Abstract
Extrahepatic nucleic acid delivery using polymers typically requires the synthesis and purification of custom monomers, post-synthetic modifications, and incorporation of additional excipients to augment their stability, endosomal escape, and in vivo effectiveness. Here, we report the development of a single-component and excipient-free, polyester-based nucleic acid delivery nanoparticle platform comprised of ionizable N-methyldiethanolamine (MDET) and various hydrophobic alkyl diols (Cp) that achieves lung-selective nucleic acid transfection in vivo. PolyMDET and polyMDET-Cp polyplexes displayed high serum and enzymatic stability, while delivering pDNA or mRNA to “hard-to-transfect” innate immune cells. PolyMDET-C4 and polyMDET-C6 mediated high protein expression in lung alveolar macrophages and dendritic cells without inducing tissue damage or systemic inflammatory responses. Improved strategies using readily-available starting materials to produce a simple, excipient-free, non-viral nucleic acid delivery platform with lung-selective and innate immune cell tropism has the potential to expedite clinical deployment of polymer-based genetic medicines.
Keywords: Ionizable Polyesters, Innate Immune Cells, Transfection, Extrahepatic, Excipient-free
Graphical Abstract
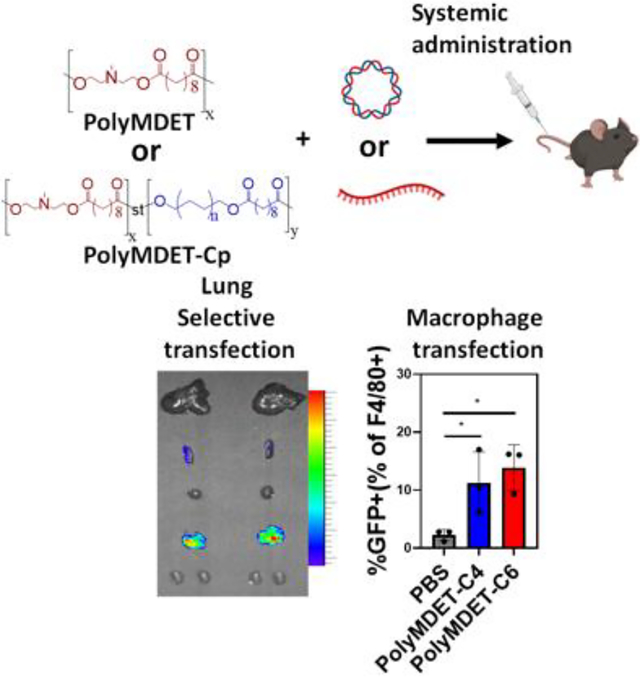
INTRODUCTION:
Nucleic acid delivery to immune cells has the potential to revolutionize therapeutic outcomes for various diseases.1 However, the delivery of nucleic acids to primary immune cells, particularly macrophages, presents several challenges that limit transfection efficiency. First, macrophages contain degradative enzymes that destroy nucleic acids after transfection and limit expression of exogenous genes.2 Second, the low-proliferative nature of primary cells restricts the entry of extracellular DNA molecules into the nucleus, which leads to marginal transfection.3 Viral and non-viral vectors have been developed to enhance nucleic acid delivery. Viral vectors (adenoviruses, adeno-associated viruses, lentiviruses) are highly efficient and have been used successfully for several preclinical and clinical applications4 but these vectors present potential safety concerns including mutagenesis, immunogenicity, and cytotoxicity.5–8 Non-viral gene delivery methods, on the other hand, offer the ability to overcome many of these limitations, while enabling a wide chemical space to be accessed to develop libraries of materials to investigate structure-function relationships though tuning of composition, surface functionality, and other physicochemical properties.8–11 Recently, significant advancements have been made in the design of non-viral nucleic acid delivery platforms including lipid nanoparticles (LNPs) and polymer-based systems.12–20 LNPs have received significant interest due to their remarkable success as a delivery platform for COVID-19 mRNA vaccines.20 These LNPs consist of a mixture of multiple components and excipients including ionizable lipid, helper lipid, PEG-lipid, and cholesterol to enable adequate qualities for mRNA delivery. Despite the advantages of LNPs, lipid-based molecules are less stable and difficult to synthesize and purify, thus limiting their cost-effectiveness.21 To facilitate the wide implementation of nucleic acid-based therapeutics, identification of alternative non-viral delivery platforms with reduced complexity that do not compromise delivery efficacy is required. Off the shelf cationic polymers such as polyethylenimine (PEI), poly(l-lysine) (PLL), and others have been thoroughly examined for nucleic acid delivery applications; yet their in vitro efficacy often does not effectively translate to in vivo applications. Strategies such as increasing the amine to phosphate (N:P) ratio offers the possibility to improve the transfection abilities of these platforms but compromises their toxicity profile.22 Functional polyester-based carriers containing ionizable subunits are advantageous due to their biodegradability, modularity, adaptability to high throughput synthesis, and low toxicity.23 For example, poly(β-amino ester)s (PBAE) are a common platform used for plasmid DNA (pDNA) and mRNA transfection; however, successful delivery to immune cells typically requires functional modification of the polymer backbone or the terminal groups in addition to the incorporation of excipients such as lipids24–26 or other polymers27 for surface coating to enable efficient transfection.
Here, we report the one-pot synthesis of hydrophobic ionizable polyesters as a single-component nucleic acid delivery platform with high serum and enzymatic stability capable of efficient transfection of “hard-to-transfect” innate immune cells with lung-selective mRNA transfection in vivo. To identify the impact of polymer design parameters on transfection, a set of ionizable polyesters (polyMDET and polyMDET-Cp) was synthesized via a polycondensation reaction from commercially-available N-methyldiethanolamine (MDET), four alkyl diols (p = 4, 6, 8, 10), and sebacoyl chloride. Polyplex stability was determined and transfection efficiency using immortalized RAW 264.7 cells and primary macrophages derived from bone marrow (BMDM) was assessed in vitro without the incorporation of any excipients or targeting ligands. Systemic administration of polyplexes by intravenous injection showed over 23-fold lung-selective protein expression and efficient transfection of lung alveolar macrophages and dendritic cells with no detectable organ toxicity or systemic inflammation. Given the current focus and challenges associated with non-viral nucleic acid therapeutics, nanoparticles prepared from polymers synthesized using simple, one-pot reactions from readily-accessible starting materials, and in the absence of any additional excipients for formulation offer great promise for translation of gene delivery technologies.
EXPERIMENTAL SECTION
Materials:
N-methyldiethanolamine (MDET), 1,4-butanediol, 1,6-hexanediol, 1,8-octanediol, 1,10-decanediol, sebacoyl chloride, pyridine, anhydrous dichloromethane (DCM, >99.8%), methanol (>99.9%), diethyl ether, chloropromazine hydrochloride (CHP), 5-[N-ethyl-N-isopropyl]-amiloride (EIPA), methyl-β-cyclodextrin (MBCD), cytochalasin D (CytD), and filipin were purchased from MilliporeSigma (St. Louis; MO). Dimethylsulfoxide-d6 and chloroform-d were purchased from Cambridge Isotope Labs Inc (Tewksbury, MA). Maxiprep Endotoxin-free kit was purchased from Qiagen (Germantown, MD). Unless otherwise noted, any additional reagents were purchased from MilliporeSigma.
RAW 264.7 Cell Culture:
RAW 264.7 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (MilliporeSigma; St. Louis, MO) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (VWR; Radnor, PA) and 1% penicillin/streptomycin (P/S) (Invitrogen Corporation; Carlsbad, CA) at 37°C and 5% CO2 atmosphere.
Bone Marrow-Derived Macrophage (BMDM) Cell Culture:
Bone marrow from the tibia and femurs of C57BL/6 mice was harvested to obtain a primary population of macrophages as previously described by our group.28 Cell media consisted of RPMI 1640 supplemented with L-glutamine (Life Technologies, Carlsbad, CA), penicillin (100 units/mL), streptomycin (100 μg/mL), and 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen Corporation, Carlsbad, CA) at 37°C and 5% CO2. The media was further supplemented with 20% L929 (ATCC, Manassas, VA) cell-conditioned medium (containing M-CSF). BMDMs were differentiated for 8 days, with cell conditioned media changes on days 3 and 6. Versene (Invivogen, San Diego, CA) was used for BMDM cell lifting. Cell count and viability were obtained using a trypan blue exclusion dye and the EVE™ automated cell counter (NanoEntek, Waltham, MA).
GFP Plasmid Preparation:
DH5α Escherichia coli competent cells (Invitrogen Corporation; Carlsbad, CA) were transformed with eGFP (GFP) pDNA (courtesy of the National Center for Toxicological Research, FDA, Jefferson, AR), which encoded for kanamycin resistance. Transformed cells were expanded in an overnight liquid LB culture at 37°C under vigorous shaking, lysed, and purified using a Qiagen Maxiprep endotoxin-free kit. The concentration of pDNA was verified using a SpectraMax iD3 microplate reader and a SpectraDrop microvolume microplate (Molecular Diagnostics; San Jose, CA) by measuring absorbance at 260 and 280 nm and agarose gel electrophoresis. The pDNA was stored at −20°C until further use.
Polymer Synthesis:
The synthesis of diethanolamine-based ionizable polyester (polyMDET): The diethanolamine-based ionizable polyester (polyMDET) was synthesized by one-pot condensation polymerization using N-methyldiethanolamine (MDET) and sebacoyl chloride in the presence of pyridine as an HCl scavenger. Initially, 595 μL of MDET (5 mmol) was dissolved in 5 mL DCM and added to a 50 mL round bottom flask. A total of 808.79 μL (10 mmol) of pyridine was added and stirred under an Ar atmosphere for 15 minutes. Next, 1 mL of sebacoyl chloride (5 mmol) was dissolved in 5 mL of DCM and slowly added to the reaction mixture for 30 minutes. After complete addition, the reaction was further carried out overnight (16–18 hours). The white precipitate, pyridine hydrochloride, was removed by centrifugation, and the supernatant containing DCM was removed using a rotary evaporator. Next, the resulting solid product was dissolved in a minimum amount of methanol. Finally, the polymer was purified by precipitation into diethyl ether to remove unreacted monomers and oligomers. This process was repeated for a total of three times. The product was dried for two days at room temperature, and 1H NMR spectra were obtained. Similarly, other polymers were also synthesized, maintaining the same polyester backbone, but integrating hydrophobic aliphatic chains (C4, C6, C8, C10) via mixing respective alkyl diols into the reaction mixture. The respective polymers were termed polyMDET-Cp, with p = the number of carbon atoms present in the diols used during the reaction. For example, polyMDET-C10 was prepared by mixing 1 mmol of 1,10-decanediol with 5 mmol of MDET and 10 mmol of pyridine in 5 mL DCM. As described earlier, the reaction was carried out by adding 6 mmol of sebacoyl chloride dropwise to the reaction mixture under Ar atmosphere.
Polymer Characterization:
The polymers were characterized by 1H NMR after dissolving the samples in DMSO-d6 or CDCl3. 1H NMR spectra was acquired using a 400 MHz Varian spectrometer. The diffusion measurement was carried out by observing the attenuation of the 1H NMR signals during a pulsed field gradient experiment using a 500 MHz Varian spectrometer in DMSO-d6. The molecular weight and polydispersity index (PDI) of the polymers was determined using MALDI-TOF mass spectrometry. The polymer and 2,5 dihydroxybenzoic acid (DHB) matrix were dissolved in methanol at 1 mg/mL and mixed. The mixture was dried on a metal sample plate and placed in the high vacuum source chamber for measurement by analyzing “time of flight” of the produced sample ions. The most probable peak was calculated from the spectra, which was reported as the molecular weight of the polymer.
pH Titration of Polymers:
1 mg/mL of each polymer was dissolved in DMSO and acidified with HCl solution (pH~2). This solution was titrated with 0.1 M NaOH by adding dropwise to the solution and final pH was measured using a pH meter. pKa was calculated from the inflection point of the obtained titration curve, which was identified as the pH at half of the neutralization point.
