Abstract
Objective
Hepatocellular carcinoma (HCC) often develops in patients with alcohol-related cirrhosis at an annual risk of up to 2.5%. Some host genetic risk factors have been identified but do not account for the majority of the variance in occurrence. This study aimed to identify novel susceptibility loci for the development of HCC in people with alcohol related cirrhosis.
Design
Patients with alcohol-related cirrhosis and HCC (cases: n=1214) and controls without HCC (n=1866), recruited from Germany, Austria, Switzerland, Italy and the UK, were included in a two-stage genome-wide association study using a case–control design. A validation cohort of 1520 people misusing alcohol but with no evidence of liver disease was included to control for possible association effects with alcohol misuse. Genotyping was performed using the InfiniumGlobal Screening Array (V.24v2, Illumina) and the OmniExpress Array (V.24v1-0a, Illumina).
Results
Associations with variants rs738409 in PNPLA3 and rs58542926 in TM6SF2 previously associated with an increased risk of HCC in patients with alcohol-related cirrhosis were confirmed at genome-wide significance. A novel locus rs2242652(A) in TERT (telomerase reverse transcriptase) was also associated with a decreased risk of HCC, in the combined meta-analysis, at genome-wide significance (p=6.41×10−9, OR=0.61 (95% CI 0.52 to 0.70). This protective association remained significant after correction for sex, age, body mass index and type 2 diabetes (p=7.94×10−5, OR=0.63 (95% CI 0.50 to 0.79). Carriage of rs2242652(A) in TERT was associated with an increased leucocyte telomere length (p=2.12×10−44).
Conclusion
This study identifies rs2242652 in TERT as a novel protective factor for HCC in patients with alcohol-related cirrhosis.
Keywords: hepatocellular carcinoma, genetic polymorphisms
What is already known on this subject?
Hepatocellular carcinoma (HCC) is the most common primary malignancy of the liver, responsible for ~0.8M deaths per year worldwide. Most alcohol-related HCCs develop in patients with established alcohol-related cirrhosis (ArC).
Older age, male sex, obesity and type 2 diabetes are risk factors for the development of HCC in people with ArC.
Only three genetic loci—PNPLA3, TM6SF2 and WNT3A-WNT9A—have been associated with the development of alcohol-related HCC, at genome-wide significance, to date. Other risk loci are likely to exist.
What are the new findings?
We identify the rs2242652 germline variant in TERT as a novel susceptibility locus for HCC development in ArC.
Specifically, the rs2242652 A allele is associated with an decreased risk of HCC development in ArC.
Carriage of rs2242652 in TERT is not associated with the risk for developing ArC.
HOW MIGHT IT IMPACT ON CLINICAL PRACTICE IN THE FORESEEABLE FUTURE?
Exploration of the functional significance of TERT variants could provide important insights into the pathogenesis of HCC in people with ArC.
Genetic profiling of patients with ArC might inform HCC screening programmes.
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver malignancy worldwide and is responsible for ~0.8 million deaths per annum.1 The global incidence of HCC is rising and may surpass 1 million cases annually by 2025.2 Alcohol-related liver disease (ArLD) is a leading underlying cause of HCC in Europe and Northern America.3 4 Most cases of alcohol-related HCC develop in patients with established cirrhosis. Cohort studies indicate that the cumulative incidence of HCC approaches 2.5% per annum for alcohol-related cirrhosis (ArC) patients attending specialist care centres.3 4 Clinical risk factors for the development of HCC in people with ArC include older age, male sex, type 2 diabetes and obesity2 5—but explain only a fraction of the total variability in HCC occurrence.6 7
In recent years, interest has focused on dissecting the underlying host genetics of HCC through candidate gene association studies. In the studies undertaken to date, loci in the genes coding for patatin-like phospholipase domain containing 3 (PNPLA3; rs738409) and transmembrane 6 superfamily member 2 (TM6SF2; rs58542926) were robustly confirmed to increase the risk of developing HCC in ArC,8 while loci, rs72613567:TA in hydroxysteroid 17-beta dehydrogenase 13 (HSD17B13) and rs429358:C in apolipoprotein E (APOE), were found to attenuate risk.9–11 As the products of these genes are involved in lipid turnover and processing, it is not surprising that the same loci also modulate the risk for HCC development in people with non-alcoholic fatty liver disease (NAFLD).12
The variants currently identified as associated with HCC risk in ArC only account for a small proportion of the heritability risk, suggesting the existence of additional genetic modulators.7 8 Also, the genetic risk loci recognised hitherto do not relate to genes considered pivotal to hepatocarcinogenesis.13 Identifying these additional, potential genetic modulators of hepatocarcinogenesis requires large genome-wide association studies (GWASs) in which cases are defined as people with ArC with HCC and controls as people with ArC who have no evidence of HCC. These definitions are critical to enable the detection of risk loci with a direct molecular link to hepatocarcinogenesis per se, rather than to the development of alcohol-related steatosis, inflammation or fibrosis.
A European GWAS of HCC in ArLD, while not conforming to this exact design, was recently undertaken by Trépo et al.14 In their discovery analysis comparing 775 HCC cases (80% with F3/F4 fibrosis) against 1332 non-HCC controls (94% with F3/F4 fibrosis), a genome-wide significant association was identified between the rs708113:T allele locus near WNT3A-WNT9A and a reduction in the risk for developing alcohol-related HCC.14
The aim of his study was to undertake a GWAS in patients with HCC against a background of ArC comprising 1066 cases and 844 controls using a case–control design.
Methods
Patient cohorts
Germany/Switzerland/Austria Alcohol Cohort (discovery cohort)
The diagnosis of ArC was established based on a history of long-term, sustained alcohol intake of a minimum of 40 g/day in women and 60 g/day in men, together with histological examination of liver tissue; or compatible historical, clinical, laboratory, radiological and endoscopic features. Patients were excluded if they had any other potential cause of liver injury, specifically if they were positive for hepatitis B surface antigen (HBsAg), anti-hepatitis C IgG (anti-hepatitis C virus (HCV) IgG), antinuclear antibodies (titre >1:80) or antimitochondrial antibodies (titre >1:40), had elevated serum ferritin levels with a transferrin saturation of >50%, a serum ceruloplasmin of <20 mg/dL (0.2 g/dL), a serum alpha-1 antitrypsin of <70 mg/dL (13 µmol/L) or were morbidly obese. The diagnosis of HCC was made following on histological examination of tumour tissue or based on criteria applied to images obtained using multiphasic CT or dynamic contrast-enhanced MRI15 16 (online supplemental methods A).
gutjnl-2022-327196supp001.pdf (3.4MB, pdf)
UK Alcohol Cohort (replication cohort 1)
The UK Biobank (UKB) is a large-scale biomedical database containing in depth genetic and health information from a prospective study of approximately half a million middle-aged individuals from the UK recruited in 2006–2010.17 Participants have been deeply phenotyped and are linked to UK hospital in-patient, cancer and mortality registries. A nested case–control dataset (n=860) was created using this resource. Cases were defined as participants with a hospital admission for ArC International Classification of Diseases 10 (ICD10:K70.3), and an HCC diagnosis (ICD10:C22.0 or ICD9:155.0). Controls were participants with a hospital admission for ArC but with no record of an HCC diagnosis. Analyses were restricted to participants of white British ancestry. These nested case–control data were then pooled with 306 patients recruited from the Centre for Hepatology at the Royal Free Hospital, London who had histologically proven ArC with or without HCC, as described previously18 (online supplemental methods B).
Germany and Italy Alcohol Cohort (replication cohort 2)
The replication cohort included 238 patients with ArC (42 with HCC) from the University of Bonn, and 72 patients with ArC (36 with HCC) from the University of Milan.
Validation cohorts
Patients with a history of alcohol misuse (AM) but without evidence of significant alcohol-related liver injury were recruited from psychiatric units in Germany (n=1080)19 20 and from Hepatology Centres in Heidelberg, Germany (n=99) and London, UK (n=341)18 (online supplemental methods C).
Genotyping and imputation
Discovery cohort
Genotyping was performed using genomic DNA extracted from peripheral blood samples as described previously.18 The GWAS (stage 1) included 1910 patients with ArC genotyped on the InfiniumGlobal Screening Array (V.24v2, Illumina) (table 1) (online supplemental methods D). Genotype imputation was performed with Minimac4 to the Haplotype Reference Consortium reference panel (HRC r1.1)21 using the Michigan Imputation Server22 (online supplemental methods E).
Table 1.
