ABSTRACT
Despite the deployment of combination tuberculosis (TB) chemotherapy, efforts to identify shorter, nonrelapsing treatments have resulted in limited success. Recent evidence indicates that GSK2556286 (GSK286), which acts via Rv1625c, a membrane-bound adenylyl cyclase in Mycobacterium tuberculosis, shortens treatment in rodents relative to standard of care drugs. Moreover, GSK286 can replace linezolid in the three-drug, Nix-TB regimen. Given its therapeutic potential, we sought to better understand the mechanism of action of GSK286. The compound blocked growth of M. tuberculosis in cholesterol media and increased intracellular cAMP levels ~50-fold. GSK286 did not inhibit growth of an rv1625c transposon mutant in cholesterol media and did not induce cyclic AMP (cAMP) production in this mutant, suggesting that the compound acts on this adenylyl cyclase. GSK286 also induced cAMP production in Rhodococcus jostii RHA1, a cholesterol-catabolizing actinobacterium, when Rv1625c was heterologously expressed. However, these elevated levels of cAMP did not inhibit growth of R. jostii RHA1 in cholesterol medium. Mutations in rv1625c conferred cross-resistance to GSK286 and the known Rv1625c agonist, mCLB073. Metabolic profiling of M. tuberculosis cells revealed that elevated cAMP levels, induced using either an agonist or a genetic tool, did not significantly affect pools of steroid metabolites in cholesterol-incubated cells. Finally, the inhibitory effect of agonists was not dependent on the N-acetyltransferase MtPat. Together, these data establish that GSK286 is an Rv1625c agonist and sheds light on how cAMP signaling can be manipulated as a novel antibiotic strategy to shorten TB treatments. Nevertheless, the detailed mechanism of action of these compounds remains to be elucidated.
KEYWORDS: TB, adenylyl cyclase, antibiotic, antitubercular
INTRODUCTION
Tuberculosis (TB), caused by Mycobacterium tuberculosis, remains a prevalent infectious disease claiming ~1.6 million lives and causing new disease in ~10.6 million individuals annually (1). A major factor that contributes to our failure to control the disease is the inefficiency and long duration of current multidrug treatment regimens. The lengthy treatment duration and severe side effects results in treatment non-compliance, leading to the development of drug resistant strains. The emergence of multidrug resistance and extensive drug resistance in M. tuberculosis is especially threatening to disease control with ~450,000 new cases of drug resistance in over 100 countries in 2021 alone (1). These discouraging statistics exemplify the urgent need for novel antibacterial agents for the treatment of TB. Shorter treatment regimens with more efficacious antibiotics will likely be essential to limit the spread of drug-resistant strains and lower the global burden of TB.
Nix-TB, the recently approved three-drug regimen that consists of bedaquiline (B), pretomanid (Pa), and linezolid (L), produces durable cures against drug-resistant TB within 6 months of treatment (2). While this treatment is clinically effective, the prolonged exposure to L is associated with adverse side effects such as myelosuppression, anemia, peripheral neuropathy, and impaired vision (2). Importantly, recent studies in a BALB/c relapsing mouse model for TB treatment indicated that administering GSK286 with BPa achieved a similar proportion of nonrelapsing mice as the BPaL regimen when administered across the same duration (3). This suggests that GSK286 could potentially replace linezolid in the Nix-TB regimen without loss of treatment efficacy. Thus, understanding the mechanism of action for GSK286 could inform the development of the next generation of TB combination treatments.
GSK286 was identified from a screen for compounds that inhibit intracellular growth of M. tuberculosis in THP-1, human macrophage-like cells (3). Importantly, the standard growth conditions routinely used for the cultivation of M. tuberculosis in the laboratory do not recapitulate the growth substrates of the bacterium during infection and, when used for screening compound libraries, can identify compounds that lack efficacy in animal models (4). For example, cholesterol is a substrate for M. tuberculosis during intracellular growth (5), cholesterol catabolism contributes to the virulence of M. tuberculosis in various infection models, and the catabolic pathway has emerged as a potential drug target (6–10). Catabolism of cholesterol in M. tuberculosis generates acetyl coenzyme A (acetyl-CoA), propionyl-CoA, succinyl-CoA, and pyruvate, all of which feed into central metabolism (11). Previous studies characterizing macrophage-active compounds found that a subset of these compounds retains activity against extracellular M. tuberculosis in the presence of cholesterol and that the activity of these compounds requires Rv1625c (5, 12, 13). Two of these compounds, V-59 and mCLB073, are chemically related to each other, but GSK286 has a distinct chemical scaffold (see Fig. S1 in the supplemental material). Interestingly, GSK286 also inhibits the growth of M. tuberculosis on cholesterol, and mutations in Rv1625c confer resistance to GSK286 (3). It is therefore possible that, despite the structural difference, GSK286 may have a mechanism similar to that of these Rv1625c chemical agonists.
Here, we characterize M. tuberculosis’s response to GSK286, specifically investigating the compound’s effect on cyclic AMP (cAMP) levels, its requirement for Rv1625c in its mechanism of action, and its effect on cholesterol catabolism in M. tuberculosis. We investigated the effect of overexpression of Rv1625c on M. tuberculosis’s susceptibility to GSK286, and the effect of GSK286 treatment on cholesterol-derived propionyl-CoA pools in M. tuberculosis during infection in macrophages. We used metabolite profiling to evaluate the pools of cholesterol metabolites during growth on cholesterol and cross-resistance of spontaneous resistance mutants to compare the mechanism of action of GSK286 with that of the known Rv1625c agonist, mCLB073. Finally, given the potentially easy path of drug resistance to GSK286 through Rv1625c mutation, we investigated whether M. tuberculosis mutants lacking Rv1625c have a fitness defect in the lungs of mice. We discuss the findings with respect to GSK286’s mechanism of action, as well as the linezolid-replacing potential of Rv1625c agonists in the Nix-TB regimen.
RESULTS
GSK286 acts on Rv1625c to stimulate cAMP production.
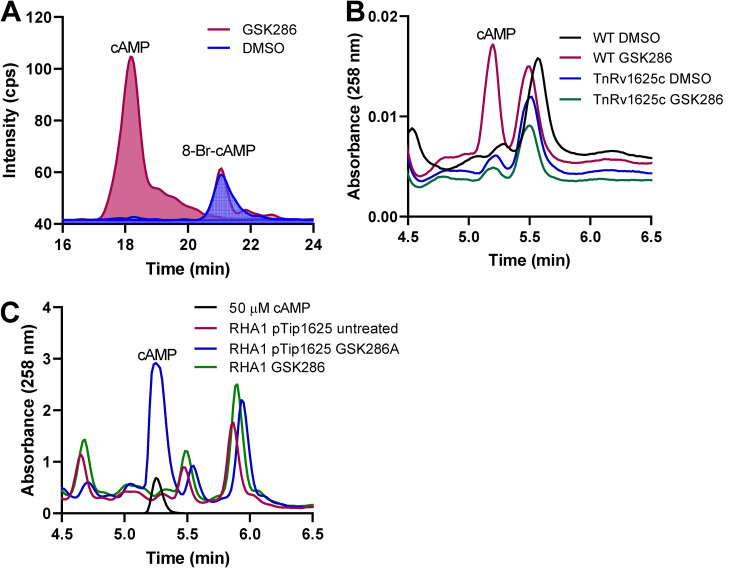
To investigate the necessity of Rv1625c in the mode of action of GSK286, M. tuberculosis Erdman was grown on cholesterol or glycerol to an optical density at 600 nm (OD600) of ~0.6 and then treated with the GSK286 or dimethyl sulfoxide (DMSO) for 24 h. The intracellular cAMP levels, analyzed by LC-MS/MS, were ~50-fold higher in treated versus untreated cells (Fig. 1A). cAMP production was not induced in a M. tuberculosis CDC1551 rv1625c transposon mutant, which suggests that the compound-induced cAMP is produced solely by Rv1625c and not one of the other 10 biochemically confirmed adenylate cyclase proteins encoded by the M. tuberculosis genome (Fig. 1B) (14, 15). Furthermore, when a plasmid carrying rv1625c or an empty plasmid control was transformed into Rhodococcus jostii RHA1, an actinobacterium with a homologous cholesterol catabolic pathway but lacking a homolog of Rv1625c (Fig. 1C), cAMP production was induced by GSK286 treatment only when Rv1625c was expressed (Fig. 1C).
