ABSTRACT
We report that ribavirin exerts an inhibitory and mutagenic activity on SARS-CoV-2-infecting Vero cells, with a therapeutic index higher than 10. Deep sequencing analysis of the mutant spectrum of SARS-CoV-2 replicating in the absence or presence of ribavirin indicated an increase in the number of mutations, but not in deletions, and modification of diversity indices, expected from a mutagenic activity. Notably, the major mutation types enhanced by replication in the presence of ribavirin were A→G and U→C transitions, a pattern which is opposite to the dominance of G→A and C→U transitions previously described for most RNA viruses. Implications of the inhibitory activity of ribavirin, and the atypical mutational bias produced on SARS-CoV-2, for the search for synergistic anti-COVID-19 lethal mutagen combinations are discussed.
KEYWORDS: SARS-CoV-2, COVID-19, viral quasispecies, ribavirin, lethal mutagenesis
INTRODUCTION
Lethal mutagenesis (defined as extinction by an excess of mutations) is a broad-spectrum antiviral design which is increasingly applied to treat viral infections. Its conceptual foundation was formulated as a corollary of quasispecies theory developed by Eigen and Schuster (1). Such a corollary stated that for a replicative system, there is a maximum value of the error rate for template copying, which is compatible with maintenance of the inheritable information conveyed by the system (1–3). Support came also from population genetics, with the prediction of extinction of asexual populations by accumulation of mutations, or mutational meltdown, as a consequence of the operation of Muller’s ratchet (4–6). Experimental proof of the concept was provided by showing the detrimental effect on RNA virus survival of replication in the presence of mutagenic agents (7–11). To date, at least 25 different RNA viruses have been suppressed or extinguished in cell culture or in vivo assays by base and nucleoside analogues, which exert at least part of their antiviral activity through viral mutagenesis by the corresponding nucleoside-triphosphates (examples of specific studies in references 12 to 15; recent reviews in references 16 and 17).
Lethal mutagenesis is one of the mechanisms of antiviral activity of the purine analogue ribavirin (1-β-d-ribofuranosyl-1-H-1,2,4-triazole-3-carboxamide) (Rib), an antiviral agent used since the 1970s (18). For decades, there was no awareness that its multiple mechanisms of action (depletion of GTP levels through inhibition of cellular inosine-monophosphate dehydrogenase, enhancement of the antiviral Th1 immune response, inhibition of mRNA cap formation, polyamine depletion, etc.) included also a mutagenic activity in viral RNA (13, 19). Rib has proven effective against several viruses (20–22). It induced lethal mutagenesis of hantaviruses (23–25) and West Nile virus (WNV) (26). It exerted a mutagenic activity on lymphocytic choriomeningitis virus (LCMV) (27), 6B virus B (28), hepatitis E virus (29), and hepatitis C virus (HCV) in cell culture and in vivo (30–35). Serial passages in cell culture of Zika virus (ZIKV) and Usutu virus in the presence of Rib correlated with increases in mutation frequency and resulted in virus extinction (36). Curing of cells persistently infected with foot-and-mouth disease virus (FMDV) involved mutagenesis by Rib (37).
The emergence of COVID-19, and the partial efficacy of the vaccines available to date, has triggered interest in the search for antiviral agents against coronaviruses, in particular for SARS-CoV-2 infections (38). Several anti-SARS-CoV-2 inhibitors are currently in use, including molnupiravir, the isopropyl ester prodrug of β-d-N4-hydroxycytidine (NHC), which acts as a lethal mutagen (39–42). Concerning Rib, infection of Vero E6 cells with nCoV-2019 (BetaCoV/Wuhan/WIV04/20192) at a multiplicity of infection (MOI) of 0.05 PFU/cell in the presence of the analogue resulted in a half-maximal effective concentration (EC50) of 109.50 μM, half cytotoxic concentration (CC50) of >400 μM, and a selectivity index (SI) of >3.65 (43). A retrospective study that involved 19 critically ill (intensive care unit-admitted) COVID-19 patients who were treated with Rib suggested a benefit in virus clearance and patient survival following ribavirin treatment (44). The analogue mediated a cell type-dependent suppression of several SARS-CoV-2 and host proteins (45).
Despite the widespread use of Rib as an antiviral agent, administered either alone or in combination with other inhibitors, quantifications of its inhibition of SARS-CoV-2, and possible alterations of the mutational spectrum it evokes on the virus, are lacking. We have adapted a deep sequencing protocol that allows examination of point mutations and deletions in mutant spectra of SARS-CoV-2 with a mutant frequency cutoff of 0.1%, which has greatly expanded the potential to reliably detect low-frequency mutations in mutant spectra of diagnostic samples from COVID-19 patients, relative to using a 0.5% cutoff (46). These high-resolution deep-sequencing data provide a solid basis for comparison of the corresponding mutational spectra during a single infection of SARS-CoV-2 in Vero E6 cells in the presence of Rib. Here, we show that ribavirin displays a good therapeutic index for SARS-CoV-2 under cell culture conditions and that it induces an unusual mutational bias in the virus even after a single round of infection. The results encourage extending preclinical trials to consider the use of ribavirin, despite its recognized side effects (47), in combination therapies for severe COVID-19 infections.
RESULTS
Inhibition of SARS-CoV-2 replication in Vero E6 cells by ribavirin.
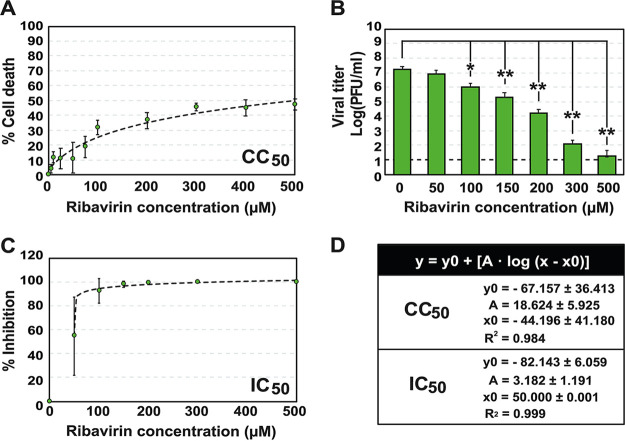
The cytotoxicity of Rib for Vero E6 cells was quantified by measuring cell viability, following exposure of subconfluent cell monolayers for 72 h to different drug concentrations. The Rib concentration that reduced cell viability by 50% (CC50) was >500 μM (Fig. 1A and D). SARS-CoV-2, isolate USA-WA1/2020 (GenBank accession number MT246667), was used to infect 2 × 105 Vero E6 cells at a multiplicity of infection (MOI) of 0.001 PFU/cell, in the absence or the presence of increasing concentrations of Rib, and the infections were allowed to proceed for 48 h. The inhibition was dose dependent (Fig. 1B), and the Rib concentration that produced a 50% decrease in infectious progeny production (IC50) of SARS-CoV-2 was 50.30 ± 24.01 μM (Fig. 1C and D). Both CC50 and IC50 exhibited a good fit with a logarithmic function, y = y0 + [A × log (x − x0)] (R2 ≥ 0.98) (Fig. 1D). These values yield a therapeutic index (TI = CC50/IC50) higher than 10, suggesting a good inhibitory activity of Rib on SARS-CoV-2, very close to the value calculated for hepatitis C virus (HCV) in cell culture (35).
FIG 1.
Cytotoxicity for Vero E6 cells and inhibition of SARS-CoV-2 progeny production by Rib. (A) Determinations of cytotoxic concentration 50 (CC50); percent cell death is plotted as a function of Rib concentration. (B) Inhibition of SARS-CoV-2 infectious progeny production by Rib. The experiments and titrations were performed in triplicate; values and standard deviations were calculated using the program SigmaPlot. (C) Determinations of inhibitory concentration 50 (IC50); percent inhibition is plotted as a function of Rib concentration. (D) Function and values that correspond to the discontinuous lines depicted in panels A and C. Mock infections were carried out in parallel (negative control). Experimental conditions for cell growth, SARS-CoV-2 infection, determination of cell viability, and SARS-CoV-2 infectivity are described in Materials and Methods.
Mutant spectrum of SARS-CoV-2 in the absence or presence of ribavirin.
To examine if SARS-CoV-2 inhibition by Rib was associated with increased mutagenesis of the viral RNA, total RNA of the cell culture supernatants at 48 h postinfection in the absence or presence of 100 and 150 μM Rib was extracted. Viral RNA corresponding to the nsp12 (polymerase)- and spike (S)-coding region was amplified using specific oligonucleotide primers (Materials and Methods; see Table S1 posted at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX), and subjected to ultradeep sequencing (UDS) analyses using MiSeq platform (Illumina) as previously described (46, 48, 49). These genomic regions were chosen because the amplification and bioinformatics data processing provided a high resolution of minority mutations, covering the important functional domains of the proteins. Four amplicons (A1 to A4) covering nucleotides 14,534 to 16,054 of the nsp12 (polymerase)-coding region and two amplicons (A5 and A6) covering nucleotides 22,872 to 23,645 of the S-coding region were analyzed with a 0.1% cutoff frequency for point mutations and deletions (see Fig. S1 posted at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX). The total number of clean reads was 2,041,147, which yielded an average of 113,397 reads per amplicon (see Table S2 posted at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX). The use of a 0.1% mutation frequency cutoff for a reliable representation of SARS-CoV-2 mutant spectra is justified by (i) the clean read coverage, (ii) the observation that the mutations found with a 0.5% mutant frequency cutoff were also included among the set identified with the 0.1% cutoff, and (iii) because, with the two cutoff levels, there was an identical mutation composition, amino acid representation, and predicted functional effects (46) (see also Materials and Methods).
