectra were aligned by the Pearson PAFFT method (peak alignment by fast Fourier transform) and then normalised by probabilistic quotient normalisation (PQN).
Six biological replicates were used for this analysis.
Metabolite identification and quantification
2D spectra were used on one of the samples for identification purposes, consisting of a Total Correlation SpectroscopY ([1H–1H] TOCSY), using the Bruker pulse sequence “dipsi2gpphzs”, slightly modified to include presaturation, with eight scans, 256 t1 increments, a spectral width of 13.7 ppm in both dimensions and a relaxation delay of 2 s, along with a Heteronuclear Single-Quantum Correlation (([1H–13C] HSQC) spectrum, using the Bruker pulse sequence “hsqcetgpsisp”, with 16 scans, 256 t1 increments, a spectral width of 170 ppm in the 13C dimension and 12 ppm in the 1H dimension, and a relaxation time of 2 s. Metabolites were assigned using information acquired from the human metabolome database (HMDB)32 and the biological magnetic resonance data bank (BMRB)33. Relative quantification of metabolites was achieved by integration of peaks to calculate peak area, which is proportional to the number of nuclei that give rise to the signal. Mean ± SD values of peak areas were calculated for each metabolite.
Statistical analysis using MetaboAnalyst
After identification of the metabolites, a list of representative peaks was chosen for each of the identified metabolites (Supplementary Table 1). Their peak area for each spectrum was taken and stored in a table which, as a .csv file, was then loaded onto MetaboAnalyst35 for the statistical analysis. For multivariate analyses, spectra were scaled by the pareto method74. To determine whether spectra cluster into groups, PCA and PLS-DA with k-fold cross-validation were performed, along with a one-way ANOVA to see which metabolites were statistically different for specific group comparison.
Metabolic pathways enrichment analysis using MetaboAnalyst
The signal integrals corresponding to the identified metabolites were used to perform a Quantitative Enrichment Analysis employing MetaboAnalyst 5.035 platform with the KEGG library, a database of relevant metabolic pathways found in humans75–77. Significant pathways were those with p-value < 0.05. The top 25 enriched pathways were shown.
Supplementary Information
Acknowledgements
We are grateful to Associate Professor JI Johnsen, Karolinska Institutet, for generously sharing the MB cell lines. This research was supported by the Swedish Research Council, the Swedish Cancer Society, the Swedish Childhood Cancer Fund, Radiumhemmet Research Funds, and Karolinska Institutet to MAH. NMR experiments and analysis were performed in the Centre for Biomolecular Spectroscopy, King’s College London, which was established with funds to MRC from Wellcome and British Heart Foundation, ref. 202767/Z/16/Z and BHF-IG/16/2/32273. VG was supported by a scholarship from Radiumhemmet, Stockholm and ARG by a postdoctoral position from the Swedish Cancer Society.
Author contributions
V.G., M.A.H. and M.R.C. conceived the study. V.G. designed/performed the experiments and interpreted the results. A.R.G. and L.S.A. carried out the cellular assays. A.L.G. ran and processed the metabolomics spectra. V.G. and A.L.G. performed the metabolomics analysis. M.A.H. and M.R.C. supervised the study. V.G., M.A.H. and M.R.C. wrote the manuscript and all authors edited and approved the final version.
Funding
Open access funding provided by Karolinska Institute.
Data availability
A large part of data generated or analysed during this study are included in this published article and its supplementary information files. Some of the datasets used and/or analysed during the current study have not been included and are available from the corresponding authors on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Marie Arsenian Henriksson, Email: marie.arsenian.henriksson@ki.se.
Maria R. Conte, Email: sasi.conte@kcl.ac.uk
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-27375-z.
References
- 1.Louis DN, et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Huang GH, et al. Medulloblastoma stem cells: Promising targets in medulloblastoma therapy. Cancer Sci. 2016;107:583–589. doi: 10.1111/cas.12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, P. & Cushing, H. A classification of the tumours of the glioma group on a histogenetic basis, with a correlated study of prognosis. By Percival Bailey and Harvey Cushing. Medium 8 vo. Pp. 175, with 108 illustrations. 1926. Philadelphia, London, and Montreal: J. B. Lippinco. Br. J. Surg.14, 554–555 (1927).
