Abstract
Purpose
Choroidal melanoma (CM) and ciliary body melanoma (CBM) are the two most common subtypes of uveal melanoma. Starting from the observation that CBM tends to have a higher metastatic potential than CM, we hypothesized that specific cytogenetic abnormalities could be associated with tumor location – reflecting distinct genetic signatures that would drive the risk of distant spread.
Methods
Chromosomal alterations were investigated by molecular cytogenetic techniques in 217 and 97 patients with CM and CBM, respectively. Cox proportional hazards regression analysis was used to identify the independent predictors of distant metastasis.
Results
Patients with CBM had larger tumor sizes (P < 0.001), higher disease stages (P < 0.001), and more frequently showed distant metastasis (P = 0.002) than those with CM. On analyzing the entire study cohort, we found that specific chromosomal alterations – including chromosome 8p loss (P < 0.001), 1p loss (P < 0.001), and monosomy 3 (P < 0.005) – were independent predictors of distant metastasis. Based on a decision-tree learning algorithm, we identified three specific subgroups of patients with uveal melanoma at high risk of distant spread. Monosomy 3 occurred significantly more frequently in patients with T3 CBM tumors.
Conclusions
Specific cytogenetic abnormalities – including chromosome 8p loss, 1p loss, and monosomy 3 – are independent risk factors for distant metastasis in uveal melanoma. Larger tumor size at presentation and monosomy 3 contribute to a higher metastatic risk in patients with CBM.
Keywords: uveal melanoma, distant metastasis, choroidal melanoma (CM), ciliary body melanoma (CBM), cytogenetics
Uveal melanoma is the most common primary ocular malignancy and can be classified according to tumor location. Choroidal melanoma (CM) and ciliary body melanoma (CBM) – which account for 85% and 10% of all uveal melanomas, respectively – are the two most prevalent subtypes, followed by iris melanoma (approximately 5%).1,2 Despite recent advances in local tumor control, uveal melanoma has the potential to metastasize, predominantly to the liver, in approximately 50% of the affected patients. The presence of distant metastasis has been associated with a mortality rate of 50% at 10 to 15 years after primary treatment.3–5 Compared with patients with CM, those with CBM generally have an increased risk of distant metastasis and less favorable survival figures.4–7 However, the underlying reasons are complex and only partially understood. In general, the main characteristics of CBM include larger tumor sizes at diagnosis, a trabecular meshwork growth pattern, and a high burden of chromosomal aberrations.4–7
Recent advances in uveal melanoma cytogenetics have fostered our ability to offer a more patient-tailored approach through an improved prognostic stratification.
Monosomy 3 and gain of chromosome 8q have been associated with an increased risk of distant metastasis, whereas chromosome 6q gain predicts a favorable prognosis.4–7 Compared with patients with CM, those with CBM have been reported to show more adverse outcomes. This has been attributed to a generally larger tumor size at diagnosis and a higher burden of cytogenetic abnormalities – including chromosome 1p loss, monosomy 3, and 8q gain. Although the prognostic accuracy can be improved when the American Joint Commission on Cancer (AJCC) stage is combined with information on cytogenetic abnormalities, the genomic landscape of CBM has not been entirely elucidated.4,8,9 In light of the complexity and heterogeneity of uveal melanoma, this study was undertaken to characterize the main differences in terms of chromosomal alterations between CM and CBM and to analyze their associations with the risk of distant metastasis. Starting from the observation that melanomas originating from the choroid and ciliary bodies behave differently,4–7 it can be hypothesized that tumor location could be related to specific cytogenetic abnormalities – reflecting distinct genetic signatures that drive a different risk of distant metastasis.
