Abstract
Protocadherin-19 (PCDH19) mutations cause early-onset seizures and cognitive impairment. The PCDH19 gene is on the X-chromosome. Unlike most X-linked disorders, PCDH19 mutations affect heterozygous females (PCDH19HET♀), but not hemizygous males (PCDH19HEMI♂); however, why remains to be elucidated. We demonstrate PCDH19, a cell-adhesion molecule, is enriched at hippocampal mossy-fiber synapses. Pcdh19HET♀, but not Pcdh19HEMI♂, mice show impaired mossy-fiber synaptic structure and physiology. Consistently, Pcdh19HET♀, but not Pcdh19HEMI♂, mice exhibit reduced pattern completion and separation abilities, which require mossy-fiber synaptic function. Furthermore, PCDH19 appears to interact with N-cadherin at mossy-fiber synapses. In Pcdh19HET♀ conditions, mismatch between PCDH19 and N-cadherin diminishes N-cadherin-dependent signaling and impairs mossy-fiber synapse development. Indeed, N-cadherin overexpression rescues Pcdh19HET♀ phenotypes. These results reveal novel molecular and cellular mechanisms underlying the female-specific PCDH19 disorder phenotype.
One Sentence Summary:
In PCDH19 disorder, PCDH19–N-cadherin mismatch causes female-specific synaptic dysfunction and cognitive impairment.
Mutations of the X-linked Protocadherin-19 (PCDH19) gene cause PCDH19 disorder with epilepsy (Epilepsy and Mental Retardation Limited to Females). PCDH19 disorder is also often associated with cognitive impairment: about 70% of patients have intellectual disabilities (1, 2). Typically, X-linked disorders exhibit more severe phenotypes in males due to X-linked recessive inheritance. In contrast, symptoms of PCDH19 disorder manifest in heterozygous females, while hemizygous males are largely asymptomatic. Since heterozygous females have a mixture of cells expressing either the wild-type (WT) or the mutant form of PCDH19, due to random X-inactivation, the mosaic expression of PCDH19 is proposed to cause pathogenic symptoms (3). The gene responsible for this disorder, PCDH19, encodes a single transmembrane protein that belongs to the δ2-protocadherin family (4) (Fig. 1A). PCDH19 mediates calcium-dependent cell-cell adhesion by homophilic binding via extracellular cadherin (EC) domains (5, 6). A majority of PCDH19 mutations, including nonsense and missense mutations found in PCDH19 disorder, alter sites in the extracellular domain of the PCDH19 protein (2), suggesting that the mutations may affect PCDH19 homophilic interactions, and as a result, cell-cell adhesion. Inactivation of PCDH19 in zebrafish or mice results in changes in neuronal migration, cortical neuron sorting, network activity, and behaviors such as stress responses (7–10). However, the precise roles of PCDH19 in the brain, and the molecular, synaptic, and circuit bases of female-specific disease phenotypes remain to be elucidated.
Fig. 1. PCDH19 is localized at mossy-fiber synapses during synapse development, but is dispensable for dentate granule cell migration and mossy-fiber targeting.
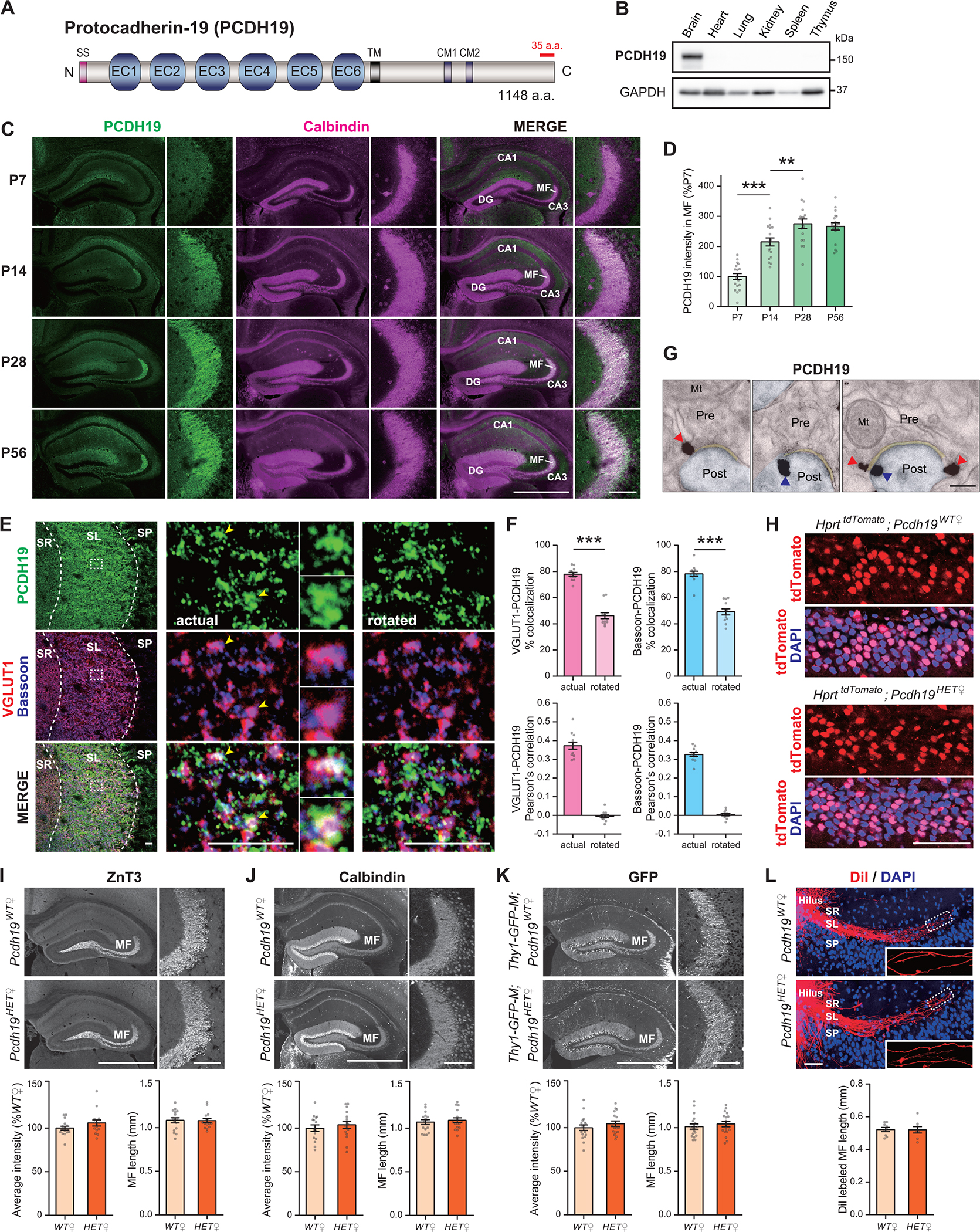
(A) Schematic representation of the mouse PCDH19 protein structure (NP_001098715.1). The epitope for our anti-PCDH19 antibody is indicated in red (C-terminal 35 a.a.). SS, signal sequence; EC, extracellular cadherin-like domain; TM, transmembrane domain; CM1/2, conserved motifs among the δ-protocadherin family; a.a., amino acid.
(B) Immunoblot showing the brain-specific expression of PCDH19 in P10 mouse tissues. GAPDH was used as a loading control.
(C and D) Double-staining for PCDH19 and Calbindin, a mossy-fiber (MF) marker, of hippocampal sections prepared from P7, P14, P28, and P56 female mice. Images are from the hippocampus (low magnification) and MFs (high magnification). PCDH19 is highly expressed along the MF pathway during synapse development. Quantification of the PCDH19 staining intensity within the MF pathway is shown in (D) (% P7). n = 18 fields from 3 female mice per age. DG, dentate gyrus.
(E and F) Triple-staining for PCDH19, VGLUT1, and Bassoon in the CA3 SL layer of P28 female mice. Left: lower magnification images. Middle: higher magnification images; puncta indicated by the arrowheads are magnified. Right: the PCDH19 image was rotated 180° for correlation analysis. Percentages of VGLUT1/Bassoon puncta colocalized with PCDH19 puncta and Pearson’s correlation coefficients between VGLUT1/Bassoon and PCDH19 are quantified (F). These analyses indicate that VGLUT1 and Bassoon puncta are associated with PCDH19 puncta. n = 12 fields from 4 female WT mice for actual and rotated images. SR, stratum radiatum; SL, stratum lucidum; SP stratum pyramidale.
(G) Pre-embedding immunogold electron microscopy for PCDH19 in the CA3 SL layer of P28 female mice. PCDH19 is localized at both pre- (red arrowheads) and post- (blue arrowheads) synaptic sites of MF synapses. Experiments were repeated 3 times with 3 female WT mice. Mt, mitochondria.
