Abstract
Genomics revealed the sequence of 3924 genes of the H37Rv strain of Mycobacterium tuberculosis. Proteomics complements genomics in showing which genes are really expressed, and here we show the expression of six genes not predicted by genomics, as proved by two-dimensional electrophoresis and matrix-assisted laser desorption ionization and nano-electrospray mass spectrometry.
Each year eight million new cases and two million deaths are caused by tuberculosis (5). Therefore, the World Health Organization (WHO) declared tuberculosis to be a global emergency, and new strategies toward the prevention and therapy are urgently required. Six years after the first publication of a complete bacterial genome (3), the complete genomes of 38 microorganisms have been sequenced (http://www-fp.mcs.anl.gov/∼gaasterland/genomes.html and http://www.tigr.org/tdb/mdb/mdbcomplete.html), including Mycobacterium tuberculosis strain H37Rv (1). The sequencing of the genome of a clinical isolate of M. tuberculosis, CDC1551, is also nearly complete (http://www.tigr.org/tdb/CMR/gmt/htmls/SplashPage.html). The proteome reflects the functional status of a cell in response to environmental stimuli and thus serves as a valuable complement to genomics. In searching for novel strategies for immune intervention, we have initiated a systematic proteome investigation by comparing the protein compositions of virulent M. tuberculosis strains with attenuated vaccine strains (4). Approximately 1,800 protein spots were separated by two-dimensional electrophoresis (2-DE) and, despite the similarity of the overall patterns, distinct and reproducible differences were detected between the strains. Only +/− variants were accepted, which occurred in all gels of independent preparations of six virulent and six attenuated strains. A total of 263 proteins were identified by Matrix-assisted laser desorption ionization (MALDI) mass spectrometry (MS) and a bioinformatics platform was constructed to store our data and connect it by hyperlinks with the genomics data (10) (http://www.mpiibberlin.mpg.de/2D-PAGE/). Using this proteome approach, namely, a combination of 2-DE (6) and MS, we detected six genes previously not predicted in the genome of M. tuberculosis H37Rv. Our data demonstrate the value of proteomics in identifying gene products undetected by the genomics approach.
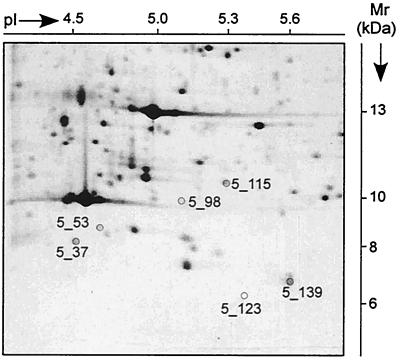
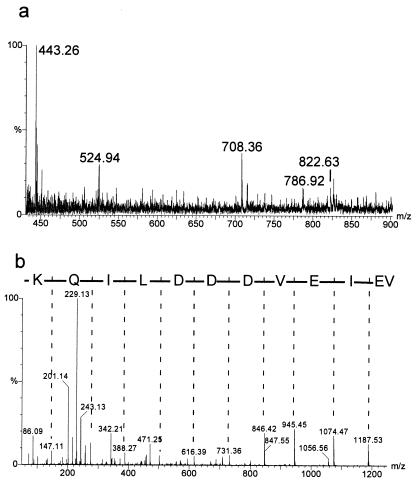
M. tuberculosis H37Rv was grown in Middlebrook medium for 6 to 8 days to a cell density of 1 × 108 to 2 × 108 cells/ml. The cells were washed and sonicated in the presence of proteinase inhibitors, and the proteins were treated with urea, dithiothreitol, and Triton X-100 to obtain final concentrations of 9 M, 70 mM, and 2%, respectively (4). Up to 900 μg of proteins were separated in preparative 2-DE gels (23 by 30 cm) and stained with Coomassie brilliant blue (CBB) G-250 (2). Spot positions were assigned to the standard 2-DE pattern, in which proteins are detected by silver staining. Given that proteins are detectable by CBB, the sequence coverage is superior when CBB-stained spots are the starting material compared to the use of silver-stained spots (11). Therefore, we started identification with CBB-stained spots. Peptide mass fingerprints were obtained by tryptic in-gel digestion and MALDI MS (Voyager Elite; Perseptive Biosystems, Framingham, Mass.) (7). Sequence information resulted from nanoelectrospray-tandem MS (nano-ESI-MS/MS) (Q-TOF; Micromass, Manchester, United Kingdom). The sequence tag method (8) was used to search the proteins in a translated protein sequence database (http://195.41.108.38/PA_PeptidePatternForm.html). If no protein matched, de novo sequencing was performed. Then the tBLASTN program of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov:80/blast.cgi?Jform=1) and the sequence search program of the Institute for Genome Research (TIGR) (http://www.tigr.org/tdb/CMR/gmt/htmls/SeqSearch.html) were applied to search within the entire genome of M. tuberculosis H37Rv and the clinical isolate CDC1551. Detailed investigations were focused on 190 spots in the pI range from 4 to 6 and the Mr range from 6 to 15 kDa representing about one-sixth of the whole 2-DE gel and one-tenth of all spots of the complete gel (9). Sixty-two 2-DE spots were identified by their peptide mass fingerprints, and ten further spots needed sequence information by n-ESI-MS/MS for their identification. Eleven spots