Patients and Methods
Study Setting and Participants
All procedures and visits occurred at the Princess Margaret Cancer Centre (Toronto, Ontario, Canada). We retrospectively reviewed the clinical charts of all consecutive adult (>18 year old) patients diagnosed with uveal melanoma who consented to undergo fine needle aspiration biopsy for prognostic stratification between February 2015 and October 2021. All biopsy specimens were collected through a trans-scleral or transretinal approach – either before radiotherapy or after enucleation. Patients with inadequate quality or insufficient amount of tumor samples and those with iris melanomas were excluded, as were cases lost to follow-up. The study followed the tenets of the Declaration of Helsinki and was granted ethical approval by the local institutional review board. Written informed consent was obtained from all participants.
Data Collection
Patients were divided into two groups according to the presence or absence of ciliary body involvement using the AJCC TNM classification system. Therefore, ciliary body tumors were defined by the presence of ciliary body involvement. Variables collected from the study patients were sex, age at diagnosis, date of metastatic disease, and duration of follow-up. The following tumor data were recorded: largest basal diameter, height, involved quadrant at the time of treatment, and presence of extra-scleral extension. Patients were staged according to the AJCC TNM staging system for uveal melanoma.10,11 The T category (T1−T4) was assigned according to the measured tumor size. Distant metastases were defined as the spread of uveal melanoma to distant organs confirmed by biopsy or imaging investigations.
Cytogenetic Analysis
All cytogenetic analyses were carried out by an independent company (Impact Genetics Inc., Bowmansville, Ontario, Canada).12 Chromosomal aberrations in tumor specimens were detected using multiplex ligation-dependent probe amplification using a uveal melanoma kit (MRC Holland) that contain probes for chromosomes 1, 3, 6, and 8 (Supplementary Materials). Microsatellite analysis was applied in smaller samples to detect loss of an allele copy or loss of heterozygosity of chromosome 3.12–14
Data Analysis
The general characteristics of the study patients are presented using descriptive statistics. Groups were compared on baseline variables by Fisher's exact test (categorical data) or Student's t-tests (continuous data). The cumulative incidence of distant metastasis was plotted using the Kaplan-Meier method, and differences were compared with the log-rank test. Censoring was performed on the date of the last follow-up. Cox proportional hazards regression analysis was applied to identify the independent predictors of distant metastasis. A multivariate forward selection procedure was implemented, and the results were expressed as hazard ratios (HRs) with 95% confidence intervals (CIs). A decision-tree learning algorithm was used to explore how the study variables influenced the risk of distant metastasis. Statistical calculations were performed using SPSS, version 22.0 (IBM, Armonk, NY, USA). All tests were 2-sided, and statistical significance was set as a P value of < 0.05.
Results
Patient Characteristics
After the exclusion of patients with inadequate quality or an insufficient amount of tumor samples, those with iris melanoma, and cases with a follow-up interval shorter than 6 months, a total of 314 patients (168 men and 146 women) who had undergone cytogenetic analysis were included in the study. A total of 217 (69.1%) and 97 (30.9%) cases were diagnosed with CM and CBM, respectively. The median follow-up time was 29.2 months (range = 6–80.2 months), without significant differences between patients with CM (median = 30.26 months, range = 6–80.2 months) and CBM (median = 28.2 months, range = 6–80.0 months). The clinical characteristics and cytogenetic findings of the study participants are summarized in Table 1. There were no significant intergroup differences in terms of age, although the proportion of men was higher in patients in CM. Cases with CBM had significantly larger tumor sizes, higher disease stages (P < 0.001), and more frequently developed distant metastasis (P = 0.002). On analyzing cytogenetic abnormalities, monosomy 3 and chromosome 8q gain were significantly more common in patients with CBM than in those with CM (P < 0.001).
Table 1.