Polyplex Preparation and Characterization:
PolyMDET/pDNA or polyMDET-Cp/pDNA polyplexes were prepared by mixing polyMDET or polyMDET-Cp polymers with pDNA at different weight ratios (55:1, 110:1, or 220:1). Briefly, polymer solutions were prepared at a concentration of 50 mg/mL in DMSO. Next, different amounts of polymer solutions were diluted in 50 μL acetate buffer (25 mM pH 5), and 2 μg pDNA encoding GFP was added to the solution and incubated for 10 minutes at room temperature. Similarly, polyMDET/mRNA or polyMDET-Cp/mRNA polyplexes were also prepared by mixing polyMDET and polyMDET-Cp polymers with 1 μg GFP encoded mRNA (CleanCap® EGFP mRNA, TriLink Biotechnologies, San Diego, CA) at 55:1 weight ratio in the presence of 25 mM acetate buffer (pH 5). The encapsulation efficiency of pDNA and mRNA polyplexes was determined using 4’,6-diamidino-2-phenylindole (DAPI) or RediPlate 96 RiboGreen RNA quantitation kit (Invitrogen Corporation; Carlsbad, CA), respectively.29, 30 The polyplexes were prepared as described above, centrifuged, and the supernatant was collected for analysis. DAPI binds with the double stranded DNA and gives rise to 20-fold enhancement of fluorescence intensity. The RiboGreen fluorescent dye binds to mRNA and produces a fluorescent signal that is proportional to mRNA content. The fluorescence signal was measured using a SpectraMax iD3 fluorescence plate reader and the pDNA and mRNA content was determined by comparing to respective standard curves.
The polyplexes were characterized by dynamic light scattering (DLS) and zeta potential analysis. Fresh polyplex solutions were prepared with 2 μg of GFP plasmid or mRNA in three different buffer solutions (pH 5, pH 6, and pH 7.4). One milliliter of each sample was added into a disposable cuvette, and the size was measured using a Zetasizer Nano ZSP (Malvern Instruments Inc., Westborough, MA). Fifteen runs were performed in triplicate for each sample. Subsequently, samples were transferred to a folded capillary cell (Malvern), and zeta potential measurements were performed in triplicate for each sample using the Zetasizer Nano ZSP.
The stability of polyplexes was assessed by gel electrophoresis before or after incubating with 55% FBS, a physiologically relevant serum concentration, for 30–60 min. The success of a nucleic acid delivery platform relies on protecting the genetic material from enzymatic degradation, for example, DNase I present in serum, extracellular matrices, and mucosal surfaces.31 Therefore, the ability of the polyplexes to protect DNA was investigated using agarose gel electrophoresis. First, polyplexes were prepared, as described earlier, with polyMDET or polyMDET-Cp, and incubated with 20/K unit DNase I for 1 hour at room temperature. After that, the integrity of DNA was assessed by agarose gel electrophoresis (150 V, 20–40 min).
In Vitro Transfection of pDNA in RAW 264.7 Macrophages and Primary BMDMs:
PolyMDET/pDNA or polyMDET-Cp/pDNA polyplexes were prepared at a weight ratio of 110:1 containing 2 μg of pDNA encoding GFP. RAW 264.7 cells were seeded in a 24 well plate at 2 × 105 density and treated with polyplexes (2 μg pDNA/220 μg polyMDET or polyMDET-Cp) for 4 hours in serum-containing media, followed by washing to remove the excess complex prior to overnight incubation in complete media. The excess complex was removed by washing with DPBS and replaced with complete DMEM. Similarly, primary BMDMs were incubated with polyplexes for 4 hours in serum-containing RPMI media. After that, the cells were washed with DPBS and incubated with fresh complete RPMI media for 24 hours. GFP expression was visualized using a revolve fluorescence microscope (ECHO, San Diego, CA) 24 hours post-transfection. Cationic polymers PEI and jetOPTIMUS were used as positive controls for pDNA transfection. PEI/pDNA polyplexes (PEI30) were prepared at N/P ratio 30, as PEI30 showed significantly higher transfection in serum-containing media according to our previous study.22 Transfection with jetOPTIMUS was achieved by following the manufacturer’s protocol.
In Vitro Transfection of mRNA in RAW 264.7 Macrophages and Primary Bone Marrow-Derived Macrophages:
PolyMDET/mRNA or polyMDET-Cp/mRNA polyplexes were prepared by mixing polymer solution with GFP mRNA at a 110:1 weight ratio (1 μg mRNA/110 μg polyMDET or polyMDET-Cp). Next, RAW 264.7 macrophages or primary BMDMs were treated with the polyplexes (24 well plate, 2 × 105 density) and incubated for 4 hours in serum-containing media. After that, the cells were washed with DPBS and further incubated with fresh complete media for 24 hours. GFP expression was visualized using a revolve fluorescence microscope at 24 hours post-transfection. PEI30 and jetOPTIMUS were also used as positive controls for mRNA transfection.
Endosomal Escape of Polyplexes:
To examine the endosomal escape, particles were prepared by mixing PolyMDET-C6 with Cy5.5 dye at a 110:1 weight ratio by mixing at pH 5 (acetate buffer). The particles were dialyzed against DPBS buffer (pH 7.4) for 1 hour to remove free Cy5.5. Next, RAW 264.7 cells were incubated with the particles for 3 h followed by washing and incubation with fresh complete DMEM media for 24 h. The cells were then treated with 100 nM of lysotracker green for 15 minutes and washed with DPBS buffer (pH 7.4). The cells were imaged using a revolve fluorescence microscope to visualize the colocalization of polyplexes with lysotracker.
Flow Cytometry:
RAW 264.7 or primary BMDMs were seeded in a 24 well plate at a density of 2 × 105 cells/well. Next, cells were transfected with polyplexes made with different polymers and GFP encoded pDNA/mRNA in serum-containing media for 4 hours. Subsequently, cells were washed and incubated for another 24 hours in fresh serum-containing media. After 24 hours, cells were collected using a cell scraper, followed by centrifugation at 500 × g for 5 min, and resuspended in fresh flow cytometry buffer (DPBS, 5% FBS and 2% EDTA). For analysis of live cells only, DAPI was used as an exclusion dye to determine cell viability. Data were collected using an LSR II (Becton-Dickinson, San Jose, CA) flow cytometer and analyzed by FCS Express 7 (De Novo Software, Pasadena, CA). Transfection efficiency was measured as the percentage of live cells, which were GFP+ compared to non-transfected controls.
Uptake Mechanism Study using Endocytosis Inhibitors:
RAW 264.7 cells were seeded in a 24 well plate and cultured for 24 hours in complete DMEM media at a 2 × 105 cell density/well. The cells were preincubated in presence or absence of chlorpromazine (CHP) (50 μM), methyl-β-cyclodextrin (MBCD) (10 mM), Filipin (1 μg/μL), (5-[N-ethyl-N-isopropyl] amiloride) (50 μM) (EIPA), Cytochalasin D (CytD) (4 μM) for 30 min. The cells were then treated with polyplexes polyMDET/pDNA or polyMDET-Cp/pDNA for another 3 hours. The cells were subjected to flow cytometry analysis after being washed with DPBS and incubated with fresh DMEM for 24 hours. The cellular uptake of the polyplexes was evaluated by measuring the mean fluorescence intensity (MFI) of GFP signals.
Cell Viability:
The cytotoxicity of polyplexes was evaluated using an MTS assay (Abcam; Cambridge, MA). RAW 264.7 cells were seeded in a 24 well plate at a density of 2 × 105 cells/well overnight prior to treatment with polyMDET/pDNA and polyMDET-Cp/pDNA polyplexes for 4 hours in serum-containing media. Following the incubation, cells were washed and incubated with fresh complete DMEM media and incubated for 24 hours. Next, 50 μL of MTS solution was added to each well and incubated for an additional 3 hours. The optical density (OD) of the solution was measured using a SpectraMax iD3 microplate reader at 570 nm, and the percentage of cell viability was measured as the ratio of OD at 570 nm and compared to untreated control.
In Vivo mRNA Transfection:
Female C57BL/6 (6–8 weeks old) were purchased from the Jackson Laboratories. The mice were housed under specific pathogen-free conditions in the School of Medicine, University of Maryland, Baltimore animal facilities. All animal procedures were performed according to the guidelines and protocols of the University of Maryland, Baltimore Animal Care and use committee and approved by the Institutional Animal Care and use Committee (IACUC; protocol #0721010). Polyplexes were prepared in acetate buffer (25 mM; pH 5) by mixing different polymers (50 mg/mL) and FLuc mRNA (1 mg/mL) (CleanCap® FLuc mRNA (5moU), TriLink Biotechnologies, San Diego, CA) at 110:1 weight ratio. After the preparation, the polyplexes were dialyzed against DPBS buffer (pH 7.4) for 30–60 minutes to remove DMSO and acetate buffer. All the polymers were stable after dialysis except polyMDET-C10/mRNA. The polyplexes were administered to mice via tail vein injection (10 μg mRNA/injection). After 24 hours, the mice were injected with D-luciferin (300 μL, 15 mg/mL) intraperitoneally. After 15 minutes, the mice were euthanized, and various organs were collected (liver, spleen, heart, lung, kidneys) and imaged using the Xenogen IVIS® Spectrum Imaging System (Alameda, CA).
Biodistribution Study of mRNA Polyplexes In Vivo:
The polyplexes were prepared by mixing the polymers and Fluc mRNA at a 110:1 weight ratio. The polyplexes were then mixed with 10 μg of Cy5.5 (1 mg/mL) and incubated for 60 minutes at room temperature. The polyplexes were then dialyzed against DPBS buffer (pH 7.4) for 30 minutes to remove excess Cy5.5 dye. Next, the polyplexes were injected in mice intravenously by tail vein injection. After 24 hrs, the localization and transfection of polyplexes were evaluated by IVIS® imaging as described above. Cy5.5 fluorescence indicates the organ trafficking of polyplexes whereas luminescence signal indicates the luciferase expression.
Flow Cytometry For In Vivo Studies:
For the evaluation of immune cell transfection, female C57BL/6 (6–8 weeks old) were intravenously injected with GFP mRNA polyMDET-C4/mRNA and polyMDET-C6/mRNA polyplexes (10 μg mRNA/injection). After 24 hours, the mice were euthanized, and lung and spleen were collected. Single cell suspensions were prepared using a standardized protocol. First, tissue samples were placed in a petri dish and injected with Liberase (Liberase TM for lungs and Liberase TL for spleen; MilliporeSigma) followed by incubation for 10 minutes at 37°C. After that, cells were isolated by mashing the lung and spleen through a 70 μm cell strainer (Thermo Fisher) and treated ACK lysis buffer (Thermo Fisher). Cells were then pelleted by centrifuging at 500 × g for 5 minutes, followed by resuspension in DPBS supplemented with 10% FBS. Cell staining was conducted according to BioLegend protocols. All antibodies were purchased from BioLegend (San Diego, CA). Flow cytometric data were collected using a BD LSR II flow cytometer. FcR blocking was performed with anti-CD16/32 (clone 98) antibody prior to staining with extracellular antibodies: PerCP anti-mouse CD45 (clone 30-F11), PE/Cy7 anti-mouse F4/80 (clone BM8), and BV605 anti-mouse CD11c (clone N418). Viability was assessed using DAPI. Data analysis was performed using FCS Express 7 (De Novo, Glendale, CA).
Histopathological Analysis:
Organs were harvested after intravenous administration of DPBS or mRNA polyplexes prepared with PolyMDET, PolyMDET-C4, and PolyMDET-C6. After 24 h, the mice were euthanized and the liver, spleen, heart, lungs, and kidneys were collected and fixed in 4% paraformaldehyde in DPBS (pH 7.4) for 24 h for sectioning. The organ tissues were dehydrated and embedded in paraffin before being sectioned and stained with haematoxylin and eosin (H&E) and observation using the revolve microscope under brightfield.
Detection of Cytokines by Enzyme Linked Immunosorbent Assay (ELISA):
Polyplexes containing Fluc mRNA were prepared as described above and administered via tail vein injection. After 24 h, the blood was collected by cardiac puncture and stored in EDTA-coated tubes. Within 30 minutes of blood collection, the blood was centrifuged at 1000 × g to separate the cellular fraction from the plasma. The plasma was then utilized to assess the proinflammatory cytokine levels for tumor necrosis factor-alpha (TNFα) and interleukin-6 (IL-6) using ELISA following the manufacturer’s protocols (BioLegend, San Diego, CA).