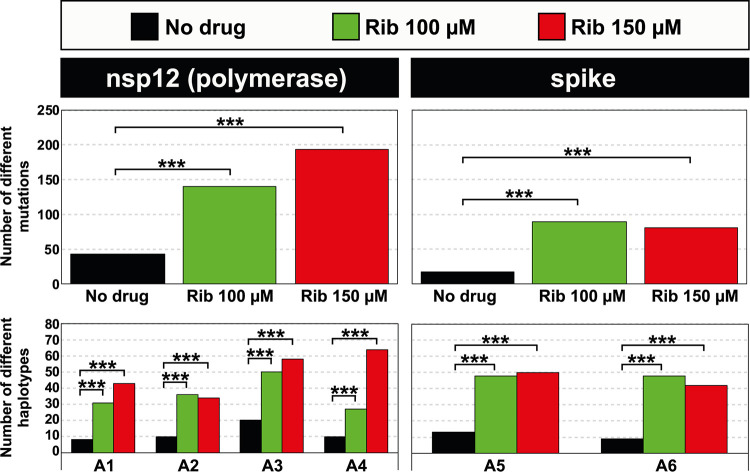
A heatmap was constructed to represent the point mutations and their frequencies within the mutant spectrum of the populations that resulted from the infection in the absence or presence of Rib (Fig. 2 and Table S3 posted at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX). Mutations were counted using, as reference, the genomic sequence of the parental Wuhan isolate (GenBank accession no. NC_045512). Most point mutations were found at frequencies in the range of 0.1% to 0.49% in each mutant spectrum, with the exception of 5 and 13 mutations in the nsp12 (polymerase)- and S-coding regions, respectively (color coded in Fig. 2). There was a statistically significant increase in the number of different mutations and haplotypes associated with Rib treatment (P < 0.0001 in all comparisons; proportion test) (Fig. 3).
FIG 2.
Heatmap of SARS-CoV-2 point mutations in the mutant spectrum of SARS-CoV-2 populations grown in the absence (no drug) or presence of ribavirin (Rib), with a cutoff mutation frequency of 0.1%. Frequency values are color coded, as depicted in the upper box. Data are presented in two blocs, one for the nsp12 (polymerase)-coding region (genomic residues 14,534 to 16,054) and another for the S-coding region (genomic residues 22,872 to 23,645). Only positions with a mutation are represented; mutations have been identified relative to GenBank accession no. NC_045512. The complete set of mutations, their frequency and type, codon position, amino acid substitutions, tolerability, and potential functional effect are given in Table S3 at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX. Procedures are detailed in Materials and Methods.
FIG 3.
Number of different point mutations and haplotypes in mutant spectra of SARS-CoV-2 populations grown in the absence (no drug) or presence of ribavirin (Rib) (color code in the top box). The genomic region is indicated at the top of each panel group, and the amplicons (A) are depicted in abscissa. The number of mutations per amplicon has not been presented due to the overlapping regions among successive amplicons (Fig. S1 at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX) that contain mutations that would be counted twice. The statistical significance of the differences was calculated using the proportion test (***, P < 0.001). The complete list of mutations and amino acid substitutions is given in Table S3 posted at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX.
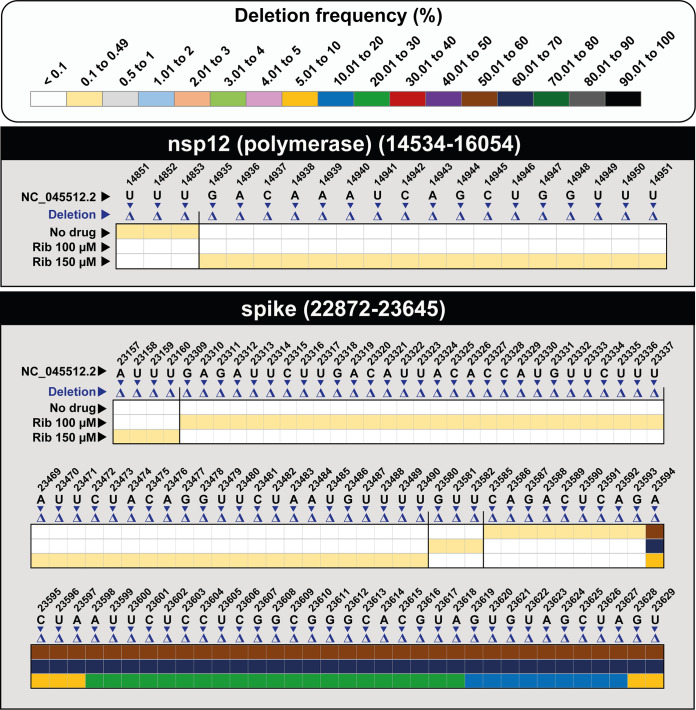
Deletions in the mutant spectra were more abundant in the S- than in the nsp12 (polymerase)-coding region (P = 0.01; proportion test). Rib did not induce an increase in the number of deletions (P = 0.657 and P = 0.668 for Rib 100 μM and Rib 150 μM, respectively; proportion test) (Fig. 4 and Fig. S2 posted at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX). Contrary to the majority of point mutations, which were found at frequencies below 0.49%, 46% of the deletions were present at a frequency higher than 0.49% (Fig. 4). The deletions affected 3 to 36 nucleotides, and they did not occur at homopolymeric tracts or recognizable sequence signatures; in 36% of them, the reading frame was altered and originated a downstream stop codon (Fig. S2 posted at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX).
FIG 4.
Heatmap of SARS-CoV-2 deletions in SARS-CoV-2 populations grown in the absence (no drug) or presence of ribavirin (Rib), with a cutoff deletion frequency of 0.1%. Frequency values are color coded, as depicted in the top box. Data are presented in two blocks, one for the nsp12 (polymerase)-coding region (genomic residues 14,534 to 16,054) and another for the S-coding region (genomic residues 22,872 to 23,645). Only positions within a deletion are represented; deletions have been identified relative to NCBI reference sequence, GenBank accession no. NC_045512. The position, length, sequence context, frequency, and the number of affected haplotypes for each deletion are given in Fig. S2 at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX. Procedures are detailed in Materials and Methods.
Therefore, the main effect associated with Rib inhibition of SARS-CoV-2 was to increase the number of mutations and corresponding haplotypes, but not the number of deletions.
Effect of ribavirin on SARS-CoV-2 mutant spectrum complexity.
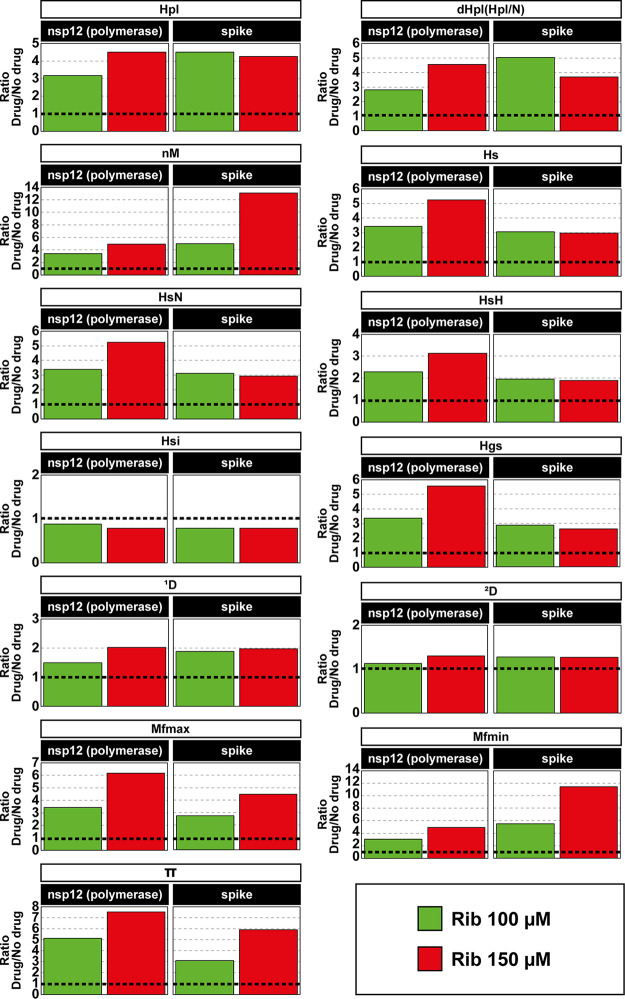
The SARS-CoV-2 mutant spectra produced in the infections in the absence or presence of Rib were further analyzed by determining the diversity indices that were proposed by Gregori and colleagues to characterize viral population complexity (50). Indices were calculated on the basis of the number of point mutations derived from clean reads, and deletions enter the calculation by considering each different deletion as one point mutation. The results, which are presented as the ratio of each index obtained in the presence to absence of Rib (Fig. 5; numerical values shown in Table S4 posted at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX), show a statistically significant increase of the majority of diversity indices, in agreement with a mutagenic activity of Rib. An exception to the increase was the Simpson index (Hsi) because this index represents the probability that two randomly chosen genomes from a population belong to the same haplotype (50); such probability is not expected to increase as a result of mutagenesis (see also Discussion). No significant differences were observed in the effect of Rib on the complexity of the nsp12 (polymerase)- and the S-coding regions, and the index variations were not affected by counting mutations relative to the Wuhan reference isolate or relative to the consensus sequence of the corresponding populations.