Clinical Data and Cytogenetic Abnormalities of 314 Patients With Uveal Melanoma
| Characteristics | Full Sample (n = 314) n (%) | Choroidal Melanoma (n = 217) n (%) | Ciliary Body Involved Melanoma (n = 97) n (%) | P Value |
|---|---|---|---|---|
| Age, y (mean, SD) | 60.9 (11.6) | 60.8 (11.5) | 61.2 (11.9) | 0.754 |
| Gender | 0.029 | |||
| F | 146 (46.5) | 92 (42.4) | 54 (55.7) | |
| M | 168 (53.5) | 125 (57.6) | 43 (44.3) | |
| AJCC tumor size | <0.001 | |||
| T1 | 51 (16.2) | 40 (18.4) | 11 (11.3) | |
| T2 | 116 (36.9) | 99 (45.6) | 17 (17.5) | |
| T3 | 127 (40.4) | 70 (32.3) | 57 (58.8) | |
| T4 | 20 (6.4) | 8 (3.7) | 12 (12.4) | |
| AJCC stage | <0.001 | |||
| I | 40 (12.7) | 40 (18.4) | 0 (0) | |
| II | 196 (62.4) | 166 (76.5) | 30 (30.9) | |
| III | 78 (24.8) | 11 (5.1) | 67 (69.1) | |
| Monosomy 3 | <0.001 | |||
| No | 161 (51.3) | 129 (59.4) | 32 (33) | |
| Yes | 153 (48.7) | 88 (40.6) | 65 (67) | |
| 8q gain | <0.001 | |||
| No | 165 (52.5) | 129 (59.4) | 36 (37.1) | |
| Yes | 149 (47.5) | 88 (40.6) | 61 (62.9) | |
| 6p gain | 0.360 | |||
| No | 212 (67.5) | 143 (65.9) | 69 (71.1) | |
| Yes | 102 (32.5) | 74 (34.1) | 28 (28.9) | |
| 8p gain | 0.989 | |||
| No | 288 (91.7) | 199 (91.7) | 89 (91.8) | |
| Yes | 26 (8.3) | 18 (8.3) | 8 (8.2) | |
| 6q loss | 0.091 | |||
| No | 251 (79.9) | 179 (82.5) | 72 (74.2) | |
| Yes | 63 (20.1) | 38 (17.5) | 25 (25.8) | |
| 8p loss | 0.920 | |||
| No | 271 (86.3) | 187 (86.2) | 84 (86.6) | |
| Yes | 43 (13.7) | 30 (13.8) | 13 (13.4) | |
| 1p loss | 0.131 | |||
| No | 261 (83.1) | 185 (85.3) | 76 (78.4) | |
| Yes | 53 (16.9) | 32 (14.7) | 21 (21.6) | |
| 6p loss | 0.309 | |||
| No | 313 (99.7) | 217 (100) | 96 (99) | |
| Yes | 1 (0.3) | 0 (0) | 1 (1) | |
| 6q gain | 0.729 | |||
| No | 304 (96.8) | 209 (96.3) | 95 (97.9) | |
| Yes | 10 (3.2) | 8 (3.7) | 2 (2.1) | |
| 8q loss | >0.999 | |||
| No | 308 (98.4) | 212 (98.1) | 96 (99) | |
| Yes | 5 (1.6) | 4 (1.9) | 1 (1) | |
| Metastatic | 0.002 | |||
| No | 247 (78.7) | 181 (83.4) | 66 (68) | |
| Yes | 67 (21.3) | 36 (16.6) | 31 (32) |
AJCC, American Joint Committee on Cancer; SD, standard deviation.
Independent Predictors of Distant Metastasis
In multivariate analyses (Table 2), the presence of T3 to T4 tumors (adjusted HR = 2.7, 95% CI = 1.6–4.7, P < 0.001), loss of chromosome 1p (adjusted HR = 2.7, 95% CI = 1.6–4.7, P = 0.031), and loss of chromosome 8p (adjusted HR = 3.4, 95% CI = 2.0–5.9, P < 0.001) were independently associated with the occurrence of distant metastasis in the entire study cohort, whereas the presence of disomy 3 (adjusted HR = 0.4, 95% CI = 0.2–0.8, P = 0.005) showed an inverse relation. Based on the measured tumor size, chromosomal aberrations were analyzed in different T subgroups. After applying the Holm-Bonferroni method, we found that monosomy 3 occurred more frequently in T3 CBM tumors (Table 3).
