Abstract
Aims
Heart failure with preserved ejection fraction (HFpEF) and non‐alcoholic fatty liver disease (NAFLD) are related conditions with an increasing incidence. The mechanism of their relationship remains undefined. Here, we aimed to explore the potential mechanisms, diagnostic markers, and therapeutic options for HFpEF and NAFLD.
Methods and results
HFpEF and NAFLD datasets were downloaded from the Gene Expression Omnibus (GEO) database. Common differentially expressed genes (DEGs) were screened for functional annotation. A protein–protein interaction network was constructed based on the STRING database, and hub genes were analysed using GeneMANIA annotation. ImmuCellAI (Immune Cell Abundance Identifier) was employed for analysis of immune infiltration. We also used validation datasets to validate the expression levels of hub genes and the correlation of immune cells. To screen for diagnostic biomarkers, we employed the least absolute shrinkage and selection operator and support vector machine‐recursive feature elimination. Drug signature database was used to predict potential therapeutic drugs. Our analyses identified a total of 33 DEGs. Inflammation and immune infiltration played important roles in the development of both diseases. The data showed a close relationship between chemokine signalling pathway, cytokine–cytokine receptor interaction, calcium signalling pathway, neuroactive ligand–receptor interaction, osteoclast differentiation, and cyclic guanosine monophosphate‐protein kinase G signalling pathway. We demonstrated that PRF1 (perforin 1) and IL2RB (interleukin‐2 receptor subunit beta) proteins were perturbed by the diseases and may be the hub genes. The analysis showed that miR‐375 may be a potential diagnostic marker for both diseases. Our drug prediction analysis showed that bosentan, eldecalcitol, ramipril, and probucol could be potential therapeutic options for the diseases.
Conclusions
Our findings revealed common pathogenesis, diagnostic markers, and therapeutic agents for HFpEF and NAFLD. There is need for further experimental studies to validate our findings.
Keywords: Heart failure with preserved ejection fraction, Non‐alcoholic fatty liver disease, Differentially expressed genes, Immune infiltration, Diagnostic markers, Therapeutics
Introduction
Heart failure (HF) is a major public health problem. A 2017 report showed that the global burden of HF was approximately 64.34 million. 1 Compared with HF with reduced ejection fraction (HFrEF), the prevalence of HF with preserved ejection fraction (HFpEF) was closely associated with increase in age. 2 Therefore, as the global aged population increases, HFpEF has become the most prevalent form of HF. 3 Patients with HFpEF require more nursing care, are more likely to be hospitalized, and have longer hospital stays compared with those with HFrEF. 4 Although HFpEF mortality varies with study type, study design, and inclusion criteria, the overall risk of death increases with the intensity of comorbidities, and the 5 year mortality rate of HFpEF is often >50%. 5
Non‐alcoholic fatty liver disease (NAFLD), which includes non‐alcoholic fatty liver (NAFL) and non‐alcoholic steatohepatitis (NASH), contributes significantly to the burden of chronic liver disease in the world. 6 According to a previous meta‐analysis that included 8 515 431 samples, the prevalence of NAFLD is approximately 25.24% worldwide, 7 with rates as high as 29.62% in Asia and 42.04% in Southeast Asia. 8 The NAFLD disease burden is directly related to both liver disease and cardiovascular disease (CVD). 9 Recent data have demonstrated an association between HF and NAFLD. For instance, a previous retrospective cohort study that comprised 870 535 Medicare beneficiaries revealed a higher risk of developing HF in patients with NAFLD compared with those without NAFLD and a stronger association with HFpEF than HFrEF. 10 Similarly, a cohort study involving 10 422 Swedish adults showed that NAFLD was closely associated with new occurrence of HF. 11 This interaction might be associated with many potential mechanisms. For instance, a retrospective study including 886 patients with a left ventricular ejection fraction (LVEF) ≥ 40%, without obstructive epicardial coronary artery disease, showed that NAFLD patients experienced a higher incidence of coronary microvascular dysfunction and lower coronary blood flow reserve compared with non‐NAFLD patients. 12 Besides, a meta‐analysis involving 280 645 participants from 12 studies showed a significant association between NAFLD and diastolic cardiac dysfunction. 13 However, data on the relationship between HF and NAFLD remain scant, despite the fact that there is a strong correlation between the two, especially HFpEF and NAFLD.
Analysis of enormous volumes of data available in public databases reveals new insights into pathological processes mediating many diseases. For instance, Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo), a comprehensive database that harbours high‐throughput gene expression data, contains datasets from international institutions that were generated using high‐throughput sequencing and microarray technologies. 14 In this study, we used the GEO database to evaluate potential mechanisms, diagnostic markers, and possible therapeutic agents for HFpEF and NAFLD.
Methods
Downloading and processing of datasets
We searched the GEO database using the mesh ‘heart failure with preserved ejection fraction’ and ‘non‐alcoholic fatty liver disease’. Datasets with gene expression profiles including case and control and those with a sample size ≥ 5 in each group were included in this study. The included datasets had data from tissue samples, and those used to analyse diagnostic markers were peripheral blood samples. Datasets containing data on reanalysis of processed or raw data and those lacking drug intervention were also included in our analysis. The normalized representation matrix was shown in boxplot whereas the principal component analysis (PCA) was used to visualize distribution patterns between samples.
Identification of differentially expressed genes
R package ‘limma’ was used to identify differentially expressed genes (DEGs) in the datasets; |log2Fold change| > 0.5 and P < 0.05 were used as the standard. Venn diagrams were used to identify common DEGs in the two diseases.
Construction of protein–protein interaction networks and screening of hub genes
A protein–protein interaction (PPI) network of common DEGs was created using STRING (https://www.string‐db.org). The nodes in the network with a confidence score ≥ 0.4 were screened and disconnected nodes were hidden. Cytoscape3.8.2 software was employed for visualization, whereas Cytohubba plug‐in was used to screen for top 10 genes with node scores. MCODE plug‐in was used to identify key modules in the PPI network, and the screening criterion included a degree cutoff of 2, a node score cutoff of 0.2, a K‐score of 2, and a max depth of 100. The module with the highest score was used as the final functional module, whereas the intersection of the two was used as the hub genes of the network. To better understand the biological characteristics of these hub genes, GeneMANIA was used to construct and analyse the network as well as their co‐expressed genes.
Functional enrichment analysis
The R package ‘clusterProfiler’ was used to perform the Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) enrichment analysis of the common DEGs. Gene Set Enrichment Analysis (GSEA) is a knowledge‐based method used to interpret genome‐wide expression profiles. In our study, we employed the GSEA for analysis of HFpEF and NAFLD datasets, to identify potential biological pathways. An adjusted P‐value < 0.05 was considered to be statistically significant.
Analysis of immune infiltration
Immune Cell Abundance Identifier (ImmuCellAI; http://bioinfo.life.hust.edu.cn/ImmuCellAI) was used to estimate the abundance of 24 immune cells, which included 18 T cell subtypes and 6 other immune cells, such as B cells, natural killer (NK) cells, monocytes, macrophages, neutrophils, and dendritic cells. Histograms demonstrating the ratio of immune cells, violin diagrams showing the difference between immune cells in groups, and the correlation between immune cells were drawn by R and related packages.
Validation of the expression of hub genes and correlation of immune infiltration
The expression profiles of the hub genes and the immune infiltration results were analysed using separate datasets. A T‐test was used to compare the two groups of data. A P < 0.05 was considered to be statistically significant.
Screening of biomarkers
The screening criteria used for DEGs were employed to screen for differentially expressed miRNAs. The least absolute shrinkage and selection operator (LASSO) and support vector machine‐recursive feature elimination (SVM‐RFE) were used to screen for diagnostic biomarkers in the differentially expressed miRNAs. Besides, we verified the data by performing searches in PubMed (https://www.ncbi.nlm.nih.gov/). Receiver operating characteristic (ROC) and area under the curve (AUC) were used to estimate the diagnostic value of the screened biomarkers.
Prediction of potential therapeutic drugs
Common DEGs were imported into the drug signature database (DSigDB) to identify potential drug candidates. The top 10 drugs with the highest scores were used as drug candidates.
Results
Evaluation of datasets
We utilized four GEO datasets: GSE192886, GSE126848, GSE89632, and GSE185062, as summarized in Supporting Information, Table S1 . Our data showed linear distribution trend of the expression matrix of the datasets, indicating that there was no obvious effect of the data batch (Supporting Information, Figure S1 A–D). PCA results showed good repeatability of the data (Supporting Information, Figure S1 E–H). We further used the GSE192886 and GSE126848 datasets as test sets for DEGs analysis, whereas GSE89632 pairing was used as the validation set for the DEGs analysis. On the other hand, GSE185062 was used to identify potential biomarkers in peripheral blood. NASH was divided into test set whereas NAFL was divided into validation set.
Identification of differentially expressed genes
Compared with the control group, there were a total of 194 DEGs (83 up‐regulated and 111 down‐regulated), 4404 DEGs (1958 up‐regulated and 2446 down‐regulated), and 4911 DEGs (2603 up‐regulated and 2308 down‐regulated) in the HFpEF, NAFL, and NASH groups, respectively (Figure 1 A–C ). Heatmap results showed top 50 smallest genes as demonstrated by the P‐values (Figure 1 E–G ). HFpEF and NAFL had 37 common DEGs (6 up‐regulated and 31 down‐regulated), whereas HFpEF and NASH had 42 common DEGs (8 up‐regulated and 34 down‐regulated). There were 33 common DEGs in the HFpEF, NAFL, and NASH groups (4 up‐regulated and 29 down‐regulated; Figure 1 D ).
