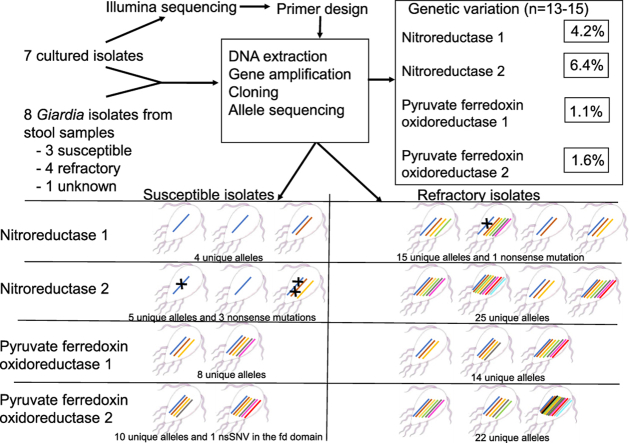
Abstract
The effectiveness of metronidazole against the tetraploid intestinal parasite Giardia lamblia is dependent on its activation/inactivation within the cytoplasm. There are several activating enzymes, including pyruvate ferredoxin reductase (PFOR) and nitroreductase (NR) 1 which metabolize metronidazole into toxic forms, while NR2 on the other hand inactivates it. Metronidazole treatment failures have been increasing rapidly over the last decade, indicating genetic resistance mechanisms. Analyzing genetic variation in the PFOR and NR genes in susceptible and refractory Giardia isolates may help identify potential markers of resistance.
Full length PFOR1, PFOR2, NR1 and NR2 genes from clinical culturable isolates and non-cultured clinical Giardia assemblage B samples were cloned, sequenced and single nucleotide variants (SNVs) were analyzed to assess genetic diversity and alleles.
A similar ratio of amino acid changing SNVs per gene length was found for the NRs; 4.2% for NR1 and 6.4% for NR2, while the PFOR1 and PFOR2 genes had less variability with a ratio of 1.1% and 1.6%, respectively. One of the samples from a refractory case had a nonsense mutation which caused a truncated NR1 gene in one out of six alleles. Further, we found three NR2 alleles with frameshift mutations, possibly causing a truncated protein in two susceptible isolates. One of these isolates was homozygous for the affected NR2 allele. Three nsSNVs with potential for affecting protein function were found in the ferredoxin domain of the PFOR2 gene. The considerable variation and discovery of mutations possibly causing dysfunctional NR proteins in clinical Giardia assemblage B isolates, reveal a potential for genetic link to metronidazole susceptibility and resistance.
Keywords: Nitroreductase, Pyruvate ferredoxin oxidoreductase, Metronidazole, Genetic diversity, Allele, Resistance, SNP, SNV, Nonsense mutation
Abbreviations: MTZ, Metronidazole; SNV, Single nucleotide variant; nsSNV, non-synonymous SNV; NR, nitroreductase; PFOR, pyruvate ferredoxin oxidoreductase
Graphical abstract
Highlights
-
•
Mutations in ferredoxin domains of thr nitroreductases and pyruvate ferredoxin oxidoreductases may alter protein function.
-
•
Nonsense mutation in nitroreductase 1 alleles may contribute towards metronidazole tolerance in Giardia.
-
•
Susceptible Giardia isolates had frameshift mutations in nitroreductase 2 alleles, possibly affecting metronidazole toxicity.
1. Introduction
Giardia lamblia is a flagellated microaerophilic protozoan parasite causing asymptomatic or symptomatic intestinal infection worldwide with almost 300 million clinical cases of Giardia being reported annually (Ankarklev et al., 2010; Kirk et al., 2015). The species G. lamblia is divided into eight distinctive assemblages, where assemblages A and B are known to infect humans, the latter being more frequently reported in human infections (Caccio and Ryan, 2008; Ankarklev et al., 2010). Assemblage B isolates usually show a higher genetic diversity and allelic sequence heterozygosity (ASH) than assemblage A isolates (0.5% vs. <0.01–0.04%) (Yu et al., 2002; Franzen et al., 2009; Ankarklev et al., 2012).
The first-line treatment against giardiasis, in most countries, is the prodrug metronidazole (MTZ), with an estimated efficacy rate ranging from 60% to 100% (Argüello-García et al., 2020). However, over the past decade, treatment failures with MTZ have been reported more frequently and has become a growing concern (Lalle and Hanevik, 2018), with 15–70% of the cases not cured with a standard 5–7-day course of MTZ treatment (Munoz Gutierrez et al., 2013; Nabarro et al., 2015; Requena-Mendez et al., 2017; Cañete et al., 2020; Ydsten et al., 2021; Neumayr et al., 2021).
MTZ needs to be activated intracellularly by a partial reduction at its nitro group, in order to create highly reactive toxic intermediates (Leitsch, 2015). MTZ has previously been demonstrated to be activated by three enzymes in Giardia, namely the nitroreductase (NR)1 (Pal et al., 2009), pyruvate-ferredoxin oxidoreductase (PFOR)1 (Leitsch et al., 2011) and the thiol-cycling associated enzyme thioredoxin reductase (Leitsch et al., 2016). NR2 inactivates MTZ by fully reducing it to an inert and non-toxic form of the antibiotic (Muller et al., 2013, 2015).
There are two PFOR genes in Giardia, named PFOR1 and PFOR2. The PFORs together with the co-factor ferredoxin serve as initial enzymes in electron transport reactions in Giardia i.e., activation of MTZ (Townson et al., 1996; Leitsch et al., 2011; Ansell et al., 2017). PFOR1 has previously been inhibited by a hammerhead ribozyme, showing that reduced expression of PFOR1 rendered Giardia more resistant towards MTZ and caused better growth in the presence of oxygen (Dan et al., 2000). Even if lower levels of PFOR1 gene expression have been linked to MTZ resistance, it has not yet been found to be significantly different between susceptible and resistant isolates, and thus not deemed a good marker of MTZ resistance (Begaydarova et al., 2015).
The NRs are essential metabolic enzymes that most likely have been acquired by lateral transfer from anaerobic bacteria or archaebacteria (Nixon et al., 2002; Muller and Muller, 2019). The two different NRs in Giardia, NR1 and NR2 are considered to be paralogs with different MTZ metabolizing capacities (Muller et al., 2013, 2015; Ansell et al., 2017). NR1 expression in MTZ-resistant laboratory Giardia lines has been found to be downregulated, whereas NR2 has been found to be upregulated (Leitsch, 2019). Further, E. coli expressing both NR1 and NR2 from Giardia has been shown not to be susceptible to MTZ, indicating that NR2's MTZ inactivating capabilities surpass NR1's activating ones (Muller et al., 2015).
There are currently no known genetic markers of MTZ resistance in Giardia, and genetic diversity represented by allele identification and characterization by cloning of the whole CDS has only been carried out for a handful of studies (Lasek-Nesselquist et al., 2009; Kosuwin et al., 2010; Siripattanapipong et al., 2011; Choy et al., 2015; Aguiar et al., 2016; Gabin-Garcia et al., 2017; Mizuno et al., 2020). Most studies of MTZ resistance in Giardia have examined cultivable and historical strains of sub-assemblage AI (WB) which rarely infect humans and may not be representative for sub-assemblage AII and assemblage B normally infecting humans (Caccio and Ryan, 2008). In these studies, MTZ tolerance has been induced by slowly increasing MTZ concentrations in growth medium, meaning they are not naturally resistant. Various potential adaptive mechanisms have been identified (Ansell et al., 2017; Müller et al., 2018; Muller et al., 2008). However, these adaptations seem to be lost after one en-/excystation cycle, indicating that such acquired resistance is not necessarily passed on from one infection to the next (Muller et al., 2008). Still, the rapid increase of treatment refractory Giardia infections, especially those seen over the last decade in South Asia, point towards heritable traits enabling Giardia to resist MTZ treatment.
The aim of this study was to analyze genetic variation and allelic composition of Giardia PFOR1 and 2 and NR1 and 2 in recently axenized and in non-cultured assemblage B clinical isolates. Some of the isolates were obtained from MTZ treatment refractory patients. The identified genetic variants were evaluated in relation to clinical information where this was available and for their potential effect on protein function.
2. Materials and methods
2.1. Giardia samples, purification, DNA extraction and PCR
Two different sets of Giardia clinical samples were used in the present study. One set represents trophozoite cultures of a Giardia biobank at the Robert Koch-Institute (RKI) in Berlin, with seven Giardia assemblage B isolates that were cultured as previously described (Keister, 1983). The other set represents DNA from non-cultured MTZ susceptible or MTZ refractory clinical samples of Giardia assemblage B infections collected from 2004 to 2017 at Haukeland University Hospital, Bergen, Norway. Trophozoites from the RKI culturable isolates were collected followed by DNA extraction and concentration measurements before Illumina whole genome sequencing was carried out according to (Saghaug et al., 2019). No clinical data were available for these Giardia isolates.
The cysts from the non-cultured isolates were obtained from 19 returning travelers. The cysts were purified from patient stool samples by sucrose flotation alone or in combination with immunomagnetic separation (IMS) (Dynabeads® G-C combo kit, Life technologies) according to (Hanevik et al., 2015) but omitting the final steps releasing cysts from beads. Cysts from a sample originating from the 2004 Bergen outbreak, VA, was obtained by salt flotation and IMS and DNA was extracted according to (Robertson et al., 2006).
DNA from the isolated Giardia cysts was extracted using the kit MagAttract® HMW DNA (Qiagen). Giardia cysts with beads from previous IMS steps, were frozen and thawed three times before adding proteinase K followed by AL buffer and RNase A from the MagAttract® kit. The samples were then incubated at 56 °C for 1 h according to (Ogren et al., 2016), followed by 2 freeze-thaw cycles before a final heat incubation at 98 °C for 15 min. Remaining beads and cyst wall debris were pelleted and removed by centrifugation at 11000xG for 2 min before following the manufacturer's instructions.
qPCR detection of Giardia was carried out for the 19 non-cultured, purified cyst samples with the Light Cycler 480 II instrument (Roche Diagnostics GmbH, Mannheim, Germany), according to (Yang et al., 2014), except for using a Giardia assemblage B-specific forward primer, FAM probe was used instead of Joe 670 and a standard curve using 10 fold dilutions of a quantified pUC57 based plasmid construct (Genscript, NJ, USA) containing the target sequence of glutamate dehydrogenase (gdh), accession number MT108431.1. The primers and probes are listed in Table 1.
