Abstract
Alcohol dehydrogenase (ADH) is important for preventing alcohol toxicity and developmental disorders, and may be involved in other diseases including neurodegenerative diseases. We found that the major acceptor protein of polyADP-ribosylation in a model organism of neurodegeneration using a Drosophila melanogaster mutant lacking poly(ADP-ribose) glycohydrolase, was ADH. Thus we postulated that human ADH activity might be regulated by polyADP-ribosylation, a post-translational modification. The radioactivity of [32P]NAD+ was incorporated into human ADH1 by human poly(ADP-ribose) polymerase 1 in vitro, but was not incorporated when heat-inactivated PARP1 or a PARP inhibitor, 3-aminobenzamide, was used. The incorporated radioactivity was not released from ADH1 protein in the presence of excess amount of ADP-ribose or poly(ADP-ribose) as competitors. However, it was released by incubation with 1 M neutral NH2OH or 0.1 N NaOH, but was not with 0.1 N HCl, suggesting the bond between ADH1 and poly(ADP-ribose) is an ester linkage. When HepG2 cells, a human hepatoma cell line, were cultured in the presence of another PARP inhibitor, olaparib, ADH activity of the cell was significantly increased. These results suggest that polyADP-ribosylation could regulate ADH activity in vivo and might be involved in neurodegeneration.
Keywords: ADH1, Drosophila melanogaster, Human hepatoma cells, Neurodegeneration, PARP1
1. Introduction
Alcohol dehydrogenases (ADH) can metabolize various substrates including alcohols, aldehydes, retinoids and lipid peroxidation-derived carbonyls [1]. Among 5 classes of ADH genes in human and rodents, most studies on ADH have focused on three classes, class I ADH (ADH1), class III ADH (ADH3) and class IV ADH (ADH4), which are highly conserved in all mammalian species [1]. ADH1 is mainly distributed in the liver and catalyzes oxidation of ethanol and retinol and also reduction of lipid peroxidation products [2–4]. ADH3 is found in every tissues and from kinetic studies it catalyzes omega-hydroxy fatty acids in addition to function as formaldehyde dehydrogenase [3]. ADH4 is present in the stomach, the esophagus and the other mucosa [2], and can also catalyze oxidation of ethanol and retinol and also reduction of the lipid peroxidation products [3,4]. ADH is important for preventing alcohol toxicity that causes human diseases [5,6]. It is reported that Adh1 and Adh1/4 gene knockout mice showed Parkinson’s disease-like phenotypes such as reduction of locomotion, reduced sensitivity of smell, reduced monoamine levels and decreased body weight [7]. Different kinds of toxic aldehydes could be formed from peroxidation of polyunsaturated fatty acids through lipid peroxidation [4]. These aldehydes could be reduced by ADH 1 in vivo from enzymatic constants [3]. So if ADH1 and ADH1/4 gene are knocked out, accumulation of toxic aldehydes might cause deleterious effects. It is known that acetaldehyde can react with dopamine and form salsolinol [8], a neurotoxin with an effect similar to parkinsonism-inducing product 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrimidine (MPTP) [9]. Thus, if the compound reaches within the brain, it can inhibit the mitochondrial respiratory chain complex I, causing degeneration of dopamine neurons in the substantia nigra [10]. A truncating mutation in ADH1C (G78Stop) showed significant association with Parkinson disease from a large international survey [11]. Parkinson’s disease is characterized by accumulation of α-synuclein as fibrils in the brain. However it is not clear how the accumulated α-synuclein fibrils drives neuronal cell death. Recently Kam et al. reported that neurotoxicity induced by α-synuclein preformed fibrils was dependent on polyADP-ribosylation in mouse primary cortical neuron [12].
PolyADP-ribosylation is a posttranslational modification of proteins and is suggested to be involved in regulation of various biological events including cell death through addition of ADP-ribose molecules to various acceptor proteins [13]. From recent approach using mass spectroscopy, various acceptor proteins of polyADP-ribosylation were reported including ADH3 (class III) (ADH5 from old nomenclature) from a human breast cancer cell line [14] and Adh3 (class III) (Adh5 from old nomenclature) from β-amyloid peptide-treated mouse BV2 microglial cells [15]. Thus, we hypothesized that human ADH1 protein is polyADP-ribosylated in human hepatoma cells and investigated whether the modification could regulate ADH activity in vivo.
2. Materials and methods
2.1. Cells and reagents
Human liver cancer cell lines, HepG2 cells (Riken, Tsukuba, Japan) and Li7 cells, were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin (100 U/ml) and streptomycin (100 μg/ml) in a humidified incubator with 5% CO2 at 37.0 °C. The other human liver cancer cell lines, HLE cells, HuH6 cells, HuH7 cells and PLC/PRF/5 cells, were cultured in DMEM supplemented with 10% heat-inactivated FBS, penicillin (100 U/ml) and streptomycin (100 μg/ml) in a humidified incubator with 5% CO2 at 37.0 °C. 3-Aminobenzamide (3AB) was purchased from Tokyo Chemical Industry Co. (Tokyo, Japan). [32P]NAD+ was purchased from PerkinElmer (Massachusetts, USA), olaparib from ChemScene LLC (New Jersey, USA), recombinant human ADH1 protein (ab123147) from Abcam (Cambridge, UK), and recombinant human PARP1 from Trevigen (Maryland, USA). Poly(ADP-ribose) was prepared as described [16]. Anti-poly(ADP-ribose) IgG column was prepared by binding of monoclonal antibody against poly(ADP-ribose) (10H) [17] to CNBr-activated Sepharose 4B (GE Healthcare).
2.2. Drosophila melanogaster mutant that has increased polyADP-ribosylation
Drosophila melanogaster mutant, parg27.1/Y, that lacks poly(ADP-ribose) glycohydrolase [18], a major enzyme to degrade poly(ADP-ribose), was established as described [19].
Extraction and purification of polyADP-ribosylated proteins was as described by Nodono et al. [17]. The purified proteins were separated with 2D gel electrophoresis. The proteins appearing as new spots on the gel after alkaline treatment of the proteins (Fig. 1, right panel) were digested with trypsin and subjected to mass spectrometer (LCMS-IT-TOF, Shimazu Co., Kyoto, Japan). The data were analyzed using LCMS solution (Shimazu Co.) and MASCOT Version 2.2 (Matrix Science K.K., Tokyo) to find out proper proteins from data base from all organism by comparing with NCBInr and SwissProt. Significance threshold was set as p < 0.05.
Fig. 1.