Table 2.
Univariate and Multivariate Analyses of Metastatic Risk
| Univariate | Multivariate (Stepwise) | ||||||
|---|---|---|---|---|---|---|---|
| Characteristics | N | HR | 95% CI | P Value | HR | 95% CI | P Value |
| Group | |||||||
| Choroidal melanoma | 217 | Ref. | |||||
| Ciliary body involved melanoma | 97 | 2.0 | 1.3–3.3 | 0.004 | |||
| AJCC tumor size | |||||||
| T1 and T2 | 167 | Ref. | Ref. | ||||
| T3 and T4 | 147 | 3.4 | 2.0–5.7 | 0.000 | 2.7 | 1.6–4.7 | 0.000 |
| Disomy 3 | |||||||
| No | 182 | Ref. | Ref. | ||||
| Yes | 132 | 0.3 | 0.1–0.5 | 0.000 | 0.4 | 0.2–0.8 | 0.005 |
| Monosomy 3 | |||||||
| No | 161 | Ref. | |||||
| Yes | 153 | 3.0 | 1.8–5.2 | 0.000 | |||
| Isodisomy 3 | |||||||
| No | 285 | Ref. | |||||
| Yes | 29 | 0.8 | 0.4–2.0 | 0.697 | |||
| 8q gain | |||||||
| No | 165 | Ref. | |||||
| Yes | 149 | 4.1 | 2.4–7.3 | 0.000 | |||
| 6p gain | |||||||
| No | 212 | Ref. | |||||
| Yes | 102 | 0.7 | 0.4–1.2 | 0.250 | |||
| 8p gain | |||||||
| No | 288 | Ref. | |||||
| Yes | 26 | 2.2 | 1.2–4.1 | 0.013 | |||
| 6q loss | |||||||
| No | 251 | Ref. | |||||
| Yes | 63 | 2.9 | 1.8–4.8 | 0.000 | |||
| 8p loss | |||||||
| No | 271 | Ref. | Ref. | ||||
| Yes | 43 | 4.9 | 2.9–8.2 | 0.000 | 3.4 | 2.0–5.9 | 0.000 |
| 1p loss | |||||||
| No | 261 | Ref. | Ref. | ||||
| Yes | 53 | 3.2 | 1.9–5.4 | 0.000 | 2.7 | 1.6–4.6 | 0.000 |
| 6p loss | |||||||
| No | 313 | Ref. | |||||
| Yes | 1 | NA | |||||
| 6q gain | |||||||
| No | 304 | Ref. | |||||
| Yes | 10 | 1.0 | 0.2–4.2 | 0.981 | |||
| 8q loss | |||||||
| No | 308 | Ref. | |||||
| Yes | 5 | 3.2 | 1–10.3 | 0.049 | |||
CI, confidence interval; HR, hazard ratio; Ref., reference; NA, not applicable.
Table 3.