- 4.Juraschka K, Taylor MD. Medulloblastoma in the age of molecular subgroups: a review. J. Neurosurg. Pediatr. 2019;24:353–363. doi: 10.3171/2019.5.PEDS18381. [DOI] [PubMed] [Google Scholar]
- 5.Shih DJH, et al. Cytogenetic prognostication within medulloblastoma subgroups. J. Clin. Oncol. 2014;32:886. doi: 10.1200/JCO.2013.50.9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menyhárt O, Győrffy B. Molecular stratifications, biomarker candidates and new therapeutic options in current medulloblastoma treatment approaches. Cancer Metastasis Rev. 2020;39:211–233. doi: 10.1007/s10555-020-09854-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gajjar A, Pizer B. Role of high-dose chemotherapy for recurrent medulloblastoma and other CNS primitive neuroectodermal tumors. Pediatr. Blood Cancer. 2010;54:649–651. doi: 10.1002/pbc.22378. [DOI] [PubMed] [Google Scholar]
- 8.Moxon-Emre I, et al. Intellectual outcome in molecular subgroups of medulloblastoma. J. Clin. Oncol. 2016;34:4161–4170. doi: 10.1200/JCO.2016.66.9077. [DOI] [PubMed] [Google Scholar]
- 9.Pérez-Salvia M, Esteller M. Bromodomain inhibitors and cancer therapy: From structures to applications. Epigenetics. 2017;12:323–339. doi: 10.1080/15592294.2016.1265710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mertz JA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moyer MW. First drugs found to inhibit elusive cancer target. Nat. Med. 2011;17:1325. doi: 10.1038/nm1111-1325. [DOI] [PubMed] [Google Scholar]
- 15.Li F, et al. Metabolism of JQ1, an inhibitor of bromodomain and extra terminal bromodomain proteins, in human and mouse liver microsomes. Biol. Reprod. 2020;103:427–436. doi: 10.1093/biolre/ioaa043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vázquez R, et al. Promising in vivo efficacy of the BET bromodomain inhibitor OTX015/MK-8628 in malignant pleural mesothelioma xenografts. Int. J. Cancer. 2017;140:197–207. doi: 10.1002/ijc.30412. [DOI] [PubMed] [Google Scholar]
- 17.Pasqua, A. E., Wilding, B., Cheeseman, M. D. & Jones, K. Targeting protein synthesis, folding, and degradation pathways in cancer. in Comprehensive Medicinal Chemistry III. Vol. 5–8 (Elsevier, 2017).
- 18.Odore E, et al. Phase I population pharmacokinetic assessment of the oral bromodomain inhibitor OTX015 in patients with haematologic malignancies. Clin. Pharmacokinet. 2016;55:397–405. doi: 10.1007/s40262-015-0327-6. [DOI] [PubMed] [Google Scholar]
- 19.Moreno V, et al. Phase I study of CC-90010, a reversible, oral BET inhibitor in patients with advanced solid tumors and relapsed/refractory non-Hodgkin’s lymphoma. Ann. Oncol. 2020;31:780–788. doi: 10.1016/j.annonc.2020.03.294. [DOI] [PubMed] [Google Scholar]
- 20.Pérez-Salvia M, Esteller M. Bromodomain inhibitors and cancer therapy: From structures to applications. Epigenetics. 2017 doi: 10.1080/15592294.2016.126571012,323-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berenguer-Daizé C, et al. OTX015 (MK-8628), a novel BET inhibitor, displays in vitro and in vivo antitumor effects alone and in combination with conventional therapies in glioblastoma models. Int. J. Cancer. 2016;139:2047–2055. doi: 10.1002/ijc.30256. [DOI] [PubMed] [Google Scholar]
- 22.Vohidov F, et al. Design of BET inhibitor bottlebrush prodrugs with superior efficacy and devoid of systemic toxicities. J. Am. Chem. Soc. 2021;143:4714–4724. doi: 10.1021/jacs.1c00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: Identification and analysis of c-MYC target genes. Proc. Natl. Acad. Sci. 2002;99:6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kress TR, Sabo A, Amati B. MYC: Connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer. 2015;15:593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- 25.Carroll PA, Freie BW, Mathsyaraja H, Eisenman RN. The MYC transcription factor network: balancing metabolism, proliferation and oncogenesis. Front. Med. 2018;12:412–425. doi: 10.1007/s11684-018-0650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.vanderHeiden MG, DeBerardinis RJ. Understanding the Intersections between metabolism and cancer biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dejure FR, Eilers M. MYC and tumor metabolism: chicken and egg. EMBO J. 2017;36:3409–3420. doi: 10.15252/embj.201796438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanov DP, Coyle B, Walker DA, Grabowska AM. In vitro models of medulloblastoma: Choosing the right tool for the job. J. Biotechnol. 2016;236:10–25. doi: 10.1016/j.jbiotec.2016.07.028. [DOI] [PubMed] [Google Scholar]
- 29.Di C, et al. Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans retinoic acid. Cancer Res. 2005;65:919. doi: 10.1158/0008-5472.919.65.3. [DOI] [PubMed] [Google Scholar]