Overview of the study populations included in the discovery and replication cohorts
| Variable | Discovery (GWAS stage 1)* (n=1910) |
Replication (stage 2)* (n=1170) |
Validation† Patients with alcohol misuse (n=1520) |
|||||||||||
| Germany-Switzerland-Austria (n=1910) |
UK (cohort 1) (n=860) |
Germany (cohort 2) (n=238) |
Italy (cohort 3) (n=72) |
Germany (n=1179) |
UK (n=341) |
|||||||||
| Cases (n=1066) |
Controls (n=844) |
‡P value | Cases (n=70) |
Controls (n=790) |
‡P value | Cases (n=42) |
Controls (n=196) |
P value‡ | Cases (n=36) |
Controls (n=36) |
‡P value | Non-cirrhosis controls |
Non-cirrhosis controls |
|
| Age (yr) | 64.8 (8.5) (100%) |
57.1 (9.7) (100%) |
<0.0001 | 60.2 (5.9) (100%) |
56.3 (8.9) (100%) |
<0.0001 | 67.1 (9.1) (100%) |
58.5 (9.7) (100%) |
>0.05 | 72.7 (8.0) (100%) |
53.3 (8.9) (100%) |
<0.0001 | 42.7 (10.4) (100%) |
48.6 (10.5) (100%) |
| Proportion male (n: %) |
968 (90.8) | 624 (73.9) | <0.0001 | 67 (95.7) | 577 (73.0) | <0.0001 | 35 (83.3) | 131 (66.8) | <0.05 | 32 (88.9) | 31 (86.1) | >0.05 | 1148 (97.4) | 263 (77.1) |
| BMI (kg/m2) § | 28.1 (4.8) (69%) |
26.5 (5.3) (91%) |
<0.0001 | 29.3 (4.4) (100%) |
27.5 (4.8) (92%) |
<0.05 | 24.8 (3.5) (57%) |
26.1 (5.8) (52%) |
>0.05 | 27.0 (3.8) (64%) |
27.0 (6.8) (75%) |
>0.05 | 25.3 (4.5) (81%) |
24.7 (2.3) (53%) |
| BMI kg/m2; (n: %) <25 |
183 (24.7) | 308 (40.3) | <0.0001 | 13 (18.6) | 227 (31.3) | <0.05 | 13 (54.2) | 52 (51.0) | >0.05 | 6 (26.1) | 13 (48.1) | >0.05 | 505 (52.6) | 94 (52.2) |
| 25–30 | 333 (45.0) | 295 (38.6) | 27 (38.6) | 288 (39.7) | 8 (33.3) | 27 (26.5) | 12 (52.2) | 7 (25.9) | 345 (35.9) | 86 (47.8) | ||||
| >30 | 224 (30.3) | 162 (21.2) | 30 (42.9) | 211 (29.1) | 3 (12.5) | 23 (22.5) | 5 (21.7) | 7 (25.9) | 110 (11.5) | 0 (0) | ||||
| Diabetes type II + (n: %)§ |
337 (47.1) (67%) |
136 (30.1) (54%) |
<0.0001 | 20 (28.6) (100%) |
102 (12.9) (100%) |
<0.0001 | 14 (36.8) (90%) |
36 (19.7) (93%) |
<0.05 | 16 (44.4) (100%) |
5 (13.9) (100%) |
<0.05 | 58 (5.7) (86%) |
8 (3.6) (66%) |
*Cases and controls were assigned to groups as detailed in the Methods section.
†Validation cohorts were used in post hoc risk assessment.
‡P values were calculated from Student’s t-test for quantitative variables and as Pearson’s χ2 test for categorical variables.
§Data are reported as mean±SD or as number (%); Completeness of phenotypic information for age, BMI and type 2 diabetes status are reported as percentage of subjects with available information below the mean value.
BMI, body mass index; GWAS, Genome-Wide Association Study.
Replication and validation samples
Patients from the Royal Free Hospital, London and Germany were genotyped using the OmniExpress array (24v1-0a, Illumina).12 The replication (stage 2) included 1170 patients with ArC (table 1). Patients from Italy were genotyped on the InfiniumGlobal Screening Array (24v2, Illumina) (online supplemental methods D). Genotypic data were imputed for each cohort to the HRC reference (online supplemental methods E). Imputed genotypic data from 606 patients were obtained from the UKB Resource.23
Statistical analyses
GWAS analysis
Association analyses for 7 946 762 variants were performed using Plink V.2.024 with allele dosages obtained after imputation (imputation info score >0.3, minor allele frequency >1%). The lambda inflation factor λGC for the unadjusted GWAS analysis was 1.085 indicative of subtle population stratification. To account for the observed inflation, the top 20 principal components (PCs) on the LD-pruned data set were calculated and the top 15 PCs of genetic ancestry included as covariates in the regression models.25 The corrected λGC was 1.03. Two discovery GWAS analyses were performed: GWAS 1 (primary GWAS analysis): included only the top 15 PCs as covariates in the regression model. The p value threshold for lead single-nucleotide polymorphism (SNPs) for replication follow-up was set to p<5×10−6 to allow loci with suggestive association to be included at the replication stage. GWAS 2 (sensitivity GWAS analysis): included sex, age and the top 15 PCs as covariates; the top 15 independent loci were follow-up at Stage 2.
Loci discovery and annotation
Independent genomic risk loci and lead variants (for p<5×10−6) were derived from FUMA (V.1.3.1)26 based on GWAS summary statistics, as previously described.27 For a locus to be defined as independent it had to be separated from other loci by at least 500 kb of genomic distance; the top-ranking SNPs were deemed potential lead markers.
Power analysis
The expected power to identify a true association between a SNP and HCC development in ArC was calculated using the GAS Power Calculator.28 The power for SNPs with minor allele frequencies of >20% was estimated to be 49% for alleles with a relative risk of 1.5, increasing to 81% for a relative risk of 1.6, for a p value threshold of 5×10−8 (online supplemental table 1).
Replication analysis
In stage 2, the selected SNPs were validated in independent samples from the UK, Germany and Italy. Study-specific β estimates and standard errors were further analysed using fixed-effect meta-analysis. Two criteria were required to demonstrate replication: (1) p<5.55×10−3 (corresponding to p<0.05 after Bonferroni correction for nine tests in the primary analysis); or p<3.33×10−3 (corresponding to p<0.05 after Bonferroni correction for 15 tests in the sensitivity analysis) (2) and consistency of allelic effect direction between discovery and replication samples (online supplemental methods F).
Additional replication analyses
The association between novel risk loci and HCC/liver cancer were also assessed using: (1) publicly available summary statistics from a recent alcohol-related HCC GWAS performed by Trépo et al 14; (2) data from two large population-based cohorts (Finngen and BioBank Japan) and (3) data from a UK cohort of patients with HCV-related cirrhosis (STOP-HCV) (online supplemental methods F).
Association with other cancers (pleiotropy)
Moreover, we assessed if novel risk loci were associated with selected cancers unrelated to the liver in both the UKB and FinnGen population-based cohorts. Each cancer phenotype was defined by ICD codes present in hospital admissions, death records and cancer registry records. In addition, the NHGRI-EBI Catalogue of human GWAS was searched for association of novel risk loci with cancer phenotypes (online supplemental methods F).
Meta-analysis GWAS
A fixed-effect meta-analysis restricted to markers present in all data sets (n=5 552 382) was performed using METAL29 to: (1) use the total study sample (n=3080) for the discovery stage and (2) to determine the combined effect size of replicated loci across stages 1 and 2 datasets.
eQTL analysis
Variants at novel loci were tested for cis-eQTL effect on gene expression in: (1) liver tissue (n=266) using the database of the Genotype-Tissue Expression Project (GTEx) release V830 and (2) whole-blood (n=24 376) using the database of the eQTLGen Consortium.31
SNP Heritability Analysis
The proportion of phenotypic variance explained by the additive genetic effect of common genome-wide significant SNPs (h²SNP: SNP heritability) was estimated using a Genomic relatedness matrix REstricted Maximum Likelihood analysis implemented in GCTA32 (online supplemental methods G).
Association with HCC-related phenotypes
Replicating loci were tested in the total UKB for association with two HCC-related phenotypes: leucocyte telomere length33 and liver fat content.34 Leucocyte telomere length was available for 474 074 participants in UKB (Field ID: 22191), while liver fat content was available for 8315 imaging substudy participants (Field ID: 22436) (online supplemental methods H).
Patient and public involvement
There was no patient and public involvement in the design and conduct of this study.
Results
GWAS and validation of the loci
After imputation a total of 7 946 762 variants with a MAF >0.01 were tested for association with HCC in 1066 cases with ArC and HCC and 844 controls with ArC but with no evidence of HCC (table 1).
Associations with HCC were observed at genome-wide significance (p<5×10−8) for two independent genomic loci viz PNPLA3 and TM6SF2 (table 2; figure 1A, online supplemental figure 1). The strongest signal was at rs2294915, located in PNPLA3 (p=6.21×10−15) which encodes 1-acylglycerol-3-phosphate O-acyltransferase. This tag SNP rs2294915, located in intron 8 of PNPLA3, is in strong linkage disequilibrium (LD) (r2=0.92) with the functional variant rs738409 C>G p.I148M in exon 3 of PNPLA3 that yielded a similar p value at the discovery stage (p=7.23×10−15, OR (95% CI)=1.71 (1.49 to 1.96)).
Table 2.