Chromosome Aberrations in Different Subgroups of Uveal Melanoma (Full Sample, n = 314 Patients)
| Choroidal Melanoma (n = 217) | Ciliary Body Involved Melanoma (n = 97) | |||
|---|---|---|---|---|
| Variables | n (%) | n (%) | P Value | Adjusted P Value |
| Monosomy 3, n (%) | ||||
| T1 | 12/40 (30) | 5/11 (45.5) | 0.336 | 0.584 |
| T2 | 42/99 (42.4) | 12/17 (70.6) | 0.032 | 0.096 |
| T3 | 29/70 (41.4) | 38/57 (66.7) | 0.005 | 0.020 |
| T4 | 5/8 (62.5) | 10/12 (83.3) | 0.292 | 0.584 |
| Chromosome 8q gain | ||||
| T1 | 7/40 (17.5) | 4/11 (36.4) | 0.178 | 0.712 |
| T2 | 34/99 (34.3) | 7/17 (41.2) | 0.586 | 0.759 |
| T3 | 41/70 (58.6) | 39/57 (68.4) | 0.253 | 0.759 |
| T4 | 6/8 (75) | 11/12 (91.7) | 0.306 | 0.759 |
| Chromosome 8p loss | ||||
| T1 | 3/40 (7.5) | 1/11 (9.1) | 0.862 | 1.000 |
| T2 | 8/99 (8.1) | 2/17 (11.8) | 0.617 | 1.000 |
| T3 | 16/70 (22.9) | 8/57 (14) | 0.207 | 0.828 |
| T4 | 3/8 (37.5) | 2/12 (16.7) | 0.292 | 0.876 |
| Chromosome 1p loss | ||||
| T1 | 1/40 (2.5) | 2/11 (18.2) | 0.050 | 0.200 |
| T2 | 14/99 (14.1) | 3/17 (17.6) | 0.706 | 1.000 |
| T3 | 14/70 (20) | 13/57 (22.8) | 0.701 | 1.000 |
| T4 | 3/8 (37.5) | 3/12 (25) | 0.550 | 1.000 |
| Chromosome 6p gain | ||||
| T1 | 6/40 (15) | 2/11 (18.2) | 0.797 | 1.000 |
| T2 | 36/99 (36.4) | 3/17 (17.6) | 0.131 | 0.524 |
| T3 | 28/70 (40) | 17/57 (29.8) | 0.233 | 0.699 |
| T4 | 4/8 (50) | 6/12 (50) | 1.000 | 1.000 |
| Metastasis | ||||
| T1 | 1/40 (2.5) | 2/11 (18.2) | 0.050 | 0.200 |
| T2 | 14/99 (14.1) | 2/17 (11.8) | 0.793 | 1.000 |
| T3 | 16/70 (22.9) | 20/57 (35.1) | 0.128 | 0.384 |
| T4 | 5/8 (62.5) | 7/12 (58.3) | 0.852 | 1.000 |
The P values were adjusted by the Holm–Bonferroni method.
Decision-Tree Learning Algorithm
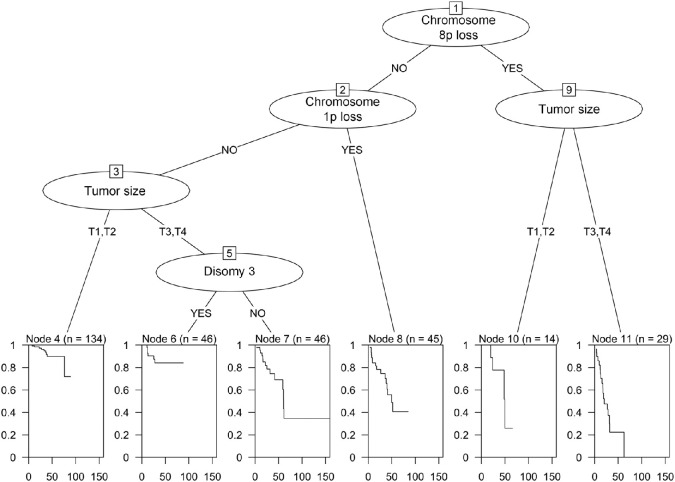
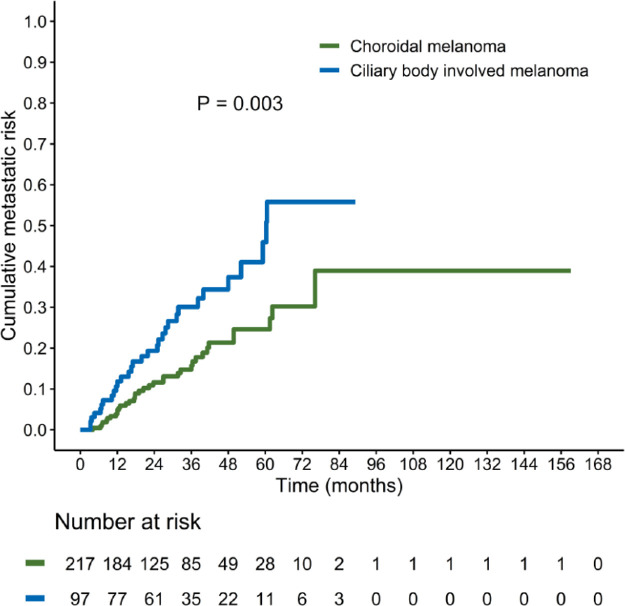
By taking into account the four independent predictors of distant metastasis identified in multivariate analysis, a decision-tree learning process was implemented to identify specific subgroups of patients with uveal melanoma at high risk of distant spread (Fig. 1). The following high-risk subgroups were identified: (1) patients with loss of chromosome 8p (regardless of tumor size); (2) patients with loss of chromosome 1p; and (3) patients with T3 to T4 tumors who did not harbor disomy 3. Kaplan-Meier plots revealed that the cumulative incidence of distant metastasis was significantly higher in the CBM group (Fig. 2).