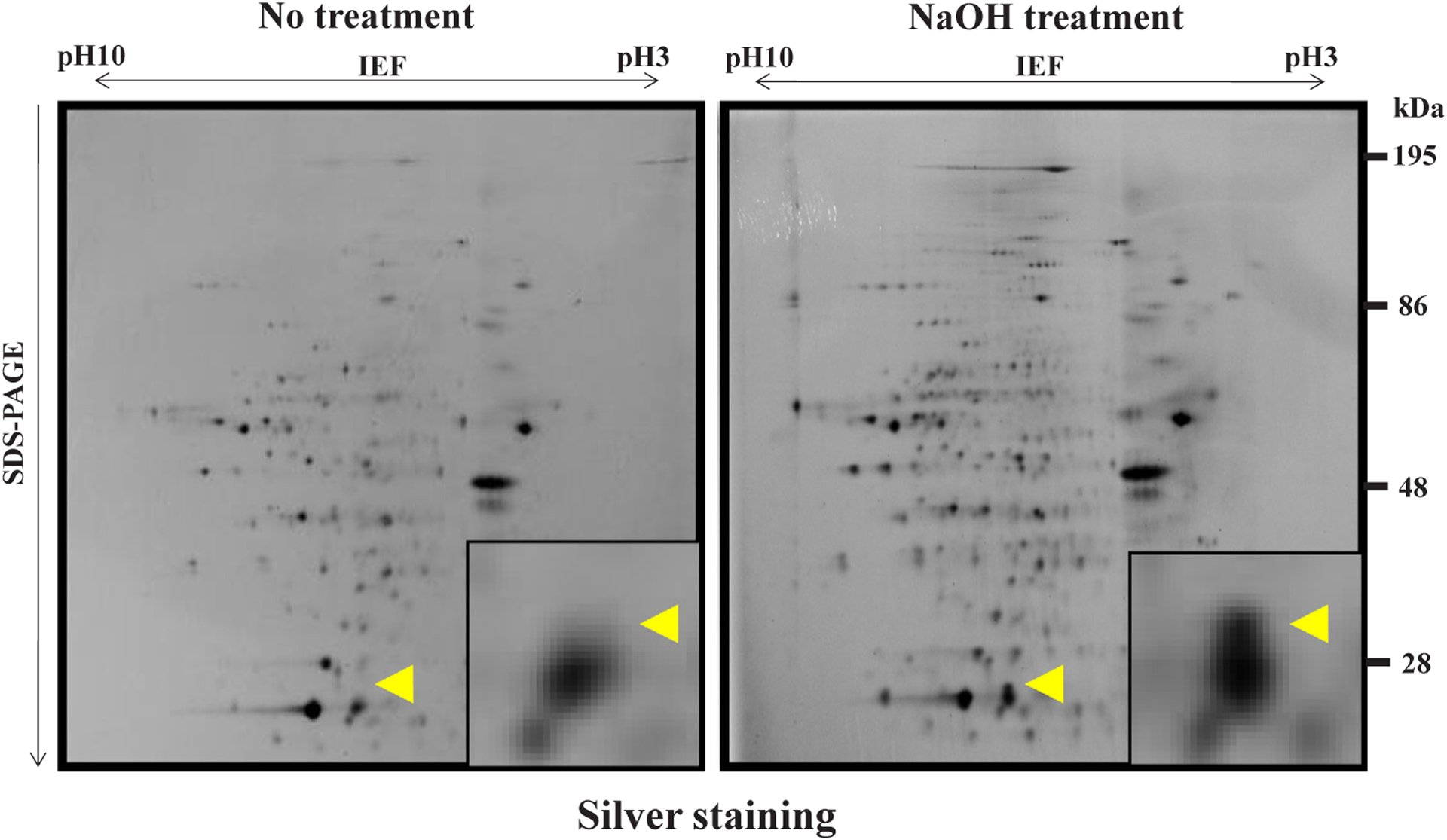
Separation on 2-D gel electrophoresis of affinity purified proteins without or with alkaline treatment Silver staining of 2-D gel electropherograms, each derived from fifteen thousand of Drosophila mutant flies. The most prominent protein spot, which appeared after NaOH treatment (right panel) but not without NaOH treatment (left panel), is marked with an arrowhead. The insets represent magnified vision. The representative data from two independent experiments are presented [reproduced from ref. 17 with the permission from Springer Nature, copyright 2011].
2.3. PolyADP-ribosylation reaction with [32P]NAD+
In vitro polyADP-ribosylation was performed as described previously [20]. Human ADH1 protein was incubated in 50 μl polyADP-ribosylation reaction buffer (100 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 1 mM DTT) with 20 U of PARP1, 1 μg of sonicated salmon sperm DNA, and 12.5 μCi (16 pmol, 10.6 ng) [32P]NAD+ for 30 min at 37 °C. The reaction was stopped by adding sample buffer (0.35 M Tris-HCl, pH 6.8, 10% SDS, 0.84 M 2-mercaptoethanol, 36% glycerol, 0.03% bromophenol blue). One millimolar of 3AB was included as a negative control for polyADP-ribosylation. As another negative control, human ADH1 protein was incubated in 50 μl polyADP-ribosylation reaction buffer with 20 U of PARP1, which had been heat-inactivated for 5 min at 100 °C. The reaction products were resolved by SDS-12.5% polyacrylamide gel electrophoresis. The polyacrylamide gels were dried at 65 °C for 2 h by vacuum using Electrophoresis RapiDry (ATTO Corporation, Japan) and exposed to X-ray film to detect radioactive materials at −80 °C for 3 h in the dark.
2.4. Characterization of binding of poly(ADP-ribose) to ADH1
To characterize the mode of binding between ADP-ribose and ADH1 protein, the products after incubation in the polyADP-ribosylation reaction mix were incubated further with 100 μg of ADP-ribose or 300 ng of poly(ADP-ribose) as competitors, for 30 min at 37 °C before addition of sample buffer, then incubated for 5 min at 100 °C and analyzed with SDS-12.5% polyacrylamide gel electrophoresis.
The reaction products after completion of the polyADP-ribosylation reaction were treated for 1 h at 25 °C with either 1 M neutral NH2OH, 0.1 M NaOH or 0.1 M HCl. NaCl at 1 M or 0.1 M was used as negative controls. The reaction products were neutralized and analyzed by SDS-12.5% polyacrylamide gel electrophoresis. The radioactive materials were detected as described in 2.3.
2.5. Cell lysate preparation for ADH activity assay
HepG2 cells (1.0 × 106) were plated in 100-mm cell culture dishes. After one day incubation, cells were treated with 30 μM olaparib or 0.15% dimethyl sulfoxide (DMSO) as a vehicle, for 30 min in a humidified incubator with 5% CO2 at 37.0 °C. Cells were washed with PBS with 30 μM olaparib or 0.15% DMSO, then harvested by treatment with a 0.025% trypsin/EDTA solution containing 30 μM olaparib or 0.15% DMSO. After centrifugation at 100 × g for 3 min, the precipitates were washed with PBS containing 30 μM olaparib or 0.15% DMSO. The cell pellets were collected by centrifugation at 100 × g for 3 min and dissolved in ice-cold RIPA buffer with 30 μM olaparib or 0.15% DMSO and sonicated for 3 sec × 10 times at a setting of 2 with a HUX-XL2000 sonicator (Misonix, USA). Protein concentration was determined using BCA kit (Thermo Fisher Scientific, USA).
2.6. Assay for ADH activity
HepG2 cell lysates were incubated in 100 μl of reaction mixture containing 1 M ethanol, 1 mM NAD+, and 100 mM sodium phosphate buffer (pH 8.5). ADH activity was measured for 30 min at 25.0 °C by the increase of absorbance at 340 nm following the reduction of NAD+ to NADH. Reaction mixtures containing HepG2 cell lysates but without 1 M ethanol were used as a blank.
2.7. Statistics
Statistical significance of the difference between the control group and PARP inhibitor group was evaluated by Student’s t-test.
3. Results
3.1. The major acceptor protein of polyADP-ribosylation in a Drosophila mutant was ADH
A Drosophila mutant, which is defective in poly(ADP-ribose) glycohydrolase (PARG) [18] that degrades poly(ADP-ribose), was used. They were processed as described [17]. The acceptor proteins of polyADP-ribosylation were isolated from the extract of Drosophila mutant and were purified with affinity chromatography using an anti-poly(ADP-ribose) IgG column. Since the acceptor proteins are bound with poly (ADP-ribose) of varying chain length, they could not be detected as distinct bands in gel electrophoresis, but could appear as converged bands after cleaving off poly(ADP-ribose) chain. Therefore, the purified acceptor proteins were treated without or with 1 M NaOH for 15 min on ice to cleave the ester bond between the acceptor proteins and poly (ADP-ribose). Then, they were separately resolved with 2D gel electrophoresis (Fig. 1). 2D gel electrophoresis was repeated 8 times and the newly appeared spots (arrowhead) derived from 28,800 Drosophila mutant flies were collected and were subjected to mass spectrometry as described in Materials and Methods. The most prominent spot as shown in Fig. 1 (right panel) was identified as Drosophila ADH protein (Uni-ProtKB/SwissProt – P00334 (ADH_DROME); Score 458, Mass 27 kDa; pI = 7.74; coverage 51%). We conclude that the major acceptor protein of polyADP-ribosylation in Drosophila melanogaster is ADH.