RESULTS AND DISCUSSION:
Design and Synthesis of polyMDET and polyMDET-Cp:
Biodegradable polymers can overcome several challenges associated with non-viral gene delivery, including in vivo stability, susceptibility to enzymatic degradation, poor cellular uptake, and inefficient endosomal escape. Ionizable polyesters as DNA/RNA delivery platforms containing tertiary amines and synthesized from custom monomers have gained attention due to their abilities to facilitate a pH-dependent, charge-altering characteristic based on the pKa of the polymer. Further, post-synthetic modifications of side chains or end groups allow for tunable hydrophobicity or alternative charge characteristics to be engineered for optimal nucleic acid delivery.
Recognizing the need for less complex non-viral nucleic acid delivery platforms, we hypothesized that we could utilize commercially-available monomers in a one-pot reaction to create a single-component nucleic acid delivery platform for efficient pDNA and mRNA transfection in vitro and in vivo. We synthesized a set of ionizable polyesters containing tertiary amines and various hydrophobic alkyl diols by condensation polymerization (Fig. 1A and B). The main components of the polymers were MDET and sebacoyl chloride (e.g. polyMDET) and alkyl diols that varied in chain length (e.g. polyMDET-Cp) (p = 4, 6, 8, 10). Following this methodology, five types of polymers with varying degrees of hydrophobicity were synthesized and no post-synthetic modifications were performed. Statistical incorporation of hydrophobic alkyl chains into the polymer backbone produced polymers with greater hydrophobicity than the parent polyMDET polymer. Importantly, statistical polymers have shown a unique advantage in gene delivery applications, such as enhanced gene transfection and reduced toxicity in the presence of serum compared to common block copolymers.32, 33 1H NMR characterization of the synthesized polymers confirmed the presence of characteristic protons in the polymer backbone indicating successful polymerization (Fig. 1C and S1). The peak at 4.2 ppm confirmed the methylene protons of MDET (-OCH2-), and the peak around 2.2–2.3 ppm represented the methylene protons of sebacoyl chloride (-OCOCH2-) at an integration ratio of 1:1, confirming conjugation. Similarly, the incorporation of alkyl chains (Cp) generated an additional peak at 4.0 ppm for the methylene protons (-OCH2-). The integration ratio of the methylene protons of MDET and Cp was 1:0.3, implying that the percentage of Cp was 30% of the total MDET present in the polymer backbone. To confirm the absence of any unreacted monomers or oligomers, we performed diffusion ordered NMR spectroscopy (DOSY) of polyMDET and polyMDET-Cp polymers (Fig. S2). The DOSY spectrum displays the chemical shifts of NMR resonances against their translational diffusion coefficient, where signals along the same horizontal line belong to the same polymer. Analysis of the DOSY spectrum for individual polymers revealed signals with similar diffusion coefficients consistent with an efficient polymerization and absence of any monomers or oligomers.
Figure 1.
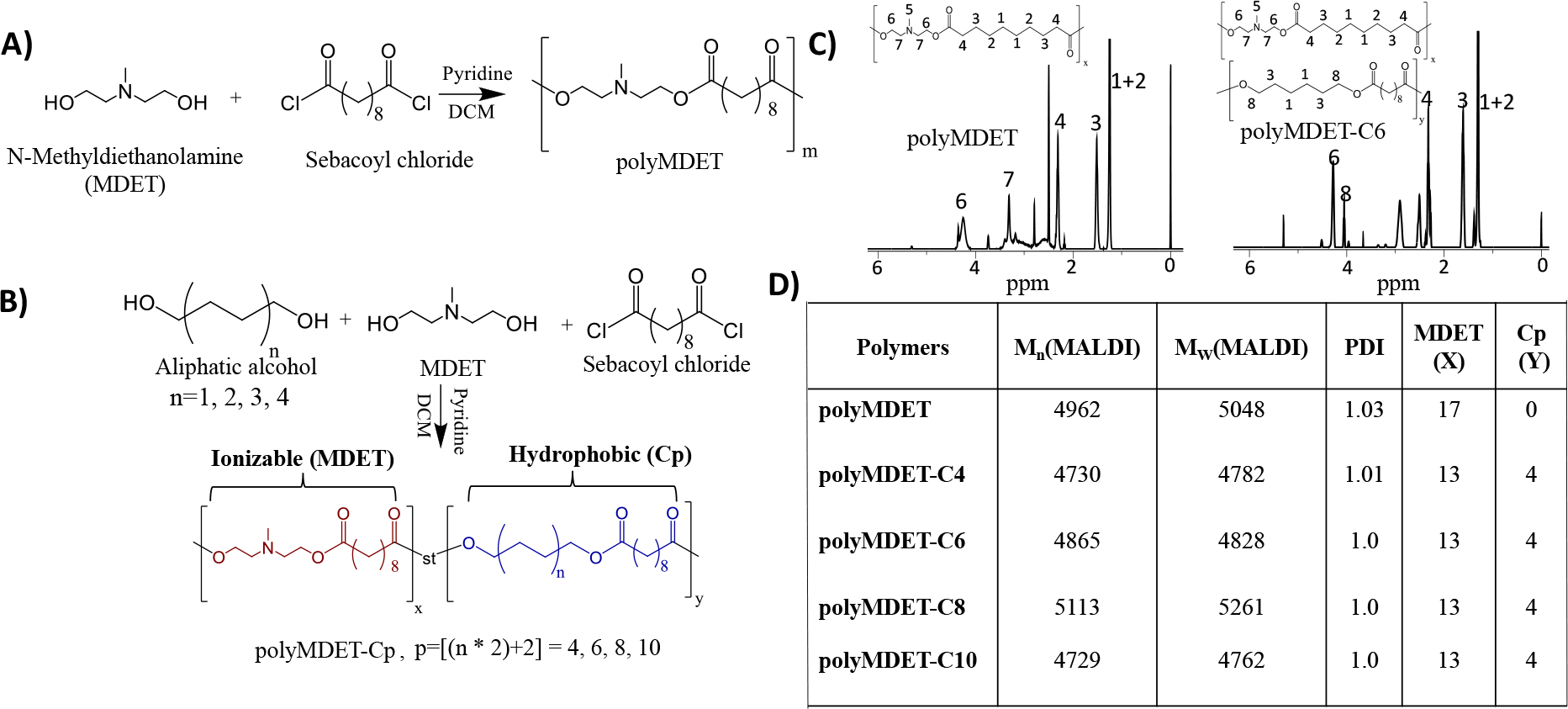
Synthesis and characterization of ionizable polyesters. Synthetic schemes for (A) polyMDET and (B) polyMDET-Cp polyesters, respectively. The polyesters were synthesized via a polycondensation reaction between N-methyldiethanolamine (MDET) and sebacoyl chloride for polyMDET or using MDET, sebacoyl chloride, and various alkyl diols for polyMDET-Cp, where “p” stands for the total number of carbons in the aliphatic chains. (C) Representative 1H NMR spectra of polyMDET and polyMDET-C6. The peak at 4.2 ppm and 1.2–1.5 ppm signifies the protons for MDET and sebacoyl chloride. The peak around 4 ppm (8) indicates the protons of the hexyl group, demonstrating successful incorporation into the polymer backbone. (D) Molecular weight and quantification of MDET and Cp determined by MALDI-TOF MS and 1H NMR spectra.
The molecular weight and polydispersity index (PDI) of the polymers was determined by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) spectrometry using 2,5 dihydroxybenzoic acid (DHB) as the matrix (Fig. 1D and S3). The m/z difference (285 Da) between the peaks corresponded to MDET blocks. The number average molecular weight (Mn), weight average molecular weight (Mw), and PDI were calculated using the following equations, where Ni and Mi represent the abundance and mass of the ith oligomer, respectively.34, 35
The molecular weight of the polymers was similar and varied between 4–5 kDa demonstrating that the incorporation of different alkyl diols minimally affected the polymerization. While we did not explore the reaction conditions that affect the molecular weight of the polymers synthesized, the same molar ratio for all the starting materials was maintained and the molecular weight of the polymers was found to be similar as shown by MALDI-TOF (Fig. S3).36
Formation of Polyplexes and Their Physicochemical Characterization:
The assembly of macromolecules is governed by the attractive forces operating between them. Cationic polymers interact electrostatically with negatively charged nucleic acids to form condensed structures called polyplexes. Studies have suggested that electrostatic interaction alone is insufficient to offer stability of the polyplexes under physiological conditions due to competition with other electrolytes present.37 Incorporation of hydrophobic ligands can enhance the cooperative binding with the nucleic acids, which facilitates increases in encapsulation efficiency, reduces the size of polyplexes, and offers improved stability. Further, hydrophobic moieties increase cell membrane interactions, increase dissociation of polyplexes, and release nucleic acid into the cytosol.38 PolyMDET and polyMDET-Cp formed polyplexes with both pDNA (polyMDET/pDNA and polyMDET-Cp/pDNA) and mRNA (polyMDET/mRNA and polyMDET-Cp/mRNA) as shown in Fig. 2A. The stability of polyMDET/pDNA or polyMDET-Cp/pDNA polyplexes at three different weight ratios of polymer to pDNA (55:1, 110:1, and 220:1) was assessed using gel electrophoresis (Fig. 2B). Generally, the polyplexes were stable at 55:1 weight ratio except polyMDET-C10, where a slight band indicating incomplete complexation was observed. PolyMDET-C6 polyplexes prepared at weight ratios lower than 55:1 were unable to stably condense pDNA (Fig. S4). Therefore, we utilized the 110:1 ratio to prepare polyMDET/pDNA and polyMDET-Cp/pDNA polyplexes. PolyMDET and polyMDET-Cp also formed stable polyplexes with mRNA yet a lower weight ratio (55:1) was required (Fig. 2C). We measured the encapsulation efficiency of pDNA and mRNA (Fig. S5), which was found to vary between 72–84% and 92–97%, respectively.
Figure 2.
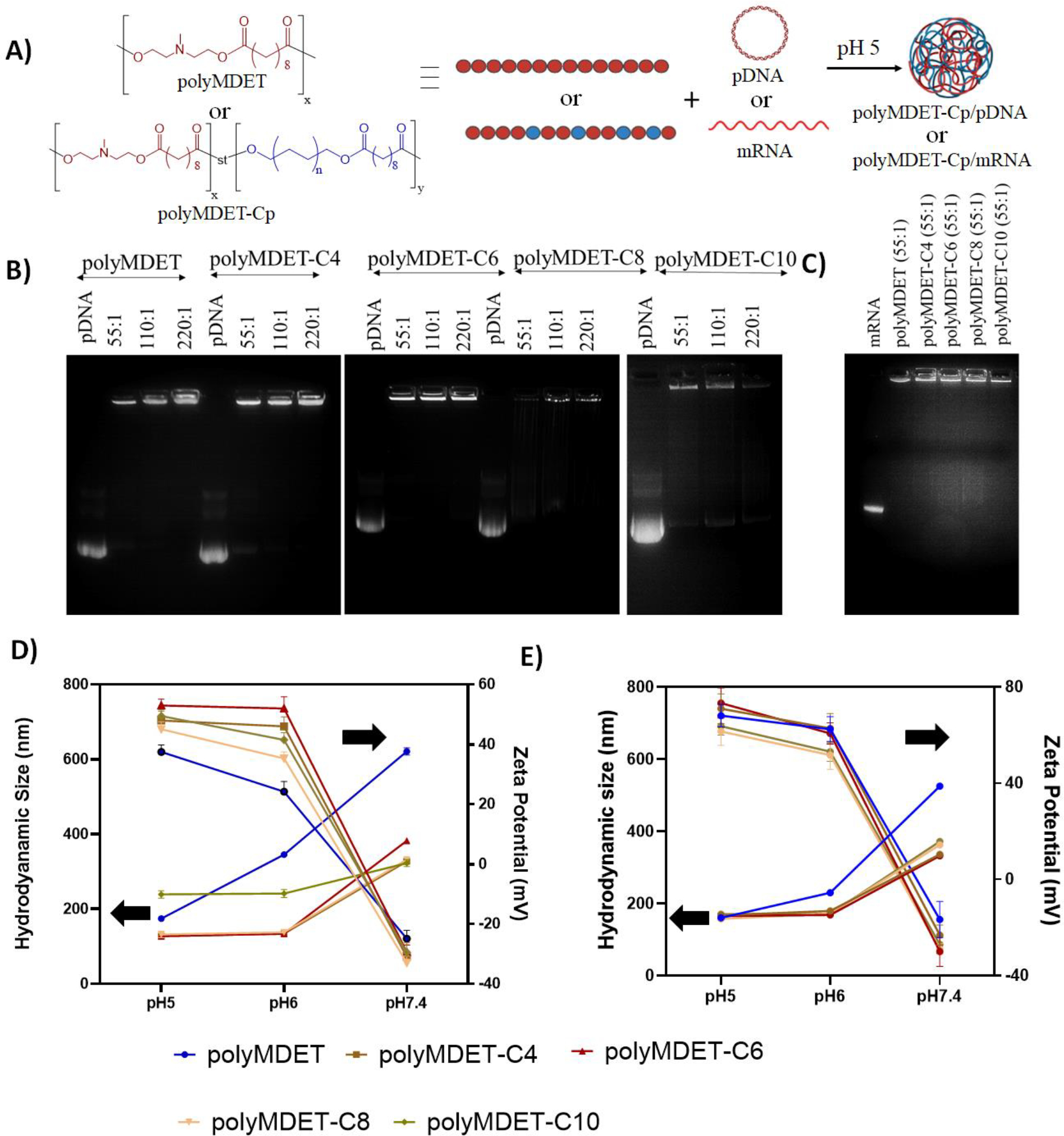
Polyplex preparation and characterization. (A) Schematic representation of the polyplex preparation with polymers and pDNA or mRNA. (B) Agarose gel electrophoresis showing the stability of polyplexes made with various polymers and pDNA at different weight ratios (55:1, 110:1, and 220:1). (C) Agarose gel electrophoresis showing the stability of polyplexes prepared using various polymers and mRNA at 55:1 weight ratio. Hydrodynamic size and zeta potential of polyplexes prepared with (D) pDNA and (E) mRNA, respectively. Data are representative of n=3 experiments. Errors bars represent standard deviation. Left arrow indicates the hydrodynamic diameter of the polyplexes, whereas the right arrow indicates the zeta potential.
