ABSTRACT
Here, this report presents two genomes of Vibrio cholerae O1 serotype Ogawa, recovered from cholera cases in Australia linked to travel to Pakistan in 2022. Their multidrug-resistant genotype represents the current activity of cholera within the seventh pandemic. One of the genome sequences was assembled using both short- and long-read sequences.
ANNOUNCEMENT
Seven distinct pandemics associated with Vibrio cholerae O1 have been described since 1896 (1). The current seventh pandemic arose in 1961 with global spread via three separate waves and remains a public health burden for many countries experiencing localized outbreaks (2–5). There has been a recent upsurge of cholera cases in Pakistan amid the global COVID-19 pandemic (4, 6), and WHO recognized a risk of the outbreak spread associated with international travel (7). This report provides high-quality genome sequences recovered from human cases diagnosed with cholera linked to that region during this upsurge. Two isolates of V. cholerae O1 serotype Ogawa which were cultured from two unrelated cases of cholera in Australian residents in 2022 were subjected to whole-genome sequencing. Case 1 (Vch-N1252) was diagnosed in New South Wales (NSW) and had an indirect link to a recent traveler from Pakistan. Case 2 (Vch-Q4233) was diagnosed in Queensland (QLD) and reported recent travel to Pakistan. A nonresearch determination for activities of Vch-N1252 was granted by Health Protection New South Wales as it was performed under the NSW Public Health Act. An analysis of Vch-Q4233 was performed under the QLD Public Health Act, and analysis received Forensic and Scientific Services (FSS) Human Ethics Committee Clearance (HEC 22–26).
Both isolates were first identified as V. cholerae O1 biotype El Tor serotype Ogawa in NSW and QLD. Parameters from isolation through to whole-genome sequencing are listed in Table 1. Default parameters were used for all software unless otherwise specified. Short reads of both isolates were trimmed (SLIDINGWINDOW: 4:20, LEADING: 3, TRAILING: 3 MINLEN: 36 [Vch-Q4233] or 100 [Vch-N1252]) using Trimmomatic version 3.04 (8), and Vch-Q4233 was assembled using SPAdes version 3.12.0 with the inclusion of the “--careful” flag (9). Live base-calling and demultiplexing (“Trim barcodes” and “Mid read barcode filtering” enabled) were performed on a GridION device running MinKNOW version 21.10. Nanopore reads were filtered (--min_length 1000, --keep_percent 95) using Filtlong version 0.2.1 (https://github.com/rrwick/Filtlong). Hybrid assembly, along with the circularization and genomic rotation of Vch-N1252, was performed using Unicycler version 0.4.8 (10). Chromosome one was rotated to start with dnaA, while chromosome two was not rotated. Circularity was confirmed visually using Bandage version 0.9.0 (11). Genes conferring antimicrobial resistance (AMR) were detected using Abricate version 1.0.1 (https://github.com/tseemann/abricate) against AMRFinderPlus (12).
TABLE 1.