FIG 5.
Effect of ribavirin mutagenesis on the complexity of mutant spectra of SARS-CoV-2 populations. The diversity indices are those previously defined (50). Abbreviations (written at the top empty, elongated boxes) are as follows: Hpl, number of haplotypes; dHpl (Hpl/N), number of haplotypes normalized to the number of reads; nM, number of mutations; Hs, Shannon entropy; HsN, Shannon entropy normalized to the number of reads; HsH, Shannon entropy normalized to the number of haplotypes; Hsi, Simpson index; Hgs, Gini-Simpson index; 1D, Hill number for q = 1; 2D, Hill number for q = 2; Mfmax, maximum mutation frequency; Mfmin, minimum mutation frequency; and π, nucleotide diversity. Bars represent the average ratios of the four nsp12 (polymerase) amplicons or the two spike amplicons (indicated in the filled boxes) (value obtained in the presence of Rib divided by the corresponding value in the absence of drug). The statistical significance of the increase of the values (Rib versus no drug) was a P value of <0.05 for all comparisons, except for the difference in Mfmax and π for Rib 100 μM and in Hsi that did not reach statistical significance (Wilcoxon test).
Mutations evoked by ribavirin, as well as mutation site preferences.
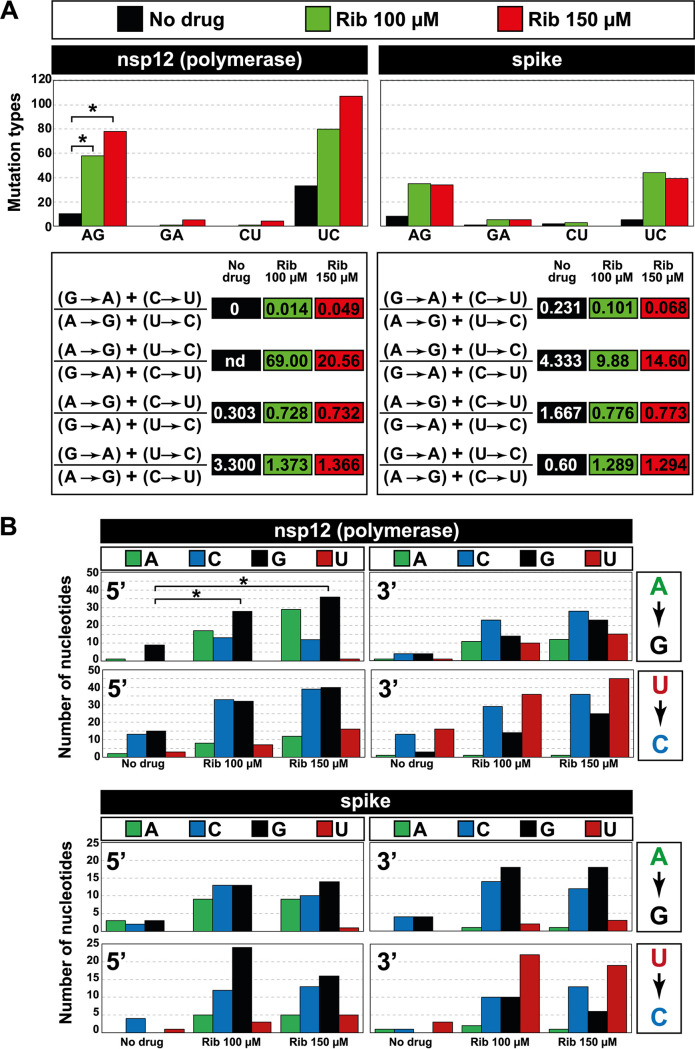
The types of different mutations at the nsp12 (polymerase) and S-coding regions analyzed in the virus grown in the presence of Rib (Fig. 6 and Table S3 posted at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX) indicate a predominance of A→G and U→C transitions over other mutation types, relative to the virus grown in the absence of Rib (Fig. 6A). In particular, the ratio [(A→G) + (U→C)]/[(G→A) + (C→U)] reached values that ranged between 9.88 and 69.00 (Fig. 6A).
FIG 6.
Mutation types evoked by ribavirin on SARS-CoV-2 and neighbor preferences at the mutation types. (A) Mutation types evoked by ribavirin. The absence or presence of Rib is color coded in the top box. Left panels give data for the nsp12 (polymerase)-coding region, and right panels give data for the spike-coding region. Of the observed biases, the preference for A→G transitions reached statistical significance for the nsp12 (polymerase)-coding region (*, P < 0.05; proportion test). Mutation bias is reflected in transition type ratios, as depicted in the bottom panels; nd, not determined because the denominator for that ratio was zero. (B) Analysis of nucleotide preference at the 5′ side or 3′ side of the mutation sites. Coding region and nucleotide type are indicated in the top boxes, and mutation types are displayed in the boxes at the right of each panel. Absence or presence of ribavirin is given in the abscissae. The number of each type of nucleotide located at the 5′ side or 3′ side of the mutation site is written in ordinate; note the different scales for the two coding regions. The complete list of mutations on which the calculations are based is given in Table S3 in the supplemental material at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX.
To determine if there was some preference for mutation sites, haplotypes were aligned without considering their abundance. Quantification of the nucleotide type at the 5′ and 3′ sides of the A→G or U→C mutation sites indicated a statistically significant preference for the presence of a G residue at the 5′ side of A→G transitions at the nsp12 (polymerase)-coding region (P = 0.035 and P = 0.023 for Rib 100 μM and Rib 150 μM, respectively; proportion test). Other additional neighbor preferences were noted, but they did not reach statistical significance (Fig. 6B). Thus, Rib is a mutagenic agent for SARS-CoV-2 that greatly accentuated a bias in favor of A→G and U→C transitions. At least in the case of A→G transitions in the nsp12 (polymerase)-coding region, the mutated sites were not random.
DISCUSSION
In the present study, we have quantified the inhibitory effect of Rib in a SARS-CoV-2-Vero E6 infection model and analyzed, by high-resolution ultradeep sequencing, the mutation and deletion repertoire that resulted from virus replication in the presence of Rib in a single infection initiated at a low MOI. The therapeutic index (TI > 10) is comparable to that calculated for Rib on hepatitis C virus (TI = 12.8 [35]). Rib increased the number of point mutations but not of deletions compared with virus grown in parallel in the absence of drug. Four of the 11 different deletions present in the mutant spectra of virus replicated in the absence or presence of ribavirin affected the furin-like cleavage site, as an expected outcome of SARS-CoV-2 replication in Vero cells (51, 52).
The diversity indices also reflected the mutagenic activity of ribavirin. In addition to the expected lack of increase of Hsi in populations replicated in the presence of ribavirin, it is noteworthy that indices 1D and 2D (Hill numbers for q = 1 and q = 2, respectively) show a decreasing trend in the effect of Rib (Fig. 5). As q increases, the D value becomes progressively less sensitive to rare haplotypes, whose frequency is a good marker of lethal mutagenesis (53). The index comparisons confirm also, for a mutagenized viral population, the correlation of 1D and 2D with Shannon entropy (Hs) and Hsi, respectively (50). This was revealed by a minimal (or absent) effect of Rib on Hsi and 2D, in contrast with a significant increase of Hs and 1D. Thus, nonuniform effects on diversity indices constitute a complementary diagnosis of mutagenic effects of drugs on viral populations. The 2- to 11-fold Rib-mediated increase of maximum mutation frequency (Mfmax) and minimum mutation frequency (Mfmin) on SARS-CoV-2 (Fig. 5 and Table S4 posted at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX) is comparable to the increases observed with mutagenic base and nucleoside analogues acting on other RNA viruses for which early molecular cloning and Sanger sequencing data are available (such values are reviewed in reference 16).
