Abstract
Helicobacter hepaticus causes disease in the liver and lower intestinal tract of mice. It is strongly urease positive, although it does not live in an acidic environment. The H. hepaticus urease gene cluster was expressed in Escherichia coli with and without coexpression of the Helicobacter pylori nickel transporter NixA. As for H. pylori, it was difficult to obtain enzymatic activity from recombinant H. hepaticus urease; special conditions including NiCl2 supplementation were required. The H. hepaticus urease cluster contains a homolog of each gene in the H. pylori urease cluster, including the urea transporter gene ureI. Downstream genes were homologs of the nik nickel transport operon of E. coli. Nongastric H. hepaticus produces urease similar to that of H. pylori.
Helicobacter hepaticus is a gram-negative, microaerophilic, urease-positive spiral rod (13). It is a pathogen of mice and causes chronic active hepatitis, hepatic tumors, and proliferative typhlocolitis (40, 53). Although it was first identified in the liver, the primary site of H. hepaticus colonization is the intestinal tract; it has not been found in the stomach. After the initial identification of H. hepaticus in 1992, this bacterium was found to infect large numbers of rodents used in biomedical research (39, 44). Since that time, additional Helicobacter species, including H. bilis, H. cholecystus, H. rodentium, and “H. typhlonicus” have been identified in laboratory rodents with disease of the hepatobiliary or intestinal tracts (14–18, 21).
The best-known and most-studied member of the genus Helicobacter is H. pylori, which causes peptic ulcer and gastric cancers in humans (8). Nongastric helicobacters also cause human illness. H. pullorum, H. canis, H. fennelliae, and H. cinaedi are associated with enteritis; in addition, proctocolitis and bacteremia have been reported for some Helicobacter species (11, 49).
Urease is an important virulence factor in H. pylori and in H. mustelae, the gastric pathogen of ferrets. In those species, urease is required to colonize the gastric mucosa of laboratory animals (3, 10, 51). Urease catalyzes the hydrolysis of urea to ammonia and carbon dioxide (33). Ammonium ion causes a pH increase that allows Helicobacter cells to survive and grow in a highly acidic niche (43). Urease contributes to disease by both direct and indirect mechanisms. Urease itself activates phagocytes, induces cytokine production, and enhances gastric inflammation (22). Ammonia can be used as a nitrogen source for protein synthesis (19), and ammonium ion is toxic to gastric epithelial cells (47).
Urease is a heteromultimer nickel-containing metalloenzyme (33). H. pylori urease contains 12 copies each of structural subunits, UreA and UreB, encoded by the genes ureA and ureB (20). Production of enzymatically active urease requires these structural genes and four accessory genes, ureE, ureF, ureG, and ureH, which are essential for assimilation of nickel ions into the apoenzyme. An additional gene, ureI, encodes an integral membrane protein that transports urea to the cytoplasm under acidic conditions (42, 55).
Urease is enzymatically active only when nickel ions are incorporated during assembly of the mature enzyme (33). Escherichia coli carrying the H. pylori urease gene cluster is only weakly active except under specific culture conditions (24). The E. coli host must be grown in medium supplemented with NiCl2 and devoid of amino acids which chelate nickel ions, thus making them unavailable for intracellular transport. H. pylori possesses redundant mechanisms for nickel acquisition, so that active urease is produced even in amino acid-rich medium. One method of transport is via the high-affinity nickel transport protein, NixA (32). Providing a copy of nixA in E. coli carrying H. pylori urease genes leads to greatly enhanced urease activity by improving nickel transport into the cell (30, 32). An additional method of nickel transport in H. pylori may be via an ATP-binding cassette (ABC) transporter composed of the genes abcABCD (23).
While urease is an important virulence factor for gastric helicobacters which inhabit a highly acidic environment, the function of urease in the nongastric helicobacters, whose environment is not acidic, is unclear. Recently, a partial sequence of the H. hepaticus urease structural genes was published (45). We have extended that information by sequencing, cloning, and expressing the entire H. hepaticus urease gene cluster. This knowledge will be useful for understanding comparative aspects of the role of urease in the pathogenesis of gastric versus nongastric helicobacters.
Bacterial strains, plasmids, and media.
H. hepaticus strain MU94-1 was isolated from the liver of a naturally infected mouse and grown on chocolate agar as previously described (13). H. pylori ATCC 49503 was purchased from the American Type Culture Collection (Rockville, Md.) and grown on 10% sheep blood agar. Escherichia coli DH5α (Gibco BRL Life Technologies, Gaithersburg, Md.) was grown on Luria-Bertani (LB) agar or in LB broth (41). Kanamycin (50 μg/ml) and/or chloramphenicol (20 μg/ml) were added to media when needed to maintain plasmids.
The plasmid pHP8080 (30) was digested with the NruI and AvaI restriction endonucleases. A 1.2-kb fragment containing nixA was ligated into corresponding sites in pACYC184 and designated pACYC184-nixA (Table 1).
TABLE 1.