(H) Random distribution of Pcdh19+ (nuclear-tdTomato+) and Pcdh19− (nuclear-tdTomato–) cells in the DGC layer of 2- to 4-month-old Pcdh19HET♀ mice, visualized by mating with the HprttdTomato line. DAPI is in blue. Distribution patterns of tdTomato+ and tdTomato– DGCs are similar between Pcdh19WT♀ and Pcdh19HET♀ mice. Experiments were repeated 3 times.
(I and J) Immunostaining for ZnT3, a MF bouton marker, and for Calbindin showing normal MF axonal projections in P28 Pcdh19HET♀ mice. Quantification of the staining intensities in the MF region (% WT) and MF lengths (mm) is shown in the graphs. n = 15–16 fields from 4 mice per genotype.
(K) MF projections are visualized by mating Pcdh19WT♀ and Pcdh19HET♀ mice with the Thy1-GFP-M line. MF projections in P28 Pcdh19HET♀ mice are normal. Quantification is shown in the graphs. n = 17–18 fields from 4 mice per genotype.
(L) DiI labeling of MF axons in P28 Pcdh19WT♀ and Pcdh19HET♀ mice. Quantification of the lengths of DiI-labeled axons shows that MF lengths are not significantly different between Pcdh19WT♀ and Pcdh19HET♀ mice. n = 8–9 sections.
The scale bars represent 1 mm (low magnification) and 100 μm (high magnification) (C and I to K), 10 μm (E), 200 nm (G) and 50 μm (H and L). Data are mean ± SEM. ***P < 0.005 by Student’s t-test (F, I to L). ***P < 0.005 by one-way ANOVA followed by the Tukey test (D).
PCDH19 is localized at mossy fiber synapses
To understand the roles of PCDH19 in the brain, we generated Pcdh19 knockout mice using the CRISPR-Cas9 system, which were confirmed by genomic PCR and RT-PCR (fig. S1, A to C). We then developed antibodies to the PCDH19 protein (fig. S1, D to F) and verified the specificity of the antibodies using the Pcdh19 knockout mice (fig. S1, G to I). Using these antibodies, we first examined the expression and localization of PCDH19 in mice. PCDH19 is selectively expressed in the brain (Fig. 1B). Since in situ hybridization data showed that Pcdh19 mRNA is highly expressed in the cortex and hippocampus (11, Allen Brain Atlas), we examined PCDH19 protein distributions in these two areas. In the cortex, the PCDH19 protein is localized to layer 5A (fig. S2A), and its expression levels decrease postnatally (fig. S2, B and C). In the hippocampus, the PCDH19 protein is abundant in the mossy-fiber pathway (Fig. 1C; Calbindin labels mossy fibers), where, in contrast to the cortex, its expression levels increase postnatally during the time of synapse development (from P7 to P28; 12) (Fig. 1D). The mossy-fiber pathway terminates in synapses formed by dentate granule cells onto CA3 pyramidal neurons. Pcdh19 mRNA is expressed by both dentate granule cells and CA3 pyramidal neurons (fig. S3), indicating that PCDH19 is expressed by both the pre- and postsynaptic neurons of mossy-fiber synapses. It appears that there is some discrepancy between the mRNA expression and protein expression (fig. S3 and Fig. 1C), suggesting that there may be region-specific regulations of mRNA translation for Pcdh19. Immunostaining for PCDH19, VGLUT1 (marker of glutamatergic synaptic vesicles), and Bassoon (marker of active zones) shows that PCDH19 colocalizes with VGLUT1 and Bassoon at mossy-fiber synapses (Fig. 1, E and F), indicating that PCDH19 localizes to mossy-fiber synapses. To further confirm the synaptic localization of PCDH19, we performed immuno-electron microscopy for PCDH19. PCDH19 is localized at both pre- and postsynaptic sites of mossy-fiber synapses (Fig. 1G). These results demonstrate that PCDH19 is in position to regulate mossy-fiber synapse development, stabilization/maintenance, and function.
PCDH19 is dispensable for mossy-fiber targeting
We next investigated the phenotype of Pcdh19 mutant mice. The expression of PCDH19 in the mossy-fiber pathway is about half in heterozygous female (Pcdh19HET♀) mice relative to WT mice, and is not detectable in hemizygous male (Pcdh19HEMI♂) mice (fig. S1, H and I). The weight of Pcdh19HET♀ mice is normal, the mice do not die early, and the mice are fertile. Furthermore, the gross appearance of the brains from Pcdh19HET♀ mice is normal (fig. S4A). Overall cellular structure in the hippocampus and cortex, as assessed by DAPI staining, is also normal (fig. S4, B and C). We then examined the distribution of Pcdh19+ and Pcdh19− cells in Pcdh19HET♀ mice by crossing them with the HprttdTomato line (13), in which nuclear-localized tdTomato is expressed from one of the X chromosomes. In Pcdh19HET♀ mice, the Pcdh19− and HprttdTomato alleles are on different X-chromosomes, and thus, Pcdh19+ cells will be tdTomato positive, and Pcdh19− cells will be tdTomato negative. We found that Pcdh19HET♀ mice show abnormal aggregation of Pcdh19+ and Pcdh19− cortical neurons (fig. S5, A and B), which is consistent with previous reports (9, 10). Unlike the cortex, dentate granule cells do not show clumped aggregations (Fig. 1H). We next asked if PCDH19 regulates mossy-fiber targeting. Mossy-fiber axonal projections, as visualized by staining for ZnT3 and Calbindin, appear normal in Pcdh19HET♀ mice (Fig. 1, I and J). Furthermore, we utilized the Thy1-GFP-M line (14), in which GFP is expressed by a subpopulation of dentate granule cells, to label mossy fibers, then examined their axons. We found that GFP-labeled mossy-fiber axons are normal in Pcdh19HET♀ mice (Fig. 1K). Finally, we performed sparse labeling of individual mossy-fiber axons with DiI and quantified the lengths of DiI labeled axons. The quantification showed that the lengths of mossy-fiber axons were not significantly different between Pcdh19WT♀ and Pcdh19HET♀ mice (Fig. 1L). These results indicate that PCDH19 does not play a role in mossy-fiber targeting.
Pcdh19HET♀-specific defects in mossy-fiber presynaptic development
We next analyzed the morphology of mossy-fiber terminals in the CA3 stratum lucidum (SL) layer (Fig. 2A) of Pcdh19 mutant mice, utilizing the Thy1-GFP-M line. We found that Pcdh19HET♀, but not Pcdh19HEMI♂, mice showed a significant decrease in the size of mossy-fiber boutons compared to control (Pcdh19WT♀ or Pcdh19WT♂, respectively) mice, without changes in mossy-fiber bouton density (Fig. 2, B and C). We then performed electron microscopic analysis to examine the ultrastructure of mossy-fiber synapses, which consists of a large and complex presynaptic bouton that contains multiple synapses (15). In the mossy-fiber synapse, Pcdh19HET♀ mice showed a significant decrease in the number of synaptic vesicles (SVs) relative to Pcdh19WT♀ mice (Fig. 2D). In contrast, Pcdh19HEMI♂ mice had a comparable number of SVs to Pcdh19WT♂ mice (Fig. 2E). The density and length of postsynaptic densities were similar between all genotypes examined (Fig. 2, D and E). To address the morphology of postsynaptic CA3 pyramidal neurons, we again utilized the Thy1-GFP-M line. In the Thy1-GFP-M line, a small population of CA3 pyramidal neurons is also labeled with GFP. Thus, we examined dendritic branching, spine density, and spine maturation in GFP+ CA3 pyramidal neurons. The results indicate that dendritic branching and spine development are not affected in Pcdh19HET♀ mice (Fig. 2F). To further evaluate postsynaptic development, we performed staining for PSD95 and glutamate receptors (GluR1) in the CA3 SL layer. The densities and sizes of PSD95 and GluR1 puncta were similar between Pcdh19WT♀ and Pcdh19HET♀ mice (Fig. 2, G and H), suggesting that postsynaptic development is not impaired in Pcdh19HET♀ mice. These results indicate that female Pcdh19HET♀ mice show presynaptic defects at mossy-fiber synapses.
Fig. 2. Pcdh19HET♀, but not Pcdh19HEMI♂, mice show impaired mossy-fiber presynaptic development and function.