The dominant number of A→G and U→C transitions (Fig. 6 and Table S3 at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX) coincides with the ranking of mutation preferences quantified in SARS-CoV-2 mutant spectra of COVID-19 patients [(U→C) > (A→G) ≥ (C→U)] (48). Thus, Rib forced an increase in the mutational types which are the most frequent also in diagnostic samples of the virus. The percentage of nonsynonymous mutations in the nsp12 (polymerase)-coding region was 53.5% in nonmutagenized (control) populations and 63.7% in Rib-mutagenized populations. The corresponding values for the spike-coding region were 64.7% and 57.2%, respectively. Only one nonsense point mutation was detected in the spike-coding region in the population passaged in the presence of 150 μM Rib. The complete information on mutation types is given in Table S3 posted at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX.
The major mutation types (A→G and U→C) induced by Rib on SARS-CoV-2 are opposite to those preferentially induced by Rib on other RNA viruses. A dominance of G→A and C→U transitions induced by Rib (comparing mutant spectra with those of the corresponding viruses passaged in the absence of drug) has been documented for Hantaan virus (54), Usutu virus (36), West Nile virus (26), poliovirus (19), foot-and-mouth disease virus (37, 55, 56), lymphocytic choriomeningitis virus (27), and HCV (35). In contrast, Rib produced an excess of A→G and U→C on Zika virus RNA, with a ratio (A→G) + (C→U)/(G→A) + (C→U) of around 2, lower than the range of 9.88 to 69.00 obtained for SARS-CoV-2 (Fig. 6). The mutational pattern renders unlikely that the Rib-mediated mutagenesis was the result of activated cellular editing activities. APOBEC enzymes evoke an excess of C→U transitions (57). The frequency of A→G transitions is unlikely to be explained by ADAR-promoted viral genome variation (58). Moreover, contrary to the observed preference of a G residue at the 5′ side of the A→G mutated sites found in our analysis (Fig. 6B), G at the 5′ side is not a preferred residue for the ADAR-edited sites (59, 60). The preference for A→G and U→C mutations suggests that during SARS-CoV-2 RNA synthesis, Rib-triphosphate competed more favorably with ATP than with GTP for incorporation. In line with our mutational analysis, a predictive in silico study of molecular docking suggested a higher affinity of Rib-triphosphate than ATP for the SARS-CoV-2 polymerase (61).
The molecular basis of the different mutational preferences for the same mutagen acting on different viruses is unknown. Competitive preferences between standard and modified nucleotides are influenced by several structural and environmental factors (reviewed in reference 62), which include the electronic structures of the analogues (63), and by relative affinities for standard and modified nucleotides dictated by the active site of the polymerases and neighbor residues (64, 65). Whatever their origin, such preferences may inform of potential synergies between lethal mutagens, as those described for HCV (66), Hantaan virus (67), and Junin virus (12). Synergy may be favored by the individual nucleotide analogues acting on nonidentical steps of cellular metabolism or the virus replication cycle, despite the analogues sharing a mutagenic activity, resulting in reinforcement of the inhibition. Synergy increase may also come about by different types of induced mutations, for example, as a result of combining pyrimidine and purine analogues, or different preferred template sites where mutations are introduced in the RNA product. For example, despite ribavirin and favipiravir sharing a preference to produce G→A and C→U transitions in HCV, favipiravir, but not ribavirin, favored the introduction of G→A mutations when A was present at the 5′ side of the mutation site and C→U mutations when U was present at the 3′ side of the mutation site; such a difference might have contributed to the strong synergy between the two analogues (66). A mutation bias—not only the increase in mutation load—is, by itself, a factor of vulnerability to lethal mutagenesis. This was documented with a triple FMDV polymerase mutant that conferred resistance to ribavirin through modulation of mutation types, which counteracted the mutational bias produced by ribavirin (56). A mutation-type modulation operated also as a mechanism of FMDV resistance to 5-fluorouracil (68). Therefore, detailed deep sequencing-based screening of mutational biases and mutation site preferences may be informative of potential synergism between lethal mutagens.
The effective mutagenic activity of Rib suggests that ExoN of SARS-CoV-2 might be less effective than ExoN of other coronaviruses in excising Rib incorporated into nascent RNA (69). The SARS-CoV-2 ExoN is active in vitro in removing misincorporated nucleotides and some analogues (70–73). The Rib-enhanced SARS-CoV-2 mutation frequency in our cell culture system suggests that the ExoN was ineffective in removing terminally incorporated Rib-monophosphate or that it was insufficient to cope with all incorporated Rib-monophosphate residues. Other possibilities are that Rib or some of its metabolites inhibit ExoN activity or the interaction between nsp14 and the nsp12-nsp7-nsp8 polymerase complex. Additional work is needed to clarify this point.
Molnupiravir appears to evoke mainly G→A and C→U in murine hepatitis virus (MHV) and MERS-CoV (39, 74). The same mutational bias was predicted for molnupiravir-mutagenized SARS-CoV-2 by combining a biochemical and structural approach (42, 75). Furthermore, C→U transitions were the most frequent mutation type in virus from swab nasopharyngeal samples of Omicron BA.2.2-infected patients that were treated with molnupiravir (76). These data suggest the possibility of synergistic activity between molnupiravir and Rib for SARS-CoV-2.
Administration of lower doses of an individual mutagenic analogue, which becomes possible when synergisms operate, may reduce the side effects which are well established for ribavirin and other polymerase inhibitors used in the clinic. For example, contradictory information has been reported for a potential mutagenic activity of molnupiravir (or its metabolites) on the host cells (77–79). In favor of pursuing clinical trials with ribavirin against COVID-19 are the promising results with Rib monotherapy administered to critically ill patients (44) and the potential of finding synergistic combinations suggested by the mutational spectrum revealed by our study. Additional supportive considerations are the balance between risk and benefit and the transient exposure to inhibitors in the case of acute respiratory infections (17, 79). An anti-COVID-19 strategy of synergistic combinations with inclusion of ribavirin would follow the paths of successful treatments of other RNA viral pathogens, such as HCV, for which Rib continues being part of some direct-acting drug combinations (80).
MATERIALS AND METHODS
Cells and virus.
Vero E6 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Merck) supplemented with 1 mM sodium pyruvate (Merck), 1% nonessential amino acids (Merck), 4 mM l-glutamine (Merck), 50 μg/mL gentamicin (PanReac), 0.2 μg/mL antifungal (Sigma), and 10% fetal bovine serum (FBS; Sigma). Cells were cultured at 37°C and 5% CO2, and they were periodically thawed from a large frozen stock and passaged a maximum of 30 times at a split ratio of 1:6 to 1:8.
The virus used in the experiments was USA-WA1/2020, NR-52281 (deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH) (SRA accession no. NR-52281_70036318). To prepare a virus stock, 3 × 106 Vero E6 cells were infected with the virus at a multiplicity of infection (MOI) of 0.001 PFU/cell in DMEM supplemented with 25 mM HEPES (PanReac) and 2% FBS, and the infection was allowed to proceed for 48 h at 37°C. The titer of the viral stock was 1 × 107 PFU/mL. Virus infections were performed following standard procedures using closed flasks. To control for the absence of contamination, the supernatants of mock-infected cells, which were maintained in parallel with the infected cultures, were titrated; no infectivity in the mock-infected cultures was detected in any of the experiments.
Virus titration.
Virus titrations were performed in Vero E6 cells following standard procedures using plates sealed in plastic bags. For titration of infectious SARS-CoV-2, cell culture supernatants were serially diluted and applied to 1 × 106 Vero E6 cells. After 1 h adsorption with gentle stirring every 15 min, the inoculum was removed, and medium containing DMEM 2×, agar 1% (Gibco), 1% FBS, and 1% DEAE-Dextran (Sigma) was added to the plates. After 72 h, cells were fixed with 10% formaldehyde (PanReac) for 1 h and then stained with 2% crystal violet (Merck) in formaldehyde.
Infections in the presence of ribavirin.
Rib (Sigma) solutions were prepared in phosphate-buffered saline (PBS), sterilized by filtration, and stored at −70°C. Prior to use, the stock solutions were diluted in DMEM to reach the desired concentration. Vero E6 cells were pretreated with the appropriate concentrations (or with DMEM without drug) for 16 h prior to infection. Then, 2 × 105 Vero E6 cells were infected (or mock infected) with USA-WA1/2020 (MOI of 0.001 PFU/cell); the adsorption time was 1 h, and the infection continued for 48 h in the absence or presence of the drug.
Drug toxicity and inhibitory activity assays.
Rib toxicity was measured in Vero E6 cells by seeding 1 × 104 cells in 96-well plates and adding different drug concentrations to the cells for 72 h. Then, cells were incubated for 4 h with MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] (Sigma) at a final concentration of 500 μg/mL. Then, 100 μL of dimethyl sulfoxide (DMSO) (Merck) was added to the cells, and the optical density at 550 nm was measured. The drug concentration required for 50% cell killing (CC50) was calculated as previously described (81).
Rib inhibitory activity was determined by seeding 1 × 105 Vero E6 cells in 24-well plates, and cells were treated with Rib for 16 h before the infection. Then, the cells were infected with SARS-CoV-2 at an MOI of 0.001 PFU/cell. After 1 h of incubation at 37°C, the inoculum was removed, and medium with or without different concentrations of Rib was added to each well. The infections were maintained for 48 h. Viral titers were determined, and the drug concentration required for 50% inhibition of infectious SARS-CoV-2 yield (IC50) was calculated using the program CompuSyn. CC50 and IC50 values were calculated as the average of the results of at least three determinations.
