Abstract
Certain strains of Clostridium difficile produce the ADP-ribosyltransferase CDT, which is a binary actin ADP-ribosylating toxin. The toxin consists of the binding component CDTb, which mediates receptor binding and cellular uptake, and the enzyme component CDTa. Here we studied the enzyme component (CDTa) of the toxin using the binding component of Clostridium perfringens iota toxin (Ib), which is interchangeable with CDTb as a transport component. Ib was used because CDTb was not expressed as a recombinant protein in Escherichia coli. Similar to iota toxin, CDTa ADP-ribosylates nonmuscle and skeletal muscle actin. The N-terminal part of CDTa (CDTa1–240) competes with full-length CDTa for binding to the iota toxin binding component. The C-terminal part (CDTa244–263) harbors the enzyme activity but was much less active than the full-length CDTa. Changes of Glu428 and Glu430 to glutamine, Ser388 to alanine, and Arg345 to lysine blocked ADP-ribosyltransferase activity. Comparison of CDTa with C. perfringens iota toxin and Clostridium botulinum C2 toxin revealed full enzyme activity of the fragment Ia208–413 but loss of activity of several N-terminally deleted C2I proteins including C2I103–431, C2I190–431, and C2I30–431. The data indicate that CDTa belongs to the iota toxin subfamily of binary actin ADP-ribosylating toxins with respect to interaction with the binding component and substrate specificity. It shares typical conserved amino acid residues with iota toxin and C2 toxin that are suggested to be involved in NAD-binding and/or catalytic activity. The enzyme components of CDT, iota toxin, and C2 toxin differ with respect to the minimal structural requirement for full enzyme activity.
Clostridium difficile is the causative pathogen of antibiotic-associated diarrhea and pseudomembranous colitis (10, 14). It produces two major protein toxins (toxins A and B), which are the prototypes of the family of large clostridial cytotoxins. Both toxins inhibit the functions of Rho GTPases by monoglucosylation. Beside toxins A and B, some strains of C. difficile produce a binary ADP-ribosylating toxin (CDT) which modifies actin (22). The pathogenic role of CDT in diseases induced by C. difficile is not clear, but about 6 to 12.5% of strains isolated from patients with enteritis contained CDT genes (20, 26).
The binary CDT is composed of the enzymatic component, CDTa (48 kDa), and the binding and translocation component, CDTb (94 kDa), which mediates cell entry of CDTa (19). CDT belongs to the group of binary actin ADP-ribosylating toxins (4), which can be divided into C2 toxin and iota toxin subfamilies (2, 5, 21). The subfamilies differ with respect to their actin substrate specificities (24). Clostridium botulinum C2 toxin ADP-ribosylates only β/γ-nonmuscle actin and γ-smooth muscle actin, whereas iota-like toxins ADP-ribosylate all actin isoforms, including α-actin (16). Furthermore, whereas the binding components of the iota toxin subfamily, including those of iota toxin, Clostridium spiroforme toxin, and CDTa, are interchangeable, no functional complementation between the binding components and the enzymatic components of C2 toxin and the toxins of the iota subfamily was observed (12, 21).
Here we studied the enzyme component of CDT and characterized its enzyme activity. We show that the N-terminal part of CDTa is responsible for interaction with the binding component, whereas the C-terminal part harbors transferase activity. For delivery of CDTa into cells we used the Ib binding and translocation component from iota toxin, because CDTb was not expressed as a recombinant protein in Escherichia coli. Recently, we characterized the catalytic center of the ADP-ribosyltransferase C2I and identified several amino acid residues essential for the transferase activity (8). Here we studied the functional roles of several amino acid residues of CDTa suggested to be conserved among the actin ADP-ribosylating toxins. Moreover, we compared the minimal structural requirements of CDTa with those of the enzyme components of iota toxin (Ia) and C2 toxin (C2I).
MATERIALS AND METHODS
Materials.
Cell culture medium was purchased from Biochrom (Berlin, Germany), fetal calf serum was purchased from PAN Systems (Aidenbach, Germany), and cell culture materials were purchased from Falcon (Heidelberg, Germany). Platelet cytosol was prepared as described previously (23). G-actin from rabbit skeletal muscle was prepared as described previously (17). The binding component (Ib) from Clostridium perfringens iota toxin was purified and activated with chymotrypsin as described previously (9). Protein A coupled to peroxidase and the enhanced chemiluminescence detection kit were from Amersham (Braunschweig, Germany). The nitrocellulose blotting membrane was obtained from Schleicher and Schuell (Dassel, Germany). The molecular weight protein marker was from Bio-Rad (Hercules, Calif.). Oligonucleotides were purchased from MWG (Ebersberg, Germany), the pGEX2T vector (included in the glutathione S-transferase [GST] Gene Fusion System) and glutathione Sepharose 4B were obtained from Pharmacia Biotech (Uppsala, Sweden), the DNA molecular weight marker (lambda HindIII) and restriction enzymes were from Roche Diagnostics (Mannheim, Germany), and T4 ligase and competent E. coli cells were from Stratagene (Heidelberg, Germany). The Topo-TA-Cloning kit was from Invitrogen (Groningen, The Netherlands). Taq polymerase was purchased from Roche Diagnostics. The JETsorb DNA extraction kit was from Genomed GmbH (Bad Oeynhausen, Germany). PCR was performed in the Gene Amp PCR System 2400 from Perkin-Elmer (Langen, Germany), and DNA sequencing was done with the Cycle Sequencing Ready Reaction Kit (ABI PRISM) from Perkin-Elmer. Thrombin and phalloidin-rhodamine were obtained from Sigma (Deisenhofen, Germany). [32P]NAD (30 Ci/mmol) was purchased from DuPont NEN (Bad Homburg, Germany).
Construction of CDTa.
