Abstract
Purpose:
Predictive biomarkers for capecitabine benefit in triple-negative breast cancer (TNBC) have been recently proposed using samples from phase III clinical trials, including non-basal phenotype and biomarkers related to angiogenesis, stroma, and capecitabine activation genes. We aimed to validate these findings on the larger phase III GEICAM/CIBOMA clinical trial.
Experimental Design:
Tumor tissues from patients with TNBC randomized to standard (neo)adjuvant chemotherapy followed by capecitabine versus observation were analyzed using a 164-gene NanoString custom nCounter codeset measuring mRNA expression. A prespecified statistical plan sought to verify the predictive capacity of PAM50 non-basal molecular subtype and tested the hypotheses that breast tumors with increased expression of (meta)genes for cytotoxic cells, mast cells, endothelial cells, PDL2, and 38 individual genes benefit from adjuvant capecitabine for distant recurrence-free survival (DRFS; primary endpoint) and overall survival.
Results:
Of the 876 women enrolled in the GEICAM/CIBOMA trial, 658 (75%) were evaluable for analysis (337 with capecitabine and 321 without). Of these cases, 553 (84%) were profiled as PAM50 basal-like whereas 105 (16%) were PAM50 non-basal. Non-basal subtype was the most significant predictor for capecitabine benefit [HRcapecitabine, 0.19; 95% confidence interval (CI), 0.07–0.54; P < 0.001] when compared with PAM50 basal-like (HRcapecitabine, 0.9; 95% CI, 0.63–1.28; P = 0.55; Pinteraction<0.001, adjusted P value = 0.01). Analysis of biological processes related to PAM50 non-basal subtype revealed its enrichment for mast cells, extracellular matrix, angiogenesis, and features of mesenchymal stem-like TNBC subtype.
Conclusions:
In this prespecified correlative analysis of the GEICAM/CIBOMA trial, PAM50 non-basal status identified patients with early-stage TNBC most likely to benefit from capecitabine.
Translational Relevance.
Recent evidence has demonstrated a significant survival benefit when capecitabine is added to standard adjuvant chemotherapy in patients with triple-negative breast cancer (TNBC) compared with non-TNBC. However, TNBC identifies a heterogeneous group and predictive biomarkers that define the subset of TNBC deriving the most benefit from adjuvant capecitabine are necessary in clinical practice. In this hypothesis-testing study, we examine the capacity of candidate RNA biomarkers to predict benefit from extended adjuvant capecitabine using materials from the phase III CIBOMA/2004–01_GEICAM/2003–11 clinical trial. Following a prospective–retrospective prespecified study design per REMARK criteria, we report that PAM50 non-basal subtype defines the TNBC subset most likely to benefit from adjuvant capecitabine. These findings still require a confirmation in a second similar prospective–retrospective clinical trial series to reach level 1B evidence. In the context of other approved options in clinical practice, such as immunotherapy, our findings may guide the selection of patients with TNBC who may still benefit from adjuvant capecitabine and could be extended to inform study designs for patients with TNBC with residual disease after neoadjuvant therapy.
Introduction
Chemotherapy is an important component of the treatment of triple-negative breast cancer (TNBC), with anthracycline and taxane-based regimens as the most frequently administered agents in the adjuvant and neoadjuvant settings (1). Early-stage TNBC with tumors larger than 1–2 cm and/or with positive axillary lymph nodes are often treated with neoadjuvant chemotherapy (2), with those having residual disease after surgery often receiving additional capecitabine as an extended adjuvant therapy (3). However, TNBC identifies a heterogenous group (4–9) and the recent introduction of additional targeted therapy options, including immunotherapy and PARP inhibitors (10–12), highlights the need to identify biomarkers for the subset of patients who still achieve the greatest benefit from adjuvant cytotoxic chemotherapies, including capecitabine.