FIG 1.
cAMP production by M. tuberculosis and Rhodococcus jostii RHA1 expressing Rv1625c treated with GSK286 (A) LC-MS total ion chromatogram from M. tuberculosis lysates following GSK286 treatment. 8-Br-cAMP (8-bromoadenosine 3′,5′-cyclic monophosphate) was supplied to M. tuberculosis as a control and run as a standard for LC-MS/MS analysis. (B) WT and an rv1625c transposon mutant of M. tuberculosis CDC1551 were grown in 7H9 medium supplemented with 0.2% glycerol, concentrated, and then incubated in 7H9 medium supplemented with 0.5 mM cholesterol with or without 5 μM GSK286. Cells were harvested after 24 h, and cell lysates were analyzed by HPLC. (C) R. jostii RHA1 carrying pTip1625 or empty pTip plasmid was grown in M9 medium supplemented with 0.5 mM cholesterol with or without 5 μM GSK286. Cell lysates were analyzed by HPLC. Data represent the means of three biological replicates.
GSK286 blocks cholesterol metabolism in M. tuberculosis in liquid culture and macrophages.
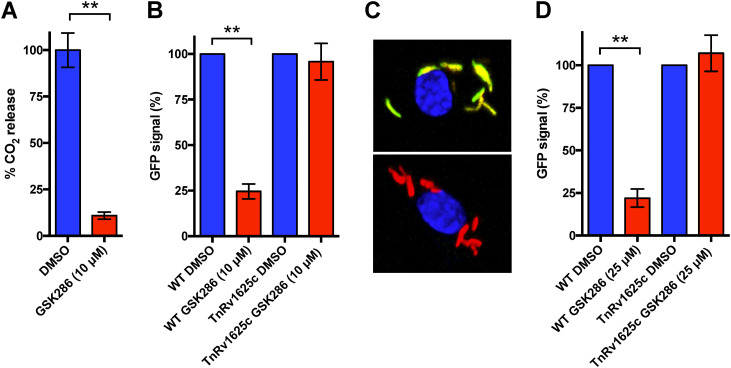
Chemically activating Rv1625c blocks cholesterol metabolism (5, 13). Therefore, we quantified the effect of GSK286 on cholesterol catabolism by monitoring the degradative release of 14CO2 from 14C-cholesterol in wild-type (WT) M. tuberculosis Erdman. Treatment with GSK286 reduced the amount of 14CO2 released from 14C-cholesterol by 90% (Fig. 2A), consistent with the inhibition of cholesterol catabolism. Catabolic degradation of cholesterol by M. tuberculosis releases significant amounts of propionyl-CoA (16). Thus, we used a GFP reporter (prpD′::GFP) to quantify propionyl-CoA levels in M. tuberculosis following treatment with GSK286 in liquid culture and during infection in macrophages. The prpD′::GFP reporter expresses GFP when propionyl-CoA pools increase following cholesterol degradation in the bacterium. We found that GSK286 treatment decreased the green fluorescent protein (GFP) signal in M. tuberculosis when the bacteria were cultured in cholesterol media supplemented with acetate (Fig. 2B). In these experiments, cholesterol medium was supplemented with short-chain fatty acid to facilitate GFP measurements by rescuing the growth inhibition from Rv1625c agonists (13). Importantly, the transposon mutant (TnRv1625c) containing a nonfunctional copy of rv1625c did not respond to the compound treatment in cholesterol-acetate media (Fig. 2B). The compound also reduced the GFP signal in WT M. tuberculosis during infection in macrophages, and this reduction in signal is dependent on Rv1625c (Fig. 2C and D).
FIG 2.
GSK286 treatment reduces cholesterol-derived propionyl-CoA pools in M. tuberculosis. (A) GSK286 blocks the degradative release of 14CO2 from 14C-cholesterol in wild-type M. tuberculosis during growth in cholesterol-acetate media. (B) The GFP signal from the prpD′::GFP reporter decreases in wild-type M. tuberculosis in an Rv1625c-dependent manner following treatment with GSK286 during growth in cholesterol-acetate media. (C) Representative images of macrophages infected with wild-type M. tuberculosis carrying the prpD′::GFP reporter (green), the constitutive M. tuberculosis mCherry signal (red), and DAPI stain (blue). DMSO-treated (top panel) and GSK286-treated (bottom panel) samples are shown. (D) Flow cytometric quantification of the prpD′::GFP reporter signal M. tuberculosis isolated from infected macrophages. The median GFP signal was quantified from 10,000 bacteria and calculated relative to the DMSO control for each strain of bacteria. Bars represent means ± the standard deviations (SD; n = 4) from two independent replicates with two technical replicates each. (A) **, P < 0.05 (Student t test); (B and D) **, P < 0.05 (ordinary one-way ANOVA with Dunnett’s multiple-comparison test).
GSK286 lowers levels of cholesterol catabolic gene transcripts.
In light of the effect of GSK286 on the rate of cholesterol catabolism by M. tuberculosis, we next determined the effect of the compound on the expression of cholesterol catabolic genes using the reverse transcription-quantitative PCR (RT-qPCR). In this experiment, we evaluated the transcript levels of kshA and echA20 since they are part of the KstR and KstR2 regulons, respectively. In cells growing on cholesterol, treatment with GSK286A or V-59, a previously described Rv1625c agonist (5), reduced the levels of kshA transcripts to 23 and 20% and echA20 transcripts to 28 and 30% of the levels in untreated cells for GSK286 and V-59 treatment, respectively (see Fig. S2). This is consistent with the effect of these compounds on the rate of cholesterol catabolism, as well as the effect of V-59 on the transcriptome of M. tuberculosis growing on cholesterol (5).
Overexpression of rv1625c increases the potency of GSK286 in M. tuberculosis.