For construction of pGEX2T-CDTa, the translated region of CDTa (1,393 bp) was amplified by PCR using genomic DNA from C. difficile strain 75 (a clinical isolate from a patient with an unusual extragastrointestinal C. difficile infection of the hip; kindly provided by Gisela Schallehn, Bonn, Germany) as a template. PCR was performed using the primer pair CDTAs (5′-GGATCCATGAAAAAATTTAGGAAAC-3′) and CDTAa (5′-GAATTCTTAAGGTATCAATGTTGC-3′). Amplification was performed by denaturation at 94°C for 10 s, primer annealing at 48°C for 30 s, and extension at 68°C for 3.5 min, which was repeated for 30 cycles. The PCR product (2 μl) was cloned into the Topo cloning vector pCR2.1 (Invitrogen) and transformed into E. coli TOP10 F′cells according to the manufacturer's instructions. The plasmid was isolated and digested with BamHI-EcoRI for mobilization of CDTa. After separation on a 1% agarose gel, the bands were excised and extracted using the JETsorb DNA extraction kit (Genomed) according to the manufacturer's instructions and further subcloned in the BamHI-EcoRI-digested pGEX2T plasmid.
Construction of CDTa mutants.
The CDTa mutants were constructed by site-directed mutagenesis, using the pGEX2T-CDTa plasmid as a template and the respective oligonucleotides. The Quikchange kit was used according to the manufacturer's instructions. For each mutant, two complementary synthetic oligonucleotides were needed (only one of the two complementary oligonucleotides is indicated): R345K, 5′-CT AAT TTA ACT GTA TAT AAA AGA TCT GGT CCT CAA G-3′; R346K, 5′-CT GTA TAT AGA AAA TCT GGT CCT CAA GAA TTT GG-3′; S388A, 5′-GCA CTG TCT TAT CCA AAC TTT ATT GCT ACT AGT ATT GGT AGT GTG-3′; T389V, 5′-CCA AAC TTT ATT AGT GTT AGT ATT GGT AGT GTG-3′; S390A, 5′-CCA AAC TTT ATT AGT ACT GCT ATT GGT AGT GTG AAT ATG AGT GC-3′; E428Q, 5′-CCA GGT TAT GCA GGT CAA TAT GAA GTG CTT TTA AAT CAT GG-3′; and E430Q, 5′-GGT TAT GCA GGT GAA TAT CAA GTG CTT TTA AAT CAT GG-3′. The mutated plasmids were transformed into E. coli TG1 pACYC-IRL 10 competent cells (Promega Corporation), and the respective mutations were confirmed by DNA sequencing.
Construction of CDTa244–463 and CDTa1–240.
CDTa1–240 was amplified from chromosomal DNA of C. difficile strain 75 with Taq polymerase (Roche Diagnostics) and the oligonucleotide primers CdribAs (5′-GGA TCC ATG AAA AAA TTT AGG AAA C-3′) and CDTa240a-BamHI (5′-GGA TCC AAT TTT ATC TAT TTT TAT AC-3′), both containing a BamHI site. Amplification was performed by denaturation at 94°C for 10 s, primer annealing at 48°C for 30 s, and extension at 68°C for 1 min, which was repeated for 25 cycles. The resulting PCR product (4 μl) was cloned into the Topo cloning vector pCR2.1 and transformed into E. coli TOP10F′ cells according to the manufacturer's instructions. The plasmids were isolated, digested with BamHI, and separated on a 1% agarose gel. The CDTa1–240 band was excised and extracted, using the JETsorb DNA extraction kit (Genomed) according to the manufacturer's instructions, and cloned into the BamHI-digested pGEX2T plasmid. CDTa244–463 was amplified from plasmid pGEX2T-CDTa with Taq polymerase and the oligonucleotide primers CDTA244s-BamHI (5′-GGA TCC GTT ATA GAT GGG AAG CAC-3′) and CdribAa (5′-GAA TTC TTA AGG TAT CAA TGT TGC-3′). Amplification was performed by denaturation at 94°C for 10 s, primer annealing at 52°C for 30 s, and extension at 68°C for 1 min, which was repeated for 25 cycles. The resulting PCR product (4 μl) was cloned into the Topo cloning vector pCR2.1 and transformed into E. coli TOP10F′ cells according to the manufacturer's instructions. The plasmids were isolated, digested with BamHI-EcoRI, and separated on a 1% agarose gel. The CDTa244–463 band was extracted with the JETsorb kit according to the manufacturer's instructions and cloned into the BamHI-EcoRI-digested pGEX2T plasmid.
Construction of N-terminally truncated C2I and iota toxin a.
Truncations of the enzyme components C2I and iota toxin a were obtained by PCR using the pGEX-2T-C2I and pGEX-2T-Ia plasmids, respectively, as templates with the following primers. The 5′ primers harbor a BglII restriction site (underlined), as follows: C2I103–431, AGA TCT AAT TTT ACA CCT AAA GAG CTT G; C2I190–431, AGA TCT GGT TTT TCT ATA ATT CCT GAG; Ia101–413, AGA TCT GCG TTT AAT AAA GAA ATA AGA AC; Ia177–413, AGA TCT ACA TTA ATA GAA CAA GAC TAT AGC; Ia208–413, AGA TCT GTA AAC AGT CTT GAT TTT AAA G; and Ia270–413, AGA TCT GAA CTA GAC TCT AAA GTA AAT AAC. The 3′ primer for the C2I constructs contains a BglII site (underlined): AGA TCT CTA AAT CTC TTT ATT TTG TAT AC. The 3′ primer for iota toxin a truncations has a BamHI site (underlined): GGA TCC TTA ATT TAT CAA TGT TGC ATC CAA. The PCR program was as follows: denaturation for 1 min at 94°C; 25 cycles of denaturation for 10 s at 94°C, annealing at 30 s at 51°C for pGEX-2T-C2I or at 50°C for pGEX-2T-Ia, and elongation with Taq polymerase for 1 min at 68°C; and a final elongation for 10 min at 68°C. The PCR products were cloned into the pCR2.1 vector. The fragments containing C2I truncations were excised by digestion with BglII. For iota toxin a truncations, a double digest with BglII and BamHI was performed. Subsequently, the DNA was ligated into a BamHI-digested and dephosphorylated pGEX-2T plasmid. The correct orientation of the insert was confirmed by sequencing.