The size and zeta potential of the prepared polyplexes was found to be pH dependent, owing to the ionizable tertiary amines present in the polymer backbones. pH titration curves revealed the charge-altering characteristics of the polymers as shown by pKa values between 4–5 (Fig. S6). Polyplexes were prepared at pH 5 and subsequently dialyzed against DPBS at pH 7.4. During the initial polyplex formation, the zeta potential was greater than 40 mV, however neutralizing the pH resulted in a reversal of the zeta potential to less than −20 mV. Furthermore, the sizes of polyMDET/pDNA, polyMDET-Cp/pDNA, polyMDET/mRNA, and polyMDET-Cp/mRNA were similar ~200 nm at pH 5 and increased as a function of pH (Fig. 2D, Fig. 2E, Table S1), although the increase in size was not the same for all the polyplexes. The increased hydrophobicity of polyMDET-Cp versus polyMDET polymers likely contributed to the less significant increase in size at higher pH. Interestingly, the size of polyMDET-C10/pDNA and polyMDET-C10/mRNA was similar at pH 7.4. However, at lower pH, we measured a slight (~80 nm) difference in size between polyMDET-C10/pDNA and polyMDET-C10/mRNA that could be related to the size of the pDNA compared to mRNA (~5-fold greater nucleotides). Over the course of 7 days, we observed differential levels of stability for various polyplexes (Fig. S7) and all polyplexes displayed similar size characteristics to their initial formulations for at least 2 days except for polyMDET-C10. Gel electrophoresis also showed that all polyplexes displayed a reduced ability to fully condense pDNA after 7 days (Fig. S8), which can possibly be explained by the degradation of the polymers over time. In fact, another study found that similar MDET containing polyesters that were modified with cholesterol side chains p(MDS-co-CES) lost approximately 54% of their weight in PBS pH 7.4 and 37°C over the course of 8 weeks.39
Polyplex Stability in the Presence of Serum and DNase I:
The ability for polyplexes to protect nucleic acid cargoes under physiological conditions was evaluated by incubating polyplexes in a physiologically relevant concentration (55% FBS) of serum or DNase I for 30 min or 1 hour, respectively followed by performing gel electrophoresis.22, 40–43 Serum incubation revealed partial destabilization of pDNA polyplexes prepared from polyMDET and polyMDET-C10 polyplexes, whereas the other polyplexes remained stable (Fig. S9). Fig. S10 shows that all polyplexes protected DNA from DNase I degradation.
Physicochemical Properties of Polymers Results in Differential In Vitro Transfection of pDNA and mRNA:
To assess the structure-property relationships between polyplexes and in vitro transfection efficiency, a monocyte/macrophage-like cell line, RAW 264.7 was used. All transfection experiments were performed in the presence of 10% FBS. Fluorescence microscopy and flow cytometry revealed that the transfection efficiency of polyMDET/pDNA polyplexes prepared at 110:1 ratio was similar to PEI (at N/P ratio 30) (Fig. 3A). The N/P ratio of PEI was chosen based on our previous study that showed PEI30 was able to produce higher transfection in macrophages in the presence of serum.22 MTS assay showed that the polyMDET and polyMDET-Cp polyplexes evaluated did not reduce cell viability at the concentrations used, whereas PEI30 displayed a 40% reduction in viability (Fig. S11). The toxicity of PEI is caused by its high positive charge density, which can lead to strong interactions with cell surfaces and subsequent damage.29 The absence of a high positive charge density and pH-dependent charge properties of polyMDET and polyMDET-Cp are likely reasons for the lack of toxicity observed for these polyesters. Hydrophobic modification enhanced the transfection efficiency and reached a maximum for polyMDET-C6/pDNA (3-fold higher compared to PEI30). Fig. 3B demonstrated that 62% of RAW 264.7 cells were GFP+ compared to untreated cells and the transfection efficiency of polyMDET-C6/pDNA was comparable to another commercially-available transfection reagent, jetOPTIMUS (Fig. 3C, 3D, and Fig. S12).
Figure 3.
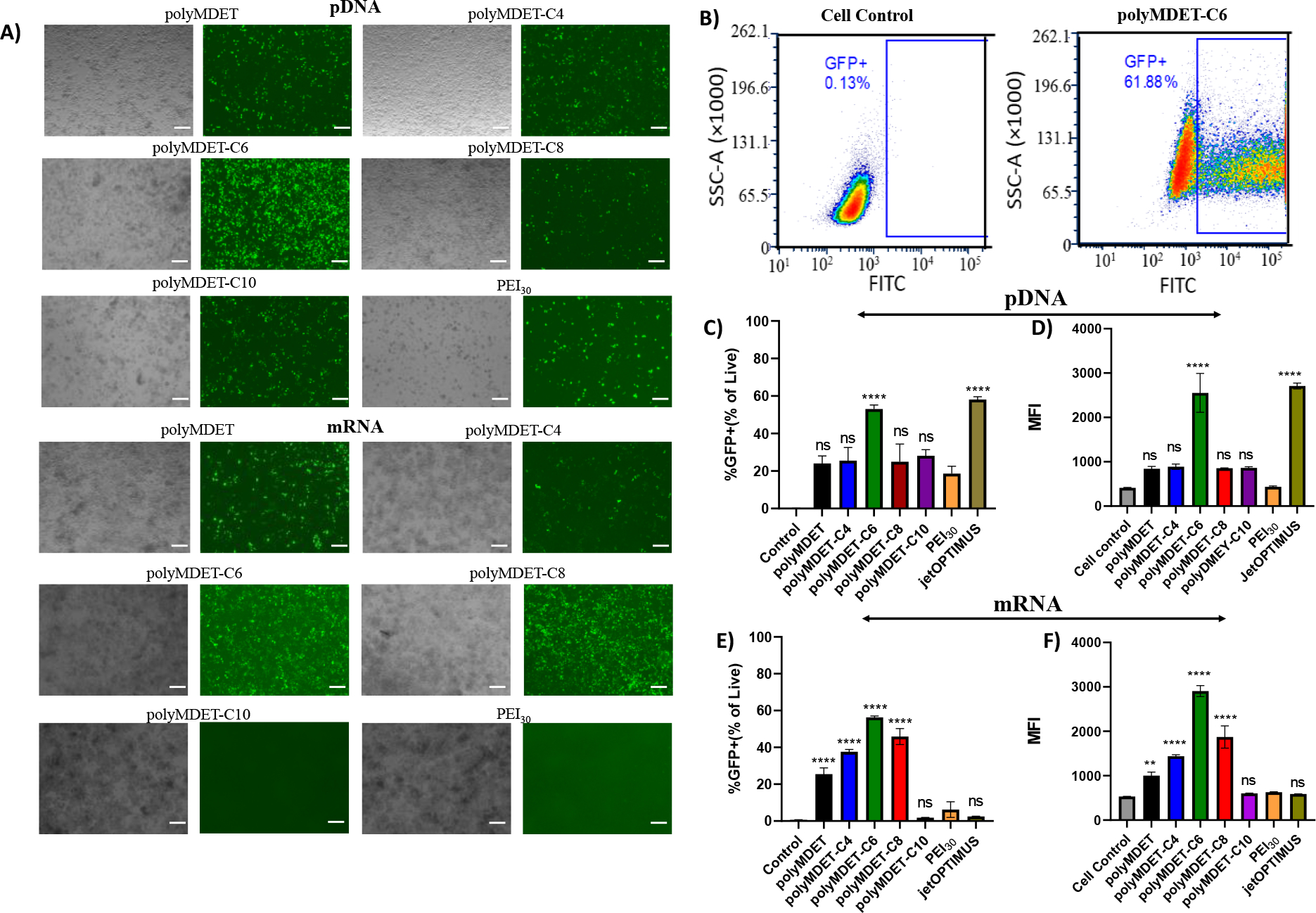
Transfection of GFP encoded pDNA and mRNA in RAW 264.7 cells. (A) RAW 264.7 cells were transfected with polyMDET/pDNA or polyMDET-Cp/pDNA and polyMDET/mRNA or polyMDET-Cp/mRNA polyplexes prepared at a weight ratio of 110:1. GFP was used to assess for the transfection efficiency of the polyplexes. Scale bar 100 μm. (B) Representative gating strategy for the quantification of transfection by flow cytometry. Flow cytometry-based quantification of transfection efficiency of (C) polyMDET/pDNA and polyMDET-Cp/pDNA and (E) polyMDET/mRNA, polyMDET-Cp/mRNA polyplexes, respectively. Mean fluorescence intensity (MFI) of GFP expression after transfection of (D) GFP-encoded pDNA and (F) mRNA, respectively. PEI30 corresponds to polyplexes prepared at N/P ratio 30. The amount of GFP pDNA and GFP mRNA used was 2 μg and 1 μg, respectively per 2 × 105 RAW 264.7 cells. Data are representative of n=3 experiments. Statistical differences between groups were determined by performing a one-way ANOVA and Tukey’s post-hoc test (**p<0.01, ****p<0.0001). Errors bars represent standard deviation.
Transfection of immune cells using mRNA is advantageous compared to pDNA, as it does not require nuclear entry and allows for efficient protein expression in lesser proliferative primary cells. Hence, mRNA transfection has become a mainstay for developing various biomedical applications, including vaccines,44 protein replacement therapy,45 and immunotherapies.46 To monitor the mRNA transfection efficiency of the various polyplexes, we utilized GFP reporter mRNA. Fig. 3A shows representative fluorescence microscopy images of the GFP signal. We further quantified the transfection efficiency using flow cytometry (Fig. 3E, 3F, and Fig. S12). Conversely to pDNA transfection, all mRNA-containing polyplexes efficiently transfected RAW264.7 cells except for polyMDET-C10. Notably, the transfection efficiency for the best performing polyMDET-C6/mRNA polyplex was over 10-fold greater than PEI30 and jetOPTIMUS that were used as controls.
The pDNA and mRNA transfection of polyMDET and polyMDET-Cp polyplexes was next evaluated using primary bone marrow-derived macrophages (BMDM) and compared with PEI30 and jetOPTIMUS as controls (Fig. 4). The transfection efficiency of polyMDET and polyMDET-Cp was significantly higher (3–4 fold) compared to the controls with polyMDET-C6 performing the best followed by polyMDET-C4 and polyMDET-C8, which was further validated using fluorescence microscopy (Fig. S13). Interestingly, there were no differences in the trend for transfection between pDNA and mRNA, although mRNA transfection was 50% more efficient than pDNA (30% versus 20% for polyMDET-C6). Importantly, these results were similar to other efficient PBAE polyplexes that utilized a poly(glutamic acid)-dimannose targeting ligand to enhance the uptake of polyplexes for the transfection of BMDMs.27
Figure 4.