Figure 1.
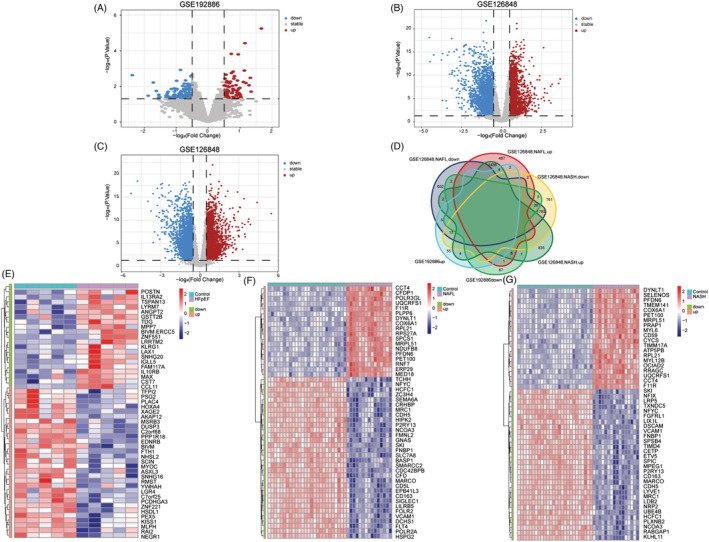
Identification of differentially expressed genes (DEGs). (A) The volcano plot of GSE192886. (B) The volcano plot of non‐alcoholic fatty liver (NAFL) in GSE126848. (C) The volcano plot of non‐alcoholic steatohepatitis (NASH) in GSE126848. (D) Co‐expressed genes in heart failure with preserved ejection fraction (HFpEF) and non‐alcoholic fatty liver disease (NAFLD). (E) The top 50 genes with the most remarkable expression changes of GSE192886. (F) The top 50 genes with the most remarkable expression changes of NAFL in GSE126848. (G) The top 50 genes with the most remarkable expression changes of NASH in GSE126848.
Protein–protein interaction network and hub genes
We then imported 33 common DEGs into STRING to create an 18‐node and 28‐edge network (Figure 2 A ). Using the Cytohubba plug‐in, we showed the top 10 important targets in the PPI network (Figure 2 B ). The MCODE plug‐in clustering found two functional modules as shown in Supporting Information, Table S2 and Figure 2 C,D . Besides, GZMB, GATA3, IL2RB (interleukin‐2 receptor subunit beta), CXCR3, and PRF1 (perforin 1) were the hub genes that intersected with the above findings in Module 1.
Figure 2.
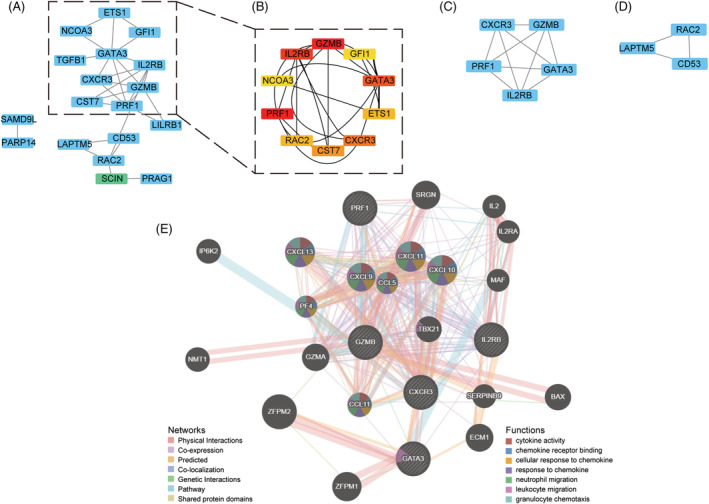
Protein–protein interaction (PPI) network and hub genes. (A) PPI networks of the differentially expressed genes (DEGs). (B) The top 10 important targets in the PPI network found by Cytohubba plug‐in. (C) Module 1 found by MCODE plug‐in. (D) Module 2 found by MCODE plug‐in. (E) Hub genes and their co‐expression genes were analysed using GeneMANIA.
GeneMANIA built a network of 20 related targets, for a total of 25 targets and 4888 total links. Five hub genes revealed a complex PPI network with 77.64% physical interactions, 8.01% co‐expression, 5.37% prediction, 3.63% co‐localization, 2.87% genetic interactions, 1.88% pathway, and 0.60% shared protein domains. Functional analyses of the network indicated targets related to cytokine activity, chemokine receptor binding, cellular response to chemokine, response to chemokine, neutrophil migration, leukocyte migration, and granulocyte chemotaxis (Figure 2 E ).
Functional enrichment analysis
Functional enrichment analysis was performed on 33 common genes. According to the GO, 84 biological processes (BPs) were involved where the predominant association was positive regulation of immune effector process (Figure 3 A ). KEGG results showed 12 related pathways, which were mainly related to T helper 17 (Th17) cell differentiation and NK cell‐mediated cytotoxicity (Figure 3 B ). These findings point to the importance of inflammation and immune infiltration in HFpEF and NAFLD. What's more, GSEA results showed that HFpEF, NAFL, and NASH datasets were enriched in 22, 68, and 40 signalling pathways, respectively. The intersection in the Venn diagram (Figure 3 C ) demonstrated a total of seven pathways. Further analysis revealed that chemokine signalling pathway, cytokine–cytokine receptor interaction, calcium signalling pathway, neuroactive ligand–receptor interaction, osteoclast differentiation, and cyclic guanosine monophosphate‐protein kinase G (cGMP‐PKG) signalling pathway may be related to the diseases (Figure 3 D–F ).
Figure 3.
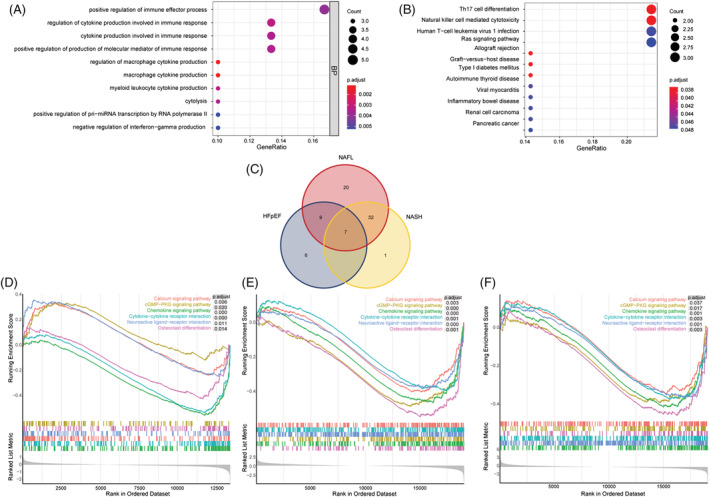
Functional enrichment analysis. (A) Gene Ontology (GO) function analysis results on common differentially expressed genes (DEGs). (B) Kyoto Encyclopaedia of Genes and Genomes (KEGG) function analysis results on common DEGs. (C) The co‐signalling pathways in heart failure with preserved ejection fraction (HFpEF) and non‐alcoholic fatty liver disease (NAFLD). (D) Signalling pathways found by Gene Set Enrichment Analysis (GSEA) in HFpEF. (E) Signalling pathways found by GSEA in non‐alcoholic fatty liver (NAFL). (F) Signalling pathways found by GSEA in non‐alcoholic steatohepatitis (NASH).
Immune infiltration
The composition of immune cells was different between the HFpEF group and the control group (Figure 4 A ). CD8_T and nTreg were enriched in the HFpEF group (Figure 4 B ). In addition, we calculated the correlations between each immune cell type to explore their relationships and possible interactions. The results showed that there was a strong positive correlation between Tfn and Tr1, Tfh and iTreg, or Tr1 and iTreg, whereas CD4_T and macrophage had the highest negative correlation (Figure 4 C ).
Figure 4.
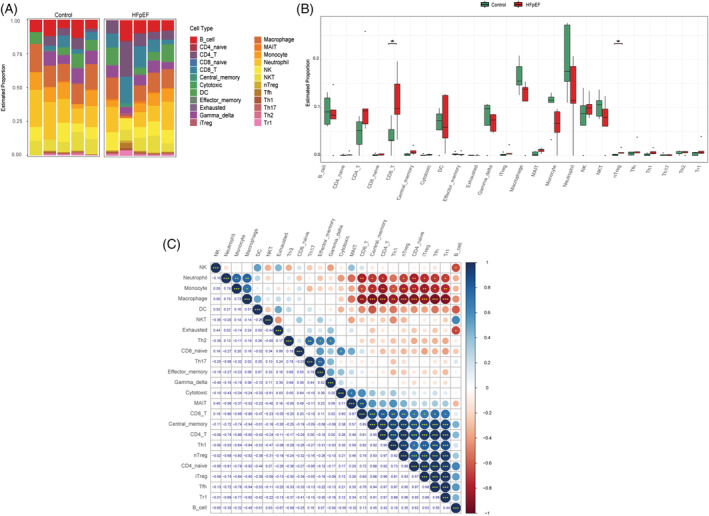
Immune infiltration of heart failure with preserved ejection fraction (HFpEF). (A) The fraction of 24 subsets of immune cells in HFpEF and control samples. (B) Bar chart of the ratio of immune cells. *P < 0.05, **P < 0.01, ***P < 0.001 vs. controls. (C) Correlation matrix among 24 immune cell subtype fractions.