Expression and purification of recombinant proteins.
Recombinant wild-type CDTa (rCDTa) or various CDTa mutant proteins were expressed as recombinant GST fusion proteins in E. coli TG1 cells. Bacteria were grown at 37°C in Luria-Bertani medium containing 100 μg of ampicillin per ml and 25 μg of chloramphenicol per ml to an optical density (at 600 nm) of 0.8. At this time, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.1 mM. The cultures were incubated at 29°C for another 24 h. Bacteria were sedimented at 7,000 × g (10 min, 4°C) and resuspended in lysis buffer (10 mM NaCl, 20 mM Tris [pH 7.4], and 1% Triton X-100). The GST fusion proteins were cleaved directly from the beads with thrombin, and the proteins were stored in phosphate buffer containing 0.5% Tween 20 and 0.5% Triton X-100. C2I-C and Ia-C proteins were expressed in E. coli. The proteins were purified as described in detail elsewhere (8).
SDS-PAGE and immunoblotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the methods of Laemmli (15). The gels were stained with Coomassie brilliant blue R-250. The amounts of the CDTa proteins were determined by Coomassie blue staining. Immunoblot analysis was done with an antiserum against C. spiroforme toxin as described previously (7).
ADP-ribosylation assay.
In vitro [32P]ADP-ribosylation of β/γ-nonmuscle actin was performed with platelet cytosol (50 μg of protein) as the substrate and the various toxins as described previously (8). For [32P]ADP-ribosylation of α-actin, purified rabbit skeletal muscle actin (1 μg) was incubated with [32P]NAD and the various toxins at 37°C. Proteins were subjected to SDS–12.5% PAGE, and radiolabeled actin was visualized by phosphorimaging. For in vitro [32P]ADP-ribosylation of lysate proteins from Vero cells, cells were lysed and 50 μg of lysate protein was incubated for 15 min at 37°C in the presence of [32P]NAD.
Cell culture and cytotoxicity assays.
Vero cells were cultivated in tissue culture flasks at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium containing 5% heat-inactivated (30 min, 56°C) fetal calf serum, 2 mM l-glutamate, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Cells were routinely trypsinized and reseeded twice a week. For cytotoxicity assays, Vero cells were grown as subconfluent monolayers in 12-well plates and treated with chymotrypsin-activated Ib together with rCDTa or the mutant CDTa protein in complete medium. For competition assays, subconfluent Vero cells were incubated with Ib plus CDTa together with a 400-fold excess of CDTa1–240.
RESULTS
Cloning and characterization of rCDTa and CDTa mutants.
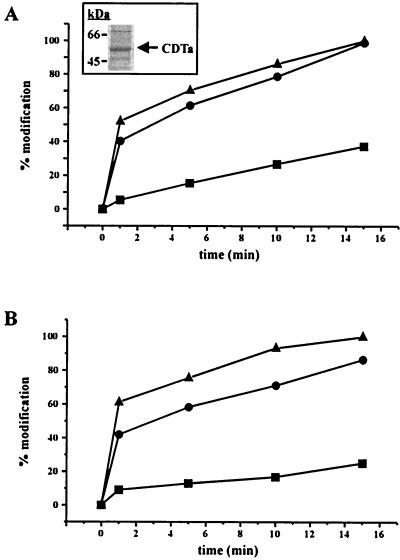
The cDTa gene was amplified from C. difficile strain 75 and ligated into the pGEX plasmid for expression of GST fusion proteins in E. coli. The purified rCDTa was analyzed by SDS-PAGE. The Coomassie blue-stained protein is shown in Fig. 1. rCDTa activity was tested in an in vitro ADP-ribosylation assay with [32P]NAD and β/γ-actin from platelet cytosol or α-actin purified from rabbit skeletal muscle as the substrate. As shown in Fig. 1, rCDTa caused the ADP-ribosylation of platelet actin and of skeletal muscle actin, indicating the same substrate specificity as for iota toxin (11).
FIG. 1.
Characterization of CDTa. In vitro ADP-ribosylation of actin with 1 (▪), 5 (●), and 10 ng (▴) of CDTa was performed for 15 min at 37°C with cytosolic actin from platelets (50 μg of protein) (A) or purified rabbit skeletal muscle actin (1 μg of protein) (B) and [32P]NAD as a cosubstrate. Labeled proteins were detected by SDS-PAGE and phosphorimaging. The percentage of modified actin is shown. The 15-min value for 10 ng of CDTa was taken as 100%. The inset in panel A shows the Coomassie blue staining of purified recombinant CDTa.
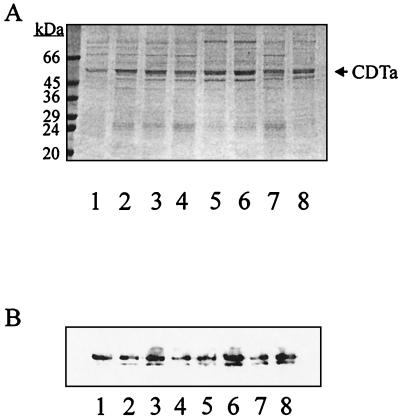
Sequence comparison of CDTa with the C2I ADP-ribosyltransferase from C. botulinum revealed the positions of the highly conserved amino acid residues in CDTa suggested to be involved in NAD-binding and/or enzyme activity (Fig. 2A). Site-directed mutagenesis was performed with the pGEX-CDTa plasmid as a template, and mutations were confirmed by sequencing. Figure 2B summarizes the sequences of mutated CDTa proteins. Proteins were expressed in E. coli and cleaved with thrombin to remove the GST part. The purified CDTa proteins were subjected to SDS-PAGE, and the stained gel is shown in Fig. 3A. Furthermore, CDTa proteins were analyzed by immunoblot analysis with an antibody raised against the C. spiroforme ADP-ribosyltransferase, which is immunologically related to CDTa (20) (Fig. 3B). CDTa and the mutant proteins were recognized by the antibody, showing the expected molecular mass of ≈48 kDa. This antibody did not recognize E. coli proteins and showed no cross-reaction with C2I transferase from C. botulinum (not shown). The lower migrating band was present in all preparations of recombinant CDTa proteins and most likely represents a degradation product.