Table 1.
Primer pairs for glutamate dehydrogenase qPCR, nitroreductase 1 and 2 genes, pyruvate ferredoxin oxidoreductase 1 and 2 amplification PCR and pyruvate ferredoxin sequencing primers. The gdh primers obtained from (Yang et al., 2014), with adapted assemblage B specific forward primer and a different quencher for the probe. The primers for the NR genes were designed using Geneious prime, and specificity was checked using the NCBI blast tool. Primer design was based on pre-and post-CDS conserved regions derived from eight previously Illumina sequenced cultured assemblage B isolates. For the NR1, two different primer sets were used in order to obtain positive PCR reactions for all isolates. All primers ordered from TIB Molbiol, Berlin, Germany.
| Gene | Direction | Primer Sequence (5’ – 3′) | Bp | [Tm]a | Position related to CDS | Amplicon length | Reference |
|---|---|---|---|---|---|---|---|
| gdh | Forward | GGGCAAGTCGGACAACGA | 18 | 61 | −434 | 262 bp |
(Yang et al., 2014) and this study |
| gdh | Reverse | GTCTACTTCCTGGAGGAGATGTGC | 24 | 62.2 | −696 | Yang et al., (2014) | |
|
gdh |
Probe |
6FAM-TCATGCGCTTCTGCCAG-BBQ |
17 |
62.3 |
−454 |
Yang et al., (2014) |
|
| NR1 | Forward | GTGATGGAGCAAAGTCGC | 18 | 64 °C | −162 | 1135 bp |
This study |
| NR1 | Reverse1 | GTGGATGGGGCTCTTGAATA | 20 | 64 °C | +178 | This study | |
|
NR1 |
Reverse2 |
GTGGATGAGGCTCTTGAATA |
20 |
61 °C |
+178 |
This study |
|
| NR2.1 | Forward | ATCTACATAAGATCCGCGCACT | 22 | 65 °C | −174 | 1212 bp | This study |
| NR2.1 | Reverse | TACTCTGCACCTCATCGCCG | 20 | 69 °C | +171 | This study | |
| NR2.2 | Forward | GACTCACAGAGTGGCAACGA | 20 | 67 °C | −251 | 1271 bp |
This study |
|
NR2.2 |
Reverse |
CGCCGAGCAATGTAGTGGTT |
20 |
68 °C |
+156 |
This study |
|
| PFOR1 | Forward | CACAGTCCCCAATCACAGAC | 20 | 59 °C | −143 | 3987 bp | This study |
| PFOR1 | Reverse | GGAGGACATGGAGAGCAAGG | 20 | 61 °C | +82 | This study | |
| PFOR1 | Sequencing1 | TGCCAAGTGGAGGAGCGAAA | 20 | 59 °C | NA | NA | This study |
| PFOR1 | Sequencing2 | AGCACACGCCACATCGAG | 18 | 58 °C | NA | NA | This study |
| PFOR1 | Sequencing3 | GACAAGTACGTCAAGGACATCA | 22 | 58 °C | NA | NA | This study |
|
PFOR1 |
Sequencing4 |
CACTGAGAGCTGCAACCTC |
19 |
58 °C |
NA |
NA |
This study |
| PFOR2 | Forward | CGCATAATAGCTTTGACCGTT | 21 | 55 °C | −66 | 4018 bp | This study |
| PFOR2 | Reverse | TTCCAGCTTCTGTCGCTAC | 19 | 56 °C | +152 | This study | |
| PFOR2 | Sequencing1 | TCCGATCTGGATGCAGGC | 18 | 58 °C | NA | NA | This study |
| PFOR2 | Sequencing2 | GTCTCTTCTTCGGAATGGGATCC | 23 | 62 °C | NA | NA | This study |
| PFOR2 | Sequencing3 | AAGGTCGTCAACATGAACCTTG | 22 | 58 °C | NA | NA | This study |
| PFOR2 | Sequencing4 | TGCCAGGTCTGCTCCCAG | 18 | 61 °C | NA | NA | This study |
NA= Not applicable.
Tm is estimated from NEB Tm calculatorwith 400 nM primer concentration and Q5® polymerase.
Isolates with ∼900 gene copies per μl sample, or higher, were selected for downstream experiments, resulting in a total of eight samples. The eight samples included one sample from an individual where treatment result was not known, three samples from individuals clinically MTZ susceptible and four from MTZ refractory individuals. Sample overview can be found in the Supplementary Table 1. Samples with fractionated DNA, contaminants, inhibitors or low concentrations of Giardia DNA were not included in the analysis.
2.2. Cloning the genes
Genomic Giardia DNA was used in polymerase chain reactions (PCR) for obtaining the PFOR1, PFOR2, NR1 and NR2 genes. All gene-PCRs were carried out using the Q5® High-Fidelity DNA Polymerase (catalog nr M0491L, New England BioLabs (NEB) Ipswich, MA, USA) and reaction setup for 25 μl according to manufacturer. Negative controls with nuclease free water were included for each experiment. Primers used to obtain full length genes are shown in Table 1. The gene names of the NRs in the current study are based on two publications by Müller et al. from 2007 to 2013, where NR1 (GSB_22677) is also known as GlNR1 or Fd-NR2 in the GiardiaDB database, while NR2 (GSB_153178) is also known as GlNR2 or Fd-NR1 (Muller et al., 2007, 2013).
The following PCR conditions were used: an initial denaturation of 98 °C for 30 s, 35–40 cycles of 98 °C for 10 s, 62–64 °C for 20 s and 72 °C for 31 s, followed by a final extension of 72 °C for 2 min. 20 μl PCR products were run on a 1% agarose gel pre-stained with GelRed® Nucleic Acid Stain (catalog nr: 41003, Biotium, San Francisco, CA, USA) and positive bands were excised from the gel using a LED-based imaging system: iBright (Thermo Fisher Scientific, A44240). The PCR products were purified from the agarose gel using the Wizard® SV Gel and PCR Clean-Up System (Promega, A9282), following the manufacturer's instructions, with the exception of using 70 °C nuclease free water for the final elution. The concentrations of purified PCR products were measured using NanoDrop.
Cloning methods have been described previously (Saghaug et al., 2020), with the only exception that Escherichia coli DH5α competent cells (cat nr: C2987U, NEB) were used for the transformation. Vector inserts were Sanger sequenced by Genewiz (Leipzig, Germany).
2.3. Data analysis of sequences and single nucleotide variation
The reference genome files of G. lamblia assemblage B (GS_B version 26, AHHH) were downloaded from Giardiadb.org (Aurrecoechea et al., 2009) and imported into the genome analysis software Geneious Prime ® 2020 (Biomatter Ltd., version 2.4, Auckland, New Zealand). One FASTA-file and one general feature format (GFF) file were combined to achieve annotated reference genomes. Illumina sequenced isolates (7 previously described cultured assemblage B isolates (Saghaug et al., 2019)) were imported into Geneious and used to validate corresponding cloned sequences. Forward and reverse sequenced gene chromatograms from clones were aligned and compared to the reference genes of interest, PFOR1; GSB_114609, PFOR2; GSB_17063, NR1; GSB_22677 (Fd-NR2) and NR2; GSB_153178 (Fd-NR1).
For cultured, previously Illumina sequenced, isolates we validated the SNVs in the same way as described before (Saghaug et al., 2020), whereas in clinical non-cultured isolates we considered any SNV present in more than one of all clone sequences from any isolate as valid. The consensus sequences from all the clones from one isolate were aligned together and sorted into excel files with SNV positions inspired by (Lecova et al., 2019), and then sorted into haplotypes. Phylogenetic trees of the genes were made in Geneious using the Tamura-Nei with method UPGMA.
Haplotype hybrids, better known as chimeric products created during PCR amplification (Haas et al., 2011), were identified using the programs Bellerophon (Huber et al., 2004), Geneious and Excel. The consensus sequences from all the clones from one isolate were aligned and sorted into unique alleles.
2.4. Analysis of amino acid changes
The universal resource databank, Uniprot.org, was used to locate the ferredoxin (fd) domains. Single nucleotide variants (SNVs) were identified in Geneious. Our analysis included amino acid (aa) changes that could lead to potential rearrangements of the secondary structure, truncation of the proteins (Grantham, 1974), or aa changes located in the ferredoxin (fd) domains or in close proximity to the domain.
Open reading frames of NR1 (GSB_22677) and NR2 (GSB_153178) genes were modeled with simplified AlphaFold 2 (Jumper et al., 2021) using Google Colab implementation (colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb), and visualized in Chimera (Pettersen et al., 2004).
3. Results
3.1. Genetic variation in clinical isolates of Giardia
The PFOR1, PFOR2, NR1 and NR2 full length genes from up to 15 recent clinical Giardia lamblia assemblage B isolates were successfully amplified and cloned into appropriate bacterial vector system and sequenced for characterization of single nucleotide variants (SNVs) and characterization of allelic variation. Two of the isolates failed to produce PCR products for the PFOR genes (VA and Ag10). The overall genetic variation, defined as the total number of combined SNV positions for all isolates per gene length, was similar for the NR genes: NR1 (10.9%), and NR2 (10.7%), while a little lower for the PFORs; 6.4% for PFOR1 and 8.1% for PFOR2. The percentage of total SNV positions for individual isolates was an average of 3.5% (0.3–5.9%) for NR1, 2.6% (0.7–5.1%) for NR2, 0.2%(0–0.4%) for PFOR1 and 0.2% (0–0.4%) for PFOR2. However, the higher percentage found in NR1 is due to exceptionally high numbers of SNVs (>40) in six isolates compared to two isolates for NR2. The percentages of nsSNV per CDS were found to be slightly less for the NR1 gene, 4.2% with 33 variable positions compared to 6.4% and 57 positions for the NR2 gene (Table 2 and Supplementary Tables 3 and 4). Both PFOR genes had lower numbers of nsSNVs per gene length, where PFOR1 was found to have 1.1% with 43 variable positions and PFOR2 had 1.6% with 59 variable positions.