Further analysis showed that gamma delta T cells, macrophages, monocytes, Th17, and Th2 were abundant in the NAFL group compared with the control group, whereas B cells and neutrophils were enriched in the control group (Figure 5 A,B ). The correlation analysis of immune cells revealed a strong positive correlation between CD8_T and exhausted T cells, followed by the strongest negative correlation between gamma_delta T cells and B cells (Figure 5 C ). When compared with the control group, the NASH group had higher levels of CD8_T, cytotoxic T cells, gamma_delta T cells, mucosal‐associated invariant T (MAIT) cells, monocyte, Th17, and Th2, whereas B cells and neutrophils were enriched in the control group (Figure 5 A,B ). Our immunocell correlation analyses showed the highest positive correlation between the Tfn and CD4_T cells and the strongest negative correlation between monocytes and neutrophils (Figure 5 D ).
Figure 5.
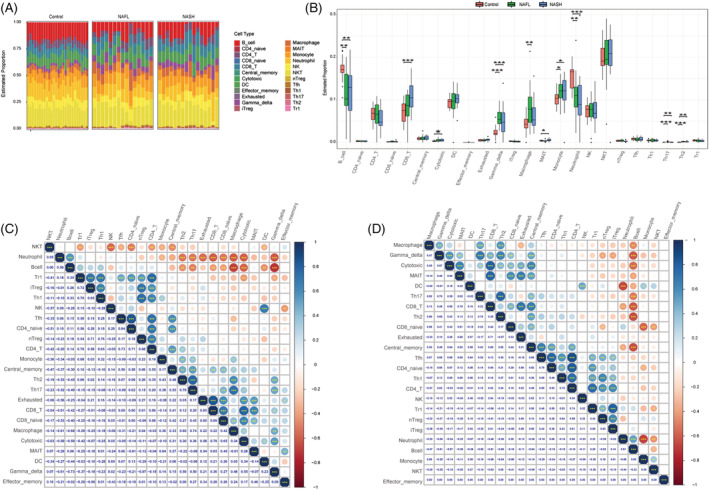
Immune infiltration of non‐alcoholic fatty liver disease (NAFLD). (A) The fraction of 24 subsets of immune cells in NAFLD and control samples. (B) Bar chart of the ratio of immune cells. *P < 0.05, **P < 0.01, ***P < 0.001 vs. controls. (C) Correlation matrix of 24 immune cell subtype fractions in non‐alcoholic fatty liver (NAFL). (D) Correlation matrix of 24 immune cell subtype fractions in non‐alcoholic steatohepatitis (NASH).
Validation of the hub genes expression and correlation of immune infiltration
To evaluate the reliability and accuracy of the bioinformatics results, the validation dataset was used to verify the expression of hub genes. The results showed that the expression of PRF1 and IL2RB in five hub genes was significantly higher in NAFLD at different stages compared with the control group (Figure 6 A ). We then examined the relationship between PRF1, IL2RB, and immune cells. Tr1, NK, and neutrophils were negatively associated with IL2RB, whereas Th2, Th17, Th1, monocyte, iTreg, gamma_delta, exhausted, effector_memory, CD8_T, CD8_naive, and CD4_T were favourably connected with IL2RB (Figure 6 B ). PRF1 was negatively correlated with Tr1, NK, and neutrophil, whereas Th2, Th17, Th1, nTreg, monocyte, iTreg, effector_memory, CD8_T, CD8_naive, and CD4_T had a positive correlation in NAFL (Figure 6 C ). In NASH, our findings demonstrated that IL2RB was negatively correlated with Tr1, NK, neutrophils, and B cells and positively correlated with Th2, Th17, Th1, monocyte, gamma_delta, effector_memory, CD8_T, CD8_naive, and CD4_T cells. PRF1 was positively correlated with Th2, Th17, Th1, nTreg, monocyte, MAIT, iTreg, effector_memory, CD8_T, and CD4_T and negatively correlated with NK, neutrophils, and B cells (Figure 6 D,E ).
Figure 6.
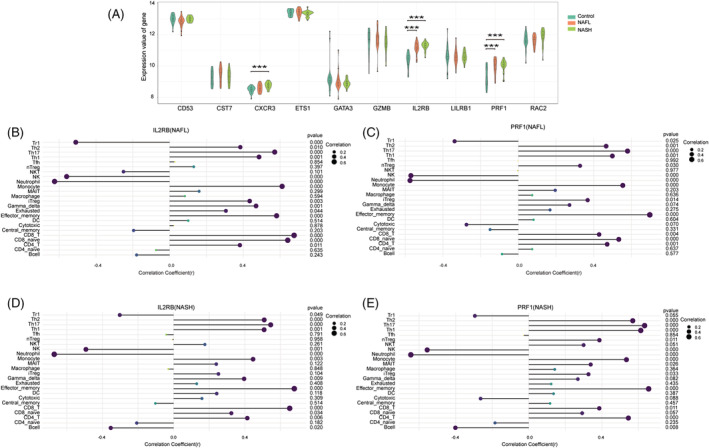
Validation of hub genes expression and correlation of immune infiltration in GSE89632. (A) The expression levels of hub genes in non‐alcoholic fatty liver disease (NAFLD). *P < 0.05, **P < 0.01, ***P < 0.001 vs. controls. (B) Analysis of correlation between IL2RB (interleukin‐2 receptor subunit beta) and immune cells in non‐alcoholic fatty liver (NAFL). (C) Analysis of correlation between PRF1 (perforin 1) and immune cells in NAFL. (D) Analysis of correlation between IL2RB and immune cells in non‐alcoholic steatohepatitis (NASH). (E) Analysis of correlation between PRF1 and immune cells in NASH.
Screening of biomarkers
A total of 351 differentially expressed miRNAs (167 up‐regulated and 184 down‐regulated) were found in the NASH group compared with the control group (Figure 7 A,B ). The LASSO algorithm identified 16 differential miRNAs with non‐zero regression coefficients, with a lambda.min of 0.01357128 (Figure 7 C,D ). Five key miRNAs were screened out by SVM‐RFE algorithm (Figure 7 E ). The analysis identified a total of 19 possible miRNAs, where only miR‐375 was shown to be associated with HFpEF when searched in PubMed (Figure 7 F ). 15 The AUC of miR‐375 in NASH was 0.859 (Figure 8 A ), and its expression level in the NAFL group was lower than that in the control group (Figure 8 C ), with an AUC of 0.873 (Figure 8 B ). These data demonstrate that miR‐375 has high diagnostic value.
Figure 7.
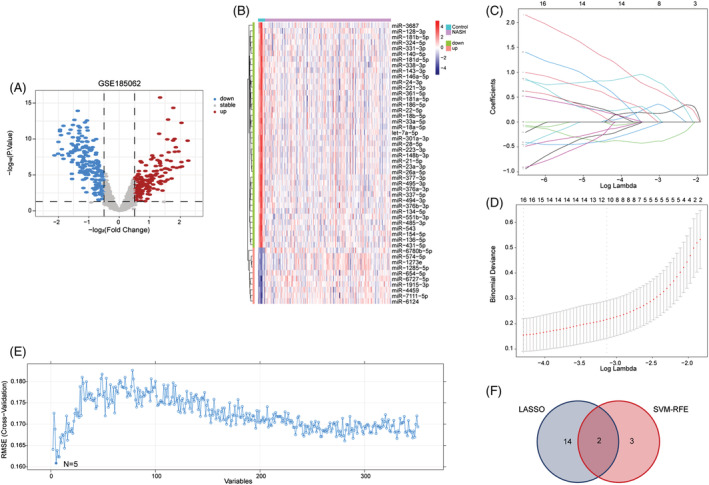
Screening of biomarkers in GSE185062. (A) The volcano plot of miRNAs in non‐alcoholic steatohepatitis (NASH). (B) The top 50 miRNAs with the most remarkable expression changes of NASH in GSE126848. (C) Ten‐time cross‐verification for tuning parameter selection in the least absolute shrinkage and selection operator (LASSO) model. (D) LASSO coefficient profiling. When 16 miRNAs were selected, the cross‐validation error is the smallest. (E) A plot of biomarkers option through support vector machine‐recursive feature elimination (SVM‐RFE) algorithm. When n = 5, the cross‐validation error is the smallest. (F) Venn diagram showed that 19 potential markers were found by LASSO and SVM‐RFE.
Figure 8.
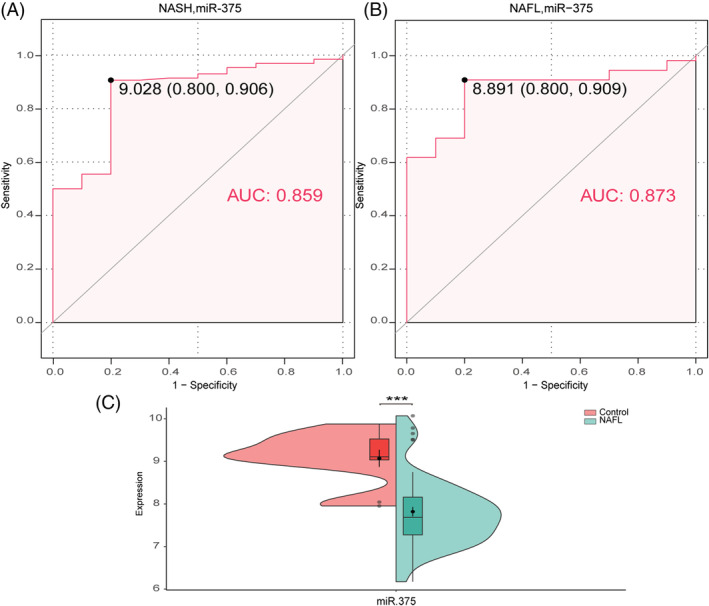
Expression of miR‐375 in non‐alcoholic fatty liver (NAFL) and its diagnostic value in non‐alcoholic fatty liver disease (NAFLD). (A) Receiver operating characteristic (ROC) curves estimating the diagnostic performance of miR‐375 in non‐alcoholic steatohepatitis (NASH). (B) ROC curves estimating the diagnostic performance of miR‐375 in NAFL. (C) The expression of miR‐375 in NAFL. *P < 0.05, **P < 0.01, ***P < 0.001 vs. controls. AUC, area under the curve.