3.2. PolyADP-ribosylation of human ADH1 by human PARP1 in vitro
Radioactivity from [32P]NAD+ was incorporated mostly into poly (ADP-ribose) polymerase 1 (PARP1), which is the canonical PARP among PARP family members [21], as known as automodification of PARP1 [22,23] (Fig. 2). But significant incorporation was observed in human ADH1 (Fig. 2). The incorporation into ADH1 was not observed when heat-inactivated PARP1 or a PARP inhibitor, 3AB, was used (Fig. 2). Histone H1, known to be polyADP-ribosylated with short chain of poly(ADP-ribose) [24], was used as a positive control. Thus, it appeared that human ADH1 was ADP-ribosylated by PARP1. In addition, the incorporated radioactivity was not removed from ADH1 protein in the presence of excess amount of ADP-ribose (100 μg, 180 nmol, 3.6 mM) or poly(ADP-ribose) (300 ng, 0.55 nmol as ADP-ribose residues, 11 μM) as competitors (Fig. 3). These results further indicated that this linkage was covalent, as was also shown with histone H1 as a positive control.
Fig. 2.
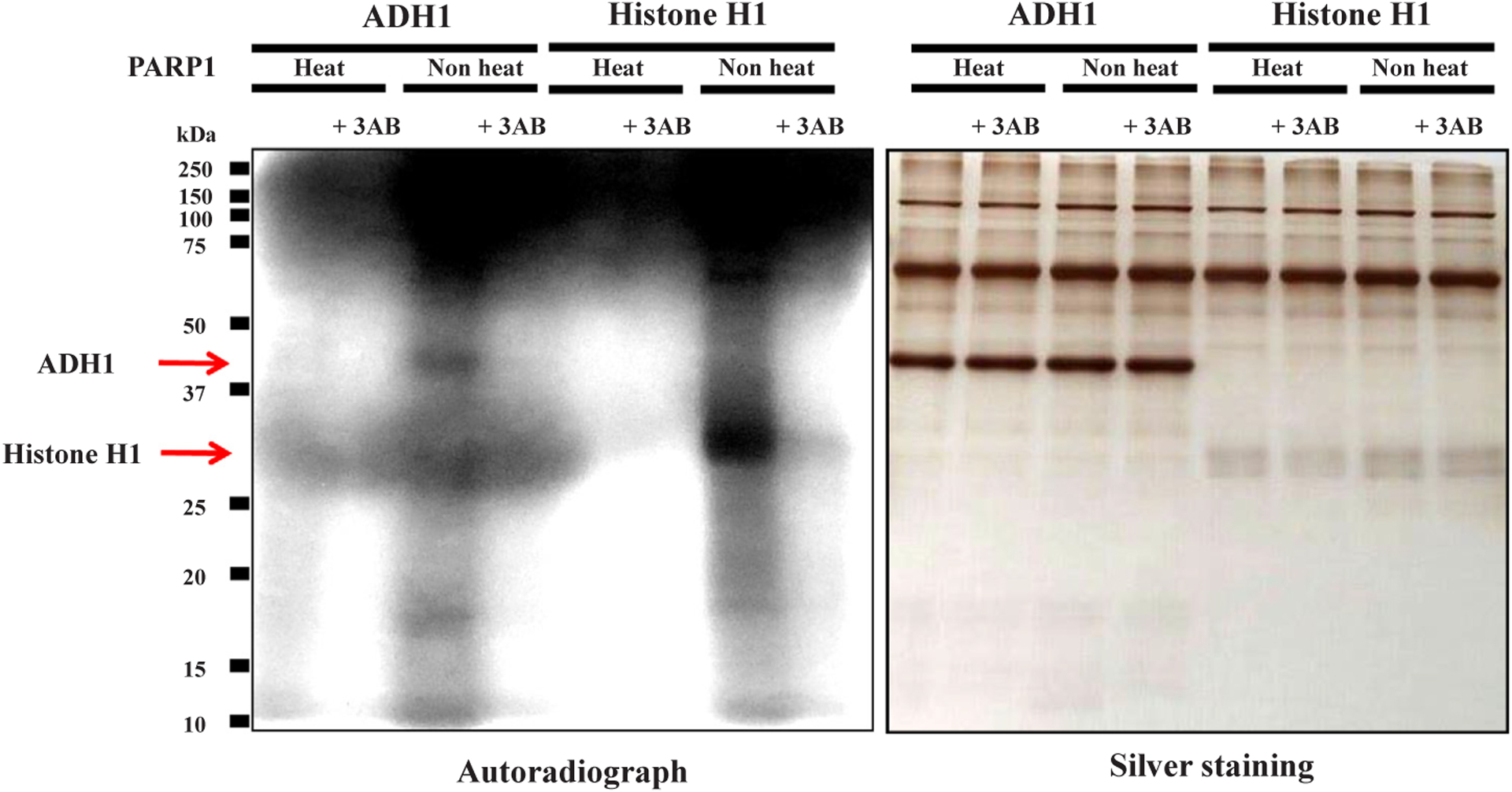
PolyADP-ribosylation of human ADH1 with [32P]NAD+ in vitro Human ADH1 protein was incubated with [32P]NAD+ under polyADP-ribosylation reaction conditions; with native or heat-inactivated PARP1 without or with 3AB. These samples were subjected to SDS-12.5% polyacrylamide gel electrophoresis followed by autoradiography (left panel) and silver staining (right panel). Histone H1 is a positive control for polyADP-ribosylation. Data are representative of three experiments.
Fig. 3.