Plasmids used in this study
| Plasmid | Vector | Insert size (kb) | Source of insert | Relevance |
|---|---|---|---|---|
| pHHuc1 | pCR-XL-TOPO | 6.1 | H. hepaticus | Urease cluster genes |
| pC2 | pCR-XL-TOPO | 7 | Irrelevant fragment | Negative control for pHHuc1 |
| pACYC184-nixA | pACYC184 | 1.2 | H. pylori | Nickel transporter gene nixA |
| pACYC184 | pACYC184 | None | None | Vector control for pACYC184-nixA |
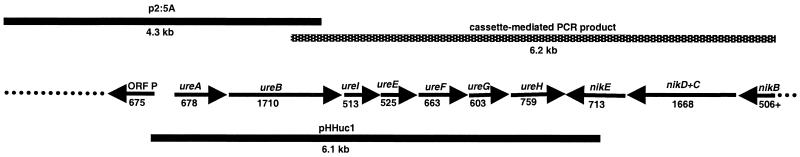
| p2:5A | pUC18 | 4.3 | H. hepaticus | Library clone |
Construction of an H. pylori ureAB probe.
A 1.6-kb PCR fragment containing the H. pylori urease genes ureA and ureB was amplified from H. pylori genomic DNA with the PCR primers Hp2794f and Hp4324r (Table 2). The PCR product was labeled with digoxigenin-11-dUTP (Roche Molecular Biochemicals, Indianapolis, Ind.) by PCR according to the manufacturer's guidelines (The DIG System User's Guide for Filter Hybridization, 2000; Roche).
TABLE 2.
Oligonucleotide primers
| Primer | Nucleotide sequence | Organism | Reference | Product |
|---|---|---|---|---|
| Hp2769f | 5′-TTTGATTAGTGCCCATATTATGGAAG | H. pylori | 48 | ureAB probe |
| Hp4346r | 5′-TGGTGGCACACCATAAGCATGTC | H. pylori | 48 | ureAB probe |
| Hh653f | 5′-CCGGAATTCGGCTTTGCATACCCTATTGACAAACa | H. hepaticus | This work | Urease cluster |
| Hh6778r | 5′-CCCGAGCTCTGCGTGGTGGAACATATAAGGATAGa | H. hepaticus | This work | Urease cluster |
| HhureB1 | 5′-ATGGAATGATTGTAGCAGCAAAAATAGGGG | H. hepaticus | This work | Cassette-mediated PCR |
| HhureB2 | 5′-GGATTCTAATGCTTCTATTCCTACTCCTGAACCTG | H. hepaticus | This work | Cassette-mediated PCR |
Underlined nucleotides indicate restriction endonuclease sites.
Hybridizations.
H. hepaticus genomic DNA was digested with the restriction endonuclease HindIII, electrophoresed, and transferred to nylon membranes according to standard techniques (4, 41). Membranes were hybridized with the H. pylori ureAB probe under stringent conditions (65°C), washed, incubated with alkaline phosphatase-conjugated anti-digoxigenin Fab fragments (Roche), and detected using the chemiluminescent substrate CSPD (disodium 3-(4-methoxyspiro{1,2- dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl) phenyl phosphate; Roche) using the manufacturer's protocol. A single band of 2.8 kb was identified, indicating that a single copy of the structural urease genes is present in H. hepaticus (data not shown).
An H. hepaticus plasmid library (29) harbored in E. coli strain DH5αMCR was screened for clones containing urease genes by colony hybridization with the H. pylori ureAB probe by standard techniques (4, 41). Membranes containing plasmid DNA were hybridized and washed under stringent conditions (65°C), incubated with alkaline phosphatase-conjugated anti-digoxigenin Fab fragments (Roche), and detected with nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl-phosphate (Roche). An E. coli clone that hybridized with the ureAB probe was selected; the corresponding plasmid was designated p2:5A.
DNA sequencing and analysis.
Vector-insert junctions of the plasmid p2:5A were sequenced by the dideoxy chain termination method using the ABI Prism BigDye Terminator Cycle Sequencing Kit (PE Applied Biosystems, Foster City, Calif.) and M13/pUC sequencing primers. The obtained sequence had strong homology with Helicobacter urease genes (BLAST; National Center for Biotechnology Information, National Library of Medicine, NIH [http://www.ncbi.nlm.nih.gov/BLAST]), so the entire 4.3-kb insert of plasmid p2:5A (Table 1) was sequenced. The insert contained one open reading frame (ORF) with homology to ureA and a partial ORF with homology to ureB (GCG software package, Wisconsin Package Version 10.1; Genetics Computer Group, Inc., Madison, Wisc.; and Omiga Version 2.0; Oxford Molecular, Ltd., Madison, Wisc.).