Prediction of potential therapeutic drugs
Five hub genes were used to screen potential drugs for HFpEF and NAFLD. We identified the top 10 candidates using combined score as shown in Supporting Information, Table S3 .
Discussion
In this study, we identified a total of 33 common DEGs in HFpEF and NAFLD. Results from the GO, KEGG, and GeneMANIA implied that inflammatory and immune mechanisms are critical in both disorders. Inflammation has been widely implicated in CVD, and increase of pro‐inflammatory cytokines is associated with poor HF outcomes. Canakinumab, an interleukin (IL)‐1 inhibitor, was shown to significantly reduce the risk of hospitalization and death in HF patients with elevated C‐reactive protein (CRP ≥ 2 mg/L) and a history of myocardial infarction (MI). 16 Inflammation of HFpEF is more systemic and serves as a bridge between a series of extracardiac complications, including NAFLD, and cardiac structural changes. 17 NAFLD increases the production of triglyceride‐enriched very low‐density lipoprotein particles that are transported to peripheral tissues, resulting in abnormal plasma lipoprotein levels. Apolipoprotein C3 and apolipoprotein B can lead to endothelial dysfunction and toll‐like receptor (TLR) activation. 18 TLR, an important part of the innate immune system, contains nucleotide‐binding oligomerization domain‐like pyrin domain containing protein 3 (NLRP3), which can fuel downstream release of a variety of cytokines and chemokines such as IL‐1, IL‐6, IL‐8, tumour necrosis factor (TNF)‐α, and IL‐12. 19 Other studies showed that NAFLD‐induced low‐grade chronic systemic inflammation can damage coronary microvascular functions, generate oxidative stress in endothelial cells, and eventually develop into cardiac hypertrophy, increased heart hardness, and collagen deposition. 20
The KEGG analysis showed that the Th17 cells may be associated with the two disorders. Th17 is a T cell subpopulation that mainly produces IL‐17. 21 In a transverse aortic constriction (TAC)‐induced HF rat model, there was also significant increase in the plasma and heart tissue levels of IL‐17, which led to impaired cardiac function through the nuclear factor‐κB‐mediated calcium pathway. 22 In spontaneous hypertension rats, inhibition of IL‐17 signalling pathway can reduce myocardial collagen deposition and improve cardiac diastolic function. 23 A study involving 77 patients with HF showed an increase in the IL‐17 plasma level, which was negatively correlated with LVEF and fractional shortening. 22 In 187 hypertensive adults and 70 healthy controls, an increased percentage of Th17 was observed in hypertensive patients, particularly in adults with left ventricular (LV) hypertrophy. 24 Th17 is associated with the progression of NAFLD from NAFL to NASH to liver fibrosis. 25 Previous studies have shown that the liver microenvironment increases macrophage infiltration and hepatocyte balloon‐like degeneration by changing the features of the liver Th17 cells, and the mechanism is related to the activation of the C‐X‐C motif chemokine receptor (CXCR)3/C‐X‐C motif chemokine ligand (CXCL)9/10 axis. 26
Immune infiltration analysis revealed enhanced CD8_T cells in both conditions. Increased infiltration of CD8_T cells in myocardium was observed in HFpEF mouse models established by N‐nitro‐l‐arginine methyl ester and a high‐fat diet. 27 Furthermore, CD8_T cells were shown to affect cardiac macrophages, which are crucial in compensatory cardiac hypertrophy in mouse TAC models, and their absence promoted compensatory hypertrophy and inhibited HF. 28 The development of NASH is significantly fuelled by CD8_T cells infiltration, which is also associated with eventual development of hepatocellular carcinoma. 29 Analysis of NAFLD mice generated with conditional lack of mineralocorticoid receptor in myeloid cells using methionine‐choline‐deficient (MCD) diet showed reduced degree of steatosis, and the mechanism may be connected to decreased activation of CD8_T cells. 30
KEGG analysis also showed the role of NK cells. In the TAC mouse model, two distinct NK cell clusters (1 and 12) were identified in the heart. Both clusters were shown to significantly express interferon (IFN)‐γ, whereas the smaller and expanding NK cells clusters 12 were most likely to undergo activation with the disease occurrence and were significantly correlated with immune activation and cytokine production pathways. 31 Activated NK cells can facilitate the growth of NASH by modulating the Janus kinase‐signal transducer and activator of transcription 1/3 axis, according to three distinct animal models, including MCD, choline‐deficient high‐fat diet, and high‐fat diet with streptozotocin injection. 32 NK cells‐derived IFN‐γ can maintain the inflammatory balance of the liver and thus limit the progression of NASH to liver fibrosis in MCD model mice. 33 The identification of peripheral blood immune cells from 42 NAFLD patients and 13 healthy volunteers revealed changes in the phenotype and function of NK cells in NAFLD patients, which are closely associated with sialic acid‐binding immunoglobulin‐like lectin 7, CD57, and PD‐1 expression. 34
Results from the GSEA suggested that chemokine signalling pathway, cytokine–cytokine receptor interaction, calcium signalling pathway, neuroactive ligand–receptor interaction, osteoclast differentiation, and cGMP‐PKG signalling pathway mediate the development of the two diseases. The role of chemokines in both diseases has been demonstrated. For instance, in an HFpEF mouse model constructed by salty drinking water/unilateral nephrectomy/aldosterone or macrophages, major infiltrating inflammatory cells in the heart expressed CXCR4 in large numbers. The high expression of CXCR4 can inhibit peroxisome proliferator‐activated receptor γ activity, enhance the pro‐inflammatory state of macrophages, and also fuel CXCL3 expression to promote myofibroblast differentiation and fibrosis. 35 Inhibition of the C‐C motif chemokine ligand (CCL)2/C‐C motif chemokine receptor (CCR)2 axis can improve cardiac dysfunction and delay fibrosis in the TAC mouse model. 36 Peripheral blood analysis of 30 HFPEF patients, 30 asymptomatic LV diastolic dysfunction patients, and 23 asymptomatic hypertension patients showed increased CCL17, CCL18, and CXCL10. 37 A previous meta‐analysis showed increased CCL2 and CXCL8 in NAFL and increased CCL3, CCL4, CCL20, CXCL8, and CXCL10 in NASH. 38 CCR2 and its ligand monocyte chemoattractant protein‐1 play a central role in obesity‐related insulin resistance, 39 and insulin resistance is closely related to NAFLD and HFpEF. 40 Furthermore, many studies have reported the involvement of the CCR9/CCL25 axis in NAFLD and carcinogenesis in humans and mice. 41
The cytokines include IL, IFN, TNF, colony‐stimulating factor, growth factor, and chemokines and are synthesized, secreted, or released by immune cells and non‐immune cells after stimulation. 42 In animal models, with increased mitochondrial over‐acetylation and NLPR3 inflammasome synthesis leading to increased IL‐1β/IL‐18 production and myocardial fibrosis in HFpEF mouse models by combining age, long‐term high fat diet and desoxycorticosterone. 43 Circulating IL‐1β and IL‐12 were higher whereas TNF‐α was lower in HFpEF rat models constructed by high‐salt diet compared with HFrEF constructed by ligation of the left coronary artery. 44 Adiponectin is an adipocyte‐derived cytokine that prevents progression of aldosterone‐induced HFpEF, and its mechanism may be associated with the reduction of oxidative stress and regulation of intracellular calcium‐processing regulatory proteins. 45 Analysis of 6814 participants in the multi‐ethnic study of atherosclerosis showed that IL‐2 is an important component of suboptimal inflammation in the pathogenesis of HFpEF. 46 On the other hand, the PREVEND (Prevention of Renal and Vascular End‐Stage Disease) study showed a significant association between IL‐6 levels and HFpEF development. 47 Elevated plasma levels of growth differentiation factor 15 (GDF15) are another significant independent predictor of complex outcomes in HFpEF patients. 48 A meta‐analysis of 51 studies involving 36 074 patients and 47 052 controls showed that elevated levels of CRP, IL‐1β, IL‐6, TNF‐α, and intercellular cell adhesion molecule‐1 were significantly associated with increased risk of NAFLD. 49 Plasma GDF15 concentration was measured in 175 overweight/obese adolescents and showed that GDF15 changed with changes in intrahepatic fat content. 50 Ectodysplasin A, a liver secreted protein, is elevated in NAFL and NASH and is associated with progression of steatosis and fibrosis. 51 Moreover, a Korean study involving 2735 participants showed an inverse association between adiponectin and the prevalence of NAFLD and could even serve as a novel biomarker for NAFLD. 52