Capecitabine is an orally available nucleoside analogue, a prodrug that exerts its antitumoral effect following conversion to its active metabolite of 5-fluorouracil (5-FU) in tumor tissue (13, 14). The incorporation of capecitabine in the adjuvant setting of TNBC has been evaluated in several clinical trials for its capacity to improve breast cancer outcomes (3, 15–21). Most of these trials have tested the concurrent administration of capecitabine with standard chemotherapy and have reported inconsistent survival benefits with an increase in side effects (15–19). However, an important recent meta-analysis of individual patient data from 12 randomized clinical trials, evaluating the benefit of capecitabine in either neoadjuvant or adjuvant setting, demonstrated that there is a significant improvement in both disease-free survival (DFS) and overall survival (OS) when capecitabine is added to standard chemotherapy in patients with TNBC compared with non-TNBC (22).
The CIBOMA/2004–01_GEICAM/2003–11 phase III clinical trial (ref. 21; referred herein as GEICAM/CIBOMA) evaluated another approach: The sequential addition of capecitabine after standard chemotherapy. This trial randomized 876 patients with early-stage TNBC from Spain and Latin America treated with surgery and standard (neo)adjuvant chemotherapy, to receive either capecitabine or observation (21). Original results did not show a significant improvement in survival with capecitabine across all TNBC cases, but a pre-planned analysis of an IHC-defined stratum for the basal-like subtype of TNBC showed that although those expressing the basal biomarkers cytokeratin 5/6 (CK5/6) or EGFR (23) did not benefit from capecitabine, those classified as non-basal by IHC did have a highly significant benefit from capecitabine with an OS hazard ratio (HR) of 0.43 versus 1.23 (Pinteraction = 0.005) and a pronounced trend toward a better DFS with an HR of 0.53 versus 0.94 (Pinteraction = 0.06) for IHC non-basal versus basal (21).
Candidate-predictive molecular biomarkers for capecitabine benefit in TNBC have been recently identified analyzing samples from the Finland Capecitabine phase III clinical trial (FinXX; refs. 17, 18), using a NanoString-based technology to assess RNA expression of 800 genes representing 37 biologically important signatures on standard formalin-fixed, paraffin embedded (FFPE) excision specimens. Genes and metagenes related to angiogenesis, mast cells, cytotoxic cells, PDL2 and capecitabine activation were predictive for capecitabine benefit. These results are discovery-based and require validation (24).
Using a focused 164-gene NanoString custom nCounter codeset, applied to breast tumors obtained from patients in the phase III GEICAM/CIBOMA randomized clinical trial, we designed a formal prospective–retrospective hypothesis-testing analysis following Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) criteria. We sought to (1) verify the capacity of the PAM50 non-basal molecular subtype to predict capecitabine benefit, as previously found by IHC in the original GEICAM/CIBOMA analysis, and (2) test the hypotheses that breast tumors with increased expression of genes and metagene signatures for mast cells, endothelial cells, cytotoxic cells, PDL2 and 38 individual genes previously identified to be predictive for capecitabine benefit in TNBC in FinXX, would predict benefit from adjuvant capecitabine in GEICAM/CIBOMA.
Materials and Methods
Study population
GEICAM/CIBOMA (ClinicalTrials.gov identifier: NCT00130533) is a multicenter, open label, randomized phase III clinical trial that was conducted in 80 centers across 8 countries (Spain, Brazil, Chile, Colombia, Ecuador, Mexico, Peru, and Venezuela) between October 2006 and September 2011 (21). A total of 876 patients with breast cancer were recruited; these included women, at age ≥18 and ≤70 years old, with a histologically centrally confirmed invasive breast adenocarcinoma and TNBC status defined by IHC as negative for estrogen receptor (<1%), progesterone receptor (<1%), and Her2. Eligible patients were those with ipsilateral axillary lymph node involvement classified as pN1a, pN2a, or pN3a (excluding metastatic infraclavicular lymph nodes) or those without axillary node involvement (N0) with a primary tumor size ≥1 cm.