Association results for lead markers of regions entering the validation stage of the primary and sensitivity GWAS analysis
| Lead SNPs |
Locus | Chr | SNP ID | EA, ED |
Discovery (stage 1) | Replication (stage 2) | Eff. Dir. |
I2 | p heterog. | Combined (stage 1 & 2)‡ | |||||
| P value*† | OR (95% CI) | EAF Ca|Co|Eur |
Meta P value+*† |
OR (95% CI) | Meta P value+*† | OR(95% CI) | I2 | ||||||||
| GWAS analysis 1 (pc adjusted)* | |||||||||||||||
| SNP 1 | PNPLA3§ | 22 | rs2294915 | T+ | 6.21×10−15 | 1.71 (1.50 to 1.96) | .49|.36|.24 | 6.19×10-6 | 1.89 (1.44 to 2.50) | +++ | 0 | 0.517 | 2.44×10−19 | 1.75 (1.55 to 1.97) | 0 |
| PNPLA3 | 22 | rs738409 | G+ | 7.23×10−15 | 1.71 (1.49 to 1.96) | .48|.35|.22 | 9.74×10-6 | 1.89 (1.42 to 2.50) | +++ | 0 | 0.578 | 4.31×10−19 | 1.74 (1.54 to 1.97) | 0 | |
| SNP 2 | TM6SF2¶ | 19 | rs58489806 | T+ | 1.42×10−9 | 1.87 (1.53 to 2.29) | .17|.10|.08 | 5.22×10-4 | 1.91 (1.33 to 2.76) | ++− | 54 | 0.110 | 3.04×10−12 | 1.88 (1.57 to 2.25) | 32 |
| TM6SF2 | 19 | rs58542926 | T+ | 2.81×10−9 | 1.94 (1.56 to 2.42) | .15|.08|.07 | 7.58×10-5 | 2.11 (1.46 to 3.04) | ++− | 61 | 0.076 | 1.00×10−12 | 1.98 (1.64 to 2.40) | 43 | |
| SNP 3 | TERT | 5 | rs2242652 | A- | 7.87×10−7 | 0.64 (0.53 to 0.76) | .13|.19|.19 | 1.07×10-3 | 0.48 (0.31 to 0.74) | −−− | 0 | 0.814 | 6.40×10−9 | 0.61 (0.52 to 0.72) | 0 |
| SNP 4 | LINC00939 | 12 | rs12371263 | A- | 9.59×10−7 | 0.63 (0.52 to 0.76) | .16|.21|.20 | 0.332 | 0.83 (0.57 to 1.21) | −+− | 0 | 0.535 | – | – | – |
| SNP 5 | DMAC2 | 19 | rs17318596 | A- | 2.49×10−6 | 0.71 (0.61 to 0.82) | .33|.40|.37 | 0.849 | 1.03 (0.77 to 1.38) | −+− | 15 | 0.308 | – | – | – |
| SNP 6 | SP100 | 2 | rs6743289 | C- | 2.77×10−6 | 0.72 (0.62 to 0.82) | .45|.52|.47 | 0.046 | 0.75 (0.57 to 1.00) | −−− | 0 | 0.936 | – | – | – |
| SNP 7 | GPIHBP1 | 8 | rs118088203 | T- | 3.60×10−6 | 0.24 (0.13 to 0.44) | .01|.03|.02 | 0.229 | 1.64 (0.73 to 3.07) | +++ | 0 | 0.697 | – | – | – |
| SNP 8 | CNPY1 | 7 | rs12698003 | T+ | 3.65×10−6 | 1.39 (1.21 to 1.60) | .46|.39|.41 | 0.053 | 0.74 (0.55 to 1.00) | −−− | 41 | 0.179 | – | – | – |
| SNP 9 | GLYR1 | 16 | rs741692 | T+ | 4.16×10−6 | 1.58 (1.30 to 1.92) | .18|.12|.15 | 0.783 | 1.05 (0.73 to 1.51) | ++− | 0 | 0.541 | – | – | – |
| GWAS analysis 2 (pc, sex, age adjusted)† | |||||||||||||||
| SNP 1 | PNPLA3§ | 22 | rs2294915 | T+ | 6.31×10−14 | 1.76 (1.52 to 2.05) | .49|.36|.24 | 3.24×10-5 | 1.89 (1.40 to 2.54) | +++ | 0 | 0.428 | 1.06×10-17 | 1.79 (1.57 to 2.04) | 0 |
| PNPLA3 | 22 | rs738409 | G+ | 1.67×10−13 | 1.75 (1.51 to 2.03) | .48|.35|.22 | 4.17×10-5 | 1.85 (1.37 to 2.50) | +++ | 0 | 0.448 | 5.35×10-17 | 1.77 (1.55 to 2.02) | 0 | |
| SNP 2 | TM6SF2¶ | 19 | rs143988316 | T+ | 4.40×10−8 | 1.91 (1.51 to 2.41) | .16|.09|.07 | 5.17×10-2 | 1.54 (1.00 to 2.38) | ++− | 0 | 0.621 | 9.14×10-9 | 1.81 (1.51 to 2.16) | 0 |
| TM6SF2 | 19 | rs58542926 | T+ | 1.21×10−7 | 1.93 (1.51 to 2.45) | .15|.08|.07 | 1.56×10-4 | 2.16 (1.45 to 3.22) | ++− | 48 | 0.149 | 8.80×10-11 | 1.99 (1.61 to 2.44) | 26 | |
| SNP 3 | SCN5A | 3 | rs6599222 | C+ | 2.86×10−6 | 1.53 (1.28 to 1.84) | .25|.20|.21 | 0.977 | 1.01 (0.68 to 1.48) | −++ | 0 | 0.984 | – | – | – |
| SNP 4 | intergenic | 13 | rs148892410 | A- | 3.77×10−6 | 0.16 (0.07 to 0.35) | .01|.02|.01 | 0.798 | 1.45 (0.09 to 24.2) | ++− | 17 | 0.299 | – | – | – |
| SNP 5 | intergenic | 2 | rs6739777 | G- | 5.03×10−6 | 0.69 (0.59 to 0.81) | .29|.34|.30 | 0.388 | 0.86 (0.61 to 1.21) | −+− | 40 | 0.193 | – | – | – |
| SNP 6 | ENSG00000269151 | 19 | rs143660337 | A- | 5.14×10−6 | 0.41 (0.28 to 0.60) | .03|.05|.04 | 0.151 | 1.68 (0.83 to 3.41) | ++− | 0 | 0.589 | – | – | – |
| SNP 7 | LOC105374308 | 3 | rs58339845 | T- | 5.84×10−6 | 0.46 (0.33 to 0.65) | .05|.07|.07 | 0.361 | 1.34 (0.72 to 2.50) | +++ | 0 | 0.919 | – | – | – |
| SNP 8 | intergenic | 7 | rs16869539 | G+ | 5.96×10−6 | 1.48 (1.25 to 1.75) | .36|.30|.37 | 0.537 | 0.90 (0.65 to 1.25) | −−− | 0 | 0.983 | – | – | – |
| SNP 9 | CELF2 | 10 | rs2277212 | T+ | 6.84×10−6 | 1.57 (1.29 to 1.91) | .75|.70|.74 | 0.282 | 1.22 (0.85 to 1.74) | +−+ | 16 | 0.303 | – | – | – |
| SNP 10 | intergenic | 7 | rs6462611 | C+ | 7.82×10−6 | 1.41 (1.21 to 1.64) | .49|.44|.50 | 0.017 | 0.68 (0.49 to 0.93) | ++− | 0 | 0.465 | – | – | – |
| SNP 11 | ENSG00000227757 | 21 | rs2017196 | T+ | 8.73×10−6 | 1.70 (1.34 to 2.14) | .89|.85|.88 | 0.092 | 0.68 (0.44 to 1.06) | −−− | 0 | 0.513 | – | – | – |
| SNP 12 | TERT | 5 | rs2242652 | A- | 9.28×10−6 | 0.64 (0.52 to 0.78) | .13|.19|.19 | 2.60×10-4 | 0.41 (0.25 to 0.66) | ++− | 0 | 0.699 | 4.08×10-8 | 0.60 (0.50 to 0.72) | 17 |
| SNP 13 | RARB | 3 | rs7617311 | A+ | 9.32×10−6 | 1.50 (1.25 to 1.80) | .28|.21|.26 | 0.454 | 0.87 (0.61 to 1.25) | −+− | 0 | 0.751 | – | – | – |
| SNP 14 | CIAO2A | 15 | rs2922508 | T+ | 9.53×10−6 | 1.61 (1.30 to 1.99) | .17|.13|.15 | 0.153 | 1.33 (0.90 to 1.97) | +++ | 0 | 0.797 | – | – | – |
| SNP 15 | intergenic | 2 | rs56209271 | T- | 9.67×10−6 | 0.69 (0.59 to 0.82) | .28|.34|.30 | 0.168 | 0.78 (0.55 to 1.11) | −−− | 61 | 0.075 | – | – | – |
*OR and p value adjusted for top 15 PCs of genetic ancestry.
†OR and p value adjusted for sex, age and top 15 PCs of genetic ancestry.
‡The results of the combined analyses are only provided for variants meeting a Bonferroni corrected p<0.05 at the replication stage (printed in bold face).