Ca2+ is a ubiquitous second messenger that mediates various cellular processes. 53 It has been shown that, in the metabolic risk‐related model of HFpEF, the level of mitochondrial free Ca2+ is increased to compensate for the required ATP by mild dysfunction of mitochondrial complex I. However, continuous increase may lead to opening of mitochondrial permeability transition pore. 54 Increased cytoplasmic Ca2+ levels mediated by store‐operated Ca2+ channels and inositol 1,4,5‐trisphosphate receptors can inhibit autophagy and aggravate NAFLD progression. 55 Importantly, changes in the mitochondrial Ca2+ in NAFLD liver cells have been shown to lead to more lipid absorption into the mitochondria, creating an imbalance between mitochondrial metabolism and energy production. 56
The discovery of neuroactive ligand–receptor interaction suggests the role of neural signals in the two diseases. The C‐hydroxyephedrine positron emission tomography (PET) study indicated that diastolic dysfunction in patients with HFpEF was associated with impaired myocardial sympathetic innervation. 57 A longitudinal cohort study involving 33 899 Korean participants has shown that autonomic dysregulation increases the risk of NAFLD, 58 and consistent findings have been obtained in the Chinese population. 59
cGMP is generated by the action of guanylate cyclase on guanosine triphosphate. 60 The cGMP‐dependent protein kinase (PKG) is an important downstream target of cGMP, and its activation can regulate cardiac contractility and myocardial remodelling. 61 Decreased PKG activity has been observed in patients with HFpEF. 62 Soluble guanylate cyclase (sGC) activation in cardiomyocytes from hypertensive rat models induced by a high‐salt diet for 10 weeks and from patients with HFpEF inhibited cardiac hypertrophy and improved cardiac function, in part due to improvements in cGMP‐PKG signalling pathway. 63 Further research into the cGMP‐PKG signalling pathway in HFpEF is required. Resistant starch significantly reduced liver weight and inhibited lipid accumulation in liver tissues in ob/ob mouse models by modulating the cGMP‐PKG signalling pathway. 64 In a choline‐deficient, l‐amino acid‐defined, and high‐fat diet‐induced mouse NASH model, sGC stimulators prevented hepatic steatosis, inflammation, and fibrosis. 65
Aside from the pathway, the major genes involved in the development of HFpEF and NAFLD remain unclear. By screening and verification, this study revealed the core positions of PRF1 and IL2RB in the two disorders, which can control immune invasion and inflammation. In the Framingham and InCHIANTI cohorts, whole‐transcriptome analysis of blood samples from patients revealed that PRF1 was associated with chronic inflammation in old age. 66 In the NASH model constructed by MCD feeding in PRF1‐deficient mice, more severe liver injury and inflammation were observed, mainly involving CD8_T cells. More importantly, high PRF1 expression was also observed in the plasma and liver of NASH patients. 67 Aside from IL‐2, IL2RB can also act as a signalling receptor for IL‐15 and is involved in the regulation of T cell‐mediated immune responses. 68 NASH occurred in Bama miniature pigs fed on a high‐fat and high‐sucrose diet for 23 months, and IL2RB is an associated inflammatory gene. 69
We finally predicted that the potential diagnostic molecule was miR‐375. Some studies reported that miR‐375 confer cardioprotection through the 3‐phosphoinositide‐dependent protein kinase 1/protein kinase B signalling pathway in MI and doxorubicin‐induced HF models. 70 , 71 miR‐375 can also interact with IL‐10 to inhibit cardiac inflammation. 72 Our findings were consistent with those reported in earlier studies, demonstrating the important role of miR‐375 in NAFLD. 73 , 74 Interestingly, a recent retrospective study suggested that miR‐375 may be regulated by vitamin D during the pathogenesis of NAFLD. 75
Among the 10 potential drugs with the highest combined score predicted by DSigDB, the drugs closely related to two diseases may be bosentan, eldecalcitol, ramipril, and probucol. Bosentan, an endothelin receptor antagonist, is the first‐line drug for the treatment of pulmonary hypertension. 76 In a rat model of NAFLD constructed using MCD, bosentan reduced intrahepatic vascular resistance, increased intrahepatic extravascular flow, and reduced steatosis and liver injury. 77 Bosentan has also been shown to increase resting caloric expenditure in obese patients, contributing to weight loss. 78
Eldecalcitol is an oral active vitamin D analogue used for the treatment of osteoporosis. 79 Interestingly, we found in GSEA that osteoclast differentiation may be associated with both diseases. Low 25(OH)D levels were associated with poor clinical outcomes among patients with asymptomatic diastolic dysfunction or newly diagnosed HFpEF patients. 80 However, there are no studies on the efficacy of vitamin D supplements in HFpEF patients. A case–control study of 3023 subjects in a community in Nanjing, China, found that an elevated serum vitamin D level decreased the risk of NAFLD. 81 In a 12 month randomized controlled trial, low and moderate doses of vitamin D (1000 IU/day) reduced hepatic steatosis and fibrosis in NAFLD patients. 82 A meta‐analysis of 544 NAFLD subjects from 10 trials also showed that vitamin D supplementation improved glycaemic control and insulin resistance in NAFLD patients. 83 These studies demonstrate a role for vitamin D in both diseases, but further prospective and mechanistic studies should be conducted to evaluate the role of eldecalcitol in HFpEF and NAFLD.
Ramipril belongs to the family of angiotensin‐converting enzyme inhibitors (ACEI). Currently, it is not clear whether ACEI can improve HFpEF. A meta‐analysis showed that ACEI treatment reduced the risk of hospitalization for HFpEF. 84 A Hong Kong study showed that diuretics combined with ramipril slightly improved LV diastolic longitudinal function and reduced N‐terminal pro‐B‐type natriuretic peptide (NT‐proBNP) within 1 year. 85 Retrospective studies have shown that ACEI can reduce the risk of liver‐related events in NAFLD patients, particularly those with chronic kidney disease. 86 Given the subtle differences between different types of ACEI, studies related to ramipril should also be conducted to assess the role of ramipril in both diseases.
Probucol has antioxidant and low‐density lipoprotein‐lowering effects. 87 A recent prospective study involving 876 Japanese patients with coronary heart disease and dyslipidaemia found that probucol had some benefits despite decreasing high‐density lipoprotein (HDL). 88 In 26 NASH patients with dyslipidaemia, probucol treatment (500 mg/day for 48 weeks) improved insulin resistance, decreased serum transaminase levels, and decreased fatty liver activity scores, and this benefit was independent of its lipid‐lowering effects. 89 , 90
In conclusion, 33 common DEGs of HFpEF and NAFLD were obtained in this work. We investigated the roles of inflammation and immune infiltration, and chemokine signalling pathway, cytokine–cytokine receptor interaction, calcium signalling pathway, neuroactive ligand–receptor interaction, osteoclast differentiation, and cGMP‐PKG signalling pathway, total of six signalling pathways, in these two diseases. Moreover, PRF1 and IL2RB are the primary genes in both diseases. Potentially helpful medications for these diseases include bosentan, eldecalcitol, ramipril, and probucol, although additional research is required to demonstrate their effectiveness. Notably, miR‐375 was found to be a diagnostic marker for the two disorders. These findings expand our understanding of the diseases. At present, there is no suitable animal model for HFpEF, not to mention the exploration of HFpEF and NAFLD in the same model, targeting Th17, NK cells, or related inflammatory factors and intervention at the gene level may provide new ideas for the construction of the model. However, this study has some limitations. First, only one dataset was used and no validation sets for HFpEF were used. Second, although the results were validated using additional NAFLD datasets, clinical data were missing due to the retrospective nature of the study. It was scarce in the aspect of biological function verification of target genes in in vitro and in vivo experiments. We plan to resolve these issues in our future studies.
Conflict of interest
None declared.
Funding
This study was supported by the National Key Research and Development Program of China (No. 2018YFC1707410‐02) and Capital Clinical Characteristic Application Research (No. Z181100001718128).
Supporting information
Figure S1. Data evaluation. (A) Normalized expression of GSE192886. (B) Normalized expression of GSE126848. (C) Normalized expression of GSE89632. (D) Normalized expression of GSE185062. (E) PCA of GSE192886. (F) PCA of GSE126848. (G) PCA of GS E89632. (H) PCA of GSE185062.
Table S1. Summary of the four GEO datasets involving HFrEF and NAFLD.
Table S2. Cluster details based on MCODE plug‐in.
Table S3. Top 10 predicted target drugs.
Acknowledgements
We would like to acknowledge the Home for Researchers editorial team (www.home‐for‐researchers.com) for their language editing service.
Wang, A. , Li, Z. , Sun, Z. , Wang, Y. , Fu, S. , Zhang, D. , and Ma, X. (2023) Heart failure with preserved ejection fraction and non‐alcoholic fatty liver disease: new insights from bioinformatics. ESC Heart Failure, 10: 416–431. 10.1002/ehf2.14211.