Since the entire H. hepaticus urease cluster was not present in a single clone, the 3′ end of ureB and downstream genes were amplified by cassette-mediated PCR using the TaKaRa LA PCR in vitro cloning kit (PanVera Corporation, Madison, Wisc.). Specific primers HhureB1 and HhureB2 (Table 1) were designed near the 3′ end of the H. hepaticus ureB gene. H. hepaticus genomic DNA, digested with the restriction enzyme PstI and ligated to cassettes, was amplified with cassette primer C1 and HhureB1, 400 μM (each) dNTP, 10× LA BufferII, and 2.5 mM MgCl2. Samples were denatured at 94°C for 9 min before 2.0 U of TaKaRa LA Taq DNA polymerase was added. Thirty cycles of 15 s at 94°C, 2 s at 58.7°C, and 6.25 min at 72°C were completed followed by a final extension at 72°C for 10 min. The nested primers C2 and HhureB2 reamplified products from the first PCR. The approximately 6.2-kb product was directly sequenced to reduce the chance of incorporating PCR-generated mistakes commonly fixed in individual strands when the PCR product is cloned before sequencing.
Analysis of the combined sequences of the library clone and the cassette-mediated PCR product revealed seven ORFs homologous to the H. pylori urease structural genes ureA and ureB, the urea transporter gene ureI, and the accessory genes ureE, ureF, ureG, and ureH (Fig. 1). The deduced amino acid sequences were highly homologous to those corresponding to urease genes of other Helicobacter species (Table 3). H. hepaticus UreB had 100% identity to two urease signature consensus patterns (Motifs program, GCG package).
FIG. 1.
Map of H. hepaticus genes included on plasmids and PCR products. Arrows represent ORFs and directions of transcription. Solid bars represent plasmid inserts, and the patterned bar represents the cassette-mediated PCR product. Size is shown in base pairs or kilobases for genes and larger fragments, respectively. Dotted lines represent incompletely sequenced fragments.
TABLE 3.
Amino acid sequence identity of predicted products of the H. hepaticus urease gene cluster
| Comparison species | % Identity of:
|
||||||
|---|---|---|---|---|---|---|---|
| UreA | UreB | UreI | UreE | UreF | UreG | UreH | |
| H. pylori | 66 | 77 | 62 | 37 | 46 | 82 | 43 |
| H. felis | 66 | 76 | 58 | ||||
| H. mustelae | 71 | 84 | |||||
| “H. heilmannii” | 66 | 76 | |||||
ATG codons initiated each ORF of the H. hepaticus urease cluster except ureF, which began with a TTG codon. A putative ribosome-binding site preceded each ORF except the accessory gene ureE. Two intergenic regions, ureI–ureE and ureE-ureF, had overlapping start and stop codons. The intergenic region between H. hepaticus ureB and ureI contained only 9 bp in contrast to approximately 200 bp between these genes in H. pylori. The G+C content of the H. hepaticus urease cluster genes was 36%.
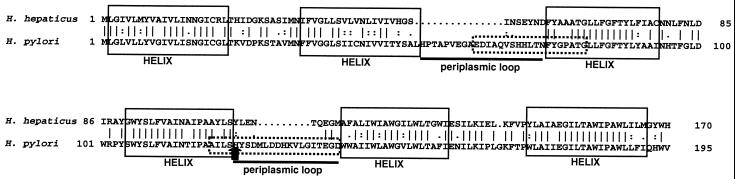
The H. hepaticus urease cluster contained a homolog of the H. pylori urea transporter gene, ureI. Kyte Doolittle hydropathy analysis of the encoded amino acid sequence predicted six hydrophobic regions separated by hydrophilic regions, very similar to H. pylori UreI. Alignment of the amino acid sequences of the two UreI proteins showed two gaps of 15 and 9 amino acids in the H. hepaticus product within extracellular loops of H. pylori UreI (55) (Fig. 2).
FIG. 2.
Alignment of UreI amino acid sequences from H. hepaticus MU94-1 and H. pylori. Identical residues are depicted by “|.” Similar residues are depicted with “:” or “.” Transmembrane helices and periplasmic loops are shown as determined by Weeks et al. (55) for H. pylori UreI. Helices are enclosed in solid boxes. Solid bars indicate periplasmic loops. Peptides used by Scott et al. (42) to develop anti-UreI antibodies are enclosed in dashed boxes. The arrowhead points to histidine 123 of H. pylori UreI.
DNA in the downstream region flanking the H. hepaticus urease cluster had homology with bacterial ABC transporters, which function as dipeptide, oligopeptide, and nickel transporters. These ORFs were on the complementary strand of the cassette-mediated PCR product with opposite polarity to genes of the urease cluster (Fig. 1) and aligned closely with the E. coli nik operon, a nickel transport system (28, 34). A partial nikB ORF was identified in H. hepaticus followed by a long ORF that encompassed domains homologous to nikC and nikD, and finally, a nikE homolog. Both the nikD and nikE homologs contained consensus patterns for the ABC transporter family signature and ATP/GTP-binding site motifs A and B (28, 31, 34, 52). A predicted ORF, ORF P, upstream of the ureA gene in the H. hepaticus clone p2:5A contained three sets of direct repeats with 51, 43, and 43 bases each and had homology to putative periplasmic proteins of Campylobacter jejuni and Neisseria meningitidis (36, 37).
