Abstract
Molecular composition is intricately intertwined with cellular function, and elucidation of this relationship is essential for understanding life processes and developing next-generational therapeutics. Technological innovations in capillary electrophoresis (CE) and liquid chromatography (LC) mass spectrometry (MS) provide previously unavailable insights into cellular biochemistry by allowing for the unbiased detection and quantification of molecules with high specificity. This chapter presents our validated protocols integrating ultrasensitive MS with classical tools of cell, developmental, and neurobiology to assess the biological function of important biomolecules. We use CE and LC MS to measure hundreds of metabolites and thousands of proteins in single cells or limited populations of tissues in chordate embryos and mammalian neurons, revealing molecular heterogeneity between identified cells. By pairing microinjection and optical microscopy, we demonstrate cell lineage tracing and testing the roles the dysregulated molecules play in the formation and maintenance of cell heterogeneity and tissue specification in frog embryos (Xenopus laevis). Electrophysiology extends our workflows to characterizing neuronal activity in sections of mammalian brain tissues. The information obtained from these studies mutually strengthen chemistry and biology and highlight the importance of interdisciplinary research to advance basic knowledge and translational applications forward.
Keywords: Single cell, mass spectrometry, functional biology, proteomics, metabolomics, cell and developmental biology, neurobiology, Xenopus laevis, zebrafish, mouse
1. INTRODUCTION
Modern ‘omics provide an unprecedented set of technologies to study connections between molecular composition and biological function. By enabling the detection and quantification of genes, transcripts, proteins, peptides, and metabolites, valuable information is gained on the molecular underpinnings of biological processes. High-throughput sequencing routine profiling of gene expression to large populations of single cells [1–3]. Identification and quantification of molecular markers from discovery ‘omics help develop hypotheses and design experiments to evaluate their biological function.
Functional biological experiments leverage diverse types of approaches and technologies. Functional tests borrow tools of molecular biology to knock down or out genes, transcripts, or proteins, e.g., by using transcription or translation blocking morpholinos and CRISPR-Cas9. Electrophysiology allows for eavesdropping on neurons. Different types of microscopies, typically optical to electron, enable characterization of cell morphology, phenotype, and anatomy (Fig. 1). Single-cell RNA-sequencing with single-molecule fluorescence in situ hybridization (sm-FISH) recently captured molecular cell heterogeneity in the brain in high spatial and molecular resolution [4], supplementing classical knowledge of brain anatomy with molecular information [5]. By comparing single thalamic neurons projecting to motor, somatosensory, and visual cortices in the mouse brain, this approach uncovered several cell types within each projection. Technologies from modern ‘omics empower classical biology and neuroscience with new investigative capabilities.
Figure 1.
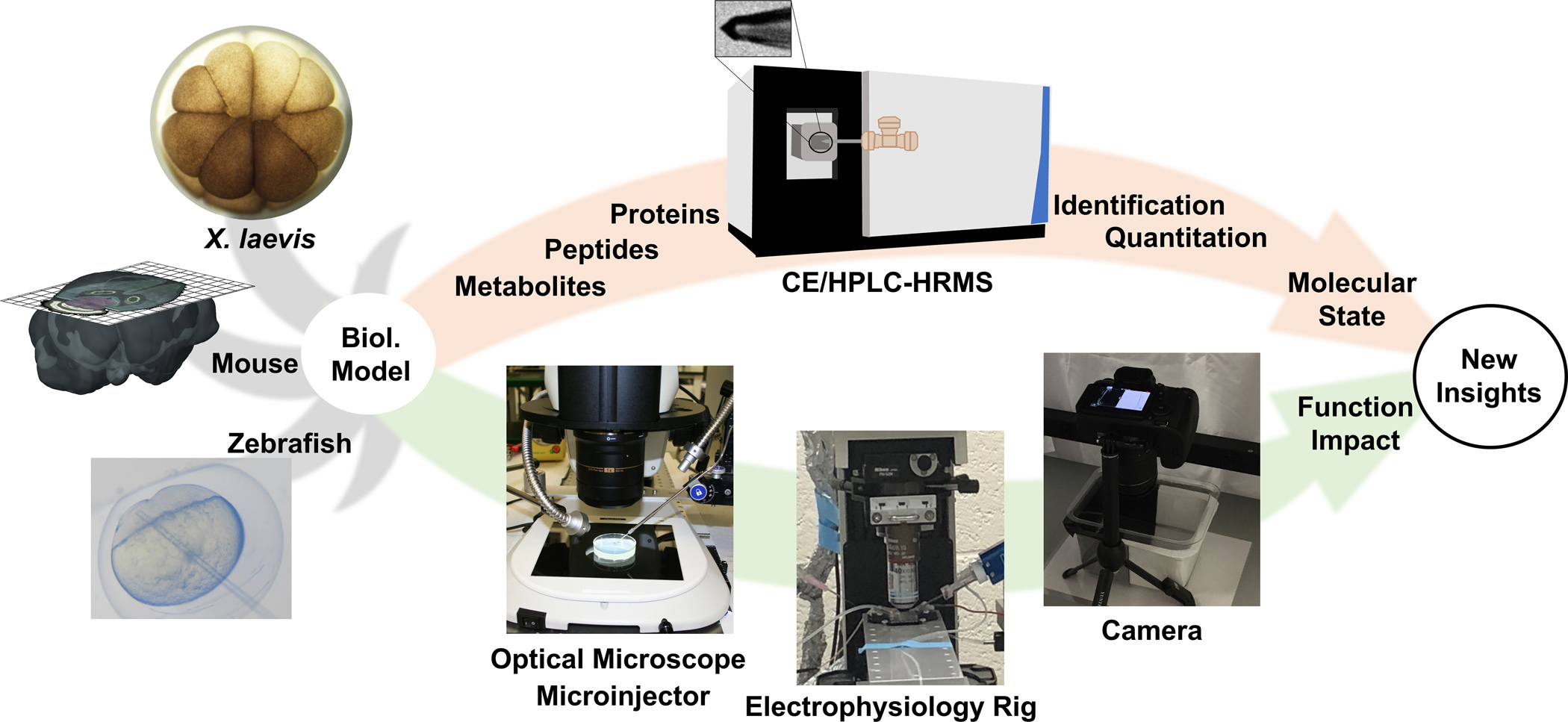
Our approach for assessing chemistry and functional in biological models. The protocols discussed in this chapter were developed and validated using embryos from the frog Xenopus laevis and zebrafish and sections from the mouse brain. (Adapted with permission from Anal. Chem. 2017, 89, 13, 7069–7076. Copyright 2017 American Chemical Society. Adapted with permission from Anal. Chem. 2019, 91, 7, 4797–4805. Copyright 2019 American Chemical Society.)
Single-cell mass spectrometry (MS) supports systems biological studies with information on the molecular state of cells. It complements single-cell transcriptomics by directly measuring proteomic, peptidomic, and metabolomic composition (Fig. 1). Detection with excellent molecular specificity facilitates identification without the requirement for functional probes. For example, MS does not necessitate antibodies for detection. High sensitivity and a broad linear dynamic range permit quantification of molecules at endogenous concentrations. Stringent reporting guidelines [6,7] and public repositories hosting data for reuse, reanalysis, and exchange (e.g., PRIDE [8] and MetabolomicsWorkbench [9]) promote scientific rigor and accountability in MS-based research. The current state of the field of single-cell MS was the focus of several reviews covering technology and application [10–19]. Single-cell MS is adaptable to broad types of molecules, can be made sufficiently sensitive to quantify physiological concentrations, and is compatible with cells of broad dimensions and types, as well as different model systems used in biology and health studies.
Optical microscopy with single-cell MS integrates morphological and molecular information. Recent reviews provide a comprehensive discussion of single-cell MS [12,18–20]. Figure 1 illustrates our protocol for optically guided single-cell MS in embryos of the South African clawed frog (X. laevis) and zebrafish and sections of the mouse brain. Cells were identified, dissected [21,22] or their contents directly microaspirated [23–25] for metabolomic and proteomic analysis using capillary electrophoresis (CE) or liquid chromatography (LC) electrospray ionization (ESI) high-resolution MS (HRMS) (reviewed in [26–29]). Microscopy with single-cell MS enabled detection of ~1,500 proteins in 2- to 50-cell X. laevis embryos and orthogonal validation using immunohistochemistry [30,31]. Single-cell proteomics by MS (SCoPE MS) quantified 3,000+ proteins from 1,490 cells [32,33], and a single-cell printer with liquid vortex capture enabled rapid metabolomics (~25 cells/min) [34]. With low attomole sensitivity, HPLC columns of narrow bore dimension or with a stationary phase supported on a monolith or a porous layer open tubular (PLOT) format allowed for the identification of ~1,300–4,000 proteins from 50–200 cells via magnetophoretic isolation from whole blood [35–37]. Nanodroplet processing in one pot for trace samples (NanoPOTS) identified over ~1,500 proteins from 10 HeLa cells and ~2,400 proteins from 100 pancreatic islet cells, supporting profiling across clinical samples [38]. These and other leaps in technology expanded the classical toolbox of cell biology, as was discussed in our recent review of the fields [26–29].
We and others built CE-MS platforms to study biomolecules and their role in cell and neurobiological processes. CE combines advantages for single-cell analyses. The physical dimensions of fused silica capillaries are amenable to the limited amounts of sample that are contained in single cells. CE provides several methods for concentration enrichment in the capillary to boost sensitivity to low-abundance molecules (e.g., reviewed in [39]). An exquisite separation power and various data alignment strategies permit reproducible identifications [40,41]. CE-ESI interfaces offer various designs to help hyphenate CE with HRMS for sensitivity, robustness, and reproducibility [42,43].
These custom-built CE-ESI-HRMS platforms revealed previously unknown details on cellular biochemistry. Proteins or metabolites were measured in single cells of X. laevis and zebrafish embryos (Fig. 1 and 2) [21–23,25], single neurons dissected from Aplysia californica [44,45] and mouse [46,40], and single HeLa [47] cells. Our CE-ESI interface enabled the identification of hundreds of cationic and anionic metabolites (Fig. 3) [48] and ~700 proteins from ~5 ng protein digest from single X. laevis cells [25]. Targeted neuropeptides were detected in record sensitivity in the subfornical organ (SFO) and the paraventricular nucleus (PVN) of the mouse hypothalamus (Fig. 3D) [49]. With a 200-zmol lower limit of detection, this technology also identified ~500 proteins from ~1 ng and ~225 proteins from ~500 pg protein digest, which estimates to a single neuron [50,46,51]. Our second-generation CE-ESI HRMS design employing a microprobe capillary enabled the in situ and in vivo analysis of single identified cells in live embryos [25,23,52] and the mouse brain [53,54,50] (Fig. 2). As shown in Figure 4, its integration with cell labeling and stereomicroscopy permitted the tracing of tissue developmental trajectory. These instrumental capabilities revealed differences in the proteomic [22,25,55] and metabolic [21,23] state of cells in X. laevis and zebrafish embryos, including those occupying the dorsal-ventral [21,22,56], animal-vegetal [25,57], and left-right [58,59] developmental axes in the frog. They also led to discovering metabolite-induced cell fate changes [21,60] and metabolic communication between neighboring cells in X. laevis [61]. Further, the approach can be extended to patch-clamp electrophysiology, permitting the metabolomic [62] and proteomic [63,53,54] characterization of identified neurons in the mouse brain. Most recently, we also integrated single-cell CE MS with assays measuring background color preference and swim pattern to assess the impact of metabolic perturbation on organismal behavior [61,52].