TNBC status confirmation and an IHC-defined preplanned stratum for basal versus non-basal were performed centrally by the GEICAM Spanish breast cancer group. IHC basal status was defined as TNBC with any staining for CK5/6+ or EGFR+; patients with TNBC negative for both these biomarkers were classified as IHC non-basal (23). Patients treated with surgery and standard (neo)adjuvant chemotherapy were randomly assigned to capecitabine versus observation. Patients assigned to adjuvant capecitabine received 8 cycles of oral capecitabine 1,000 mg/m2 twice daily on days 1 to 14 of each 21-day cycle. Full details on the GEICAM/CIBOMA study treatment protocols have been reported (21).
Study design and endpoints
The primary endpoint of the GEICAM/CIBOMA trial was DFS defined as time from random assignment to locoregional or distant recurrence, second primary malignancy, or death, whichever occurred first. OS was one of the secondary endpoints of the original analysis, defined as the time from the date of randomization to the date of death from any cause. Given that the DFS definition included second primary malignancies (non-breast) reported to be higher on the observation arm compared with capecitabine (3% vs. 1.3%) in GEICAM/CIBOMA patients and that the reduction in DFS events with capecitabine was mainly due to distant relapses among IHC non-basal cases, the current correlative study uses the primary endpoint of distant recurrence-free survival (DRFS) to avoid a potential reporting bias (21). DRFS is defined as time from randomization to distant recurrence of breast cancer (documented deaths due to breast cancer without distant recurrence were also considered as a distant recurrence event; local recurrence, regional recurrence, and contralateral second primary or secondary breast cancer in the ipsilateral breast are not considered distant recurrence events). OS was used as a secondary endpoint in the current correlative study. DFS (the primary endpoint of the GEICAM/CIBOMA trial) is included as a Supplementary Analysis. Exploratory analyses in the current study investigated the predictive capacity in relation to treatment effect for (i) categorical expression of biomarkers, and (ii) continuous expression of metagenes for CD8 T cells, exhausted CD8 cells and other single genes included in the codeset. Additional exploratory analyses assessed the prognostic capacity of continuous biomarker expression.
The current study follows a formal prospective–retrospective design per REMARK criteria (25), and per the guidelines for use of archived clinical trial specimens for predictive biomarker evaluation on clinical trials (26). An analysis plan was prespecified in writing and agreed to by the Vancouver group (who generated the RNA expression data but had no access to the clinical data) and the GEICAM statistical office (who executed the analysis) before performing any outcome analyses.
Ethics approval and consent
All patients signed a written informed consent to participate in the GEICAM/CIBOMA trial that allows the use of their tumor tissue for study-related research purposes. The subsequent use of patients’ specimens without disclosure of patient identifiers met waiver of informed consent policy criteria in accordance with the Declaration of Helsinki ethical guidelines. The study was approved by the Ethics Committee of the GEICAM Spanish Breast Cancer Group, the University of British Columbia, and the Clinical Research Ethics Board of BC Cancer (approval number: H17–01207).
Procedures
Archival FFPE tumor tissue samples were assembled from patients enrolled in the GEICAM/CIBOMA trial, who all received their allocated treatments. Hematoxylin and eosin slides for these FFPE samples were reviewed by pathologists (F. Rojo and D. Gao) who marked areas with viable invasive tumor cells. These areas guided macro-dissections of 10-μm unstained sections to obtain tissue for RNA extraction as previously published (27). Samples were analyzed on the nCounter NanoString system using a 164-gene custom codeset, comprising 18 housekeeping genes and 146 target genes that allow calculation of scores for metagene signatures for PAM50 subtypes, mast cells, endothelial cells, cytotoxic cells, CD8 T cells, exhausted CD8 cells, PDL2 and 38 individual genes postulated to be associated with capecitabine sensitivity (Supplementary Table S1). A minimum of 20 ng/μL was required as input following the manufacturer's protocol recommendations. 7 μL of RNA per sample was used for the hybridization reaction with the NanoString codeset performed overnight using the high-sensitivity protocol.