Cloning the H. hepaticus urease cluster.
To express recombinant H. hepaticus urease, the entire cluster of genes was cloned as a single fragment amplified by long PCR. Primers Hh653f and Hh6778r (Table 2) amplified the urease structural genes, accessory genes, and approximately 300 bases of flanking sequence from H. hepaticus DNA. Long PCR reactions contained 100 to 200 ng of H. hepaticus genomic DNA, 0.3 μM (each) primer, 300 μM (each) dNTP, 1 mM MgSO4, 2.5 U of Platinum Pfx polymerase, and Pfx Amplification Buffer (Gibco BRL Life Technologies) in a 50-μl volume. Reactions were denatured for 3 min followed by 20 cycles of 94°C for 15 s, 58.2°C for 2 s, and 68°C for 6 min 20 s with a final 7-min extension at 68°C. A 6.1-kb product was obtained.
Terminal 3′ deoxyadenosine overhangs were added to the PCR product by incubation with 200 μM dNTPs, 1 U of Taq DNA polymerase, and 10× PCR buffer (Roche) at 72°C for 15 min. The fragment was ligated to pCR-XL-TOPO (TOPO XL PCR cloning kit; Invitrogen, Carlsbad, Calif.) according to the manufacturer's recommendations and was designated pHHuc1 (Table 1). The ligation reaction was electroporated into DH5α that had been previously transformed with pACYC184-nixA to facilitate identification of urease-positive clones. After cloning, the nucleotide sequence of the insert in pHHuc1 was redetermined and compared to sequence of the cassette-mediated PCR product and plasmid p2:5A to ensure that no base errors were introduced during PCR amplification.
A 7-kb control DNA fragment supplied with the TOPO XL PCR cloning kit was amplified, ligated to the vector, and designated pC2. The plasmid pC2 was cotransformed into DH5α carrying pACYC184-nixA as a urease-negative control strain. A second DH5α control strain was prepared with pHHuc1 and insert-free pACYC184 (Table 1). DH5α was also transformed with pHHuc1 and pC2 as single plasmids.
Qualitative tests for urease activity.
E. coli DH5α cotransformants carrying pHHuc1 plus pACYC184-nixA and control strains were screened for enzymatic activity on modified urea segregation agar (30). Only cotransformants carrying pHHuc1 and pACYC184-nixA turned the medium pink as the pH rose because of urea hydrolysis.
Antigenic cross-reactivity of H. hepaticus urease structural subunits.
Cultured bacteria were centrifuged (10,000 × g, 10 min, 4°C), washed twice in 50 mM HEPES, pH 7.5, and frozen at −20°C until used. Pellets were resuspended in 50 mM HEPES, sonicated 3 times for 30 s, and centrifuged at 12,000 × g, and the supernatant was retained. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed as described by Laemmli (27). Proteins were electrophoresed, transferred to an ImmobilonP membrane (Millipore, Bedford, Mass.), and detected using the Lumi-LightPLUS Western Blotting Kit (Roche) using the manufacturer's protocol. Antibodies to H. pylori UreA and UreB (24) identified immunoreactive bands at approximately 27 and 60 kDa, respectively, in H. pylori, H. hepaticus, and DH5α containing plasmid pHHuc1 (data not shown). No bands were identified for DH5α transformed with pC2.
Enzymatic activity of recombinant H. hepaticus urease.
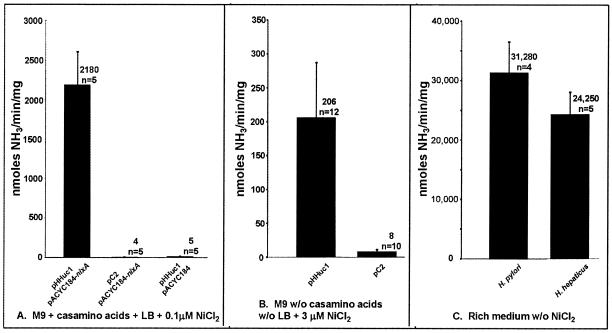
Growth media for DH5α strains varied based on which plasmids were carried. DH5α cotransformed with pHHuc1 and pACYC184-nixA, or corresponding control constructs (Table 1), was grown in M9 minimal medium (per liter, 6 g of Na2HPO4, 3 g of KH2PO4, 0.5 g of NaCl, 1 g of NH4Cl, 0.4% glucose, 1 mM MgSO4, 0.1 mM CaC l2, and 1.68 μM thiamine-HCl) supplemented with 0.5% casamino acids, 1% LB, 0.1 μM NiCl2, 50 μg of kanamycin/ml, and 20 μg of chloramphenicol/ml (30). Bacteria were centrifuged, washed, and sonicated as described for sodium dodecyl sulfate-polyacrylamide gel electrophoresis Urease activities were determined by the phenol-hypochlorite assay, a spectrophotometric assay that measures ammonia production, as previously described (30, 54). Data were statistically analyzed using Sigmastat for Windows (Version 2.03; SPSS, Inc., San Rafael, Calif.). High levels of urease activity were measured only when the H. hepaticus urease cluster and the nickel transporter gene were both present in the same strain (pHHuc1 and pACYC184-nixA) (Fig. 3A). When either of these plasmids was present in combination with its negative control, only negligible ammonia was produced.