Figure 2.
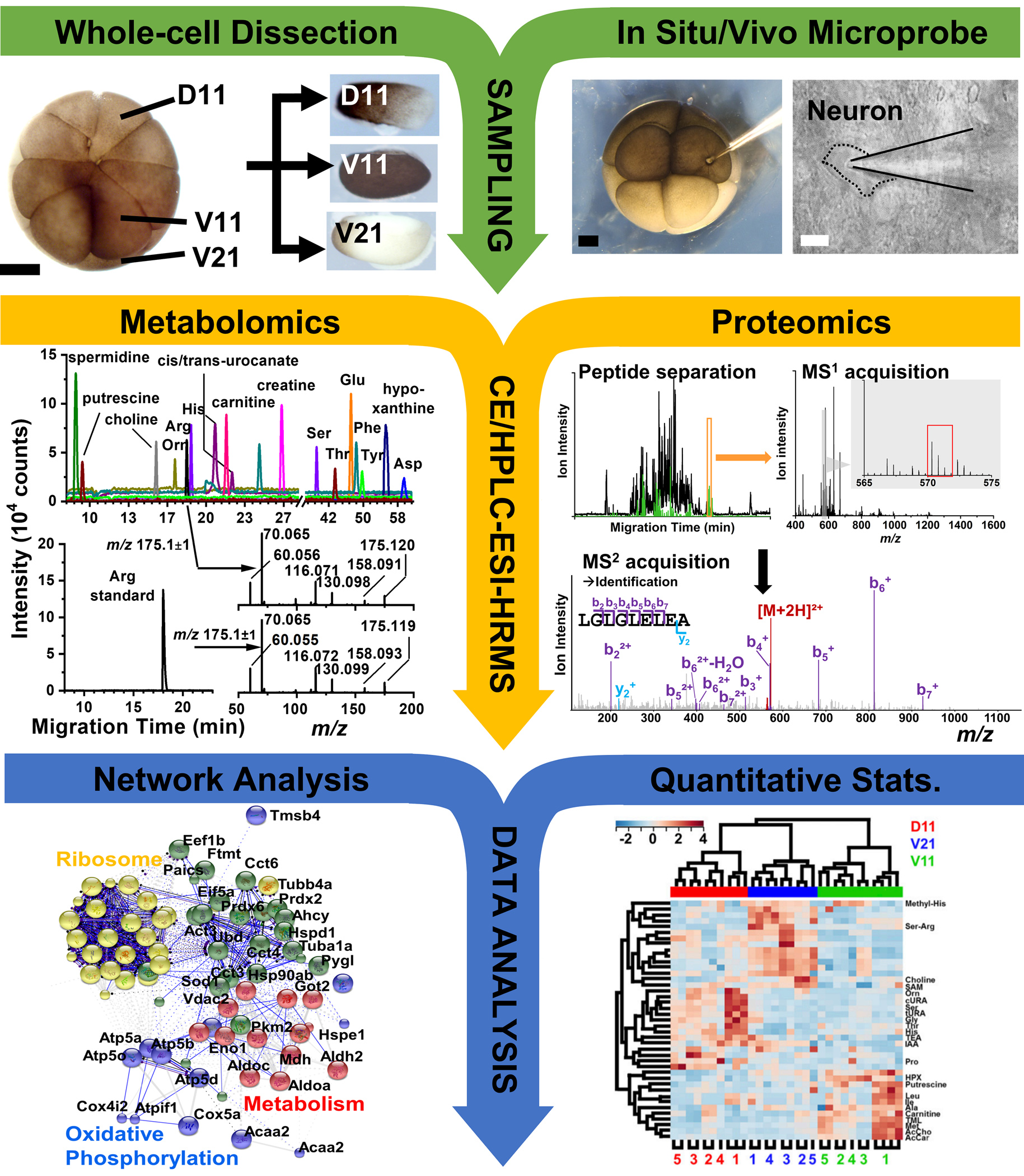
Our general single-cell MS workflow enabling the analysis of metabolites and proteins in single cells of Xenopus laevis or zebrafish and single neurons in a section of the mouse brain. Scale bars: 250 μm (black), 20 μm (white). (Adapted with permission from Ref. [57]; Adapted with permission from Ref. [21]. Copyright 2015 National Academy of Sciences; Adapted with permission from Anal. Chem. 2017, 89, 13, 7069–7076. Copyright 2017 American Chemical Society; Adapted with permission from Ref. [26].)
Figure 3.
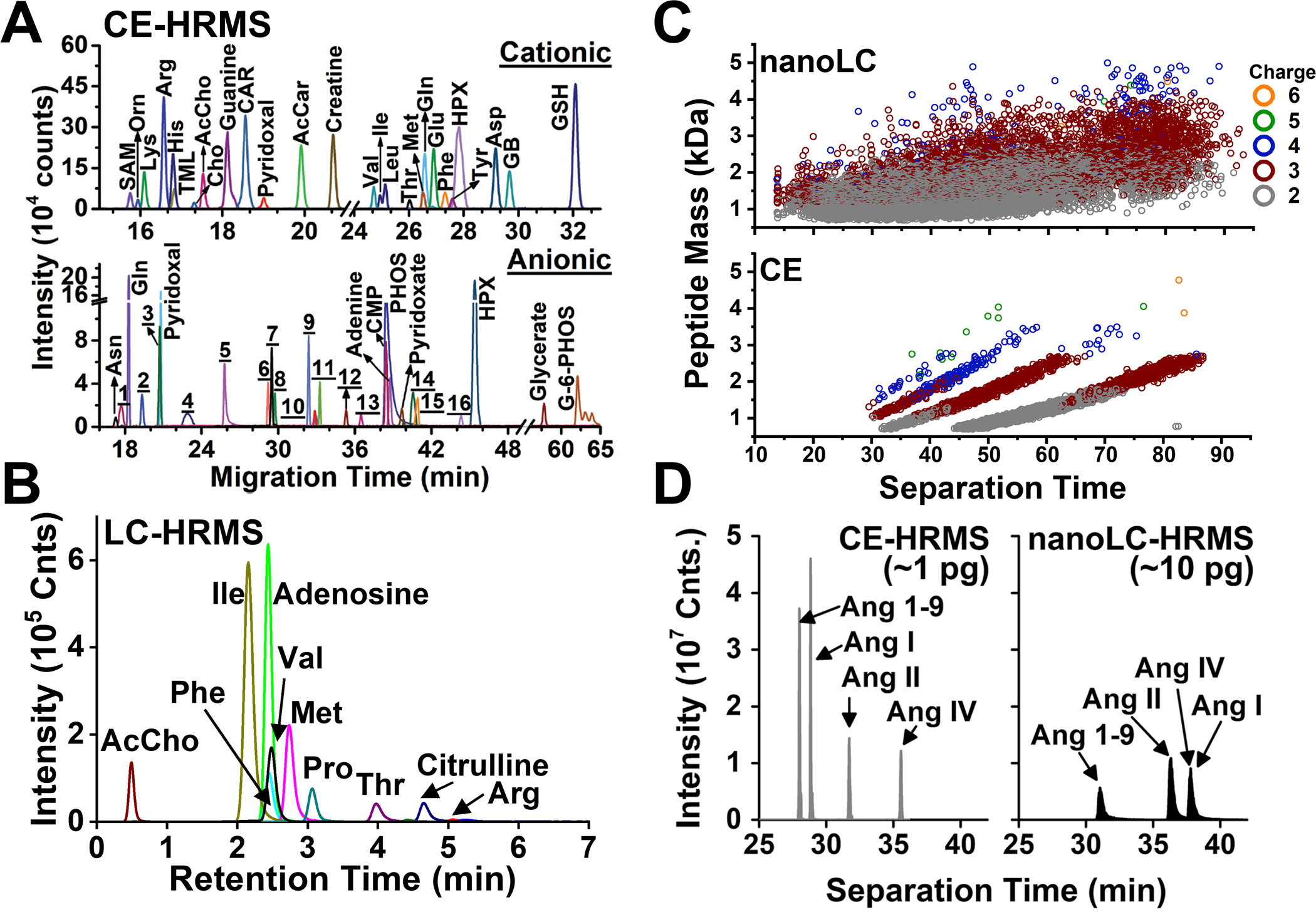
Representative detection of metabolites and proteins by CE and HPLC ESI-HRMS. (A) Chemical profiling of anionic and cationic metabolites in a single X. laevis cell using CE-ESI-MS. (B) HILIC LC-MS of polar metabolites from limited populations of cells. (C) Comparison of peptide identifications by CE-ESI-HRMS and nanoLC-nanoESI-HRMS. (D) Targeted detection of angiotensin peptides in the PVN and SFO of the mouse hypothalamus using CE and nanoLC. (Reproduced from Ref. [48,49] with permission from The Royal Society of Chemistry and Springer Nature, Copyright 2019.)
Figure 4.