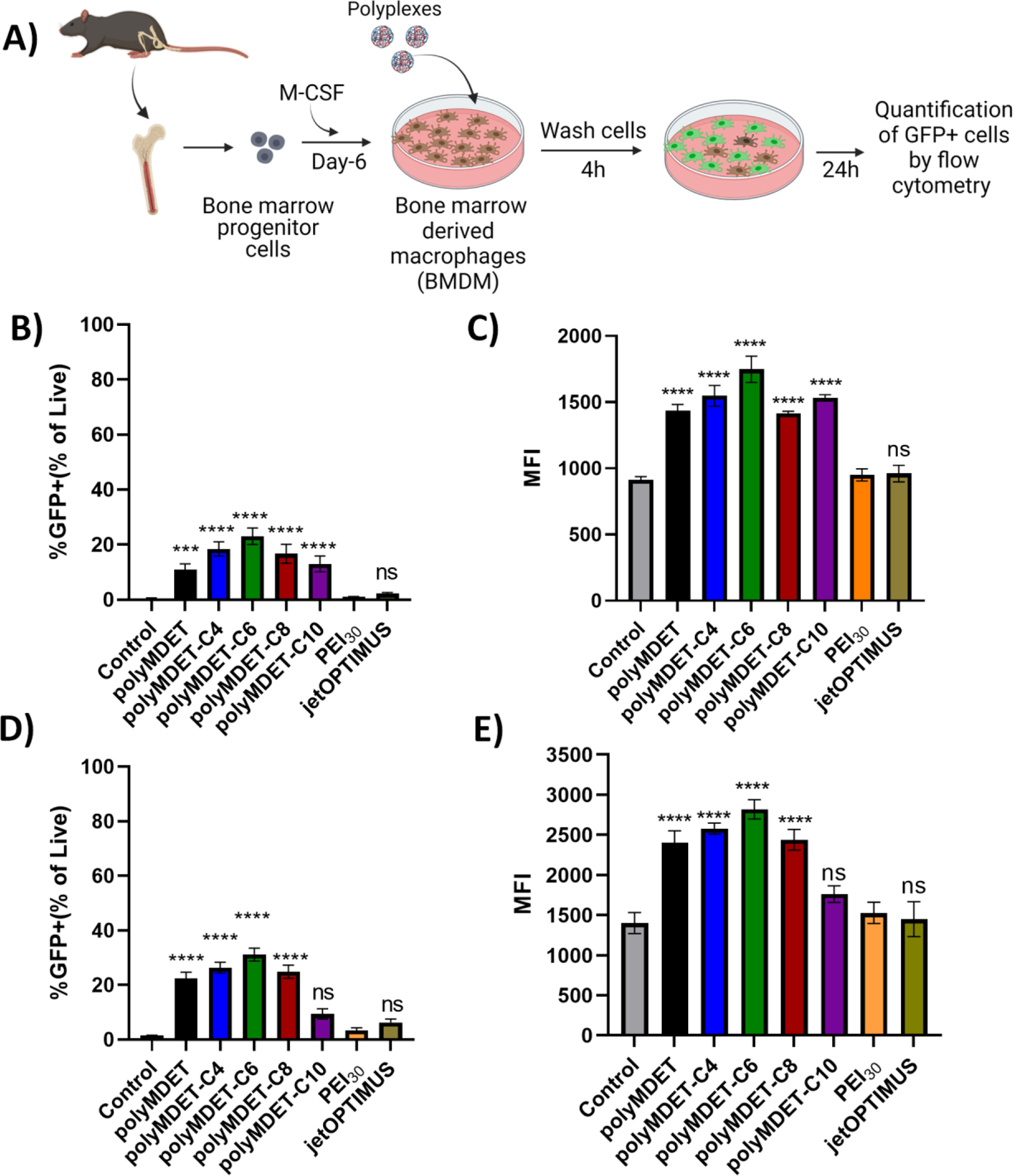
Flow cytometry-based quantification of transfection efficiency of polyMDET and polyMDET-Cp polyplexes in primary BMDM cells. (A) Representative schematic for extraction of bone marrow from the femur and tibia of C57BL/6 mouse, differentiation into bone marrow-derived macrophages (BMDM), and polyplex treatment. Created with BioRender. (B and D) Flow cytometry-based quantification of transfection efficiency of polyMDET/pDNA, polyMDET-Cp/pDNA and polyMDET/mRNA, polyMDET/mRNA polyplexes, respectively, in BMDMs. (C and E) Mean fluorescence intensity (MFI) of GFP expression after transfection of GFP encoded pDNA and mRNA, respectively. The amount of GFP pDNA and GFP mRNA used was 2 μg and 1 μg, respectively per 2 × 105 BMDMs. PEI30 corresponds to polyplexes prepared at N/P ratio 30. Data are representative of n=3 experiments. Statistical differences between groups were determined by performing a one-way ANOVA and Tukey’s post-hoc test (***P<0.001 ****P<0.0001). Errors bars represent standard deviation.
The enhancement of transfection in both RAW 264.7 cells and BMDMs for polyMDET and polyMDET-CP polyplexes without the need to incorporate additional excipients, surface coatings or targeting ligands could be due to multiple reasons. It was reported that the balance between hydrophobicity and cationic charge can significantly affect nucleic acid delivery.38 The hydrophobicity of the polymers can enhance cell membrane interactions, resulting in increased cellular uptake of polyplexes, and improved gene expression.16–18 The increased expression of GFP could also be attributed to the efficient endosomal escape of the polymers, where a previous study showed that interplay between the alkyl chain length and the monomer ratio can lead to a significant change in nucleic acid loading, endosomal escape, nucleic acid delivery, and eventually therapeutic outcomes.47 Although there are minor differences in the hydrophobic alkyl chains among PolyMDET-C6, PolyMDET-C8, and PolyMDET-C10, it could lead to significant differences in transfection efficiency due to impairment of one or multiple intracellular processes as previously shown.48–50 The pKa of the polymers is anticipated to be protonated at endosomal pH (Fig. S6), which could allow for polyplexes to utilize the proton sponge effect to escape endosomes. We confirmed the ability of polyMDET-C6 polyplexes to escape endosomal trafficking by encapsulating Cy5.5 and co-staining using lysotracker for fluorescence imaging (Fig. S14). After 24 h, the observation of polyplexes co-localized and not co-localized with lysotracker staining, in combination with transfection data demonstrating GFP expression in various cell types (Fig. 3 and Fig. 4), supports that the polyplexes were able to escape endosomes. Overall, these results demonstrated that single-component, excipient-free polyMDET and polyMDET-Cp polyplexes were highly efficient in transfecting both pDNA and mRNA in “hard-to-transfect” innate immune cells, including RAW 264.7 cells and primary BMDMs. The hydrophobicity of the polymers was a driving factor for efficient transfection, with moderately hydrophobic polyMDET-C6 offering the highest transfection compared to the more hydrophobic polyMDET-C8 and polyMDET-C10 polyplexes.
Cellular Uptake Mechanism of Polyplexes:
The cellular uptake mechanism of polyplexes was evaluated using RAW 264.7 cells. The intensity of the GFP fluorescence signal from nucleic acid delivery by polyplexes was used to determine the impact of various uptake inhibitors known to alter clathrin-dependent endocytosis (chlorpromazine (CHP)), lipid raft and caveolin-dependent endocytosis (methyl-β-cyclodextrin (MBCD)), caveolin-dependent endocytosis (filipin), micropinocytosis (5-(N-ethyl-N-isopropyl)- amiloride (EIPA), and phagocytosis (cytochalasin D (CytD)).44, 51 Fig. 5 shows CHP and MBCD treatment completely inhibited polyMDET and polyMDET-Cp induced GFP expression. Filipin treatment reduced GFP expression by polyMDET-C6 by approximately 40%, whereas no significant reductions were observed for other polyplexes. EIPA treatment partially reduced GFP expression from polyMDET, polyMDET-C6, and polyMDET-C8 polyplexes. Lastly, CytD treatment significantly reduced GFP expression induced by all polyplexes with the greatest inhibition noted for polyMDET polyplexes. Taken together, these results indicated that the polyplexes mostly relied on both clathrin- and lipid raft-dependent endocytosis mechanisms to induce gene expression and that blocking one of the pathways was sufficient to eliminate gene expression. All other inhibitors showed variable levels of inhibition that was dependent on the type of polyplex. For example, CytD treatment completely inhibited gene expression induced by polyMDET polyplexes but affected other polyplexes to a lesser, but significant extent. This effect could partially be explained by the differences in the size of polyplexes at pH 7.4. Nanoparticle size plays a crucial role in the endocytosis pathways, which is known to affect their uptake efficiency.52 Phagocytosis is a pathway by which cells uptake various bacteria, viruses, apoptotic, and necrotic cells. This pathway is known to allow for internalization of particles that are larger in size (0.5–10 μm), which correlated with the DLS results for polyMDET polyplexes being larger than other variants tested.53, 54
Figure 5.
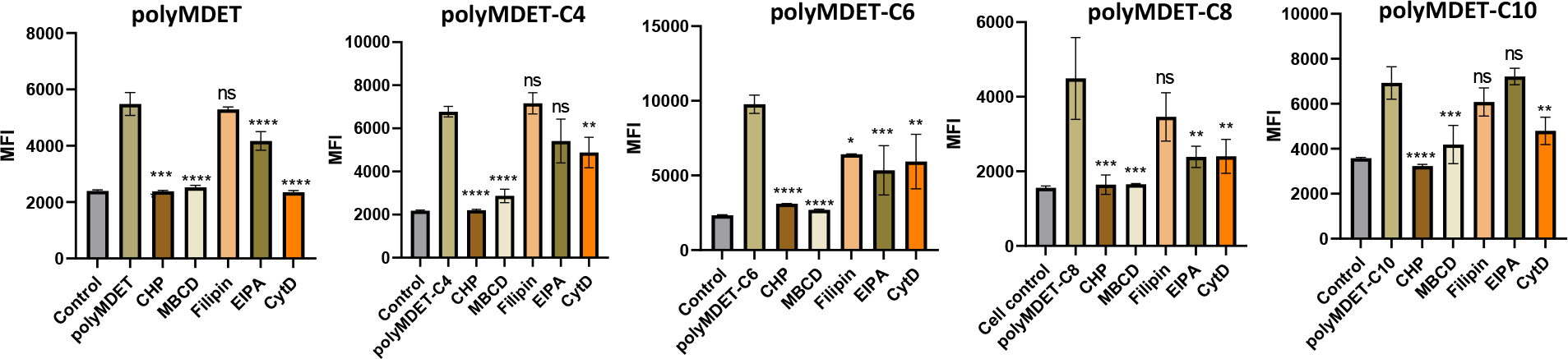
Uptake mechanism study of polyMDET/pDNA and polyMDET-Cp/pDNA polyplexes in RAW 264.7 cells. RAW 264.7 macrophages were incubated with various endocytosis inhibitors for 1 hour before treatment with the polyplexes. The MFI of GFP expression was analyzed by flow cytometry after 24 hours. The amount of GFP pDNA used was 2 μg per 2 × 105 RAW 264.7 cells. Data are representative of n=3 experiments. Statistical differences between groups were determined by performing a one-way ANOVA and Tukey’s post-hoc test (*P<0.05, **P<0.01, ***P<0.001 ****P<0.0001). Errors bars represent standard deviation.
In Vivo mRNA Delivery and Lung-Selective Transfection:
After successful in vitro transfection of pDNA and mRNA, we next explored the efficiency of in vivo transfection with these polyplexes in C57BL/6 mice. Since mRNA was more effective to transfect BMDMs (Fig. 4), we performed these studies using mRNA. Mice were intravenously administered polyMDET/mRNA or polyMDET-Cp/mRNA polyplexes containing 10 μg of luciferase mRNA (FLuc). After 24 hours, the mice were injected with D-luciferin solution intraperitoneally, euthanized, and the luminescence signal from various organs (liver, spleen, heart, lungs, kidneys) was measured using IVIS® (Fig. 6A). A strong luminescence signal was observed in the lungs for certain polyplexes, with the highest signal for polyMDET-C4/mRNA (Fig. 6B and 6C). PolyMDET showed a strong but equal distribution of protein expression in the lung and spleen with minimal expression in the liver, heart, and kidneys. Interestingly, polyMDET-C4 and polyMDET-C6 displayed a significant increase in the ratio of protein expression in the lung versus the spleen, 23-fold and 12-fold, respectively (Fig. 6D). PolyMDET-C8 polyplexes only showed minor transfection in the spleen. Other organs besides the lung and spleen were minimally transfected.