§The tag SNP rs2294915 in PNPLA3 is in LD (r2= 0.92) with the functional variant rs738409 previously reported at the PNPLA3 locus58 59.
¶The intergenic tag SNP rs143988316 is in strong LD (r2= 0.88) with the functional variant rs58542926 previously reported at the TM6SF2 locus.60
+, Significance derived from a fixed effect meta-analysis; Ca, Cases (Cirrhosis with HCC); Chr, chromosome; Co, Controls (Cirrhosis without HCC); EA, effect allele; EAF, allele frequency of the effect allele; ED, effect direction; HCC, hepatocellular carcinoma; I2, percentage of between cohort heterogeneity; LD, linkage disequilibrium; pheterog, heterogeneity p value of the meta-analysis; SNP, single-nucleotide polymorphism.
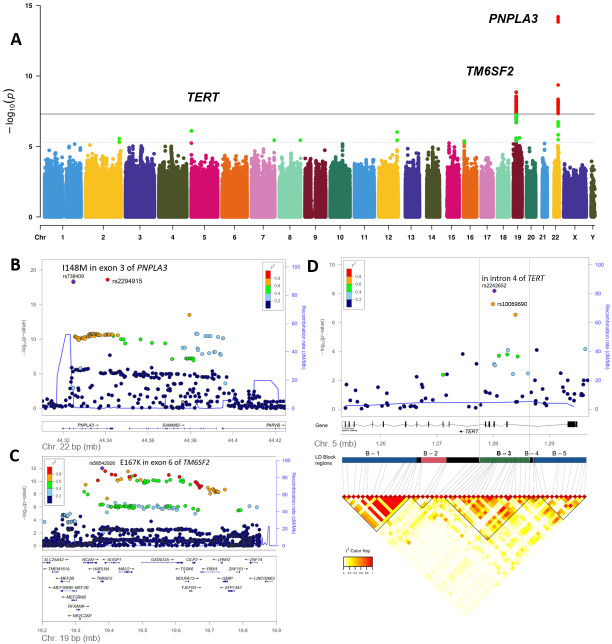
Figure 1.
Genome-wide association study (Discovery GWAS) results. Principal findings of genetic analyses. (A): Manhattan plot of genome-wide association results for alcohol-related hepatocellular carcinoma (HCC) in the primary discovery cohort. P values (−log10) are shown for SNPs that passed quality control. The genome-wide significance threshold (5×10−8) is shown as a black line. The threshold for replication follow-up (p<5×10−6) is shown as a dashed line. Gene names for replicating loci (table 2) are shown. Variants with significance p<5×10−8 are highlighted in red, those with p<5×10−6 are highlighted in green. (B) Locus plot for HCC risk locus PNPLA3. The −log10 (p values, meta-analysis of discovery and replication samples) are plotted against SNP genomic position based on NCBI Build 37, with the names and location of nearest genes shown at the bottom. The variant with the lowest p value (lead variant) in the discovery analysis in the region is marked by a purple diamond. SNPs are coloured to reflect correlation with the most significant SNP, with red denoting the highest LD (r2 >0.8) with the lead SNP. The top association signal is located in exon 3 of PNPLA3. Estimated recombination rates from the 1000 Genomes Project (hg19, EUR population) are plotted in blue to reflect the local LD structure. (C) Locus plot for HCC risk locus TM6SF2. The top association signal is located in exon 6 of TM6SF2. (D) Locus plot for HCC risk locus TERT. Fine-mapping analysis of the TERT association signals. Annotated LD-Blocks are clusters of strong pairwise LD SNPs and reflect the LD pattern in the Discovery GWAS cohort. The lead association signal is located in intron 4 of the TERT gene (annotated on the reverse strand), located in LD block B-3 spanning from intron 4 to intron 2 of TERT. NCBI, National Center for Biotechnology Information; SNP, single-nucleotide polymorphism.
The other signal associated with HCC at genome-wide significance was rs58489806, located in intron 1 of MAU2 (p=1.49×10−9) encoding MAU2 sister chromatid cohesion factor; 49 additional genome-wide significant SNPs were mapped to this locus. The variant rs58489806 is in strong LD (r2=0.80) with the coding variant rs58542926 p.E167K at the TM6SF2 locus (encoding transmembrane 6 superfamily member 2) that yielded (p=2.81×10−9, OR (95% CI)=1.94 (1.56 to 2.42)) at the discovery stage.
In stage 2, the nine lead SNPs from HCC associated loci were validated in independent cohorts from the UK, Germany and Italy in fixed-effect meta-analysis (table 1; online supplemental tables 2–4). In addition to rs2294915 in PNPLA3 (p=6.19×10−6) and rs58489806 in TM6SF2/MAU2 (p=5.22×10−4), disease association was replicated for the minor allele in rs2242652:A (p=1.07×10−3) in TERT (telomerase reverse transcriptase) (table 2). In the combined analysis of all stage 1 and stage 2 samples, the association of rs2242652:A in TERT with alcohol-related HCC attained genome-wide significance (p=6.41×10−9, OR (95% CI)=0.61 (0.52 to 0.72) (table 2). The protective effect associated with carriage of TERT rs2242652:A remained significant after correction for sex, age, body mass index (BMI), type 2 diabetes and the top 15 PCs of genetic ancestry, but did not reach genome-wide significance (p=7.94×10−5; OR (95% CI)=0.63 (0.50 to 0.79) (online supplemental table 5) reflecting the loss of power associated with the high number of missing BMI and diabetes data points in the analysis (table 1).
A sensitivity analysis in which the genome-wide analysis was additionally adjusted for sex and age also showed genome-wide significant association with HCC for two independent genomic loci PNPLA3 and TM6SF2 with HCC and suggestive evidence of association for TERT (p=9.28×10−6). (table 2; online supplemental figures 2 and 3). Of the top 15 associated loci, only the variants in PNPLA3, TM6SF2 and TERT were replicated (table 2).
The combined GWAS meta-analyses of stage 1 and 2 data sets of the primary and the sensitivity analyses confirmed genome-wide significant association with HCC for genomic loci in rs738409 in PNPLA3, rs58542926 in TM6SF2 and rs2242652 in TERT. No additional risk locus attained genome-wide significance p<5.0×10−8 (online supplemental table 6). Forest plots showing the association between genomic loci in PNPLA3, TM6SF2, TERT and HCC are shown in online supplemental figures 4–6. Regional association plots of these three loci are shown in figure 1B–1D and in online supplemental figures 7–9.
Previously reported associations of HCC in the context of ArC with variants of HSD17B13 rs72613567:TA (p=8.95×10−3; OR=0.81 (95% CI 0.69 to 0.95) and APOE rs429358:C (p=5.44×10−3; OR=0.74 (95% CI 0.60 to 0.91) were nominally significant in this study, but did not achieve genome-wide significance in the discovery cohort (online supplemental tables 5 and 7). In contrast, a recently reported association between rs708113:T near WNT3A was not confirmed (online supplemental tables 5 and 7). Other previously described HCC risk loci, for example, DEPDC5 in HCV-related HCC35 or STAT4 and HLA-DQ 36 were not significantly associated with ArC-related HCC in this study (online supplemental table 7).
Allelic and genotypic associations for TERT were highly significant, in the univariate analyses, for the comparisons HCC vs ArC (Pallelic=2.81×10−11, Pgenotypic 2.32×10−10) and HCC versus alcohol misuse but not for ArC vs alcohol misuse using combined genotype counts from the stage 1 and 2 data sets (online supplemental table 8; figure 2). The protective effect for HCC was greater in homozygous carriers of TERT rs2242652:A (OR=0.41 (95% CI 0.25 to 0.67)) than in heterozygous carriers (OR=0.61 (95% CI 0.51 to 0.72)). In contrast, variants in PNPLA3 and TM6SF2 were strongly associated both with ArC and ArC-related HCC (online supplemental tables 9 and 10, figure 2).
Figure 2.
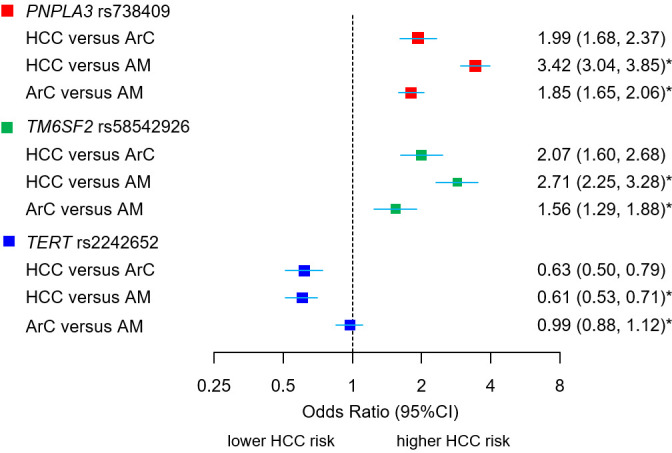
Association between novel (TERT) and confirmed loci (PNPLA3, TM6SF2) with HCC and cirrhosis phenotypes. ORs and 95% CIs for the susceptibility loci for alcohol-related HCC and alcohol-related cirrhosis (ArC) in comparison to alcohol misusers without cirrhosis (AM). The comparison HCC vs ArC displays allelic ORs of combined stage 1 and 2 samples (meta-analysis), derived from allele dosage data, adjusted for age, sex, BMI, type 2 diabetes status and top 15 principal components of genetic ancestry. *The comparison HCC versus AM and ArC versus AM display unadjusted allelic ORs derived from 2×2 contingency tables of allele counts observed in the total cohort, provided in online supplemental tables 2–4. BMI, body mass index; HCC, hepatocellular carcinoma.