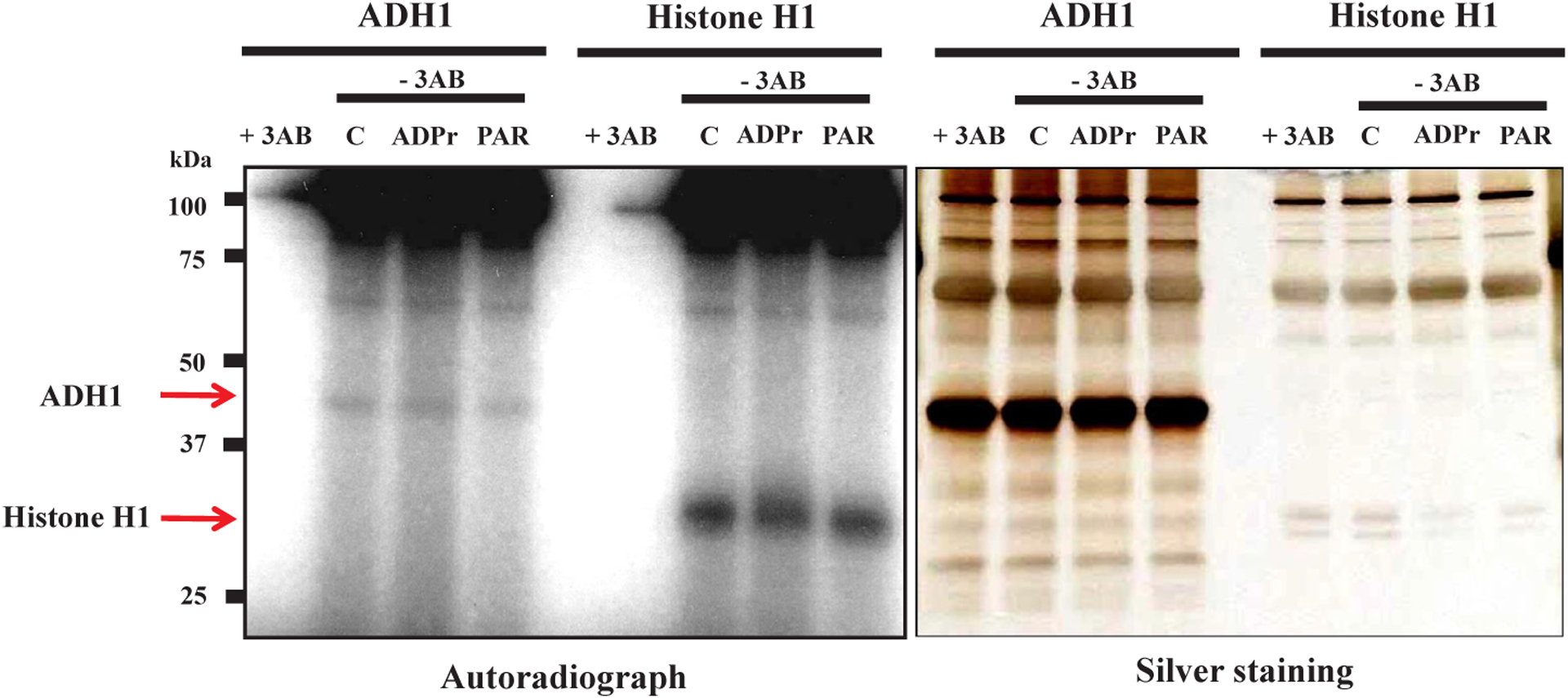
Covalent linkage between human ADH1 and poly(ADP-ribose) in vitro Human ADH1 protein was incubated with [32P]NAD+ (12.5 μCi, 16 pmol, 10.6 ng) under polyADP-ribosylation reaction conditions. The reaction was stopped by addition of 1 mM 3AB and then distilled water (Control, C), 100 μg (180 nmol) ADP-ribose (ADPr) or 300 ng (0.55 nmol as ADP-ribose residues) poly(ADP-ribose) (PAR). These samples were subjected to SDS-12.5% polyacrylamide gel electrophoresis followed by autoradiography (left panel) and silver staining (right panel). Histone H1 is a positive control for polyADP-ribosylation. Data are representative of three experiments.
3.3. The major chemical bond between ADH1 and poly(ADP-ribose) is an ester linkage
To know the nature of the chemical bond between the acceptor amino acid and poly(ADP-ribose), the chemical stability of the bond was analyzed. The bond between ADH1 and poly(ADP-ribose) was mostly cleaved with either 1 M neutral NH2OH or 0.1 M NaOH for 1 h at 25 °C, but not with 0.1 M HCl, suggesting the presence of an ester bond as the major linkage [25,26] (Figs. 4 and 5). The chain length of incorporated ADP-ribose into ADH did not appear to be sufficiently long to change the electrophoretic mobility, similar to the findings with the histone H1 acceptor in vitro, suggesting that it might be < 10 ADP-ribose moieties. Thus, we conclude that human ADH1 could be polyADP-ribosylated in vitro with an ester bond as the major linkage.
Fig. 4.
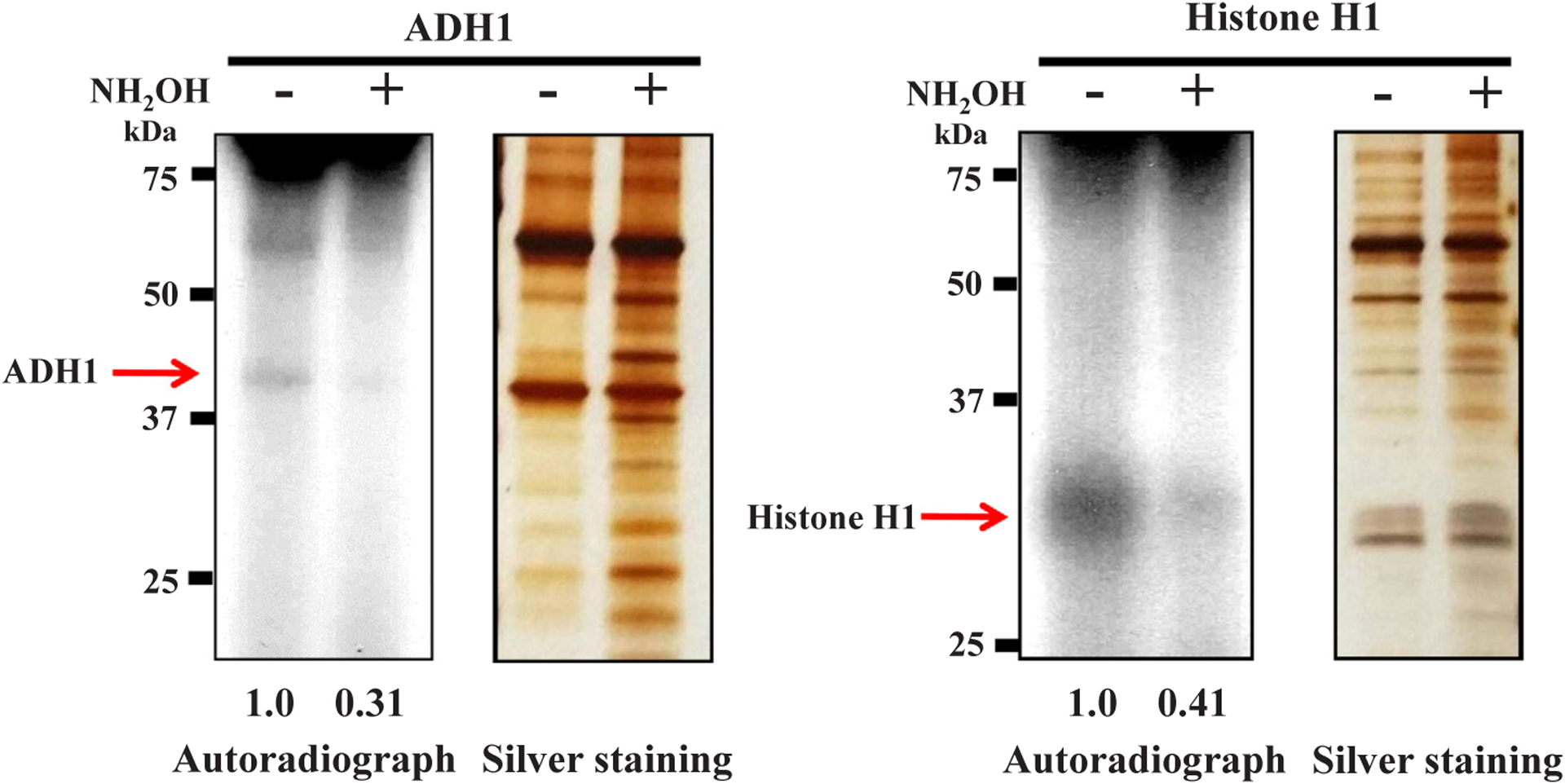
Chemical susceptibility to NH2OH of the bond between human ADH1 and poly(ADP-ribose) PolyADP-ribosylated ADH1 was treated with 1 M NH2OH (pH 7.5) for 1 h at 25 °C. The sample was subjected to SDS-12.5% polyacrylamide gel electrophoresis followed by autoradiography and silver staining (left panel). Histone H1 is a positive control for polyADP-ribosylation (right panel). Data are representative of three experiments. The levels of radiolabeled ADH1 and histone H1 proteins were also calculated by Image-J and were digitized.
Fig. 5.