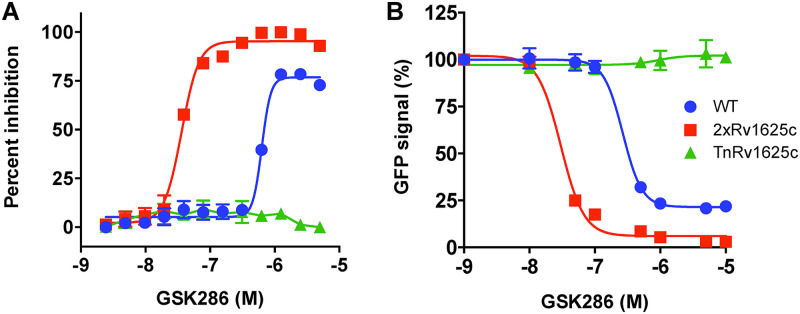
Given that the M. tuberculosis genome encodes 16 distinct adenylyl cyclases, we sought to further confirm that GSK286 specifically activates Rv1625c. Accordingly, we overexpressed rv1625c in M. tuberculosis, hypothesizing that increasing the copy number of the protein would increase the susceptibility of M. tuberculosis to GSK286 and thereby decrease the MIC of GSK286. Overexpressing rv1625c in wild-type M. tuberculosis decreased the MIC of GSK286 in cholesterol media ~10-fold (Fig. 3A). Importantly, a M. tuberculosis mutant with a nonfunctional Rv1625c (TnRv1625c) was resistant to the compound across all concentrations tested. Furthermore, we leveraged the prpD′::GFP reporter to test the hypothesis that the concentration of GSK286 required to inhibit the GFP signal in cholesterol/acetate media would also decrease when rv1625c is overexpressed in wild-type M. tuberculosis. Indeed, overexpressing rv162c5 decreased the concentration of GSK286 required to inhibit the GFP signal ~10-fold in this assay (Fig. 3B). Similar to our previous results (Fig. 2B), the GFP signal was unchanged in the TnRv1625c mutant treated with GSK286 across all concentrations, confirming that Rv1625c is required for the compound-dependent inhibition of cholesterol catabolism in the bacterium. Together, these data indicate that GSK286 specifically stimulates Rv1625c to inhibit cholesterol catabolism in M. tuberculosis and the bacterial growth of this lipid.
FIG 3.
Rv1625c is required for GSK286-dependent growth inhibition and block of cholesterol metabolism in M. tuberculosis. (A) MIC curves for GSK286 in M. tuberculosis grown in cholesterol media. Inhibition was calculated using alamarBlue fluorescence with 100% inhibition established using rifampicin treatment (10 μM). All of the inhibition values are normalized to this value. The x-axis values are logarithms of concentrations expressed as a molar unit. (B) GFP fluorescence inhibition curves for GSK286 using the prpD′::GFP reporter in cholesterol-acetate media. The strains included WT (wild-type M. tuberculosis), 2×Rv1625c (WT strain carrying a plasmid expressing rv1625c from an hsp60 promoter), and TnRv1625c (a transposon-disrupted mutant with a nonfunctional Rv1625c). Each point in the fluorescence inhibition curve was obtained by quantifying the GFP signal from 10,000 bacteria, and error bars indicate means ± the SD (n = 4) from two independent replicates with two technical replicates for each.
Mutations in M. tuberculosis confer cross-resistance to Rv1625c agonists.
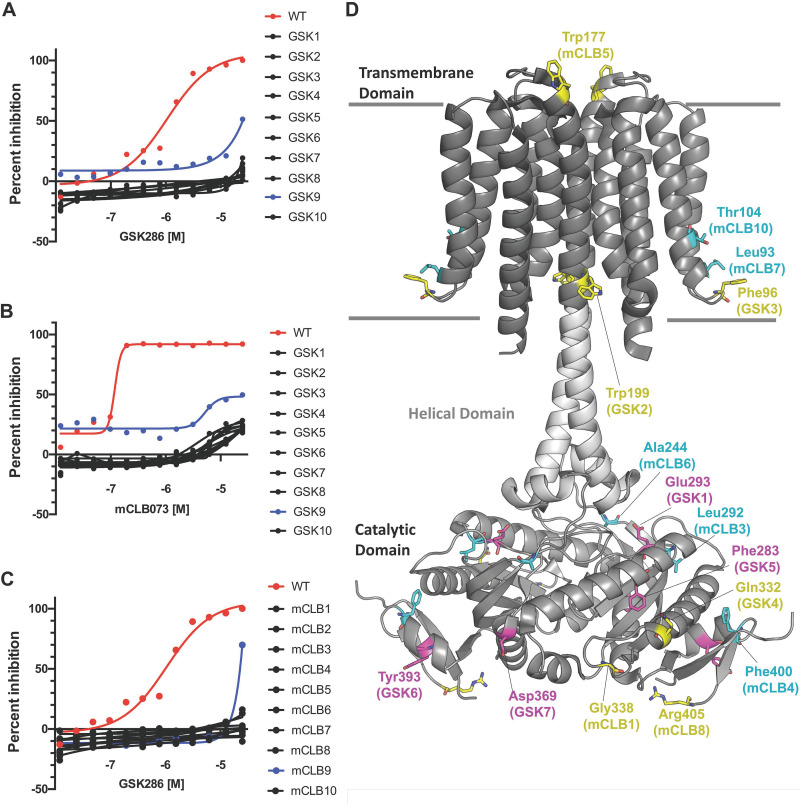
We next used cross-resistance to investigate the similarity in the modes of action of GSK286 and another Rv1625c agonist, mCLB073. We found that all spontaneous resistant mutants raised against GSK286 (this study) and mCLB073 (previous work) conferred resistance to both compounds. Briefly, we plated 7 × 105 bacteria onto cholesterol-based agar plates containing GSK286 or mCLB073 (13) at concentrations ranging from 25 to 100 μM. Under these conditions, resistant mutants occurred with a frequency of ~2.2 × 10−3 regardless of the concentration of the compound. We isolated 10 spontaneous resistant mutants with GSK286 (Table 1), these mutations conferred resistance to GSK286 in cholesterol media with MIC90’s to exceeding 50 μM for each strain with a resazurin-based MIC assay (Fig. 4A). For this assay, we set 100% inhibition using the resazurin fluorescence values from bacteria treated with 10 μM rifampicin and express all inhibition values as a percentage of this value. Moreover, each of these mutants were also resistant to mCLB073 in cholesterol media (Fig. 4B) with similar MIC shifts. Finally, each of the 10 spontaneous resistant mutants we had previously isolated against mCLB073 (13) were cross resistant to GSK286 in cholesterol media (Fig. 4C).
TABLE 1.
Spontaneous mutations that confer resistance to GSK286
| Mutant | Location, gene | Mutation/result |
|---|---|---|
| GSK1 | Glu293 (GAG), rv1625c | Gly (GGG) |
| GSK2 | Trp199 (TGG), rv1625c | STOP (TGA) |
| GSK3 | After Phe96, rv1625c | Extra G (frameshift to stop at 204) |
| GSK4 | 14-bp insert Glu332, rv1625c | Frameshift to stop at 338 |
| GSK5 | Phe283 (TTC), rv1625c | Ser (TCC) |
| GSK6 | Tyr393 (TAC), rv1625c | Asp (GAC) |
| GSK7 | Asp369 (GAC), rv1625c | Tyr (TAC) |
| GSK8 | Unknown | Unknown |
| GSK9 | Unknown | Unknown |
| GSK10 | Unknown | Unknown |
FIG 4.
Spontaneous resistance mutations confer cross-resistance to the Rv1625c agonists. (A) GSK286 MIC assay in cholesterol media with resistant mutants raised against GSK286 on cholesterol. (B) mCLB073 MIC assay in cholesterol media with resistant mutants raised against GSK286 on cholesterol. (C) GSK286 MIC assay in cholesterol media with resistant mutants raised against mCLB073 on cholesterol. The data are from one experiment with two technical replicates (n = 2). Symbols indicate means, and curves display the nonlinear fit of dose response. Inhibition was calculated using alamarBlue fluorescence with 100% inhibition established using rifampicin treatment (10 μM); all inhibition values are normalized to this value. The x-axis values are logarithms of concentrations expressed as a molar unit. (D) Mutations mapped onto the structure of Rv1625c. The overall fold (PDB ID 7YZK) is shown as a cartoon representation with the transmembrane, helical, and catalytic domains shown as different shades of gray with the approximate boundaries of the membrane drawn as gray lines. The location of missense mutations arising from GSK286 treatment in this study (Table 1) are highlighted as magenta sticks. The locations of cross-resistant missense mutations arising from mCLB073 treatment are shown as cyan sticks. The location of all other mutations, including nonsense, insertion, and frameshift mutations conferring resistance to GSK286, are highlighted as yellow sticks.
