Abstract
The cancer stem cell (CSC) model states that heterogeneous tumor cell populations are organized in a hierarchical manner, with a small population of CSCs at the apex. These CSCs are capable of self-renewal and giving rise to other cancer cell populations, conceptually analogous to the function of normal adult stem cells present in almost all organs. However, there has been significant controversy regarding the existence and identification of CSCs. We argue that technical differences in experimentation and CSC assays, CSC niche-dependency and plasticity, and CSC heterogeneity itself may explain some of the differences observed.
Introduction
Generally speaking, to acquire cancer stem cell (CSC) properties, specific signaling pathways and transcriptional programs need to be active to instruct stemness. Thus, there must be differences between CSCs and non-CSC cells, be it proteins, noncoding RNAs or epigenetic differences, to explain their different functional properties. Thus, if one subscribes to the notion that heterogeneity and functional differences between cancer cells exist, from this directly follows that there must also exist markers that characterize these differences. It is worth noting that molecules specifically expressed in CSCs arguably are more likely to have an important function in CSC biology. Indeed, some of the best characterized CSC markers regulate pathways known to be crucial for cancer stemness, the WNT receptor component Lgr5 being a prime example (1).
However, while these general considerations may suggest that markers for CSCs should exist, in many tumor types a plethora of different markers have been proposed, without a clear consensus on what the functional differences between the different suggested CSC populations might be. There is thus considerable controversy on delineating the “true” CSC population.
Conceptual and Technical Considerations
The first description of a CSC population was in acute myeloid leukemia. Using the FACS technique combined with xenotransplantation assays, Bonnet and Dick identified a subpopulation of leukemic cell with leukemia-initiating capacity (2). Consequently, CSCs were defined functionally for their ability to initiate tumors when transplanted. This has become the main experimental approach to measure cancer stemness, which explains the synonymous name for CSCs, namely tumor-initiating cells.
While FACS coupled with xenotransplantation assays is the “gold standard” for identifying CSCs, technical factors need to be considered. First, the immunodeficient status of the recipient mice, highly immunocompromised NOD/SCID/Il2rg−/− animals or less immunocompromised mouse models, can affect the xenografting efficiency and CSC read-out. Second, the variable protocols for tumor cell dissociation, staining and sorting can influence the viability and fitness of cancer cells queried. Third, the injection sites (heterotopic or orthotopic) and the usage of Matrigel and other co-injected matrices can also bias the xenografting efficiency. All these parameters may cause discrepancies of CSC functional read-out and frequencies for a given cancer.
It is important to consider that CSCs reside within the context of the tumor microenvironment. Tumor cells are surrounded by a diverse array of different nontumor stromal cells, e.g., cancer-associated fibroblasts and various types of lymphocytes, and the cancer cells interact closely with their stromal environment. Thus, CSC function might be an intrinsic cellular property independent of any interaction with surrounding cells, or, alternatively, cancer stemness may depend on paracrine interactions with niche cells.
Cancer stemness of a particular tumor cell subpopulation may depend on instructive signals from a niche. In these scenarios, non-CSC cells might acquire CSC properties once a niche is vacated. For example, in colon cancer, after Lgr5-positive CSCs had been eliminated, some remaining cancer cells regained stemness in the primary tumor, but not in liver metastases, demonstrating plasticity of non-CSCs to acquire CSC properties and suggesting that a specific niche is required for the acquisition of stemness (3).
Importantly, some of these experimental limitations are particularly relevant for the characterization of human CSCs by transplantation. The characterization of human CSCs is hampered by the unavoidable interspecies differences inherent to this experimental approach. Some human CSCs might depend on paracrine signaling from stromal niche cells. These CSCs will only grow if these murine signaling proteins are functionally homologous to their human counterparts. Murine niche factors required for CSC propagation might not be functional on transplanted human cells, implying that only a subset of human CSC populations might be able to be identified using xenotranplantation (4).