Table 2.
Overview of NR1, NR2, PFOR1 and PFOR2 single nucleotide variants (SNVs) relative to the assemblage B reference genome GS_B (AHHH) and alleles.
| Gene | Gene length (bp) | Number of SNV positions | SNVs per gene length (%) | Number of nsSNV positions | nsSNVs per gene length (%) | Distinct alleles found | Alleles leading to truncated proteins |
|---|---|---|---|---|---|---|---|
| NR1 | 795 | 87 | 10.9 | 33 | 4.2 | 69 | 1a |
| NR2 | 884 | 95 | 10.7 | 57 | 6.4 | 90 | 7b |
| PFOR1 | 3762 | 239 | 6.4 | 43 | 1.1 | 42 | 0 |
| PFOR2 | 3600 | 293 | 8.1 | 59 | 1.6 | 70 | 0 |
Nonsense mutation in one sample's alleles.
Single nucleotide deletions causing frameshifts and nonsense mutations in three Giardia samples.
Table 3, Table 4 show that the number of SNVs per isolate in Giardia assemblage B NR and PFOR genes of clinical samples are variable, ranging from as low as two SNVs up to as many as 46 SNVs for the NRs and 136 for the much larger PFOR genes. SNVs and nsSNVs in the ferredoxin (fd) domains of NR1, NR2, PFOR1 and PFOR2 and number of identified alleles are presented in Table 3, Table 4 The NR1 fd domains reside at positions 10–99 and 106–195, while the NR2 fd domains are found at position 37–126 and 130–222. The fd domains of PFOR1 are located at positions 715–749 and 778–807, while the fd domains of PFOR2 are located at positions 697–726 and 756–790. Both cultured and non-cultured sample groups were found to contain a spectrum in the number of variants, from highly variable (reflecting potentially mixed infections), to homozygous samples. The degree of variability in NR1 was associated with similar variability in NR2. For the NR1 gene, the average number of nsSNVs per CDS length and number of nsSNVs in the fd domain were similar in the cultured and non-cultured isolates (5.3 vs 5.1 and 1.71 vs 1.63). For the NR2 gene, a higher number of nsSNVs per CDS length and number of nsSNVs in the fd region was found in the non-cultured isolates compared to the cultured ones (14.3 vs 11.4 and 2.5 vs 1.5). The average numbers of nsSNVs for the non-cultured isolates of PFOR1 and PFOR2 were higher than for the cultured isolates (10.6 vs 4.1 and 6.4 vs 9.7).
Table 3.
Number of validated cloned sequences, single nucleotide variants and alleles of NR1 and NR2 genes in Giardia assemblage B isolates.
| Nitroreductase 1 |
Nitroreductase 2 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sequenced clonesa | SNVs | nsSNVs | nsSNVs fd | Alleles | Sequenced clonesa | SNVs | nsSNVs | nsSNVs fd | Alleles | |
| Cultured samples, no clinical data available | ||||||||||
| P344 | 29 | 46 | 12 | 3 | 21 | 19 | 45 | 31 | 5 | 14 |
| P387 | 26 | 42 | 9 | 4 | 8 | 14 | 31 | 16 | 2 | 5 |
| P413 | 20 | 15 | 4 | 1 | 1 | 20 | 9 | 6 | 1 | 1 |
| P424 | 19 | 14 | 3 | 1 | 1 | 20 | 9 | 6 | 1 | 1 |
| P427 | 28 | 44 | 9 | 3 | 9 | 15 | 35 | 17 | 5 | 8 |
| P433 | 19 | 2 | 0 | 0 | 3 | 16 | 6 | 2 | 0 | 4 |
| P458 | 24 | 3 | 0 | 0 | 4 | 20 | 6 | 2 | 0 | 3 |
| Fecal sample, no clinical data available | ||||||||||
| 0099 | 21 | 42 | 10 | 2 | 5 | 15 | 41 | 25 | 3 | 4 |
| Fecal samples, clinically susceptible | ||||||||||
| Ag30 | 16 | 18 | 1 | 1 | 1 | 18 | 6 | 2 | 0 | 1 |
| VA | 20 | 23 | 3 | 1 | 1 | 19 | 16 | 12 | 4b | 1 |
| Ag15 | 6 | 31 | 4 | 1 | 2 | 16 | 29 | 16 | 3b | 3 |
| Fecal samples, clinically refractory | ||||||||||
| Ag10 | 16 | 42 | 8 | 2 | 4 | 14 | 28 | 13 | 2 | 6 |
| Ag13 | 15 | 44 | 8 | 3 | 6 | 16 | 25 | 15 | 3 | 9 |
| Ag20 | 8 | 25 | 2 | 1 | 2 | 6 | 19 | 10 | 2 | 3 |
| Ag22 | 19 | 28 | 6 | 2 | 3 | 17 | 37 | 21 | 3 | 7 |
The number of sequenced clones corresponds to the number of clones obtained after removal of chimeras (see Supplementary Table 2 for the total number of clones obtained).
Single nucleotide deletions are included in the number.
Table 4.
Number of validated cloned sequences, single nucleotide variants and alleles of PFOR1 and PFOR2 genes in Giardia assemblage B isolates.
| Sequenced clonesa | Pyruvate ferredoxin oxidoreductase 1 |
Pyruvate ferredoxin oxidoreductase 2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNVs | nsSNVs | nsSNVs fd | Alleles | Sequenced clonesa | SNVs | nsSNVs | nsSNVs fd | Alleles | ||
| Cultured samples, no clinical data available | ||||||||||
| P344 | 6 | 52 | 8 | 4 | 20 | 117 | 15 | 1 | 4 | |
| P387 | 13 | 115 | 16 | 5 | 19 | 123 | 10 | 1 | 14 | |
| P413 | 15 | 11 | 1 | 1 | 16 | 35 | 10 | 7 | ||
| P424 | 19 | 11 | 1 | 1 | 14 | 26 | 1 | 1 | ||
| P427 | 15 | 11 | 1 | 1 | 14 | 119 | 9 | 8 | ||
| P433 | 15 | 12 | 2 | 2 | 20 | 0 | 0 | 1 | ||
| P458 | 12 | 4 | 0 | 3 | 15 | 0 | 0 | 1 | ||
| Fecal sample, no clinical data available | ||||||||||
| 0099 | 8 | 114 | 13 | 7 | 6 | 119 | 13 | 4 | ||
| Fecal samples, clinically susceptible | ||||||||||
| Ag30 | 7 | 36 | 5 | 3 | 7 | 33 | 4 | 4 | ||
| VA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| Ag15 | 11 | 103 | 10 | 5 | 11 | 115 | 8 | 2 | 6 | |
| Fecal samples, clinically refractory | ||||||||||
| Ag10 | NA | NA | NA | NA | NA | NA | NA | NA | ||
| Ag13 | 11 | 88 | 10 | 3 | 8 | 125 | 13 | 1 | 6 | |
| Ag20 | 4 | 97 | 14 | 4 | 16 | 115 | 8 | 5 | ||
| Ag22 | 10 | 72 | 12 | 7 | 13 | 129 | 12 | 11 | ||
NA=Not available seuences due to not enough DNA or failure to produce PCR products from samples.
No chimera detected for the two PFOR genes.