FIG. 2.
(A) Highly conserved amino acid residues in the catalytic centers of the ADP-ribosyltransferases C2I from C. botulinum, Ia from C. perfringens, and CDTa from C. difficile. Numbers indicate the positions of amino acid residues. The conserved residues are given in boldface. (B) Amino acid sequences of rCDTa (wild type) and the respective mutants. The exchanged amino acid residues are given in boldface.
FIG. 3.
SDS-PAGE and immunoblot analysis of mutant CDTa proteins. CDTa proteins were expressed in E. coli as GST fusion proteins and purified as described in the text. Proteins (500 ng of each) were subjected to SDS–12.5% PAGE and stained with Coomassie blue (A) or blotted onto nitrocellulose and detected with an antiserum against C. spiroforme ADP-ribosyltransferase and peroxidase-coupled protein A (B). Lanes 1, rCDTa; lanes 2, E428Q; lanes 3, E430Q; lanes 4, S388A; lanes 5, T389V; lanes 6, S390A; lanes 7, R345K; lanes 8, R346K.
In vitro ADP-ribosyltransferase activity of CDTa mutants.
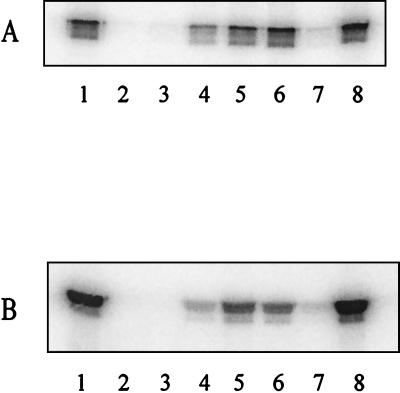
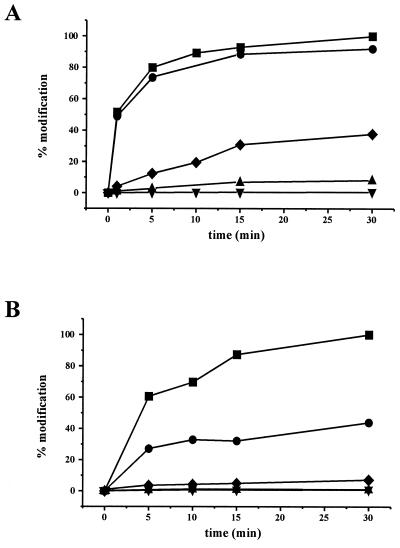
We tested the ADP-ribosyltransferase activities of rCDTa and mutant CDTa in an in vitro ADP-ribosylation assay with [32P]NAD and either platelet cytosol actin or rabbit skeletal muscle actin as a substrate. The autoradiogram of [32P]ADP-ribosylated actin is shown in Fig. 4. Because rCDTa and the CDTa mutant proteins T389V, S390A, and R346K labeled actin from platelet cytosol (Fig. 4A) and skeletal muscle actin (Fig. 4B), these proteins were enzymatically active. By contrast, E428Q, E430Q, S388A, and R345K mutants of CDTa showed no or significantly reduced labeling of actin, indicating that these mutants had no or largely reduced ADP-ribosyltransferase activity. To test the CDTa mutant proteins in more detail, a time course study of the ADP-ribosylation of actin from platelet cytosol (Fig. 5A) and of actin from rabbit skeletal muscle (Fig. 5B) was performed.
FIG. 4.
In vitro ADP-ribosylation of actin with rCDTa and mutant CDTa proteins. ADP-ribosylation of cytosolic actin from platelets (50 μg of protein) (A) and purified rabbit skeletal muscle actin (1 μg of protein) (B) was performed for 15 min at 37°C with [32P]NAD and the various CDTa proteins (100 ng of each). Labeled proteins were detected by SDS-PAGE and phosphorimaging. The autoradiogram is shown. Lanes 1, rCDTa; lanes 2, E428Q; lanes 3, E430Q; lanes 4, S388A; lanes 5, T389V; lanes 6, S390A; lanes 7, R345K; lanes 8, R346K.
FIG. 5.
Time courses of actin ADP-ribosylation by rCDTa (▪) and CDTa mutant proteins E428Q (▾), E430Q (▴), S388A (●), and R345K (♦). ADP-ribosylation was done with platelet cytosol (1 μg of protein) (A) and with purified rabbit muscle actin (1 μg of protein) (B). Proteins were incubated with [32P]NAD, rCDTa (100 ng of CDTa in panel A and 25 ng of CDTa in panel B), and 1 μg of CDTa mutant protein for 1, 5, 10, 15, and 30 min at 37°C. Labeled proteins were detected by SDS-PAGE, and the amount of radioactively labeled actin was quantified by phosphorimaging. The value for the 30-min incubation of the actin with rCDTa was taken as 100%. Shown are results from a representative experiment, which was repeated at least three times with similar results.
Cytotoxic effects of rCDTa and mutant proteins on Vero cells.