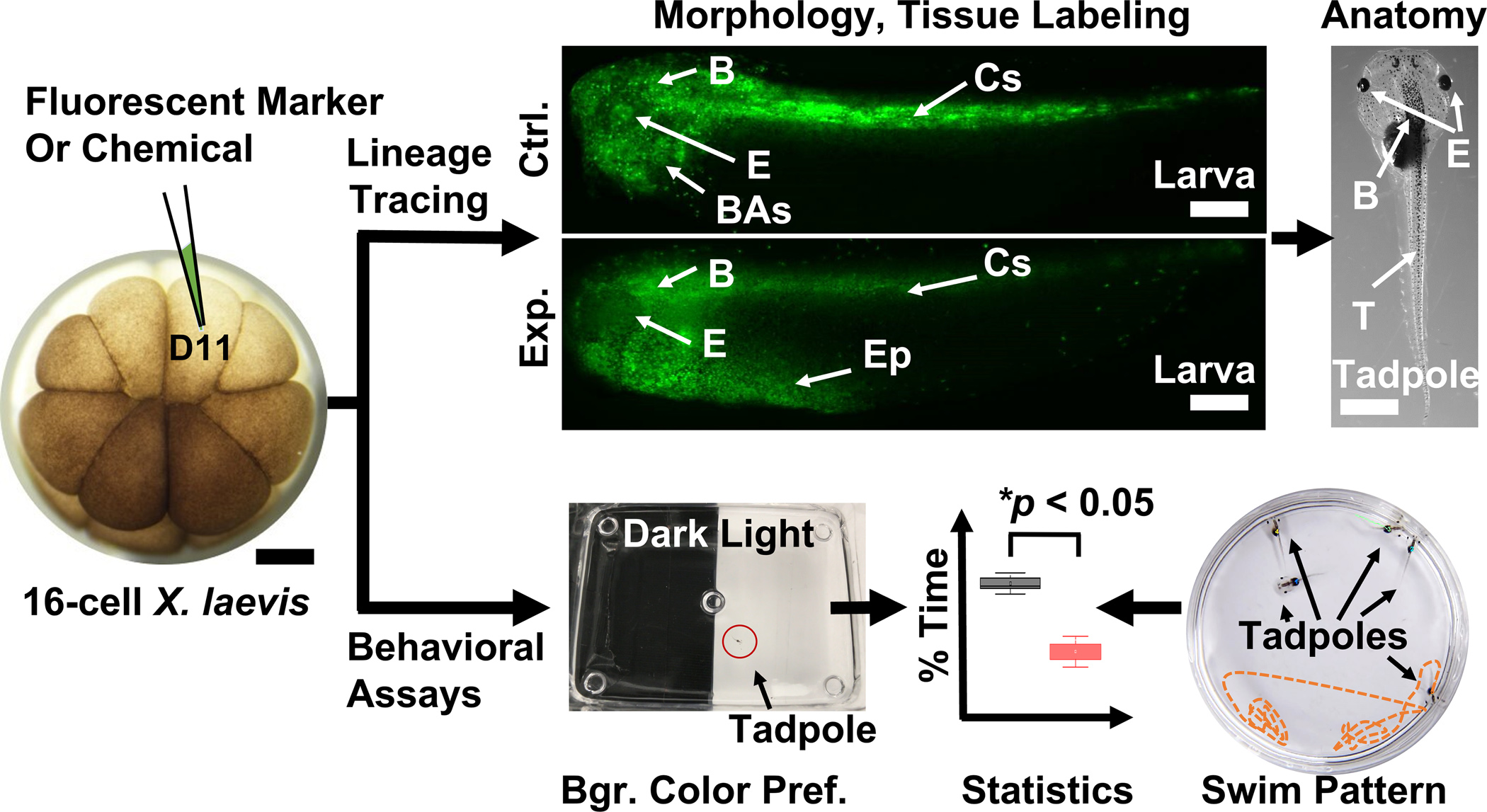
Techniques to investigate chemistry and function during development. (Top panel) Analysis of cell fate, morphology, and anatomy following fluorescence lineage tracing of the left dorsal-animal midline cell (D11) in control (Ctrl.) and experimental (Exp.) X. laevis larvae. (Bottom panel) Background color preference and swim patter assays evaluating behavior in X. laevis tadpoles. Key: B, brain; BAs, branchial arches; Cc; central somites; E, eye; Ep; epidermis. Scale bars = ~250 μm (embryo, larvae), ~1.5 mm (tadpole). (Adapted with permission from Anal. Chem. 2017, 89, 13, 7069–7076. Copyright 2017 American Chemical Society.)
This chapter presents our protocol enabling functional biological studies with insights to the proteomic and metabolomic state of cells in chordate embryos and mammalian neurons (Fig. 1). We overview required consumables and instruments (see Materials and Methods) and discuss the experimental workflow (Fig. 2). After identifying cells based on optical microscopy or electrophysiology and cell sampling by dissection or microprobe aspiration, the collected material is processed, and the resulting metabolites, peptides, or proteins are measured using LC and CE. Statistical analysis of signal abundances detected by HRMS–MS/MS allowed us to identify compounds for biological investigations. As an example, we present our approaches to test the effect of select compounds on tissue development (Fig. 4). Representative examples are discussed with references to data showing the integration of single-cell ‘omics (Fig. 3) with functional biology (Fig. 4). The Note section advises on troubleshooting from the vantage point of an experimentalist, thus hoping to promote the combined use of single-cell HRMS-based proteomics/metabolomics with functional biology in other cell types and biological models.
2. MATERIALS
2.1. Culturing Embryos and Neurons
Animals: Adult male and female Xenopus laevis frogs (e.g., from Nasco, Fort Atkinson, WI); adult male and female zebrafish (e.g., from Zebrafish International Resource Center, Eugene, OR); Adult male mouse (e.g., from Charles River Laboratories, Wilmington, MA) (see Note 1).
Equipment: Incubators set to 14 °C and 18 °C; Stereomicroscope.
-
Solutions:
Dejellying solution (2% cysteine): Dissolve 4 g of cysteine in 200 mL deionized (DI) water. Add 10 M sodium hydroxide dropwise to adjust pH to 8.
100% Steinberg’s solution (SS): Mix 3.4 g sodium chloride (NaCl), 0.05 g potassium chloride (KCl), 0.08 g calcium nitrate (Ca(NO3)2 × 4 H2O), 0.205 g magnesium sulfate (MgSO4 × 7 H2O), 0.66 g Trizma hydrochloride, 0.075 g Trizma base in 1 L of DI water. Adjust the pH to 7.4 by adding Trizma base and autoclave the solution. Store at 4–14 °C.
Anesthetic solution (ketamine, 20 mg/mL and dexmedetomidine, 0.1 mg/mL): Prepare by mixing 200 μL of ketamine stock solution (100 mg/mL) and 200 μL of dexmedetomidine stock solution (0.5 mg/mL) with 800 μL of 0.9% saline solution for injection (e.g., part no. NDC0409-4888-06, Hopira, Inc., Lake Forest, IL).
HEPES ringer solution: Prepare by mixing the following reagents to the following final concentrations: 86 mM NaCl, 2.5 mM KCl, 1.2 mM sodim phosphate (NaH2PO4), 35 mM sodium bicarbonate (NaHCO3), 20 mM HEPES, 25 mM glucose, 5 mM sodium ascorbate, 2 mM thiourea, 3 mM sodium pyruvate, 1 mM MgSO4, 2 mM calcium chloride (CaCl2).
Perfusion solution for mouse brain slices: Prepare by mixing the following reagents to final concentration: 126 mM NaCl, 21.4 mM NaHCO3, 2.5 mM KCl, 1.2 mM NaH2PO4, 2.4 mM CaCl2, 1.0 mM MgSO4, and 11.1 mM glucose.
2.2. Embryology
-
Equipment:
Stereomicroscope for embryology to identify, inject, and microsample single cells (e.g., SMZ1270, Nikon, Melville, NY) and phenotype embryos, larvae, and tadpoles (e.g., SMZ18, Nikon).
Microinjector to inject into or aspirate contents from identified cells (e.g., PLI-100A, Warner Instruments, Hamden, CT).
Micromanipulator (e.g., MM-33, Warner Instruments)
Capillary pullers (e.g., P-1000 for fused silica capillaries and P-2000 for borosilicate capillaries, Sutter Instruments, Novato, CA)
Ancillary equipment: Heat block; Centrifuge (e.g., Refrigerated 5430R, Eppendorf)
-
Materials and Solutions:
Fine sharp forceps (e.g., Dumont #5, Fine Science Tools)
Centrifuge vials (e.g., 0.2–0.5 mL LoBind tubes, Eppendorf)
Borosilicate glass capillary (e.g., 0.5/1.0 mm inner/outer diameter, Sutter Instruments)
Hair loop: Place both ends of a fine hair (~10 cm length) into a 6-inch Pasteur pipet to form a 2 mm loop. Secure it in place with melted paraffin. Sterilize before use by dipping in 70% ethanol and air-drying.
Dissection dish: Cover the bottom of a Petri dish (35- or 60-mm) with non-toxic modeling clay (e.g., Claytoon was tested in Xenopus labs [64]). Make 3–5 wells of ~1.5 mm diameter using a cool glass bead to hold the embryos in place.
Injection dish: Cover the bottom of a Petri dish (35- or 60-mm) with non-toxic modeling clay. Make several ~1.5 mm wells using a cool glass bead across the dish to hold ~20–30 embryos.
50% Steinberg’s solution: Dilute 500 mL of 100% SS with 500 mL of DI water.
20% Steinberg’s solution: Dilute 200 mL of 100% SS with 800 mL of DI water.
2.3. Sample Processing
Equipment: Vacuum concentrator (e.g., CentriVap, LabConco, Kansas City, MO)
Reagents: All reagents are LC-MS grade to reduce chemical interference during MS detection. Methanol, anhydrous acetonitrile (ACN), water, acetic acid, 100 mM TEAB, 5% hydroxylamine, isobaric labeling kit (e.g., TMTsixplex, ThermoFisher Scientific or iTRAQ, AB Sciex)
-
Solutions:
Metabolite extraction solvent: 50% (v/v) methanol in water containing 0.5% acetic acid.
Proteomic digestion buffer: 50 mM ammonium bicarbonate containing protease inhibitor (1 protease inhibitor cocktail tablet per 10 mL)
Trypsin (0.5 μg/μL) in 50 mM acetic acid
Patch-clamp solution: We use 50 mM ammonium bicarbonate in water as a compromise between sensitivity and function.
2.4. Mass Spectrometry
-
Instrument and Materials:
CE system (e.g., laboratory-built following [21,40] or CESI, AB Sciex, Toronto, Canada)
High-resolution tandem mass spectrometer (e.g., quadrupole time-of-flight, Impact HD or TimsTOF, Bruker Scientific, Billerica, MA or quadrupole orbitrap Q-Exactive Plus or Orbitrap Fusion Lumos, Thermo Scientific, Fig. 1)
HPLC (e.g., Acquity I-class UPLC, Waters, Milford, MA and Dionex Ultimate 3000, ThermoScientific)
Separation CE capillary (e.g., 40/100 μm inner/outer diameter fused silica, Polymicro Technologies, Phoenix, AZ)
LC column (e.g., Acquity UPLC BEH Amide Column, 1.7 μm, 1 mm × 100 mm, and Acclaim PepMap C18 column, 3 μm, 0.075mm × 250 mm, Waters)
-
Solutions: All solvents and reagents are LC-MS grade.
CE background electrolyte solution (BGE): 1% (v/v) formic acid in water for metabolomics; 25% (v/v) acetonitrile in water with 1 M formic acid for proteomics.