Seven of the ten mutants raised on GSK286 carried mutations in rv1625c, and none of these mutations were previously described (3). Mapping the cross-resistant point mutations arising against GSK286 and mCLB073 to the Rv1625c cryo electron microscopy (cryo-EM) structure (PDB ID 7YZK) (17) reveals a remarkable distribution of amino acids spanning Thr104 (mutant mCLB10) in the transmembrane domain to Tyr393 (GSK6) in the catalytic domain of Rv1625c (Fig. 4D). Mutations that disrupt folding, dimerization or catalytic activity could give rise to resistance. However, the fact that all mutations displayed cross-resistance suggests the agonists may have a similar mechanism of action.
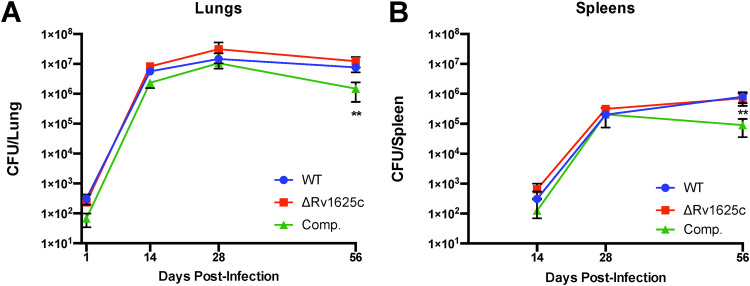
Rv1625c is dispensable in the TB mouse model of infection.
The major mechanism by which M. tuberculosis develops resistance to Rv1625c agonists is through mutations in the rv1625c gene. It was unknown whether M. tuberculosis lacking Rv1625c displays a fitness defect in vivo that could potentially reduce the likelihood of M. tuberculosis acquiring these resistance mutations during infection. To test this possibility, we infected BALB/c mice by the aerosol route with the WT, a Rv1625c mutant (ΔRv1625c), and a complemented mutant that overexpresses Rv1625c (Comp). The bacterial load was quantified from the lungs and spleens at 2, 4, and 8 weeks postinfection with five mice per treatment group at each time point. Loss of Rv1625c did not confer a fitness cost to M. tuberculosis during infection. The complemented strain exhibited a slight reduction in bacterial numbers in the lungs (~0.75-log10 reduction) and a decrease in bacterial burden in the spleens (~1-log10 reduction) of mice selectively during the chronic stage of infection (Fig. 5). The slight cost associated with overexpression of Rv1625c is similar to previously published infection assays (13).
FIG 5.
M. tuberculosis lacking Rv1625c has no fitness defect in a mouse model. (A and B) Bacterial burdens in the lungs (A) and spleens (B) of BALB/c mice infected with ~200 CFU of WT, ΔRv1625c, or complemented strain by the aerosol route. The data are from one experiment with five mice per group for each time point. The data are means ± the SD. **, P < 0.05 (ordinary one-way ANOVA with Dunnett’s multiple-comparison test).
Effect of agonists and elevated cAMP on pools of cholesterol-derived metabolites.
To determine which step(s) of the cholesterol catabolic pathway might be inhibited by Rv1625c agonists, extracellular and intracellular cholesterol-derived metabolites were analyzed by liquid chromatography-mass spectrometry (LC-MS). Briefly, cells were grown in 7H9 medium containing glycerol to generate biomass and were then concentrated and incubated in cholesterol-containing medium with or without V-59 for 48 h (13). Metabolites throughout the cholesterol catabolic pathway were subjected to high-resolution LC-QTOF (liquid chromatography-quadrupole time of flight mass spectrometry) analysis and validated using commercial standards or standards generated in vitro (11, 18). In addition, M. tuberculosis carrying a genetic construct that produces cAMP following the addition of anhydrotetracycline (19) was used to evaluate the impact of cAMP increases on cholesterol metabolism independent of the Rv1625c agonist (13). The metabolites readily detected in both treatments include side-chain degradation products, AB ring degradation intermediates, CD ring degradation intermediates, and cAMP, as well as the metabolic end products propionyl-, succinyl-, and acetyl-CoA (see Fig. S3). This broad coverage allows us to assess changes in the distribution of metabolite pools through the entire degradation pathway. Overall, there were small reductions in the abundance of cholesterol-specific metabolites from the treated and untreated cells, suggesting an early block in cholesterol catabolism in M. tuberculosis. Importantly, no specific metabolites accumulated in treated cells, suggesting the pathway is not inhibited at one particular step, either by treatment with the agonist or cAMP overproduction (see Fig. S3).
The GSK286 mechanism of action does not involve MtPat-mediated acetylation.
In M. tuberculosis, cAMP regulates numerous metabolic pathways via MtPat, an N-acetyltransferase. Briefly, the binding of cAMP to MtPat activates the latter to posttranslationally downregulate activity of key enzymes by acetylation (20–24). Therefore, we hypothesized that an increase in intracellular cAMP in M. tuberculosis upon treatment with GSK286 may lead to MtPat activation and deactivation of cholesterol catabolic enzymes by protein acetylation. To test this hypothesis, we deleted the genes encoding MtPat (rv0998) and the corresponding deacetylase (rv1151c), respectively, and tested the sensitivity of these strains to GSK286. The Δmtpat and the Δrv1151c strains are both sensitive to GSK286 (see Fig. S4). These findings suggest that the primary mechanism of action of GSK286 does not involve the MtPat-mediated acetylation of cholesterol catabolic enzymes.
DISCUSSION
This study establishes that GSK286 acts as an agonist of Rv1625c. Specifically, treatment of cells with GSK286 increased intracellular cAMP levels in an Rv1625c-dependent manner in M. tuberculosis and a related, nonpathogenic mycolic-acid-producing actinobacterium (Fig. 1). GSK286 also lowered the levels of transcripts of cholesterol catabolic genes under transcriptional control of the KstR and KstR2 regulators, respectively (see Fig. S2), and blocked the growth of M. tuberculosis on cholesterol in an Rv1625c-dependent manner (Fig. 3). Compounds with similar mode of action to GSK286 were identified by VanderVen et al. (5) and were further characterized by Wilburn et al. (13), demonstrating that Rv1625c agonists such as mCLB073 and its analog V-59, stimulate cAMP production and block cholesterol metabolism in M. tuberculosis. Importantly, spontaneous mutants that were resistant to GSK286 were also resistant to mCLB073 and vice versa (Fig. 4). Overall, the identification of several compounds targeting Rv1625c in independent macrophage-based screens combined with the data demonstrating the promising in vivo activity these compounds suggests that this enzyme is a useful target for the development of TB therapeutics.