Next, we tested whether the various recombinant CDTa proteins caused a breakdown of the actin cytoskeleton in Vero cells. The binding component of iota toxin, Ib, was used to deliver CDTa into the cells. Subconfluently growing Vero cells were incubated with Ib (300 ng/ml) together with the CDTa protein (200 ng/ml) in complete medium at 37°C. For controls, cells were treated without any toxin, with Ib alone, or with the various CDTa proteins alone. After 4.5 h, pictures were taken (Fig. 6). The CDTa proteins (200 ng/ml) which were active in the in vitro ADP-ribosylation assay were also able to induce cell rounding in the presence of Ib. By contrast, Ib plus the CDTa mutant proteins E428Q, E430Q, S388A, and R345K did not cause cell rounding, even when they were added at a higher concentration (1 μg/ml). Ib alone and the CDTa proteins alone had no effect on cell morphology. To confirm that cell rounding was caused by ADP-ribosylation of cellular actin, cells were incubated with Ib together with rCDTa and each CDTa mutant protein. After 4.5 h, cells were lysed and lysate proteins were subjected to an in vitro ADP-ribosylation assay with [32P]NAD. Actin from cells which were intoxicated with active CDTa was already ADP-ribosylated and did not serve as a substrate for radiolabeling. In Fig. 7 the autoradiogram of labeled actin is shown. The actin from cells which were rounded up after toxin treatment was only weakly labeled, whereas the actin from cells which were previously treated with the CDTa mutant proteins E428Q, E430Q, S388A, and R345K showed strong actin labeling.
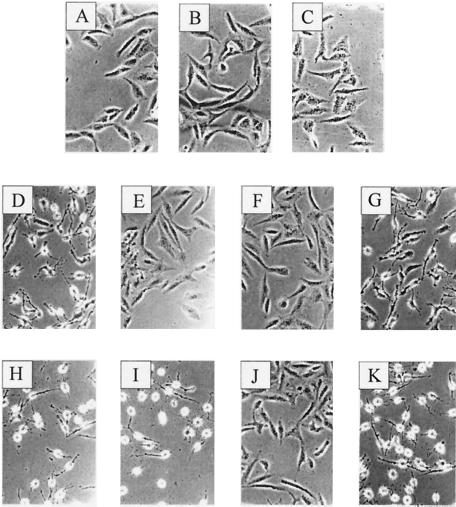
FIG. 6.
Cytotoxic effects of rCDTa and mutant CDTa on Vero cells. Subconfluent Vero cells were incubated in complete medium with the indicated protein at 37°C, and pictures were taken after 4.5 h. For controls, cells were incubated without any toxin (A), with Ib (300 ng/ml) alone (B), or with rCDTa (500 ng/ml) alone (C). The toxin treatments were as follows: rCDTa (D), E428Q (E), E430Q (F), S388A (G), T389V (H), S390A (I), R345K (J), and R346K (K). Samples in panels D to K contained 200 ng of each protein per ml together with 300 ng of Ib per ml.
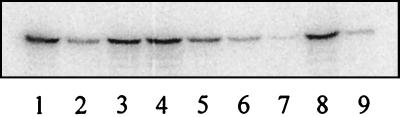
FIG. 7.
In vitro [32P]ADP-ribosylation of Vero cell lysates after treatment of intact cells with Ib and rCDTa and mutant CDTa proteins. Cells were treated for 4.5 h with the toxins (300 ng of Ib per ml and 200 ng of the CDTa protein per ml) at 37°C and lysed. Lysates (50 μg of protein each) were incubated with 50 ng of CDTa and [32P]NAD for 15 min at 37°C. Proteins were subjected to SDS–12.5% PAGE, and labeled actin was detected by phosphorimaging. Lane 1, control cells; lane 2, cells treated with rCDTa; lane 3, cells treated with E428Q; lane 4, cells treated with E430Q; lane 5, cells treated with S388A; lane 6, cells treated with T389V; lane 7, cells treated with S390A; lane 8, cells treated with R345K; lane 9, cells treated with R346K.
In vitro ADP-ribosyltransferase activity of CDTa244–463.
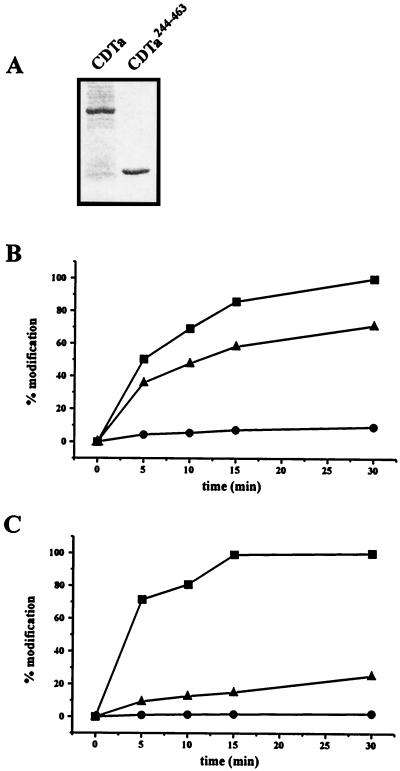
The catalytic center of the CDTa appears to be located at the C-terminal part of the protein. We tested whether the C terminus of CDTa represents a functionally independent domain carrying an ADP-ribosyltransferase activity. An N-terminally truncated fragment, CDTa244–463, was constructed and expressed in E. coli. Figure 8A shows the stained recombinant CDTa and CDTa244–463 proteins after cleavage with thrombin and SDS-PAGE. Both proteins were tested in an in vitro ADP-ribosylation assay with actin from platelet cytosol (Fig. 8B) and from rabbit skeletal muscle (Fig. 8C). In the presence of either actin substrate, the fragment CDTa244–463 was much less active than the full-length CDTa protein.
FIG. 8.
Characterization of CDTa and CDTa244–463 proteins. Proteins were expressed in E. coli, purified as described in the text, and cleaved with thrombin. (A) Proteins were subjected to SDS–12.5% PAGE and stained with Coomassie blue. (B) Time course of ADP-ribosylation of actin from platelet cytosol (50 μg of protein) by full-length CDTa (5 ng) (▪) and CDTa244–463 (100 ng [●] and 1 μg [▴]). (C) Time course of ADP-ribosylation of actin from rabbit skeletal muscle (1 μg of protein) by full-length CDTa (5 ng) and CDTa244–463 (100 ng and 1 μg). The value for the 15-min incubation with full-length CDTa was taken as 100%. Panels B and C show results from representative individual experiments. The experiments were done three times.
Enzymatic activities and substrate specificities of CDTa, Ia, and C2I.