Figure 1.
Cumulative incidence of distant metastasis in patients with uveal melanoma, stratified according to tumor location (choroidal melanoma versus ciliary body melanoma).
Figure 2.
Patients at high risk of distant metastasis were identified through a decision-tree learning process. The first classification tree distinguished the presence versus absence of chromosome 8p loss. When this alteration was lacking, the risk of distant spread was trained according to chromosome 1p. When 1p was also lacking, the classification tree in patients harboring T3 to T4 tumors and monosomy 3 as consecutive decision node to classify the risk of distant metastasis. The high-risk group comprised (1) patients with chromosome 8p loss (regardless of tumor size); (2) patients harboring chromosome 1p loss; and (3) patients harboring T3 to T4 tumors and monosomy 3.
Discussion
In the current study, we were able to confirm that CBM portends a higher risk of metastatic dissemination compared with CM.3,10 The high metastatic burden associated with CBM is generally explained by its late detection, which not only results in greater tumor growth but also increases the likelihood of systemic spread. As expected, we found that patients with CBM had a larger tumor size than those with CM. Interestingly, we also observed that both monosomy 3 and chromosome 8q gain, 2 well-known unfavorable prognostic biomarkers in uveal melanoma,14–16 were over-represented in patients with CBM. This prompted us to further dissect the role of cytogenetic abnormalities in the prognostic stratification of uveal melanoma in relation to its anatomic location. In general, we can highlight three principal findings from our study. First, specific chromosomal alterations (chromosome 8p loss, 1p loss, and the absence of disomy 3) were identified as independent predictors of distant metastasis in the entire cohort. Second, based on the combined analysis of tumor size and chromosomal alterations through a decision-tree learning algorithm, we identified three specific subgroups of patients with uveal melanoma at high risk of distant spread. Third, monosomy 3 was significantly more frequent in patients with T3 CBM tumors.