- 30.Portwine C, et al. Absence of germline and somatic p53 alterations in children with sporadic brain tumors. J. Neuro-Oncol. 2001;52:227–235. doi: 10.1023/A:1010661831335. [DOI] [PubMed] [Google Scholar]
- 31.Zhu F, et al. Mouse models for the p53 R72P polymorphism mimic human phenotypes. Cancer Res. 2010;70:5851–5859. doi: 10.1158/0008-5472.CAN-09-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wishart DS, et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ulrich EL, et al. BioMagResBank. Nucleic Acids Res. 2008;36:402–408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bijlsma S, et al. Large-scale human metabolomics studies: A strategy for data (pre-) processing and validation. Anal. Chem. 2006 doi: 10.1021/ac051495j. [DOI] [PubMed] [Google Scholar]
- 35.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 36.Bandopadhayay P, et al. Neuronal differentiation and cell-cycle programs mediate response to BET-bromodomain inhibition in MYC-driven medulloblastoma. Nat. Commun. 2019;10:2400. doi: 10.1038/s41467-019-10307-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrow JJ, et al. Bromodomain inhibitors correct bioenergetic deficiency caused by mitochondrial disease complex I mutations. Mol. Cell. 2016;64:163. doi: 10.1016/j.molcel.2016.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deeney JT, Belkina AC, Shirihai OS, Corkey BE, Denis GV. BET Bromodomain proteins Brd2, Brd3 and Brd4 selectively regulate metabolic pathways in the pancreatic β-cell. PLoS ONE. 2016;11:1–16. doi: 10.1371/journal.pone.0151329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat. Rev. Cancer. 2004;4:551–561. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- 40.Han J, Li Q, Chen Y, Yang Y. Recent metabolomics analysis in tumor metabolism reprogramming. Front. Mol. Biosci. 2021;8:1–16. doi: 10.3389/fmolb.2021.763902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alarcon-Barrera JC, Kostidis S, Ondo-Mendez A, Giera M. Recent advances in metabolomics analysis for early drug development. Drug Discov. Today. 2022;27:1763–1773. doi: 10.1016/j.drudis.2022.02.018. [DOI] [PubMed] [Google Scholar]
- 42.Mizisin AP, et al. Polyol pathway and osmoregulation in JS1 Schwann cells grown in hyperglycemic and hyperosmotic conditions. Am. J. Physiol. 1996 doi: 10.1152/ajprenal.1996.270.1.F90. [DOI] [PubMed] [Google Scholar]
- 43.Bizzarri M, Dinicola S, Bevilacqua A, Cucina A. Broad spectrum anticancer activity of myo-inositol and inositol hexakisphosphate. Int. J. Endocrinol. 2016;2016:5616807. doi: 10.1155/2016/5616807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bizzarri M, Dinicola S, Cucina A. Myoinositol and inositol hexakisphosphate in the treatment of breast cancer. Mol. Mech. 2018 doi: 10.1007/978-3-319-63540-8_20. [DOI] [Google Scholar]
- 45.Chhetri DR. Myo-inositol and its derivatives: Their emerging role in the treatment of human diseases. Front. Pharmacol. 2019;10:1172. doi: 10.3389/fphar.2019.01172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garg G, et al. Targeted metabolomic profiling of low and high grade serous epithelial ovarian cancer tissues: A pilot study. Metabolomics. 2018;14:1–13. doi: 10.1007/s11306-018-1448-3. [DOI] [PubMed] [Google Scholar]
- 47.Cisternas P, et al. The oxidized form of vitamin C, dehydroascorbic acid, regulates neuronal energy metabolism. J. Neurochem. 2014;129:663–671. doi: 10.1111/jnc.12663. [DOI] [PubMed] [Google Scholar]
- 48.Uetaki M, Tabata S, Nakasuka F, Soga T, Tomita M. Metabolomic alterations in human cancer cells by vitamin C-induced oxidative stress. Sci. Rep. 2015;5:1–9. doi: 10.1038/srep13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang B, et al. BRCA1 deficiency sensitizes breast cancer cells to bromodomain and extra-terminal domain (BET) inhibition. Oncogene. 2018;37:6341–6356. doi: 10.1038/s41388-018-0408-8. [DOI] [PubMed] [Google Scholar]
- 50.Putney JW, Tomita T. Phospholipase C signaling and calcium influx. Adv. Biol. Regul. 2012;52:152. doi: 10.1016/j.advenzreg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas DMP, Mills DSJ, Potter PDBVL. The, “other” inositols and their phosphates: Synthesis, biology and medicine (with recent advances in myo-inositol chemistry) Angew. Chem. Int. Ed. Engl. 2016;55:1614. doi: 10.1002/anie.201502227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsui MM, York JD. Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv. Enzyme Regul. 2010;50:324. doi: 10.1016/j.advenzreg.