contained more than one protein. Ten genes gave rise to more than one protein species. Within this sector of the gel (Fig. 1) sequences of six proteins could not be assigned to genes of M. tuberculosis H37Rv. As an example for the MS analysis, the identification of spot 5_98 is shown in Fig. 2a with the MS spectrum of the peptide mixture after digestion with trypsin, and in Fig. 2b with the MS/MS spectrum obtained by fragmentation of one peptide. Open reading frames (ORFs) were found in the genome of the strain CDC1551 for five spots, and no ORF was found for one spot (Table 1). A search in the genome of M. tuberculosis H37Rv revealed the presence of these DNA sequences, suggesting that the ORFs were not recognized by the search algorithms used by Cole et al. (1). The predicted Mr values from theoretical gene sequences are in the same range as the ones estimated by 2-DE. Three of the gene sequences are completely identical between H37Rv and CDC1551 (5_53, 5_139, and 5_37). The reasons for the failure of detection of these ORFs in H37Rv remain elusive. In contrast, the exchange of methionine in position 1 in 5_98, 5_123, and 5_115 by leucine, valine, and proline-valine, respectively, may have prevented the detection of the starting codon. Spot 5_53 contains two further proteins: 14-kDa antigen (SwissProt: 14KD_MYCTU) and hypothetical protein Rv2626c (PIR: A70573). The protein of spot 5_37 was predicted neither in the H37Rv nor CDC1551 genome so far. A hypothetical M. leprae protein (SwissProt: Y525_MYCLE) shows 83.5% similarity to the new ORF. Recently, a sequence as part of an U.S. patent was published (EMBLNEW: AX023830) identical to the sequence of spot 5_53 without the residues 1 to 7 and methionine instead of valine as residue 8.
FIG. 1.
Sector 5 of M. tuberculosis H37Rv 2-DE pattern. Proteins were stained with silver nitrate. The Mr range between 6 and 15 kDa and the pI range between 4 and 6 are shown. The spots numbered were sequenced de novo by nanospray MS/MS and revealed ORFs not predicted previously.
FIG. 2.
MS analysis of spot 5_98. (a) Spectrum of the trypsinized protein. Labeled peptides were fragmented to obtain sequence information. (b) fragmentation pattern of the peptide with an m/z of 708.36 identified as VEIEVDDDLIQK.
TABLE 1.
Protein identification by n-ESI-MS/MS (boldface residues) and MALDI MS (underlined residues) of previously unpredicted ORFs of M. tuberculosis H37Rv
| Spot | H37Rv Sequence | Sanger EMBL accession no. | ORF detected in CDC1551 | Comparison of H37Rv and CDC1551 | Mr | pI |
|---|---|---|---|---|---|---|
| 5_37 | GGAPVARVVV HVMPKAEILD | Z80226 (32260–32508) | 100% identity | 8,872 | 4.5 | |
| POGQAIVGAL GRLGHLGISD | ||||||
| VRQGKRFELE VDDTVDDTTL | ||||||
| AEIAESLLAN TVIEDWTISR | ||||||
| DPQ | ||||||
| 5_53 | MPMEGATVEV KIGITDSPRE | Z95120 (5517–5311) | 03128 | 100% identity | 10,118 | 4.9 |
| LVFSSAQTPS EVEELVSNAL | ||||||
| RDDSGLLTLT DERGRRFLIH | ||||||
| TARIAYVEIG VADARRVGFG | ||||||
| VGVDAAAGSA GKVATSG | ||||||
| 5_98 | LGSDCGCGGY LWSMLKRVEI | Z92772 (17111–17359) | D0043 | Leu-1→Met-1 in CDC1551 | 9,403 | 4.9 |
| EVDDDLIQKV IRRYRVKGAR | ||||||
| EAVNLALRTL LGEADTAEHG | ||||||
| HDDEYDEFSD PNAWVPRRSR | ||||||
| DTG | ||||||
| 5_115 | PVTVYRRGMA VLTDEQVDAA | Z95584 (24791–24486) | 06120 | Pro-1-Val-2→Met-1 in CDC1551 | 11,309 | 5.9 |
| LHDLNGWQRA GGVLRRSIKF | ||||||
| PTFMAGIDAV RRVAERAEEV | ||||||
| NHHPDIDIRW RTVTFALVTH | ||||||
| AVGGITENDI AMAHDIDAMF | ||||||
| GA | ||||||
| 5_123 | VQEGGPQETM SARSTQHDAA | AL021646 (44673–44494) | 03103 | Val-1→Met-1 in CDC1551 | 7,253 | 4.9 |
| DALFRAIIET LDKHRNERTL | ||||||
| TEDVLDTLAR AYASISTNVP | ||||||
| EQGRLG | ||||||
| 5_139 | MSNHTYRVIE IVGTSPDGVD | Z79701 (17944–17735) | 00401 | 100% identity | 7,629 | 5.8 |
| AAIQGGLARA AQTMRALDWF | ||||||
| EVQSIRGHLV DGAVAHFQVT | ||||||
| MKVGFRLEDS |
MALDI MS proved highly effective in the rapid identification of the main components of a 2-DE gel, if the proteins are known in a sequence database. A more detailed analysis of spots in 2-DE gels by nano-ESI-MS/MS elucidated additional proteins per spot and additional genes not predicted from genome investigations. Our findings illustrate the value of proteomics in complementing genomics in both functional and genomic analyses. Proteomics is a further building block to unravel the molecular network in bacterium-host interactions, a prerequisite for the development of new vaccines to fight against infectious diseases like tuberculosis.