In an effort to improve the prognostic stratification in patients with uveal melanoma, the identification of chromosomal alterations independently associated with clinical outcomes has recently gained momentum.13–18 Apart from the absence of disomy 3, losses of chromosomes 8p and 1p were identified as being independently associated with distant metastasis in our study. Surprisingly, loss of chromosome 8p was the strongest predictor for the occurrence of distant metastasis. In addition, the results of our decision-tree learning algorithm revealed that the presence of chromosome 8p loss was sufficient to identify a subgroup of patients at high risk of distant metastasis, regardless of tumor size. The prognostic significance of loss of chromosome 8p has been rarely reported in the published literature and is in need of verification via further confirmatory research. For example, on analyzing 356 patients with uveal melanoma with data on chromosome 3 and chromosome 8, Damato and coworkers found no evidence of this aberration.15 However, it is worth noting that Onken et al.19 identified the leucine zipper tumor suppressor-1 (LZTS1) gene as a potential metastasis-suppressor located on chromosome 8p. Interestingly, Yavuzyigitoglu et al.20 have recently reported that chromosome 1p loss and 8p loss were significantly more frequent in primary uveal melanomas of patients who eventually developed miliary metastases compared with those who developed single solitary hepatic metastases. Collectively, this evidence should prompt further research on the potential role of chromosome 8p loss in influencing the risk of distant spread in patients with uveal melanoma.
By applying a decision-tree learning process, three subgroups at high risk of distant metastasis were identified. The first classification tree distinguished the presence versus absence of chromosome 8p loss regardless of tumor size. When this alteration was lacking, the risk of distant spread was trained according to chromosome 1p loss. Finally, the classification tree in patients harboring T3 to T4 tumors was based on the presence or absence of disomy 3 as the main decision node (see Fig. 1).
Several caveats of this study should be acknowledged. First, its single-center design may have limited the external validity of the findings. Second, the study participants represented a convenience sample. The prevalence of CBM in our study cohort (31%) was higher than that reported in the literature (10%).1,2 Therefore, prospective confirmation is required to assert the generalizability of our findings. Third, the multiplex ligation-dependent probe amplification (MLPA) assay used for this study did not comprise probes for 1q, although 7 probes for 1p were available. Fourth, it would have been interesting to include cytogenetic findings related to the BAP1 status, which is related to metastatic risk. Unfortunately, we did not have the data. Finally, the median follow-up of both groups was relatively short to detect a long-term effect of cytogenetic abnormalities on metastatic outcomes. Despite these limitations, this study provides initial evidence that the prognostic significance of cytogenetic abnormalities and tumor size in uveal melanoma outweighs that of tumor location.
Conclusions
The risk of distant metastasis in uveal melanoma is independently associated with tumor size and the presence of specific cytogenetic abnormalities (absence of disomy 3 and losses of chromosomes 8p and 1p). The combined analysis of lesion size and chromosomal alterations through a decision-tree learning algorithm allowed identifying specific subgroups at high risk of distant spread, which should be prioritized for periodic systemic surveillance.
Supplementary Material
Acknowledgments
Disclosure: A.-N. Chao, None; K. Rose, None; H. Racher, None; F. Altomare, None; H. Krema, None
References
- 1. Egan KM, Seddon JM, Glynn RJ, Gragoudas ES, Albert DM.. Epidemiologic aspects of uveal melanoma. Surv Ophthalmol . 1988; 32: 239–251. [DOI] [PubMed] [Google Scholar]