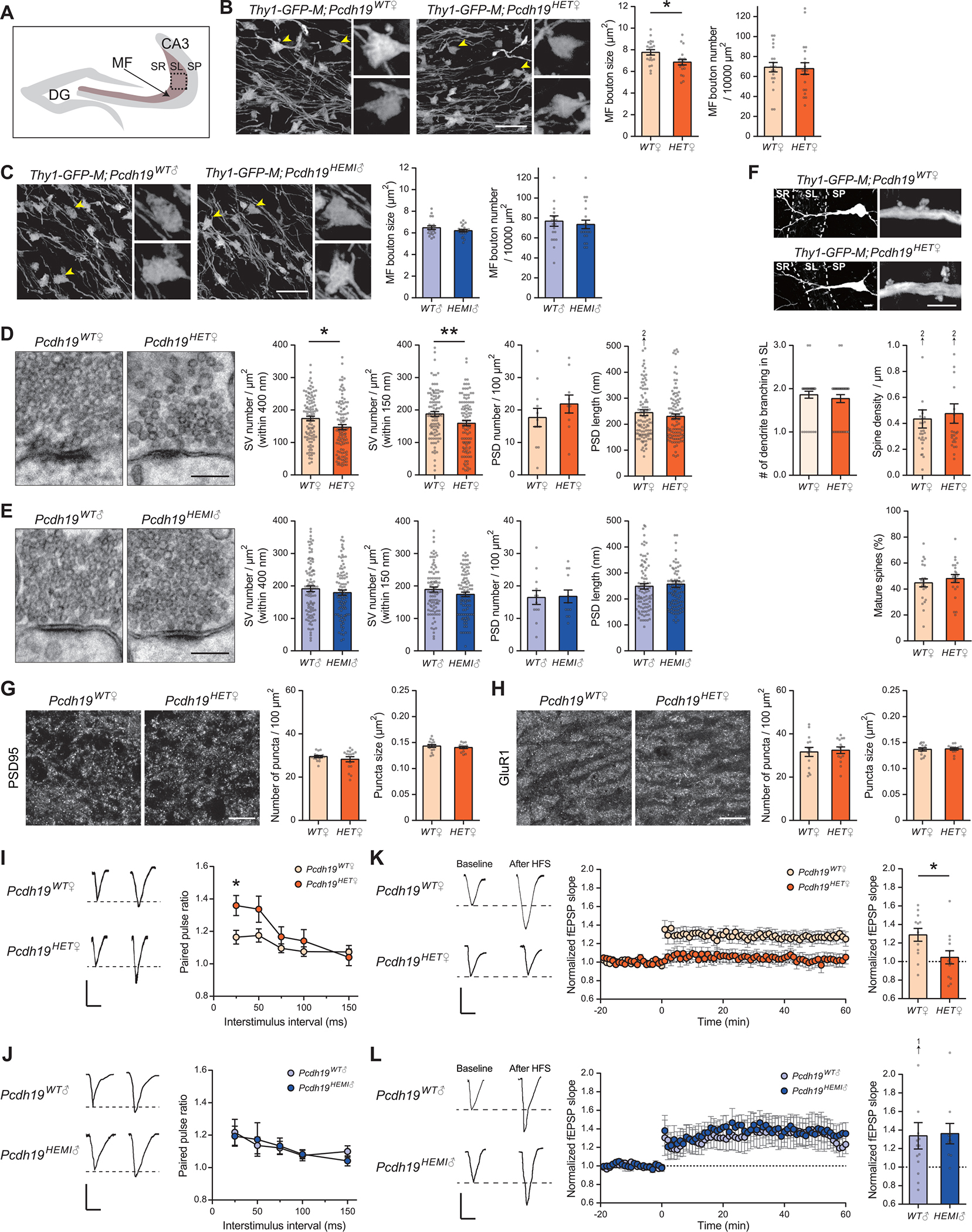
(A) Illustration indicating the pictured area in B, C, G, and H. Images were taken from the CA3 SL layer (MF layer; boxed).
(B and C) Analysis of MF terminals in 4-week-old female (Pcdh19WT♀ and Pcdh19HET♀; B) and male (Pcdh19WT♂ and Pcdh19HEMI♂; C) mice, visualized by mating them with the Thy1-GFP-M line. MF terminals indicated by the arrowheads are magnified. MF bouton size is decreased in Pcdh19HET♀, but not in Pcdh19HEMI♂, mice compared to controls. MF bouton density is similar between control and mutant. n = 20 fields from 4 mice per genotype.
(D and E) Electron microscopic analysis of MF synapses in 4-week-old female (D) and male (E) mice. Densities of synaptic vesicles (SVs) within 400 nm and 150 nm from the active zone are decreased in Pcdh19HET♀, but not in Pcdh19HEMI♂, mice compared their respective controls. The density and length of PSDs are comparable between all genotypes. n = 93–103 synapses from 10–12 fields from 2 mice per genotype.
(F) Analysis of dendrites and spines of CA3 pyramidal neurons in 4-week-old Pcdh19WT♀ and Pcdh19HET♀ mice, visualized by mating them with the Thy1-GFP-M line. Low and high magnification images are shown. Number of dendritic branches in the SL layer (n = 35–36 dendrites from 5 mice per genotype), spine density (n = 25–28 fields from 5 mice per genotype), and % of mature spines (n = 25–28 fields from 5 mice per genotype) are similar between Pcdh19WT♀ and Pcdh19HET♀ mice.
(G and H) Immunostaining for postsynaptic proteins PSD95 and GluR1 in the CA3 SL layer in 4-week-old Pcdh19WT♀ and Pcdh19HET♀ mice. Density and size of PSD95 (G) and GluR1 (H) puncta are comparable between Pcdh19WT♀ and Pcdh19HET♀ mice. n = 14–16 fields from 4 mice per genotype.
(I and J) Paired-pulse ratio (PPR) of evoked EPSCs at MF synapses from 5-week-old female (I) and male (J) mice (left, sample traces with a 25 ms inter-stimulus interval; right, PPR across a range of inter-stimulus intervals). PPR is significantly increased in Pcdh19HET♀, but not in Pcdh19HEMI♂, mice compared to controls. n = 21 slices for Pcdh19WT♀, 14 for Pcdh19HET♀, 11 for Pcdh19WT♂, and 14 for Pcdh19HEMI♂ from 4 mice each.
(K and L) MF-LTP induced by high-frequency stimulation (2.5 s train at 25 Hz × 25 stimulations) in 5-week-old female (K) and male (L) mice (Left, sample traces before and after the LTP induction; Middle, LTP time course; Right, fEPSP slopes after the LTP induction normalized to baseline). MF-LTP is abolished in Pcdh19HET♀, but not in Pcdh19HEMI♂ mice. n = 12 slices for Pcdh19WT♀, 13 for Pcdh19HET♀, 14 for Pcdh19WT♂, and 10 for Pcdh19HEMI♂ from 4 mice each.
The scale bars represent 10 μm (B, C, F: low magnification, G, and H), 200 nm (D and E), 5 μm (F: high magnification), and 100 μV and 25 ms (I to L). Data are mean ± SEM. *P < 0.05, **P < 0.01 by Student’s t-test (B to H). *P < 0.05 by two-way ANOVA followed by the Sidak test (I and J). *P < 0.05 by Mann-Whitney U test (K and L).
To determine whether mossy-fiber synapses are functionally impaired in Pcdh19 mutant mice, we performed electrophysiological recordings. We first performed whole-cell recordings from CA3 pyramidal neurons in Pcdh19WT♀ and Pcdh19HET♀ mice. The frequency (indicative of presynaptic function), but not the amplitude (indicative of postsynaptic function), of miniature excitatory postsynaptic current (mEPSC) seemed decreased in Pcdh19HET♀ mice relative to in Pcdh19WT♀ mice, although the decrease was not significant (fig. S6A). We then evaluated the connectivity between mossy fibers and CA3 pyramidal neurons. The input-output curves generated by mossy-fiber stimulation were similar between Pcdh19WT♀ and Pcdh19HET♀ mice (fig. S6B), suggesting that PCDH19 does not regulate basal mossy-fiber strength. To assess potential presynaptic changes in Pcdh19HET♀ mice, we next analyzed the paired-pulse ratio (PPR) of evoked EPSCs at mossy-fiber synapses. The PPR was significantly increased in Pcdh19HET♀ as compared to Pcdh19WT♀ mice (Fig. 2I), while it was similar between Pcdh19WT♂ and Pcdh19HEMI♂ mice (Fig. 2J), suggesting that the neurotransmitter release probability is decreased in female Pcdh19HET♀ and not in male Pcdh19HEMI♂ mice. Finally, we examined long-term potentiation (LTP) at mossy fiber-CA3 synapses, which is a presynaptic form of synaptic plasticity (16, 17). We found that mossy-fiber LTP was completely abolished in Pcdh19HET♀ mice (Fig. 2K). In contrast, mossy-fiber LTP was normal in Pcdh19HEMI♂ mice (Fig. 2L). Taken together with our histological studies (Fig. 1, I to L and Fig. 2, B to H), these results indicate that Pcdh19HET♀ mice specifically show impairment in presynaptic function and plasticity at mossy fiber-CA3 synapses.