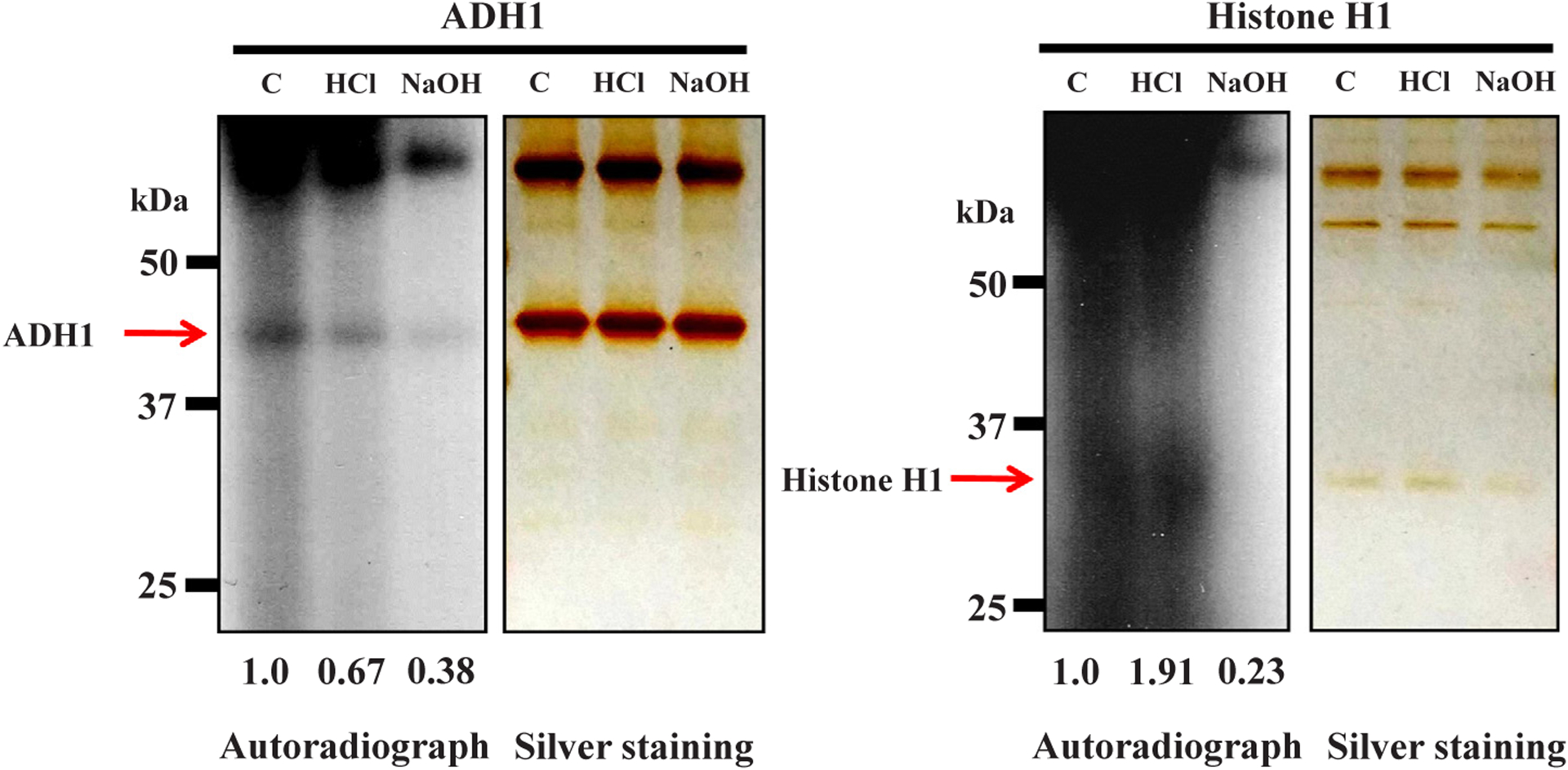
Chemical susceptibility of the bond between human ADH1 and poly(ADP-ribose) to NaOH but not to HCl PolyADP-ribosylated ADH1 was treated with 0.1 M NaCl (Control, C), 0.1 M HCl or 0.1 M NaOH for 1 h at 25 °C. The sample was subjected to SDS-12.5% polyacrylamide gel electrophoresis followed by autoradiography and silver staining (left panel). Histone H1 is a positive control for polyADP-ribosylation (right panel). Data are representative of three experiments. The levels of radiolabeled ADH1 and histone H1 proteins were also calculated by Image-J and were digitized.
3.4. PARP inhibitor increased the enzymatic activity of ADH1 in HepG2 cells
To know how ADP-ribosylation affects ADH1 activity in vivo, we measured ADH1 activity in human liver cell lines using PARP inhibitor. First, we checked the expression levels of ADH1, PARP1 and PARG proteins. ADH1 and PARP1 protein levels were similar in human liver cell lines except for HuH-6 and HuH-7 cells. PARG protein level was similar in human liver cell lines except for Li7 (Fig. 6). We chose HepG2 cells, which expressed a significant amount of ADH1, PARP1 and PARG proteins for determination of ADH1 activity. When HepG2 cells were cultured in the presence of olaparib, a potent inhibitor of polyADP-ribosylation, there was no significant cytotoxicity below 50 μM within 72 h (Fig. 7). It was expected that if ADP-ribosylation of ADH1 inhibited its enzymatic activity, then the PARP inhibitor would increase ADH activity. In fact, when olaparib was added to HepG2 cell culture at 30 μM for 30 min, ADH activity of the cell lysate (1 mg) was significantly increased as compared to the control (Fig. 8). Therefore, we conclude that ADH activity of a human hepatoma cell line, HepG2 cells, could be regulated through polyADP-ribosylation.
Fig. 6.
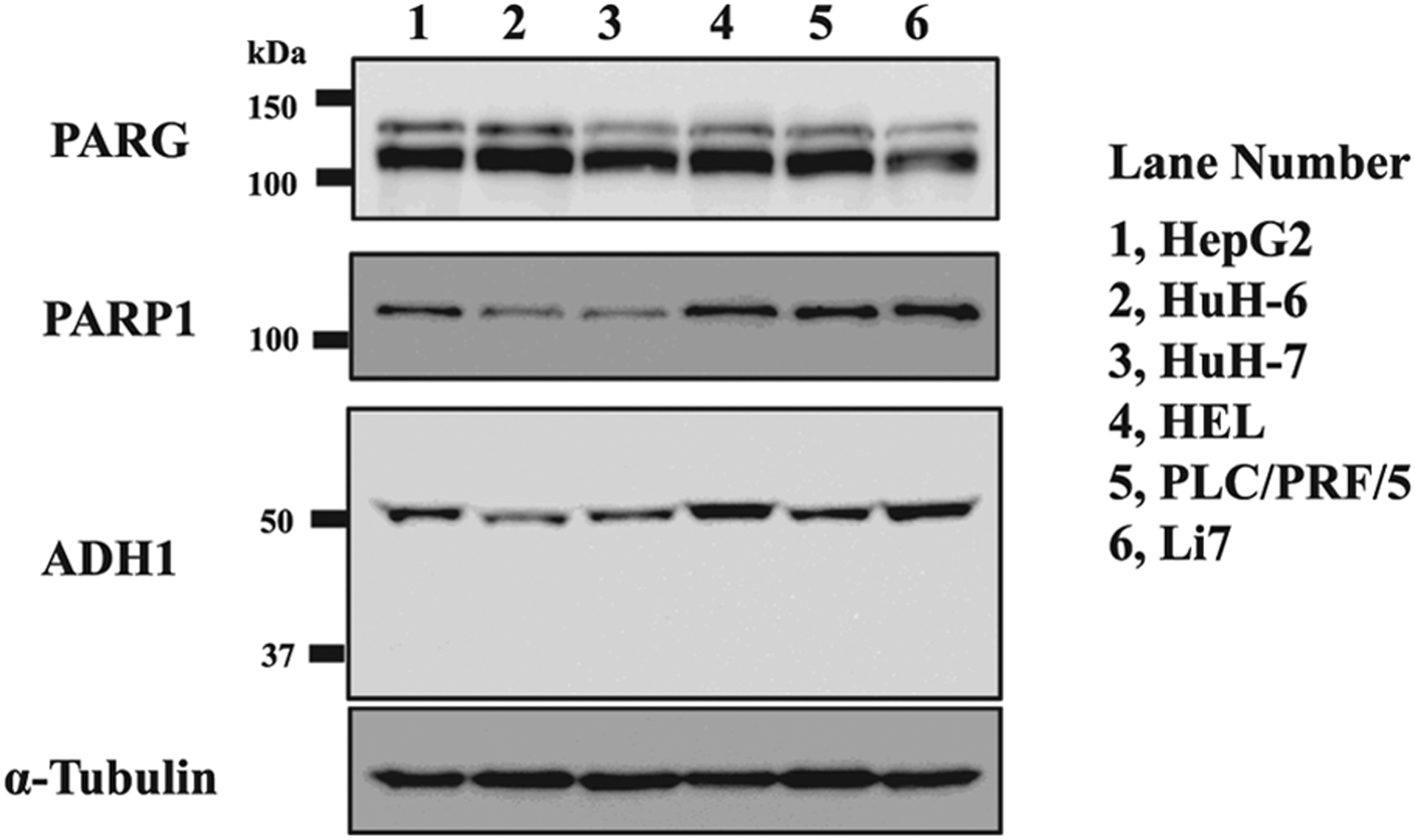
ADH1 protein levels in human liver cell lines Lysates from proliferating human liver cell lines were subjected to SDS-10% polyacrylamide gel electrophoresis followed by Western blotting using antibodies against ADH1, PARG, PARP1 and α -tubulin.