Samples were analyzed on the nCounter and data from the NanoString output files were analyzed using the nSolver software package and R statistical software. Gene expression analysis was performed following prespecified established algorithms developed by NanoString technologies consistent with methods previously used for the Breast Cancer 360 NanoString 770-gene panel (24, 28). These established algorithms were trained using datasets from The Cancer Genome Atlas (TCGA) and validated on immunotherapy datasets for different immune cell populations’ abundances as previously described (28). In brief, the training datasets originally included 9,986 samples from 32 tumor types in TCGA and algorithms were developed to include a small subset of candidate genes that had the most highly specific and stable expression for each cell type, and that showed similar and reproducible performance across the different TCGA datasets. These prespecified algorithms were then validated on independent immunotherapy datasets as predicting response to checkpoint inhibitor therapy in patients with metastatic melanoma (28).
The normalization in the custom codeset was carried out using both housekeeping genes and panel standards (consisting of a 16fM synthetic oligonucleotide pool corresponding to all panel gene targets) to control for run-to-run variation. Following the NanoString Gene Expression Data Analysis Guidelines, normalization was automatically generated in nSolver by calculating the geometric mean of housekeeping genes for each lane compared with the geometric mean across all sample lanes. Data were then log2 transformed, and the average of the log2 transformed counts was calculated for each gene across the 32 lanes of panel standard included in this study. The average values across the panel standard lanes for each gene were subtracted from the housekeeper normalized data, and this dataset was then used for gene expression analysis. PAM50-intrinsic subtype analysis was performed to identify the prototypical luminal A, luminal B, Her2-Enriched, basal-like, and normal-like breast cancer subtypes as published (29).
Statistical analysis
The prespecified statistical hypothesis tested whether breast tumors with increased continuous expression of genes and metagene signatures for mast cells, endothelial cells, cytotoxic cells, PDL2 and 38 individual genes previously identified to be predictive for capecitabine benefit in TNBC in FinXX would also predict benefit from adjuvant capecitabine in GEICAM/CIBOMA. On the basis of the significant previous findings observed on 229 cases classified as IHC non-basal in the original GEICAM/CIBOMA, we estimated that we required at least 226 cases to identify a significant predictive benefit for capecitabine with 95% power (type I error of 0.05). On the basis of the RNA immune biomarker prevalence and the survival rates observed in the FinXX trial for the genes/metagenes previously identified to be predictive for capecitabine benefit, we estimated that we required at least 363 total cases to identify a significant predictive benefit for capecitabine with 95% power (type I error of 0.05). Considering the large sample size of 876 cases accrued in the original GEICAM/CIBOMA cohort, we projected we did have an adequate number of cases in the translational cohort to test our study hypotheses with >95% power (type I error of 0.05).