In solid tumors, the ability to form cancer organoids in culture has recently been established as a surrogate read-out of cancer stemness (5). Cancer organoids are three-dimensional (3D) self-organized assemblies of neoplastic cells derived from murine or cancer patient tissue samples that mimic key histopathologic, genetic, and phenotypic features of the parental tumor. While the in vitro 3D culture systems are being actively developed, this experimental system has the potential to combine CSC culture with niche cells, thereby allowing a more complete identification of CSCs with different growth requirements.
Arguably, the most stringent experimental test of CSC function is analysis in the context of the intact growing tumor. Lineage tracing is the identification of all progenies of a single cell, in cancer mouse models typically done by cre/LoxP-mediated activation of expression of a fluorescent marker protein. Lineage tracing of CSCs has demonstrated that CSCs are the subpopulation capable of generating large hierarchically organized tumor cell clones, suggesting that CSCs underlie tumor growth. Conversely, CSC-specific expression of suicide genes such as herpes simplex virus thymidine kinase or diphtheria toxin receptor, can be used to specifically eliminate the CSC subpopulation, which is expected to cause decreased tumor growth and reduced cellular heterogeneity (3). Advances in CRISPR-mediated genome editing should enable lineage tracing and cell ablation approaches also in human cancer organoids and xenografts, by introducing an inducible cre or suicide gene into a putative CSC marker locus (6).
Thus, because of experimental limitations and the availability of more relevant functional assays, murine CSCs tend to be much better characterized than their human counterparts. Given its robustness, we speculate that lineage tracing and cell ablation will be increasingly used in studies of human CSCs in patient-derived organoid models and patient-derived xenograft models. In parallel, next-generation transplantation assays that include cotransplantation with stromal cells to better recapitulate the native tumor microenvironment will lead to a more rigorous assessment of CSCs as well as niche cell function. In addition, advanced single-cell RNA sequencing and spatial transcriptomics will also increasingly contribute to delineating the heterogeneity of CSCs as well as their localization and niche.
CSC Markers: Functional Overlap and Plasticity
While general considerations mentioned above suggest the existence of definitive markers identifying cancer cell subpopulations including CSCs, in practice the picture is often rather unclear. For example, in pancreatic ductal adenocarcinoma, several markers have been reported to identify CSCs, including CD44, CD133, c-MET, ALDH1, Dclk1, CD90, Msi1, Msi2, and CD9, with the functional relationship between these cellular subsets being unresolved (7). Similar differences in CSC characterization have also been seen in other tumor types. There are several possible scenarios to conceptualize these differences.
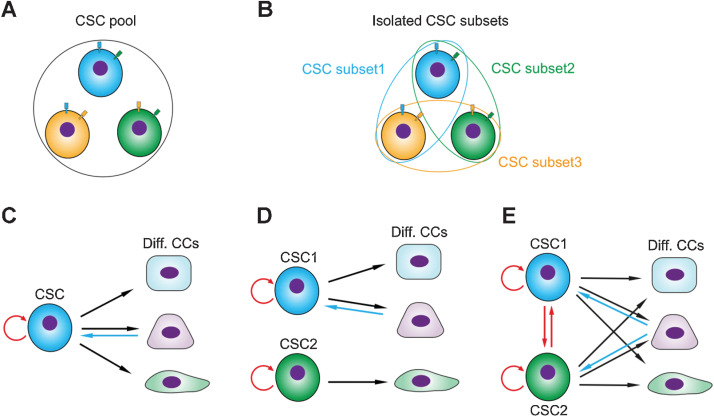
While in principle it might be possible to identify CSC markers that precisely cover the CSC population, in practice it is much more likely that any given CSC marker will only isolate a fraction of all CSCs. Thus, different CSC markers may isolate CSCs with slightly different molecular characteristics, but which are all part of the same functional cellular group (Fig. 1A and B). Secondly, it is assumed that typically there exists only one single CSC population, however this is not necessarily the case. It is conceivable that a tumor harbors functionally distinct CSC populations, possibly in distinct tumor subclones, which both contribute to tumor growth (Fig. 1C and D; ref. 8). Thirdly, the same CSC population may exist in different “states” with distinct molecular characteristics. A recent example is the description of Lgr5-positive matrix-dependent “dormant” colon cancer CSCs, which however can be reactivated upon chemotherapy and transplantation (Fig. 1E; ref. 9). It is noteworthy that tumor cells appear to have substantial plasticity, with non-CSCs being capable of acquiring CSC function, e.g., upon depletion of the original CSC pool, or non–stem cells (non-SC) dedifferentiating to stem cells being capable of initiating tumors upon an inflammatory microenvironmental clue (10).