3.2. Alleles in the nitroreductase 1 and 2 genes
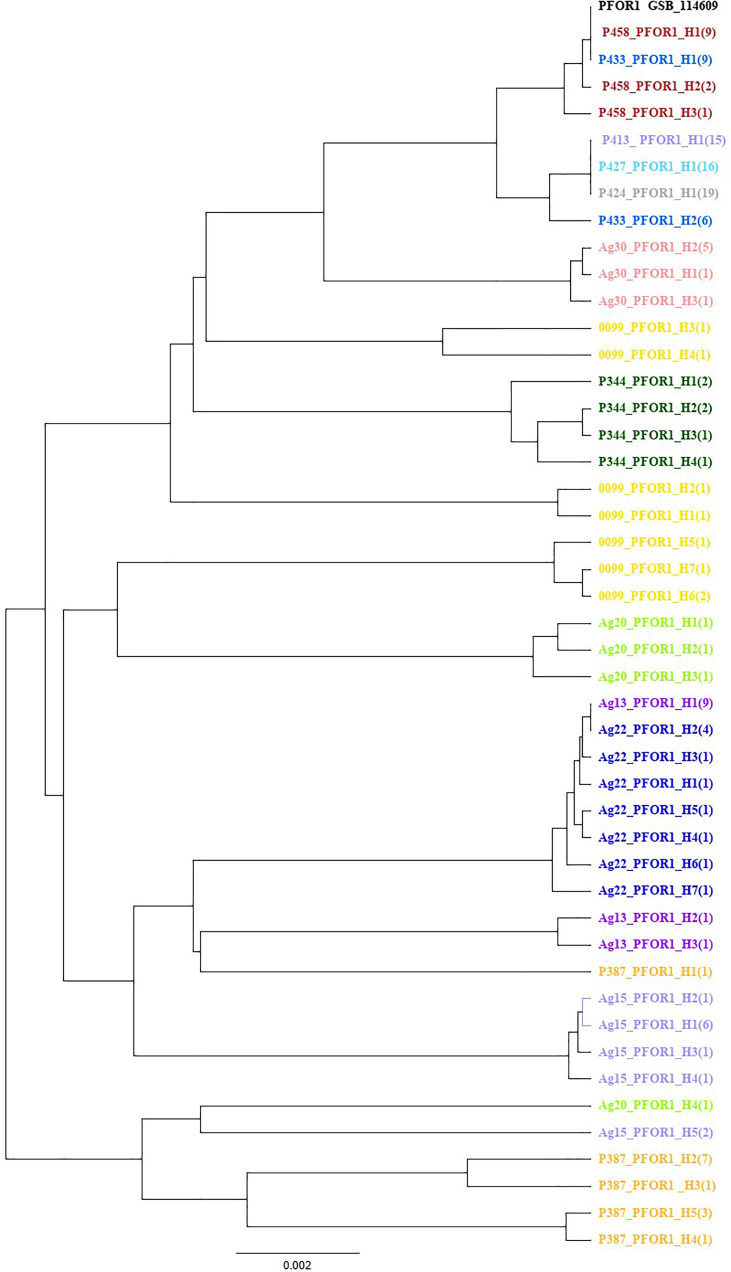
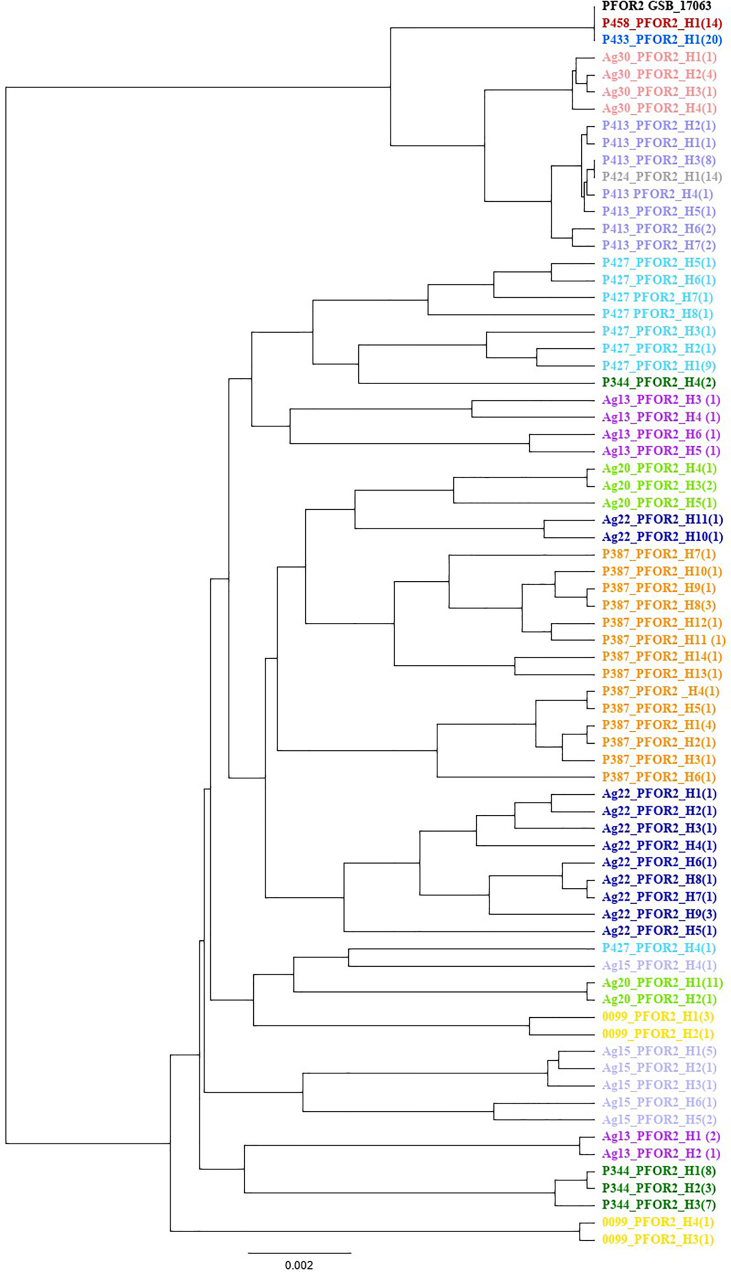
A total of 69 different alleles were found in the NR1 gene, while 90 different alleles were found in the NR2 gene in all 15 isolates of Giardia analyzed in the present study (Table 3). We found a total of 42 alleles for the PFOR1 gene and 70 alleles for the PFOR2 gene in 13 isolates. A total of three isolates (P413, P424 and P427) were found to be homozygous for PFOR1 (see Table 4). One isolate was homozygous for both PFOR genes (P424), while the two isolates P433 and P58 were homozygous for PFOR2. Four of the isolates were homozygous at both NR genes:P413, P424, VA and Ag30, Table 3. Chimeric sequences occurred for the NR genes in most of the heterozygous samples. 33 chimeric NR1 sequences were found in nine samples (range 0%–26%), and 27 NR2 chimeric sequences in ten samples (range 4%–29%) (see Supplementary Table 2). Chimeric sequences were removed from the main analysis. Even after removal of chimeras in the NR genes, the number of distinct alleles and genetic variation remained high in three cultured isolates, P344, P427 and P387, and also in the non-cultured Ag10, Ag13 and 0099 (see Table 3, Supplementary Table 2 and supplementary text 1). The number of distinct alleles was higher in the PFOR2 gene compared to the PFOR1 gene (see Table 4). With the number of distinct alleles exceeding four for at least one of the genes, these were considered to be possibly isolates from infections by more than one strain. For the isolates with 2–4 alleles, we found allele distributions compatible with all possible combinations (1:3, 2:2, 1:2:1, 1:1:1:1).
For the NR1 gene, only two alleles were found to be present in more than one of the cultured isolates. The two isolates P433 and P458 had two alleles in common: one of them being identical to the reference NR1 gene sequence, GSB_22677. No shared alleles were found in any of the non-cultured isolates.
One NR2 allele was found to be present in three cultured isolates (P458, P344 and P433), while five other NR2 alleles were found to be present in two of the isolates examined (see Supplementary Figs. S1 and S2 for allele distribution in phylogenetic trees). One of these alleles was found in the Ag13 isolate from a treatment refractory case and in the Ag15 isolate from a treatment-susceptible case. Another of the shared alleles was found in both a cultured isolate (P427) and a non-cultured isolate from a treatment-susceptible case (VA).
Some of the alleles from the cultured clinical isolates of Giardia were found in more than one isolate. For PFOR1 the two isolates P458 and P433 had the same allele as the reference sequence GSB_114609. One other allele was found to be present in the three isolates P413, P427 and P424. Two of the MTZ refractory isolates, Ag13 and Ag22 was also found to have the same PFOR1 allele. For PFOR2 the two isolates P458 and P433 were found to have the same allele as the GSB_17063 reference. One other allele was found in the two clinical isolates P413 and P424 (see Supplementary Figs. S3 and S4).
3.3. Evaluating the putative nsSNV effects on the protein function
The number of nsSNVs found in each gene for every isolate is listed in Table 2, while the nucleotide positions and changes in the fd domain and changes causing potentially important alterations are listed in Table 5, Table 6. The rest of the SNVs, nsSNVs and deletions are presented in the Supplementary Tables 3 and 4 In NR1 a common nsSNV at codon 264 leading to a change from the basic amino acid lysine (K) to arginine (R) was found in eight of the 15 isolates (see Table 5) A nsSNV at codon 8 resulted in the large and bulky phenylalanine (F) being changed to the hydrophobic leucine (L) in the fd domain of three isolates (see Table 5). Further, a nsSNV change from the acidic aspartic acid (D) to the smallest amino acid glycine (G) at codon 101 was identified in several of the isolates from all three groups (cultured, and non-cultured isolates from MTZ susceptible and refractory cases). Isolate Ag13 was found to have one allele with a nonsense mutation at codon 195. This caused a truncated, most likely dysfunctional NR1 protein, encoded by one of the alleles of this isolate (represented by 6 out of 15 cloned sequences).
Table 5.
Non-synonymous SNVs causing potentially important alterations in NR1, including all that were found in the ferredoxin domains.
| Position Nucl. ref | 10 |
11 |
17 |
22 |
40/42 |
59 |
76 |
88 |
91 |
145 |
173 |
302 |
583 |
786 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | G | C | T | G-G | T | A | G | G | G | G | A | C | C | |
| Cultured samples, no clinical data available | ||||||||||||||
| P344 | A/C | G/A | A/G | A/G | C/T | |||||||||
| P387 | C/G | G/A | A/G | A/G | A/G | C/T | ||||||||
| P413 | C | |||||||||||||
| P424 | C | |||||||||||||
| P427 | C/G | G/A | A/G | A/G | C/T | |||||||||
| P433 | ||||||||||||||
| P458 | ||||||||||||||
| Fecal sample, no clinical data available | ||||||||||||||
| 0099 | A-A/G-G | A/G | G | C/T | ||||||||||
| Fecal samples, clinically susceptible | ||||||||||||||
| Ag30 | C | |||||||||||||
| VA | A | G | T | |||||||||||
| Ag15 | A/G | G | C/T | |||||||||||
| Fecal samples, clinically refractory | ||||||||||||||
| Ag10 | A/G | A | G | C/T | ||||||||||
| Ag13 | G/A | A/G | A/G | A/G | C/T | C/T | ||||||||
| Ag20 | A | |||||||||||||
| Ag22 |
C/T |
A/G |
A/G |
|||||||||||
| Amino acid change (codon position) | G > S (4) | G > A (4) | P > Q (6) | F > L (8) | V > I (14) | V > A (20) | M > V (26) | V > M (30) | D > N (31) | G > R (49) | S > N (57) | D > G (101) | Q >* (195) | K > R(264) |
Table 6.
Non-synonymous SNVs causing potentially important alterations in NR2, including all that were found in the ferredoxin domains.
| Position Nucl. ref | 42 |
52 |
99 |
123 |
124 |
126 |
131 |
156 |
169 |
184 |
220 |
381 |
565 |
751 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | G | C | G | A | G | G | A | T | T | A | C | A | T | |
| Cultured samples, no clinical data available | ||||||||||||||
| P344 | A/G | T/G | G/A | G/C | C/T/A | A/G | T/C | |||||||
| P387 | G/A | C/A | A/G | C | ||||||||||
| P413 | G | |||||||||||||
| P424 | G | |||||||||||||
| P427 | Del | G | A/G | C | C | G/A | A/G | C | ||||||
| P433 | T/C | |||||||||||||
| P458 | T/C | |||||||||||||
| Fecal sample, no clinical data available | ||||||||||||||
| 0099 | . | . | . | . | G/A | G/C | C/T/A | C | ||||||
| Fecal samples, clinically susceptible | ||||||||||||||
| Ag30 | C | |||||||||||||
| VA | Del | G | A | C | G | C | ||||||||
| Ag15 | Del | G | C | Del | G | C | ||||||||
| Fecal samples, clinically refractory | ||||||||||||||
| Ag10 | G/A | C | G | C | ||||||||||
| Ag13 | G | C | C/T | A/G | C | |||||||||
| Ag20 | G | C | A/G | C | ||||||||||
| Ag22 |
G |
G/C. |
C/T |
A/G |
C |
|||||||||
| Amino acid change (codon position) | (14) | D > N (18) | (33) | K > N (41) | K > E (42) | K > Da (42) | R > K (44) | E > D (52) | C > Rb (57) | S > P (62) | I > V (74) | (127) | R > G(189) | S > P(251) |
Combination of two nsSNVs with a G at position 124 and a C at position 126 changes K to D.