Pcdh19HET♀-specific impairment in mossy-fiber dependent cognitive behaviors
Given the functional mossy-fiber synaptic deficits in female Pcdh19HET♀ mice, we tested several cognitive behavioral paradigms associated with hippocampal function, both those associated with mossy-fiber function, and those associated with other areas of the hippocampus. Previous reports have demonstrated that mice with mossy-fiber synaptic dysfunction show cognitive impairments in pattern completion, a process to restore an engram and recall/complete the pattern of memories, and pattern separation, a process to separate similar patterns of activities (18, 19). In contrast, the mice with mossy-fiber synaptic dysfunction show normal contextual and cued learning, tasks associated with other areas of the hippocampus (18, 20). We first examined contextual and cued learning of Pcdh19 mutant mice using contextual and cued fear conditioning tests (Fig. 3A). We found that there were no significant differences in terms of freezing responses during conditioning (Fig. 3, B and E), contextual tests (Fig. 3, C and F), and cued tests (Fig. 3, D and G) between Pcdh19 mutant (Pcdh19HET♀ and Pcdh19HEMI♂) and control mice. Freezing responses tested 28 days after conditioning were also normal (fig. S7, A and B), indicating normal memory retention. Thus, hippocampal learning not associated with mossy-fiber function was normal.
Fig. 3. Pcdh19HET♀, but not Pcdh19HEMI♂, mice lack pattern completion and pattern separation abilities.
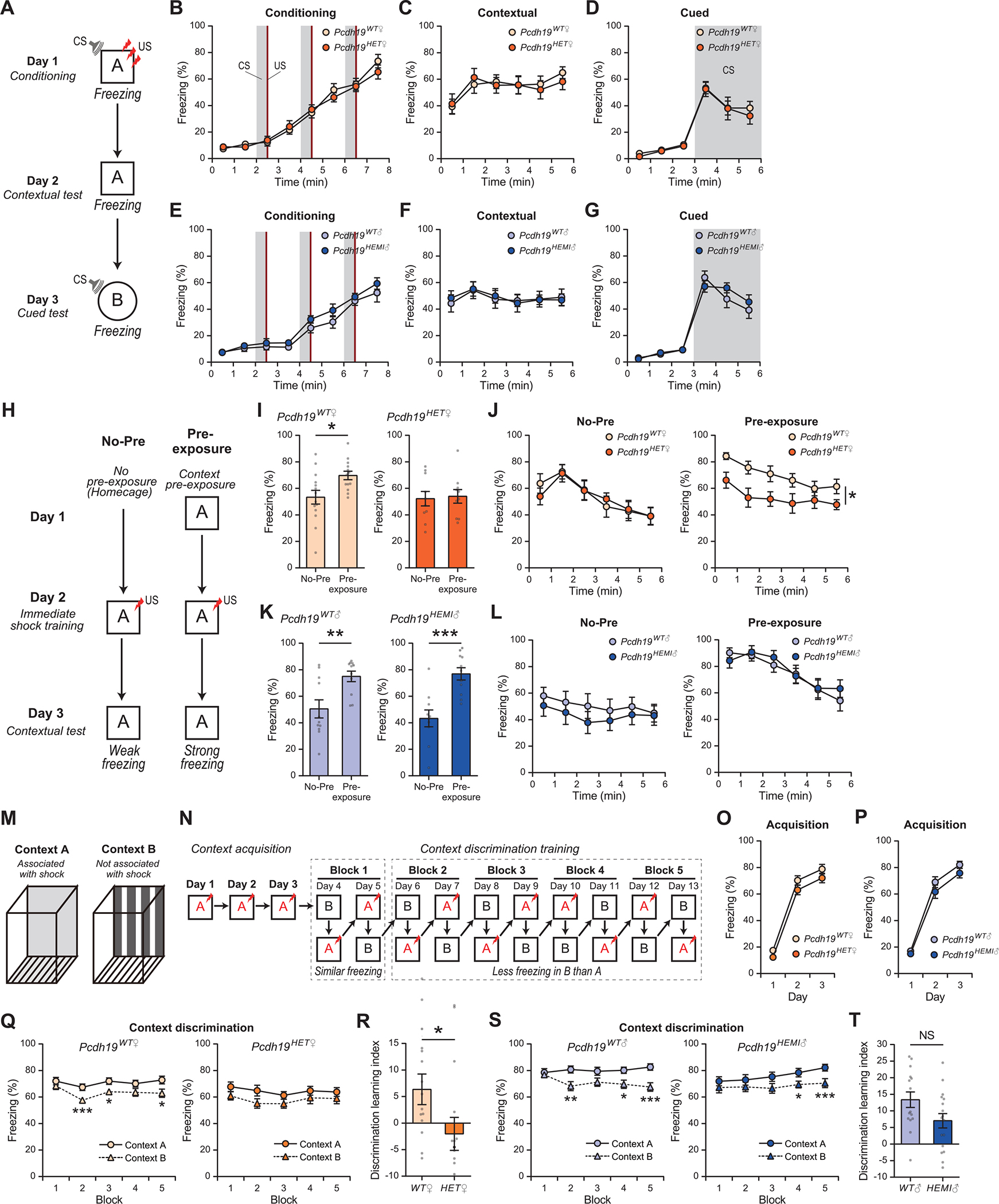
(A to G) Fear conditioning test. (A) Schematic of the contextual and cued fear conditioning test. Context A is different from context B. CS, white noise; US, 0.3 mA foot shock. (B to G) Freezing responses of female (B to D) and male (E to G) mice. (B and E) Freezing responses on day 1. CS was presented for 30 s, 3 times, (gray shadow) and at the end of each CS, a US was given (red). (C and F) Freezing responses on day 2 (contextual test). (D and G) Freezing responses on day 3 (cued test). CS was presented at 3–6 min (gray shadow). Freezing responses during the conditioning, contextual and cued tests were similar between Pcdh19 mutant and control mice. n = 14 mice for Pcdh19WT♀, 10 for Pcdh19HET♀, 12 for Pcdh19WT♂, and 12 for Pcdh19HEMI♂.
(H to L) The pattern completion test. (H) Schematic of the pre-exposure dependent contextual fear task (pattern completion-mediated contextual task). On day 1, mice were either left in their home cage (No-Pre) or pre-exposed to context A for 10 min (Pre-exposure). On day 2, the mice received an immediate shock in context A (a 0.5 mA, 2 sec foot shock during a 42 sec stay). On day 3, freezing responses in context A were monitored. (I to L) Pcdh19WT♀, Pcdh19WT♂, and Pcdh19HEMI♂, but not Pcdh19HET♀, mice show increased freezing responses when pre-exposed to the context on day 1. n = 14 (No-Pre) and 13 (Pre-exposure) mice for Pcdh19WT♀, 10 and 10 for Pcdh19HET♀, 11 and 11 for Pcdh19WT♂, and 11 and 11 for Pcdh19HEMI♂. (J and L) Time course of the freezing responses.
(M to T) The pattern separation test. (M) Context A and B are very similar but slightly different in terms of a floor and walls. (N) Schematic of the contextual discrimination task (pattern separation-mediated contextual task). Only in context A, a foot shock is given to mice. Days 1–3 are context acquisition, and days 4–13 are context discrimination training. After the discrimination training, mice can separate context A (showing strong freezing) from context B (weak freezing). (O and P) Normal freezing responses during context acquisition in Pcdh19WT♀ and Pcdh19HET♀ (O) and in Pcdh19WT♂ and Pcdh19HEMI♂ mice (P). (Q and S) Freezing responses in context A (solid line) and B (dashed line) during discrimination training. Pcdh19WT♀ (Q, left), Pcdh19WT♂ (S, left), and Pcdh19HEMI♂ (S, right), but not Pcdh19HET♀ (Q, right), mice showed less freezing responses in context B than in context A after training. (R and T) Discrimination learning index of % freezing = (A – B in Block5) – (A – B in Block1). Pcdh19WT♀, Pcdh19WT♂, and Pcdh19HEMI♂, but not Pcdh19HET♀, mice learned to discriminate the two contexts. n = 17 mice for Pcdh19WT♀, 15 for Pcdh19HET♀, 15 for Pcdh19WT♂, and 17 for Pcdh19HEMI♂.