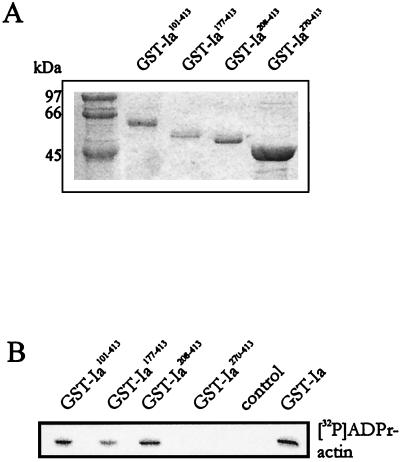
The iota-like toxins CDTa and Ia both ADP-ribosylate β/γ-nonmuscle actin as well as α-actin isoforms (24). By contrast, C2I accepts β/γ-actin but not α-actin isoforms as a substrate (16). Because the enzyme activity of CDTa244–463 was reduced compared to that of the full-length protein, we tested various C-terminal fragments of Ia and C2I for their ADP-ribosyltransferase activities. Various fragments of Ia and C2I were constructed and expressed as GST fusion proteins. For Ia (413 amino acid residues), the fragments GST-Ia101–413, GST-Ia177–413, GST-Ia208–413, and GST-Ia270–413 were purified and analyzed by SDS-PAGE (Fig. 9A). These fragments and GST-Ia were used for in vitro ADP-ribosylation of platelet cytosol. The autoradiogram of 32P-labeled actin is shown in Fig. 9B. GST-Ia, GST-Ia101–413, GST-Ia177–413, and GST-Ia208–413 were enzymatically active, but the fragment GST-Ia270–413 did not ADP-ribosylate actin. Similar results were obtained when thrombin-cleaved Ia was used instead of GST-Ia (not shown).
FIG. 9.
ADP-ribosylation of actin by C-terminal fragments of Ia. (A) Coomassie blue-stained GST-Ia fragments after SDS-PAGE. The numbers give the amino acid residues of Ia. (B) In vitro ADP-ribosylation of actin from platelet cytosol (50 μg of protein) with various GST-Ia proteins. Labeled proteins were detected by SDS-PAGE and phosphorimaging. The autoradiogram is shown.
Next, we tested the C-terminal domain of C2I for ADP-ribosyltransferase activity. Because C2I226–431 was enzymatically inactive, we made further C2I truncations to test which part of the N terminus of this protein can be removed without a loss of ADP-ribosyltransferase activity. All of these N-terminally truncated C2I fragments (C2I103–431 and C2I190–431) were enzymatically inactive, including the fragment C2I30–431 (not shown).
CDTa1–240 mediates the interaction of CDTa with Ib.
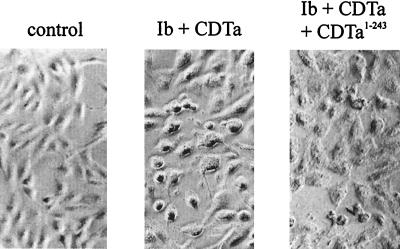
To study the function of the N-terminal domain of CDTa, we expressed the C-terminally truncated CDTa1–240 fragment (amino acids 1 to 240) in E. coli and tested its ability to interact with activated Ib by performing a competition assay with CDTa. Vero cells were incubated with Ib or CDTa in the presence of a 400-fold excess of CDTa1–240. As shown in Fig. 10, after an 8-h incubation period, fewer cells were rounded up after treatment with CDTa plus Ib in the presence of CDTa1–240 than after treatment with CDTa plus Ib. To exclude any nonspecific effects caused by the large amount of CDTa1–240, bovine serum albumin (BSA) and C3 transferase from Clostridium limosum were tested for inhibition of cytotoxic effects induced by CDTa plus Ib. Vero cells were incubated with Ib and CDTa in the presence of BSA (120 μg/ml) or C3 transferase (120 μg/ml). Neither BSA nor C3 transferase showed any effect on the intoxication of the cells by CDTa plus Ib (not shown). This observation suggests that the competition of CDTa1–240 with CDTa was specific and that the N-terminal domain of CDTa interacts with Ib.
FIG. 10.
Influence of CDTa1–240 on cytotoxic effects of Ib plus CDTa on Vero cells. Subconfluently growing Vero cells were incubated in complete medium with 300 ng of Ib per ml with CDTa (300 ng/ml) and with or without a 400-fold excess of CDTa1–240. For a control, cells were treated without toxin. After 8 h, pictures were taken.
DISCUSSION
Here, we characterized the actin ADP-ribosyltransferase CDTa from C. difficile. CDT is a member of the iota family of binary ADP-ribosylating toxins, which also includes iota toxin from C. perfringens and the ADP-ribosyltransferase from C. spiroforme (3, 5, 20, 21, 25). The enzymatic components of these proteins (CDTa, Ia, and Sa) show high sequence homology to the enzymatic component of the C2 toxin from C. botulinum and to VIP2, the enzymatic component of the vegetative insecticidal protein (VIP) from Bacillus cereus. Although it has been directly shown only for C2 toxin (28) and iota toxin (27), all of these proteins are suggested to ADP-ribosylate G-actin at Arg177 to block actin polymerization and to induce the breakdown of the actin cytoskeleton (1).