Acknowledgments
This work was supported by Chiron Behring, Marburg, Germany, and the WHO (Global Programme for Vaccines and Immunization–Vaccine Research and Development).
REFERENCES
- 1.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G. Deciphering the biology of Mycobacterium tuberculosisfrom the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 2.Doherty N S, Littman B H, Reilly K, Swindell A C, Buss J M, Anderson N L. Analysis of changes in acute-phase plasma proteins in an acute inflammatory response and in rheumatoid arthritis using two-dimensional gel electrophoresis. Electrophoresis. 1998;19:355–363. doi: 10.1002/elps.1150190234. [DOI] [PubMed] [Google Scholar]
- 3.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, Mckenney K, Sutton G, Fitzhugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Venter J C, et al. Whole-genome random sequencing and assembly of Haemophilus influenzaeRD. Science. 1995;269:496–511. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 4.Jungblut P R, Schaible U E, Mollenkopf H-J, Zimny-Arndt U, Raupach B, Mattow J, Halada P, Lamer S, Hagens K, Kaufmann S H E. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovisBCG strains: towards functional genomics of microbial pathogens. Mol Microbiol. 1999;33:1103–1117. doi: 10.1046/j.1365-2958.1999.01549.x. [DOI] [PubMed] [Google Scholar]
- 5.Kaufmann S H E. Is the development of a new tuberculosis vaccine possible? Nat Med. 2000;6:955–960. doi: 10.1038/79631. [DOI] [PubMed] [Google Scholar]
- 6.Klose J, Kobalz U. Two-dimensional electrophoresis of proteins: an updated protocol and implications for a functional analysis of the genome. Electrophoresis. 1995;16:1034–1059. doi: 10.1002/elps.11501601175. [DOI] [PubMed] [Google Scholar]
- 7.Lamer S, Jungblut P R. Matrix-assisted laser desorption-ionization mass spectrometry peptide mass fingerprinting for proteome analysis: identification efficiency after on-blot or in-gel digestion with and without desalting procedures. J Chromatogr B. 2001;752:311–322. doi: 10.1016/s0378-4347(00)00446-1. [DOI] [PubMed] [Google Scholar]
- 8.Mann M, Wilm M. Error tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 9.Mattow J, Jungblut P R, Müller E-C, Kaufmann S H E. Identification of acidic, low molecular mass proteins of Mycobacterium tuberculosisstrain H37Rv by MALDI- and ESI-mass spectrometry. Proteomics. 2001;1:494–507. doi: 10.1002/1615-9861(200104)1:4<494::AID-PROT494>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 10.Mollenkopf H-J, Jungblut P R, Raupach B, Mattow J, Lamer S, Zimny-Arndt U, Schaible U E, Kaufmann S H E. A dynamic two-dimensional polyacrylamide gel electrophoresis database: the mycobacterial proteome via the internet. Electrophoresis. 1999;20:2172–2180. doi: 10.1002/(SICI)1522-2683(19990801)20:11<2172::AID-ELPS2172>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 11.Scheler C, Lamer S, Pan Z, Li X-P, Salnikow J, Jungblut P. Peptide mass fingerprint sequence coverage from differently stained proteins on 2-DE patterns by MALDI-MS. Electrophoresis. 1998;19:918–927. doi: 10.1002/elps.1150190607. [DOI] [PubMed] [Google Scholar]