Data are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005 by Student’s t-test (I, K, Q, and S). *P < 0.05; two-way repeated-measures ANOVA followed by the Sidak test (B to G, J, L, O, and P). *P < 0.05 by Mann-Whitney U test (R and T). NS: not significant.
We next evaluated the mossy fiber-dependent pattern completion ability using the pre-exposure-dependent contextual fear facilitation test (18, 21). For this, we prepared and compared two groups of mice, No-Pre and Pre-exposure, as diagramed in Figure 3H. The No-Pre group of mice were in the home cage on day 1, received a single foot shock immediately after being placed in a context on day 2, and tested for freezing in the same context on day 3. Because of the short stay in the context, only on day 2, it is difficult for the No-Pre group of mice to associate the context with the foot shock, resulting in only weak freezing on day 3. On the other hand, the Pre-exposure group of mice were pre-exposed to the context on day 1, followed by a foot shock in the context on day 2, and tested for freezing on day 3. Because the Pre-exposure group of mice was already exposed to the context before the shock training, they can recall the context during the training and associate it with the foot shock (21). As a result of this “pattern completion”, the Pre-exposure group of mice show strong freezing on day 3 (Fig. 3H). Indeed, the Pre-exposure group of Pcdh19WT♀ mice did show significantly more freezing responses than the No-Pre group, demonstrating pattern completion ability in control mice (Fig. 3, I and J). However, Pcdh19HET♀ mice exhibited similar levels of freezing responses between the Pre-exposure and the No-Pre groups, showing a failure to demonstrate pattern completion (Fig. 3, I and J). Pcdh19WT♂ and Pcdh19HEMI♂ mice both showed normal pattern completion ability (Fig. 3, K and L). These results indicate that pattern completion ability is impaired selectively in Pcdh19HET♀ mice, which is consistent with their mossy-fiber synaptic defects (Fig. 2).
We then evaluated pattern separation ability using the contextual discrimination fear conditioning test (18, 22). In this test, mice were placed in one of two boxes that have slightly different contexts: context A and context B (Fig. 3M). The mice received a foot-shock in context A, but not in context B. During the pattern separation test, mice were first placed in context A for three days in a row and received a foot shock (context acquisition; Fig. 3, N to P). From day 4 to day 13, mice were placed in either context A or B, where context A is associated with a foot shock and B is not (context discrimination training). Initially, mice cannot separate these two closely related contexts, and thus, they show similar freezing levels in both contexts (Fig. 3, N, Q and S, days 4 and 5; Block 1). However, after a few days, normal mice can separate the two contexts and show less freezing in context B than in context A (Fig. 3, N, Q and S, day 6 and later; Blocks 2 to 5). In our test, both Pcdh19WT♀ and Pcdh19HET♀ mice showed normal contextual acquisition (Fig. 3O). Pcdh19WT♀ mice then started to show reduced freezing responses in context B after two days of context discrimination training (Fig. 3Q, after Block 2). However, Pcdh19HET♀ mice never demonstrated differences in freezing responses between context A and B, indicating that Pcdh19HET♀ mice could not distinguish context B from context A. This failure is depicted by the discrimination learning index (Fig. 3R), suggesting that Pcdh19HET♀ mice lack the contextual pattern separation ability. Pcdh19WT♂ and Pcdh19HEMI♂ mice showed normal context acquisition and pattern separation ability (Fig. 3, P, S and T).
To exclude the possibility that vision impairment of Pcdh19HET♀ mice might underlie their defects in cognitive behaviors, we performed the distinct context discrimination test, which requires visually discriminating different context (fig. S7, C to H). There was no difference in this test between Pcdh19WT♀ and Pcdh19HET♀ mice, suggesting that the vision of Pcdh19HET♀ mice is normal. Altogether, Pcdh19HET♀ mice, a model of PCDH19 disorder, show defects in cognitive behaviors associated with mossy-fiber function, but not other cognitive tasks, which, again, is consistent with their morphological and physiological mossy-fiber synaptic defects (Fig. 2).
PCDH19–N-cadherin complexes at mossy-fiber synapses
What are the molecular bases of female heterozygote (Pcdh19HET♀)-specific mossy-fiber synaptic dysfunction and cognitive impairment? Since male hemizygotes (Pcdh19HEMI♂) do not show mossy-fiber synaptic dysfunction, lack of homophilic interactions between PCDH19 alone cannot explain the defects. Therefore, we looked for additional molecular players that may interact with PCDH19. To this end, we focused on the interaction between PCDH19 and N-cadherin (Ncad; also called as Cadherin2 [CDH2]), another cell adhesion molecule localized at synapses (23, 24). PCDH19 has been proposed to form a cis-complex with Ncad, which may mask the homophilic binding ability of Ncad (25). If this is the case, the PCDH19–N-cad cis-complex may send inter-cellular signals through the PCDH19-mediated interactions, but not through the Ncad-mediated interactions (see Fig. 4O). In the absence of PCDH19 (Pcdh19HEMI♂), Ncad would be unmasked and could send inter-cellular signals through Ncad–Ncad interactions. However, if the PCDH19 expression is mosaic (Pcdh19HET♀), the PCDH19–N-cad cis-complex could not mediate inter-cellular signals between PCDH19-positive and negative cells. This PCDH19–Ncad mismatch would result in reduced downstream intracellular signals and cellular defects. We tested this possibility at mossy-fiber synapses.
Fig. 4. PCDH19–N-cad mismatch impairs Ncad signaling and presynaptic development at mossy-fiber synapses.
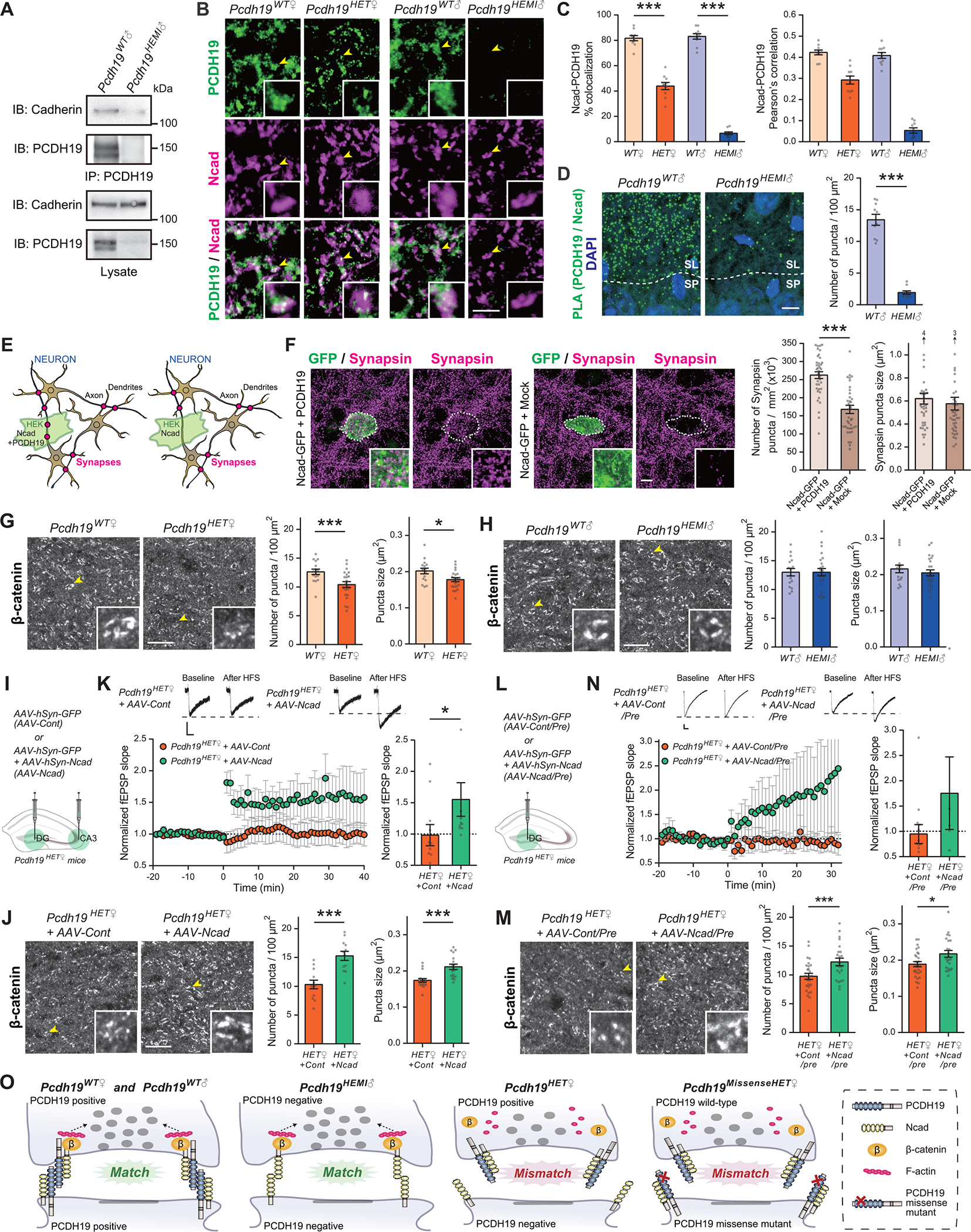
(A) PCDH19 interacts with Cadherin. Immunoprecipitates with anti-PCDH19 antibodies from hippocampal lysates prepared from 4-week-old Pcdh19WT♂ and Pcdh19HEMI♂ mice were immunoblotted for Cadherin or PCDH19 (upper panels). Lysates were also tested for the presence and absence of Cadherin and PCDH19 (lower panels). Anti-Cadherin antibody: Cat# sc-59876. The two bands for PCDH19 are splice variants due to alternative splicing in the cytoplasmic domain (NM_001105245.1 and NM_001105246.1).