Figure 1.
Models of CSC heterogeneity. A, A functionally homogeneous CSC pool in a tumor. B, Experimental marker–based isolation of CSC subsets from a functionally homogeneous CSC pool. Blue, green, and orange lines indicate marker proteins. C, Classical model of CSCs. D, Two distinct subpopulations of CSCs contributing to different tumor cell lineages. E, Two distinct interconvertible CSC states. In C, D, and E, circle arrows indicate self-renewal, red arrows indicate interconversion, black arrows indicate cellular differentiation, and blue arrows indicate dedifferentiation (plasticity from non-CSCs to CSCs). Diff. CCs, differentiated cancer cells.
Together with differences in techniques used to experimentally characterize CSCs, these mechanisms may underlie some of the conflicting results obtained.
Conclusions
CSCs have attracted significant attention as it has been postulated that CSC eradication is a prerequisite for long-term cancer cure. While the cellular heterogeneity of tumors is undisputed, an unambiguous characterization of CSCs has often been controversial. Better experimental systems and approaches will be required to further advance the CSC field.
Acknowledgments
The authors apologize to colleagues whose work could not be cited due to the brevity of the format. The authors thank D. Bonnet and V.M.Y. Wang for critical reading. L. Lan and A. Behrens are funded by the Breast Cancer Now Toby Robins Research Centre at the Institute of Cancer Research (CTR-Q5-Y2).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authors' Disclosures
No disclosures were reported.
References
- 1. de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signaling. Nature 2011;476:293–7. [DOI] [PubMed] [Google Scholar]
- 2. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997;3:730–7. [DOI] [PubMed] [Google Scholar]
- 3. de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, et al. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature 2017;543:676–80. [DOI] [PubMed] [Google Scholar]
- 4. Abarrategi A, Foster K, Hamilton A, Mian SA, Passaro D, Gribben J, et al. Versatile humanized niche model enables study of normal and malignant human hematopoiesis. J Clin Invest 2017;127:543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science 2019;364:952–5. [DOI] [PubMed] [Google Scholar]
- 6. Cortina C, Turon G, Stork D, Hernando-Momblona X, Sevillano M, Aguilera M, et al. A genome editing approach to study cancer stem cells in human tumors. EMBO Mol Med 2017;9:869–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evan T, Wang VM, Behrens A. The roles of intratumor heterogeneity in the biology and treatment of pancreatic ductal adenocarcinoma. Oncogene 2022;41:4686–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Boer B, Prick J, Pruis MG, Keane P, Imperato MR, Jaques J, et al. Prospective isolation and characterization of genetically and functionally distinct AML subclones. Cancer Cell 2018;34:674–89. [DOI] [PubMed] [Google Scholar]
- 9. Ohta Y, Fujii M, Takahashi S, Takano A, Nanki K, Matano M, et al. Cell-matrix interface regulates dormancy in human colon cancer stem cells. Nature 2022;608:784–94. [DOI] [PubMed] [Google Scholar]
- 10. Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem cell–like properties. Cell 2013;152:25–38. [DOI] [PubMed] [Google Scholar]