Fig. 7.
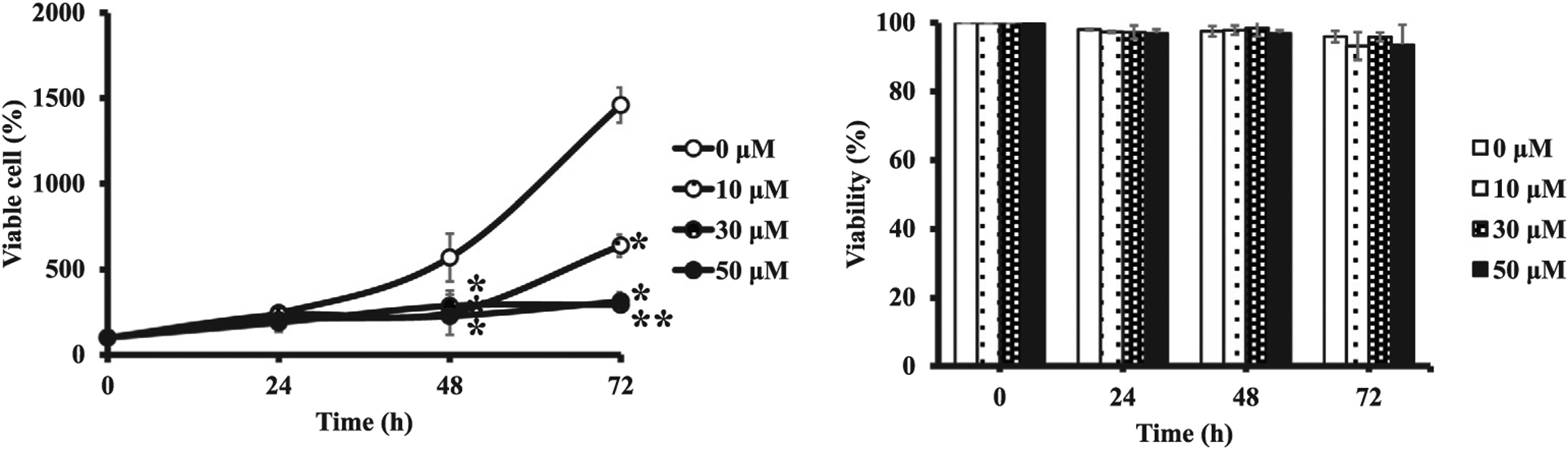
Effect of PARP inhibitor (olaparib) on proliferation and viability of HepG2 cells (A) Proliferation of HepG2 cells was determined at 24 h, 48 h and 72 h without or with olaparib. (B) Cell viability was calculated by dividing the number of cells that did not stain with trypan blue by the total number of cells. Cells were counted with hemocytometer. The result is the means ± standard deviations from two independent experiments. Start; 1.0 × 105 cells/3.5 cm dish (represented as 100%). Significances were 0 μM vs. 10 μM olaparib, 0 μM vs. 30 μM olaparib, 0 μM vs. 50 μM olaparib. *p < 0.05, **p < 0.01.
Fig. 8.
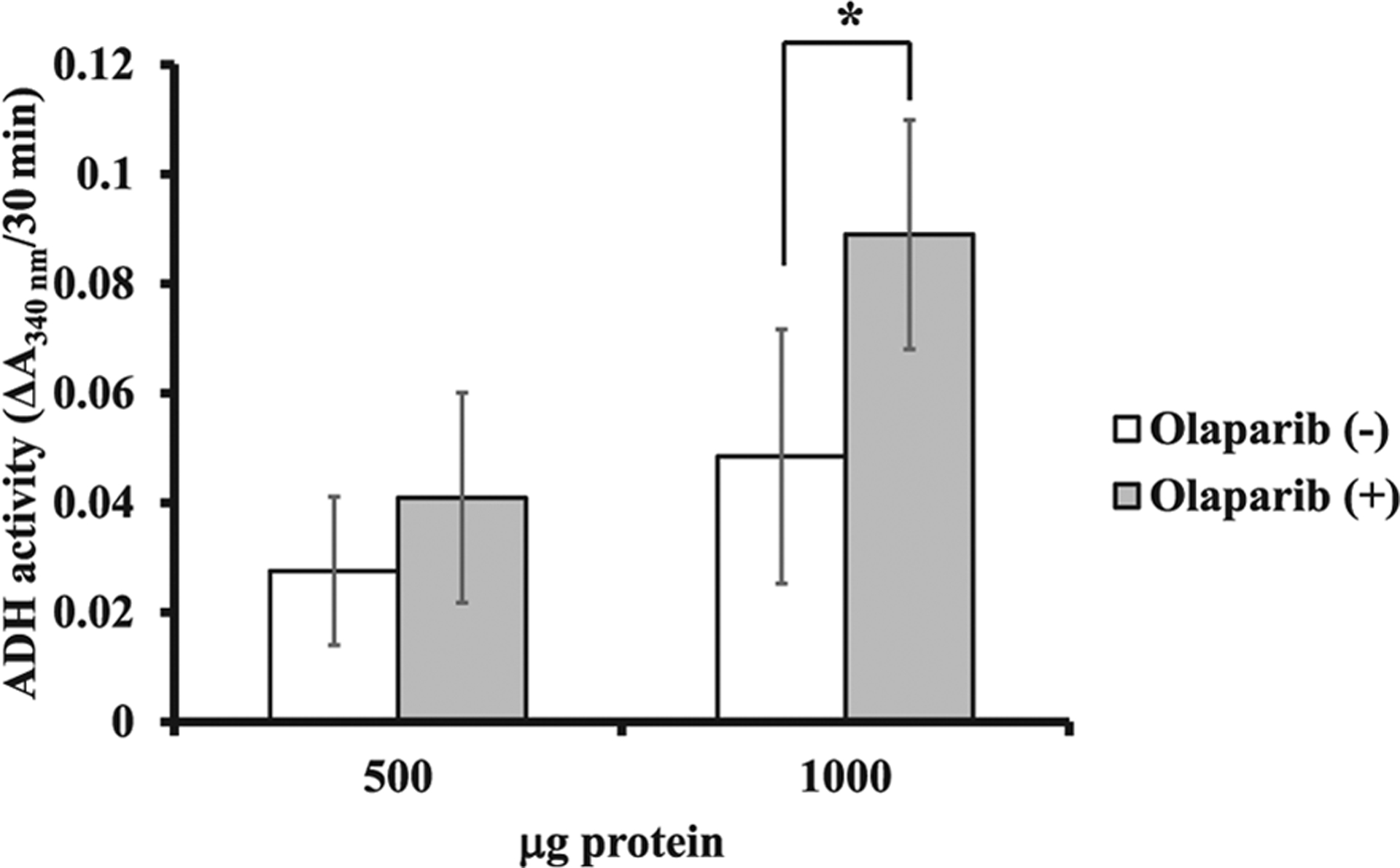
Effect of PARP inhibitor on ADH1 activity ADH1 activity was assayed as described in “Materials and Methods”. The result is the means ± standard deviations from three independent experiments. Cell lysates containing 500 μg and 1000 μg protein were used. Significances were olaparib (−) vs. olaparib (+). *p < 0.05.