(B and C) Double staining for PCDH19 and Ncad in the CA3 SL layer of 4-week-old Pcdh19WT♀, Pcdh19HET♀, Pcdh19WT♂, and Pcdh19HEMI♂ mice. Insets are higher-magnification views of a MF bouton, indicated by arrowheads. Percentages of Ncad puncta colocalized with PCDH19 puncta and Pearson’s correlation coefficients between Ncad and PCDH19 are quantified (C). n = 10 fields from 3 mice per genotype. Anti-Ncad antibody: Cat# 610920.
(D) Proximal ligation assay (PLA) detecting PCDH19 and Ncad interaction in situ in hippocampal sections from 4-week-old Pcdh19WT♂ and Pcdh19HEMI♂ mice. PLA signals (green puncta) are detected in the CA3 SL layer of Pcdh19WT♂, but not of Pcdh19HEMI♂, mice. n = 10 fields from 3 mice per genotype. Anti-Ncad antibody: Cat# 610920.
(E and F) HEK cell–neuron co-culture assay. (E) Schematic of the co-culture assay to examine the synaptogenic activity. HEK cells may organize presynaptic differentiation (red dots) in contacting neuronal axons only when HEK cells are expressing synapse organizing molecules (in this case, Ncad + PCDH19; left panel). If HEK cells only express Ncad, they cannot organize presynaptic differentiation (right panel). In the adjacent areas, synapses may form where neuronal axons and dendrites contact with each other. (F) HEK cells overexpressing Ncad-GFP only or Ncad-GFP + PCDH19 were co-cultured with hippocampal neurons for 2 days and stained for Synapsin (to label presynaptic terminals) and GFP (to label HEK cells) at DIV18. Insets show high magnification views of staining on HEK cells. The density of Synapsin puncta formed on HEK cells overexpressing Ncad only was significantly less than that on HEK cells overexpressing Ncad + PCDH19. n = 39 cells from 3 independent experiments. The cDNA constructs used for overexpression were human NCAD (CDH2) and mouse Pcdh19.
(G and H) β-catenin clustering in the CA3 SL layer in 4-week-old female (G) and male (H) mice. Insets are higher-magnification view of a MF bouton indicated by arrowheads. Density and size of β-catenin puncta were significantly decreased in Pcdh19HET♀, but not in Pcdh19HEMI♂, mice compared to controls. n = 16–24 fields from 3 mice per genotype.
(I to K) Overexpression of Ncad in DG and CA3 regions of Pcdh19HET♀ mice restored β-catenin clustering and MF-LTP. (I) AAV expressing GFP only (AAV-Cont) or GFP together with Ncad (AAV-Ncad) was injected into both DG and CA3 regions of P4 Pcdh19HET♀ mice by stereotaxic injection. (J) β-catenin clustering in the CA3 SL layer of 4-week-old female Pcdh19HET♀+AAV-Cont and Pcdh19HET♀+AAV-Ncad mice. Insets are higher-magnification views of a MF bouton indicated by arrowheads. Density and size of β-catenin puncta are significantly increased in Pcdh19HET♀+AAV-Ncad compared to Pcdh19HET♀+AAV-Cont. n = 18 fields from 3 mice per condition. (K) MF-LTP induced by high-frequency stimulation in 5-week-old female Pcdh19HET♀+AAV-Cont and Pcdh19HET♀+AAV-Ncad mice (upper panel, sample traces before and after the LTP induction; lower left panel, LTP time course; lower right panel, fEPSP slope after the LTP induction normalized to baseline). n = 11 slices for Pcdh19HET♀+AAV-Cont and 11 for Pcdh19HET♀+AAV-Ncad from 4 mice each.
(L to N) Overexpression of Ncad in the DG region of Pcdh19HET♀ mice restored β-catenin clustering and MF-LTP. (L) AAV expressing GFP or GFP together with Ncad was injected into the DG region of P4 Pcdh19HET♀ mice by stereotaxic injection (AAV-Cont/Pre or AAV-Ncad/Pre). (M) β-catenin clustering in the CA3 SL layer of 4-week-old female Pcdh19HET♀+AAV-Cont/Pre and Pcdh19HET♀+AAV-Ncad/Pre mice. Insets are higher-magnification views of a MF bouton indicated by arrowheads. Density and size of β-catenin puncta are significantly increased in Pcdh19HET♀+AAV-Ncad/Pre compared to Pcdh19HET♀+AAV-Cont/Pre. n = 23–27 fields from 3–4 mice per condition. (N) MF-LTP induced by high-frequency stimulation in 5-week-old female Pcdh19HET♀+AAV-Cont/Pre and Pcdh19HET♀+AAV-Ncad/Pre mice (upper panel, sample traces before and after the LTP induction; lower left panel, LTP time course; lower right panel, fEPSP slope after the LTP induction normalized to baseline). n = 13 slices for Pcdh19HET♀+AAV-Cont/Pre and 5 for Pcdh19HET♀+AAV-Ncad/Pre from 3–4 mice each.
(O) A proposed model. At Pcdh19WT♀, Pcdh19WT♂, and Pcdh19HEMI♂ synapses, PCDH19 and/or Ncad homophilic matching mediates β-catenin signaling and organizes presynaptic terminals. However, at Pcdh19HET♀ synapses, PCDH19–N-cad mismatching diminishes β-catenin signaling and impairs presynaptic development. PCDH19 with missense mutations, which may not bind to WT PCDH19, could also lead to synaptic impairment (Pcdh19MissenseHET♀).
The scale bars represent 5 μm (B), 10 μm (D, F, G, H, J, M), and 100 μV and 25 ms (K and N). Data are mean ± SEM. *P < 0.05, ***P < 0.005; Student’s t-test (D, F, G, H, J, M). ***P < 0.005 by one-way ANOVA followed by the Tukey test (C). *P < 0.05 by Mann-Whitney U test (K and N).
We first examined the physical interactions between PCDH19 and Ncad in the hippocampus. Co-immunoprecipitation analysis showed that PCDH19 does interact with Cadherins in the hippocampus (Fig. 4A). We next analyzed the localization of PCDH19 and Ncad in mossy-fiber terminals. Both PCDH19 and Ncad are localized at mossy-fiber synapses: a majority (~80%) of Ncad puncta was colocalized with PCDH19 puncta in Pcdh19WT♀ and Pcdh19WT♂ mice (Fig. 4, B and C). ~40% of Ncad was colocalized with PCDH19 in Pcdh19HET♀ mice, and almost none in Pcdh19HEMI♂ mice. Pearson’s correlation analysis also supported the colocalization between PCDH19 and Ncad (Fig. 4C). At the electron microscopic level, PCDH19 is localized perisynaptically (Fig. 1G), which is similar to known Ncad localization (23, 24, 26). To directly detect the possible interaction between PCDH19 and Ncad in situ, we performed a proximity ligation assay for PCDH19 and Ncad in hippocampal sections (27). Signals were detected in the CA3 SL layer of Pcdh19WT♂ mice, but not Pcdh19HEMI♂ mice (Fig. 4D), indicating that PCDH19 and Ncad are near each other, likely in a complex, at mossy-fiber synapses.