The prespecified approved statistical analysis plan was independently executed by the GEICAM central office, testing the predictive capacity of gene and metagene expression by treatment arm. Univariate and multivariate survival analyses were performed for continuous expression scores for genes and metagenes using Cox regression models. The associations between HR and 95% confidence intervals (CI) with unit increase in each signature score were calculated in each treatment arm. Multivariate analysis was adjusted for age at randomization (continuous), menopausal status (postmenopausal vs. premenopausal), histological grade (G1 vs. G2 vs. G3 vs. GX), tumor size (T1 vs. T2 vs. T3), stage (I vs. II vs. III), breast surgery (lumpectomy vs. mastectomy), region (Spain vs. Latin America), nodal status (negative vs. 1–3 vs. ≥4), chemotherapy regimen (anthracyclines and taxanes vs. anthracyclines without taxanes), and phenotype by IHC (basal vs. non-basal). Interaction tests of heterogeneity that assess the associations of biomarker expression with clinical outcomes between treatment arms were used. Primary and secondary analyses for the prespecified hypotheses tested were adjusted for multiplicity using the Benjamini–Hochberg (BH) method. Exploratory analyses evaluating the predictive capacity of categorical expression of genes/metagenes used Kaplan–Meier curves to display survival outcomes according to gene or metagene expression status using the median as a cutoff point. Cox proportional hazard regression models were used to estimate HR and 95% CI by the gene/metagene-defined group and differences in survival outcome were compared using the log-rank test. Exploratory analyses evaluating the prognostic significance of continuous increases in the scores of genes/metagenes in relation to clinical outcomes were performed in the entire cohort, including both treatment arms. Application of genes/metagenes to basal versus non-basal groups was performed to explore whether findings could be related to PAM50 subtype. The χ2 test was used to assess associations between treatment arms and clinicopathological (categorical) variables. Differential gene expression analysis between PAM50 basal versus non-basal was performed on the log2 transformed data using the t test. All tests were 2-sided, at a significance level of 0.05 using the R statistical software.
Data availability
Clinical data for the patients included in this study are not publicly available per the GEICAM Spanish Breast Cancer Group policy to protect patient privacy. Any queries for data access used in this study should be directed to the corresponding author.
Results
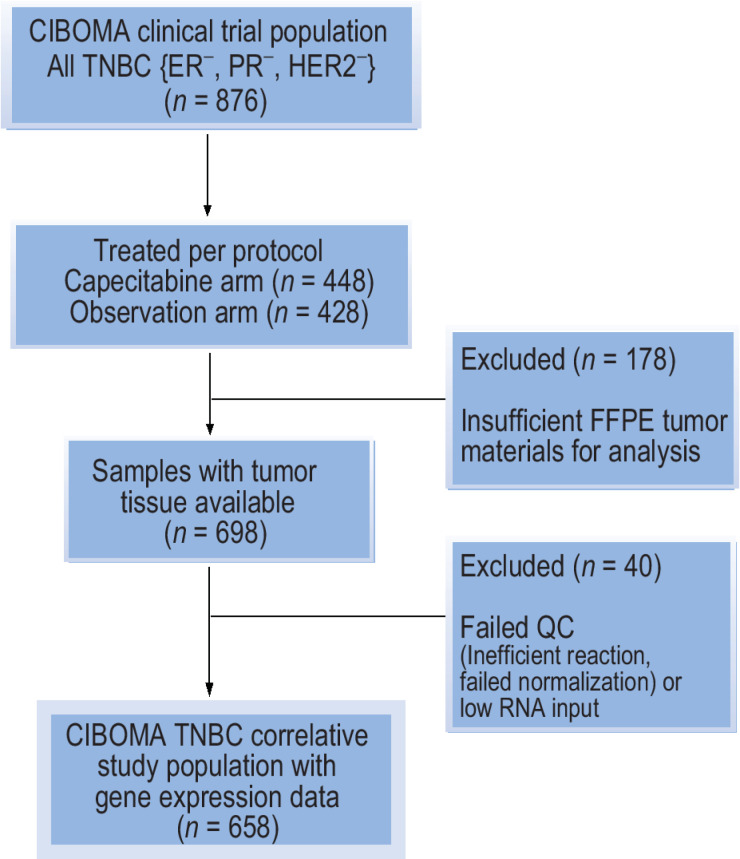
Of the 876 women enrolled in the GEICAM/CIBOMA trial, 698 (80%) had tumor tissue samples available and of these, 658 (75%) were evaluable for RNA analysis (Fig. 1). These cases defined the translational study cohort and were more available from patients treated with prior adjuvant than neoadjuvant therapy (Supplementary Table S2). Other baseline characteristics were similar between the translational cohort relative to the intention-to-treat population (Supplementary Table S2). Among the 658 evaluable cases, 337 patients were treated with capecitabine whereas 321 were assigned to the observation arm, and there were no imbalances in clinicopathological characteristics between the two study populations (Table 1).