Rv1625c agonists appear to inhibit cholesterol catabolism indirectly. The prpD′::GFP reporter, which provides a relative measure of the propionyl-CoA that is liberated during cholesterol breakdown in M. tuberculosis, revealed that these compounds reduce cholesterol catabolism (Fig. 2 and 3). In agreement with this, the degradative release of 14CO2 from 14C-cholesterol is also inhibited by these compounds (Fig. 2). Interestingly, LC-MS analysis indicated that treatment with an Rv1625c agonist or induction of cAMP synthesis through a genetic tool did not result in the significant accumulation of any cholesterol-derived metabolite(s) in M. tuberculosis (see Fig. S3). This suggests that the production of cAMP does not inhibit one specific cholesterol catabolic enzyme. The apparent discrepancy between the metabolite profiles and the 14CO2 and prpD′::GFP readouts may be because the former only indicate the size of the metabolite pools and not the turnover of those pools or the flux through the pathway. The 50% effective concentration (EC50) of the Rv1625c agonists varied in the different assays described here. Therefore, we used them at concentrations between 5 and 20 times their EC50 values in each of the respective assays, and it is unlikely that differences in potency account for any of the observations described here. Overall, the data indicate that the agonists may inhibit cholesterol uptake and/or downregulate the pathway as a whole. Clearly, further studies are needed to elucidate the precise mechanism involved in blocking cholesterol metabolism in a cAMP-dependent manner.
The inhibitory effect following Rv1625c stimulation and cAMP production appears to contradict some previously reported findings. Shleeva et al. demonstrated that overexpression of the M. tuberculosis adenylyl cyclase encoded by rv2212 led to increased intracellular cAMP concentrations and that this led to enhanced virulence in a mouse model of infection (25). The authors postulated that the increased pathogenicity was due to the cAMP-mediated survival and recovery from dormancy. Furthermore, cAMP levels have been shown to increase ~50-fold in M. tuberculosis upon infection in macrophages, and this increase in cAMP does not correspond to bacterial cell death in the absence of any Rv1625c agonists (26). Finally, M. tuberculosis is capable of excreting cAMP and has been shown to do so into the culture medium and into infected macrophages, possibly as a mechanism of immune modulation (26, 27). Whether M. tuberculosis is able to mitigate the effects of Rv1625c stimulation through cAMP secretion or turnover via a cAMP-phosphodiesterase remains to be determined (15). Recent reports have implicated cAMP in regulating Mce1-mediated import of fatty acids in M. tuberculosis through the activity of the adenylyl cyclase, Rv3645 (28) and the cAMP-phosphodiesterase, Rv1339 (28, 29). Given the shared homology of the four Mce transporters in M. tuberculosis (30) and that these individual transporters also share protein subunits (29, 31, 32), cAMP could plausibly be a universal regulator of Mce transporters in M. tuberculosis. It will be interesting to determine whether fluctuating levels of cAMP regulate Mce4-mediated import of cholesterol.
It remains unclear how or whether the structurally different Rv1625c agonists directly engage Rv1625c. GSK286 induces cAMP production in R. jostii RHA1 expressing recombinant Rv1625c (Fig. 1) and the other Rv1625c agonists (V-58, V-59, and mCLB073) induce cAMP production in an Escherichia coli cya mutant strain expressing Rv1625c (12, 13). To date, the only evidence that the agonists bind directly to Rv1625c is from nuclear magnetic resonance studies demonstrating that the chemical environment of V-58 changes upon interacting with E. coli cya mutant strain expressing Rv1625c (12). The recent cryo-EM and X-ray structures of the full-length, dimeric Rv1625c provide at least two important insights into understanding the potential interactions between the agonists and the enzyme. First, activation of the adenylyl cyclase domains of the Rv1625c dimer are coordinated through conformational changes in the transmembrane regions of the protein complex (17). Second, the transmembrane domain contains multiple putative substrate-binding clefts, any one of which could potentially bind native signaling molecules or small molecule compounds (17). It will be important to determine whether Rv1625c agonists interact with these putative binding sites and to elucidate the molecular interactions between these compounds and the enzyme. However, the fact that all mutations displayed cross-resistance suggests the agonists may have a similar mechanism of action. Finally, it remains to be determined whether a signaling molecule binds to Rv1625c to promote cAMP-dependent signaling and cholesterol utilization during some stage of the M. tuberculosis life cycle.
Specific combinations of drugs that shorten TB treatment are highly desired, and yet mechanisms that help shorten TB treatment are poorly understood. Given the treatment shortening potential of GSK286 in mice (3), it will be exciting to determine whether Rv1625c agonists similarly contribute to TB treatment shortening in humans and to discover the treatment-shortening mechanism provided by Rv1625c agonists. Nevertheless, one potential limitation of Rv1625c agonists as therapeutics is that Rv1625c itself does not appear to be essential: M. tuberculosis mutants lacking this enzyme have no apparent fitness defect in the mouse infection model (Fig. 5). This nonessentiality potentially increases the likelihood of resistance mutations during infection given that inactivating mutations that confer resistance could be selected without any fitness cost to the bacterium. However, it is unclear how frequently Rv1625c mutations arise during infection in the presence of combination therapy. Given the unusual mechanism for Rv1625c agonists and their potential to eliminate bacteria that are dependent on cholesterol and/or residing in macrophages, these compounds present an interesting potential component of novel regimens for the treatment of drug-sensitive and drug-resistant TB.
MATERIALS AND METHODS
Chemicals and reagents.
Cholesterol (>99%), 5α-cholestane, 8-Br-cAMP, and resazurin were purchased from Sigma-Aldrich. Restriction enzymes were purchased from Thermo Fisher Scientific, Inc. T7 DNA ligase and Gibson assembly master mix were purchased from New England Biolabs. Oligonucleotides were ordered from Integrated DNA Technologies. Water for buffers was purified using a Barnstead Nanopure Diamond system to a resistivity of at least 18 MΩ. Reagents were of high-pressure liquid chromatography (HPLC) or analytical grade.
Bacterial strains and growth.
Bacterial strains and plasmids used in this study are provided in Tables S1 and S2, respectively. M. tuberculosis strains were cultured in 7H9 medium supplemented with either 0.05% Tween 80 or 0.5% tyloxapol and various added carbon sources. Strains were incubated at 37°C in roller bottles. Growth was measured by determining the OD600 or CFU/mL. Briefly, cells were serially diluted in saline containing 0.05% Tween 80 and plated on 7H10 agar containing oleic acid-albumin-dextrose-catalase (OADC) and 0.2% glycerol. Mutant strains were grown in media supplemented with 20 μg/mL kanamycin. RHA1 strains were cultured at 30°C in M9 minimal medium shaking at 200 rpm, as described previously (33). Growth was measured by determining the OD600. Expression from pTipQC2-based plasmids was induced using 10 μg/mL thiostrepton. For strains harboring pTip1625c and pTipQC2, chloramphenicol was included in the medium at 34 μg/mL chloramphenicol.
Mutant generation in M. tuberculosis.
Genes rv0998 and rv1151c were deleted in M. tuberculosis using homologous recombineering (34). Allelic-exchange substrates (AES) were generated using the oligonucleotides listed in Table S3 to amplify the flanking regions of rv0998 and rv1151c and cloning them on either side of the hygR cassette in pYUB854 to generate pKO0998 and pKO1151, respectively (see Table S3). The linearized AES was electroporated into M. tuberculosis harboring pJV53 (34). The rv1625c gene was deleted in M. tuberculosis Erdman by allelic exchange as described previously (35). For this, ~1,000-bp DNA fragments corresponding to the upstream and downstream region of the rv1625c locus were cloned into pYUB854+rpsL to flank the hygromycin resistance gene. The final plasmid was electroporated into streptomycin-resistant M. tuberculosis Erdman. Hygromycin-resistant and streptomycin-resistant clones were selected. Hygromycin-resistant colonies were screened for the absence of the gene of interest and the presence of the hygR cassette by PCR. The orientation of the cassette within the genome was determined using screening primers that anneal outside the AES.