RNA extraction and SARS-CoV-2 amplification.
Total RNA was extracted from 140 μL of cell culture supernatants using the QIAamp viral RNA minikit (250) (Qiagen) as specified by the manufacturer. Amplifications of nsp12 (polymerase)- and S-coding regions were performed by reverse transcriptase PCR (RT-PCR). Each region was amplified from 5 μL of the RNA preparation by RT-PCR using Transcriptor one-step RT-PCR kit (Roche Applied Science). To perform the RT-PCR, 5 μL of the preparation was mixed with 10 μL of 5× buffer, 2 μL of a solution containing the forward primer, 2 μL of a solution with the reverse primer (50 ng/μL, each) (Table S1 at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX), and 1 μL of polymerase. Reaction parameters were 50°C for 30 min for the reverse transcription, an initial denaturing step at 94°C for 7 min, followed by 35 cycles of a denaturing step at 94°C for 10 s, an annealing step at 46 to 48°C for 30 s, an extension step at 68°C for 40 s, and then a final extension at 68°C for 7 min. Negative controls (amplification reactions in the absence of RNA) were included in parallel to ascertain the absence of contamination by template nucleic acids. Amplification products were analyzed by 2% agarose gel electrophoresis, using Gene Ruler 1-kb Plus DNA ladder (Thermo Scientific) as molar mass standard. PCR products were purified (QIAquick gel extraction kit; Qiagen), quantified (Qubit double-stranded DNA [dsDNA] assay kit; Thermo Fisher Scientific), and tested for quality (TapeStation system, Agilent Technologies) prior to sequencing using the Illumina MiSeq platform. Dilutions of 1:10, 1:100, and 1:1,000 of the initial RNA preparation and subsequent amplification by RT-PCR were carried out for one patient of each disease severity. When amplification with the 1:1,000 dilution of the template produced a visible DNA band, the ultradeep sequencing analysis was performed with the undiluted template to avoid redundant copying of the same template molecules, as we have previously documented (66, 82).
Ultradeep sequencing.
PCR products were adjusted to 4 × 109 molecules/μL before generating DNA pools that were purified using Kapa pure beads (Kapabiosystems; Roche), quantified using Qubit as previously described (48, 83–85), and then adjusted at 1.5 ng/μL. Purified DNA pools were further processed using the DNA library preparation kit Kapa Hyper Prep kit (Roche), during which each pool was indexed using SeqCap adapter kit A/B (Nimblegen) (24 index). Each DNA pool was quantified by LightCycler 480 and sequenced using MiSeq sequencing platform with MiSeq reagent kit v3 (2 × 300-bp mode with the 600 cycle kit) (Illumina).
Bioinformatics processing of deep sequencing data.
To establish the reliability of deep sequencing data to characterize mutant spectra of viral populations, we previously carried out several experimental and bioinformatics controls to show that conclusions on mutant spectrum complexity from molecular cloning and Sanger sequencing were coherent with the conclusions reached by pyrosequencing (454 Life Sciences/Roche, Brandford, CT) (86). Then, the coherence was tested between results obtained by pyrosequencing and the MiSeq Illumina platform (83). In addition, controls to establish basal error rate, reproducibility of clean read composition upon subjecting different aliquots of the same sample to deep sequencing, detection of minority variants from reconstructed mixtures of mutant HCV RNAs, and read coverage requirement to attain a given mutation frequency cutoff were performed and have been previously reported (46, 83, 84, 87, 88). Of particular relevance to the present analysis of SARS-CoV-2 replicated in the absence and presence of Rib is that the number of clean reads per amplicon (A1 to A6) ranged from 83,593 to 177,211, with average values ranging from 91,759 to 128,426 (see Table S2 posted at https://saco.csic.es/index.php/s/So94ey5ECYgMZdX). The average quality score (Q) was higher than 70%. The coverage and Q attained allowed the establishment of a mutation frequency cutoff of 0.1% for read analysis (46).
In addition to read coverage, the following quantifications validated the extension from a 0.5% to a 0.1% for mutant spectrum analysis of SARS-CoV-2: (i) detection of 98.9% of the different point mutations and 100% of the deletions scored both with a 0.5% and a 0.1% cutoff; (ii) an agreement of 80% in the number of positions with or without mutations in the regions which overlap among neighbor amplicons; (iii) preservation at the two cutoff levels of the mutational bias and ranking of mutations in the three codon positions that characterize SARS-CoV-2 mutant spectra; (iv) similar preservation of acceptability scores (PAM250 metric) of the amino acid substitutions deduced from haplotype mutations determined with 0.1% and 0.5% cutoffs; (v) a similar percentage (of around 97%) of amino acid substitutions deduced with the two mutation frequency cutoffs were present in the “outbreak.info” database; and, finally, (vi) when increasing the detection capacity from 0.5% to 0.1%, there was a statistically significant increase in the number of amino acid substitutions, which, according to the SNAP2 (Screening for Non-Acceptable Polymorphisms) predictor, have a predicted functional effect (89). Such an increase is incompatible with being a consequence of sequencing errors, and it is expected from deleterious amino acid substitutions populating low-frequency (low-fitness) SARS-CoV-2 genomes (90). Data on the six points listed above to validate the 0.1% mutation frequency cutoff have been published (46, 48, 49).
FASTQ data were analyzed using the SeekDeep pipeline (91) with the following options: “--extraExtractorCmds=-- checkRevComplementForPrimers –primerNumOfMismatches 3” “—extraProcessClusterCmds=–fracCutOff0.001 --rescueExcludedOneOffLowFreqHaplotypes.”
Diversity indices.
For the study of the genetic diversity of viral populations, several indices were determined, typical of the field of ecology and common application also to virology (defined and explained in reference 50). Since no diversity index can fully capture the complexity of a viral population, a multivariate analysis was performed, examining the behavior of different indices for the same data set. These indices were Hpl, number of haplotypes; dHpl (Hpl/N), number of haplotypes normalized to the number of reads; nM, number of mutations; Hs, Shannon entropy; HsN, Shannon entropy normalized to the logarithm of the number of reads; HsH, Shannon entropy normalized to the logarithm of the number of haplotypes; Hsi, Simpson index; Hgs, Gini-Simpson index; qD, Hill number for q = 1, 2; Mfmax, maximum mutation frequency; Mfmin, minimum mutation frequency; and π, nucleotide diversity. The diversity indices were calculated according to references 50 and 92 and with an algorithm implemented in the C programming language.
Statistics.
The statistical significance of different comparisons was calculated by the proportion test. One-way analysis of variance (ANOVA) followed by Dunnett’s test was used to compare the differences in the viral titer between the control population and different concentrations of Rib using GraphPad 8.0.2. The statistical significance of differences between diversity indices was calculated with a Wilcoxon test, using R software version 4.0.2.
Ethics approval and consent to participate.
All experiments with SARS-CoV-2 were performed in biosafety level 3 (BSL-3) facilities at Centro de Biología Molecular Severo Ochoa (CBMSO) (Consejo Superior de Investigaciones Científicas-Autonomous University of Madrid [CSIC-UAM]) according to the guidelines set forth by the institution. This study was approved by the Ethics Committee 082/2021 from CSIC.
Data availability.
FASTQ files of SARS-CoV-2 samples included in this study are available in ENA under accession no. PRJEB56440.
ACKNOWLEDGMENTS
This work was supported by Instituto de Salud Carlos III, Spanish Ministry of Science and Innovation (COVID-19 Research Call COV20/00181) and cofinanced by the European Development Regional Fund, “A way to achieve Europe.” The work was also supported by grants CSIC-COV19-014 from Consejo Superior de Investigaciones Científicas (CSIC), project 525/C/2021 from Fundació La Marató de TV3, PID2020-113888RB-I00 and 202220I116 from Ministerio de Ciencia e Innovación, BFU2017-91384-EXP from Ministerio de Ciencia, Innovación y Universidades (MCIU), PI18/00210 and PI21/00139 from Instituto de Salud Carlos III, and S2018/BAA-4370 (PLATESA2 from Comunidad de Madrid/FEDER). This research work was also funded by the European Commission-NextGenerationEU (regulation EU 2020/2024) through the CSIC’s Global Health Platform (PTI Salud Global). C.P. is supported by the Miguel Servet program of the Instituto de Salud Carlos III (CP14/00121 and CPII19/00001), cofinanced by the European Regional Development Fund (ERDF). CIBERehd (Centro de Investigación en Red de Enfermedades Hepáticas y Digestivas) is funded by Instituto de Salud Carlos III. Institutional grants from the Fundación Ramón Areces and Banco Santander to the CBMSO are also acknowledged. The team at CBMSO belongs to the Global Virus Network (GVN). C.G.-C. is supported by predoctoral contract PRE2018-083422 from MCIU. P.S. is supported by postdoctoral contract Margarita Salas, CA1/RSUE/2021 from MCIU. B.M.-G. is supported by predoctoral contract PFIS FI19/00119 from ISCIII, cofinanced by Fondo Social Europeo (FSE).