CE-ESI sheath solution: 0.1% (v/v) formic acid and 50% methanol for metabolomics; 10% (v/v) acetonitrile in water with 0.05% acetic acid for proteomics.
LC mobile phase for metabolomics using hydrophilic interaction LC (HILIC): For cationic separation, mobile phase A is aqueous 0.1% formic acid and B is acetonitrile containing 0.1% formic acid; for anionic separation, mobile phase A is aqueous 5% acetonitrile with 10 mM ammonium bicarbonate (pH 9) and B is aqueous 95% acetonitrile with 10 mM ammonium bicarbonate (pH 9).
LC mobile phase for proteomics using reversed-phase LC (RPLC): Mobile phase A is aqueous 0.1% formic acid and B is acetonitrile containing 0.1% formic acid.
2.5. Functional Studies
-
Instrument and Equipment:
Epifluorescence stereomicroscope (e.g., SMZ18, Nikon, Fig. 1)
Inverted microscope (e.g., Eclipse Ti-U, Nikon)
Microinjector (e.g., PLI-100A, Warner Instruments)
Micromanipulator (e.g., MM-33, Warner Instruments)
Camera with tripod (e.g., ESO70D, Canon, Fig. 1)
Software for processing movies (e.g., Windows media player software)
Ancillary: Incubator set to 14 °C; Nutator rotator
Patch amplifier for electrophysiology (e.g., Sutter Instruments)
-
Materials:
Tadpole food (e.g., part no. SA05964, Nasco)
Transfer pipets
Black electrical tape
26 G needle (e.g., part no. BD305115, Fisher Scientific)
1/2-gallon tank (e.g., part no. SB19271M, Nasco)
Fine sharp forceps (e.g., Dumont #5)
Inoculating turntable (e.g., part no. 50809-022, VWR)
-
Reagents:
200 proof ethanol
Gentamicin antibiotic (e.g., part no. 17-528Z, Fisher Scientific)
Sylgard 184 silicone elastomer (e.g., part no. NC9285739, Fisher Scientific)
Ficoll 400 (Sigma-Aldrich, St. Louis, MO)
Benzocaine (part no. E1501-500G, Sigma-Aldrich)
1X phosphate-buffered saline (PBS) (Fisher Scientific)
Fluorescent lineage tracer (e.g., fluorescent dextran 10,000 MW lysine fixable or mRNA lineage tracer, Invitrogen, Carlsbad, CA)
-
Solutions:
3% Ficoll in 100 % SS: Prepare by mixing 3 g of Ficoll in 100 mL of 100% SS.
4% paraformaldehyde: Prepare by mixing 4 g of paraformaldehyde in 40 mL DI water at 60 °C. Add a few drops of 1 N NaOH to adjust pH to 7.4. Add DI water to a total volume of 100 mL.
DEPC water: Add 1 mL diethyl pyrocarbonate to 1 L DI water. Autoclave the solution, seal, and store at room temperature.
10% benzocaine: Weight out 2 g of benzocaine and place in a glass beaker. Add 20 mL 200 proof ethanol and stir using a magnetic bar in a stirrer.
2% benzocaine in 20% SS: Add 2 mL of 10% benzocaine to 8 mL of 20% SS dropwise, ensuring complete dissolution.
3. METHODS
3.1. Culturing
This step lays out our methodology to culture X. laevis embryos to larvae or tadpoles and primary neurons from the mouse on the basis of established protocols [65,66]. X. laevis embryos require additional dejellying step for manipulation and functional experiments, as described below.
1. Frog Embryos
Obtain fertilized eggs by gonadotropin-induced natural mating of adult X. laevis frogs or in vitro fertilization as detailed elsewhere [65]. See Note 1 on working with live vertebrate animals.
-
Remove the jelly coat from fertilized eggs as follows:
Remove excess media from dishes containing fertilized eggs.
Add dejellying solution and keep embryos unperturbed for 2 min.
Gently swirl dishes over a 2 min period and immediately decant excess dejellying solution.
Transfer the embryos to a 250 mL beaker and add 10% SS. Gently swirl for ~30 s and decant excess liquid.
-
Rinse embryos four times with 10% SS to remove remaining dejellying solution.
Transfer ~300–500 embryos into individual 100 mm Petri dishes containing 100% SS. Place dishes in the 14 °C incubator (see Note 2).
-
Collect two-cell embryos:
Under a stereomicroscope, identify 2-cell embryos that display stereotypical pigmentation to accurately mark the dorsal-ventral axis, in reference to established cell fate maps [67].
Place the selected 2-cell embryos into a 100 mm Petri dish containing 100% SS and incubate at 14 °C until the desired developmental stage.
2. Brain Section
-
Brain sections were collected following established protocols [68]:
Anesthetize male mice aged postnatal day (PND) 21–35 with an intraperitoneal injection of anesthetic solution and perfuse with ice-cold HEPES ringer solution.
After perfusion, dissect the brain rapidly in horizontal slices (220 μm) prepared in HEPES ringer solution using a vibratome.
Recover slices for 1 h at 34°C in oxygenated HEPES holding solution. Then, place slide in the same solution at room temperature until use.
3.2. Sample Collection from Single Cells
The goal of this step is to collect material from targeted single cells. In what follows, we give an example for Xenopus. The workflow starts with the identification of single cells in the X. laevis embryo in reference to established cell fate maps [69,70]. As shown in Figure 2, our laboratory established orthogonal strategies to collect single-cell samples. We dissected identified whole cells from X. laevis embryos [21,26] or used fabricated microcapillaries as microprobes to aspirate portions of single identified cells from the embryo [23,28,24] or electrophysiologically characterized mouse neurons [63,53].
Whole-Cell Dissection from X. laevis Embryos
-
Under a stereomicroscope, identify the cell of interest based on stereotypical cleavage and pigmentation, then dissect it manually as follows:
Transfer the embryo into a dissection dish containing 50% SS.
Using a hair loop, place the embryo of interest in a groove.
Gently remove the vitelline membrane surrounding the embryo using sharp forceps.
Use forceps to hold the embryo, preferably at the opposite side of the cell of interest. Lightly pull away the selected cell from the rest of the embryo.
Transfer the isolated cell using a pipet into a LoBind Eppendorf vial containing chilled 10 μL methanol (~4 °C) or digestion buffer (see Note 3).
Cool the Eppendorf vial (on ice) to preserve sample at low temperature and store samples at −80 °C until analysis and up to 3 months without detectable degradation.
1. In Situ/Vivo Microsampling
Fabricate the microprobe by pulling a borosilicate glass capillary to create a fine tip. We use a capillary puller (P-1000 Sutter Instrument) with custom settings: heat = 355, pull = 65, velocity = 80, time = 150.
Using sharp forceps, cut the needle tip to an aperture of ~10–20 μm (see Note 4).
Mount the microprobe into a capillary holder on a three-axis micromanipulator and connect its distal end to a microinjector.
Transfer X. laevis embryos into an injection dish containing 50% SS. This protocol does not require removal of vitelline membrane.
Use a stereomicroscope to aid viewing and manipulation of the embryo. Use a hair loop to immobilize the embryo of interest into a well in preparation for microsampling. Identify the cell of interest following protocols established elsewhere [69,70].
Using a micromanipulator, guide the tip of the microprobe into the targeted cell to pierce through the membrane of the cell. Withdraw ~10–15 nL (or as needed) volume from the targeted cell by applying negative pressure to the microprobe using the connected microinjector.
To end the microsampling, reduce pressure and retract the microprobe from the cell.
Transfer the collected content into a LoBind Eppendorf vial containing 4 μL of metabolite extraction solvent or digestion buffer (see Note 5). We usually inject the collected material from the capillary by inserting the tip into the solvent/buffer and applying a positive pressure pulse.
Cool the Eppendorf vial (on ice) to preserve sample at low temperature and immediately process the samples via metabolomics or proteomics workflows to prevent molecular degradation.
3.3. Sample Processing for MS-Based ‘Omics
This section discusses protocols to process the collected materials for HRMS analysis. We use LC and CE to separate biomolecules in complementary performance prior to ESI-HRMS.
1. Metabolomics Workflow
-
Extract metabolites from dissected single cells [21,23] or aspirates collected by microprobe sampling [23,48] as follows:
For dissected single cells: Retrieve the single-cell samples stored in 100% methanol from −80 °C freezer and vacuum-dry them at 4 °C. Add 10 μL metabolite extraction solvent. Vortex-mix the vials for 30 s at room temperature to facilitate cell lysis and extraction of metabolites. Sonicate the sample vials for 3 min in an ice bath, followed by vortex-mixing for 1 min at room temperature.
For microprobe aspirated samples: Retrieve single-cell samples collected in metabolite extraction solvent preserved on ice. Vortex-mix the vials for 1 min at room temperature to facilitate extraction of metabolites.
For CE-ESI-HRMS, centrifuge the samples (dissected or microsamples) for 5 min at 8,000 × g at 4 °C to pellet cellular debris. Proper pelleting is important to avoid CE capillaries from getting clogged. We usually store the aliquot together with the pelleted debris to avoid sample losses. The samples are kept at −80 °C until analysis.
For HILIC-ESI-HRMS, centrifuge the samples at 13,000 × g for 10 min at 4 °C and transfer the supernatant into a microvial and vacuum-dry the samples at 4 °C. Reconstitute the samples in 10 μL 95% (v/v) acetonitrile in water and centrifuge the samples at 13,000 × g for 10 min at 4 °C to pellet potential debris that could clog the column (see Note 6). Transfer the supernatant into an LC vial and store the sample at −80 °C until analysis.
2. Proteomics Workflow
Lyse the collected cell or aspirate by sonication for 5 min. Heat the sample to 60 °C for ~15 min to denature proteins. We usually skip reduction and alkylation steps for our single cell samples for higher sensitivity [25].
For one-step digestion, add ~50 ng of trypsin protease to the protein extract and incubate the mixture at 37 °C for 5–6 h. For neurons yielding less starting protein amounts, add ~2 ng trypsin and digest at 60 °C for 1 h.
Vacuum-dry the resulting protein digest and store it at −80 °C until analysis.
-
(Optional) To enable multiplexing relative quantification, barcode the dried protein digests. We use TMT isobaric labeling following the vendor’s protocol (see Note 7), downscaled to the total amount of protein/peptide contained in the sample:
Reconstitute the dried protein digest in 10 μL of 100 mM TEAB and tag it with 1 μL of 85 mM TMT reagent.
Incubate each sample for 1 h at room temperature.
Quench the reaction with 2 μL of 5% hydroxylamine and incubate the mixture for 15 min at room temperature.