Figure 6.
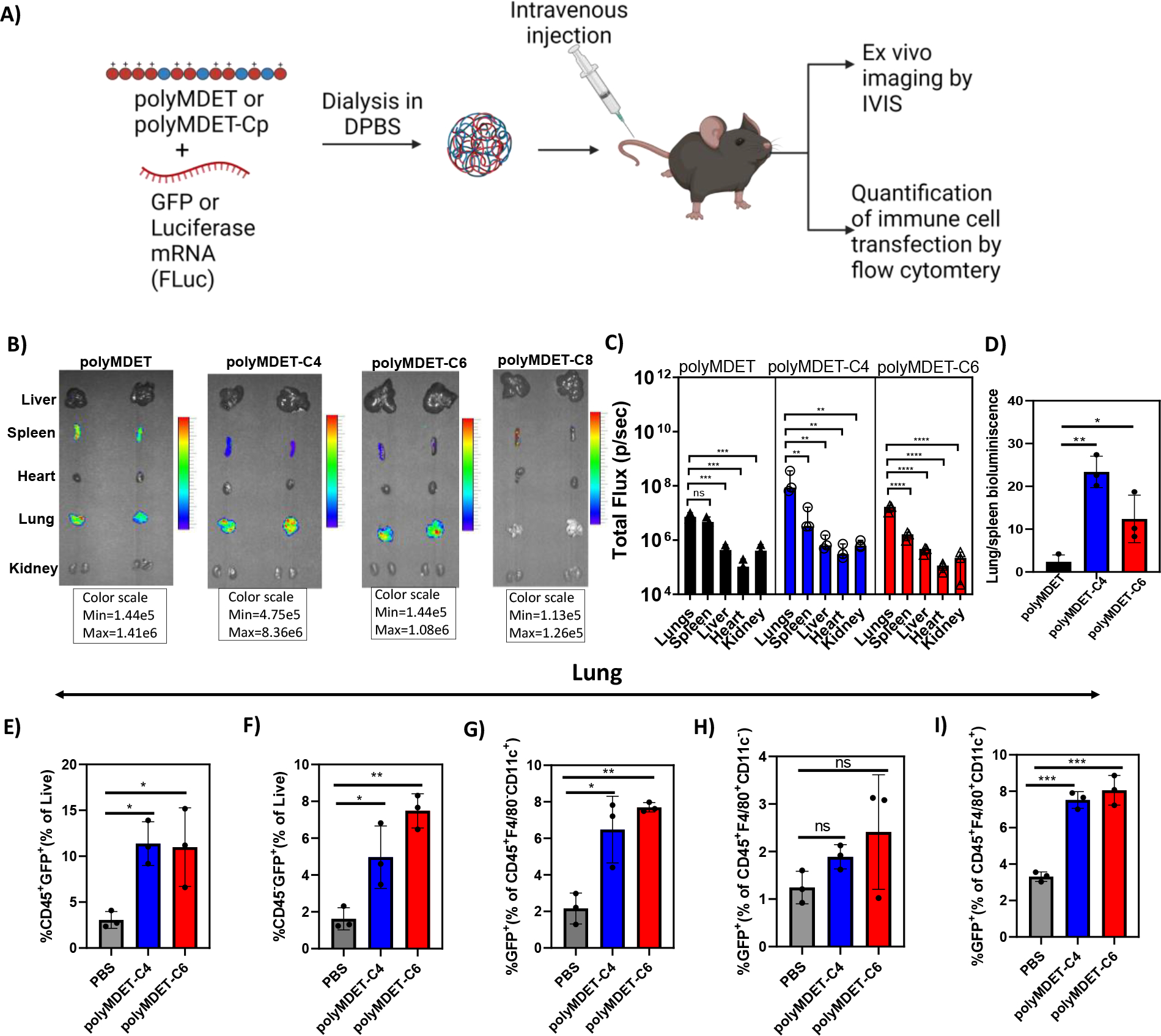
In vivo mRNA transfection in C57BL/6 mice and quantification of immune cell transfection. (A) Schematic for mRNA polyplex preparation, in vivo administration, and analysis of cell transfection. (B) Representative ex vivo bioluminescence images of organs after 24 h of intravenously injected FLuc-encoded mRNA polyplexes in C57BL/6 mice. (C) Quantification of FLuc mRNA expression in selected tissues. (D) Relative bioluminescence intensity of lung compared to spleen in PolyMDET, PolyMDET-C4, and PolyMDET-C6. Flow cytometric analysis in the lung of (E) total percentage of CD45+GFP+ cells. (F) total percentage of CD45−GFP+ cells. (G) total percentage of CD45+F4/80−CD11c+GFP+. (H) total percentage of CD45+F4/80+CD11c−GFP+ (I) CD45+F4/80+CD11c+ cells in lungs. Statistical differences between groups were determined by performing a one-way ANOVA and Turkey’s post-hoc test (*P<0.05, **P<0.01, ***P<0.001 ****P<0.0001). Errors bars represent standard deviation. N=3 mice per experimental group.
As polyMDET and polyMDET-Cp polyplexes showed slightly different mRNA translation in the lung, we studied the biodistribution of the polyplexes. As shown in Fig. S15, the most significant fluorescence signals were detected in the liver, lung, kidney, and spleen. Previous studies have shown that the pKa and hydrophobicity of polymers affected the organ selectivity of the protein expression.55 However, our results differed from previously reported results for LNPs that showed pKa values around 9 led to increased lung selectivity, whereas the pKa for the polymers used in our studies were ≤ 5 (Fig. S6). Several factors may play a role in the observed differences between our findings and LNPs, including differential uptake and trafficking mechanisms (Fig. 5), differential protein corona fingerprints, and other physicochemical properties including size and zeta potential.56, 57 Taken together, these results support that the physicochemical properties of polyMDET-C4/mRNA and polyMDET-C6/mRNA polyplexes played a major role in determining the lung selectivity of FLuc expression.
In Vivo Immune Cell Transfection:
To quantify the immune cell populations transfected in the lung and spleen, we prepared GFP mRNA-containing polyMDET-C4 and polyMDET-C6 polyplexes. Polyplexes containing 10 μg of mRNA were intravenously administered via the tail vein into C57BL/6 mice and the resulting GFP signal in the lungs and spleen was measured using flow cytometry after 24 hours. The gating strategy used for analysis is shown in Fig. S16. We enumerated GFP expression in a variety of cell populations (Fig. 6E–I), such as CD45+ (lymphocytes), CD45− (non-lymphocytes, which includes endothelial and epithelial cells, among others), CD45+F4/80+CD11c+ (alveolar macrophages), CD45+F4/80+CD11c− (interstitial macrophages), and CD45+F4/80−CD11c+ (dendritic cells).58–61 PolyMDET-C4 and polyMDET-C6 polyplexes transfected approximately 10–11% of lymphocytes, 5–8% of non-lymphocytes, 6–8% of dendritic cells, 2–3% of interstitial macrophages and 7–8% of alveolar macrophages.
Total cell transfection in spleen was less than the lung for both polyplexes evaluated, which correlated with the IVIS® images that showed a 10-fold lower luciferase signal in the spleen for both polyplexes evaluated. Lymphocyte transfection in the spleen was also lower than the lungs with 2.5–3% of CD45+CD11c+ dendritic cells and 3.5–4.6% of CD45+F4/80+ macrophages being found GFP+ (Fig. S17). The reduced transfection within the spleen may be related to distinct differences in cell populations between the spleen and lungs, mainly the significant presence of B and T cells. This corresponded with our in vitro studies evaluating these polyplexes for T cell transfection (Jurkat cells), where no transfection was observed (data not shown). The percentage of CD45+F4/80−CD11c−GFP+ cells was also calculated in both the spleen and lungs (Fig. S18), which would include immune cells besides macrophages and dendritic cells, which could include monocytes, neutrophils, B cells, and T cells, among others. The spleen did not show any significant difference compared to PBS, which corroborated the findings that few lymphocytes were transfected. Moreover, 1.5–2.5% of CD45+F4/80−CD11c− cells were GFP+ in the lungs. These studies demonstrated that appreciable levels of mRNA transfection can be achieved using polyMDET-C4 and polyMDET-C6 polyplexes in an organ-selective manner to reach “hard-to-transfect” innate immune cells in vivo. Future studies aimed to further distinguish lymphocyte and non-lymphocyte populations within the lung and spleen may better classify differences between polyMDET-C4 and polyMDET-C6 polyplexes.
Analysis of organ tissue histology and plasma cytokines following polyplex treatment:
To assess the biocompatibility of polyMDET and polyMDET-Cp polyplexes used in these studies, we performed histological examination and plasma cytokine analyses. Mice were injected intravenously with polyplexes containing 10 μg mRNA via tail vein injection. After 24 h several major organs (liver, spleen, heart, lung, and kidney) were collected, fixed, and stained using hematoxylin and eosin (H&E). This timepoint was chosen for evaluation as it corresponded to the timepoint used for in vivo transfection experiments in Fig. 6. Examination of the tissue sections did not indicate induction of inflammation nor alterations from normal tissue architecture after polyplex treatment (Fig. 7A). LNP administration has been shown to induce proinflammatory responses, owing to the use of ionizable lipids, and strategies have been developed to reduce this effect.62, 63 We analyzed the plasma proinflammatory cytokine levels using an enzyme-linked immunosorbent assay (ELISA) following polyplex treatment. Interleukin-6 (IL-6) and tumor necrosis factor α (TNFα) and were not significantly increased compared to the PBS control, which demonstrated the lack of proinflammatory responses generated by the polyplexes (Fig. 7B). Taken together, these results support the biocompatibility, anti-inflammatory properties, and safety of the polyMDET and polyMDET-Cp polyplexes for in vivo use. Future toxicology studies will be useful to comprehensively evaluate the dose- and time-dependent responses following polyplex administration in a therapeutically-relevant model.
Figure 7.