Fine-mapping of TERT locus
In the primary meta-analysis of stage 1 and stage 2 samples, the strongest association signal was obtained for the minor allele in rs2242652:A (p=6.40×10−09; OR=0.61 (95% CI 0.52 to 0.72)), although the alternative allele in rs10069690:T was similarly associated (p=5.19×10−08, OR=0.66 (95% CI 0.57 to 0.77)). Both variants are located in intron 4 of TERT and are correlated (r2=0.70; online supplemental table 11). The analysis of LD structure at the TERT locus showed that the association signal spans a narrow range from intron 2 to intron 6 of TERT—here termed LD block B-3 region (online supplemental table 11 and figure 7). The conditional analysis on allele dosage of rs2242652:A or rs10069690:T on each of the 20 SNPs from the B-3 region confirmed rs2242652 to be the lead locus (online supplemental table 11 and 12). Indeed, none of the other variants within the B-3 block, including rs10069690 was associated with HCC after conditioning on rs2242652 (online supplemental table 11).
Replication of the TERT variant’s association with HCC
Significant associations were observed between rs2242652:A and HCC in patients with HCV-related cirrhosis (p=0.047; OR=0.72 (95% CI 0.53 to 0.99) and in the population-based FinnGen, UKB and BioBank Japan cohorts (table 3, online supplemental figure 10 and table 13).
Table 3.
Replication of TERT variants in patients with alcohol-related and chronic HCV-related cirrhosis and in population-based cohorts
| Cohort | Controls | Cases phenotype (ICD-10) | N cases | controls |
TERT
Variant |
EA | P value | OR (95% CI) |
| Current study* | ALD cirrhosis* | C22.0 Liver cell carcinoma (HCC) in alcohol-related cirrhosis | 1214 | 1866 | rs2242652 | A | 6.40×10−9 | 0.61 (0.52 to 0.72) |
| Replication cohorts | |||||||
| Trépo et al† | ALD F0-F4 fibrosis | C22.0 HCC in alcohol-related liver disease (F0-F4 fibrosis) | 775 | 1332 | rs2242652 | A | 0.179 | 0.89 (0.75 to 1.06 |
| Stop-HCV‡ | HCV cirrhosis | C22.0 Liver cell carcinoma (HCC) in HCV-related cirrhosis | 169 | 890 | rs2242652 | A | 0.047 § | 0.72 (0.53 to 0.99) |
| Zhang et al¶ | Healthy volunteers | C22.0 Liver cell carcinoma (HCC) | 473 | 564 | rs2242652 | A | 0.004 | 0.70 (0.55 to 0.90) |
| Dong et al** | Healthy volunteers | C22.0 Liver cell carcinoma (HCC) (hepatitis-induced) | 162 | 106 | rs10069690 | T | 0.00014§ | 0.36 (0.21 to 0.63) |
| FinnGen†† | General population | C22 malignant neoplasm of liver and intrahepatic bile duct | 442 | 204 070 | rs2242652 | A | 0.007 | 0.80 (0.68 to 0.94) |
| UKBB‡‡ | General population | C22 malignant neoplasm of liver and intrahepatic bile duct | 874 | 348 465 | rs2242652 | A | 0.027 | 0.87 (0.78 to 0.97) |
| UKBB‡‡ | General population | C22.0 Liver cell carcinoma (HCC) | 383 | 348 956 | rs2242652 | A | 0.028 | 0.80 (0.66 to 0.98) |
| BBJ Japan§§ | BBJ population¶¶ | C22.0 Liver cell carcinoma (HCC) | 1866 | 195 745 | rs72709458 | T | 0.00031 | 0.84 (0.76 to 0.92) |
*Combined effect estimates of stage 1 and 2 samples of current study as shown in table 1 (for comparison).
†Cases: patients with ALD (80% with F3-4 fibrosis; 20% F0-2 fibrosis) and HCC, controls: patients with ALD (90% with F3-4 fibrosis, 10% F0-2 fibrosis) from Trépo et al.14
‡Cases: patients with HCV related cirrhosis and HCC, controls: patients with HCV related cirrhosis without HCC (online supplemental methods F).
§Allelic ORs were calculated from 2×2 tables on allele counts. Significance was calculated as 1 df χ2 test.
¶Zhang et al,52 Huang et al 51 Han Chinese patients with HCC (individuals were excluded from the study if they had HCV).
**Dong et al 61 62 male Han Chinese patients with viral hepatitis-induced primary hepatocellular carcinoma (r2=0.85 between rs10069690:T and rs2242652:A, both variants are in high linkage disequilibrium).
††General population controls (excluding all cancers).
‡‡As UKB data were incorporated into our discovery analysis, further interrogation of liver cancer phenotypes from UKB does not constitute independent validation.
§§Variants rs2242652 and rs10069690 were not available in the summary GWAS data from Ishigaki et al 63 (PMID: 32514122, publicly available from http://jenger.riken.jp/en/result) (rs72709458 is the closest proxy to rs2242652 (r2=0.973)).
¶¶Removed diseases from control samples (biliary tract cancer, oesophageal cancer, gastric cancer, colorectal cancer and pancreatic cancer).
ALD, alcohol liver disease; BBJ, BioBank Japan; EA, effect allele; FinnGen, FinnGen Biobank; GWAS, Genome-Wide Association Studies; HCV, hepatitis C virus; ICD, International Classification of Diseases;; UKB, UK Biobank.
Association of TERT variants with non-liver cancers
Associations between TERT rs2242652:A and the 10 most frequent cancers were explored in the UKB and FinnGen (FG) cohorts (online supplemental figure 10). Significant associations were observed with bladder cancer (FG: p=6.10 × 10-6, OR=0.83 (95% CI 0.67 to 0.90)), UKB: p=5.82 × 10−7, OR=0.84 (95% CI 0.79 to 0.90)), and prostate cancer (FG: p=5.11 × 10−11, OR=0.87 (95% CI 0.84 to 0.90)), UKB: p=6.16 × 10−16; OR=0.86 (95% CI 0.83 to 0.89)) while weaker associations were observed for lung and skin cancer. The effect sizes for prostate and bladder cancer were smaller than those for HCC/primary liver cancer in these cohort (UKB: HCC: p=0.028; OR=0.80 (95% CI 0.66 to 0.89), FG: primary liver cancer: p=0.009; OR=0.81 (95% CI 0.69 to 0.95)). These effect sizes are broadly consistent with those reported in the NHGRI-EBI Catalogue of human GWASs (online supplemental table 14).
Additive effect of risk variants
The proportions of patients with ArC, in the discovery and validation cohorts, who developed HCC increased with cumulative carriage of the risk increasing alleles rs738409:G in PNPLA3, rs58542926:T in TM6SF2 and rs2242652:G in TERT (online supplemental figure 11). In the discovery cohort, the OR for alcohol-related HCC was 2.12 (95% CI 1.76 to 2.56) in patients carrying three to four risk alleles, and 5.24 (95% CI 2.82 to 9.77) in patients carrying five to six risk alleles (online supplemental table 15). In the UK replication cohort, the ORs for carriage of three to four risk alleles and five to six risk alleles were even higher at 3.25 (95% CI 1.84 to 5.73) and 17.8 (95% CI 6.38 to 49.6), respectively (online supplemental figure 12 and table 15).