FIG. 3.
Urease activities of sonicated bacterial proteins in the phenol-hypochlorite assay. Scales differ for the three panels. (A) E. coli DH5α cotransformed with H. hepaticus urease genes (pHHuc1) and the H. pylori nickel transporter (pACYC184-nixA), or control plasmids (P = 0.008; Kruskal-Wallis one-way analysis of variance on ranks). (B) E. coli DH5α singly transformed with H. hepaticus urease genes (pHHuc1) or control plasmid (pC2) (P = <0.001; Mann-Whitney rank sum test). (C) Wild-type H. pylori and H. hepaticus. Differences are not statistically significant (t test).
Starter cultures of DH5α singly transformed with pHHuc1 or pC2 were grown as for cotransformed DH5α. Cultures were then diluted 1:500 into M9 minimal medium with 3 μM NiCl2 and kanamycin (no casamino acids or LB). These cultures were incubated at 37°C with shaking until optical density measurements were stable, approximately 44 h. Casamino acids and LB were not used to prevent amino acid chelation of nickel ions (24). DH5α(pHHuc1) had consistent urease activity (Fig. 3B), while DH5α(pC2) had negligible activity. Sonicated wild-type H. hepaticus MU94-1 had an average urease activity similar to that for H. pylori (Fig. 3C).
Nucleotide sequence accession numbers.
The sequence of pHHuc1 containing the urease gene cluster of H. hepaticus MU94-1 was deposited in GenBank under accession number AF3322656. Sequence of the 5′ and the 3′ flanking regions was deposited under accession numbers AF332654 and AF332655, respectively. The latter two sequences were determined in one direction only.
Discussion.
This is the first report of the cloning and sequencing of a complete urease gene cluster for a Helicobacter species other than H. pylori. The urease gene cluster of H. hepaticus is similar to the urease cluster of H. pylori in many ways. As in other helicobacters, two structural subunit genes are present (Fig. 1), in contrast to the more common bacterial pattern of a three-subunit urease (33). Both structural subunits of H. hepaticus cross-react with immune sera directed against H. pylori urease subunits, indicating that recombinant H. hepaticus urease is stable and important antigenic epitopes are conserved. Indeed, protein sequence alignment of the UreA and UreB structural subunits of H. pylori and H. hepaticus confirm this high degree of relatedness (Table 3).
Despite overall similarities of the H. hepaticus and H. pylori urease gene clusters, there are notable differences. In H. hepaticus, the ureB–ureI intergenic distance is 9 bp, compared to approximately 200 bp (strain dependent) in H. pylori; in H. pylori, the sequence contains a promoter for ureI and downstream accessory genes (1, 26). This sequence difference suggests that the two species differ in regulation of ureI and the accessory genes. Although the overall sequence of UreI in H. hepaticus and H. pylori is well conserved, alignment shows gaps in the H. hepaticus product (Fig. 2). This may explain why antibodies to H. pylori UreI failed to detect products in Western blots of H. hepaticus and other nongastric species (42). The antibodies used by Scott et al. (42) were directed against peptides within extracellular loops of H. pylori UreI which are truncated in H. hepaticus UreI. H. hepaticus UreI also lacks the critical histidine 123 residue, important for acid activation of urea transport in H. pylori (55). A similar histidine residue is also present in UreI of gastric H. felis (GenBank accession no. A41012) (46). The presence of such a histidine residue in gastric helicobacters and its absence in nongastric H. hepaticus may represent specific adaptations of these organisms to acidic versus nonacidic environments.
The significant urease activity of DH5α(pHHuc1) proves that all of the H. hepaticus genes essential for urease activity are present on this plasmid (Fig. 3B). Without a specific nickel transporter, urease activity was obtained only when E. coli cells were grown in medium devoid of amino acids which chelate nickel ions and prevent their assimilation into the apoenzyme (24, 32). When the NixA nickel transporter was coexpressed with the H. hepaticus urease gene cluster in DH5α, much higher levels of urease activity were obtained (Fig. 3A). These levels were similar to urease activities of cloned H. pylori urease genes and emphasize the importance of nickel acquisition to urease activity. In the presence of a nickel transporter, urease activity was high even when host cells were grown in medium supplemented with amino acids. Urease activity of DH5α cotransformed with pHHuc1 and pACYC184-nixA remained about 10-fold lower than that of wild-type H. hepaticus (Fig. 3C). This is similar to findings with comparable clones carrying H. pylori genes and suggests that other factors in addition to urease genes and nickel transport may be necessary for full wild-type levels of urease activity.