Contributor Information
Esteban Domingo, Email: edomingo@cbm.csic.es.
Celia Perales, Email: celia.perales@cnb.csic.es.
REFERENCES
- 1.Eigen M, Schuster P. 1979. The hypercycle. In A principle of natural self-organization. Springer, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 2.Nowak MA, Schuster P. 1989. Error thresholds of replication in finite populations mutation frequencies and the onset of Muller's ratchet. J Theor Biol 137:375–395. 10.1016/S0022-5193(89)80036-0. [DOI] [PubMed] [Google Scholar]
- 3.Schuster P. 2016. Quasispecies on fitness landscapes. Curr Top Microbiol Immunol 392:61–120. 10.1007/82_2015_469. [DOI] [PubMed] [Google Scholar]
- 4.Lynch M, Gabriel W. 1990. Mutation load and the survival of small populations. Evolution 44:1725–1737. 10.1111/j.1558-5646.1990.tb05244.x. [DOI] [PubMed] [Google Scholar]
- 5.Lynch M, Burger R, Butcher D, Gabriel W. 1993. The mutational meltdown in asexual populations. J Hered 84:339–344. 10.1093/oxfordjournals.jhered.a111354. [DOI] [PubMed] [Google Scholar]
- 6.Jensen JD, Stikeleather RA, Kowalik TF, Lynch M. 2020. Imposed mutational meltdown as an antiviral strategy. Evolution 74:2549–2559. 10.1111/evo.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holland JJ, Domingo E, de la Torre JC, Steinhauer DA. 1990. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J Virol 64:3960–3962. 10.1128/JVI.64.8.3960-3962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeb LA, Essigmann JM, Kazazi F, Zhang J, Rose KD, Mullins JI. 1999. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc Natl Acad Sci USA 96:1492–1497. 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sierra S, Dávila M, Lowenstein PR, Domingo E. 2000. Response of foot-and-mouth disease virus to increased mutagenesis: influence of viral load and fitness in loss of infectivity. J Virol 74:8316–8323. 10.1128/jvi.74.18.8316-8323.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eigen M. 2002. Error catastrophe and antiviral strategy. Proc Natl Acad Sci USA 99:13374–13376. 10.1073/pnas.212514799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biebricher CK, Eigen M. 2005. The error threshold. Virus Res 107:117–127. 10.1016/j.virusres.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Zadeh VR, Afowowe TO, Abe H, Urata S, Yasuda J. 2022. Potential and action mechanism of favipiravir as an antiviral against Junin virus. PLoS Pathog 18:e1010689. 10.1371/journal.ppat.1010689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crotty S, Maag D, Arnold JJ, Zhong W, Lau JYN, Hong Z, Andino R, Cameron CE. 2000. The broad-spectrum antiviral ribonucleotide, ribavirin, is an RNA virus mutagen. Nat Med 6:1375–1379. 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 14.Arias A, Thorne L, Goodfellow I. 2014. Favipiravir elicits antiviral mutagenesis during virus replication in vivo. Elife 3:e03679. 10.7554/eLife.03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guedj J, Piorkowski G, Jacquot F, Madelain V, Nguyen THT, Rodallec A, Gunther S, Carbonnelle C, Mentre F, Raoul H, de Lamballerie X. 2018. Antiviral efficacy of favipiravir against Ebola virus: a translational study in cynomolgus macaques. PLoS Med 15:e1002535. 10.1371/journal.pmed.1002535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perales C, Gallego I, de Avila AI, Soria ME, Gregori J, Quer J, Domingo E. 2019. The increasing impact of lethal mutagenesis of viruses. Future Med Chem 11:1645–1657. 10.4155/fmc-2018-0457. [DOI] [PubMed] [Google Scholar]
- 17.Hadj Hassine I, Ben M’hadheb M, Menéndez-Arias L. 2022. Lethal mutagenesis of RNA viruses and approved drugs with antiviral mutagenic activity. Viruses 14:841. 10.3390/v14040841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidwell OW, Simon LN, Witkowski JT, Robins RK. 1974. Antiviral activity of virazole: review and structure-activity relationships. Prog Chemotherapy 2:889–903. [Google Scholar]
- 19.Crotty S, Cameron CE, Andino R. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci USA 98:6895–6900. 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beaucourt S, Vignuzzi M. 2014. Ribavirin: a drug active against many viruses with multiple effects on virus replication and propagation. Molecular basis of ribavirin resistance. Curr Opin Virol 8:10–15. 10.1016/j.coviro.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Clercq E, Li G. 2016. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev 29:695–747. 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geraghty RJ, Aliota MT, Bonnac LF. 2021. Broad-spectrum antiviral strategies and nucleoside analogues. Viruses 13:667. 10.3390/v13040667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Severson WE, Schmaljohn CS, Javadian A, Jonsson CB. 2003. Ribavirin causes error catastrophe during Hantaan virus replication. J Virol 77:481–488. 10.1128/jvi.77.1.481-488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonsson CB, Milligan BG, Arterburn JB. 2005. Potential importance of error catastrophe to the development of antiviral strategies for hantaviruses. Virus Res 107:195–205. 10.1016/j.virusres.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Chung DH, Vastermark A, Camp JV, McAllister R, Remold SK, Chu YK, Bruder C, Jonsson CB. 2013. The murine model for Hantaan virus-induced lethal disease shows two distinct paths in viral evolutionary trajectory with and without ribavirin treatment. J Virol 87:10997–11007. 10.1128/JVI.01394-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Day CW, Smee DF, Julander JG, Yamshchikov VF, Sidwell RW, Morrey JD. 2005. Error-prone replication of West Nile virus caused by ribavirin. Antiviral Res 67:38–45. 10.1016/j.antiviral.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 27.Moreno H, Gallego I, Sevilla N, de la Torre JC, Domingo E, Martin V. 2011. Ribavirin can be mutagenic for arenaviruses. J Virol 85:7246–7255. 10.1128/JVI.00614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanford RE, Chavez D, Guerra B, Lau JY, Hong Z, Brasky KM, Beames B. 2001. Ribavirin induces error-prone replication of GB virus B in primary tamarin hepatocytes. J Virol 75:8074–8081. 10.1128/jvi.75.17.8074-8081.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Todt D, Gisa A, Radonic A, Nitsche A, Behrendt P, Suneetha PV, Pischke S, Bremer B, Brown RJ, Manns MP, Cornberg M, Bock CT, Steinmann E, Wedemeyer H. 2016. In vivo evidence for ribavirin-induced mutagenesis of the hepatitis E virus genome. Gut 65:1733–1743. 10.1136/gutjnl-2015-311000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Contreras AM, Hiasa Y, He W, Terella A, Schmidt EV, Chung RT. 2002. Viral RNA mutations are region specific and increased by ribavirin in a full-length hepatitis C virus replication system. J Virol 76:8505–8517. 10.1128/jvi.76.17.8505-8517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asahina Y, Izumi N, Enomoto N, Uchihara M, Kurosaki M, Onuki Y, Nishimura Y, Ueda K, Tsuchiya K, Nakanishi H, Kitamura T, Miyake S. 2005. Mutagenic effects of ribavirin and response to interferon/ribavirin combination therapy in chronic hepatitis C. J Hepatol 43:623–629. 10.1016/j.jhep.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 32.Cuevas JM, Gonzalez-Candelas F, Moya A, Sanjuan R. 2009. Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo. J Virol 83:5760–5764. 10.1128/JVI.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lutchman G, Danehower S, Song BC, Liang TJ, Hoofnagle JH, Thomson M, Ghany MG. 2007. Mutation rate of the hepatitis C virus NS5B in patients undergoing treatment with ribavirin monotherapy. Gastroenterology 132:1757–1766. 10.1053/j.gastro.2007.03.035. [DOI] [PubMed] [Google Scholar]
- 34.Dietz J, Schelhorn SE, Fitting D, Mihm U, Susser S, Welker MW, Fuller C, Daumer M, Teuber G, Wedemeyer H, Berg T, Lengauer T, Zeuzem S, Herrmann E, Sarrazin C. 2013. Deep sequencing reveals mutagenic effects of ribavirin during monotherapy of hepatitis C virus genotype 1-infected patients. J Virol 87:6172–6181. 10.1128/JVI.02778-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortega-Prieto AM, Sheldon J, Grande-Perez A, Tejero H, Gregori J, Quer J, Esteban JI, Domingo E, Perales C. 2013. Extinction of hepatitis C virus by ribavirin in hepatoma cells involves lethal mutagenesis. PLoS One 8:e71039. 10.1371/journal.pone.0071039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bassi MR, Sempere RN, Meyn P, Polacek C, Arias A. 2018. Extinction of Zika virus and Usutu virus by lethal mutagenesis reveals different patterns of sensitivity to three mutagenic drugs. Antimicrob Agents Chemother 62:e00380-18. 10.1128/AAC.00380-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Airaksinen A, Pariente N, Menendez-Arias L, Domingo E. 2003. Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis. Virology 311:339–349. 10.1016/s0042-6822(03)00144-2. [DOI] [PubMed] [Google Scholar]