The ADP-ribosyltransferases C2I, Ia, and CDTa are composed of N- and C-terminal domains which divide the proteins into two sections of similar size. Han et al. reported for the enzyme component (VIP2) of the related VIP toxin that the N- and C-terminal domains share the same topology (13) and may have developed by duplication from a common ancestral ADP-ribosyltransferase gene. We were interested in studying whether the N- and C-terminal parts of ADP-ribosyltransferases represent functionally independent domains that possess adapter and/or enzyme activities when expressed as separated fragments. Our studies with CDTa1–240 revealed that the N-terminal part of the binary ADP-ribosyltransferases is involved in the interaction with the binding component. For C2I, we showed previously that the N terminus (amino acid residues 1 to 225), which is enzymatically inactive, binds to the C2II binding component and mediates uptake of C2I as well as that of the C2I1–225-C3 fusion toxin (6). Even the truncation C2I1–87, consisting of amino acid residues 1 to 87, was able to mediate uptake of the C3 fusion toxin into cells (H. Barth et al., submitted for publication). This strongly suggests that the N-terminal part of C2I represents a functional domain which interacts with C2II. Also, the N terminus of CDTa acts as an adapter and mediates the interaction of the enzyme component with the binding component. Here, we report that the N-terminal domain of CDTa (amino acids 1 to 240) competed with full-length CDTa for the binding component. In our experiments, we used Ib, the binding component of the iota toxin, to deliver CDTa into cells, because the native CDTb binding component was not well expressed as a recombinant protein in E. coli.
The catalytic sites of Ia and C2I, which are located within the C-terminal part of the proteins, were previously characterized in detail. In the enzyme component Ia of C. perfringens iota toxin, Glu380 was identified as the catalytic glutamate that is essential for enzyme activity (18). For the enzyme component C2I of C. botulinum C2 toxin, we reported earlier that Glu387, Glu389, Ser348, and Arg299 are essential for ADP-ribosyltransferase activity, with Glu389 being the catalytic glutamic acid residue (8). In the present study, we show that the equivalent amino acid residues in CDTa are E428, E430, S388, and R345. Mutation of one of these residues inhibited or reduced the enzyme activity of CDTa and prevented intoxication of cells. As deduced from the crystal structure analysis of the related VIP2, the catalytic glutamic acid residue of CDTa (E430) is most likely involved in stabilization of the oxocarbenium transition state by forming a hydrogen bond with the 2′-hydroxyl group of the nicotinamide ribose. Glu428, which is equivalent to Glu426 of VIP2, is suggested to participate in the interaction with the incoming substrate and increases the nucleophilicity of the ADP-ribose acceptor amino acid residue Arg177 of actin. Ser388 is part of the STS motif and is probably involved in stabilization of the NAD-binding pocket. Similarly, Arg345 is likely to be involved in NAD binding (13).
We tested whether the C-terminal domains of the actin ADP-ribosylating toxins CDTa, Ia, and C2I were enzymatically active when they were expressed without their N-terminal parts. Whereas the enzyme domain of iota toxin was fully active compared to the full-length enzyme, CDTa244–463 was markedly less active than the full-length protein. Skeletal muscle actin was less effectively ADP-ribosylated by the CDTa244–463 fragment than cytosolic actin from platelets. One reason might be that nonmuscle actin is a preferred substrate for the toxin. However, it is also plausible that the presence of additional proteins in the cytosolic preparation which prevent polymerization of G-actin to F-actin facilitates ADP-ribosylation, because G-actin but not F-actin is a substrate for ADP-ribosylation (1, 27, 28).
All N-terminally truncated C2I fragments tested were unable to catalyze ADP-ribosylation of actin. Surprisingly, even C2I30–431, from which amino acid residues 1 to 29 were deleted, did not exhibit ADP-ribosyltransferase activity at all. We suggest that amino acid residues 1 to 29 of C2I are necessary for proper folding of the C-terminal part of this enzyme. Thus, although various conserved amino acid residues that may be similarly involved in the enzyme activity of the toxins could be identified, the various actin ADP-ribosylating toxins appear to differ in their minimal structural requirements for proper folding and/or stability.
So far the clinical relevance and pathogenetic role of the actin ADP-ribosylating toxin CDTa are not clear. Only about 7 to 12.5% of clinical isolates contain CDT (20, 26). Diseases induced by C. difficile differ greatly in severity. The spectrum includes mild and trivial diarrhea, pseudomembranous colitis, toxic megacolon, and even fulminant life-threatening colitis. Because CDT is, like other actin ADP-ribosylating toxins, a highly potent cytotoxin, it is plausible that damage of the mucosal barrier function by CDT prepares the way for the action of typical clostridial cytotoxins. Thus, it remains to be clarified whether CDTa plays a specific role in one of the severe clinical manifestations induced by C. difficile.
ACKNOWLEDGMENTS
We thank Otilia Wunderlich for expert technical assistance and B. Stiles (USAMRIID, Frederick, Md.) for the antibody against C. spiroforme ADP-ribosyltransferase.
This study was supported by the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 388).