DNA manipulation.
DNA was propagated, digested, and ligated using standard protocols (36). To construct pTip1625, Rv1625c was amplified from genomic DNA using Rv1625-F and Rv1625-R (see Table S3) and digested with the indicated restriction enzymes. Oligonucleotides were purchased from Integrated DNA Technologies (San Diego, CA). The nucleotide sequence of pTip1625 was verified prior to their use.
GSK286 effect on MtPat-mediated acylation.
WT and mutant M. tuberculosis Erdman were grown in 7H9 medium containing 0.5% tyloxapol supplemented with 0.2% glycerol or 0.5 mM cholesterol in 150 μL in 96-well plates at 37°C with increasing concentrations of GSK286 or DMSO alone, with each concentration in triplicate. Strains were incubated for 7 days. Growth was measured by adding 50 μM resazurin, incubating the sample for 24 h at 37°C, and measuring the fluorescence at 530-nm excitation and 590-nm emission. Data were analyzed by GraphPad Prism to generate a nonlinear regression model to fit the normalized results of the dose-response curves and EC50.
Analysis of cAMP.
For WT M. tuberculosis Erdman, WT M. tuberculosis CDC1551, and Tn1625c M. tuberculosis CDC1551, the strains were grown in 200 mL of 7H9 medium containing 0.5 mM cholesterol to an OD600 of 0.4. GSK286 was added at a concentration of 5 μM, and strains were incubated for 16 h at 37°C. Cells were harvested by centrifugation (4,000 × g for 10 min) at room temperature, suspended in 4 mL od acetonitrile-methanol-water (2:2:1 [vol/vol/vol]). Then, 1.5 μM 8-Br-cAMP was added to samples, followed by LC-MS analysis. Cells were lysed by bead beating (three cycles at 6 m/s for 30 s each with 5 min on ice between each run). Lysates were cleared by centrifugation at 16,000 × g for 10 min at 4°C. Samples were filtered through a 0.2-μm-pore size filter, removed from containment level 3, and then dried under nitrogen. For LC-MS analysis, the dried extracts were suspended in 100 mM ammonium acetate (pH 4.5). Samples were analyzed on an Agilent 6460 Triple Quadrupole LC-MS apparatus operated in positive-ion mode at the Proteomics Core Facility at the University of British Columbia, connected to an 80 × 0.25 mm Luna 3 μm PFP(2) analytical column (prepared in-house). For HPLC analysis, the samples were suspended in 100 mM ammonium acetate (pH 4.5). Samples were analyzed using a Waters 2695 Separation HPLC module (Milford, MA) equipped with a Waters 2996 photodiode array detector and a Luna 3-μm PFP(2) 50 × 4.6-mm column (Phenomenex, Torrance, CA). The column was equilibrated with 0.1 M ammonium acetate (pH 4.5). CoA thioesters were eluted using a 20-min linear gradient of 0 to 90% methanol in 0.1 M ammonium acetate (pH 4.5). The eluate was monitored at 258 nm.
Macrophage culture.
Murine J774 macrophage cells were maintained in DMEM supplemented with 10% heat-inactivated fetal calf serum, 2.0 mM l-glutamine, and 1.0 mM sodium pyruvate (100 U/mL penicillin and 100 mg/mL streptomycin) at 37°C and 6.0% CO2. Media without antibiotics were used for infections with M. tuberculosis.
Propionyl-CoA reporter assays.
Fluorescent prpD′::GFP assays in liquid media were conducted in 7H12 medium containing cholesterol (100 μM) and acetate (100 μM) as described previously (31). Bone marrow-derived macrophages (BMDMs) were infected at a multiplicity of infection (MOI) of 5. Extracellular M. tuberculosis was removed after 2 h and replaced with fresh medium containing DMSO or GSK286. After 24 h, infected BMDMs were scraped and fixed in paraformaldehyde. Fixed BMDMs were suspended in lysis buffer (0.1% SDS and 0.1 mg/mL proteinase K in H2O) and lysed by passage through a 25-gauge needle. Pellets were retained for analysis. The GFP mean fluorescence intensity was quantified from 10,000 bacteria by flow cytometry and analyzed by using FlowJo (Becton Dickinson).
14CO2 release experiments.
Catabolism of [4-14C]cholesterol to 14CO2 was quantified as described previously, with minor modifications (31). Briefly, M. tuberculosis cultures were pregrown in 7H12 medium containing cholesterol (100 μM) and acetate (100 μM) for 1 week and adjusted to an OD600 of 0.5. DMSO or GSK286 was added 45 min prior to adding [4-14C]cholesterol. 14CO2 released from the vented M. tuberculosis culture flasks was collected as described previously (31).
RT-qPCR.
RT-qPCR was performed on sigA, kshA, and echA20 in WT Erdman M. tuberculosis. Briefly, four 100-mL cultures in 7H9 with 0.5 mM cholesterol were grown to an OD of 0.6. Cultures were split into 25-mL portions, and 10 μM GSK286, V-59, or DMSO alone was added to cultures in triplicate; the cultures were then incubated at 37°C for 3 h. Next, 10 mL of each culture was pelleted, suspended in 750 μL of TRIzol (Invitrogen) as described by the manufacturer, and processed by bead beating at 6 m/s for 30 s. RNA was precipitated using alcohol, briefly dried, suspended in 200 μL of RNase-free water, and filtered. Samples were treated twice with DNase I and cDNA were synthesized using Invitrogen VILO cDNA synthesis kit according to manufacturer protocols. Samples were assessed for gDNA contamination, and then RT-qPCR was performed in biological duplicate using gene-specific primers and TaqMan probes to sigA, echA20, and kshA as previously described (37). Data were analyzed using the ΔΔCT method, and statistical analyses were performed using one-way analysis of variance (ANOVA) and Tukey’s multiple-comparison test.
Generating spontaneous resistance mutants.
The frequency of spontaneous resistance to GSK286 in vitro was estimated by plating M. tuberculosis on cholesterol agar plates containing GSK286 (25 to 100 μM). Cholesterol was dissolved in 500 mM methyl-β-cyclodextrin and added to 7H10 agar at 100 μM. Then, 7 × 105 CFU of WT Erdman M. tuberculosis was spread per 150-mm plate, and colonies were enumerated. Mutant clones were isolated and subjected to the inhibition assay. The rv1625c region was amplified by PCR and sequenced to determine the location of each mutation.
Inhibition assay.
To determine compound potency against M. tuberculosis in liquid culture, an alamarBlue reduction assay was used. For inhibition assays in medium containing cholesterol as the main carbon source, M. tuberculosis was first cultured to an (OD600 0.4) in 7H12 acetate media (7H9 base, 0.1% Casamino Acids, 100 mM 2-morpholinoethanesulfonic acid [pH 6.6]), 0.1% (wt/vol) acetate, and 0.05% tyloxapol, as described previously (5). The bacteria were washed three times in sterile PBS containing 0.05% tyloxapol and added to 7H12 cholesterol media (7H9 base, 0.1% Casamino Acids, 100 mM 2-morpholinoethanesulfonic acid [pH 6.6]), 100 μM cholesterol, and 0.05% tyloxapol. Cholesterol was added to the culture media as ethanol/tyloxapol micelles. For the inhibition assay, 1.0 × 106 bacteria were added to 96-well microplates to a final volume of 200 μL containing the experimental compounds. The microplates were incubated for 10 days in humidified, sealed plastic bags at 37°C. To quantify bacterial proliferation, 40 μL of an alamarBlue solution (50%) was added to each well, and the plates were reincubated at 37°C for 16 h. alamarBlue reduction was quantified by using an Envision Multilabel plate reader (Perkin-Elmer) with λex = 492 nm and λem = 595 nm.