Cysteine responsible for making Fe–S clusters.
For NR2, most of the samples had the nucleotide G at position 124 in the fd domain translating to glutamic acid (E) instead of the basic lysine (K) by the respective codon of the reference (see Table 6). The larger and basic arginine (R) was changed to the small glycine (G) due to an SNV in codon 189 for all three groups of samples and the aliphatic amino acid serine (S) was replaced with the ring structured proline (P) at codon position 251 in 13 out of 15 isolates. A total of seven distinct NR2 alleles had three different single nucleotide deletions causing a frameshift mutation resulting in a nonsense mutation, hence, truncated versions of NR2. A nonsense mutation at position 33, was found in two isolates: the MTZ susceptible non-cultured isolate VA and the cultured and potentially non-clonal isolate P427 (Table 6). For the homozygous VA isolate, this means all NR2 proteins most likely would be dysfunctional. Stop codons were also found at two different positions codon 14 and 127, in two of the three alleles of the isolate Ag15, from a MTZ-susceptible case, potentially causing dysfunctionality in the majority of NR2 proteins in Ag15 (see Supplementary Table 4). In two of the alleles of NR2 in the isolate from a MTZ-refractory case, Ag13, a nsSNV in the fd domain caused a switch from cysteine (C) to arginine (R) at codon position 57, likely disrupting this protein variant's ability to stably bind an iron-sulfur (Fe–S) cluster at this site.
For the PFOR genes, A total of four amino acid changes were found in the fd domains of the PFOR2 gene (see Table 7). The first one at codon position 241 a switch from Lysine (K) to arginine (R) where both amino acids have positively charged side chains were found in two alleles from the non-cultured isolate Ag13. In the cultured isolate P344 one allele was found to have a switch at codon 255 from the small amino acid glycine (G) to the other small amino acid cysteine (C). One allele from the MTZ-susceptible isolate Ag15 was found to have a switch from the negatively charged aspartic acid (D) to the uncharged asparagine at codon 262. Finally, a switch from the hydrophobic alanine (A) to the polar and uncharged threonine at codon 264 was found in seven alleles of the cultured isolate P387. The nsSNVs outside of the fd domains were mostly conservative, meaning that important properties of the substitute amino acids were not changed (see Supplementary Tables 5 and 6 for overview).
Table 7.
Non-synonymous SNVs causing potentially important alterations in PFOR2 in the ferredoxin domains.
| Position Nucl. ref | 722 |
763 |
784 |
790 |
|---|---|---|---|---|
| A | G | G | G | |
| Cultured samples, no clinical data available | ||||
| P344 | G/T | |||
| P387 | G/A | |||
| P413 | ||||
| P424 | ||||
| P427 | ||||
| P433 | ||||
| P458 | ||||
| Fecal sample, no clinical data available | ||||
| 0099 | . | . | . | . |
| Fecal samples, clinically susceptible | ||||
| Ag30 | ||||
| VA | ||||
| Ag15 | G/A | |||
| Fecal samples, clinically refractory | ||||
| Ag10 | ||||
| Ag13 | G/A | |||
| Ag20 | ||||
| Ag22 |
||||
| Amino acid change (codon position) | K > R(241) | G > C (255) | D > N (262) | A > T (264) |
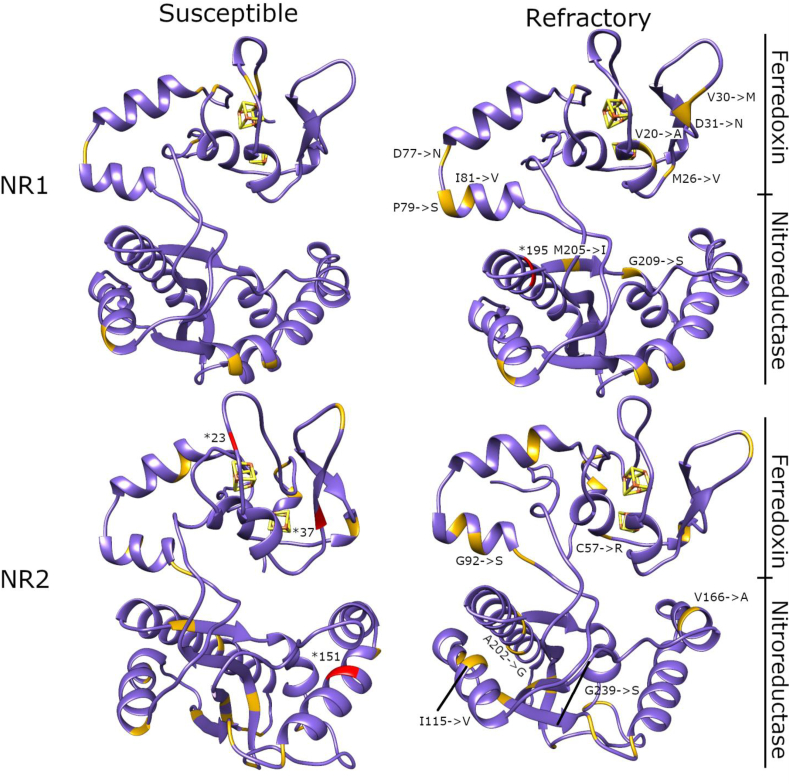
To gain further insight into the potential importance of different mutations and their locations in the NR1 and NR2 subdomains, we constructed models using AlphaFold 2 and mapped the identified nsSNV in these (Fig. 1). Giardia NR1 and 2 have a unique domain organization with an fd domain and NR domain together. No structural homologs exist with the same domain organization, based on a sequence search in Protein Data Bank. Some segments within the models score high, and these are interspersed with low scoring segments.
Fig. 1.
Cartoon presentations of NR1 and NR2 homology models. The ferredoxin (fd) domain is above, and iron-sulfur cluster is shown as a red-yellow cage within the fd domain. The larger NR domain is below. nsSNVs identified in the present study are depicted with orange color. Red color indicates nonsense mutations at positions 23, 37 and 151 from susceptible NR2 isolates, and a nonsense mutation leading to a premature stop codon at position 195 from a refractory NR1 isolate (positions highlighted by *). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
First, we compared our homology model to other structurally related NRs and fd containing proteins using the DALI server (Holm, 2020). Structural sequence alignments showed that both subdomains had conserved residues, and these were also retained in our homology models. This suggests that nsSNVs do not obstruct activity of NR1 and NR2. nsSNVs unique to clinical isolates from treatment refractory cases in NR1 were mostly found in the fd domain (Fig. 1). Mutations in NR2 refractory and susceptible samples are mostly in overlapping positions. Unlike in NR1, mutations in NR2 are spatially more distributed along the protein, and we found only one mutation in the fd domain.
4. Discussion
In the present study, we were able to investigate in a select collection of non-cultured clinical samples of both, MTZ susceptible and refractory Giardia assemblage B isolates, and axenized isolates derived by limiting dilution, the genetic variation and allelic diversity of the four full length MTZ-metabolism genes, NR1,NR2, PFOR1 and PFOR2.
4.1. Genetic variation and alleles of the genes
For some of the non-cultured isolates, such as Ag20 and Ag15, we obtained a limited number of sequences for both of the NR genes. The number of alleles found in these two isolates must be interpreted with caution.
The number of distinct alleles and genetic variation was high in the NR genes for three of the cultured isolates, P344, P427 and P387 and also in the non-cultured samples Ag10 and Ag13 (Table 3). The number of distinct alleles was also high for PFOR genes where P387, Ag15, Ag22 had more than four alleles. The presence a high number of distinct alleles could indicate that these isolates potentially could represent non-clonal isolates. This is puzzling since the cultured isolates were originally derived by limiting dilution. There is a possibility that they are co-existing lineages with some interdependency, or some new mutations could have occurred to adapt to in vitro culture, or a combination of both. For the isolate with the highest number of distinct alleles for NR1, P344, a total of 36 clones were sequenced and 21 alleles were found, while 20 clones were obtained for NR2 resulting in 14 alleles (see Supplementary Table 2). For the PFOR2 gene from the isolate P387 19 clones were sequenced and a total of 14 distinct alleles were identified. Several studies have concluded that the presence of more than four alleles in single isolates most likely are due to mixed infections or perhaps intragenic recombination (Lalle et al., 2005; Kosuwin et al., 2010; Siripattanapipong et al., 2011; Waldram et al., 2017; Lecova et al., 2019). This may happen if an infected host is subsequently reinfected, with a different Giardia strain (Sprong et al., 2009). An isolate may still originate from one clone but be unstable and generate many variants. While the number of isolates examined in this study is small, especially for the PFOR genes of non-cultured isolates, we nevertheless noted that only one of the four isolates, Ag22 for the PFOR1 gene, from treatment-refractory cases was homozygous, and two of the non-cultured MTZ susceptible samples were homozygous for the NR1 and NR2 loci. One of the samples that were homogenous for both of the NR genes, VA, is a sample obtained from the Bergen outbreak in 2004. A previous study investigated the sequences of glutamate dehydrogenase (gdh) and beta-giardin (Robertson et al., 2007). It was observed that some of the gene sequences found to be non-dominant in the beginning of the study were predominating at a later time point. The authors suggest that these sequences may be associated with a higher potential of transmission including virulence, environmental resistance, increased proliferation, or even a combination of factors.
Whether homozygous or heterozygous NR and PFOR alleles may decrease or increase resilience against MTZ, should be assessed in further studies.