- 2. Aronow ME, Topham AK, Singh AD.. Uveal Melanoma: 5-Year Update on Incidence, Treatment, and Survival (SEER 1973-2013). Ocul Oncol Pathol . 2018; 4: 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shields CL, Say EAT, Hasanreisoglu M. , et al. Personalized Prognosis of Uveal Melanoma Based on Cytogenetic Profile in 1059 Patients over an 8-Year Period: The 2017 Harry S. Gradle Lecture. Ophthalmology . 2017; 124: 1523–1531. [DOI] [PubMed] [Google Scholar]
- 4. Dogrusöz M, Bagger M, Van Duinen SG. , et al. The prognostic value of AJCC staging in uveal melanoma is enhanced by adding chromosome 3 and 8q status. Med Image Comput Comput Assist Interv. 2017; 58: 833–842. [DOI] [PubMed] [Google Scholar]
- 5. Singh AD, Turell ME, Topham AK.. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology . 2011; 118: 1881–1885. [DOI] [PubMed] [Google Scholar]
- 6. Damato BE, Heimann H, Kalirai H, Coupland SE.. Age, survival predictors, and metastatic death in patients with choroidal melanoma: tentative evidence of a therapeutic effect on survival. JAMA Ophthalmol . 2014; 132: 605–613. [DOI] [PubMed] [Google Scholar]
- 7. Mooy CM, De Jong PT.. Prognostic parameters in uveal melanoma: a review. Surv Ophthalmol . 1996; 41: 215–228. [DOI] [PubMed] [Google Scholar]
- 8. Cunha Rola A, Taktak A, Eleuteri A. , et al. Multicenter External Validation of the Liverpool Uveal Melanoma Prognosticator Online: An OOG Collaborative Study. Cancers (Basel) . 2020; 12: 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cassoux N, Rodrigues MJ, Plancher C. , et al. Genome-wide profiling is a clinically relevant and affordable prognostic test in posterior uveal melanoma. Br J Ophthalmol . 2014; 98: 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kujala E, Damato B, Coupland SE. , et al. Staging of ciliary body and choroidal melanomas based on anatomic extent. J Clin Oncol . 2013; 31: 2825–2831. [DOI] [PubMed] [Google Scholar]
- 11. Bagger M, Andersen MT, Andersen KK. , et al. The prognostic effect of American Joint Committee on Cancer staging and genetic status in patients with choroidal and ciliary body melanoma. Invest Ophthalmol Vis Sci . 2014; 56: 438–444. [DOI] [PubMed] [Google Scholar]
- 12. Singh AD, Aronow ME, Sun Y. , et al. Chromosome 3 status in uveal melanoma: a comparison of fluorescence in situ hybridization and single-nucleotide polymorphism array. Invest Ophthalmol Vis Sci . 2012; 53: 3331–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vaarwater J, van den Bosch T, Mensink HW. , et al. Multiplex ligation-dependent probe amplification equals fluorescence in-situ hybridization for the identification of patients at risk for metastatic disease in uveal melanoma. Melanoma Res . 2012; 22: 30–37. [DOI] [PubMed] [Google Scholar]
- 14. Aalto Y, Eriksson L, Seregard S, Larsson O, Knuutila S.. Concomitant loss of chromosome 3 and whole arm losses and gains of chromosome 1, 6, or 8 in metastasizing primary uveal melanoma. Invest Ophthalmol Vis Sci . 2001; 42: 313–317. [PubMed] [Google Scholar]
- 15. Damato B, Duke C, Coupland SE. , et al. Cytogenetics of uveal melanoma: a 7-year clinical experience. Ophthalmology . 2007; 114: 1925–1931. [DOI] [PubMed] [Google Scholar]
- 16. Sisley K, Rennie IG, Parsons MA. , et al. Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes Chromosomes Cancer . 1997; 19: 22–28. [DOI] [PubMed] [Google Scholar]
- 17. Ewens KG, Kanetsky PA, Richards-Yutz J. , et al. Genomic profile of 320 uveal melanoma cases: chromosome 8p-loss and metastatic outcome. Invest Ophthalmol Vis Sci . 2013; 54: 5721–5729. [DOI] [PubMed] [Google Scholar]
- 18. Kilic E, van Gils W, Lodder E. , et al. Clinical and cytogenetic analyses in uveal melanoma. Invest Ophthalmol Vis Sci . 2006; 47: 3703–3707. [DOI] [PubMed] [Google Scholar]
- 19. Onken MD, Worley LA, Harbour JW.. A metastasis modifier locus on human chromosome 8p in uveal melanoma identified by integrative genomic analysis. Clin Cancer Res . 2008; 14: 3737–3745. [DOI] [PubMed] [Google Scholar]
- 20. Yavuzyigitoglu S, Tang MCY, Jansen M. , et al. Radiological Patterns of Uveal Melanoma Liver Metastases in Correlation to Genetic Status. Cancers (Basel) . 2021; 13: 5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.