REFERENCES
- 1.Aktories K, Bärmann M, Ohishi I, Tsuyama S, Jakobs K H, Habermann E. Botulinum C2 toxin ADP-ribosylates actin. Nature. 1986;322:390–392. doi: 10.1038/322390a0. [DOI] [PubMed] [Google Scholar]
- 2.Aktories K, Just I. GTPases and actin as targets for bacterial toxins. In: Dickey B F, Birnbaumer L, editors. GTPases in biology I. Berlin-Heidelberg, Germany: Springer; 1993. pp. 87–112. [Google Scholar]
- 3.Aktories K, Koch G. Modification of actin and of Rho proteins by clostridial ADP-ribosylating toxins. In: Iglewski B, Moss J, Tu A T, Vaughan M, editors. Microbial toxins and virulence factors in disease. New York, N.Y: Marcel Dekker; 1995. pp. 491–520. [Google Scholar]
- 4.Aktories K, Sehr P, Just I. Actin-ADP-ribosylating toxins: cytotoxic mechanism of Clostridium botulinum C2 toxin and Clostridium perfringens iota toxin. In: Aktories K, editor. Bacterial toxins. Weinheim, Germany: Chapman & Hall; 1997. pp. 93–101. [Google Scholar]
- 5.Aktories K, Wille M, Just I. Clostridial actin-ADP-ribosylating toxins. Curr Top Microbiol Immunol. 1992;175:97–113. doi: 10.1007/978-3-642-76966-5_5. [DOI] [PubMed] [Google Scholar]
- 6.Barth H, Hofmann F, Olenik C, Just I, Aktories K. The N-terminal part of the enzyme component (C2I) of the binary Clostridium botulinum C2 toxin interacts with the binding component C2II and functions as a carrier system for a Rho ADP-ribosylating C3-like fusion toxin. Infect Immun. 1998;66:1364–1369. doi: 10.1128/iai.66.4.1364-1369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barth H, Olenik C, Sehr P, Schmidt G, Aktories K, Meyer D K. Neosynthesis and activation of Rho by Escherichia coli cytotoxic necrotizing factor (CNF1) reverse cytopathic effects of ADP-ribosylated Rho. J Biol Chem. 1999;274:27407–27414. doi: 10.1074/jbc.274.39.27407. [DOI] [PubMed] [Google Scholar]
- 8.Barth H, Preiss J C, Hofmann F, Aktories K. Characterization of the catalytic site of the ADP-ribosyltransferase Clostridium botulinum C2 toxin by site-directed mutagenesis. J Biol Chem. 1998;273:29506–29511. doi: 10.1074/jbc.273.45.29506. [DOI] [PubMed] [Google Scholar]
- 9.Blöcker D, Behlke J, Aktories K, Barth H. Cellular uptake of the binary Clostridium perfringens iota-toxin. Infect Immun. 2001;69:2980–2987. doi: 10.1128/IAI.69.5.2980-2987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borriello S P. Clostridial disease of the gut. Clin Infect Dis. 1995;20(Suppl. 2):242–250. doi: 10.1093/clinids/20.supplement_2.s242. [DOI] [PubMed] [Google Scholar]
- 11.Braun M, Herholz C, Straub R, Choisat B, Frey J, Nicolet J, Kuhnert P. Detection of the ADP-ribosyltransferase toxin gene (cdtA) and its activity in Clostridium difficile isolates from equidae. FEMS Microbiol Lett. 2000;184:29–33. doi: 10.1111/j.1574-6968.2000.tb08985.x. [DOI] [PubMed] [Google Scholar]
- 12.Considine R V, Simpson L L. Cellular and molecular actions of binary toxins possessing ADP-ribosyltransferase activity. Toxicon. 1991;29:913–936. doi: 10.1016/0041-0101(91)90076-4. [DOI] [PubMed] [Google Scholar]
- 13.Han S, Craig J A, Putnam C D, Carozzi N B, Tainer J A. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat Struct Biol. 1999;6:932–936. doi: 10.1038/13300. [DOI] [PubMed] [Google Scholar]
- 14.Kelly C P, Pothoulakis C, LaMont J T. Clostridium difficile colitis. N Engl J Med. 1994;330:257–262. doi: 10.1056/NEJM199401273300406. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Mauss S, Chaponnier C, Just I, Aktories K, Gabbiani G. ADP-ribosylation of actin isoforms by Clostridium botulinum C2 toxin and Clostridium perfringens iota toxin. Eur J Biochem. 1990;194:237–241. doi: 10.1111/j.1432-1033.1990.tb19448.x. [DOI] [PubMed] [Google Scholar]
- 17.Pardee J D, Spudich J A. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 18.Perelle S, Domenighini M, Popoff M R. Evidence that Arg-295, Glu-378, and Glu-380 are active-site residues of the ADP-ribosyltransferase activity of iota toxin. FEBS Lett. 1996;395:191–194. doi: 10.1016/0014-5793(96)01035-6. [DOI] [PubMed] [Google Scholar]
- 19.Perelle S, Gibert M, Bourlioux P, Corthier G, Popoff M R. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infect Immun. 1997;65:1402–1407. doi: 10.1128/iai.65.4.1402-1407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popoff M. Molecular biology of actin-ADP-ribosylating toxins. In: Aktories K, Just I, editors. Bacterial protein toxins. Berlin-Heidelberg, Germany: Springer; 2000. pp. 275–306. [Google Scholar]
- 21.Popoff M R, Boquet P. Clostridium spiroforme toxin is a binary toxin which ADP-ribosylates cellular actin. Biochem Biophys Res Commun. 1988;152:1361–1368. doi: 10.1016/s0006-291x(88)80435-2. [DOI] [PubMed] [Google Scholar]
- 22.Popoff M R, Rubin E J, Gill D M, Boquet P. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect Immun. 1988;56:2299–2306. doi: 10.1128/iai.56.9.2299-2306.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenberg S, Stracher A, Lucas R C. Isolation and characterization of actin and actin-binding protein from human platelets. J Cell Biol. 1981;91:201–211. doi: 10.1083/jcb.91.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schering B, Bärmann M, Chhatwal G S, Geipel U, Aktories K. ADP-ribosylation of skeletal muscle and non-muscle actin by Clostridium perfringens iota toxin. Eur J Biochem. 1988;171:225–229. doi: 10.1111/j.1432-1033.1988.tb13780.x. [DOI] [PubMed] [Google Scholar]
- 25.Simpson L L, Stiles B G, Zepeda H, Wilkins T D. Production by Clostridium spiroforme of an iotalike toxin that possesses mono(ADP-ribosyl)transferase activity: identification of a novel class of ADP-ribosyltransferases. Infect Immun. 1989;57:255–261. doi: 10.1128/iai.57.1.255-261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stubbs S, Rupnik M, Gibert M, Brazier J, Duerden B, Popoff M. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol Lett. 2000;186:307–312. doi: 10.1111/j.1574-6968.2000.tb09122.x. [DOI] [PubMed] [Google Scholar]
- 27.Vandekerckhove J, Schering B, Bärmann M, Aktories K. Clostridium perfringens iota toxin ADP-ribosylates skeletal muscle actin in Arg-177. FEBS Lett. 1987;225:48–52. doi: 10.1016/0014-5793(87)81129-8. [DOI] [PubMed] [Google Scholar]
- 28.Vandekerckhove J, Schering B, Bärmann M, Aktories K. Botulinum C2 toxin ADP-ribosylates cytoplasmic β/γ-actin in arginine 177. J Biol Chem. 1988;263:696–700. [PubMed] [Google Scholar]