Association with leucocyte telomere length and liver fat content in the UKB
The minor allele of the lead variant in TERT rs2242652:A (p=2.12×10−44) was significantly associated with an increase in LTL, as was rs10069690:T which is in strong LD with the lead variant (p=4.08×10−84) (online supplemental table 16). Additional variants located in the tested interval, that is, rs7726159, showed even stronger association with LTL (p=1.16×10−219) despite weak LD with rs2242652 (r2=0.354) (online supplemental table 11). The main association signals for HCC and LTL were both located in the LD block B-3 region, but a direct correlation in the strength of association was not observed (online supplemental figure 13 and table 11). Lead variants in PNPLA3 and TM6SF2 were not significantly associated with LTL—rs738409 (p=0.458) and rs58542926 (p=0.475), but showed significant associations with liver fat content viz. rs738409 (p=3.39×10−61), rs58542926 (p=5.94×10−45), respectively (online supplemental table 16); rs2242652 in TERT was not significantly associated with liver fat content (p=0.144).
eQTL Analysis
Carriage of rs2242652:A was associated with increased expression of TERT in blood leucocytes (p=1.39×10−5) (online supplemental table 11). However, no significant eQTLs were found for rs2242652 in liver using the GTEx data base or in any other tissues.30
SNP Heritability Analysis
The percentage heritability for ArC- HCC explained by additive genome-wide SNPs expressed as h 2 was 29.6% on the observed scale (GWAS cohort) and either 20.4% or 25.7% on the liability scale assuming a disease prevalence of 1% or 2.5%, respectively (online supplemental table 20 20). The proportion of phenotypic variation due to the underlying genetic variation in the PNPLA3/TM6SF2/TERT LD regions, expressed as h 2, was 7.5% on the observed scale and 4.2% or 5.3% on the liability scale, assuming the same disease prevalence (online supplemental table 17). The proportion of the total SNP heritability due to variance component 1 (PNPLA3/TM6SF2/TERT variants) was 25.5% for model 1, adjusted for 15 PCs, and 22.2% for model 2 adjusted for sex, age and 15 PCs. After adjustment of variance component 1 for lead variants rs738409 in PNPLA3/rs58542926 in TM6SF2/rs2242652 in TERT h2 was reduced to 0.000001%, indicating that the genetic risk of variance component 1 was fully captured by the three identified lead variants.
Discussion
In this study, associations at genome-wide significance were identified between HCC in ArC and previously recognised variants in PNPLA3 and TM6SF2, and with a variant in TERT (telomerase reverse transcriptase) on chromosome 5 not previously associated with this phenotype. In combination, these three loci may explain up to 25% of the total SNP heritability in HCC in patients with ArC.
The identification of host genetic risk factors for alcohol-related HCC has been largely undertaken using a candidate gene approach. Candidate genes have invariably been selected because of their association with progression of alcohol-related liver injury and positive robust associations for variants rs738409 in PNPLA3, and rs58542926 in TM6SF2 have been identified.8 9 These variants are known to modify liver fat content and signalling, but how they influence the mechanisms leading to tumour initiation or promotion is largely unknown.37 38 In this study, the increased risk associations between HCC in ArC and rs738409 in PNPLA3 and rs58542926 in TM6SF2 were confirmed, at genome-wide significance.
Significant associations have also been identified between rs72613567 in HSD17B13 and rs429358 in APOE and a reduced risk for developing HCC in ArC.9–11 In this study, these protective associations were confirmed but failed to reach a detectable genome-wide significance level (online supplemental table 5).
Further insights into the genetic landscape of HCC in the context of ArLD were recently provided by Trépo et al 14 who undertook a discovery GWAS of HCC in people with a spectrum of ArLD in a French-Belgian collaborative effort. Similar to this study, they confirmed genome-wide significant associations with an increased risk for developing alcohol-related HCC and variants in PNPLA3 and TM6SF2. In addition, they found an equally significant association with rs708113 in the WNT3A-WNT9A region on chromosome 1q42, which was associated with a reduced risk for development of alcohol-related HCC. The presence of this variant was associated with increased immune cell infiltration of tumour tissues and a lower frequency of beta-catenin mutations (CTNNB1) which frequently precede HCC occurrence.39 This protective effect of rs708113 was not observed in people with HCC on a background of chronic HCV infection or NAFLD.14
In this study, rs708113 in the WNT3A-WNT9A region was not significantly associated with the development of HCC, possibly reflecting differences in the cohort composition between the two studies although both comprised of participants of European descent. This assumption of population diversity is supported, to some extent, by the fact that in the French-Belgian cohorts the effect size of rs58542926 in TM6SF2 surpassed that of rs738409 in PNPLA3 which has been the strongest single genetic risk locus for ArLD in previous candidate gene association studies.40
The key finding in this study was the identification of a risk locus in TERT, that is not related to lipid turnover, inflammation or fibrogenesis but appears to be highly influential in HCC development.41 Like any cancer, HCC arises when healthy hepatocytes acquire mutations in specific genes regulating cell division. In HCC, TERT is the most commonly mutated gene, with mutations (mainly in the promoter region) present in up to 60% of tumours.42 This lends clear plausibility to the association reported in this study between inherited polymorphisms in TERT and alcohol-related HCC. Similar relationships between germline and somatic variants have been identified for other cancers types.43 The biology of telomere regulation is still being unravelled and remains incompletely understood. TERT encodes the catalytic subunit (hTERT) of the enzyme telomerase, which maintains telomeres, the repeated DNA segments found at the ends of chromosomes. In most cells telomeres progressively shorten as the cells repeatedly divide and this eventually triggers the cell to stop dividing or to undergo apoptosis. Telomerase counteracts the shortening of telomeres by adding small repetitive DNA segments to the ends of the chromosomes during each cell division cycle.44 Telomerase is also abnormally active in most cancer cells.45 TERT expression levels significantly affect telomerase activity in various cells and tissues.46 Previous studies show that older age, male gender and cirrhosis (all classic risk factors for HCC) are associated with shorter telomere length in liver tissue.47 Thus, this study, showing that rs2242652:A reduces HCC risk while at the same time increasing telomere length, is directionally concordant with this previous work. From a mechanistic perspective, it could be that shorter telomeres leave cells more vulnerable to mutations in driver genes, thus accelerating hepatocarcinogenesis.47 It is important to point out however that the association between rs2242652 and HCC may not be entirely mediated through telomere length alone. Indeed, for variants in TERT, we found that there was not a good correlation between strength of association with telomere length and strength of association with HCC. Thus, rs2242652 is not simply acting as a surrogate for telomere length. Relevant to this point is that, as part of its non-canonical functions TERT also regulates the WNT/β-catenin pathway.48 49 This signalling pathway is suggested to play a role in alcohol-induced fibrogenesis and hepatocarcinogenesis, too.14 50 However, regarding the risk of alcohol-induced fibrosis/cirrhosis, our data unequivocally show no association with rs2242652 in TERT.
There is also some support for the findings in this study in previous publications. In the GWAS undertaken by Trépo et al 14 rs2242652:A in TERT was associated with a reduced risk of HCC, but the OR was weaker than in this study and did not reach statistical significance (p=0.179; OR=0.89 (95% CI 0.75 to 1.06)). However, carriage of rs10069690:T in TERT—the nearest available proxy to rs2242652—was associated with a significantly reduced risk of HCC development (p=0.036; OR=0.84 (95% CI 0.71 to 0.99)). The significant association between rs2242652:A in TERT with liver and intrahepatic bile duct carcinoma in the population-based FinnGen cohort and with HCC in the BioBank Japan cohort additionally substantiate this study’s findings. A case–control study in Han Chinese involving 473 patients with HCC and 564 healthy volunteers, which is reported in two separate publications (Huang et al and Zhang et al 51 52), also identified associations between variants in TERT and the development of HCC; carriage of rs2242652:A in TERT was associated with a reduced risk for HCC development (OR =0.70, 95% CI 0.55 to 0.90, p =0.004), as was carriage of rs10069690:T (OR =0.75, 95% CI 0.59 to 0.96, p=0.021). Patients with chronic HCV infection were excluded from this study but otherwise it is unclear whether the patients with HCC had underlying chronic liver disease and, if so, its aetiology.
A number of HCC risk loci have been identified in patients developing HCC on a background of chronic HCV35 and chronic HBV,36 but none was significantly associated with ArC-related HCC in this study. However, there is some evidence that variants in TERT may predispose to HCC in other types of chronic liver disease. Thus, a significant association between rs2242652:A and the reduced risk for developing HCC was observed in patients with HCV-related cirrhosis in this study following reanalysis of the STOP-HCV53 data. Also, in a small study Dong et al 54 showed that carriage of the common allele T in rs10069690 is associated with an increased risk of developing HCC on a background of chronic viral hepatitis (OR = 2.78, 95% CI 1.62 to 4.78, p=0.00014). Thus, the association between rs2242652 and HCC may extend beyond its relationship with ArC. Further work is warranted to assess if a similar association applies to patients with NAFLD. A previous study showed that rare loss of function germline mutations in TERT are enriched in patients with NAFLD-HCC relative to controls—however, the specific relevance of the rs2242652 locus in this patient group is unknown.55
TERT rs2242652 has also been implicated in the susceptibility for developing other cancers but the direction of association seems to vary between cancer types (online supplemental table 14). In this study, rs2242652:A was significantly associated with reduced risks for developing bladder cancer and prostate cancer in the UKB and FinnGen cohorts. Kote-Jarai et al 56 found that carriage of TERT rs2242652:A: was associated with a lower risk for developing prostate cancer and with increased TERT expression which has been reported to improves survival, in prostate cancer. Further large studies involving diverse populations are clearly needed.