It is likely that wild-type H. hepaticus possesses a specialized system for nickel transport, but it is not known whether that system is a nixA homolog, another transporter gene, or perhaps multiple redundant means of nickel transport as is found in H. pylori. ORFs flanking the downstream end of the H. hepaticus urease gene cluster are most closely related to the nik operons of E. coli (34) and Brucella suis (25). Both of these operons were documented to mediate nickel transport, and mutation of B. suis nikA led to decreased urease activity. The closest H. pylori match to the H. hepaticus nik homologs is the dpp operon, a putative dipeptide ABC transporter (2, 50). A role for dpp genes in nickel transport has not yet been tested. Another set of H. pylori ABC transporter genes, abcABCD, appears to be necessary for full urease activity, since mutation of abcD led to decreased urease activity (23). That system, however, has not been proven to be specific for nickel.
Identification of a putative nickel transporter flanking the urease gene cluster in H. hepaticus points to a difference in genome organization between H. hepaticus and H. pylori. Documented nickel transporter genes in H. pylori are located at separate sites on the chromosome distant from the urease gene cluster (2, 50). In contrast, the H. pylori urease cluster is flanked by lspA upstream and cdrA downstream, respectively (2, 50).
H. hepaticus inhabits a biological niche where the pH is nearly neutral, yet it produces an amount of urease activity similar to that of gastric H. pylori (Fig. 3C). It is not apparent why such high levels of urease activity would be necessary in the lower bowel and liver. Possible roles for urease in H. hepaticus include improving survival during passage through the stomach, as for Yersinia enterocolitica (7), and producing ammonia as a source of nitrogen for protein biosynthesis (6, 19). Urease activity could significantly contribute to pathology, since ammonia damages host cells (47) and urease itself stimulates phagocyte chemotaxis, activates immune cells, and induces cytokine production (9).
Among the nongastric helicobacters, no clear pattern can be discerned correlating urease activity with virulence or site of colonization. Both urease-positive and urease-negative Helicobacter species have been identified in the liver and/or biliary tracts of various animal species in association with disease (14–18, 49). Some reports link helicobacters with human diseases of the liver and biliary tract (5, 12, 35, 38). Ultimately, understanding the role of urease in the pathogenesis of the enterohepatic helicobacters may contribute to a better understanding of some human hepatobiliary tract diseases. Future studies will clarify properties of the specific gene products and their roles in colonization and pathogenesis of H. hepaticus.
Acknowledgments
This work was supported in part by NIH PHS grant AI10098 (D.J.M.) and AI25567 (H.L.T.M.).
We thank Robert S. Livingston for the use of the H. hepaticus library and Howard Wilson for computer graphics assistance.
REFERENCES
- 1.Akada J K, Shirai M, Takeuchi H, Tsuda M, Nakazawa T. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol Microbiol. 2000;36:1071–1084. doi: 10.1046/j.1365-2958.2000.01918.x. [DOI] [PubMed] [Google Scholar]
- 2.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 3.Andrutis K A, Fox J G, Schauer D B, Marini R P, Murphy J C, Yan L, Solnick J V. Inability of an isogenic urease-negative mutant stain of Helicobacter mustelae to colonize the ferret stomach. Infect Immun. 1995;63:3722–3725. doi: 10.1128/iai.63.9.3722-3725.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and John Wiley & Sons; 1995. [Google Scholar]
- 5.Avenaud P, Marais A, Monteiro L, Le Bail B, Bioulac Sage P, Balabaud C, Megraud F. Detection of Helicobacter species in the liver of patients with and without primary liver carcinoma. Cancer. 2000;89:1431–1439. [PubMed] [Google Scholar]
- 6.Burne R A, Chen Y Y. Bacterial ureases in infectious diseases. Microbes Infect. 2000;2:533–542. doi: 10.1016/s1286-4579(00)00312-9. [DOI] [PubMed] [Google Scholar]
- 7.De Koning-Ward T F, Robins-Browne R M. Contribution of urease to acid tolerance in Yersinia enterocolitica. Infect Immun. 1995;63:3790–3795. doi: 10.1128/iai.63.10.3790-3795.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn B E, Phadnis S H. Structure, function and localization of Helicobacter pylori urease. Yale J Biol Med. 1998;71:63–73. [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton K A, Krakowka S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect Immun. 1994;62:3604–3607. doi: 10.1128/iai.62.9.3604-3607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox J. Enterohepatic helicobacters: natural and experimental models. Ital J Gastroenterol Hepatol. 1998;30:S264–S269. [PubMed] [Google Scholar]
- 12.Fox J G, Dewhirst F E, Shen Z, Feng Y, Taylor N S, Paster B J, Ericson R L, Lau C N, Correa P, Araya J C, Roa I. Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology. 1998;114:755–763. doi: 10.1016/s0016-5085(98)70589-x. [DOI] [PubMed] [Google Scholar]