- 38.Lai CC, Chao CM, Hsueh PR. 2021. Clinical efficacy of antiviral agents against coronavirus disease 2019: a systematic review of randomized controlled trials. J Microbiol Immunol Infect 54:767–775. 10.1016/j.jmii.2021.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agostini ML, Pruijssers AJ, Chappell JD, Gribble J, Lu X, Andres EL, Bluemling GR, Lockwood MA, Sheahan TP, Sims AC, Natchus MG, Saindane M, Kolykhalov AA, Painter GR, Baric RS, Denison MR. 2019. Small-molecule antiviral beta-d-N (4)-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J Virol 93:e01348-19. 10.1128/JVI.01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J, Guo S, Yi D, Li Q, Ma L, Zhang Y, Wang J, Li X, Guo F, Lin R, Liang C, Liu Z, Cen S. 2021. A cell-based assay to discover inhibitors of SARS-CoV-2 RNA dependent RNA polymerase. Antiviral Res 190:105078. 10.1016/j.antiviral.2021.105078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Painter WP, Holman W, Bush JA, Almazedi F, Malik H, Eraut N, Morin MJ, Szewczyk LJ, Painter GR. 2021. Human safety, tolerability, and pharmacokinetics of molnupiravir, a novel broad-spectrum oral antiviral agent with activity against SARS-CoV-2. Antimicrob Agents Chemother 65. 10.1128/AAC.02428-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon CJ, Tchesnokov EP, Schinazi RF, Gotte M. 2021. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J Biol Chem 297:100770. 10.1016/j.jbc.2021.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. 2020. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30:269–271. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Y, Li M, Zhou L, Liu D, He W, Liang W, Sun Q, Sun H, Li Y, Liu X. 2021. Ribavirin treatment for critically ill COVID-19 patients: an observational study. Infect Drug Resist 14:5287–5291. 10.2147/IDR.S330743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Unal MA, Bitirim CV, Summak GY, Bereketoglu S, Cevher Zeytin I, Besbinar O, Gurcan C, Aydos D, Goksoy E, Kocakaya E, Eran Z, Murat M, Demir N, Aksoy Ozer ZB, Somers J, Demir E, Nazir H, Ozkan SA, Ozkul A, Azap A, Yilmazer A, Akcali KC. 2021. Ribavirin shows antiviral activity against SARS-CoV-2 and downregulates the activity of TMPRSS2 and the expression of ACE2 in vitro. Can J Physiol Pharmacol 99:449–460. 10.1139/cjpp-2020-0734. [DOI] [PubMed] [Google Scholar]
- 46.Martinez-Gonzalez B, Soria ME, Vazquez-Sirvent L, Ferrer-Orta C, Lobo-Vega R, Minguez P, de la Fuente L, Llorens C, Soriano B, Ramos-Ruiz R, Corton M, Lopez-Rodriguez R, Garcia-Crespo C, Somovilla P, Duran-Pastor A, Gallego I, de Avila AI, Delgado S, Moran F, Lopez-Galindez C, Gomez J, Enjuanes L, Salar-Vidal L, Esteban-Munoz M, Esteban J, Fernandez-Roblas R, Gadea I, Ayuso C, Ruiz-Hornillos J, Verdaguer N, Domingo E, Perales C. 2022. SARS-CoV-2 mutant spectra at different depth levels reveal an overwhelming abundance of low frequency mutations. Pathogens 11:662. 10.3390/pathogens11060662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muller WE, Maidhof A, Taschner H, Zahn RK. 1977. Virazole (1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide; a cytostatic agent. Biochem Pharmacol 26:1071–1075. 10.1016/0006-2952(77)90246-5. [DOI] [PubMed] [Google Scholar]
- 48.Martinez-Gonzalez B, Soria ME, Vazquez-Sirvent L, Ferrer-Orta C, Lobo-Vega R, Minguez P, de la Fuente L, Llorens C, Soriano B, Ramos R, Corton M, Lopez-Rodriguez R, Garcia-Crespo C, Gallego I, de Avila AI, Gomez J, Enjuanes L, Salar-Vidal L, Esteban J, Fernandez-Roblas R, Gadea I, Ayuso C, Ruiz-Hornillos J, Verdaguer N, Domingo E, Perales C. 2022. SARS-CoV-2 point mutation and deletion spectra and their association with different disease outcomes. Microbiol Spectr 10:e00221-22. 10.1128/spectrum.00221-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez-Gonzalez B, Vazquez-Sirvent L, Soria ME, Minguez P, Salar-Vidal L, Garcia-Crespo C, Gallego I, Avila A, Llorens C, Soriano B, Ramos-Ruiz R, Esteban J, Fernandez-Roblas R, Gadea I, Ayuso C, Ruiz-Hornillos J, Perez-Jorge C, Domingo E, Perales C. 2022. Vaccine-breakthrough infections with SARS-CoV-2 Alpha mirror mutations in Delta Plus, Iota and Omicron. J Clin Invest 132:e157700. 10.1172/JCI157700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gregori J, Perales C, Rodriguez-Frias F, Esteban JI, Quer J, Domingo E. 2016. Viral quasispecies complexity measures. Virology 493:227–237. 10.1016/j.virol.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 51.Hossain MG, Tang YD, Akter S, Zheng C. 2022. Roles of the polybasic furin cleavage site of spike protein in SARS-CoV-2 replication, pathogenesis, and host immune responses and vaccination. J Med Virol 94:1815–1820. 10.1002/jmv.27539. [DOI] [PubMed] [Google Scholar]
- 52.Sasaki M, Uemura K, Sato A, Toba S, Sanaki T, Maenaka K, Hall WW, Orba Y, Sawa H. 2021. SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog 17:e1009233. 10.1371/journal.ppat.1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gregori J, Soria ME, Gallego I, Guerrero-Murillo M, Esteban JI, Quer J, Perales C, Domingo E. 2018. Rare haplotype load as marker for lethal mutagenesis. PLoS One 13:e0204877. 10.1371/journal.pone.0204877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung DH, Sun Y, Parker WB, Arterburn JB, Bartolucci A, Jonsson CB. 2007. Ribavirin reveals a lethal threshold of allowable mutation frequency for Hantaan virus. J Virol 81:11722–11729. 10.1128/JVI.00874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sierra M, Airaksinen A, González-López C, Agudo R, Arias A, Domingo E. 2007. Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: implications for error catastrophe. J Virol 81:2012–2024. 10.1128/JVI.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agudo R, Ferrer-Orta C, Arias A, de la Higuera I, Perales C, Perez-Luque R, Verdaguer N, Domingo E. 2010. A multi-step process of viral adaptation to a mutagenic nucleoside analogue by modulation of transition types leads to extinction-escape. PLoS Pathog 6:e1001072. 10.1371/journal.ppat.1001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ratcliff J, Simmonds P. 2021. Potential APOBEC-mediated RNA editing of the genomes of SARS-CoV-2 and other coronaviruses and its impact on their longer term evolution. Virology 556:62–72. 10.1016/j.virol.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mourier T, Sadykov M, Carr MJ, Gonzalez G, Hall WW, Pain A. 2021. Host-directed editing of the SARS-CoV-2 genome. Biochem Biophys Res Commun 538:35–39. 10.1016/j.bbrc.2020.10.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dawson TR, Sansam CL, Emeson RB. 2004. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J Biol Chem 279:4941–4951. 10.1074/jbc.M310068200. [DOI] [PubMed] [Google Scholar]
- 60.Eggington JM, Greene T, Bass BL. 2011. Predicting sites of ADAR editing in double-stranded RNA. Nat Commun 2:319. 10.1038/ncomms1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Celik I, Erol M, Duzgun Z. 2022. In silico evaluation of potential inhibitory activity of remdesivir, favipiravir, ribavirin and galidesivir active forms on SARS-CoV-2 RNA polymerase. Mol Divers 26:279–292. 10.1007/s11030-021-10215-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Domingo E, Garcia-Crespo C, Lobo-Vega R, Perales C. 2021. Mutation rates, mutation frequencies, and proofreading-repair activities in RNA virus genetics. Viruses 13:1882. 10.3390/v13091882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jena NR. 2020. Role of different tautomers in the base-pairing abilities of some of the vital antiviral drugs used against COVID-19. Phys Chem Phys 22:28115–28122. 10.1039/d0cp05297c. [DOI] [PubMed] [Google Scholar]