Key parameters from isolation through to draft assembly of Vch-Q4233 and the hybrid assembly of Vch-N1252
| Parameter | Information for: |
||
|---|---|---|---|
| Vch-Q4233 | Vch-N1252 |
||
| Short reads | Long reads | ||
| Isolation | |||
| Specimen type | Isolate was referred via a pathology provider | Feces | |
| Initial detection | Allplex-GIa-bacteria (Seegene) | ||
| Enrichment conditions | Alkaline peptone water at 37°C for 8 h | ||
| Isolation conditions | Thiosulfate-citrate-bile salts-sucrose agar at 37°C for 48 h | ||
| Isolate identification | MALDI-TOF MSb (Bruker) and ID 32E (bioMérieux) | ||
| Isolate maintenance | Thiosulfate-citrate-bile salts-sucrose agar at 37°C for 24 h | ||
| DNA extraction | |||
| Growth conditions | Horse blood agar at 37°C for 20 ± 2 h | ||
| DNA extraction kt | QiaSymphony DSP DNA minikit (Qiagen) | DNeasy blood and tissue kit (Qiagen) | DNeasy UltraClean microbial kit (Qiagen) |
| Type of extraction | Spin column | Spin column | Spin column |
| Library prepn and sequencing | |||
| Library prepn kit | Nextera XT (Illumina) | Nextera XT (Illumina) | Rapid barcoding kit (Oxford Nanopore Technologies) |
| Reagent kit/flow cell | Midoutput kit, 150 cycles | Midoutput kit, 150 cycles | R9.4.1 |
| Sequencing platform | NextSeq 500 | NextSeq 500 | GridION |
| DNA input | N/Ac | N/A | 500 ng |
| Sequencing runtime | N/A | N/A | 20 h 32 min |
| Sequencing and assembly statistics | |||
| No. of readsd | 2,439,076 | 3,559,816 | 111,064 |
| Minimum read length (no. of bases) | 36 | 100 | 1,000 |
| Maximum read length (no. of bases) | 151 | 151 | 36,440 |
| Read length N50 (no. of bases) | 150 | 150 | 5,310 |
| Cumulative read length (no. of bases) | 341,317,784 | 521,169,415 | 444,176,625 |
| Sequencing depth (×)e | 82.48 | 125.93 | 107.33 |
| No. of contigs | 176 | N/A | 2 |
| Assembly length (no. of bases) | 3,989,030 | N/A | 4,095,848 |
| Assembly N50 (no. of bases) | 77,137 | N/A | 3,046,907 |
| Size of chromosome 1 (no. of bases) | N/A | N/A | 3,046,907 |
| Size of chromosome 2 (no. of bases) | N/A | N/A | 1,048,941 |
| G+C content (%) | 47.61 | N/A | Chr. 1, 48.39; Chr. 2, 47.88 |
| Detected AMR genes | dfrA1, sul2, strA, strB, floR, varG, catB9 | dfrA1, sul2, strA, strB, floR, varG, catB9 | dfrA1, sul2, strA, strB, floR, varG, catB9 |
| Accession no. | |||
| Sequence Read Archive | SRR21074467 | SRR21074466 | SRR21074465 |
| GenBankf | JANQBL000000000 | N/A | CP102927, CP102928 |
| NCBI RefSeqg | N/A | N/A | NZ_CP102927.1, NZ_CP102928.1 |
GI, gastrointestinal.
MALDI-TOF MS, matrix-assisted laser desorption ionization–time of flight mass spectrometry.
N/A, not available.
Counts of trimmed reads and filtered reads for short reads and long reads, respectively.
Calculated using the assembly length of ASM836960v1.
Assembly of Vch-Q4233 was annotated by the NCBI Prokaryotic Genome Annotation Pipeline version 6.3 [15].
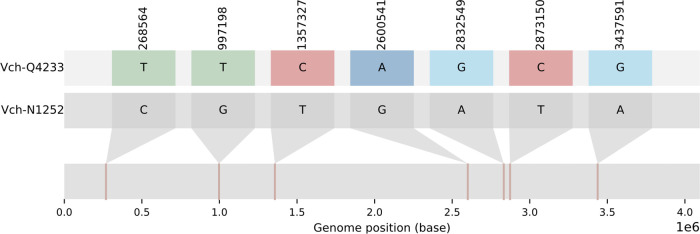
Both genomes were related genetically with only seven single nucleotide polymorphisms (SNPs) separating them (Fig. 1), and both harbored the ctxAB (ctxB7 allele) genes. The same set of AMR genes was detected in the assemblies of both isolates based on short reads, and the hybrid assembly of Vch-N1252 confirmed that drfA1, sul2, strA, strB, and floR were carried within an integrative and conjugative element, as reported previously (13). This submission is inspired by the call for a timely sharing of international genomic surveillance data (14) and may be helpful for monitoring the spread and control of the ongoing outbreak in Pakistan during the seventh pandemic of cholera and beyond.
FIG 1.
Genomic similarity between genomes of Vch-N1252 and Vch-Q4233. Graphical representation of a pair-wise alignment between Vch-N1252 and a pseudomolecule of Vch-N1252 with identified SNPs of Vch-Q4233 incorporated. The pseudomolecule was generated from an output from snippy version 4.6.0 (https://github.com/tseemann/snippy). The image was generated using snipit (https://github.com/aineniamh/snipit). Default settings were used for both programs.