4.2. Putative effect of SNV induced mutations
Several enzymes are involved in the activation of MTZ, and several metabolic systems may be involved in protection against the reactive toxic metabolites and contribute to resistance (Leitsch, 2019). The MTZ resistance mechanisms in Giardia is likely pleiotropic and may be a combination of several aspects such as a resistance mechanism where MTZ-activating enzymes become downregulated, or MTZ inactivating enzymes become up-regulated, and/or genetic variation and post-transcriptional changes (Ansell et al., 2015, 2017; Leitsch, 2017; Müller et al., 2018). Possibly, different Giardia strains may evolve distinct resistance strategies that result from combinations of these mechanisms.
There are at least three different enzymes, NR1, PFOR1 (possibly PFOR2) and thioredoxin reductase that contribute to the activation of MTZ in Giardia, whereas only one enzyme, NR2, has been associated with inactivation. If the parasite is either producing less NR2 (downregulation of the gene itself) or the NR2 is dysfunctional, the parasite would most likely be more susceptible to MTZ. In the non-cultured samples for which clinical information was available for all patients, we found two susceptible samples with frameshift mutations, leading to potentially truncated and dysfunctional NR2 proteins. Interestingly, one of the susceptible isolates (VA) was homozygous, indicating viability without NR2 function. There may be redundancy in NR2 function, allowing higher tolerance for genetic variability.
Mutations that would make the NR1 enzyme less efficient at activating MTZ or dysfunctional, could lead to reduced levels of toxic metabolites of the drug, and make the parasite more tolerant to MTZ. A study by Ansell et al., (2017) described the same nonsense mutation at nucleotide position 583 (codon position 195) as identified in our study. This mutation was found in some transcripts of NR1 in the laboratory induced MTZ-resistant isolate, 106-MtzR (Ansell et al., 2017). Further, two studies have reported that NR1 was downregulated in three resistant Giardia strains (Ansell et al., 2017; Emery et al., 2018), supporting the notion that a dysfunctional NR1 could be protective against MTZ, as seen in the refractory Ag13 sample in the present study. Indeed, mutational inactivation of a gene coding for a NR, rdxA, in Helicobacter pylori has previously been associated with resistance towards MTZ (Goodwin et al., 1998; Jenks and Edwards, 2002). Additionally, mutations and a nonsense mutation have been found in the two NR genes ntr4TV and ntr6TV of MTZ resistant T. vaginalis (Paulish-Miller et al., 2014). Nonsense mutations should therefore be further explored in samples from clinically MTZ-refractory Giardia infections to assess whether these mutations could be potential resistance markers.
It has been observed that laboratory strains of WB Giardia may lose its resistance during one en-/excystation cycle (Muller et al., 2008). This loss of resistance was found be relevant for the two drugs MTZ and nitazoxanide, and could potentially mean that resistance may not be carried through generations. Still, the authors debate whether this could be due to factors such as acquired mutations rendering the cells unable to encyst or that encystation may have eliminated the resistant population or selective pressure may have affected the resistant cells in a negative manner. This may as well mean that the drug resistant cells were not able to grow properly in the new environment after encystation. Loss of resistance during cell cycles may be linked to epigenetic and post-transcriptional changes than specific mutations being responsible for this temporary resistance pattern (Loderstädt and Frickmann, 2021). MTZ is most likely activated through several different pathways and will therefore exhibit a pleiotropic mode of action (Holmes et al., 2016). The resistance could also manifest differently in laboratory strains than in clinical isolates of Giardia. The mutations and deletions found in our study should therefore be tested in vivo to properly understand whether potential markers of resistance may be hereditary. It is clear that further research is needed for understanding how Giardia may pass on resistance.
Both NR and PFOR proteins contain a fd domain. The fd domain contains cysteine residues with iron-sulfur (Fe–S) cluster forming abilities which are part of redox-active centers responsible for biological electron transport (Johnson et al., 2005; Ansell et al., 2017). One of the cysteines in NR2 was changed to an arginine in the fd domain in the refractory isolate Ag13. However, this refractory isolate had a total of nine distinct alleles, probably due to a mixed infection, making the effect of this mutation difficult to interpret. In the PFOR2 an arginine was found to be switched to a cysteine at codon 255 in the cultured clinical isolate P344. This extra cysteine could potentially affect the electron transfer abilities of the domain. We also observed a switch from the negatively charged aspartic acid (D) to the uncharged asparagine at codon 262 for the MTZ-susceptible isolate Ag15, which potentially could influence the domain and even so the MTZ metabolizing capacity of the PFOR2. One other switch that could have impact on the fd domain of PFOR2 is the change of the hydrophobic alanine (A) to the polar and uncharged threonine at codon 264 found in alleles of the cultured isolate P387. Further research is needed to interpret how amino acid changes within the fd domains potentially can alter the function and MTZ metabolizing capacities of the proteins.
4.3. Strengths and limitations
A strength of the current study is the inclusion of non-cultured Giardia samples from relatively recent clinical patient samples. The reference strains used in most other laboratory MTZ resistance studies were isolated decades ago. The MTZ tolerance slowly induced in these isolates may not reflect the mechanisms behind the increase in MTZ treatment refractory infections observed in recent years (Munoz Gutierrez et al., 2013; Nabarro et al., 2015; Cañete et al., 2020).
For the three genes, NR2, PFOR1 ane PFOR2, we found presence of more nsSNVs in the non-cultured isolates compared to the cultured ones (see Table 3, Table 4). We cannot exclude that this could be due to mixed infections as they were not subject to the same limiting dilution procedures done for the cultured isolates.
Since no specific resistance mechanism has yet been discovered in Giardia, and treatment failure can result not only from resistant isolates, but also immune deficiency, poor treatment compliance and reinfection (Nash et al., 2001), the term refractory is used here to refer to treatment failure. A further strength of this study is that no signs of immunodeficiency or poor drug compliance were present in the cases with MTZ treatment refractory infections, and, because the risk of reinfection is small in Norway, these factors can be discounted as confounders and, as a consequence, the clinically MTZ refractory presentation is likely linked to the Giardia genotype. These refractory cases were later cured using secondary or tertiary drug regimens according to Morch et al., 2008) (Morch et al., 2008), with a lab confirmed Giardia negative stool sample after successful treatment.
A limitation was that some of the collected clinical samples could not be included or obtained in the study, especially for the two PFOR genes where only 4–6 clones were obtained from some of the isolates, due to low Giardia DNA content, making the total number of susceptible and refractory samples low, and the results need to be interpreted with caution. We also noted that the quality sequenced PFOR clones varied, even if a high number of clones was selected for sequencing, As both of the PFOR genes are 3–4 times bigger than the NR genes, several sequencing primers are needed (5–6). If one or more of these primers fail to produce a sequence, the clone cannot be analyzed. Samples with low concentrations of DNA may be analyzed in future studies by doing multiple IMS purifications and combining them before DNA extraction is carried out.
Errors induced by many amplification cycles should also be noted as a potential limitation, even if several measures were taken to avoid them (i.e., using high fidelity polymerase). It is our experience that designing and optimizing specific primers help lower the number of amplification cycles.
The eventual importance of the nsSNVs identified in this study needs to be explored in functional studies. Expression of the identified alleles could be tested in vitro (Müller et al., 2021) or in vivo in transformed E. coli susceptibility assays such as agar disk diffusion assays, as previously demonstrated by Müller et al. (Muller et al., 2007, 2013, 2015; Muller and Muller, 2019), or in a model system of genetically modified Giardia trophozoites (Jex et al., 2020). Although SNVs in full-length genes were analyzed in the present study, SNVs in potential promoter regions or before/after the CDS could potentially affect transcription of NR1 and NR2 and could be analyzed in future studies.
4.4. Conclusion
The considerable genetic variation in the two MTZ-metabolizing genes NR1 and NR2 described in this collection of Giardia assemblage B samples show the potential for genetic alterations affecting MTZ susceptibility and resistance. Frameshift mutations potentially leading to truncated NR2 proteins in susceptible Giardia isolates may affect MTZ tolerance in the parasite, as NR2 is important for detoxification of MTZ. On the other hand, a nonsense mutation in NR1, possibly leading to a truncated enzyme in a refractory Giardia isolate may protect the parasite due to NR1 MTZ-activating capabilities. Further, amino acid changes found in the ferredoxin domains may alter protein function and possibly help mediate increased MTZ tolerance.
We identifed a total of three nsSNVs in the ferredoxin domain of PFOR2 that potentially could affect the electron transfer capacity of the protein. The PFOR genes were found to be more conserved with fewer nsSNVs per gene length and generally fewer distinct alleles than the NR genes.
Ethical aspects
No patient data was collected and used from the RKI laboratory cultured isolates presented in the current study; any link between individual parasite data and patient information was removed before WGS of trophozoites and cloning experiments were carried out. For the eight non-cultured clinical isolates obtained from Haukeland University Hospital, the study was approved by the Regional Committee for Medical Research Ethics (REC) of Western Norway (2013/1285/REK vest). The VA sample was obtained from the combined research and clinical biobank of the infectious diseases department (REK vest 165.04) (Infeksjonsseksjonenes kombinerte forskning-og kliniske biobank).
Data availability
The sequences for the identified NR1, NR2, PFOR1 and PFOR2 alleles have been submitted to Genbank and have the following accession numbers: BankIt2664166: OQ267781-OQ268066.
Funding
This study has been funded by a grant from the Norwegian Surveillance System for Antimicrobial Drug Resistance (NORM), a grant from the Centre for Pharmacy, University of Bergen, a grant from Helse-Vest (grant number 912245) and The National Graduate School in Infection Biology and Antimicrobials (IBA). All of the experiments, data analysis and evaluation of the results in the present study were performed independently by the authors, without any interference from any of the funding institutions.
Declaration of competing interest
No conflict of interest has been reported by any of the authors or funding institutions.
Acknowledgements
The personnel at the Genomics Core Facility at Oslo University Hospital (oslo.genomics.no) are acknowledged due to their work and assistance with the Illumina whole-genome sequencing of the cultured clinical isolates of Giardia. Further, the personnel at the Department of Clinical Science, University of Bergen should be thanked for providing technical assistance for the sequencing process and for the assembly and filtration of files for the genome data. Petra Gosten-Heinrich, Robert Koch-Institute Berlin should be thanked for providing excellent technical assistance during culturing experiments. Ralf Ignatius, Institute of Tropical Medicine and International Health, Charité, Berlin, Germany, is acknowledged for kindly providing biological samples. Ole-Morten Morvik, Department of Clinical Science, University of Bergen is acknowledged for PFOR primer design and obtaining sequences of the PFOR genes. Lucy Robertson, Norwegian University Life Science, NMBU, Oslo and Helene Sandnes, Department of Clinical Science, University of Bergen are acknowledged for cysts purification and extraction of DNA from clinical samples. Audun Nerland, Department of Clinical Science, University of Bergen and Christel Gill Haanshuus, department of medicine, Haukeland University Hospital have provided help and methodology assistance in PCR, cloning and sequencing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2022.12.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
figs1.
figs2.
figs3.
figs4.