4. Discussion
Here we found that human ADH1 is polyADP-ribosylated in vitro by showing the incorporation of radioactivity from [32P]NAD+ with human PARP1. The chemical bond between ADH1 and poly(ADP-ribose) suggests the presence of mainly ester linkages; the acceptor amino acid could be either glutamic acid or aspartic acid as was already reported for histone H1 [25]. About 30% reduction of the radioactivity after HCl treatment of ADH might be due to the presence of a bond between serine residues of ADH1 [27] and a small amount of the radioactivity remaining after NH2OH or NaOH treatment of ADH1 might be explained by the presence of a bond between lysine residues of ADH1 [28]. Of note, when PARP inhibitor was added to cultured HepG2 cells, a human hepatoma cell line, ADH activity of the cells was significantly increased, suggesting that ADH activity is regulated by polyADP-ribosylation in vivo. In the liver, since alcohol is primarily metabolized through ADH, how liver diseases induced by alcohol are regulated through polyADP-ribosylation would be an interesting question to be studied in the future.
There is only a single ADH gene in Drosophila melanogaster in contrast to 5 classes of genes in man and rodents. Fruit flies feed on decaying fruit, which contains ethanol concentrations as high as 6–7% (v/v). When larvae of fruit flies are exposed to ethanol, they maintain high concentrations of ethanol in their body to protect against parasitoid wasp infection, since high ethanol concentrations exterminate larvae of parasitoid wasp [29]. Indeed, ADH might be one of the essential factors for survival from alcohol intoxication [30]. When larvae of fruit flies were exposed to medium containing 10% ethanol, about 60–70% of wild-type larvae reached adulthood, while 100% of ADH-negative larvae did not [31]. The Parg knockout Drosophila melanogaster mutant, which showed accumulation of poly(ADP-ribose), revealed neurodegenerative disease-like phenotypes including reduced locomotion, inability to fly and became moribund and died within 10 days [19]. Additionally, we found that the major acceptor protein of polyADP-ribosylation was ADH by mass spectroscopy after purification of the extract from the Drosophila mutant. The results suggest polyADP-ribosylation of ADH could be one of the mechanisms of neurodegeneration in Drosophila. Concerning ADP-ribosylation of ADH, Fuentes et al. previously reported that the ADH from Entamoeba histolytica was mono-ADP-ribosylated, implying the involvement of a different enzyme with respect to PARP-induced polyADP-ribosylation, and that the mono-ADP-ribosylation of ADH did not have any effect on enzymatic activity in Entamoeba histolytica [32].
The involvement of polyADP-ribosylation in the neurodegeneration of neuronal cells was recently reported [12]. Preformed α-synuclein fibrils induced PARP1 activation in mouse primary cortical neurons, leading to cell death. Intrastriatal injection of preformed α-synuclein fibrils induced loss of dopamine neurons and these pathological changes were reduced by addition of inhibitors of PARP or in PARP1 knocked out mice [12]. They also found the elevated poly(ADP-ribose) level was observed in cerebrospinal fluid of the patients with Parkinson’s disease [12]. It is not clear at present whether PARP inhibitors could be used to prevent or treat the neurodegenerative diseases in humans. Further studies are needed to clarify the involvement of ADH in neurodegeneration and the effect of PARP inhibitors for clinical use.
In conclusion, human ADH1 was polyADP-ribosylated in vitro by human PARP1. The chemical bond between human ADH1 and poly (ADP-ribose) suggests the presence of mainly ester linkages. When HepG2 cells, derived from human hepatoma, were cultured in the presence of a PARP inhibitor, ADH activity of the cells was significantly increased, suggesting that polyADP-ribosylation could regulate ADH activity in vivo.
Acknowledgments
We thank Dr. Richi Nakatake, Department of Surgery, for kindly providing HuH-6, HuH-7, HEL, PLC/PRF/5 and Li7 liver cancer cell lines and Dr. Jun-Ichi Fujisawa, Department of Microbiology, Kansai Medical University, for generous support of the work. We are indebted to Dr. F.-S. Che, Dr. K. Kamemura, and Dr. T. Komiya for their valuable comments and to the members of the laboratory in Nagahama Institute of Bio-Science and Technology for their continuous support. This work was supported in part by a Grant-in-Aid for Cancer Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan. J. M. was supported by the Intramural Research Program (National Institutes of Health [NIH], National Heart, Lung, and Blood Institute [NHLBI]).
Abbreviations:
- 3AB
3-aminobenzamide
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- FBS
fetal bovine serum
- NAD+
nicotinamide adenine dinucleotide
- NADH
reduced nicotinamide adenine dinucleotide
- PARPs
poly(ADP-ribose) polymerases
- PARG
poly(ADP-ribose) glycohydrolase
- PBS
phosphate-buffered saline
- SDS
sodium dodecyl sulfate
Footnotes
Declarations of interest
None.