Data availability.
Reads and assemblies were deposited in SRA and GenBank, respectively. Accession numbers are provided in Table 1.
ACKNOWLEDGMENTS
We acknowledge the staff of both Q-PHIRE Genomics and Public Health Microbiology, Queensland Health Forensic and Scientific Services and Centre for Infectious Diseases and Microbiology Laboratory Services, NSW Health Pathology for their assistance in confirmatory testing and sequencing. We acknowledge the case follow-up performed by Queensland Health Metro South Public Health Unit and Health Protection New South Wales. We also thank The Sydney Informatics Hub along with The University of Sydney for the provision of high-performance computing capacity.
Contributor Information
Eby M. Sim, Email: eby.sim@sydney.edu.au.
David Rasko, University of Maryland School of Medicine.
REFERENCES
- 1.Hu D, Liu B, Feng L, Ding P, Guo X, Wang M, Cao B, Reeves PR, Wang L. 2016. Origins of the current seventh cholera pandemic. Proc Natl Acad Sci USA 113:E7730–E7739. doi: 10.1073/pnas.1608732113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, Niyogi SK, Kim EJ, Ramamurthy T, Chun J, Wood JLN, Clemens JD, Czerkinsky C, Nair GB, Holmgren J, Parkhill J, Dougan G. 2011. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganesan D, Gupta SS, Legros D. 2020. Cholera surveillance and estimation of burden of cholera. Vaccine 38:A13–A17. doi: 10.1016/j.vaccine.2019.07.036. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed SH, Shaikh TG, Waseem S, Hasan MM, Bardhan M, Mukerjee N. 2022. Rise in cholera amid COVID-19: spotlight on Pakistan and Bangladesh. Lancet Reg Health Southeast Asia 4:100041. doi: 10.1016/j.lansea.2022.100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin C-S, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, Paxinos EE, Yamaichi Y, Calderwood SB, Mekalanos JJ, Schadt EE, Waldor MK. 2011. The origin of the Haitian cholera outbreak strain. N Engl J Med 364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan HA, Masood W, Siddiqui A, Ahmad S, Salman Y, Essar MY. 2022. The cholera outbreak in Karachi, Pakistan: challenges, efforts and recommendations. Ann Med Surg (Lond) 78:103873. doi: 10.1016/j.amsu.2022.103873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization. 2022. Cholera—Pakistan. World Health Organization, Geneva, Switzerland. https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON391. [Google Scholar]
- 8.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes de novo assembler. Curr Protoc Bioinformatics 70:e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 10.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, Hoffmann M, Pettengill JB, Prasad AB, Tillman GE, Tyson GH, Klimke W. 2021. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep 11:12728. doi: 10.1038/s41598-021-91456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sjölund-Karlsson M, Reimer A, Folster JP, Walker M, Dahourou GA, Batra DG, Martin I, Joyce K, Parsons MB, Boncy J, Whichard JM, Gilmour MW. 2011. Drug-resistance mechanisms in Vibrio cholerae O1 outbreak strain, Haiti, 2010. Emerg Infect Dis 17:2151–2154. doi: 10.3201/eid1711.110720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. 2022. Global genomic surveillance strategy for pathogens with pandemic and epidemic potential, 2022–2032. World Health Organization, Geneva, Switzerland. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, O'Neill KR, Haft DH, DiCuccio M, Chetvernin V, Badretdin A, Coulouris G, Chitsaz F, Derbyshire MK, Durkin AS, Gonzales NR, Gwadz M, Lanczycki CJ, Song JS, Thanki N, Wang J, Yamashita RA, Yang M, Zheng C, Marchler-Bauer A, Thibaud-Nissen F. 2021. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res 49:D1020–D1028. doi: 10.1093/nar/gkaa1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Reads and assemblies were deposited in SRA and GenBank, respectively. Accession numbers are provided in Table 1.