This study has a number of strengths including: (1) use of a two stage GWAS approach; (2) large, carefully selected case and control samples focusing on HCC in patients with established ArC; (3) careful exclusion of confounding comorbidities; (4) uniform inclusion of Caucasians participants of European ancestry; (5) the protective effect of rs2242652:A on HCC has been confirmed in the Japanese and Chinese population, suggesting that it may be applicable to East Asian population too, and (6) although the study was confined, by design, to patients with HCC on a background of ArC a cohort of patients with HCV-related HCC was also included to assess the generalisability of our findings to other aetiologies. The study also has a number of limitations: (1) it was performed retrospectively and hence potentially important information such as the lifetime alcohol history, information on diabetes and obesity were not generally available; (2) it had comparatively low power to detect true disease associations with smaller effect sizes (OR <1.4), at the levels of significance needed for GWAS analysis, and (3) only a minority of the HCC cases had histological confirmation of the diagnosis so tissue specimen for molecular analyses were not available.
In conclusion, this study identifies TERT rs2242652:A as a novel genetic factor for HCC development in ArC and confirmed the importance of the PNPLA3 and TM6SF2 as risk factors for HCC in this population. While the association between HCC and rs2242652:A in TERT is robust, the functional implications of carriage of this protective allele remains unclear. Carriage of rs2242652:A was significantly associated with an increase in leucocyte telomere lengths, but data on its effect on TERT transcription in liver tissue were not available. Thus, the functional implications of this association require further study in this specific context since the impact of TERT transcription, telomere length and the risk of malignancy remains controversial.57
Acknowledgments
The authors would like to thank all study participants, researchers, clinicians and administrative staff who contributed to this study.
Footnotes
Twitter: @StephanB76, @lucavalenti75
SB and HI contributed equally.
PD, SM, JH and FS contributed equally.
Correction notice: This article has been corrected since it published Online First. The author's name, Sascha A Müller, has been corrected.
Contributors: SB, HI, performed bioinformatic analyses, analysed the data, interpreted results and wrote the manuscript; HI gave conceptual advice; PLL, HDN, JUM, JF, KHW, JR, AM, MK, MC, FL, FE, AV, SM, JvF, RS, SRA, AM, JN managed collection of samples, performed phenotyping and curation of data, participated in the discussions, interpretation of the results; CS coordinated and supervised collection of samples, performed phenotyping, served as scientific advisor; AF gave conceptual advice, critically reviewed the manuscript; CS, MR, HA, SS coordinated, managed collection of samples, performed phenotyping; VRT, MB, CL, RES, AC, AL, TR, MM, GS, BS, CD, SR, SGC, WLI, JRM, ING, EB, MAA, JQ, collection of samples, performed phenotyping and curation of data, participated in the discussions, interpretation of the results; LV, SAM, JF-D, JT, TB, MYM obtained samples, gave conceptual advice, participated in the discussions, interpretation of the results, editing of the manuscript; PD, SM, JH, FS conceived the experimental and analytical design, supervision, interpretation of results, wrote and reviewed the manuscript. FS is the guarantor of this study.
Funding: This work was supported by grants from LiSyMKrebs (DEEP-HCC) network funded by the German Federal Ministry for Education and Research (BMBF) to JH (BMBF grant number 031L0258A). JT was supported by the German Research Foundation (DFG) project ID 403224013- SFB 1382 (A09), by the German Federal Ministry of Education and Research (BMBF) for the DEEP-HCC project and by the Hessian Ministry of Higher Education, Research and the Arts (HMWK) for the ENABLE and ACLF-I cluster projects. This work was further supported by grants from the Swiss National Funds (SNF no. 310030_169196) and the Swiss Foundation for Alcohol Research (SSA) to FS. HDN and US were supported by a grant from the Deutsche Krebshilfe (70112169). HI is supported by a viral hepatitis fellowship from the Medical Research Foundation (C0825). JM is funded by the UK Medical Research Council (MC_UU_12014/1) and the Medical Research Foundation (C0365). EB is funded by the Medical Research Council UK, the Oxford NIHR Biomedical Research Centre and is an NIHR Senior Investigator. The STOP-HCV study was funded by a grant from the Medical Research Council, UK (grant MR/K01532X/1). This work was also supported by the Deliver study funded by Cancer Research UK (C30358/A29725). See: https://www.oxcode.ox.ac.uk/research-showcase/liver-cancer. AL is supported by the funds of European Commission through the 'European funds for regional development' (EFRE) as well as by the regional Ministry of Economy, Science and Digitalization of Saxony-Anhalt as part of the 'Autonomy in old Age' (AiA) research group for “LiLife” Project (Project ID: ZS/2018/11/95324). The MICROB-PREDICT (project ID 825694), DECISION (project ID 847949), GALAXY (project ID 668031), LIVERHOPE (project ID 731875), and IHMCSA (project ID 964590) projects have received funding from the European Union’s Horizon 2020 research and innovation programme. This research has been conducted using the UK Biobank resource: application number: 8764. This research was further supported by the BioBank Dresden resource (https://www.nct-dresden.de/forschung/core-units/biobank-dresden.html).
Disclaimer: The manuscript reflects only the authors’ views, and the European Commission is not responsible for any use that may be made of the information it contains. The funders had no influence on study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests: JT has received speaking and/or consulting fees from Versantis, Gore, Bayer, Alexion, Norgine, Grifols and CSL Behring.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request.
Ethics statements
Patient consent for publication
Consent obtained directly from patient(s).
Ethics approval
This study involves human participants and was approved by name of the ethics Committee: Ethikkommission Technische Universität Dresden (TU Dresden) Ethics name ID: EK 594122019name of the ethics Committee: Kantonale Ethikkommision Bern, Switzerland (KEK)Ethics name ID:062 /11. Participants gave informed consent to participate in the study before taking part.
References
- 1., Akinyemiju T, Abera S, et al. , Global Burden of Disease Liver Cancer Collaboration . The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol 2017;3:1683–91. 10.1001/jamaoncol.2017.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2021;7. 10.1038/s41572-020-00240-3 [DOI] [PubMed] [Google Scholar]
- 3. Mancebo A, González-Diéguez ML, Cadahía V, et al. Annual incidence of hepatocellular carcinoma among patients with alcoholic cirrhosis and identification of risk groups. Clin Gastroenterol Hepatol 2013;11:95–101. 10.1016/j.cgh.2012.09.007 [DOI] [PubMed] [Google Scholar]
- 4. Ganne-Carrié N, Nahon P. Hepatocellular carcinoma in the setting of alcohol-related liver disease. J Hepatol 2019;70:284–93. 10.1016/j.jhep.2018.10.008 [DOI] [PubMed] [Google Scholar]
- 5. Mallet V, Parlati L, Martinino A, et al. Burden of liver disease progression in hospitalized patients with type 2 diabetes mellitus. J Hepatol 2022;76:265–74. 10.1016/j.jhep.2021.09.030 [DOI] [PubMed] [Google Scholar]
- 6. El-Serag HB, Mason AC. Risk factors for the rising rates of primary liver cancer in the United States. Arch Intern Med 2000;160:3227–30. 10.1001/archinte.160.21.3227 [DOI] [PubMed] [Google Scholar]
- 7. Dragani TA. Risk of HCC: genetic heterogeneity and complex genetics. J Hepatol 2010;52:252–7. 10.1016/j.jhep.2009.11.015 [DOI] [PubMed] [Google Scholar]
- 8. Stickel F, Buch S, Nischalke HD, et al. Genetic variants in PNPLA3 and TM6SF2 predispose to the development of hepatocellular carcinoma in individuals with alcohol-related cirrhosis. Am J Gastroenterol 2018;113:1475–83. 10.1038/s41395-018-0041-8 [DOI] [PubMed] [Google Scholar]
- 9. Yang J, Trépo E, Nahon P, et al. A 17-Beta-Hydroxysteroid dehydrogenase 13 variant protects from hepatocellular carcinoma development in alcoholic liver disease. Hepatology 2019;70:231-240–40. 10.1002/hep.30623 [DOI] [PubMed] [Google Scholar]
- 10. Stickel F, Lutz P, Buch S, et al. Genetic variation in HSD17B13 reduces the risk of developing cirrhosis and hepatocellular carcinoma in alcohol Misusers. Hepatology 2020;72:88–102. 10.1002/hep.30996 [DOI] [PubMed] [Google Scholar]
- 11. Innes H, Nischalke HD, Guha IN, et al. The rs429358 locus in apolipoprotein E is associated with hepatocellular carcinoma in patients with cirrhosis. Hepatol Commun 2022;6:1213-1226. 10.1002/hep4.1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abul-Husn NS, Cheng X, Li AH, et al. A Protein-Truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med 2018;378:1096–106. 10.1056/NEJMoa1712191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nahon P, Nault J-C. Constitutional and functional genetics of human alcohol-related hepatocellular carcinoma. Liver Int 2017;37:1591–601. 10.1111/liv.13419 [DOI] [PubMed] [Google Scholar]
- 14. Trépo E, Caruso S, Yang J, et al. Common genetic variation in alcohol-related hepatocellular carcinoma: a case-control genome-wide association study. Lancet Oncol 2022;23:161–71. 10.1016/S1470-2045(21)00603-3 [DOI] [PubMed] [Google Scholar]