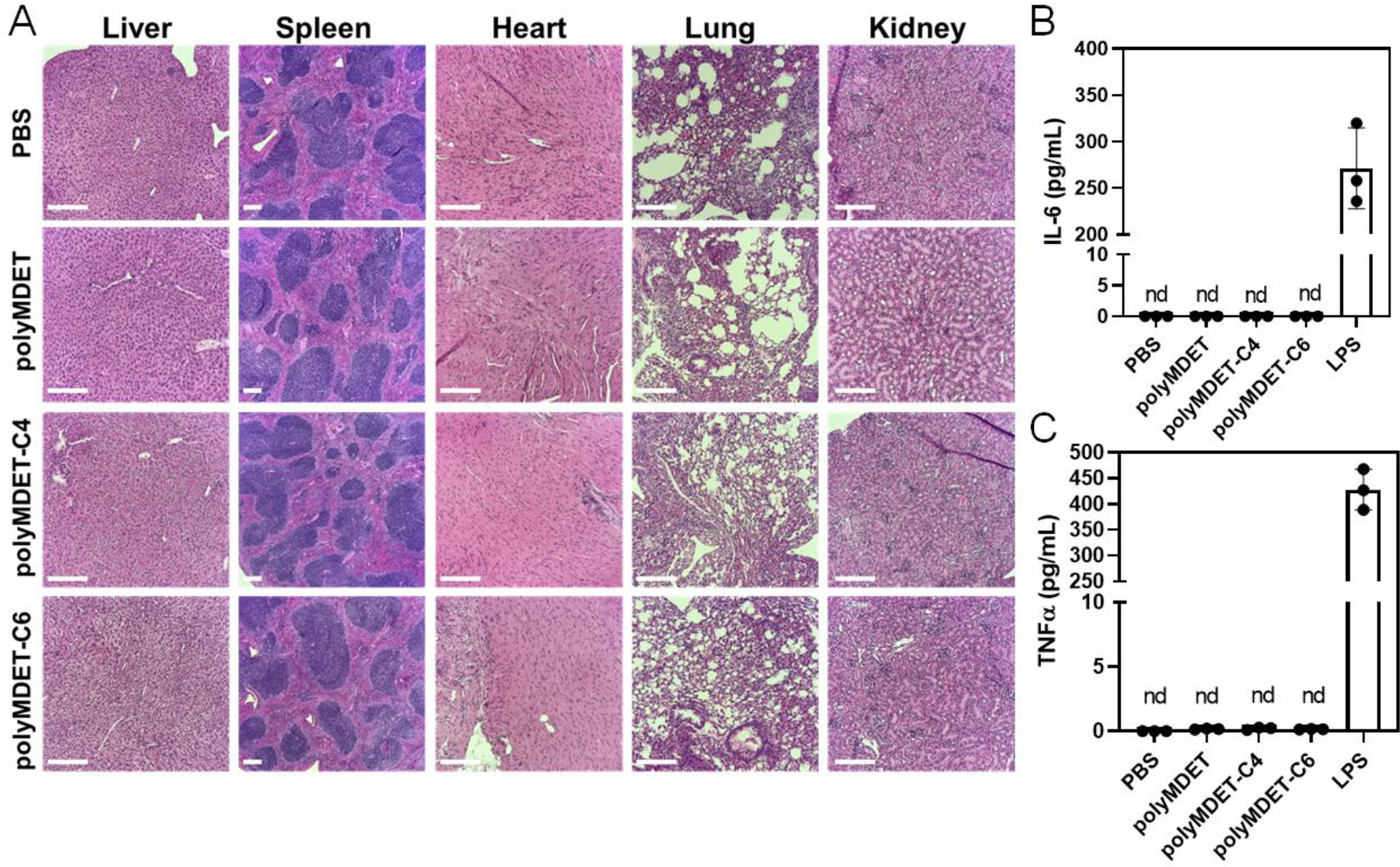
In vivo histological examination and proinflammatory cytokine levels of polyplex-treated mice. (A) Histopathological changes of various organ tissues after single dose intravenous administration of polyplexes (10 μg mRNA) in mice. Scale bar 200 μm. (B) Expression of plasma proinflammatory cytokines after intravenous administration of the polyplexes using ELISA. mRNA polyplexes did not show any proinflammatory cytokine IL-6 and TNFα expression after 24 h. Lipopolysaccharide (LPS) used as positive control induced higher amount of IL-6 and TNFα expression. nd – not detected. N=3 mice per experimental group.
Conclusion:
Designing polymer-based carriers for efficient DNA/mRNA transfection is critical for developing nucleic acid therapeutics. Here, we synthesized a set of biodegradable hydrophobic ionizable polyesters (polyMDET and polyMDET-Cp) using a one-pot synthetic methodology by incorporating variable length alkyl chains into the polymer backbone to create a single-component and excipient-free, non-viral polyplex for pDNA and mRNA delivery. Polyplexes were highly efficient to transfect pDNA and mRNA in RAW 264.7 macrophages and primary BMDMs. The hydrophobicity of the polyMDET and polyMDET-Cp was a conducive factor for efficient transfection, with the moderately hydrophobic and best performing polyMDET-C4 and polyMDET-C6 polyplexes achieving over 10-fold higher transfection than PEI30 and jetOPTIMUS in BMDMs with negligible cytotoxicity. Uptake mechanism studies using polyMDET and polyMDET-Cp polyplexes revealed that transfection was eliminated by clathrin, and lipid raft inhibitor treatment, whereas the extent of inhibition due to other inhibitors was polyplex-dependent. Intravenous administration of polyplexes identified a hydrophobicity-driven shift in the lung:spleen protein expression ratio with all protein expression being extrahepatic. Protein expression in the lungs and spleen by the more hydrophilic polyMDET polyplexes was similar, whereas polyMDET-C4 and polyMDET-C6 polyplexes displayed a lung-selective tropism for mRNA transfection of 23- and 12-fold, respectively. Flow cytometry analysis of the lungs showed 6–8% of dendritic cells and 7–8% of alveolar macrophages were transfected with mRNA, while other lymphocytes were marginally transfected (~2%) demonstrating significant opportunity to modulate innate immune cell responses.
The preferential lung transfection illustrates the importance of polyplex physicochemical properties to modulate biodistribution and immune cell uptake profiles. We envision that altering the size and surface chemistry of the polyplexes may aid in achieving transfection in hepatic or other extrahepatic tissues as previously described.64 Importantly, our data support noninflammatory mRNA delivery to the lung using polyMDET and polyMDET-Cp polyplexes, which offers significant potential to develop tolerogenic mRNA vaccines for the treatment of autoimmunity65 or allergy based on several studies utilizing polymeric nanoparticles for induction of antigen (Ag)-specific immune tolerance.66–69 Alternatively, the incorporation of nucleic acid adjuvants targeting Toll-like receptor signaling or the cGas/STING pathway may provide the opportunity to develop targeted cancer vaccines.70, 71
Supplementary Material
ACKNOWLEDGMENT
The authors acknowledge the support of the University of Maryland School of Medicine Center for Innovative Biomedical Resources, Flow Cytometry Core, Center for Translational Imaging in Research (C-TRIM), Pathology Biorepository Shared Services Core — Baltimore, Maryland for technical assistance. We thank the University of Maryland School of Pharmacy Mass Spectrometry Center for use of the MALDI-TOF instrument.
Funding Sources:
Research reported in this publication was supported by Startup funds provided by the University of Maryland, Baltimore, the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM142752, the UMGCCC P30 grant under award P30CA134274 from the National Cancer Institute; N.T. is supported by a PhRMA Foundation Predoctoral Fellowship in Drug Delivery. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the PhRMA Foundation.
Footnotes
A.C. and R.M.P. are inventors on a patent application describing the materials and polyplexes described in this article.
ASSOCIATED CONTENT
Supporting Information.
The following files are available free of charge.
NMR and MALDI-TOF characterizations of the polymers, pH titration study, agarose gel electrophoresis, cell viability, in vivo biodistribution study of the polyplexes, flow cytometry gating strategy, and percentage of in vivo immune cell transfection (PDF).
REFERENCES
- 1.Stephan MT In Empowering patients from within: Emerging nanomedicines for in vivo immune cell reprogramming, Seminars in immunology, Elsevier: 2021; p 101537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Odaka C; Mizuochi T, Role of Macrophage Lysosomal Enzymes in the Degradation of Nucleosomes of Apoptotic Cells. 1999, 163 (10), 5346–5352. [PubMed] [Google Scholar]
- 3.Maury B; Gonçalves C; Tresset G; Zeghal M; Cheradame H; Guégan P; Pichon C; Midoux P, Influence of pDNA availability on transfection efficiency of polyplexes in non-proliferative cells. Biomaterials 2014, 35 (22), 5977–85. [DOI] [PubMed] [Google Scholar]
- 4.Lee CS; Bishop ES; Zhang R; Yu X; Farina EM; Yan S; Zhao C; Zeng Z; Shu Y; Wu X; Lei J; Li Y; Zhang W; Yang C; Wu K; Wu Y; Ho S; Athiviraham A; Lee MJ; Wolf JM; Reid RR; He T-C, Adenovirus-mediated gene delivery: Potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes & Diseases 2017, 4 (2), 43–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colella P; Ronzitti G; Mingozzi F, Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol Ther Methods Clin Dev 2018, 8, 87–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shirley JL; de Jong YP; Terhorst C; Herzog RW, Immune Responses to Viral Gene Therapy Vectors. Mol Ther 2020, 28 (3), 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hacein-Bey-Abina S; Hauer J; Lim A; Picard C; Wang GP; Berry CC; Martinache C; Rieux-Laucat F; Latour S; Belohradsky B. H. J. N. E. J. o. M., Efficacy of gene therapy for X-linked severe combined immunodeficiency. 2010, 363 (4), 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van den Berg AIS; Yun C-O; Schiffelers RM; Hennink WE, Polymeric delivery systems for nucleic acid therapeutics: Approaching the clinic. Journal of Controlled Release 2021, 331, 121–141. [DOI] [PubMed] [Google Scholar]
- 9.Hong SJ; Ahn MH; Sangshetti J; Choung PH; Arote R. B. J. C. p., Sugar-based gene delivery systems: current knowledge and new perspectives. 2018, 181, 1180–1193. [DOI] [PubMed] [Google Scholar]
- 10.Arote RB; Jiang H-L; Kim Y-K; Cho M-H; Choi Y-J; Cho C.-S. J. E. O. o. D. D., Degradable poly (amido amine) s as gene delivery carriers. 2011, 8 (9), 1237–1246. [DOI] [PubMed] [Google Scholar]
- 11.Hong SJ; Ahn MH; Sangshetti J; Arote R. B. J. A. b., Sugar alcohol-based polymeric gene carriers: synthesis, properties and gene therapy applications. 2019, 97, 105–115. [DOI] [PubMed] [Google Scholar]
- 12.Lungwitz U; Breunig M; Blunk T; Göpferich A, Polyethylenimine-based non-viral gene delivery systems. European journal of pharmaceutics and biopharmaceutics : official journal of Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik e.V 2005, 60 (2), 247–66. [DOI] [PubMed] [Google Scholar]
- 13.Toncheva V; Wolfert MA; Dash PR; Oupicky D; Ulbrich K; Seymour LW; Schacht EH, Novel vectors for gene delivery formed by self-assembly of DNA with poly(l-lysine) grafted with hydrophilic polymers. Biochimica et Biophysica Acta (BBA) - General Subjects 1998, 1380 (3), 354–368. [DOI] [PubMed] [Google Scholar]
- 14.Wang J; Cooper RC; Yang HJB, Polyamidoamine Dendrimer Grafted with an Acid-Responsive Charge-Reversal Layer for Improved Gene Delivery. 2020, 21 (10), 4008–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlsson J; Rhodes KR; Green JJ; Tzeng SY, Poly(beta-amino ester)s as gene delivery vehicles: challenges and opportunities. Expert Opinion on Drug Delivery 2020, 17 (10), 1395–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao X, Cationic lipid-based gene delivery: an update. In Gene Therapy for Diseases of the Lung, CRC Press: 2020; pp 99–112. [Google Scholar]
- 17.Mohammadinejad R; Dehshahri A; Sagar Madamsetty V; Zahmatkeshan M; Tavakol S; Makvandi P; Khorsandi D; Pardakhty A; Ashrafizadeh M; Ghasemipour Afshar E; Zarrabi A, In vivo gene delivery mediated by non-viral vectors for cancer therapy. Journal of Controlled Release 2020, 325, 249–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q; Su R-C; Yi W-J; Zheng L-T; Lu S-S; Zhao Z-G, pH and reduction dual-responsive dipeptide cationic lipids with α–tocopherol hydrophobic tail for efficient gene delivery. European Journal of Medicinal Chemistry 2017, 129, 1–11. [DOI] [PubMed] [Google Scholar]
- 19.Loh XJ; Lee T-C; Dou Q; Deen GR, Utilising inorganic nanocarriers for gene delivery. Biomaterials Science 2016, 4 (1), 70–86. [DOI] [PubMed] [Google Scholar]
- 20.Hou X; Zaks T; Langer R; Dong Y, Lipid nanoparticles for mRNA delivery. Nature Reviews Materials 2021, 6 (12), 1078–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell MJ; Billingsley MM; Haley RM; Wechsler ME; Peppas NA; Langer R, Engineering precision nanoparticles for drug delivery. Nature Reviews Drug Discovery 2021, 20 (2), 101–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakraborty A; Martín Lasola JJ; Truong N; Pearson RM, Serum-Independent Nonviral Gene Delivery to Innate and Adaptive Immune Cells Using Immunoplexes. ACS Applied Bio Materials 2020, 3 (9), 6263–6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piperno A; Sciortino MT; Giusto E; Montesi M; Panseri S; Scala A, Recent Advances and Challenges in Gene Delivery Mediated by Polyester-Based Nanoparticles. Int J Nanomedicine 2021, 16, 5981–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rui Y; Wilson DR; Tzeng SY; Yamagata HM; Sudhakar D; Conge M; Berlinicke CA; Zack DJ; Tuesca A; Green JJ, High-throughput and high-content bioassay enables tuning of polyester nanoparticles for cellular uptake, endosomal escape, and systemic in vivo delivery of mRNA. Science Advances 2022, 8 (1), eabk2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaczmarek JC; Kauffman KJ; Fenton OS; Sadtler K; Patel AK; Heartlein MW; DeRosa F; Anderson DG, Optimization of a Degradable Polymer–Lipid Nanoparticle for Potent Systemic Delivery of mRNA to the Lung Endothelium and Immune Cells. Nano Letters 2018, 18 (10), 6449–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J; Eygeris Y; Gupta M; Sahay G, Self-assembled mRNA vaccines. Advanced drug delivery reviews 2021, 170, 83–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F; Parayath NN; Ene CI; Stephan SB; Koehne AL; Coon ME; Holland EC; Stephan MT, Genetic programming of macrophages to perform anti-tumor functions using targeted mRNA nanocarriers. Nature Communications 2019, 10 (1), 3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Truong N; Black SK; Shaw J; Scotland BL; Pearson RM, Microfluidic-Generated Immunomodulatory Nanoparticles and Formulation-Dependent Effects on Lipopolysaccharide-Induced Macrophage Inflammation. The AAPS Journal 2021, 24 (1), 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zintchenko A; Philipp A; Dehshahri A; Wagner E, Simple Modifications of Branched PEI Lead to Highly Efficient siRNA Carriers with Low Toxicity. Bioconjugate Chemistry 2008, 19 (7), 1448–1455. [DOI] [PubMed] [Google Scholar]