PCDH19–N-cad mismatch impairs mossy-fiber presynaptic development
To investigate the function of potential PCDH19–N-cad complexes, we designed experiments to test the effects of PCDH19–Ncad mismatch. For this, we utilized the HEK cell–neuron co-culture system (28, 29). When HEK cells express synaptogenic molecules, they induce presynaptic differentiation in contacting axons (Fig. 4E, left panel), but if HEK cells do not express synaptogenic molecules, there will be no presynaptic differentiation induced at the contact sites between neurons and HEK cells (Fig. 4E, right panel). WT hippocampal neurons endogenously express both PCDH19 and Ncad, but HEK cells only express Ncad (human Ncad, which is known to interact with mouse Ncad; 30) and do not endogenously express PCDH19. Into HEK cells, we overexpressed Ncad together with PCDH19 (Ncad was fused with GFP) or Ncad only, so that when the HEK cells are co-cultured with hippocampal neurons, they would create a PCDH19–Ncad matched or a mismatched situation, respectively. We then evaluated the presynaptic development of hippocampal neurons at the contact sites with HEK cells by staining for Synapsin, a synaptic vesicle protein. We found that the number of Synapsin puncta formed on HEK cells overexpressing Ncad-only (which mimics the Pcdh19HET♀ condition) was significantly lower than that on HEK cells overexpressing Ncad together with PCDH19 (which mimics the Pcdh19WT♀ control condition) (Fig. 4F), suggesting that, indeed, PCDH19–Ncad mismatch impairs presynaptic development.
We next investigated the effects of PCDH19–Ncad mismatch on downstream signaling mediated by PCDH19–N-cad in vivo. Activation of Ncad-mediated signaling induces the recruitment of β-catenin toward synapses and regulates presynaptic development and function (23, 24, 31). We therefore examined the clustering of β-catenin at mossy-fiber synapses in Pcdh19 mutant mice. We found that the β-catenin puncta density and size were significantly decreased in Pcdh19HET♀ relative to Pcdh19WT♀ mice (Fig. 4G). Pcdh19HEMI♂ mice did not show any differences from Pcdh19WT♂ mice (Fig. 4H). These results suggest that PCDH19–Ncad mismatch (Pcdh19HET♀) impairs β-catenin signaling at mossy-fiber synapses.
Finally, to investigate whether impaired Ncad signaling indeed underlies defects in β-catenin clustering and mossy-fiber function in Pcdh19HET♀ mice, we overexpressed Ncad in Pcdh19HET♀ mice and examined whether it can rescue the defects in Pcdh19HET♀ mice. Using AAV, we first overexpressed GFP (AAV-Cont) or GFP together with Ncad (AAV-Ncad) in neurons in the dentate gyrus and CA3 of Pcdh19HET♀ mice (Fig. 4I). Overexpression of Ncad in Pcdh19HET♀ mice not only restored β-catenin clustering in the CA3 SL layer (Fig. 4J), but also synaptic function, as demonstrated by restored mossy fiber-LTP (Fig. 4K). We next performed rescue experiments with Ncad overexpressed only in the presynaptic compartment, i.e., dentate granule cells (Fig. 4L). The presynaptic overexpression of Ncad was sufficient to restore β-catenin clustering (Fig. 4M) and mossy fiber-LTP (Fig. 4N) in Pcdh19HET♀ mice. These results indicate that impaired Ncad function underlies the phenotype observed in Pcdh19HET♀ mutants in vivo (see Fig. 4O).
Discussion
In this paper, we showed that Pcdh19HET♀, and not Pcdh19HEMI♂, mice show mossy-fiber presynaptic dysfunction and cognitive impairments, mimicking the female specific manifestation of PCDH19 disorder. We further showed that the mismatch between PCDH19 and Ncad impairs β-catenin clustering and presynaptic development at mossy-fiber synapses. We propose the following model (Fig. 4O): At Pcdh19WT♀ and Pcdh19WT♂ synapses, PCDH19–N-cad cis-complex mediates trans-synaptic signaling through PCDH19 homophilic matching, which recruits β-catenin and organizes presynaptic terminals. At Pcdh19HEMI♂ synapses, unmasked Ncad can mediate trans-synaptic signaling through Ncad homophilic matching and recruit β-catenin. However, at Pcdh19HET♀ synapses between PCDH19+ and PCDH19− neurons, PCDH19–N-cad cis-complex cannot trans-synaptically bind Ncad due to mismatching. This mismatching diminishes β-catenin clustering, leading to impaired presynaptic development, and contributes to cognitive impairment. Our results provide insights into the molecular and cellular mechanisms underlying the female-specific phenotypes in PCDH19 disorder. In the case of missense mutations (Pcdh19MissenseHET♀), many missense mutations in PCDH19 disorder alter sites in the PCDH19 extracellular domain (2, 6). Therefore, missense mutants of PCDH19 may not interact with WT PCDH19, causing a mismatch. This mismatch in Pcdh19MissenseHET♀, like in Pcdh19HET♀, could impair Ncad–β-catenin signaling and lead to synaptic impairment.
Human PCDH19 disorder is characterized by clusters of epileptic seizures and often accompanied by cognitive impairment. While Pcdh19HET♀ mice show abnormal network activity (9), they do not exhibit apparent spontaneous seizures, perhaps due to differences in the brain complexity between humans and mice. Nevertheless, as described in this paper, cognitive impairment is a common phenotype observed in both humans and mice. This suggests that the molecular and cellular mechanisms we identified likely underlie human PCDH19 disorder. A possible translational implication of our study is that the PCDH19–N-cad cis-interaction could be a target of the treatment of certain aspects of PCDH19 disorder, including cognitive impairment. Inhibition of PCDH19–N-cad cis-interactions, e.g., with a competitive peptide, in Pcdh19HET♀ could unmask Ncad and enable Ncad homophilic matching, and thus, restore mossy-fiber presynaptic function. Unmasking of Ncad may work even in the case of missense mutations (Pcdh19MissenseHET♀): inhibition of both WT PCDH19–N-cad and missense PCDH19–N-cad cis-interactions could enable Ncad homophilic matching. Furthermore, since Ncad regulates cortical neuron migration and development (32, 33), the abnormal cortical neuron sorting phenotype in Pcdh19HET♀ mice may also involve PCDH19–Ncad mismatch, and if so, unmasking of Ncad could alleviate the cell sorting phenotype in Pcdh19HET♀ as well. Thus, our study here not only provides mechanistic insights into PCDH19 disorder, but also suggests possible strategies to treat the disorder.
Summary of Materials and Methods:
PCDH19 antibodies and cDNAs
Polyclonal anti-PCDH19 antibodies were generated by immunizing rabbits with KLH-conjugated PCDH19 peptides (mouse, a.a. 1111–1145 [NP_001098715.1]). The specificity of the antibodies was confirmed by immunoblotting and immunohistochemistry (fig. S1, D to I). The mouse Pcdh19 cDNA (NM_001105245.1), human CDH2 (NCAD) cDNA (NM_001792.5), and mouse Cdh2 (Ncad) cDNA (NM_007664.5) were obtained by RT-PCR.
Animals
Pcdh19 mutant mice were generated using the CRISPR/Cas9 system (fig. S1). Two gRNAs were designed so that the 1st exon of Pcdh19 would be deleted after cleavage. Pcdh19 mutant mice were maintained on a C57BL/6J background. Thy1-GFP-M (JAX: 007788) and HprtLSL-tdTomato (JAX: 021428) mice were previously described (13, 14). Both male and female mice were used and analyzed separately. All animal care and use was in accordance with the institutional guidelines and approved by the Institutional Animal Care and Use Committee at Boston Children’s Hospital.
Histology
Mouse brain sections and cultured cells were immunostained as described previously (12, 34). Fluorescent in situ hybridization was performed with an RNAscope Fluorescent Multiplex Kit (Advanced Cell Diagnostics). Proximity ligation assay was conducted with Duolink in situ PLA fluorescence kit (MilliporeSigma). Images were taken with a BZ-X810 fluorescence microscope (Keyence) or an LSM700 confocal microscope (Carl Zeiss). Images were obtained with the identical acquisition settings. The background intensity was subtracted from each image. The intensity of signals, the density and size of puncta, the axon length, and the density and morphology of boutons and spines were quantified using Image J or MetaMorph, or manually. For colocalization experiments, % colocalization and Pearson’s correlation were quantified using Image J. Puncta smaller than 6 pixels were excluded from analysis. Electron microscopic analysis of mossy fiber synapses and pre-embedding immunogold labeling were performed as described previously (12, 34). Quantitative analysis of mossy fiber synapses was performed using Image J or manually by an individual blind to genotype.
Electrophysiology
All electrophysiological experiments and analyses were done blind. mEPSC and fEPSP recordings were conducted with acute hippocampal slices prepared from 4- to 6-week-old mice with a Multiclamp 700B amplifier and Clampex 10.7 software (Axon Instruments). mEPSCs were analyzed using Minianalysis (Synaptosoft). The paired-pulse ratio was determined as the ratio of the second to the first peak amplitude of EPSPs at various interstimulus intervals. For LTP, fEPSPs were elicited at 0.033 Hz; once a stable baseline had been achieved for 20 minutes, LTP was evoked by a 2.5 sec train delivered at 25 Hz × 25 stimulations.