References
- Aguiar J.M., Silva S.O., Santos V.A., Taniwaki S.A., Oliveira T.M., Ferreira H.L., Keid L.B., Gregori F., Soares R.M. Evidence of heterozygosity and recombinant alleles in single cysts of Giardia duodenalis. Revista brasileira de parasitologia veterinaria = Brazilian journal of veterinary parasitology : Orgao Oficial do Colegio Brasileiro de Parasitologia Veterinaria. 2016;25:187–195. doi: 10.1590/S1984-29612016031. [DOI] [PubMed] [Google Scholar]
- Ankarklev J., Jerlström-Hultqvist J., Ringqvist E., Troell K., Svärd S.G. Behind the smile: cell biology and disease mechanisms of Giardia species. Nat. Rev. Microbiol. 2010;8:413–422. doi: 10.1038/nrmicro2317. [DOI] [PubMed] [Google Scholar]
- Ankarklev J., Svard S.G., Lebbad M. Allelic sequence heterozygosity in single Giardia parasites. BMC Microbiol. 2012;12:65. doi: 10.1186/1471-2180-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansell B.R., McConville M.J., Ma'ayeh S.Y., Dagley M.J., Gasser R.B., Svard S.G., Jex A.R. Drug resistance in Giardia duodenalis. Biotechnol. Adv. 2015;33:888–901. doi: 10.1016/j.biotechadv.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Ansell B.R., Baker L., Emery S.J., McConville M.J., Svard S.G., Gasser R.B., Jex A.R. Transcriptomics indicates active and passive metronidazole resistance mechanisms in three seminal giardia lines. Front. Microbiol. 2017;8:398. doi: 10.3389/fmicb.2017.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argüello-García R., Leitsch D., Skinner-Adams T., Ortega-Pierres M.G. Drug resistance in giardia: mechanisms and alternative treatments for giardiasis. Adv. Parasitol. 2020;107:201–282. doi: 10.1016/bs.apar.2019.11.003. [DOI] [PubMed] [Google Scholar]
- Aurrecoechea C., Brestelli J., Brunk B.P., Carlton J.M., Dommer J., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O.S., Heiges M., Innamorato F., Iodice J., Kissinger J.C., Kraemer E., Li W., Miller J.A., Morrison H.G., Nayak V., Pennington C., Pinney D.F., Roos D.S., Ross C., Stoeckert C.J., Jr., Sullivan S., Treatman C., Wang H. GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res. 2009;37:D526–D530. doi: 10.1093/nar/gkn631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begaydarova R., Yukhnevich Y., Babenko D., et al. Determination of PFOR gene expression in strains of G. intestinalis with different inhibitory concentrations of metronidazole. J. Infect. Dveloping Cuntries. 2015;9(5):519–523. doi: 10.3855/jidc.5768. [DOI] [PubMed] [Google Scholar]
- Caccio S.M., Ryan U. Molecular epidemiology of giardiasis. Mol. Biochem. Parasitol. 2008;160:75–80. doi: 10.1016/j.molbiopara.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Cañete R., Noda A.L., Rodríguez M., Brito K., Herrera E., Kofoed P.E., Ursing J. 5-Nitroimidazole refractory giardiasis is common in Matanzas, Cuba and effectively treated by secnidazole plus high-dose mebendazole or quinacrine: a prospective observational cohort study. Clin. Microbiol. Infect. : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2020;26 doi: 10.1016/j.cmi.2019.12.017. 1092.e1091-1092.e1096. [DOI] [PubMed] [Google Scholar]
- Choy S.H., Mahdy M.A., Al-Mekhlafi H.M., Low V.L., Surin J. Population expansion and gene flow in Giardia duodenalis as revealed by triosephosphate isomerase gene. Parasites Vectors. 2015;8:454. doi: 10.1186/s13071-015-1084-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan M., Wang A.L., Wang C.C. Inhibition of pyruvate-ferredoxin oxidoreductase gene expression in Giardia lamblia by a virus-mediated hammerhead ribozyme. Mol. Microbiol. 2000;36(2):447–456. doi: 10.1046/j.1365-2958.2000.01863.x. [DOI] [PubMed] [Google Scholar]
- Emery S.J., Baker L., Ansell B.R.E., Mirzaei M., Haynes P.A., McConville M.J., Svard S.G., Jex A.R. Differential protein expression and post-translational modifications in metronidazole-resistant Giardia duodenalis. GigaScience. 2018;7 doi: 10.1093/gigascience/giy024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen O., Jerlstrom-Hultqvist J., Castro E., Sherwood E., Ankarklev J., Reiner D.S., Palm D., Andersson J.O., Andersson B., Svard S.G. Draft genome sequencing of giardia intestinalis assemblage B isolate GS: is human giardiasis caused by two different species? PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabin-Garcia L.B., Bartolome C., Abal-Fabeiro J.L., Mendez S., Llovo J., Maside X. Strong genetic structure revealed by multilocus patterns of variation in Giardia duodenalis isolates of patients from Galicia (NW-Iberian Peninsula) Infect. Genet. Evol. : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2017;48:131–141. doi: 10.1016/j.meegid.2016.12.014. [DOI] [PubMed] [Google Scholar]
- Goodwin A., Kersulyte D., Sisson G., Veldhuyzen van Zanten S.J., Berg D.E., Hoffman P.S. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- Grantham R. Amino acid difference formula to help explain protein evolution. Science (New York, N.Y.) 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- Haas B.J., Gevers D., Earl A.M., Feldgarden M., Ward D.V., Giannoukos G., Ciulla D., Tabbaa D., Highlander S.K., Sodergren E., Methé B., DeSantis T.Z., Petrosino J.F., Knight R., Birren B.W. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanevik K., Bakken R., Brattbakk H.R., Saghaug C.S., Langeland N. Whole genome sequencing of clinical isolates of Giardia lamblia. Clin. Microbiol. Infect. : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2015;21 doi: 10.1016/j.cmi.2014.08.014. 192.e191-192.e193. [DOI] [PubMed] [Google Scholar]
- Holm L. DALI and the persistence of protein shape. Protein Sci. : a publication of the Protein Society. 2020;29:128–140. doi: 10.1002/pro.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A.H., Moore L.S., Sundsfjord A., et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016;387:176–187. doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- Huber T., Faulkner G., Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- Jenks P.J., Edwards D.I. Metronidazole resistance in Helicobacter pylori. Int. J. Antimicrob. Agents. 2002;19:1–7. doi: 10.1016/s0924-8579(01)00468-x. [DOI] [PubMed] [Google Scholar]
- Jex A.R., Svärd S., Hagen K.D., Starcevich H., Emery-Corbin S.J., Balan B., Nosala C., Dawson S.C. Recent advances in functional research in Giardia intestinalis. Adv. Parasitol. 2020;107:97–137. doi: 10.1016/bs.apar.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.C., Dean D.R., Smith A.D., Johnson M.K. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 596, 2021:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keister D.B. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans. R. Soc. Trop. Med. Hyg. 1983;77:487–488. doi: 10.1016/0035-9203(83)90120-7. [DOI] [PubMed] [Google Scholar]
- Kirk M.D., Pires S.M., Black R.E., Caipo M., Crump J.A., Devleesschauwer B., Döpfer D., Fazil A., Fischer-Walker C.L., Hald T., Hall A.J., Keddy K.H., Lake R.J., Lanata C.F., Torgerson P.R., Havelaar A.H., Angulo F.J. World Health organization estimates of the global and regional disease burden of 22 foodborne bacterial, Protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuwin R., Putaporntip C., Pattanawong U., Jongwutiwes S. Clonal diversity in Giardia duodenalis isolates from Thailand: evidences for intragenic recombination and purifying selection at the beta giardin locus. Gene. 2010;449:1–8. doi: 10.1016/j.gene.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Lalle M., Pozio E., Capelli G., Bruschi F., Crotti D., Cacciò S.M. Genetic heterogeneity at the beta-giardin locus among human and animal isolates of Giardiaduodenalis and identification of potentially zoonotic subgenotypes. Int. J. Parasitol. 2005;35:207–213. doi: 10.1016/j.ijpara.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Lalle M., Hanevik K. Treatment-refractory giardiasis: challenges and solutions. Infect. Drug Resist. 2018;11:1921–1933. doi: 10.2147/IDR.S141468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasek-Nesselquist E., Welch D.M., Thompson R.C., Steuart R.F., Sogin M.L. Genetic exchange within and between assemblages of Giardia duodenalis. J. Eukaryot. Microbiol. 2009;56:504–518. doi: 10.1111/j.1550-7408.2009.00443.x. [DOI] [PubMed] [Google Scholar]
- Lecova L., Tumova P., Nohynkova E. Clone-based haplotyping of Giardia intestinalis assemblage B human isolates. Parasitol. Res. 2019;118:355–361. doi: 10.1007/s00436-018-6161-7. [DOI] [PubMed] [Google Scholar]
- Leitsch D., Burgess A.G., Dunn L.A., Krauer K.G., Tan K., Duchene M., Upcroft P., Eckmann L., Upcroft J.A. Pyruvate:ferredoxin oxidoreductase and thioredoxin reductase are involved in 5-nitroimidazole activation while flavin metabolism is linked to 5-nitroimidazole resistance in Giardia lamblia. J. Antimicro. Chemot. 2011;66:1756–1765. doi: 10.1093/jac/dkr192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitsch D. Drug resistance in the microaerophilic parasite giardia lamblia. Curr.Tropical Med. Rep. 2015;2:128–135. doi: 10.1007/s40475-015-0051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitsch D., Muller J., Muller N. Evaluation of Giardia lamblia thioredoxin reductase as drug activating enzyme and as drug target. International J. Parasitol. Drugs and Drug Resistance. 2016;6:148–153. doi: 10.1016/j.ijpddr.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitsch D. A review on metronidazole: an old warhorse in antimicrobial chemotherapy. Parasitology. 2017:1–12. doi: 10.1017/S0031182017002025. [DOI] [PubMed] [Google Scholar]