- 30.Lei H; Fan DJAS, A Combination Therapy Using Electrical Stimulation and Adaptive, Conductive Hydrogels Loaded with Self‐Assembled Nanogels Incorporating Short Interfering RNA Promotes the Repair of Diabetic Chronic Wounds. 2022, 2201425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Apostolov EO; Soultanova I; Savenka A; Bagandov OO; Yin X; Stewart AG; Walker RB; Basnakian AG, Deoxyribonuclease I is Essential for DNA Fragmentation Induced by Gamma Radiation in Mice. Radiation Research 2009, 172 (4), 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan E; Lv J; Hu J; Shen W; Wang H; Cheng Y. J. J. o. M. C. B., Statistical versus block fluoropolymers in gene delivery. 2018, 6 (44), 7230–7238. [DOI] [PubMed] [Google Scholar]
- 33.Ahmed M; Narain R, The effect of polymer architecture, composition, and molecular weight on the properties of glycopolymer-based non-viral gene delivery systems. Biomaterials 2011, 32 (22), 5279–5290. [DOI] [PubMed] [Google Scholar]
- 34.Williams JB; Gusev AI; Hercules DM, Characterization of Polyesters by Matrix-Assisted Laser Desorption Ionization Mass Spectrometry. Macromolecules 1997, 30 (13), 3781–3787. [Google Scholar]
- 35.Lim YB; Han SO; Kong HU; Lee Y; Park JS; Jeong B; Kim SW, Biodegradable polyester, poly[alpha-(4-aminobutyl)-L-glycolic acid], as a non-toxic gene carrier. Pharmaceutical research 2000, 17 (7), 811–6. [DOI] [PubMed] [Google Scholar]
- 36.Eltoukhy AA; Siegwart DJ; Alabi CA; Rajan JS; Langer R; Anderson DG, Effect of molecular weight of amine end-modified poly(β-amino ester)s on gene delivery efficiency and toxicity. Biomaterials 2012, 33 (13), 3594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naeye B; Deschout H; Caveliers V; Descamps B; Braeckmans K; Vanhove C; Demeester J; Lahoutte T; De Smedt SC; Raemdonck KJB, In vivo disassembly of IV administered siRNA matrix nanoparticles at the renal filtration barrier. 2013, 34 (9), 2350–2358. [DOI] [PubMed] [Google Scholar]
- 38.Sunshine JC; Akanda MI; Li D; Kozielski KL; Green JJJB, Effects of base polymer hydrophobicity and end-group modification on polymeric gene delivery. 2011, 12 (10), 3592–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y; Wang L-S; Goh S-H; Yang Y-YJB, Synthesis and characterization of cationic micelles self-assembled from a biodegradable copolymer for gene delivery. 2007, 8 (3), 1028–1037. [DOI] [PubMed] [Google Scholar]
- 40.Burke RS; Pun SH, Extracellular Barriers to in Vivo PEI and PEGylated PEI Polyplex-Mediated Gene Delivery to the Liver. Bioconjugate Chemistry 2008, 19 (3), 693–704. [DOI] [PubMed] [Google Scholar]
- 41.Zhao K; Li W; Huang T; Luo X; Chen G; Zhang Y; Guo C; Dai C; Jin Z; Zhao YJPO, Preparation and efficacy of Newcastle disease virus DNA vaccine encapsulated in PLGA nanoparticles. 2013, 8 (12), e82648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobson ME; Becker KW; Palmer CR; Pastora LE; Fletcher RB; Collins KA; Fedorova O; Duvall CL; Pyle AM; Wilson JT, Structural Optimization of Polymeric Carriers to Enhance the Immunostimulatory Activity of Molecularly Defined RIG-I Agonists. ACS Central Science 2020, 6 (11), 2008–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naidu PSR; Denham E; Bartlett CA; McGonigle T; Taylor NL; Norret M; Smith NM; Dunlop SA; Iyer KS; Fitzgerald M, Protein corona formation moderates the release kinetics of ion channel antagonists from transferrin-functionalized polymeric nanoparticles. RSC Advances 2020, 10 (5), 2856–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Islam MA; Xu Y; Tao W; Ubellacker JM; Lim M; Aum D; Lee GY; Zhou K; Zope H; Yu M; Cao W; Oswald JT; Dinarvand M; Mahmoudi M; Langer R; Kantoff PW; Farokhzad OC; Zetter BR; Shi J, Restoration of tumour-growth suppression in vivo via systemic nanoparticle-mediated delivery of PTEN mRNA. Nature Biomedical Engineering 2018, 2 (11), 850–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kowalski PS; Rudra A; Miao L; Anderson DG, Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol Ther 2019, 27 (4), 710–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beck JD; Reidenbach D; Salomon N; Sahin U; Türeci Ö; Vormehr M; Kranz LM, mRNA therapeutics in cancer immunotherapy. Molecular Cancer 2021, 20 (1), 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foroozandeh P; Aziz AA, Insight into Cellular Uptake and Intracellular Trafficking of Nanoparticles. Nanoscale Research Letters 2018, 13 (1), 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fortune JA; Novobrantseva TI; Klibanov AM, Highly effective gene transfection in vivo by alkylated polyethylenimine. Journal of drug delivery 2011, 2011, 204058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sunshine JC; Akanda MI; Li D; Kozielski KL; Green JJ, Effects of base polymer hydrophobicity and end-group modification on polymeric gene delivery. Biomacromolecules 2011, 12 (10), 3592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santos JL; Oliveira H; Pandita D; Rodrigues J; Pêgo AP; Granja PL; Tomás H. J. J. o. C. R.V, Functionalization of poly (amidoamine) dendrimers with hydrophobic chains for improved gene delivery in mesenchymal stem cells. 2010, 144 (1), 55–64. [DOI] [PubMed] [Google Scholar]
- 51.Hussain KM; Leong KLJ; Ng MM-L; Chu JJH, The essential role of clathrin-mediated endocytosis in the infectious entry of human enterovirus 71. J Biol Chem 2011, 286 (1), 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang S; Li J; Lykotrafitis G; Bao G; Suresh S, Size-Dependent Endocytosis of Nanoparticles. Advanced materials (Deerfield Beach, Fla.) 2009, 21, 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hashimoto Y; Moki T; Takizawa T; Shiratsuchi A; Nakanishi Y. J. T. J. o. I., Evidence for phagocytosis of influenza virus-infected, apoptotic cells by neutrophils and macrophages in mice. 2007, 178 (4), 2448–2457. [DOI] [PubMed] [Google Scholar]
- 54.Martin CJ; Peters KN; Behar SM, Macrophages clean up: efferocytosis and microbial control. Current opinion in microbiology 2014, 17, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blanco E; Shen H; Ferrari M, Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol 2015, 33 (9), 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaw J; Pearson RM, Nanoparticle personalized biomolecular corona: implications of pre-existing conditions for immunomodulation and cancer. Biomaterials Science 2022, 10 (10), 2540–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearson RM; Juettner VV; Hong S, Biomolecular corona on nanoparticles: a survey of recent literature and its implications in targeted drug delivery. Frontiers in Immunology 2014, 2 (108). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abernathy LM; Fountain MD; Rothstein SE; David JM; Yunker CK; Rakowski J; Lonardo F; Joiner MC; Hillman GG, Soy Isoflavones Promote Radioprotection of Normal Lung Tissue by Inhibition of Radiation-Induced Activation of Macrophages and Neutrophils. Journal of Thoracic Oncology 2015, 10 (12), 1703–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaynagetdinov R; Sherrill TP; Kendall PL; Segal BH; Weller KP; Tighe RM; Blackwell TS, Identification of myeloid cell subsets in murine lungs using flow cytometry. American journal of respiratory cell and molecular biology 2013, 49 (2), 180–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Misharin AV; Morales-Nebreda L; Mutlu GM; Budinger GR; Perlman H, Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. American journal of respiratory cell and molecular biology 2013, 49 (4), 503–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bedoret D; Wallemacq H; Marichal T; Desmet C; Quesada Calvo F; Henry E; Closset R; Dewals B; Thielen C; Gustin P; de Leval L; Van Rooijen N; Le Moine A; Vanderplasschen A; Cataldo D; Drion P-V; Moser M; Lekeux P; Bureau F, Lung interstitial macrophages alter dendritic cell functions to prevent airway allergy in mice. The Journal of Clinical Investigation 2009, 119 (12), 3723–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H; Han X; Alameh M-G; Shepherd SJ; Padilla MS; Xue L; Butowska K; Weissman D; Mitchell MJ, Rational design of anti-inflammatory lipid nanoparticles for mRNA delivery. J. Biomed. Mater. Res. A. 2022, 110 (5), 1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ndeupen S; Qin Z; Jacobsen S; Bouteau A; Estanbouli H; Igyártó BZ, The mRNA-LNP platform’s lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. iScience 2021, 24 (12). 103479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheng Q; Wei T; Farbiak L; Johnson LT; Dilliard SA; Siegwart DJ, Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nature nanotechnology 2020, 15 (4), 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krienke C; Kolb L; Diken E; Streuber M; Kirchhoff S; Bukur T; Akilli-Öztürk Ö; Kranz LM; Berger H; Petschenka J; Diken M; Kreiter S; Yogev N; Waisman A; Karikó K; Türeci Ö; Sahin U, A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science (New York, N.Y.) 2021, 371 (6525), 145–153. [DOI] [PubMed] [Google Scholar]
- 66.Saito E; Gurczynski SJ; Kramer KR; Wilke CA; Miller SD; Moore BB; Shea LD, Modulating lung immune cells by pulmonary delivery of antigen-specific nanoparticles to treat autoimmune disease. Science advances 2020, 6 (42). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casey LM; Decker JT; Podojil JR; Rad L; Hughes KR; Rose JA; Pearson RM; Miller SD; Shea LDJB; Bioengineering, Nanoparticle Dose and Antigen Loading Attenuate Antigen-Specific T Cell Responses. Biotechnology & Bioengineering 2022. doi: 10.1002/bit.28252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Freitag TL; Podojil JR; Pearson RM; Fokta FJ; Sahl C; Messing M; Andersson LC; Leskinen K; Saavalainen P; Hoover LI; Huang K; Phippard D; Maleki S; King NJC; Shea LD; Miller SD; Meri SK; Getts DR, Gliadin Nanoparticles Induce Immune Tolerance to Gliadin in Mouse Models of Celiac Disease. Gastroenterology 2020, 158 (6), 1667–1681.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Casey LM; Hughes KR; Saunders MN; Miller SD; Pearson RM; Shea LD, Mechanistic contributions of Kupffer cells and liver sinusoidal endothelial cells in nanoparticle-induced antigen-specific immune tolerance. Biomaterials 2022, 283, 121457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miao L; Li L; Huang Y; Delcassian D; Chahal J; Han J; Shi Y; Sadtler K; Gao W; Lin J; Doloff JC; Langer R; Anderson DG, Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat Biotechnol 2019, 37 (10), 1174–1185. [DOI] [PubMed] [Google Scholar]
- 71.Van Herck S; Feng B; Tang L, Delivery of STING agonists for adjuvanting subunit vaccines. Advanced drug delivery reviews 2021, 179, 114020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