- 15. Galle PR, Forner A, Llovet JM, et al. EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2018;69:182–236. 10.1016/j.jhep.2018.03.019 [DOI] [PubMed] [Google Scholar]
- 16. Lee Y-T, Wang JJ, Zhu Y, et al. Diagnostic criteria and LI-RADS for hepatocellular carcinoma. Clin Liver Dis 2021;17:409–13. 10.1002/cld.1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sudlow C, Gallacher J, Allen N, et al. Uk Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buch S, Stickel F, Trépo E, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet 2015;47:1443–8. 10.1038/ng.3417 [DOI] [PubMed] [Google Scholar]
- 19. Treutlein J, Cichon S, Ridinger M, et al. Genome-Wide association study of alcohol dependence. Arch Gen Psychiatry 2009;66:773–84. 10.1001/archgenpsychiatry.2009.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frank J, Cichon S, Treutlein J, et al. Genome-Wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addict Biol 2012;17:171–80. 10.1111/j.1369-1600.2011.00395.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McCarthy S, Das S, Kretzschmar W, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016;48:1279-83–83. 10.1038/ng.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Das S, Forer L, Schönherr S, et al. Next-Generation genotype imputation service and methods. Nat Genet 2016;48:1284–7. 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–9. 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chang CC, Chow CC, Tellier LCAM, et al. Second-Generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4. 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Price AL, Zaitlen NA, Reich D, et al. New approaches to population stratification in genome-wide association studies. Nat Rev Genet 2010;11:459–63. 10.1038/nrg2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Watanabe K, Taskesen E, van Bochoven A, et al. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017;8:1826–11. 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schafmayer C, Harrison JW, Buch S, et al. Genome-Wide association analysis of diverticular disease points towards neuromuscular, connective tissue and epithelial pathomechanisms. Gut 2019;68:854–65. 10.1136/gutjnl-2018-317619 [DOI] [PubMed] [Google Scholar]
- 28. Johnson JL, Abecasis GR. GAS power calculator: web-based power calculator for genetic association studies. bioRxiv 2017. [Google Scholar]
- 29. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–1. 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aguet F, Barbeira AN, Bonazzola R, et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020;369:1318–30. 10.1126/science.aaz1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Võsa U, Claringbould A, Westra HJ. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv 2018. https://europepmc.org/article/PPR/PPR59262 [Google Scholar]
- 32. Yang J, Lee SH, Goddard ME, et al. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011;88:76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ningarhari M, Caruso S, Hirsch TZ, et al. Telomere length is key to hepatocellular carcinoma diversity and telomerase addiction is an actionable therapeutic target. J Hepatol 2021;74:1155–66. 10.1016/j.jhep.2020.11.052 [DOI] [PubMed] [Google Scholar]
- 34. Dongiovanni P, Stender S, Pietrelli A, et al. Causal relationship of hepatic fat with liver damage and insulin resistance in nonalcoholic fatty liver. J Intern Med 2018;283:356–70. 10.1111/joim.12719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miki D, Ochi H, Hayes CN, et al. Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers. Nat Genet 2011;43:797–800. 10.1038/ng.876 [DOI] [PubMed] [Google Scholar]
- 36. Jiang D-K, Sun J, Cao G, et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet 2013;45:72–5. 10.1038/ng.2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nischalke HD, Lutz P, Bartok E, et al. The PNPLA3 I148M variant promotes lipid-induced hepatocyte secretion of CXC chemokines establishing a tumorigenic milieu. J Mol Med 2019;97:1589–600. 10.1007/s00109-019-01836-3 [DOI] [PubMed] [Google Scholar]
- 38. Newberry EP, Hall Z, Xie Y, et al. Liver-Specific deletion of mouse TM6SF2 promotes steatosis, fibrosis, and hepatocellular cancer. Hepatology 2021;74:1203–19. 10.1002/hep.31771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Calderaro J, Ziol M, Paradis V, et al. Molecular and histological correlations in liver cancer. J Hepatol 2019;71:616–30. 10.1016/j.jhep.2019.06.001 [DOI] [PubMed] [Google Scholar]
- 40. Stickel F, Moreno C, Hampe J, et al. The genetics of alcohol dependence and alcohol-related liver disease. J Hepatol 2017;66:195–211. 10.1016/j.jhep.2016.08.011 [DOI] [PubMed] [Google Scholar]
- 41. Caruso S, O'Brien DR, Cleary SP, et al. Genetics of hepatocellular carcinoma: approaches to explore molecular diversity. Hepatology 2021;73 Suppl 1:14-26–26. 10.1002/hep.31394 [DOI] [PubMed] [Google Scholar]
- 42. Müller M, Bird TG, Nault J-C. The landscape of gene mutations in cirrhosis and hepatocellular carcinoma. J Hepatol 2020;72:990–1002. 10.1016/j.jhep.2020.01.019 [DOI] [PubMed] [Google Scholar]
- 43. Zhang P, Kitchen-Smith I, Xiong L, et al. Germline and somatic genetic variants in the p53 pathway interact to affect cancer risk, progression, and drug response. Cancer Res 2021;81:1667–80. 10.1158/0008-5472.CAN-20-0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 2010;11:171-81–81. 10.1038/nrm2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang Q, Kim N-K, Feigon J. Architecture of human telomerase RNA. Proc Natl Acad Sci U S A 2011;108:20325–32. 10.1073/pnas.1100279108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang F, Cheng D, Wang S, et al. Human specific regulation of the telomerase reverse transcriptase gene. Genes 2016;7:30. 10.3390/genes7070030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nault J-C, Ningarhari M, Rebouissou S, et al. The role of telomeres and telomerase in cirrhosis and liver cancer. Nat Rev Gastroenterol Hepatol 2019;16:544–58. 10.1038/s41575-019-0165-3 [DOI] [PubMed] [Google Scholar]
- 48. Park J-I, Venteicher AS, Hong JY, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 2009;460:66-72–72. 10.1038/nature08137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hoffmeyer K, Raggioli A, Rudloff S, et al. Wnt/β-catenin signaling regulates telomerase in stem cells and cancer cells. Science 2012;336:1549–54. 10.1126/science.1218370 [DOI] [PubMed] [Google Scholar]
- 50. Behari J, Sylvester KG. Role of the Wnt/β-catenin pathway in the pathogenesis of alcoholic liver disease. Curr Mol Pharmacol 2017;10:186–94. 10.2174/1874467208666150817111256 [DOI] [PubMed] [Google Scholar]
- 51. Huang P, Li R, Shen L, et al. Single nucleotide polymorphisms in telomere length-related genes are associated with hepatocellular carcinoma risk in the Chinese Han population. Ther Adv Med Oncol 2020;12:175883592093302. 10.1177/1758835920933029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y, Wang S, Shi Y, et al. Associations of TERT polymorphisms with hepatocellular carcinoma risk in a Han Chinese population. Int J Clin Exp Pathol 2017;10:7776. [PMC free article] [PubMed] [Google Scholar]
- 53. McLauchlan J, Innes H, Dillon JF, et al. Cohort profile: the hepatitis C virus (HCV) research UK clinical database and Biobank. Int J Epidemiol 2017;46:1391–1391h. 10.1093/ije/dyw362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dong J, Wang L, Tian Y-ping, et al. [hTERT single nucleotide polymorphism is associated with increased risks of hepatocellular carcinoma and tumor metastasis]. Nan Fang Yi Ke Da Xue Xue Bao 2011;31:49–52. [PubMed] [Google Scholar]
- 55. Donati B, Pietrelli A, Pingitore P, et al. Telomerase reverse transcriptase germline mutations and hepatocellular carcinoma in patients with nonalcoholic fatty liver disease. Cancer Med 2017;6:1930–40. 10.1002/cam4.1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kote-Jarai Z, Saunders EJ, Leongamornlert DA, et al. Fine-Mapping identifies multiple prostate cancer risk loci at 5p15, one of which associates with TERT expression. Hum Mol Genet 2013;22:2520–8. 10.1093/hmg/ddt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bojesen SE, Pooley KA, Johnatty SE, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet 2013;45:371–84. 10.1038/ng.2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2008;40:1461–5. 10.1038/ng.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu Y-L, Patman GL, Leathart JBS, et al. Carriage of the PNPLA3 rs738409 C >G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J Hepatol 2014;61:75–81. 10.1016/j.jhep.2014.02.030 [DOI] [PubMed] [Google Scholar]
- 60. Kozlitina J, Smagris E, Stender S, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2014;46:352–6. 10.1038/ng.2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. hTERT single nucleotide polymorphism is associated with increased risks of hepatocellular carcinoma and tumor metastasis. [PubMed]
- 62. Dong J, Wang L, Tian Y-ping, et al. [hTERT single nucleotide polymorphism is associated with increased risks of hepatocellular carcinoma and tumor metastasis]. Nan Fang Yi Ke Da Xue Xue Bao 2011;31:49–52 https://pubmed.ncbi.nlm.nih.gov/21269955/ [PubMed] [Google Scholar]
- 63. Ishigaki K, Akiyama M, Kanai M, et al. Large-Scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat Genet 2020;52:669–79. 10.1038/s41588-020-0640-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2022-327196supp001.pdf (3.4MB, pdf)
Data Availability Statement
Data are available on reasonable request.