Figure 1.
CONSORT flow diagram for cases included in the GEICAM/CIBOMA translational study cohort of triple-negative breast cancer. The analysis of the translational study cohort followed a prospective–retrospective design testing prespecified primary and secondary hypotheses using high-quality clinical trial materials with adherence to REMARK criteria and to the guidelines for use of archived clinical trial specimens for predictive biomarker evaluation on clinical trials.
Table 1.
Patient and baseline characteristics of GEICAM/CIBOMA translational study cohort according to treatment arm.
| Characteristic | Observation arm (n = 321) | Capecitabine arm (n = 337) | P |
|---|---|---|---|
| Age | |||
| ≤50 years | 169 (53%) | 170 (50%) | 0.63 |
| >50 years | 152 (47%) | 167 (50%) | |
| Region | |||
| Spain | 205 (64%) | 218 (65%) | 0.89 |
| Latin America | 116 (36%) | 119 (35%) | |
| Race | |||
| White | 242 (75%) | 249 (74%) | 0.48 |
| Hispanic | 66 (21%) | 70 (21%) | |
| African American | 4 (1%) | 10 (3%) | |
| Other | 9 (3%) | 8 (2%) | |
| Karnofsky performance status | |||
| 80 | 17 (5%) | 6 (2%) | 0.05 |
| 90 | 47 (15%) | 48 (14%) | |
| 100 | 257 (80%) | 283 (84%) | |
| Menopausal status at diagnosis | |||
| Postmenopausal | 108 (34%) | 102 (30%) | 0.40 |
| Premenopausal | 213 (66%) | 235 (70%) | |
| Histological type | |||
| Invasive ductal | 277 (86%) | 292 (86%) | 0.87 |
| Invasive lobular | 7 (2%) | 9 (3%) | |
| Other | 37 (12%) | 36 (11%) | |
| Histological grade | |||
| G1 | 9 (3%) | 7 (2%) | 0.71 |
| G2 | 51 (16%) | 57 (17%) | |
| G3 | 238 (74%) | 255 (76%) | |
| GX | 23 (7%) | 18 (5%) | |
| Tumor size | |||
| ≤2 cm | 118 (37%) | 130 (39%) | 0.75 |
| >2 and ≤5 cm | 178 (55%) | 183 (54%) | |
| >5 cm | 21 (7%) | 18 (5%) | |
| Unknown | 4 (1%) | 6 (2%) | |
| Phenotype by IHC | |||
| Triple-negative basal | 245 (76%) | 251 (74%) | 0.65 |
| Triple-negative non-basal | 76 (24%) | 86 (26%) | |
| Stage at diagnosis | |||
| I | 56 (17%) | 51 (15%) | 0.74 |
| II | 199 (62%) | 208 (62%) | |
| III | 65 (20%) | 72 (21%) | |
| Unknown | 1 (1%) | 6 (2%) | |
| Nodal status | |||
| Negative | 169 (52%) | 183 (54%) | 0.83 |
| 1–3 | 99 (31%) | 96 (29%) | |
| ≥4 | 52 (16%) | 54 (16%) | |
| Unknown | 1 (1%) | 4 (1%) | |
| Type of prior chemotherapy | |||
| Adjuvant | 286 (89%) | 278 (83%) | 0.04 |
| Neoadjuvant | 34 (10%) | 55 (16%) | |
| Unknown | 1 (1%) | 4 (1%) | |
| pCR in patients with neoadjuvant chemotherapy | |||