Mix the multiple tagged samples, vacuum-dry the mixture, and store it at −80 °C for up to 1 month until analysis.
3.4. High-Resolution Mass Spectrometry
In this step, biomolecules in the resulting samples are separated and detected using ESI-HRMS. We present protocols for separation based on partition chromatography (LC) and electrophoresis (CE). These separation techniques provide complementary benefits in sensitivity, throughput, and molecular coverage (Fig. 3). The resulting data are processed using established approaches in bioinformatics, including but not limited to statistics, multivariate data analysis(e.g., principal component analysis and hierarchical cluster analysis), or machine learning (e.g., Trace [71,72]). These technologies and related protocols (Fig. 2) allowed us to document metabolic and proteomic differences between cells in embryos of X. laevis and zebrafish [58,21,22,25] and single neurons in the mouse brain [53,54,63].
1. CE-ESI-HRMS
-
Construct the CE-ESI interface following protocols established by us and others [40,50,73,44]. A simplified procedure to build a blunt-tip CE-ESI interface follows:
Cleave a 1-m long piece of fused-silica CE capillary.
On the outlet end of the CE capillary, burn off ~1.5 mm of polyimide coating and clean using isopropanol. Before proceeding to the next step, ensure that the capillary end is clean of burned residues to avoid the leaching of interfering ion signals from the burned residue.
Feed the CE capillary outlet-end into a T-junction connected to a sheath-flow capillary.
Mount the CE capillary into the T-junction to feed the CE capillary through the blunt tip emitter, allowing the capillary to protrude ~40–50 μm past the emitter.
Hydrate the CE capillary by flushing with LC-MS grade water overnight.
Position the tip of the CE-ESI interface ~5 mm from the inlet orifice of the mass spectrometer.
Fill CE capillary with BGE and flush sheath-flow capillary with sheath solution.
-
Initiate the electrospray as follows:
Using a translation stage, fine-position the electrospray emitter tip ~2–3 mm from the mass spectrometer orifice to operate the electrospray in the cone-jet regime (ESI voltage 1.8–2.0 kV).
Monitor the electrospray using a stereomicroscope.
Observe the stability of total ion current (TIC) for ~10–15 min to ensure stable operation before analyzing a sample (see Note 8).
Inject ~10–15 nL from the metabolite or protein extract hydrodynamically into the CE capillary following previously described protocols [24,40].
Gradually increase CE separation voltage from ground (0 V) to ~20–22 kV. Sudden application of high voltage may break the capillary.
Load the MS method as described in Table 1 (see CE-HRMS) and start data acquisition (see Note 9).
Table 1.
Our typical instrumental settings for detecting metabolites and proteins in CE and LC ESI-HRMS using the positive (+) and negative (−) ion mode.
| Parameters | CE-ESI-HRMS | LC-ESI-HRMS | ||
|---|---|---|---|---|
| Instrument Configuration | Q-TOF | Q-OT | Q-TOF | Q-OT |
| Compounds | Metabolites | Proteins | Metabolites | Proteins |
| MS survey scan frequency | MS1 –MS2, 2 Hz | MS1, 7; MS2, 13 Hz | MS1 –MS2, 2 Hz | MS1, 7; MS2, 13 Hz |
| Mass range (m/z) and spectral resolution | 50–550 at 40,000 FWHM | 400–1,700 at 35,000 FWHM for MS1 and 17,500 FWHM for MS2 |
50–1,300 at 40,000 FWHM | 400–1,700 at 35,000 FWHM for MS1 and 17,500 FWHM for MS2 |
| Dry gas | 2 L/min (N2) | – | 4 L/min (N2) | – |
| Nebulizer gas | – | – | 0.4 bar (+); 1 bar (−) | – |
| Dry temperature | 100 °C (+); 150 °C (−) | 275 °C | 220 °C | 275 °C |
| Ion-transfer capillary voltage | –1,700 V (+); +2,100 V (−) | 2,300 V | –4,500 V (+); +4,000 V (−) | 1,800 V |
| Collision energy | 18 eV (CID) | 36% (normalized, HCD) | 15–35 eV (CID) | 36% (normalized, HCD) |
| Isolation m/z window | 1.5 Da | 1.5 Da | 1.5 Da | 1.5 Da |
| Dynamic exclusion mass tolerance | 5.0 ppm | 5.0 ppm | 5.0 ppm | 5.0 ppm |
| Dynamic exclusion | 9 s | 9 s | 13 s | 13 s |
| High intensity ion signal threshold (counts) | 1.5 × 106 | 1.5 × 106 | 1.5 × 106 | 1.5 × 106 |
| AGC target (counts) | – | 1 × 106 | – | 1 × 106 |
| Minimum AGC target (ion counts) | – | 9.2 × 102 | – | 9.2 × 102 |
| Maximum injection time (ms) | – | 50 | – | 50 |
| DDA Top N | 5 | 20 | 5 | 20 |
2. LC-ESI-HRMS
For Metabolomics:
We use the following LC parameters: column temperature, 35 °C; autosampler temperature, 4 °C; injection volume, 1 μL; flow rate, 130 μL/min. Positive ion mode gradient: 0–0.5 min 95% B, 0.5–10 min 95–40% B, 10–13 min 40% B, 13–15 min 40–95% B, 15–22 min 95% B; Negative ion mode gradient: 0–0.5 min 99% B, 0.5–2.5 min 99–82.5% B, 2.5–6.5 min 82.5–68% B, 6.5–10 min 68–30% B, 10–13 min 30% B, 13–15 min 30–99% B, 15–22 min 99% B.
Select MS method parameters described in Table 1 (see LC-HRMS).
For Proteomics:
LC parameters: 0–5 min 2% B, 5–85 min 2–35% B, 86–90 min 70% B, 91–120 min 2% B; autosampler temperature, 4 °C; injection volume, 1 μL; flow rate, 300 nL/min.
Load the MS method parameters and start data acquisition. Our typical parameters are listed in Table 1. Adjust ion source settings to get a stable nanospray. (see Note 8 and 9).
3. Data Processing
Metabolomics:
Survey the MS–MS/MS data for molecular features (signals with unique m/z and separation time) using available software packages. For example, we employ MetaboScape Version 4.0.4 (Bruker Daltonics) using the following settings: intensity threshold, 1,000 counts; minimum peak length, 5 spectra.
Annotate metabolites based on the accurate mass, isotopic distribution pattern, and tandem MS spectra against reference spectra available in MS-MS/MS databases, including but not limited to METLIN [74], EMBL (http://curatr.mcf.embl.de/), MzCloud (https://www.mzcloud.org/), MassBank of North America (https://mona.fiehnlab.ucdavis.edu/), and the Human Metabolome Database [75]. For example, we use METLIN with an annotation tolerance ≤ 10 ppm mass accuracy and MS/MS score ≥ 700–900.
Perform relative/absolute quantification using under-the-peak-areas (label-free quantification) or ion signal abundances (multiplexing quantification) serving as a proxy for metabolite abundance.
Perform statistics and multivariate data analysis to select molecules for follow-up functional studies (see Fig. 2).
Proteomics:
Identify proteins using established bioinformatics software packages broadly available for bottom-up proteomics. For example, we analyze the MS–MS/MS data in ProteomeDiscoverer (Thermo Fisher Scientific) or MaxQuant (Max Planck Institute of Biochemistry) against the mouse or Xenopus proteome (e.g., downloaded from UniProt [76] or Xenbase [77]) with the following search parameters: digestion enzyme, trypsin; missed cleavages, up to 2; variable modification, methionine oxidation; precursor mass tolerance, 20 ppm; fragment mass tolerance, 4.5 ppm; minimum peptide length, 5. Peptides are filtered to <1% false discovery rate (FDR), calculated against a reversed-sequence decoy database. The reported proteins are grouped based on the closest parsimony principle. We remove common contaminants from the final list of protein identifications by manually annotating for common contaminant proteins (downloaded from UniProt).
Employ label-free or label-based strategies and software packages from the proteomics community to compare protein levels between single cells. For example, we used MaxQuant Version 1.5.5.1 [78] or Proteome Discoverer (Thermo Scientific) to quantify the proteomic state of single embryonic cells and neurons by calculating label-free quantitative indexes (LFQ values) [57,25] or relative reporter ion signal abundances from TMTs [22].
3.5. Functional Studies
The goal of this step is to link chemistry with biological function. As an example, we describe a protocol to prepare brain tissues to record neuronal activity followed by in vitro single-cell proteomics [63,53]. In the context of cell differentiation, we trace cell fates to understand how cells divide to form specific tissues and organs. Because cell fates are reproducible in X. laevis [79,80], it is possible to inject molecules into identified cells to determine their developmental impact on tissue specification and organogenesis [21,60,61]. Figure 4 shows an example, in which the vehicle or select metabolites were injected into specific cells, while fluorescently monitoring their tissue clone via the co-injection of the green fluorescent dextran. Alternatively, fluorescent proteins can be expressed in the cell, for example, by injecting the corresponding mRNA [64,21]. In X. laevis tadpoles, it is also possible to perform behavioral assays to assess sensory (e.g., visual), muscular, cognitive, and other functions. As an example, we adopted the background color preference assay [81] to test the behavior of pre-metamorphic X. laevis tadpoles after performing in vivo single-cell MS on the precursor embryo [52].
1. Single-Neuron Electrophysiology and Capillary Microsampling
Perfuse midbrain slices continuously at 1.5–2 mL/min with perfusion solution at 28–32 °C, following established protocols [82].
Backfill the patch pipettes for recording (2–4 MΩ) with ~20 μL of 50 mM ammonium bicarbonate in water (see Note 10).
Putatively identify neuron type of interest. For example, dopaminergic neurons can be identified based on their location in the lateral portion of the substantia nigra and their size. Consult with brain anatomy atlases to improve the accuracy of tissue identifications.
Neuron identification may be aided by electrophysiology. Detection of a slow pacemaker firing pattern (> 2 ms action potential), indicates dopaminergic neurons [83].
Obtain a giga-ohm seal and record action potential for 60 s in a cell-attached configuration. A patch-clamped neuron is demonstrated in Fig. 2. We used the Sutterpatch software to control the devices [53,54,63].
Following electrophysical analysis, apply a steady negative pressure at the outlet end of pipette with a syringe to aspirate a portion of neuronal soma (see Note 11).
Under an inverted microscope (40× magnification), visually inspect the neuron during microaspiration. A slight reduction in neural soma size is anticipated from successful patching. Figure 2 shows a neuron during sampling after electrophysiological recording.
Gently withdraw the pipette from the cell then expel the collected contents into an LoBind microvial containing 5 μL protein digestion solution chilled on ice. After a 1 h digestion at 60 °C, store samples at −80 °C for up to 1 month without detectable degradation, until analysis.