- Leitsch D. A review on metronidazole: an old warhorse in antimicrobial chemotherapy. Parasitology. 2019;146:1167–1178. doi: 10.1017/S0031182017002025. [DOI] [PubMed] [Google Scholar]
- Loderstädt U., Frickmann H. Antimicrobial resistance of the enteric protozoon Giardia duodenalis - a narrative review. Eur. J. Microbiol. Immunol. (Bp) 2021;11(2):29–43. doi: 10.1556/1886.2021.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Matey E.J., Bi X., Songok E.M., Ichimura H., Tokoro M. Extremely diversified haplotypes observed among assemblage B population of Giardia intestinalis in Kenya. Parasitol. Int. 2020;75 doi: 10.1016/j.parint.2019.102038. [DOI] [PubMed] [Google Scholar]
- Morch K., Hanevik K., Robertson L.J., Strand E.A., Langeland N. Treatment-ladder and genetic characterisation of parasites in refractory giardiasis after an outbreak in Norway. J. Infect. 2008;56:268–273. doi: 10.1016/j.jinf.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Muller J., Wastling J., Sanderson S., Muller N., Hemphill A. A novel Giardia lamblia nitroreductase, GlNR1, interacts with nitazoxanide and other thiazolides. Antimicro. Agent. Chemot. 2007;51:1979–1986. doi: 10.1128/AAC.01548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J., Ley S., Felger I., Hemphill A., Muller N. Identification of differentially expressed genes in a Giardia lamblia WB C6 clone resistant to nitazoxanide and metronidazole. J. Antimicro. Chemot. 2008;62:72–82. doi: 10.1093/jac/dkn142. [DOI] [PubMed] [Google Scholar]
- Muller J., Schildknecht P., Muller N. Metabolism of nitro drugs metronidazole and nitazoxanide in Giardia lamblia: characterization of a novel nitroreductase (GlNR2) J. Antimicro. Chemot. 2013;68:1781–1789. doi: 10.1093/jac/dkt106. [DOI] [PubMed] [Google Scholar]
- Muller J., Rout S., Leitsch D., Vaithilingam J., Hehl A., Muller N. Comparative characterisation of two nitroreductases from Giardia lamblia as potential activators of nitro compounds. International J. Parasitol. Drugs and Drug Resistance. 2015;5:37–43. doi: 10.1016/j.ijpddr.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Hemphill A., Müller N. Physiological aspects of nitro drug resistance in Giardia lamblia. International J. Parasitol. Drugs and Drug Resistance. 2018;8:271–277. doi: 10.1016/j.ijpddr.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J., Muller N. Nitroreductases of bacterial origin in Giardia lamblia: potential role in detoxification of xenobiotics. Microbiol. 2019;8 doi: 10.1002/mbo3.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Heller M., Uldry A.C., Braga S., Müller N. vol. 10. Pathogens; Basel, Switzerland: 2021. (Nitroreductase Activites in Giardia Lamblia: ORF 17150 Encodes a Quinone Reductase with Nitroreductase Activity). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz Gutierrez J., Aldasoro E., Requena A., Comin A.M., Pinazo M.J., Bardaji A., Oliveira I., Valls M.E., Gascon J. Refractory giardiasis in Spanish travellers. Trav. Med. Infect. Dis. 2013;11:126–129. doi: 10.1016/j.tmaid.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Nabarro L.E., Lever R.A., Armstrong M., Chiodini P.L. Increased incidence of nitroimidazole-refractory giardiasis at the hospital for tropical diseases. Clin. Microbiol. Infect. : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2015;21:791–796. doi: 10.1016/j.cmi.2015.04.019. London: 2008-2013. [DOI] [PubMed] [Google Scholar]
- Nash T.E., Ohl C.A., Thomas E., Subramanian G., Keiser P., Moore T.A. Treatment of patients with refractory giardiasis. Clin. Infect. Dis. 2001;33:22–28. doi: 10.1086/320886. [DOI] [PubMed] [Google Scholar]
- Neumayr A., Schunk M., Theunissen C., Van Esbroeck M., Mechain M., Hatz C., Mørch K., Soriano Pérez M.J., Sydow V., Sothmann P., Kuenzli E., Rothe C., Bottieau E. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America; 2021. Efficacy and Tolerability of Quinacrine Monotherapy and Albendazole Plus Chloroquine Combination Therapy in Nitroimidazole-Refractory Giardiasis: a TropNet Study. [DOI] [PubMed] [Google Scholar]
- Nixon J.E., Wang A., Field J., Morrison H.G., McArthur A.G., Sogin M.L., Loftus B.J., Samuelson J. Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase, and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukaryot. Cell. 2002;1:181–190. doi: 10.1128/EC.1.2.181-190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogren J., Van Nguyen S., Nguyen M.K., Dimberg J., Matussek A. Prevalence of Dientamoeba fragilis, Giardia duodenalis, Entamoeba histolytica/dispar, and Cryptosporidium spp in Da Nang, Vietnam, detected by a multiplex real-time PCR. APMIS : APMIS (Acta Pathol. Microbiol. Immunol. Scand.) 2016;124:529–533. doi: 10.1111/apm.12535. [DOI] [PubMed] [Google Scholar]
- Pal D., Banerjee S., Cui J., Schwartz A., Ghosh S.K., Samuelson J. Giardia, Entamoeba, and Trichomonas enzymes activate metronidazole (nitroreductases) and inactivate metronidazole (nitroimidazole reductases) Antimicro. Agent. Chemot. 2009;53:458–464. doi: 10.1128/AAC.00909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulish-Miller T.E., Augostini P., Schuyler J.A., Smith W.L., Mordechai E., Adelson M.E., Gygax S.E., Secor W.E., Hilbert D.W. Trichomonas vaginalis metronidazole resistance is associated with single nucleotide polymorphisms in the nitroreductase genes ntr4Tv and ntr6Tv. Antimicro. Agent. Chemot. 2014;58:2938–2943. doi: 10.1128/AAC.02370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Requena-Mendez A., Goni P., Rubio E., Pou D., Fumado V., Lobez S., Aldasoro E., Cabezos J., Valls M.E., Trevino B., Martinez Montseny A.F., Clavel A., Gascon J., Munoz J. The use of quinacrine in nitroimidazole-resistant giardia duodenalis: an old drug for an emerging problem. J. Infect. Dis. 2017;215:946–953. doi: 10.1093/infdis/jix066. [DOI] [PubMed] [Google Scholar]
- Robertson L.J., Hermansen L., Gjerde B.K., Strand E., Alvsvåg J.O., Langeland N. Application of genotyping during an extensive outbreak of waterborne giardiasis in Bergen, Norway, during autumn and winter 2004. Appl. Environ. Microbiol. 2006;72:2212–2217. doi: 10.1128/AEM.72.3.2212-2217.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson L.J., Forberg T., Hermansen L., Gjerde B.K., Langeland N. Molecular characterisation of Giardia isolates from clinical infections following a waterborne outbreak. J. Infect. 2007;55:79–88. doi: 10.1016/j.jinf.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Saghaug C.S., Klotz C., Kallio J.P., Brattbakk H.R., Stokowy T., Aebischer T., Kursula I., Langeland N., Hanevik K. Genetic variation in metronidazole metabolism and oxidative stress pathways in clinical Giardia lamblia assemblage A and B isolates. Infect. Drug Resist. 2019;12:1221–1235. doi: 10.2147/IDR.S177997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghaug C.S., Klotz C., Kallio J.P., Aebischer T., Langeland N., Hanevik K. Genetic diversity of the flavohemoprotein gene of giardia lamblia: evidence for high allelic heterozygosity and copy number variation. Infect. Drug Resist. 2020;13:4531–4545. doi: 10.2147/IDR.S274543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siripattanapipong S., Leelayoova S., Mungthin M., Thompson R.C., Boontanom P., Saksirisampant W., Tan-Ariya P. Clonal diversity of the glutamate dehydrogenase gene in Giardia duodenalis from Thai isolates: evidence of genetic exchange or mixed infections? BMC Microbiol. 2011;11:206. doi: 10.1186/1471-2180-11-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprong H., Cacciò S.M., van der Giessen J.W. Identification of zoonotic genotypes of Giardia duodenalis. PLoS Neglected Trop. Dis. 2009;3 doi: 10.1371/journal.pntd.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townson S.M., Upcroft J.A., Upcroft P. Characterisation and purification of pyruvate:ferredoxin oxidoreductase from Giardia duodenalis. Mol. Biochem. Parasitol. 1996;79:183–193. doi: 10.1016/0166-6851(96)02661-8. [DOI] [PubMed] [Google Scholar]
- Waldram A., Vivancos R., Hartley C., Lamden K. Prevalence of Giardia infection in households of Giardia cases and risk factors for household transmission. BMC Infect. Dis. 2017;17:486. doi: 10.1186/s12879-017-2586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Jacobson C., Gardner G., Carmichael I., Campbell A.J., Ryan U. Development of a quantitative PCR (qPCR) for Giardia and analysis of the prevalence, cyst shedding and genotypes of Giardia present in sheep across four states in Australia. Exp. Parasitol. 2014;137:46–52. doi: 10.1016/j.exppara.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Ydsten K.A., Hellgren U., Asgeirsson H. Quinacrine treatment of nitroimidazole-refractory giardiasis. J. Infect. Dis. 2021;225:1773–1776. doi: 10.1093/infdis/jiab287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.Z., Birky C.W., Jr., Adam R.D. The two nuclei of Giardia each have complete copies of the genome and are partitioned equationally at cytokinesis. Eukaryot. Cell. 2002;1:191–199. doi: 10.1128/EC.1.2.191-199.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences for the identified NR1, NR2, PFOR1 and PFOR2 alleles have been submitted to Genbank and have the following accession numbers: BankIt2664166: OQ267781-OQ268066.