References
- 1. Disease GBD, Injury I, Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018; 392: 1789–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shah SJ, Borlaug BA, Kitzman DW, McCulloch AD, Blaxall BC, Agarwal R, Chirinos JA, Collins S, Deo RC, Gladwin MT, Granzier H. Research priorities for heart failure with preserved ejection fraction: National Heart, Lung, and Blood Institute working group summary. Circulation 2020; 141: 1001–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oktay AA, Rich JD, Shah SJ. The emerging epidemic of heart failure with preserved ejection fraction. Curr Heart Fail Rep 2013; 10: 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Steinberg BA, Zhao X, Heidenreich PA, Peterson ED, Bhatt DL, Cannon CP, Hernandez AF, Fonarow GC. Trends in patients hospitalized with heart failure and preserved left ventricular ejection fraction: prevalence, therapies, and outcomes. Circulation 2012; 126: 65–75. [DOI] [PubMed] [Google Scholar]
- 5. Gladden JD, Chaanine AH, Redfield MM. Heart failure with preserved ejection fraction. Annu Rev Med 2018; 69: 65–79. [DOI] [PubMed] [Google Scholar]
- 6. Cotter TG, Rinella M. Nonalcoholic fatty liver disease 2020: the state of the disease. Gastroenterology 2020; 158: 1851–1864. [DOI] [PubMed] [Google Scholar]
- 7. Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease—meta‐analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016; 64: 73–84. [DOI] [PubMed] [Google Scholar]
- 8. Li J, Zou B, Yeo YH, Feng Y, Xie X, Lee DH, Fujii H, Wu Y, Kam LY, Ji F, Li X, Chien N, Wei M, Ogawa E, Zhao C, Wu X, Stave CD, Henry L, Barnett S, Takahashi H, Furusyo N, Eguchi Y, Hsu YC, Lee TY, Ren W, Qin C, Jun DW, Toyoda H, Wong VWS, Cheung R, Zhu Q, Nguyen MH. Prevalence, incidence, and outcome of non‐alcoholic fatty liver disease in Asia, 1999–2019: a systematic review and meta‐analysis. Lancet Gastroenterol Hepatol 2019; 4: 389–398. [DOI] [PubMed] [Google Scholar]
- 9. Mantovani A, Csermely A, Petracca G, Beatrice G, Corey KE, Simon TG, Byrne CD, Targher G. Non‐alcoholic fatty liver disease and risk of fatal and non‐fatal cardiovascular events: an updated systematic review and meta‐analysis. Lancet Gastroenterol Hepatol 2021; 6: 903–913. [DOI] [PubMed] [Google Scholar]
- 10. Fudim M, Zhong L, Patel KV, Khera R, Abdelmalek MF, Diehl AM, McGarrah RW, Molinger J, Moylan CA, Rao VN, Wegermann K, Neeland IJ, Halm EA, das SR, Pandey A. Nonalcoholic fatty liver disease and risk of heart failure among Medicare beneficiaries. J Am Heart Assoc 2021; 10: e021654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simon TG, Roelstraete B, Hagstrom H, Sundstrom J, Ludvigsson JF. Non‐alcoholic fatty liver disease and incident major adverse cardiovascular events: results from a nationwide histology cohort. Gut 2021; 70: 1375–1382. [DOI] [PubMed] [Google Scholar]
- 12. Vita T, Murphy DJ, Osborne MT, Bajaj NS, Keraliya A, Jacob S, Diaz Martinez AJ, Nodoushani A, Bravo P, Hainer J, Bibbo CF, Steigner ML, Taqueti VR, Skali H, Blankstein R, di Carli MF, Dorbala S. Association between nonalcoholic fatty liver disease at CT and coronary microvascular dysfunction at myocardial perfusion PET/CT. Radiology 2019; 291: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wijarnpreecha K, Lou S, Panjawatanan P, Cheungpasitporn W, Pungpapong S, Lukens FJ, Ungprasert P. Association between diastolic cardiac dysfunction and nonalcoholic fatty liver disease: a systematic review and meta‐analysis. Dig Liver Dis 2018; 50: 1166–1175. [DOI] [PubMed] [Google Scholar]
- 14. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002; 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Watson CJ, Gupta SK, O'Connell E, Thum S, Glezeva N, Fendrich J, Gallagher J, Ledwidge M, Grote‐Levi L, McDonald K, Thum T. MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Eur J Heart Fail 2015; 17: 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Everett BM, Cornel JH, Lainscak M, Anker SD, Abbate A, Thuren T, Libby P, Glynn RJ, Ridker PM. Anti‐inflammatory therapy with canakinumab for the prevention of hospitalization for heart failure. Circulation 2019; 139: 1289–1299. [DOI] [PubMed] [Google Scholar]
- 17. Byrne CD, Targher G. Non‐alcoholic fatty liver disease‐related risk of cardiovascular disease and other cardiac complications. Diabetes Obes Metab 2022; 24: 28–43. [DOI] [PubMed] [Google Scholar]
- 18. Zewinger S, Reiser J, Jankowski V, Alansary D, Hahm E, Triem S, Klug M, Schunk SJ, Schmit D, Kramann R, Körbel C, Ampofo E, Laschke MW, Selejan SR, Paschen A, Herter T, Schuster S, Silbernagel G, Sester M, Sester U, Aßmann G, Bals R, Kostner G, Jahnen‐Dechent W, Menger MD, Rohrer L, März W, Böhm M, Jankowski J, Kopf M, Latz E, Niemeyer BA, Fliser D, Laufs U, Speer T. Apolipoprotein C3 induces inflammation and organ damage by alternative inflammasome activation. Nat Immunol 2020; 21: 30–41. [DOI] [PubMed] [Google Scholar]
- 19. Feldman N, Rotter‐Maskowitz A, Okun E. DAMPs as mediators of sterile inflammation in aging‐related pathologies. Ageing Res Rev 2015; 24: 29–39. [DOI] [PubMed] [Google Scholar]
- 20. Franssen C, Chen S, Unger A, Korkmaz HI, De Keulenaer GW, Tschope C, Leite‐Moreira AF, Musters R, Niessen HW, Linke WA, Paulus WJ. Myocardial microvascular inflammatory endothelial activation in heart failure with preserved ejection fraction. JACC Heart Fail 2016; 4: 312–324. [DOI] [PubMed] [Google Scholar]
- 21. Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, Sobel RA, Regev A, Kuchroo VK. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol 2012; 13: 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xue GL, Li DS, Wang ZY, Liu Y, Yang JM, Li CZ, Li XD, Ma JD, Zhang MM, Lu YJ, Li Y, Yang BF, Pan ZW. Interleukin‐17 upregulation participates in the pathogenesis of heart failure in mice via NF‐κB‐dependent suppression of SERCA2a and Cav1.2 expression. Acta Pharmacol Sin 2021; 42: 1780–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu W, Wang X, Feng W, Li S, Tian W, Xu T, Song Y, Zhang Z. Lentivirus mediated IL‐17R blockade improves diastolic cardiac function in spontaneously hypertensive rats. Exp Mol Pathol 2011; 91: 362–367. [DOI] [PubMed] [Google Scholar]
- 24. Xu L, Chen G, Liang Y, Zhou C, Zhang F, Fan T, Chen X, Zhou H, Yuan W. T helper 17 cell responses induce cardiac hypertrophy and remodeling in essential hypertension. Pol Arch Intern Med 2021; 131: 257–265. [DOI] [PubMed] [Google Scholar]
- 25. Chackelevicius CM, Gambaro SE, Tiribelli C, Rosso N. Th17 involvement in nonalcoholic fatty liver disease progression to non‐alcoholic steatohepatitis. World J Gastroenterol 2016; 22: 9096–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moreno‐Fernandez ME, Giles DA, Oates JR, Chan CC, Damen M, Doll JR, Stankiewicz TE, Chen X, Chetal K, Karns R, Weirauch MT. PKM2‐dependent metabolic skewing of hepatic Th17 cells regulates pathogenesis of non‐alcoholic fatty liver disease. Cell Metab 2021; 33: 1187–204 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang Y, Zhang K, Liu M, Su J, Qin X, Wang X, Zhang J, Li S, Fan G. An herbal preparation ameliorates heart failure with preserved ejection fraction by alleviating microvascular endothelial inflammation and activating NO‐cGMP‐PKG pathway. Phytomedicine 2021; 91: 153633. [DOI] [PubMed] [Google Scholar]
- 28. Komai K, Ito M, Nomura S, Shichino S, Katoh M, Yamada S, Ko T, Iizuka‐Koga M, Nakatsukasa H, Yoshimura A. Single‐cell analysis revealed the role of CD8+ effector T cells in preventing cardioprotective macrophage differentiation in the early phase of heart failure. Front Immunol 2021; 12: 763647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McVey JC, Green BL, Ruf B, McCallen JD, Wabitsch S, Subramanyam V, Diggs LP, Heinrich B, Greten TF, Ma C. NAFLD indirectly impairs antigen‐specific CD8+ T cell immunity against liver cancer in mice. iScience 2022; 25: 103847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Munoz‐Durango N, Arrese M, Hernandez A, Jara E, Kalergis AM, Cabrera D. A mineralocorticoid receptor deficiency in myeloid cells reduces liver steatosis by impairing activation of CD8+ T cells in a nonalcoholic steatohepatitis mouse model. Front Immunol 2020; 11: 563434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martini E, Kunderfranco P, Peano C, Carullo P, Cremonesi M, Schorn T, Carriero R, Termanini A, Colombo FS, Jachetti E, Panico C, Faggian G, Fumero A, Torracca L, Molgora M, Cibella J, Pagiatakis C, Brummelman J, Alvisi G, Mazza EMC, Colombo MP, Lugli E, Condorelli G, Kallikourdis M. Single‐cell sequencing of mouse heart immune infiltrate in pressure overload‐driven heart failure reveals extent of immune activation. Circulation 2019; 140: 2089–2107. [DOI] [PubMed] [Google Scholar]