- 13.Fox J G, Dewhirst F E, Tully J G, Paster B J, Yan L, Taylor N S, Collins M J, Jr, Gorelick P L, Ward J M. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox J G, Gorelick P L, Kullberg M C, Ge Z, Dewhirst F E, Ward J M. A novel urease-negative Helicobacter species associated with colitis and typhlitis in IL-10-deficient mice. Infect Immun. 1999;67:1757–1762. doi: 10.1128/iai.67.4.1757-1762.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox J G, Yan L L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher J C, Mendes E N. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franklin C L, Beckwith C S, Livingston R S, Riley L K, Gibson S V, Besch-Williford C L, Hook R R., Jr Isolation of a novel Helicobacter species, Helicobacter cholecystus sp. nov., from the gallbladders of Syrian hamsters with cholangiofibrosis and centrilobular pancreatitis. J Clin Microbiol. 1996;34:2952–2958. doi: 10.1128/jcm.34.12.2952-2958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franklin C L, Riley L K, Livingston R S, Beckwith C S, Besch-Williford C L, Hook R R., Jr Enterohepatic lesions in SCID mice infected with Helicobacter bilis. Lab Anim Sci. 1998;48:334–339. [PubMed] [Google Scholar]
- 18.Franklin C L, Riley L K, Livingston R S, Beckwith C S, Hook R R, Jr, Besch-Williford C L, Hunziker R, Gorelick P L. Enteric lesions in SCID mice infected with “Helicobacter typhlonicus,” a novel urease-negative Helicobacter species. Lab Anim Sci. 1999;49:496–505. [PubMed] [Google Scholar]
- 19.Garner R M, Fulkerson J, Jr, Mobley H L. Helicobacter pylori glutamine synthetase lacks features associated with transcriptional and posttranslational regulation. Infect Immun. 1998;66:1839–1847. doi: 10.1128/iai.66.5.1839-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ha N C, Oh S T, Sung J Y, Cha K A, Lee M H, Oh B H. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat Struct Biol. 2001;8:505–509. doi: 10.1038/88563. [DOI] [PubMed] [Google Scholar]
- 21.Haines D C, Gorelick P L, Battles J K, Pike K M, Anderson R J, Fox J G, Taylor N S, Shen Z, Dewhirst F E, Anver M R, Ward J M. Inflammatory large bowel disease in immunodeficient rats naturally and experimentally infected with Helicobacter bilis. Vet Pathol. 1998;35:202–208. doi: 10.1177/030098589803500305. [DOI] [PubMed] [Google Scholar]
- 22.Harris P R, Mobley H L, Perez-Perez G I, Blaser M J, Smith P D. Helicobacter pylori urease is a potent stimulus of mononuclear phagocyte activation and inflammatory cytokine production. Gastroenterology. 1996;111:419–425. doi: 10.1053/gast.1996.v111.pm8690207. [DOI] [PubMed] [Google Scholar]
- 23.Hendricks J K, Mobley H L. Helicobacter pylori ABC transporter: effect of allelic exchange mutagenesis on urease activity. J Bacteriol. 1997;179:5892–5902. doi: 10.1128/jb.179.18.5892-5902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu L T, Mobley H L. Expression of catalytically active recombinant Helicobacter pylori urease at wild-type levels in Escherichia coli. Infect Immun. 1993;61:2563–2569. doi: 10.1128/iai.61.6.2563-2569.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jubier-Maurin V, Rodrigue A, Ouahrani-Bettache S, Layssac M, Mandrand-Berthelot M A, Kohler S, Liautard J P. Identification of the nik gene cluster of Brucella suis: regulation and contribution to urease activity. J Bacteriol. 2001;183:426–434. doi: 10.1128/JB.183.2.426-434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Labigne A, Cussac V, Courcoux P. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J Bacteriol. 1991;173:1920–1931. doi: 10.1128/jb.173.6.1920-1931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Linton K J, Higgins C F. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol Microbiol. 1998;28:5–13. doi: 10.1046/j.1365-2958.1998.00764.x. [DOI] [PubMed] [Google Scholar]
- 29.Livingston R S, Riley L K, Hook R R, Jr, Besch-Williford C L, Franklin C L. Cloning and expression of an immunogenic membrane-associated protein of Helicobacter hepaticus for use in an enzyme-linked immunosorbent assay. Clin Diagn Lab Immunol. 1999;6:745–750. doi: 10.1128/cdli.6.5.745-750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGee D J, May C A, Garner R M, Himpsl J M, Mobley H L. Isolation of Helicobacter pylori genes that modulate urease activity. J Bacteriol. 1999;181:2477–2484. doi: 10.1128/jb.181.8.2477-2484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mimura C S, Holbrook S R, Ames G F. Structural model of the nucleotide-binding conserved component of periplasmic permeases. Proc Natl Acad Sci USA. 1991;88:84–88. doi: 10.1073/pnas.88.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mobley H L, Garner R M, Bauerfeind P. Helicobacter pylori nickel-transport gene nixA: synthesis of catalytically active urease in Escherichia coli independent of growth conditions. Mol Microbiol. 1995;16:97–109. doi: 10.1111/j.1365-2958.1995.tb02395.x. [DOI] [PubMed] [Google Scholar]