2. Cell Lineage Tracing
Cell Labeling by Microinjection
-
Prepare fluorescent tracer solution:
Prepare a 100 μL solution containing 0.5% fluorescent dextran in DEPC water.
(Optional) Synthesize capped mRNA using an in vitro transcription kit. Prepare a working solution containing ~50–100 pg/nL mRNA pellet in DEPC-treated water.
Prepare injection needles following steps described earlier. Calibrate the volume of injection by injecting water droplet into mineral oil and measuring the diameter of the water droplet.
Fill the injection needle with ~0.5–1 μL fluorescent tracer solution using a microinjector in “fill” mode following established protocols [64].
Transfer the embryo into an injection dish containing 3% Ficoll in 100% SS.
Under a stereomicroscope, use a hair loop to gently angle the embryo to orient the cell of interest for facile access for injection (see Fig. 4).
Using a calibrated micromanipulator, guide the tip of the injection capillary into the targeted cell. Inject ~1–5 nL of the sample by applying +40 psi on the capillary for ~300–500 ms using a microinjector (see Note 12).
Culture the injected embryos in 3% Ficoll in 100% SS for 3–4 h to allow the cell membrane to heal. Transfer the embryos to 50% SS and culture at 14–22 °C until the larval stages 32–34 (see Note 13).
Fixing and Imaging of the Larvae/Tadpoles
Anesthetize the larvae or tadpoles on ice or using 0.5% benzocaine in 100% SS. Ensure the success of anesthesia by gently touching the tadpole with a capillary, anticipating no response if successful. Increase the concentration of benzocaine if necessary. Ensure tadpoles are handled and treated humanely so that the organisms do not suffer or feel pain (see Note 1).
Fix the specimens in 4% paraformaldehyde for 1 h on a rotator.
Rinse the tadpoles with 1X PBS twice. Store the tadpoles in 1X PBS at 4 °C.
For imaging, mount the specimens in a 30 mm dish containing ~1–2 mL of 1X PBS. Image the specimens using epifluorescence microscopy (e.g., SMZ18, Nikon) (see Note 14).
Acquire images using a microscope following the manufacturer’s instructions.
Conduct lineage analysis by determining the relative contribution of fluorescent cells to tissues and organs (see protocols in References [69,70,79]).
3. Behavioral Assay
Tadpole Preparation
Obtain and culture embryos as described earlier.
Inject identified cells with the test compounds as described earlier. Prepare negative control by injecting identified cells with DEPC-treated water. Use non-injected embryos for later use as the wild-type control group.
Place the embryos in the 14 °C incubator until they reach gastrula stage. Transfer the embryos to room temperature in 90 mm Petri dishes containing 20% SS and change media every two days and culture the tadpoles until the feeding stage (Stage 45) (see Note 1).
-
Feed tadpoles every other day as follows:
Mix food with 20% SS solution to form a paste.
Place the paste in a corner of the Petri dish containing the tadpoles.
Provide more food as needed.
Maintain the tadpoles under a 12 h light/dark light cycle.
Background Color Preference Assay
The color preference assay is performed in a setup consisting of nested tanks following an established protocol [81]. The inner tank holds the tadpoles. The outer tank provides the background colors with half covered with a black tape and the other half covered with a white paper. Ensure both tanks are water-leveled to aid visual inspection of tadpole behavior.
Fill the inner test tank with 20% SS to the 5 cm water mark from the top and insert the inner tank inside the outer tank.
Mount the camera on a tripod to record the entire tank from above (see Note 15).
Transfer a single tadpole on the white background of the inner tank. Limit this experiment to one tadpole at a time to avoid interactions between tadpoles which may confound behavioral phenotypes.
Record the swim pattern of tadpole for 2 min.
After 2 min, carefully lift the outer tank, rotate the outer tank, and return the inner tank into the outer tank. This step helps minimize the impact environmental factors may have on behavior. Start recording immediately and set the timer for 2 min.
Place the tadpole back to a holding tank containing 20% SS. Record 2 trials for each of ~10–15 tadpoles.
Repeat the assay on the next day on the same tadpoles to test for reproducibility and enhance statistical evaluation of the results by obtaining more data.
Euthanasia
Place tadpoles in a 90 mm Petri dish containing 2% benzocaine in 20% SS for ~15–20 min.
Monitor the tadpole’s reflex by gently touching with a hair loop. Anesthetized tadpole cannot swim or respond to mechanical stimuli (gentle touching). Only proceed to the next step if the tadpole is anesthetized.
Freeze the larvae/tadpoles at −20 °C overnight.
Dispose of the tadpoles following protocols approved by the relevant institutional and federal authorities (see Note 1)
ACKNOWLEDGMENTS.
The work presented here was in part sponsored by an Arnold and Mabel Beckman Foundation Beckman Young Investigator award (to P.N.), COSMOS Club Foundation Awards (to E.P.P., L.P., and J.L.), the National Institute of General Medical Sciences award no. 1R35GM124755 (to P.N.), and the National Science Foundation award no. IOS-1832968 (to P.N.). The authors thank Abigail Polter (The George Washington University) for permitting to photograph the electrophysiology station in her laboratory.
4. NOTES
X. laevis, mouse, and zebrafish are sentient and vertebrate animals; therefore, protocols pertaining to the care and handling of the animals must be approved by institutional and federal agencies. The work presented in this chapter was approved by the Institutional Animal Care and Use Committee of the University of Maryland (approval numbers R-DEC-17-57, R-FEB-21-07, and R-JUN-20-31) and/or The George Washington University (approval numbers #A311 and #A283).
Development is temperature-dependent in Xenopus [77,84], thus providing a helpful tool to time biological and chemical experiments. Low temperatures slow down the speed of cell-cleavage, extending the time to select embryos for experiments.
For metabolomics, place cells in ice-cold 100% methanol immediately upon collection. Methanol denatures enzymes and low temperatures slow down chemical reactions, thus minimizing metabolic changes.
Capillaries with too large or too small diameters challenge microsampling or microinjection. In Xenopus, we find apertures larger than ~20 μm tend to cause substantial damage to the cell membrane. Without the membrane being able to heal, the cytoplasm may leak into the media and the cell may not be able to continue division for functional experiments. Conversely, apertures below ~10 μm may clog with yolk and cytoplasmic content, requiring refabrication of the microprobe.
Tailor the composition of the metabolite extraction solvent to the type of metabolites of interest in a particular study. To study polar metabolites, we use aqueous 40% (v/v) acetonitrile with 40% (v/v) methanol as the extraction solution. Theoretical predictions based on partition and distribution coefficients can help experimental design [58].
Debris may clog CE capillaries or LC columns. We find that centrifugation of samples before analysis prevents clogging. The efficiency of centrifugation depends on the field force and time of centrifugation. We typically use 13,000 × g for 10 min.
To select the spectral resolution appropriate for the multiplexing relative quantification, refer to instructions from the manufacturer of the reagents.
We consider the electrospray to be stable when the total ion current exhibits less than ~15% relative standard deviation over ~40 min of separation.
Tailor MS–MS/MS experimental settings to the chosen separation technology. For example, we adjust the number of targeted molecular features and the duration of dynamic exclusion depending on typical peak widths and the complexity of the sample in LC and CE experiments (see Table 1).
It is imperative to optimize the composition of the intracellular solution used for patch clamp electrophysiology to the osmolarity of the neuron. Although potassium gluconate is commonly used in electrophysiological recordings, we use ammonium bicarbonate to minimize spectral and ionization interferences during MS caused by involatile salts.
Aid sample collection by applying consistent negative pressure and continuously monitoring the size of the neuron under the inverted microscope. Shrinking neuronal soma is an indication of successful sampling. Be careful not to aspirate the media surrounding the neuron to avoid sample dilution and interferences due to salt during MS analysis.
Limit damage to the cell membrane by carefully withdrawing the needle tip from the cell. With negligible damage, the cell membrane heals, and the embryo continues development. Damage to the cell membrane can cause leakage of cytoplasmic content, which can result in low survival rates or lethality. Take extra care to also avoid damaging the neighboring cells to facilitate development.
Based on our experience and other protocols [80], culturing at lower temperature (14–16 °C) improves survival rates.
Clearing agents (e.g., benzyl alcohol:benzyl benzoate, BA:BB) may be used in X. laevis embryos and tadpoles to improve fluorescent imaging [85]. Tissue clearing is recommended for imaging deep in intact embryos/tadpoles and sections.
The locations of the eyes in tadpoles help accurately determine crossings between the white and black backgrounds (see Fig. 4). Therefore, the camera used to record video trials should have sufficient optical resolution and frame rate to clearly identify the eyes and monitor fast tadpole swimming. We typically use ~30 fps to monitor tadpoles during both the background color preference and swim assays.