Behavioral tests
All data acquisition and analyses were carried out by an individual blind to the genotype. 2- to 4-month-old mice were used for experiments. Contextual and cued fear conditioning tests, pre-exposure dependent fear facilitation (Pattern completion) task, contextual discrimination fear learning (Pattern separation) task, and distinct contextual fear conditioning tests were conducted in a conditioning chamber (Ugo Basile) equipped with a digital video camera and a sound-attenuating chest. An automated activity detection system in EthoVision XT software (Noldus) was used to monitor freezing responses.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software, as indicated in the figure legends.
Supplementary Material
Acknowledgments:
We thank Sivapratha Nagappan-Chettiar, Akiko Terauchi, and Masahiro Yasuda for helpful comments on the manuscript, and Jaanvi Sant for mouse colony management. We also thank Mariel Seiglie, the Mouse Gene Manipulation Core and the Viral Core at Boston Children’s Hospital, Electron Microscopy Facility at Harvard Medical School, and Neurodevelopmental Behavioral Core at Boston Children’s Hospital for technical contributions.
Funding:
This work is supported by the NIH grant DA042744, SFARI grant #568285, and Harvard Brain Science Initiative Bipolar Disorder Seed Grant, supported by Kent and Liz Dauten (H.U.). N.H. was supported by JSPS Grants-in-Aid for Scientific Research #23700411 and the Uehara Memorial Foundation overseas postdoctoral research fellowship.
Footnotes
Competing interests: Authors declare no competing interests.
Data and materials availability:
All data is available in the main text or the supplementary materials. All materials and animals are available from the corresponding author upon request.
References and Notes:
- 1.Dibbens LM et al. , X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat. Genet. 40, 776–781 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolc KL et al. , A systematic review and meta-analysis of 271 PCDH19-variant individuals identifies psychiatric comorbidities, and association of seizure onset and disease severity. Mol. Psychiatry 24, 241–251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Depienne C et al. , Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet. 5, e1000381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kahr I, Vandepoele K, van Roy F, Delta-protocadherins in health and disease. Prog. Mol. Biol. Transl. Sci. 116, 169–92 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Tai K, Kubota M, Shiono K, Tokutsu H, Suzuki ST, Adhesion properties and retinofugal expression of chicken protocadherin-19. Brain Res. 1344, 13–24 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Cooper SR, Jontes JD, Sotomayor M, Structural determinants of adhesion by Protocadherin-19 and implications for its role in epilepsy. Elife 5, e18529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas S, Emond MR, Jontes JD, Protocadherin-19 and N-cadherin interact to control cell movements during anterior neurulation. J. Cell Biol. 191, 1029–1041 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper SR et al. , Protocadherins control the modular assembly of neuronal columns in the zebrafish optic tectum. J. Cell Biol. 211, 807–814 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pederick DT et al. , Abnormal cell sorting underlies the unique X-linked inheritance of PCDH19 epilepsy. Neuron 97, 59–66 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Hayashi S et al. , Loss of X-linked Protocadherin-19 differentially affects the behavior of heterozygous female and hemizygous male mice. Sci. Rep. 7, 5801 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaarschuch A, Hertel N, Expression profile of N-cadherin and protocadherin-19 in postnatal mouse limbic structures. J. Comp. Neurol. 526, 663–680 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Terauchi A et al. , Distinct FGFs promote differentiation of excitatory and inhibitory synapses. Nature 465, 783–787 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H et al. , Cellular resolution maps of X chromosome inactivation: implications for neural development, function, and disease. Neuron 81, 103–119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng G et al. , Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Rollenhagen A, Lübke JH, The mossy fiber bouton: the “common” or the “unique” synapse? Front. Synaptic Neurosci. 2, 2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicoll RA, Schmitz D, Synaptic plasticity at hippocampal mossy fibre synapses. Nat. Rev. Neurosci. 6, 863–876 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Wiera G, Mozrzymas JW, Extracellular proteolysis in structural and functional plasticity of mossy fiber synapses in hippocampus. Front. Cell Neurosci. 9, 427 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakashiba T et al. , Young dentate granule cells mediate pattern separation, whereas old granule cells facilitate pattern completion. Cell 149, 188–201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rolls ET, The mechanisms for pattern completion and pattern separation in the hippocampus. Front. Syst. Neurosci. 7, 74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang YY et al. , A genetic test of the effects of mutations in PKA on mossy fiber LTP and its relation to spatial and contextual learning. Cell 83, 1211–1222 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Cushman JD et al. , Juvenile neurogenesis makes essential contributions to adult brain structure and plays a sex-dependent role in fear memories. Front. Behav. Neurosci. 6, 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones BW et al. , Targeted deletion of AKAP7 in dentate granule cells impairs spatial discrimination. Elife 5, e20695 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tai CY, Kim SA, Schuman EM, Cadherins and synaptic plasticity. Curr. Opin. Cell Biol. 20, 567–75 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Brigidi GS, Bamji SX, Cadherin-catenin adhesion complexes at the synapse. Curr. Opin. Neurobiol. 21, 208–214 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Emond MR, Biswas S, Blevins CJ, Jontes JD, A complex of Protocadherin-19 and N-cadherin mediates a novel mechanism of cell adhesion. J. Cell Biol. 195, 1115–1121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fannon AM, Colman DR, A model for central synaptic junctional complex formation based on the differential adhesive specificities of the cadherins. Neuron 17, 423–434 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Söderberg O et al. , Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006) [DOI] [PubMed] [Google Scholar]
- 28.Biederer T, Scheiffele P, Mixed-culture assays for analyzing neuronal synapse formation. Nat. Protoc. 2, 670–676 (2007) [DOI] [PubMed] [Google Scholar]
- 29.Toth AB et al. , Synapse maturation by activity-dependent ectodomain shedding of SIRPα. Nat. Neurosci. 16, 1417–1425 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hazan RB, Phillips GR, Qiao RF, Norton L, Aaronson SA, Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J. Cell Biol. 148, 779–790 (2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li MY et al. , A critical role of presynaptic cadherin/catenin/p140Cap complexes in stabilizing spines and functional synapses in the neocortex. Neuron 94, 1155–1172 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Kadowaki M et al. , N-cadherin mediates cortical organization in the mouse brain. Dev. Biol. 304, 22–33 (2007) [DOI] [PubMed] [Google Scholar]
- 33.Shikanai M, Nakajima K, Kawauchi T, N-cadherin regulates radial glial fiber-dependent migration of cortical locomoting neurons. Commun. Integr. Biol. 24, 326–30 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoshina N et al. , Protocadherin 17 regulates presynaptic assembly in topographic corticobasal ganglia circuits. Neuron 78, 839–854 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Umemori H, Linhoff MW, Ornitz DM, Sanes JR, FGF22 and its close relatives are presynaptic organizing molecules in the mammalian brain. Cell 118, 257–270 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Ridler TW, Calvard S, Picture thresholding using an iterative selection method. IEEE Trans. Syst. Man Cybern. 8, 630–632 (1978). [Google Scholar]
- 37.Graf ER, Zhang X, Jin S, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 119, 1013–26 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science 307, 1324–1328 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Terauchi A et al. , Retrograde fibroblast growth factor 22 (FGF22) signaling regulates insulin-like growth factor 2 (IGF2) expression for activity-dependent synapse stabilization in the mammalian brain. Elife 5, e12151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith KR et al. , Cadherin-10 maintains excitatory/inhibitory ratio through interactions with synaptic proteins. J. Neurosci. 37, 11127–11139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.EOmari S et al. , Noncanonical autophagy at ER exit sites regulates procollagen turnover. Proc. Natl. Acad. Sci. U S A 115, E10099–E10108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki K et al. , A synthetic synaptic organizer protein restores glutamatergic neuronal circuits. Science 369, eabb4853 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips GR et al. , γ-Protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J. Neurosci. 23, 5096–5104 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green MV et al. , Automated live-cell imaging of synapses in rat and human neuronal cultures. Front. Cell. Neurosci. 13, 467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson-Venkatesh EM, Khan MN, Murphy GG, Sutton MA, Umemori H, Excitability governs neural development in a hippocampal region-specific manner. Development 142, 3879–3891 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorra KE, Harris KM, Overview on the structure, composition, function, development, and plasticity of hippocampal dendritic spines. Hippocampus 10, 501–511 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available in the main text or the supplementary materials. All materials and animals are available from the corresponding author upon request.