- 33.Mobley H L, Island M D, Hausinger R P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro C, Wu L F, Mandrand-Berthelot M A. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol Microbiol. 1993;9:1181–1191. doi: 10.1111/j.1365-2958.1993.tb01247.x. [DOI] [PubMed] [Google Scholar]
- 35.Nilsson I, Lindgren S, Eriksson S, Wadstrom T. Serum antibodies to Helicobacter hepaticus and Helicobacter pylori in patients with chronic liver disease. Gut. 2000;46:410–414. doi: 10.1136/gut.46.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkhill J, Achtman M, James K D, Bentley S D, Churcher C, Klee S R, Morelli G, Basham D, Brown D, Chillingworth T, Davies R M, Davis P, Devlin K, Feltwell T, Hamlin N, Holroyd S, Jagels K, Leather S, Moule S, Mungall K, Quail M A, Rajandream M A, Rutherford K M, Simmonds M, Skelton J, Whitehead S, Spratt B G, Barrell B G. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 37.Parkhill J, Wren B W, Mungall K, Ketley J M, Churcher C, Basham D, Chillingworth T, Davies R M, Feltwell T, Holroyd S, Jagels K, Karlyshev A V, Moule S, Pallen M J, Penn C W, Quail M A, Rajandream M A, Rutherford K M, van Vliet A H, Whitehead S, Barrell B G. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 38.Ponzetto A, Pellicano R, Leone N, Cutufia M A, Turrini F, Grigioni W F, D'Errico A, Mortimer P, Rizzetto M, Silengo L. Helicobacter infection and cirrhosis in hepatitis C virus carriage: is it an innocent bystander or a troublemaker? Med Hypotheses. 2000;54:275–277. doi: 10.1054/mehy.1999.0987. [DOI] [PubMed] [Google Scholar]
- 39.Riley L K, Franklin C L, Hook R R, Jr, Besch-Williford C. Identification of murine helicobacters by PCR and restriction enzyme analyses. J Clin Microbiol. 1996;34:942–946. doi: 10.1128/jcm.34.4.942-946.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell R J, Haines D C, Anver M R, Battles J K, Gorelick P L, Blumenauer L L, Gonda M A, Ward J M. Use of antibiotics to prevent hepatitis and typhlitis in male scid mice spontaneously infected with Helicobacter hepaticus. Lab Anim Sci. 1995;45:373–378. [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Scott D R, Marcus E A, Weeks D L, Lee A, Melchers K, Sachs G. Expression of the Helicobacter pylori ureI gene is required for acidic pH activation of cytoplasmic urease. Infect Immun. 2000;68:470–477. doi: 10.1128/iai.68.2.470-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott D R, Weeks D, Hong C, Postius S, Melchers K, Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology. 1998;114:58–70. doi: 10.1016/s0016-5085(98)70633-x. [DOI] [PubMed] [Google Scholar]
- 44.Shames B, Fox J G, Dewhirst F, Yan L, Shen Z, Taylor N S. Identification of widespread Helicobacter hepaticus infection in feces in commercial mouse colonies by culture and PCR assay. J Clin Microbiol. 1995;33:2968–2972. doi: 10.1128/jcm.33.11.2968-2972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen Z, Schauer D B, Mobley H L, Fox J G. Development of a PCR-restriction fragment length polymorphism assay using the nucleotide sequence of the Helicobacter hepaticus urease structural genes ureAB. J Clin Microbiol. 1998;36:2447–2453. doi: 10.1128/jcm.36.9.2447-2453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skouloubris S, Thiberge J M, Labigne A, De Reuse H. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect Immun. 1998;66:4517–4521. doi: 10.1128/iai.66.9.4517-4521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smoot D T, Mobley H L, Chippendale G R, Lewison J F, Resau J H. Helicobacter pylori urease activity is toxic to human gastric epithelial cells. Infect Immun. 1990;58:1992–1994. doi: 10.1128/iai.58.6.1992-1994.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solnick J V, O'Rourke J, Lee A, Tompkins L S. Molecular analysis of urease genes from a newly identified uncultured species of Helicobacter. Infect Immun. 1994;62:1631–1638. doi: 10.1128/iai.62.5.1631-1638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stanley J, Linton D, Burnens A P, Dewhirst F E, On S L, Porter A, Owen R J, Costas M. Helicobacter pullorum sp. nov.—genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology. 1994;140:3441–3449. doi: 10.1099/13500872-140-12-3441. [DOI] [PubMed] [Google Scholar]
- 50.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Venter J C, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 51.Tsuda M, Karita M, Morshed M G, Okita K, Nakazawa T. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect Immun. 1994;62:3586–3589. doi: 10.1128/iai.62.8.3586-3589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ward J M, Anver M R, Haines D C, Melhorn J M, Gorelick P, Yan L, Fox J G. Inflammatory large bowel disease in immunodeficient mice naturally infected with Helicobacter hepaticus. Lab Anim Sci. 1996;46:15–20. [PubMed] [Google Scholar]
- 54.Weatherburn M. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39:971–974. [Google Scholar]
- 55.Weeks D L, Eskandari S, Scott D R, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482–485. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]