References
- [1].Duester G, Farrés J, Felder MR, Holmes RS, Höög JO, Parés X, Plapp BV, Yin SJ, Jörnvall H, Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family, Biochem. Pharmacol 58 (1999) 389–395. [DOI] [PubMed] [Google Scholar]
- [2].Crabb DW, Matsumoto M, Chang D, You M, Overview of the role of alcohol dehydrogenase and aldehyde dehydrogenase and their variants in the genesis of alcohol-related pathology, Proc. Nutr. Soc 63 (2004) 49–63. [DOI] [PubMed] [Google Scholar]
- [3].Boleda MD, Saubi N, Farrés J, Parés X, Physiological substrates for rat alcohol dehydrogenase classes: aldehydes of lipid peroxidation, omega-hydroxyfatty acids, and retinoids, Arch. Biochem. Biophys 307 (1993) 85–90, 10.1006/abbi.1993.1564. [DOI] [PubMed] [Google Scholar]
- [4].Singh M, Kapoor A, Bhatnagar A, Oxidative and reductive metabolism of lipid-peroxidation derived carbonyls, Chem. Biol. Interact 234 (2015) 261–273, 10.1016/j.cbi.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Riley EP, Infante MA, Warren KR, Fetal alcohol spectrum disorders: an overview, Neuropsychol. Rev 21 (2011) 73–80, 10.1007/s11065-011-9166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rusyn I, Bataller R, Alcohol and toxicity, J. Hepatol 59 (2013) 387–388, 10.1016/j.jhep.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Anvret A, Ran C, Westerlund M, Gellhaar S, Lindqvist E, Pernold K, Lundströmer K, Duester G, Felder MR, Galter D, Belin AC, Adh1 and Adh1/4 knockout mice as possible rodent models for presymptomatic Parkinson’s disease, Behav. Brain Res 227 (2012) 252–257, 10.1016/j.bbr.2011.10.040. [DOI] [PubMed] [Google Scholar]
- [8].Yamanaka Y, Walsh MJ, Davis VE, Salsolinol, an alkaloid derivative of dopamine formed in vitro during alcohol metabolism, Nature 227 (1970) 1143–1144. [DOI] [PubMed] [Google Scholar]
- [9].Niwa T, Maruyama W, Nakahara D, Takeda N, Yoshizumi H, Tatematsu A, Takahashi A, Dostert P, Naoi M, Nagatsu T, Endogenous synthesis of N-methylsalsolinol, an analogue of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine, in rat brain during in vivo microdialysis with salsolinol, as demonstrated by gas chromatography-mass spectrometry, J. Chromatogr 578 (1992) 109–115. [DOI] [PubMed] [Google Scholar]
- [10].Mizuno Y, Ikebe S, Hattori N, Nakagawa-Hattori Y, Mochizuki H, Tanaka M, Ozawa T, Role of mitochondria in the etiology and pathogenesis of Parkinson’s disease, Biochim. Biophys. Acta 1271 (1995) 265–274. [DOI] [PubMed] [Google Scholar]
- [11].Buervenich S, Carmine A, Galter D, Shahabi HN, Johnels B, Holmberg B, Ahlberg J, Nissbrandt H, Eerola J, Hellström O, Tienari PJ, Matsuura T, Ashizawa T, Wüllner U, Klockgether T, Zimprich A, Gasser T, Hanson M, Waseem S, Singleton A, McMahon FJ, Anvret M, Sydow O, Olson L, A rare truncating mutation in ADH1C (G78Stop) shows significant association with Parkinson disease in a large international sample, Arch. Neurol 62 (2005) 74–78, 10.1001/archneur.62.1.74. [DOI] [PubMed] [Google Scholar]
- [12].Kam TI, Mao X, Park H, Chou SC, Karuppagounder SS, Umanah GE, Yun SP, Brahmachari S, Panicker N, Chen R, Andrabi SA, Qi C, Poirier GG, Pletnikova O, Troncoso JC, Bekris LM, Leverenz JB, Pantelyat A, Ko HS, Rosenthal LS, Dawson TM, Dawson VL, Poly(ADP-ribose) drives pathologic α-synuclein neurodegeneration in Parkinson’s disease, Science 362 (2018) 6414, 10.1126/science.aat8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gupte R, Liu Z, Kraus WL, PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes, Genes Dev 31 (2017) 101–126, 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhen Y, Zhang Y, Yu Y, A Cell-Line-Specific Atlas of PARP-Mediated Protein Asp/Glu-ADP-Ribosylation in Breast Cancer, Cell Rep 21 (2017) 2326–2337, 10.1016/j.celrep.2017.10.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Correani V, Martire S, Mignogna G, Caruso LB, Tempera I, Giorgi A, Grieco M, Mosca L, Schininà ME, Maras B, d’Erme M, Poly(ADP-ribosylated) proteins in β-amyloid peptide-stimulated microglial cells, Biochem. Pharmacol (2018) https://doi.org/101016/j.bcp.2018.10.026. [DOI] [PubMed] [Google Scholar]
- [16].Miwa M, Sugimura T, Structure of poly(ADP-ribose), Methods Enzymol 106 (1984) 441–450. [DOI] [PubMed] [Google Scholar]
- [17].Nodono H, Hamada A, Kuroda Y, Kamemura K, Hasegawa M, Komiya T, Kondo M, Che FS, Hanai S, Miwa M, Approaches to separate and identify polyADP-ribosylated proteins using poly(ADP-ribose) glycohydrolase-knockout Drosophila, Methods Mol. Biol 780 (2011) 377–390, 10.1007/978-1-61779-270-0_22. [DOI] [PubMed] [Google Scholar]
- [18].Miwa M, Tanaka M, Matsushima T, Sugimura T, Purification and properties of glycohydrolase from calf thymus splitting ribose-ribose linkages of poly(adenosine diphosphate ribose), J. Biol. Chem 249 (1974) 3475–3482. [PubMed] [Google Scholar]
- [19].Hanai S, Kanai M, Ohashi S, Okamoto K, Yamada M, Takahashi H, Miwa M, Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster, Proc. Natl. Acad. Sci. U.S.A 101 (2004) 82–86, 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kanai M, Hanashiro K, Kim S-H, Hanai S, Boulares AH, Miwa M, Fukasawa K, Inhibition of Crm1–p53 interaction and nuclear export of p53 by poly(ADP-ribosyl) ation Nat, Cell Biol 9 (2007) 1175–1183, 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- [21].Amé JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase, J. Biol. Chem 274 (1999) 17860–17868. [DOI] [PubMed] [Google Scholar]
- [22].Yoshihara K, Hashida T, Yoshihara H, Tanaka Y, Ohgushi H, Enzyme-bound early product of purified poly(ADP-ribose) polymerase, Biochem. Biophys. Res. Commun 78 (1977) 1281–1288. [DOI] [PubMed] [Google Scholar]
- [23].Tanaka Y, Yoshihara K, Ohashi Y, Itaya A, Nakano T, Ito K, Kamiya T, A method for determining oligo- and poly(ADP-ribosyl)ated enzymes and proteins in vitro, Anal. Biochem 145 (1985) 137–143. [DOI] [PubMed] [Google Scholar]
- [24].Nishizuka Y, Ueda K, Honjo T, Hayaishi O, Enzymic adenosine diphosphate ribosylation of histone and poly adenosine diphosphate ribose synthesis in rat liver nuclei, J. Biol. Chem 243 (1968) 3765–3767. [PubMed] [Google Scholar]
- [25].Ogata N, Ueda K, Kagamiyama H, Hayaishi O, ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites, J. Biol. Chem 255 (1980) 7616–7620. [PubMed] [Google Scholar]
- [26].Moss J, Yost DA, Stanley SJ, Amino acid-specific ADP-ribosylation, J. Biol. Chem 258 (1983) 6466–6470. [PubMed] [Google Scholar]
- [27].Leidecker O, Bonfiglio JJ, Colby T, Zhang Q, Atanassov I, Zaja R, Palazzo L, Stockum A, Ahel I, Matic I, Serine is a new target residue for endogenous ADP-ribosylation on histones, Nat. Chem. Biol 12 (2016) 998–1000, 10.1038/nchembio.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO, Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites, Nucl. Acids Res 37 (2009) 3723–3738, 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Milan NF, Kacsoh BZ, Schlenke TA, Alcohol consumption as self-medication against blood-borne parasites in the fruit fly, Curr. Biol 22 (2012) 488–493, 10.1016/j.cub.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fry JD, Mechanisms of naturally evolved ethanol resistance in Drosophila melanogaster, J. Exp. Biol 217 (2014) 3996–4003, 10.1242/jeb.110510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Heinstra PW, Scharloo W, Thörig GE, Physiological significance of the alcohol dehydrogenase polymorphism in larvae of Drosophila, Genetics 117 (1987) 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fuentes SM, Martínez-Cadena G, Silva ME, López A, Sánchez C, Alvarez AH, Avila EE, MonoADP-ribosylation of the NAD+-dependent alcohol dehydrogenase from Entamoeba histolytica, Curr. Microbiol 51 (2005) 171–174, 10.1007/s00284-005-4538-1. [DOI] [PubMed] [Google Scholar]