| No | 28 (9%) | 44 (13%) | 1 |
| Yes | 6 (2%) | 11 (3%) | |
| Unknown | 287 (89%) | 282 (84%) | |
| Chemotherapy regimen | |||
| Anthracyclines and taxanes | 217 (68%) | 225 (67%) | 0.88 |
| Anthracyclines without taxanes | 104 (32%) | 112 (33%) | |
| Breast surgery | |||
| Conservative | 183 (57%) | 189 (56%) | 1 |
| Mastectomy | 137 (42%) | 143 (42%) | |
| Unknown | 1 (1%) | 5 (2%) | |
| Axillary surgery | |||
| ALND ± SLNB | 229 (71%) | 257 (76%) | 0.18 |
| SLNB | 92 (29%) | 80 (24%) | |
| Radiation therapy | |||
| No | 256 (79%) | 262 (78%) | 0.75 |
| Yes | 64 (20%) | 71 (21%) | |
| Unknown | 1 (1%) | 4 (1%) | |
| Distant relapse events | |||
| No | 239 (74%) | 268 (80%) | 0.15 |
| Yes | 82 (26%) | 69 (20%) | |
| Recurrence events | |||
| No | 230 (72%) | 261 (77%) | 0.11 |
| Yes | 91 (28%) | 76 (23%) | |
| Death events | |||
| No | 267 (83%) | 288 (85%) | 0.49 |
| Yes | 54 (17%) | 49 (15%) | |
Abbreviations: IHC, immunohistochemistry; pCR, pathologic complete response; ALND, axillary lymph node dissection; SLNB, sentinel lymph node biopsy.
Classification of the study cohort into different PAM50 intrinsic subtypes revealed that 553 (84%) cases were PAM50 basal-like whereas 105 (16%) profiled as PAM50 non-basal (Supplementary Table S3). Overall, most of the cases were characterized by low expression of estrogen-related genes (e.g., ESR1, PGR, FOXA1, NAT1, MAPT) and high expression for genes associated with the basal-like subtype (e.g., KRT5, KRT17, MKI67, FOXC1, and PHGDH; Supplementary Fig. S1).
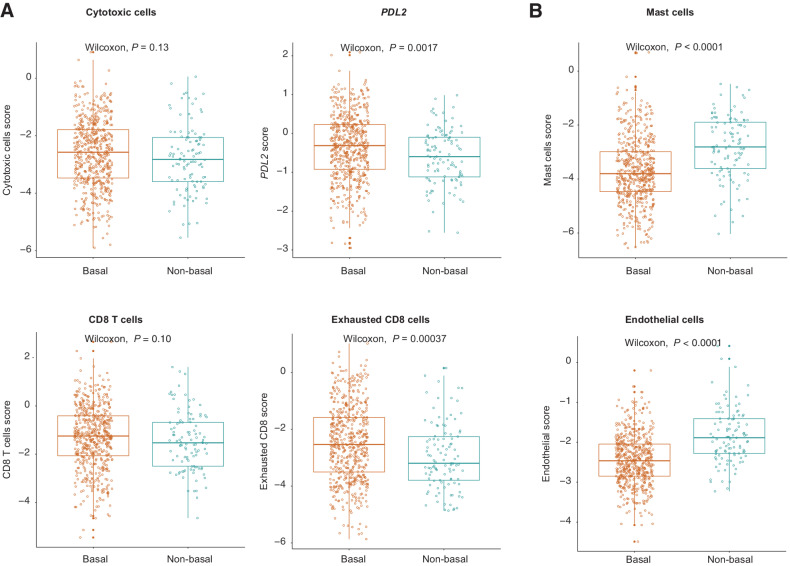
Expression levels of immune-related genes and metagenes revealed that the expression of exhausted CD8 cells and PDL2 were significantly higher among cases classified as PAM50 basal-like (Fig. 2A). In contrast, mast and endothelial cells metagenes were significantly higher in PAM50 non-basal cases (Fig. 2B).
Figure 2.
Expression levels of selected genes and metagene signatures against basal versus non-basal PAM50 status. A, Immune-related signatures. B, Mast cells and endothelial signatures. Boxplots show the median (center bar), the third (top edge), and first quartiles (bottom edge) of selected genes and metagenes. Each point represents one case. All statistical analyses were performed with the two-sided Wilcoxon rank-sum test. The gene contents for each metagene included in the NanoString custom nCounter codeset are displayed in Supplementary Table S1.