- 32. Wang F, Zhang X, Liu W, Zhou Y, Wei W, Liu D, Wong CC, Sung JJY, Yu J. Activated natural killer cell promotes nonalcoholic steatohepatitis through mediating JAK/STAT pathway. Cell Mol Gastroenterol Hepatol 2022; 13: 257–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tosello‐Trampont AC, Krueger P, Narayanan S, Landes SG, Leitinger N, Hahn YS. NKp46+ natural killer cells attenuate metabolism‐induced hepatic fibrosis by regulating macrophage activation in mice. Hepatology 2016; 63: 799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sakamoto Y, Yoshio S, Doi H, Mori T, Matsuda M, Kawai H, Shimagaki T, Yoshikawa S, Aoki Y, Osawa Y, Yoshida Y, Arai T, Itokawa N, Atsukawa M, Ito T, Honda T, Mise Y, Ono Y, Takahashi Y, Saiura A, Taketomi A, Kanto T. Increased frequency of dysfunctional Siglec‐7−CD57+PD‐1+ natural killer cells in patients with non‐alcoholic fatty liver disease. Front Immunol 2021; 12: 603133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang N, Ma Q, You Y, Xia X, Xie C, Huang Y, Wang Z, Ye F, Yu Z, Xie X. CXCR4‐dependent macrophage‐to‐fibroblast signaling contributes to cardiac diastolic dysfunction in heart failure with preserved ejection fraction. Int J Biol Sci 2022; 18: 1271–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patel B, Bansal SS, Ismahil MA, Hamid T, Rokosh G, Mack M, Prabhu SD. CCR2+ monocyte‐derived infiltrating macrophages are required for adverse cardiac remodeling during pressure overload. JACC Basic Transl Sci 2018; 3: 230–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Glezeva N, Voon V, Watson C, Horgan S, McDonald K, Ledwidge M, Baugh J. Exaggerated inflammation and monocytosis associate with diastolic dysfunction in heart failure with preserved ejection fraction: evidence of M2 macrophage activation in disease pathogenesis. J Card Fail 2015; 21: 167–177. [DOI] [PubMed] [Google Scholar]
- 38. Pan X, Chiwanda Kaminga A, Liu A, Wen SW, Chen J, Luo J. Chemokines in non‐alcoholic fatty liver disease: a systematic review and network meta‐analysis. Front Immunol 2020; 11: 1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu L, Kitade H, Ni Y, Ota T. Roles of chemokines and chemokine receptors in obesity‐associated insulin resistance and nonalcoholic fatty liver disease. Biomolecules 2015; 5: 1563–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Itier R, Guillaume M, Ricci JE, Roubille F, Delarche N, Picard F, Galinier M, Roncalli J. Non‐alcoholic fatty liver disease and heart failure with preserved ejection fraction: from pathophysiology to practical issues. ESC Heart Fail 2021; 8: 789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morikawa R, Nakamoto N, Amiya T, Chu PS, Koda Y, Teratani T, Suzuki T, Kurebayashi Y, Ueno A, Taniki N, Miyamoto K, Yamaguchi A, Shiba S, Katayama T, Yoshida K, Takada Y, Ishihara R, Ebinuma H, Sakamoto M, Kanai T. Role of CC chemokine receptor 9 in the progression of murine and human non‐alcoholic steatohepatitis. J Hepatol 2021; 74: 511–521. [DOI] [PubMed] [Google Scholar]
- 42. Dinarello CA. Historical insights into cytokines. Eur J Immunol 2007; 37: S34–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Deng Y, Xie M, Li Q, Xu X, Ou W, Zhang Y, Xiao H, Yu H, Zheng Y, Liang Y, Jiang C, Chen G, du D, Zheng W, Wang S, Gong M, Chen Y, Tian R, Li T. Targeting mitochondria‐inflammation circuit by β‐hydroxybutyrate mitigates HFpEF. Circ Res 2021; 128: 232–245. [DOI] [PubMed] [Google Scholar]
- 44. Seiler M, Bowen TS, Rolim N, Dieterlen MT, Werner S, Hoshi T, Fischer T, Mangner N, Linke A, Schuler G, Halle M, Wisloff U, Adams V. Skeletal muscle alterations are exacerbated in heart failure with reduced compared with preserved ejection fraction: mediated by circulating cytokines? Circ Heart Fail 2016; 9: e003027. [DOI] [PubMed] [Google Scholar]
- 45. Tanaka K, Wilson RM, Essick EE, Duffen JL, Scherer PE, Ouchi N, Sam F. Effects of adiponectin on calcium‐handling proteins in heart failure with preserved ejection fraction. Circ Heart Fail 2014; 7: 976–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carris NW, Mhaskar R, Coughlin E, Bracey E, Tipparaju SM, Halade GV. Novel biomarkers of inflammation in heart failure with preserved ejection fraction: analysis from a large prospective cohort study. BMC Cardiovasc Disord 2022; 22: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chia YC, Kieneker LM, van Hassel G, Binnenmars SH, Nolte IM, van Zanden JJ, van der Meer P, Navis G, Voors AA, Bakker SJL, de Borst MH, Eisenga MF. Interleukin 6 and development of heart failure with preserved ejection fraction in the general population. J Am Heart Assoc 2021; 10: e018549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chan MM, Santhanakrishnan R, Chong JP, Chen Z, Tai BC, Liew OW, Ng TP, Ling LH, Sim D, Leong KT, Yeo PS. Growth differentiation factor 15 in heart failure with preserved vs. reduced ejection fraction. Eur J Heart Fail 2016; 18: 81–88. [DOI] [PubMed] [Google Scholar]
- 49. Duan Y, Pan X, Luo J, Xiao X, Li J, Bestman PL, Luo M. Association of inflammatory cytokines with non‐alcoholic fatty liver disease. Front Immunol 2022; 13: 880298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Galuppo B, Agazzi C, Pierpont B, Chick J, Li Z, Caprio S, Santoro N. Growth differentiation factor 15 (GDF15) is associated with non‐alcoholic fatty liver disease (NAFLD) in youth with overweight or obesity. Nutr Diabetes 2022; 12: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bayliss J, Ooi GJ, De Nardo W, Shah YJH, Montgomery MK, McLean C, Kemp W, Roberts SK, Brown WA, Burton PR, Watt MJ. Ectodysplasin A is increased in non‐alcoholic fatty liver disease, but is not associated with type 2 diabetes. Front Endocrinol (Lausanne) 2021; 12: 642432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim YS, Lee SH, Park SG, Won BY, Chun H, Cho DY, Kim MJ, Lee JE, Haam JH, Han K. Low levels of total and high‐molecular‐weight adiponectin may predict non‐alcoholic fatty liver in Korean adults. Metabolism 2020; 103: 154026. [DOI] [PubMed] [Google Scholar]
- 53. Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol 2000; 1: 11–21. [DOI] [PubMed] [Google Scholar]
- 54. Miranda‐Silva D, Wust RCI, Conceicao G, Goncalves‐Rodrigues P, Goncalves N, Goncalves A, Kuster DW, Leite‐Moreira AF, van der Velden J, de Sousa Beleza JM, Magalhães J. Disturbed cardiac mitochondrial and cytosolic calcium handling in a metabolic risk‐related rat model of heart failure with preserved ejection fraction. Acta Physiol (Oxf) 2020; 228: e13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang L, Zhang Y, Jiang Y, Dou X, Li S, Chai H, Qian Q, Wang M. Upregulated SOCC and IP3R calcium channels and subsequent elevated cytoplasmic calcium signaling promote nonalcoholic fatty liver disease by inhibiting autophagy. Mol Cell Biochem 2021; 476: 3163–3175. [DOI] [PubMed] [Google Scholar]
- 56. Bhowmick S, Singh V, Jash S, Lal M, Sinha RS. Mitochondrial metabolism and calcium homeostasis in the development of NAFLD leading to hepatocellular carcinoma. Mitochondrion 2021; 58: 24–37. [DOI] [PubMed] [Google Scholar]
- 57. Aikawa T, Naya M, Obara M, Manabe O, Tomiyama Y, Magota K, Yamada S, Katoh C, Tamaki N, Tsutsui H. Impaired myocardial sympathetic innervation is associated with diastolic dysfunction in heart failure with preserved ejection fraction: 11C‐hydroxyephedrine PET study. J Nucl Med 2017; 58: 784–790. [DOI] [PubMed] [Google Scholar]
- 58. Jung I, Lee DY, Lee MY, Kwon H, Rhee EJ, Park CY, Oh KW, Lee WY, Park SW, Park SE. Autonomic imbalance increases the risk for non‐alcoholic fatty liver disease. Front Endocrinol (Lausanne) 2021; 12: 752944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sun W, Zhang D, Sun J, Xu B, Sun K, Wang T, Ren C, Li J, Chen Y, Xu M, Bi Y, Xu Q, Wang W, Gu Y, Ning G. Association between non‐alcoholic fatty liver disease and autonomic dysfunction in a Chinese population. QJM 2015; 108: 617–624. [DOI] [PubMed] [Google Scholar]
- 60. Xiao S, Li Q, Hu L, Yu Z, Yang J, Chang Q, Chen Z, Hu G. Soluble guanylate cyclase stimulators and activators: where are we and where to go? Mini Rev Med Chem 2019; 19: 1544–1557. [DOI] [PubMed] [Google Scholar]
- 61. van Heerebeek L, Hamdani N, Falcao‐Pires I, Leite‐Moreira AF, Begieneman MP, Bronzwaer JG, van der Velden J, Stienen GJ, Laarman GJ, Somsen A, Verheugt FW. Low myocardial protein kinase G activity in heart failure with preserved ejection fraction. Circulation 2012; 126: 830–839. [DOI] [PubMed] [Google Scholar]