REFERENCES
- 1.Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW, Klein AM (2018) The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360 (6392):1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan TZ, Heong V, Ye J, Lim D, Low J, Choolani M, Scott C, Tan DSP, Huang RY-J (2019) Decoding transcriptomic intra-tumour heterogeneity to guide personalised medicine in ovarian cancer. The Journal of Pathology 247 (3):305–319 [DOI] [PubMed] [Google Scholar]
- 3.Rohrback S, Siddoway B, Liu CS, Chun J (2018) Genomic mosaicism in the developing and adult brain. Dev Neurobiol 78 (11):1026–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cembrowski MS (2019) Single-cell transcriptomics as a framework and roadmap for understanding the brain. Journal of Neuroscience Methods 326:1–7 [DOI] [PubMed] [Google Scholar]
- 5.Phillips JW, Schulmann A, Hara E, Winnubst J, Liu C, Valakh V, Wang L, Shields BC, Korff W, Chandrashekar J, Lemire AL, Mensh B, Dudman JT, Nelson SB, Hantman AW (2019) A repeated molecular architecture across thalamic pathways. Nature Neuroscience 22 (11):1925–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor CF, Paton NW, Lilley KS, Binz P-A, Julian RK, Jones AR, Zhu W, Apweiler R, Aebersold R, Deutsch EW, Dunn MJ, Heck AJR, Leitner A, Macht M, Mann M, Martens L, Neubert TA, Patterson SD, Ping P, Seymour SL, Souda P, Tsugita A, Vandekerckhove J, Vondriska TM, Whitelegge JP, Wilkins MR, Xenarios I, Yates JR, Hermjakob H (2007) The minimum information about a proteomics experiment (MIAPE). Nature Biotechnology 25 (8):887–893 [DOI] [PubMed] [Google Scholar]
- 7.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TWM, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR (2007) Proposed minimum reporting standards for chemical analysis. Metabolomics 3 (3):211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic acids research 47 (D1):D442–D450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sud M, Fahy E, Cotter D, Azam K, Vadivelu I, Burant C, Edison A, Fiehn O, Higashi R, Nair SK, Sumner S, Subramaniam S (2016) Metabolomics workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic acids research 44 (D1):D463–D470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan KD, Fyrestam J, Lanekoff I (2019) Advances in mass spectrometry based single-cell metabolomics. Analyst 144 (3):782–793 [DOI] [PubMed] [Google Scholar]
- 11.Yin L, Zhang Z, Liu Y, Gao Y, Gu J (2019) Recent advances in single-cell analysis by mass spectrometry. Analyst 144 (3):824–845 [DOI] [PubMed] [Google Scholar]
- 12.Comi TJ, Do TD, Rubakhin SS, Sweedler JV (2017) Categorizing cells on the basis of their chemical profiles: Progress in single-cell mass spectrometry. Journal of the American Chemical Society 139 (11):3920–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couvillion SP, Zhu Y, Nagy G, Adkins JN, Ansong C, Renslow RS, Piehowski PD, Ibrahim YM, Kelly RT, Metz TO (2019) New mass spectrometry technologies contributing towards comprehensive and high throughput omics analyses of single cells. Analyst 144 (3):794–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Vertes A (2018) Single-cell mass spectrometry approaches to explore cellular heterogeneity. Angewandte Chemie International Edition 57 (17):4466–4477 [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Huang Y, Wu J, Liu N, Deng J, Luan T (2017) Single-cell analysis by ambient mass spectrometry. Trends in Analytical Chemistry 90:14–26 [Google Scholar]
- 16.Qi M, Philip MC, Yang N, Sweedler JV (2018) Single cell neurometabolomics. ACS Chemical Neuroscience 9 (1):40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubakhin SS, Romanova EV, Nemes P, Sweedler JV (2011) Profiling metabolites and peptides in single cells. Nature Methods 8 (4):S20–S29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evers TMJ, Hochane M, Tans SJ, Heeren RMA, Semrau S, Nemes P, Mashaghi A (2019) Deciphering metabolic heterogeneity by single-cell analysis. Analytical Chemistry 91 (21):13314–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armbrecht L, Dittrich PS (2017) Recent advances in the analysis of single cells. Analytical Chemistry 89 (1):2–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Sevinsky CJ, Davis BM, Vertes A (2018) Single-cell mass spectrometry of subpopulations selected by fluorescence microscopy. Analytical Chemistry 90 (7):4626–4634 [DOI] [PubMed] [Google Scholar]
- 21.Onjiko RM, Moody SA, Nemes P (2015) Single-cell mass spectrometry reveals small molecules that affect cell fates in the 16-cell embryo. Proc Natl Acad Sci U S A 112 (21):6545–6550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lombard-Banek C, Moody SA, Nemes P (2016) Single-cell mass spectrometry for discovery proteomics: Quantifying translational cell heterogeneity in the 16-cell frog (Xenopus) embryo. Angewandte Chemie International Edition 55 (7):2454–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onjiko RM, Portero EP, Moody SA, Nemes P (2017) In situ microprobe single-cell capillary electrophoresis mass spectrometry: Metabolic reorganization in single differentiating cells in the live vertebrate (Xenopus laevis) embryo. Analytical Chemistry 89 (13):7069–7076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onjiko RM, Portero EP, Moody SA, Nemes P (2017) Microprobe capillary electrophoresis mass spectrometry for single-cell metabolomics in live frog (Xenopus laevis) embryos. Journal of Visualized Experiments (130):e56956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lombard-Banek C, Moody SA, Manzini MC, Nemes P (2019) Microsampling capillary electrophoresis mass spectrometry enables single-cell proteomics in complex tissues: Developing cell clones in live Xenopus laevis and Zebrafish embryos. Analytical Chemistry 91 (7):4797–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lombard-Banek C, Moody SA, Nemes P (2016) High-sensitivity mass spectrometry for probing gene translation in single embryonic cells in the early frog (Xenopus) embryo. Frontiers in Cell and Developmental Biology 4 (100):1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lombard-Banek C, Portero EP, Onjiko RM, Nemes P (2017) New-generation mass spectrometry expands the toolbox of cell and developmental biology. genesis 55 (1–2):1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lombard-Banek C, Choi SB, Nemes P (2019) Single-cell proteomics in complex tissues using microprobe capillary electrophoresis mass spectrometry. In: Allbritton NL, Kovarik ML (eds) Methods in Enzymology, vol 628. Academic Press, pp 263–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onjiko RM, Portero EP, Nemes P (2018) Single-cell metabolomics with capillary electrophoresis–mass spectrometry. In: Ramautar (ed) Capillary Electrophoresis–Mass Spectrometry for Metabolomics. The Royal Society of Chemistry, pp 209–224 [Google Scholar]
- 30.Sun L, Dubiak KM, Peuchen EH, Zhang Z, Zhu G, Huber PW, Dovichi NJ (2016) Single cell proteomics using frog (Xenopus laevis) blastomeres isolated from early stage embryos, which form a geometric progression in protein content. Analytical Chemistry 88 (13):6653–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saha-Shah A, Esmaeili M, Sidoli S, Hwang H, Yang J, Klein PS, Garcia BA (2019) Single cell proteomics by data-independent acquisition to study embryonic asymmetry in Xenopus laevis. Analytical Chemistry 91 (14):8891–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Budnik B, Levy E, Harmange G, Slavov N (2018) SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biology 19 (1):1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Specht H, Emmott E, Petelski AA, Huffman G, Perlman D, Serra M, Kharchenko P, Koller A, Slavov N (2020) Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity. bioRxiv 665307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cahill JF, Riba J, Kertesz V (2019) Rapid, untargeted chemical profiling of single cells in their native environment. Analytical Chemistry 91 (9):6118–6126 [DOI] [PubMed] [Google Scholar]
- 35.Cong Y, Liang Y, Motamedchaboki K, Huguet R, Truong T, Zhao R, Shen Y, Lopez-Ferrer D, Zhu Y, Kelly RT (2020) Improved single-cell proteome coverage using narrow-bore packed nanoLC columns and ultrasensitive mass spectrometry. Analytical Chemistry 92 (3):2665–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S, Plouffe BD, Belov AM, Ray S, Wang X, Murthy SK, Karger BL, Ivanov AR (2015) An integrated platform for isolation, processing, and mass spectrometry-based proteomic profiling of rare cells in whole blood. Molecular & Cellular Proteomics 14 (6):1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanov AR, Zang L, Karger BL (2003) Low-attomole electrospray ionization MS and MS/MS analysis of protein tryptic digests using 20-μm-i.d. polystyrene–divinylbenzene monolithic capillary columns. Analytical Chemistry 75 (20):5306–5316 [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Piehowski PD, Zhao R, Chen J, Shen Y, Moore RJ, Shukla AK, Petyuk VA, Campbell-Thompson M, Mathews CE, Smith RD, Qian W-J, Kelly RT (2018) Nanodroplet processing platform for deep and quantitative proteome profiling of 10–100 mammalian cells. Nature Communications 9 (1):1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osbourn DM, Weiss DJ, Lunte CE (2000) On-line preconcentration methods for capillary electrophoresis. Electrophoresis 21 (14):2768–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nemes P, Rubakhin SS, Aerts JT, Sweedler JV (2013) Qualitative and quantitative metabolomic investigation of single neurons by capillary electrophoresis electrospray ionization mass spectrometry. Nature protocols 8 (4):783–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drouin N, van Mever M, Zhang W, Tobolkina E, Ferre S, Servais A-C, Gou M-J, Nyssen L, Fillet M, Lageveen-Kammeijer GSM, Nouta J, Chetwynd AJ, Lynch I, Thorn JA, Meixner J, Lößner C, Taverna M, Liu S, Tran NT, Francois Y, Lechner A, Nehmé R, Al Hamoui Dit Banni G, Nasreddine R, Colas C, Lindner HH, Faserl K, Neusüß C, Nelke M, Lämmerer S, Perrin C, Bich-Muracciole C, Barbas C, Gonzálvez ÁL, Guttman A, Szigeti M, Britz-McKibbin P, Kroezen Z, Shanmuganathan M, Nemes P, Portero EP, Hankemeier T, Codesido S, González-Ruiz V, Rudaz S, Ramautar R (2020) Capillary electrophoresis-mass spectrometry at trial by metabo-ring: Effective electrophoretic mobility for reproducible and robust compound annotation. Analytical Chemistry 92 (20):14103–14112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramautar R, Somsen GW, de Jong GJ (2019) CE-MS for metabolomics: Developments and applications in the period 2016–2018. Electrophoresis 40 (1):165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramautar R, Somsen GW, de Jong GJ (2017) CE–MS for metabolomics: Developments and applications in the period 2014–2016. Electrophoresis 38 (1):190–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao H-W, Rubakhin SS, Philip MC, Sweedler JV (2020) Enhanced single-cell metabolomics by capillary electrophoresis electrospray ionization-mass spectrometry with field amplified sample injection. Analytica Chimica Acta 1118:36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J-X, Aerts JT, Rubakhin SS, Zhang X-X, Sweedler JV (2014) Analysis of endogenous nucleotides by single cell capillary electrophoresis-mass spectrometry. The Analyst 139 (22):5835–5842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi SB, Lombard-Banek C, Muñoz-Llancao P, Manzini MC, Nemes P (2018) Enhanced peptide detection toward single-neuron proteomics by reversed-phase fractionation capillary electrophoresis mass spectrometry. Journal of The American Society for Mass Spectrometry 29 (5):913–922 [DOI] [PubMed] [Google Scholar]