Metabolite extraction for LC-QTOF analysis.
M. tuberculosis was cultured in 7H9 OADC to generate biomass before subculturing the bacteria in 7H9 medium supplemented with 0.5% tyloxapol and 100 μM cholesterol. The bacterial cultures were treated with V-59 (25 μM) or Anhydrotetracycline (500 ng/mL) for 48 h before metabolite isolation. To isolate metabolites, 5 × 105 bacterial cells were harvested by centrifugation. The bacterial cells were resuspended in 1 mL of ice-cold water (acetonitrile-methanol-water, 2:2:1 [vol/vol/vol]) containing the internal standard 4-chlorobenzoic acid (1.0 μM) before the cells were transferred to a screw-top tube containing silica lysis beads. The 4-chlorobenzoic acid served as a control for the extraction and all sample handling prior to LC-MS analysis, and its consistent abundance was verified in electrospray ionization (ESI)-negative mode. Bacterial cells were lysed with bead beating using three cycles at 6 m/s for 30 s each with 5 min on ice between lysis cycles. Cellular debris was removed by centrifugation, and the metabolite extract was sterilized through a 0.2-μm-pore size PTFE filter and dried to completion with a Speed-Vac.
LC-QTOF analysis of cholesterol metabolites.
Commercial standards of cholesterol, cholestenone, 3-oxocholest-4-en-26-oate, androst-4-ene-3,17-dione, and androsta-1,4-diene-3,17-dione were purchased from Sigma or Steraloids. All other standards were generated in vitro as previously described (11, 18). Chromatography was performed using an Agilent 1290 Infinity II UHPLC. For compounds with CoA moieties, an InfinityLab Poroshell 120 HILIC-Z column (100 mm by 2.1 mm by 2.7 μm) was used. Solvent A consisted of water with 10 mM ammonium acetate (pH 9.0), and solvent B consisted of 90% acetonitrile with 10 mM ammonium acetate (pH 9.0). The flow rate was 0.25 mL/min, and 2 μL of sample was injected. The column was equilibrated in 90% B and held for 2 min after injection, followed by a 10-min linear gradient from 90 to 60% B and held at 60% B for 3 min. An Agilent 6546 QTOF apparatus equipped with a dual AJS ESI source was used in negative and positive modes. The following MS parameters were used: capillary voltage, 3,500 V; nozzle voltage, 1,000 V (negative mode) or 500 V (positive mode); drying gas temperature, 250°C; drying gas flow rate, 10 L/min; sheath gas temperature, 300°C; sheath gas flow rate, 12 L/min; nebulizer pressure, 40 lb/in2; and fragmentor voltage, 100 V. All other compounds were separated using a Zorbax Eclipse Plus C18 reverse-phase column (100 mm by 2.1 mm by 1.8 μm) and run on a 16-min linear gradient from 5 to 100% solvent B at 0.45 mL/min. Solvent A was water–5 mM ammonium acetate (pH 4.8) and solvent B was 95% acetonitrile–5 mM ammonium acetate (pH 4.8). The MS parameters were the same as described above. Data were collected using MassHunter Workstation software. Untargeted molecular feature extractions were performed using MassHunter Profinder (Agilent), and data were normalized in Mass Profiler Professional (Agilent) using the total abundance for the sum of only those extracted features present in all samples. Cholesterol metabolites were identified using an m/z value and retention time matched to the standard from our in-house cholesterol metabolite library.
TB mouse model of infection.
Female BALB/cJ mice (Jackson Laboratories) aged 6 to 8 weeks were infected via an aerosol inhalation exposure system (Glass-Col) with a calibrated dose of 200 CFU of Erdman M. tuberculosis. Tissues were collected from five mice per treatment group at each time point and processed for CFU enumeration. For CFU plating, lungs were homogenized in phosphate-buffered saline–0.05% Tween 80, and serial dilutions were plated on 7H10 OADC.
Ethics statement.
Animal work was approved by Cornell University IACUC (protocol 2013-0030). All protocols conform to the USDA Animal Welfare Act, institutional policies on the care and humane treatment of animals, and other applicable laws/regulations. Euthanasia was performed via carbon dioxide.
ACKNOWLEDGMENTS
This study was supported by grants from the Canadian Institutes of Health Research (CIHR, PJT-159574) and the Canadian Lung Association to L.D.E. and from the National Institute of Allergy and Infectious Diseases (NIAID, AI130018 and AI119122) to B.C.V.
Elena Jimenez and Gareth Maher-Edwards provided valuable feedback as the manuscript was written.
Footnotes
Supplemental material is available online only.
Contributor Information
Brian C. VanderVen, Email: bcv8@cornell.edu.
Lindsay D. Eltis, Email: leltis@mail.ubc.ca.
REFERENCES
- 1.World Health Organization. 2022. Global tuberculosis report, 2022. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Conradie F, Diacon AH, Ngubane N, Howell P, Everitt D, Crook AM, Mendel CM, Egizi E, Moreira J, Timm J, McHugh TD, Wills GH, Bateson A, Hunt R, Van Niekerk C, Li M, Olugbosi M, Spigelman M, Nix TBTT, Nix-TB Trial Team . 2020. Treatment of highly drug-resistant pulmonary tuberculosis. N Engl J Med 382:893–902. 10.1056/NEJMoa1901814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nuermberger EL, Martinez-Martinez MS, Sanz O, Urones B, Esquivias J, Soni H, Tasneen R, Tyagi S, Li SY, Converse PJ, Boshoff HI, Robertson GT, Besra GS, Abrahams KA, Upton AM, Mdluli K, Boyle GW, Turner S, Fotouhi N, Cammack NC, Siles JM, Alonso M, Escribano J, Lelievre J, Rullas-Trincado J, Perez-Herran E, Bates RH, Maher-Edwards G, Barros D, Ballell L, Jimenez E. 2022. GSK2556286 is a novel antitubercular drug candidate effective in vivo with the potential to shorten tuberculosis treatment. Antimicrob Agents Chemother 66:e00132-22. 10.1128/aac.00132-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pethe K, Sequeira PC, Agarwalla S, Rhee K, Kuhen K, Phong WY, Patel V, Beer D, Walker JR, Duraiswamy J, Jiricek J, Keller TH, Chatterjee A, Tan MP, Ujjini M, Rao SPS, Camacho L, Bifani P, Mak PA, Ma I, Barnes SW, Chen Z, Plouffe D, Thayalan P, Ng SH, Au M, Lee BH, Tan BH, Ravindran S, Nanjundappa M, Lin X, Goh A, Lakshminarayana SB, Shoen C, Cynamon M, Kreiswirth B, Dartois V, Peters EC, Glynne R, Brenner S, Dick T. 2010. A chemical genetic screen in Mycobacterium tuberculosis identifies carbon-source-dependent growth inhibitors devoid of in vivo efficacy. Nat Commun 1:57. 10.1038/ncomms1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.VanderVen BC, Fahey RJ, Lee W, Liu Y, Abramovitch RB, Memmott C, Crowe AM, Eltis LD, Perola E, Deininger DD, Wang T, Locher CP, Russell DG. 2015. Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium’s metabolism is constrained by the intracellular environment. PLoS Pathog 11:e1004679. 10.1371/journal.ppat.1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pandey AK, Sassetti CM. 2008. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci USA 105:4376–4380. 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yam KC, D’Angelo I, Kalscheuer R, Zhu H, Wang JX, Snieckus V, Ly LH, Converse PJ, Jacobs WR, Jr, Strynadka N, Eltis LD. 2009. Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog 5:e1000344. 10.1371/journal.ppat.