- 62. Takimoto E. Cyclic GMP‐dependent signaling in cardiac myocytes. Circ J 2012; 76: 1819–1825. [DOI] [PubMed] [Google Scholar]
- 63. Kolijn D, Kovacs A, Herwig M, Lodi M, Sieme M, Alhaj A, Sandner P, Papp Z, Reusch PH, Haldenwang P, Falcão‐Pires I. Enhanced cardiomyocyte function in hypertensive rats with diastolic dysfunction and human heart failure patients after acute treatment with soluble guanylyl cyclase (sGC) activator. Front Physiol 2020; 11: 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang A, Liu M, Shang W, Liu J, Dai Z, Strappe P, Zhou Z. Attenuation of metabolic syndrome in the ob/ob mouse model by resistant starch intervention is dose dependent. Food Funct 2019; 10: 7940–7951. [DOI] [PubMed] [Google Scholar]
- 65. Flores‐Costa R, Alcaraz‐Quiles J, Titos E, Lopez‐Vicario C, Casulleras M, Duran‐Guell M, Rius B, Diaz A, Hall K, Shea C, Sarno R. The soluble guanylate cyclase stimulator IW‐1973 prevents inflammation and fibrosis in experimental non‐alcoholic steatohepatitis. Br J Pharmacol 2018; 175: 953–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pilling LC, Joehanes R, Melzer D, Harries LW, Henley W, Dupuis J, Lin H, Mitchell M, Hernandez D, Ying SX, Lunetta KL, Benjamin EJ, Singleton A, Levy D, Munson P, Murabito JM, Ferrucci L. Gene expression markers of age‐related inflammation in two human cohorts. Exp Gerontol 2015; 70: 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang T, Sun G, Wang Y, Li S, Zhao X, Zhang C, Jin H, Tian D, Liu K, Shi W, Tian Y, Zhang D. The immunoregulatory effects of CD8 T‐cell‐derived perforin on diet‐induced nonalcoholic steatohepatitis. FASEB J 2019; 33: 8490–8503. [DOI] [PubMed] [Google Scholar]
- 68. Smith GA, Taunton J, Weiss A. IL‐2Rβ abundance differentially tunes IL‐2 signaling dynamics in CD4+ and CD8+ T cells. Sci Signal 2017; 10: eaan4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xia J, Yuan J, Xin L, Zhang Y, Kong S, Chen Y, Yang S, Li K. Transcriptome analysis on the inflammatory cell infiltration of nonalcoholic steatohepatitis in Bama minipigs induced by a long‐term high‐fat, high‐sucrose diet. PLoS ONE 2014; 9: e113724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Garikipati VNS, Verma SK, Jolardarashi D, Cheng Z, Ibetti J, Cimini M, Tang Y, Khan M, Yue Y, Benedict C, Nickoloff E, Truongcao MM, Gao E, Krishnamurthy P, Goukassian DA, Koch WJ, Kishore R. Therapeutic inhibition of miR‐375 attenuates post‐myocardial infarction inflammatory response and left ventricular dysfunction via PDK‐1‐AKT signalling axis. Cardiovasc Res 2017; 113: 938–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang H, Tian Y, Liang D, Fu Q, Jia L, Wu D, Zhu X. The effects of inhibition of microRNA‐375 in a mouse model of doxorubicin‐induced cardiac toxicity. Med Sci Monit 2020; 26: e920557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Goswami SK, Ranjan P, Dutta RK, Verma SK. Management of inflammation in cardiovascular diseases. Pharmacol Res. 2021; 173: 105912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Guo Y, Xiong Y, Sheng Q, Zhao S, Wattacheril J, Flynn CR. A micro‐RNA expression signature for human NAFLD progression. J Gastroenterol 2016; 51: 1022–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pirola CJ, Fernandez Gianotti T, Castano GO, Mallardi P, San Martino J, Ledesma MM, Flichman D, Mirshahi F, Sanyal AJ, Sookoian S. Circulating microRNA signature in non‐alcoholic fatty liver disease: from serum non‐coding RNAs to liver histology and disease pathogenesis. Gut 2015; 64: 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang Z, Moon R, Thorne JL, Moore JB. NAFLD and vitamin D: evidence for intersection of microRNA‐regulated pathways. Nutr Res Rev 2021: 1–20. [DOI] [PubMed] [Google Scholar]
- 76. Ruopp NF, Cockrill BA. Diagnosis and treatment of pulmonary arterial hypertension: a review. JAMA 2022; 327: 1379–1391. [DOI] [PubMed] [Google Scholar]
- 77. van der Graaff D, Chotkoe S, De Winter B, De Man J, Casteleyn C, Timmermans JP, Pintelon I, Vonghia L, Kwanten WJ, Francque S. Vasoconstrictor antagonism improves functional and structural vascular alterations and liver damage in rats with early NAFLD. JHEP Rep 2022; 4: 100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Derella CC, Blanks AM, Nguyen A, Looney J, Tucker MA, Jeong J, Rodriguez‐Miguelez P, Thomas J, Lyon M, Pollock DM, Harris RA. Dual endothelin receptor antagonism increases resting energy expenditure in people with increased adiposity. Am J Physiol Endocrinol Metab 2022; 322: E508–E516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liu H, Wang G, Wu T, Mu Y, Gu W. Efficacy and safety of eldecalcitol for osteoporosis: a meta‐analysis of randomized controlled trials. Front Endocrinol (Lausanne) 2022; 13: 854439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nolte K, Herrmann‐Lingen C, Platschek L, Holzendorf V, Pilz S, Tomaschitz A, Düngen HD, Angermann CE, Hasenfuß G, Pieske B, Wachter R, Edelmann F. Vitamin D deficiency in patients with diastolic dysfunction or heart failure with preserved ejection fraction. ESC Heart Fail 2019; 6: 262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang R, Wang M, Wang M, Zhang L, Ding Y, Tang Z, Fu Z, Fan H, Zhang W, Wang J. Vitamin D level and vitamin D receptor genetic variation were involved in the risk of non‐alcoholic fatty liver disease: a case‐control study. Front Endocrinol (Lausanne) 2021; 12: 648844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lukenda Zanko V, Domislovic V, Trkulja V, Krznaric‐Zrnic I, Turk‐Wensveen T, Krznaric Z, Filipec Kanizaj T, Radic‐Kristo D, Bilic‐Zulle L, Orlic L, Dinjar‐Kujundzic P, Poropat G, Stimac D, Hauser G, Mikolasevic I. Vitamin D for treatment of non‐alcoholic fatty liver disease detected by transient elastography: a randomized, double‐blind, placebo‐controlled trial. Diabetes Obes Metab 2020; 22: 2097–2106. [DOI] [PubMed] [Google Scholar]
- 83. Guo XF, Wang C, Yang T, Li S, Li KL, Li D. Vitamin D and non‐alcoholic fatty liver disease: a meta‐analysis of randomized controlled trials. Food Funct 2020; 11: 7389–7399. [DOI] [PubMed] [Google Scholar]
- 84. Lin Y, Wu M, Liao B, Pang X, Chen Q, Yuan J, Dong S. Comparison of pharmacological treatment effects on long‐time outcomes in heart failure with preserved ejection fraction: a network meta‐analysis of randomized controlled trials. Front Pharmacol 2021; 12: 707777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yip GW, Wang M, Wang T, Chan S, Fung JW, Yeung L, Yip T, Lau ST, Lau CP, Tang MO, Yu CM. The Hong Kong diastolic heart failure study: a randomised controlled trial of diuretics, irbesartan and ramipril on quality of life, exercise capacity, left ventricular global and regional function in heart failure with a normal ejection fraction. Heart 2008; 94: 573–580. [DOI] [PubMed] [Google Scholar]
- 86. Zhang X, Wong GL, Yip TC, Tse YK, Liang LY, Hui VW, Lin H, Li GL, Lai JC, Chan HL, Wong VW. Angiotensin‐converting enzyme inhibitors prevent liver‐related events in nonalcoholic fatty liver disease. Hepatology 2022; 76: 469–482. [DOI] [PubMed] [Google Scholar]
- 87. Parthasarathy S, Young SG, Witztum JL, Pittman RC, Steinberg D. Probucol inhibits oxidative modification of low density lipoprotein. J Clin Invest 1986; 77: 641–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yamashita S, Arai H, Bujo H, Masuda D, Ohama T, Ishibashi T, Yanagi K, Doi Y, Nakagawa S, Yamashiro K, Tanabe K, Kita T, Matsuzaki M, Saito Y, Fukushima M, Matsuzawa Y, on behalf of the PROSPECTIVE Study Group . Probucol trial for secondary prevention of atherosclerotic events in patients with coronary heart disease (PROSPECTIVE). J Atheroscler Thromb 2021; 28: 103–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ishitobi T, Hyogo H, Tokumo H, Arihiro K, Chayama K. Efficacy of probucol for the treatment of non‐alcoholic steatohepatitis with dyslipidemia: an open‐label pilot study. Hepatol Res 2014; 44: 429–435. [DOI] [PubMed] [Google Scholar]
- 90. Merat S, Malekzadeh R, Sohrabi MR, Sotoudeh M, Rakhshani N, Sohrabpour AA, Naserimoghadam S. Probucol in the treatment of non‐alcoholic steatohepatitis: a double‐blind randomized controlled study. J Hepatol 2003; 38: 414–418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Data evaluation. (A) Normalized expression of GSE192886. (B) Normalized expression of GSE126848. (C) Normalized expression of GSE89632. (D) Normalized expression of GSE185062. (E) PCA of GSE192886. (F) PCA of GSE126848. (G) PCA of GS E89632. (H) PCA of GSE185062.
Table S1. Summary of the four GEO datasets involving HFrEF and NAFLD.
Table S2. Cluster details based on MCODE plug‐in.
Table S3. Top 10 predicted target drugs.