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo Y, Li S, Huang P, Zhang H, Yu C. Development of a prognostic model based on an immunogenomic landscape analysis of medulloblastoma. Biosci. Rep. 2021;41:20202907. doi: 10.1042/BSR20202907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao P, et al. BKM120 induces apoptosis and inhibits tumor growth in medulloblastoma. PLoS ONE. 2017;12:e0179948. doi: 10.1371/journal.pone.0179948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milkevitch M, et al. Increases in NMR-visible lipid and glycerophosphocholine during phenylbutyrate-induced apoptosis in human prostate cancer cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2005;1734:1–12. doi: 10.1016/j.bbalip.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 56.Krahmer N, et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP: Phosphocholine cytidylyltransferase. Cell Metab. 2011;14:504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zirath H, et al. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc. Natl. Acad. Sci. USA. 2013;110:10258–10263. doi: 10.1073/pnas.1222404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oliynyk G, et al. MYCN-enhanced oxidative and glycolytic metabolism reveals vulnerabilities for targeting neuroblastoma. iScience. 2019;21:188–204. doi: 10.1016/j.isci.2019.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benjamin DI, et al. Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proc. Natl. Acad. Sci. USA. 2013;110:14912–14917. doi: 10.1073/pnas.1310894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dickinson A, et al. Mass spectrometry-based lipidomics of oral squamous cell carcinoma tissue reveals aberrant cholesterol and glycerophospholipid metabolism—A pilot study. Transl. Oncol. 2020;13:100807. doi: 10.1016/j.tranon.2020.100807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hahnefeld L, et al. Ether lipid and sphingolipid expression patterns are estrogen receptor-dependently altered in breast cancer cells. Int. J. Biochem. Cell Biol. 2020;127:105831. doi: 10.1016/j.biocel.2020.105834. [DOI] [PubMed] [Google Scholar]
- 62.Hoy AJ, Nagarajan SR, Butler LM. Tumour fatty acid metabolism in the context of therapy resistance and obesity. Nat. Rev. Cancer. 2021;21:753–766. doi: 10.1038/s41568-021-00388-4. [DOI] [PubMed] [Google Scholar]
- 63.Dean JM, Lodhi IJ. Structural and functional roles of ether lipids. Protein Cell. 2018;9:196–206. doi: 10.1007/s13238-017-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang R, et al. LC-MS-based metabolomics reveals metabolic signatures related to glioma stem-like cell self-renewal and differentiation. RSC Adv. 2017;7:24221–24232. doi: 10.1039/C7RA03781C. [DOI] [Google Scholar]
- 65.Yang M, Vousden KH. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer. 2016;16:650–662. doi: 10.1038/nrc.2016.81. [DOI] [PubMed] [Google Scholar]
- 66.Nikiforov MA, et al. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol. Cell. Biol. 2002;22:5793. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jain M, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science (80-) 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pham, K. et al. Comprehensive Metabolic Profiling of MYC-Amplified Medulloblastoma Tumors Reveals Key Dependencies on Amino Acid, Tricarboxylic Acid and Hexosamine Pathways. Cancers (Basel). 14 (2022). [DOI] [PMC free article] [PubMed]
- 69.Cavalli FMG, et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell. 2017;31:737–754.e6. doi: 10.1016/j.ccell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Y, et al. Seryl trna synthetase cooperates with pot1 to regulate telomere length and cellular senescence. Signal Transduct. Target. Ther. 2019;4:1–11. doi: 10.1038/s41392-019-0078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khan K, Gogonea V, Fox PL. Aminoacyl-tRNA synthetases of the multi-tRNA synthetase complex and their role in tumorigenesis. Transl. Oncol. 2022;19:101392. doi: 10.1016/j.tranon.2022.101392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simpson AJ, Brown SA. Purge NMR: Effective and easy solvent suppression. J. Magn. Reson. 2005;175:340–346. doi: 10.1016/j.jmr.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 73.Clendinen CS, et al. 13C NMR metabolomics: Applications at natural abundance. Anal. Chem. 2014;86:9242–9250. doi: 10.1021/ac502346h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Worley B, Powers R. Multivariate Analysis in Metabolomics. Curr Metabolomics. Bentham Science Publishers Ltd.; 2013;1:92–107 [DOI] [PMC free article] [PubMed]
- 75.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A large part of data generated or analysed during this study are included in this published article and its supplementary information files. Some of the datasets used and/or analysed during the current study have not been included and are available from the corresponding authors on reasonable request.