- 64.Long C, Romero ME, La Rocco D, Yu J. 2021. Dissecting nucleotide selectivity in viral RNA polymerases. Comput Struct Biotechnol J 19:3339–3348. 10.1016/j.csbj.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. 2007. Sequential structures provide insights into the fidelity of RNA replication. Proc Natl Acad Sci USA 104:9463–9468. 10.1073/pnas.0700518104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gallego I, Soria ME, Gregori J, de Avila AI, Garcia-Crespo C, Moreno E, Gadea I, Esteban J, Fernandez-Roblas R, Esteban JI, Gomez J, Quer J, Domingo E, Perales C. 2019. Synergistic lethal mutagenesis of hepatitis C virus. Antimicrob Agents Chemother 63:e01653-19. 10.1128/AAC.01653-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mayor J, Engler O, Rothenberger S. 2021. Antiviral efficacy of ribavirin and favipiravir against Hantaan virus. Microorganisms 9:1306. 10.3390/microorganisms9061306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de la Higuera I, Ferrer-Orta C, de Avila AI, Perales C, Sierra M, Singh K, Sarafianos SG, Dehouck Y, Bastolla U, Verdaguer N, Domingo E. 2017. Molecular and functional bases of selection against a mutation bias in an RNA virus. Genome Biol Evol 9:1212–1228. 10.1093/gbe/evx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferron F, Subissi L, Silveira De Morais AT, Le NTT, Sevajol M, Gluais L, Decroly E, Vonrhein C, Bricogne G, Canard B, Imbert I. 2018. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci USA 115:E162–E171. 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin S, Chen H, Chen Z, Yang F, Ye F, Zheng Y, Yang J, Lin X, Sun H, Wang L, Wen A, Dong H, Xiao Q, Deng D, Cao Y, Lu G. 2021. Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity. Nucleic Acids Res 49:5382–5392. 10.1093/nar/gkab320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma Z, Pourfarjam Y, Kim IK. 2021. Reconstitution and functional characterization of SARS-CoV-2 proofreading complex. Protein Expr Purif 185:105894. 10.1016/j.pep.2021.105894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scholle MD, Liu C, Deval J, Gurard-Levin ZA. 2021. Label-free screening of SARS-CoV-2 NSP14 exonuclease activity using SAMDI mass spectrometry. SLAS Discov 26:766–774. 10.1177/24725552211008854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moeller NH, Passow KT, Harki DA, Aihara H. 2022. SARS-CoV-2 nsp14 exoribonuclease removes the natural antiviral 3'-deoxy-3',4'-didehydro-cytidine nucleotide from RNA. Viruses 14:1790. 10.3390/v14081790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, Leist SR, Schafer A, Dinnon KH, 3rd, Stevens LJ, Chappell JD, Lu X, Hughes TM, George AS, Hill CS, Montgomery SA, Brown AJ, Bluemling GR, Natchus MG, Saindane M, Kolykhalov AA, Painter G, Harcourt J, Tamin A, Thornburg NJ, Swanstrom R, Denison MR, Baric RS. 2020. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med 12:eabb5883. 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kabinger F, Stiller C, Schmitzova J, Dienemann C, Kokic G, Hillen HS, Hobartner C, Cramer P. 2021. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat Struct Mol Biol 28:740–746. 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chan WS, Law JHY, Ho MKS, Chan TL, Ma ESK, Tang BSF. 2022. Genomic characteristics and viral load dynamics of a SARS-CoV-2 Omicron BA.2.2 variant from a hospitalized patient treated with molnupiravir. Infect Genet Evol 105:105376. 10.1016/j.meegid.2022.105376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou S, Hill CS, Sarkar S, Tse LV, Woodburn BMD, Schinazi RF, Sheahan TP, Baric RS, Heise MT, Swanstrom R. 2021. Beta-d-N4-hydroxycytidine inhibits SARS-CoV-2 through lethal mutagenesis but is also mutagenic to mammalian cells. J Infect Dis 224:415–419. 10.1093/infdis/jiab247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Githaka JM. 2022. Molnupiravir does not induce mutagenesis in host lung cells during SARS-CoV-2 treatment. Bioinform Biol Insights 16:11779322221085077. 10.1177/11779322221085077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waters MD, Warren S, Hughes C, Lewis P, Zhang F. 2022. Human genetic risk of treatment with antiviral nucleoside analog drugs that induce lethal mutagenesis: the special case of molnupiravir. Environ Mol Mutagen 63:37–63. 10.1002/em.22471. [DOI] [PubMed] [Google Scholar]
- 80.Hayes CN, Imamura M, Tanaka J, Chayama K. 2022. Road to elimination of HCV: clinical challenges in HCV management. Liver Int 42:1935–1944. 10.1111/liv.15150. [DOI] [PubMed] [Google Scholar]
- 81.Vandamme A, Witvrouw M, Pannecouque C, Balzarini J, Van Laethem K, Schmit J, Desmyter J, De Clercq E. 2000. Evaluating clinical isolates for their phenotypic and genotypic resistance against anti-HIV drugs. Methods Mol Med 24:223–258. 10.1385/1-59259-245-7:223. [DOI] [PubMed] [Google Scholar]
- 82.de Avila AI, Gallego I, Soria ME, Gregori J, Quer J, Esteban JI, Rice CM, Domingo E, Perales C. 2016. Lethal mutagenesis of hepatitis C virus induced by favipiravir. PLoS One 11:e0164691. 10.1371/journal.pone.0164691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Soria ME, Gregori J, Chen Q, Garcia-Cehic D, Llorens M, de Avila AI, Beach NM, Domingo E, Rodriguez-Frias F, Buti M, Esteban R, Esteban JI, Quer J, Perales C. 2018. Pipeline for specific subtype amplification and drug resistance detection in hepatitis C virus. BMC Infect Dis 18:446. 10.1186/s12879-018-3356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soria ME, Garcia-Crespo C, Martinez-Gonzalez B, Vazquez-Sirvent L, Lobo-Vega R, de Avila AI, Gallego I, Chen Q, Garcia-Cehic D, Llorens-Revull M, Briones C, Gomez J, Ferrer-Orta C, Verdaguer N, Gregori J, Rodriguez-Frias F, Buti M, Esteban JI, Domingo E, Quer J, Perales C. 2020. Amino acid substitutions associated with treatment failure for hepatitis C virus infection. J Clin Microbiol 58:e01985-20. 10.1128/JCM.01985-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Q, Perales C, Soria ME, García-Cehic D, Gregori J, Rodríguez-Frías F, Buti M, Crespo J, Calleja JL, Tabernero D, Vila M, Lázaro F, Rando-Segura A, Nieto-Aponte L, Llorens-Revull M, Cortese MF, Fernandez-Alonso I, Castellote J, Niubó J, Imaz A, Xiol X, Castells L, Riveiro-Barciela M, Llaneras J, Navarro J, Vargas-Blasco V, Augustin S, Conde I, Rubín Á, Prieto M, Torras X, Margall N, Forns X, Mariño Z, Lens S, Bonacci M, Pérez-Del-Pulgar S, Londoño MC, García-Buey ML, Sanz-Cameno P, Morillas R, Martró E, Saludes V, Masnou-Ridaura H, Salmerón J, Quíles R, Carrión JA, Forné M, Rosinach M, Fernández I, et al. 2020. Deep-sequencing reveals broad subtype-specific HCV resistance mutations associated with treatment failure. Antiviral Res 174:104694. 10.1016/j.antiviral.2019.104694. [DOI] [PubMed] [Google Scholar]
- 86.Moreno E, Gallego I, Gregori J, Lucia-Sanz A, Soria ME, Castro V, Beach NM, Manrubia S, Quer J, Esteban JI, Rice CM, Gomez J, Gastaminza P, Domingo E, Perales C. 2017. Internal disequilibria and phenotypic diversification during replication of hepatitis C virus in a noncoevolving cellular environment. J Virol 91:e02505-16. 10.1128/JVI.02505-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Quer J, Rodriguez-Frias F, Gregori J, Tabernero D, Soria ME, Garcia-Cehic D, Homs M, Bosch A, Pinto RM, Esteban JI, Domingo E, Perales C. 2017. Deep sequencing in the management of hepatitis virus infections. Virus Res 239:115–125. 10.1016/j.virusres.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 88.Perales C, Chen Q, Soria ME, Gregori J, Garcia-Cehic D, Nieto-Aponte L, Castells L, Imaz A, Llorens-Revull M, Domingo E, Buti M, Esteban JI, Rodriguez-Frias F, Quer J. 2018. Baseline hepatitis C virus resistance-associated substitutions present at frequencies lower than 15% may be clinically significant. Infect Drug Resist 11:2207–2210. 10.2147/IDR.S172226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hecht M, Bromberg Y, Rost B. 2015. Better prediction of functional effects for sequence variants. BMC Genomics 16:S1. 10.1186/1471-2164-16-S8-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Domingo E, Schuster P, Elena SF, Perales C. 2023. Viral fitness and evolution-population dynamics and adaptive mechanisms. Current topics in microbiology and immunology, vol 439. Springer Verlag GmbH, Heidelberg, Germany. [Google Scholar]
- 91.Hathaway NJ, Parobek CM, Juliano JJ, Bailey JA. 2018. SeekDeep: single-base resolution de novo clustering for amplicon deep sequencing. Nucleic Acids Res 46:e21. 10.1093/nar/gkx1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao L, Illingworth CJR. 2019. Measurements of intrahost viral diversity require an unbiased diversity metric. Virus Evol 5:vey041. 10.1093/ve/vey041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
FASTQ files of SARS-CoV-2 samples included in this study are available in ENA under accession no. PRJEB56440.