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouellet H, Johnston JB, de Montellano PR. 2011. Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis. Trends Microbiol 19:530–539. 10.1016/j.tim.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rengarajan J, Bloom BR, Rubin EJ. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci USA 102:8327–8332. 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowe AM, Casabon I, Brown KL, Liu J, Lian J, Rogalski JC, Hurst TE, Snieckus V, Foster LJ, Eltis LD. 2017. Catabolism of the last two steroid rings in Mycobacterium tuberculosis and other bacteria. mBio 8:e00321-17. 10.1128/mBio.00321-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson RM, Bai G, DeMott CM, Banavali NK, Montague CR, Moon C, Shekhtman A, VanderVen B, McDonough KA. 2017. Chemical activation of adenylyl cyclase Rv1625c inhibits growth of Mycobacterium tuberculosis on cholesterol and modulates intramacrophage signaling. Mol Microbiol 105:294–308. 10.1111/mmi.13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilburn KM, Montague CR, Qin B, Woods AK, Love MS, McNamara CW, Schultz PG, Southard TL, Huang L, Petrassi HM, VanderVen BC. 2022. Pharmacological and genetic activation of cAMP synthesis disrupts cholesterol utilization in Mycobacterium tuberculosis. PLoS Pathog 18:e1009862. 10.1371/journal.ppat.1009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 15.Johnson RM, McDonough KA. 2018. Cyclic nucleotide signaling in Mycobacterium tuberculosis: an expanding repertoire. Pathog Dis 76:fty048. 10.1093/femspd/fty048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, McKinney JD, Bertozzi CR, Sassetti CM. 2012. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem Biol 19:218–227. 10.1016/j.chembiol.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta V, Khanppnavar B, Schuster D, Kantarci I, Vercellino I, Kosturanova A, Iype T, Stefanic S, Picotti P, Korkhov VM. 2022. Structure of Mycobacterium tuberculosis Cya, an evolutionary ancestor of the mammalian membrane adenylyl cyclases. Elife 11:470738. 10.7554/eLife.77032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casabon I, Swain K, Crowe AM, Eltis LD, Mohn WW. 2014. Actinobacterial acyl coenzyme A synthetases involved in steroid side-chain catabolism. J Bacteriol 196:579–587. 10.1128/JB.01012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rimal B, Batchelder HR, Story-Roller E, Panthi CM, Tabor C, Nuermberger EL, Townsend CA, Lamichhane G. 2022. T405, a new penem, exhibits in vivo efficacy against M. abscessus and synergy with beta-lactams imipenem and cefditoren. Antimicrob Agents Chemother 66:e0053622. 10.1128/aac.00536-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bi J, Wang Y, Yu H, Qian X, Wang H, Liu J, Zhang X. 2017. Modulation of central carbon metabolism by acetylation of isocitrate lyase in Mycobacterium tuberculosis. Sci Rep 7:44826. 10.1038/srep44826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HJ, Lang PT, Fortune SM, Sassetti CM, Alber T. 2012. Cyclic AMP regulation of protein lysine acetylation in Mycobacterium tuberculosis. Nat Struct Mol Biol 19:811–818. 10.1038/nsmb.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nambi S, Basu N, Visweswariah SS. 2010. cAMP-regulated protein lysine acetylases in mycobacteria. J Biol Chem 285:24313–24323. 10.1074/jbc.M110.118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nambi S, Gupta K, Bhattacharyya M, Ramakrishnan P, Ravikumar V, Siddiqui N, Thomas AT, Visweswariah SS. 2013. Cyclic AMP-dependent protein lysine acylation in mycobacteria regulates fatty acid and propionate metabolism. J Biol Chem 288:14114–14124. 10.1074/jbc.M113.463992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu H, Hegde SS, Blanchard JS. 2011. Reversible acetylation and inactivation of Mycobacterium tuberculosis acetyl-CoA synthetase is dependent on cAMP. Biochemistry 50:5883–5892. 10.1021/bi200156t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shleeva MO, Kondratieva TK, Demina GR, Rubakova EI, Goncharenko AV, Apt AS, Kaprelyants AS. 2017. Overexpression of adenylyl cyclase encoded by the Mycobacterium tuberculosis rv2212 gene confers improved fitness, accelerated recovery from dormancy and enhanced virulence in mice. Front Cell Infect Microbiol 7:370. 10.3389/fcimb.2017.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai G, Schaak DD, McDonough KA. 2009. cAMP levels within Mycobacterium tuberculosis and Mycobacterium bovis BCG increase upon infection of macrophages. FEMS Immunol Med Microbiol 55:68–73. 10.1111/j.1574-695X.2008.00500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agarwal N, Lamichhane G, Gupta R, Nolan S, Bishai WR. 2009. Cyclic AMP intoxication of macrophages by a Mycobacterium tuberculosis adenylate cyclase. Nature 460:98–102. 10.1038/nature08123. [DOI] [PubMed] [Google Scholar]
- 28.Wong AI, Beites T, Planck KA, Li S, Poulton NC, Rhee K, Schnappinger D, Ehrt S, Rock J. 2022. Cyclic AMP is a critical mediator of intrinsic drug resistance and fatty acid metabolism in M. tuberculosis. bioRxiv. 10.1101/2022.07.07.499113. [DOI] [PMC free article] [PubMed]
- 29.Nazarova EV, Montague CR, Huang L, La T, Russell D, VanderVen BC. 2019. The genetic requirements of fatty acid import by Mycobacterium tuberculosis within macrophages. Elife 8:43621. 10.7554/eLife.43621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casali N, Riley LW. 2007. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 8:60. 10.1186/1471-2164-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nazarova EV, Montague CR, La T, Wilburn KM, Sukumar N, Lee W, Caldwell S, Russell DG, VanderVen BC. 2017. Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. Elife 6:e26969. 10.7554/eLife.26969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perkowski EF, Miller BK, McCann JR, Sullivan JT, Malik S, Allen IC, Godfrey V, Hayden JD, Braunstein M. 2016. An orphaned Mce-associated membrane protein of Mycobacterium tuberculosis is a virulence factor that stabilizes Mce transporters. Mol Microbiol 100:90–107. 10.1111/mmi.13303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casabon I, Crowe AM, Liu J, Eltis LD. 2013. FadD3 is an acyl-CoA synthetase that initiates catabolism of cholesterol rings C and D in actinobacteria. Mol Microbiol 87:269–283. 10.1111/mmi.12095. [DOI] [PubMed] [Google Scholar]
- 34.van Kessel JC, Marinelli LJ, Hatfull GF. 2008. Recombineering mycobacteria and their phages. Nat Rev Microbiol 6:851–857. 10.1038/nrmicro2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mann FM, VanderVen BC, Peters RJ. 2011. Magnesium depletion triggers production of an immune modulating diterpenoid in Mycobacterium tuberculosis. Mol Microbiol 79:1594–1601. 10.1111/j.1365-2958.2011.07545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 37.Casabon I, Zhu SH, Otani H, Liu J, Mohn WW, Eltis LD. 2013. Regulation of the KstR2 regulon of Mycobacterium tuberculosis by a cholesterol catabolite. Mol Microbiol 89:1201–1212. 10.1111/mmi.12340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aac.01294-22-s0001.pdf, PDF file, 0.3 MB (345.7KB, pdf)