- 47.Kawai T, Ota N, Okada K, Imasato A, Owa Y, Morita M, Tada M, Tanaka Y (2019) Ultrasensitive single cell metabolomics by capillary electrophoresis–mass spectrometry with a thin-walled tapered emitter and large-volume dual sample preconcentration. Analytical Chemistry 91 (16):10564–10572 [DOI] [PubMed] [Google Scholar]
- 48.Portero EP, Nemes P (2019) Dual cationic–anionic profiling of metabolites in a single identified cell in a live Xenopus laevis embryo by microprobe CE-ESI-MS. Analyst 144 (3):892–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lombard-Banek C, Yu Z, Swiercz AP, Marvar PJ, Nemes P (2019) A microanalytical capillary electrophoresis mass spectrometry assay for quantifying angiotensin peptides in the brain. Analytical and Bioanalytical Chemistry 411 (19):4661–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi SB, Zamarbide M, Manzini MC, Nemes P (2016) Tapered-tip capillary electrophoresis nano-electrospray ionization mass spectrometry for ultrasensitive proteomics: the mouse cortex. Journal of The American Society for Mass Spectrometry 28 (4):597–607 [DOI] [PubMed] [Google Scholar]
- 51.Choi SB, Muñoz-LLancao P, Manzini MC, Nemes P (2021) A data-dependent acquisition ladder for ultrasensitive (neuro)proteomics in review [DOI] [PubMed]
- 52.Lombard-Banek C, Li J, Portero EP, Onjiko RM, Singer CD, Plotnick DO, Al Shabeeb RQ, Nemes P (2021) In vivo subcellular mass spectrometry enables proteo-metabolomic single-cell systems biology in a chordate embryo developing to a normally behaving tadpole (X. laevis). Angew Chem-Int Edit 60 (23):12852–12858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi SB, Polter AM, Nemes P Mass spectrometry-based proteomics of single dopaminergic mouse neurons identified by whole-cell electrophysiology. In: Pittsburgh Conference on Analytical Chemistry and Applied Spectroscopy, Philadelphia, PA March 17 2019. [Google Scholar]
- 54.Choi SB, Polter AM, Nemes P Single-cell mass spectrometry for proteomic analysis of patch-clamp electrophysiology identified dopaminergic neurons. In: United States Human Proteome Organization, Washington DC, March 3 2019. [Google Scholar]
- 55.Baxi AB, Lombard-Banek C, Moody SA, Nemes P (2018) Proteomic characterization of the neural ectoderm fated cell clones in the Xenopus laevis embryo by high-resolution mass spectrometry. ACS Chemical Neuroscience 9 (8):2064–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Onjiko RM, Plotnick DO, Moody SA, Nemes P (2017) Metabolic comparison of dorsal versus ventral cells directly in the live 8-cell frog embryo by microprobe single-cell CE-ESI-MS. Analytical Methods 9 (34):4964–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lombard-Banek C, Reddy S, Moody SA, Nemes P (2016) Label-free quantification of proteins in single embryonic cells with neural fate in the cleavage-stage frog (Xenopus laevis) embryo using capillary electrophoresis electrospray ionization high-resolution mass spectrometry (CE-ESI-HRMS). Molecular & Cellular Proteomics 15 (8):2756–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Onjiko RM, Morris SE, Moody SA, Nemes P (2016) Single-cell mass spectrometry with multi-solvent extraction identifies metabolic differences between left and right blastomeres in the 8-cell frog (Xenopus) embryo. Analyst 141 (12):3648–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Onjiko RM, Nemes P, Moody SA (2021) Altering metabolite distribution at Xenopus cleavage stages affects left-right gene expression asymmetries. genesis 59 (5–6):1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Portero EP, Andrews AJ, Nemes P Probing metabolic pathways during early embryonic development using stable isotope labeling and single-cell mass spectrometry. In: ASMS Conference on Mass Spectrometry and Allied Topics, Atlanta, GA, June 6 2019. [Google Scholar]
- 61.Portero EP, Pade L, Nemes P Single-cell mass spectrometry reveals cell-to-cell communication in the same vertebrate (Xenopus laevis) frog embryo. In: ASMS Conference on Mass Spectrometry and Allied Topics, Online, June 12 2020. [Google Scholar]
- 62.Aerts JT, Louis KR, Crandall SR, Govindaiah G, Cox CL, Sweedler JV (2014) Patch clamp electrophysiology and capillary electrophoresis-mass spectrometry metabolomics for single cell characterization. Anal Chem 86 (6):3203–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi SB, Polter AM, Nemes P Integration of patch-clamp electrophysiology with single-cell mass spectrometry for proteomic analysis to extend the bioanalytical toolbox of neuroscience. In: ASMS Conference on Mass Spectrometry and Allied Topics, Online, June 4 2020. [Google Scholar]
- 64.Moody SA (2018) Microinjection of mRNAs and oligonucleotides. Cold Spring Harbor protocols 2018 (12) [DOI] [PubMed] [Google Scholar]
- 65.Sive HL, Grainger RM, Harland RM (2000) Early development of Xenopus laevis: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 66.Kaech S, Banker G (2006) Culturing hippocampal neurons. Nature protocols 1 (5):2406–2415 [DOI] [PubMed] [Google Scholar]
- 67.Klein SL (1987) The 1st cleavage furrow demarcates the dorsal ventral axis in Xenopus embryos. Developmental biology 120 (1):299–304 [DOI] [PubMed] [Google Scholar]
- 68.Polter AM, Bishop RA, Briand LA, Graziane NM, Pierce RC, Kauer JA (2014) Poststress block of kappa opioid receptors rescues long-term potentiation of inhibitory synapses and prevents reinstatement of cocaine seeking. Biological Psychiatry 76 (10):785–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moody SA (1987) Fates of the blastomeres of the 32-cell-stage Xenopus embryo. Developmental biology 122 (2):300–319 [DOI] [PubMed] [Google Scholar]
- 70.Moody SA (1987) Fates of the blastomeres of the 16-cell stage Xenopus embryo. Developmental biology 119 (2):560–578 [DOI] [PubMed] [Google Scholar]
- 71.Liu ZC, Portero EP, Jian YR, Zhao YJ, Onjiko RM, Zeng C, Nemes P (2019) Trace, machine learning of signal images for trace-sensitive mass spectrometry: a case study from single-cell metabolomics. Analytical Chemistry 91 (9):5768–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie YR, Castro DC, Bell SE, Rubakhin SS, Sweedler JV (2020) Single-cell classification using mass spectrometry through interpretable machine learning. Analytical Chemistry 92 (13):9338–9347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun L, Zhu G, Zhang Z, Mou S, Dovichi NJ (2015) A third-generation electrokinetically pumped sheath-flow nanospray interface with improved stability and sensitivity for automated capillary zone electrophoresis-mass spectrometry analysis of complex proteome digests. J Proteome Res 14 (5):2312–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guijas C, Montenegro-Burke JR, Domingo-Almenara X, Palermo A, Warth B, Hermann G, Koellensperger G, Huan T, Uritboonthai W, Aisporna AE, Wolan DW, Spilker ME, Benton HP, Siuzdak G (2018) METLIN: A technology platform for identifying knowns and unknowns. Analytical Chemistry 90 (5):3156–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, Scalbert A (2018) HMDB 4.0: The human metabolome database for 2018. Nucleic acids research 46 (D1):D608–D617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bateman A, Martin MJ, Orchard S, Magrane M, Agivetova R, Ahmad S, Alpi E, Bowler-Barnett EH, Britto R, Bursteinas B, Bye-A-Jee H, Coetzee R, Cukura A, Da Silva A, Denny P, Dogan T, Ebenezer T, Fan J, Castro LG, Garmiri P, Georghiou G, Gonzales L, Hatton-Ellis E, Hussein A, Ignatchenko A, Insana G, Ishtiaq R, Jokinen P, Joshi V, Jyothi D, Lock A, Lopez R, Luciani A, Luo J, Lussi Y, Mac-Dougall A, Madeira F, Mahmoudy M, Menchi M, Mishra A, Moulang K, Nightingale A, Oliveira CS, Pundir S, Qi GY, Raj S, Rice D, Lopez MR, Saidi R, Sampson J, Sawford T, Speretta E, Turner E, Tyagi N, Vasudev P, Volynkin V, Warner K, Watkins X, Zaru R, Zellner H, Bridge A, Poux S, Redaschi N, Aimo L, Argoud-Puy G, Auchincloss A, Axelsen K, Bansal P, Baratin D, Blatter MC, Bolleman J, Boutet E, Breuza L, Casals-Casas C, de Castro E, Echioukh KC, Coudert E, Cuche B, Doche M, Dornevil D, Estreicher A, Famiglietti ML, Feuermann M, Gasteiger E, Gehant S, Gerritsen V, Gos A, Gruaz-Gumowski N, Hinz U, Hulo C, Hyka-Nouspikel N, Jungo F, Keller G, Kerhornou A, Lara V, Le Mercier P, Lieberherr D, Lombardot T, Martin X, Masson P, Morgat A, Neto TB, Paesano S, Pedruzzi I, Pilbout S, Pourcel L, Pozzato M, Pruess M, Rivoire C, Sigrist C, Sonesson K, Stutz A, Sundaram S, Tognolli M, Verbregue L, Wu CH, Arighi CN, Arminski L, Chen CM, Chen YX, Garavelli JS, Huang HZ, Laiho K, McGarvey P, Natale DA, Ross K, Vinayaka CR, Wang QH, Wang YQ, Yeh LS, Zhang J, UniProt C (2021) UniProt: the universal protein knowledgebase in 2021. Nucleic acids research 49 (D1):D480–D489. doi: 10.1093/nar/gkaa1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karimi K, Fortriede JD, Lotay VS, Burns KA, Wang DZ, Fisher ME, Pells TJ, James-Zorn C, Wang Y, Ponferrada VG, Chu S, Chaturvedi P, Zorn AM, Vize PD (2018) Xenbase: A genomic, epigenomic and transcriptomic model organism database. Nucleic acids research 46 (D1):D861–D868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y, Fonslow BR, Shan B, Baek M-C, Yates JR (2013) Protein analysis by shotgun/bottom-up proteomics. Chemical Reviews 113 (4):2343–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moody SA (2000) Cell lineage analysis in Xenopus embryos. Methods in Molecular Biology 135:331–347 [DOI] [PubMed] [Google Scholar]
- 80.Moody SA (2018) Lineage tracing and fate mapping in Xenopus embryos. Cold Spring Harbor protocols 2018 (12) [DOI] [PubMed] [Google Scholar]
- 81.Viczian AS, Zuber ME (2014) A simple behavioral assay for testing visual function in Xenopus laevis. Journal of Visualized Experiments (88):e51726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ting JT, Daigle TL, Chen Q, Feng GP (2014) Acute Brain Slice Methods for Adult and Aging Animals: Application of Targeted Patch Clamp Analysis and Optogenetics. In: Martina M, Taverna S (eds) Patch-Clamp Methods and Protocols, 2nd Edition, vol 1183. Methods in Molecular Biology. pp 221–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grace AA, Onn SP (1989) Morphology and electrophysiological properties of immunocytochemically identified rat dopamine neurons recorded in vitro. The Journal of Neuroscience 9 (10):3463–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, Papalopulu N, Smith JC, Zorn AM, Harland RM, Grammer TC (2002) Techniques and probes for the study of Xenopus tropicalis development. Developmental Dynamics 225 (4):499–510 [DOI] [PubMed] [Google Scholar]
- 85.Wallingford JB (2010) Preparation of fixed Xenopus embryos for confocal imaging. Cold Spring Harbor protocols 2010 (5):1–7 [DOI] [PubMed] [Google Scholar]