Predictive capacity of the non-basal molecular subtype of TNBC
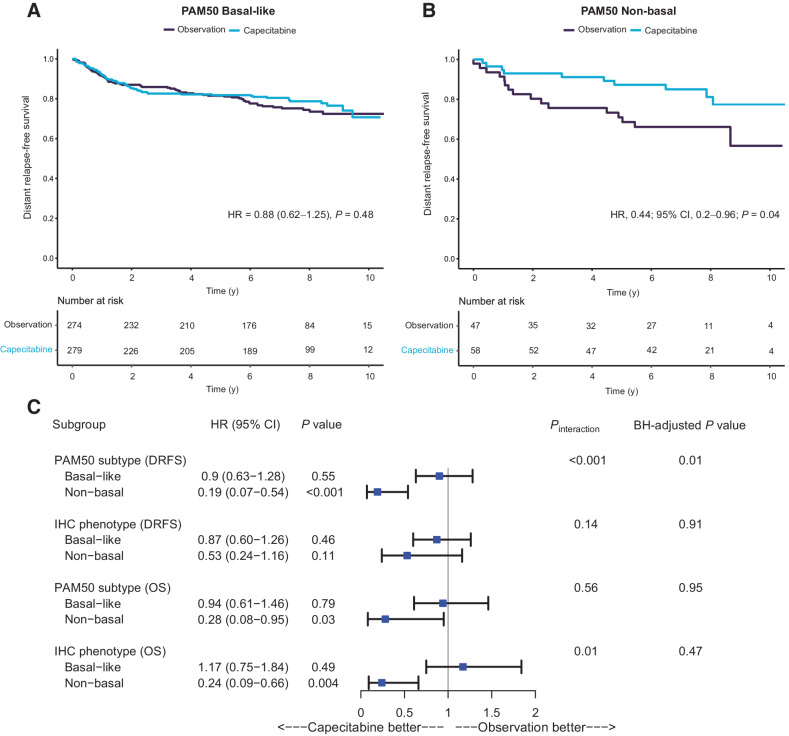
Using PAM50 subtyping, the TNBC non-basal subset predicted improved DRFS on univariate analysis (Fig. 3A and B), verifying the IHC-based result that was part of the original GEICAM/CIBOMA planned stratified analysis (21). In a multivariate DRFS analysis corrected for multiple testing, PAM50 non-basal subtype was the most significant predictor for capecitabine benefit (HRcapecitabine, 0.19; 95% CI, 0.07–0.54; P = 0<0.001) when compared with PAM50 basal-like (HRcapecitabine, 0.9; 95% CI, 0.63–1.28; P = 0.55; Pinteraction <0.001, adjusted BH P value = 0.01; Fig. 3C). The assignment of non-basal status by PAM50 showed a higher magnitude of capecitabine DRFS benefit when compared with the IHC definition (Fig. 3C). A secondary analysis for the OS endpoint suggested that cases classified as non-basal (particularly by IHC) benefitted significantly from capecitabine, although these results were not significant when adjusted for multiplicity (Fig. 3C). PAM50 non-basal subtype was found to be the most significant predictor for capecitabine benefit for the DFS endpoint (Supplementary Fig. S2).
Figure 3.
Survival analyses showing the primary endpoint of DRFS for patients randomly assigned to capecitabine or observation in the GEICAM/CIBOMA translational study cohort. A, Kaplan–Meier curves for basal patients as defined by RNA-based PAM50. B, Kaplan–Meier curves for non-basal patients as defined by RNA-based PAM50. C, Forest plot for the GEICAM/CIBOMA translational study cohort primary endpoint of DRFS and secondary endpoint of OS on the capecitabine arm versus observation arm. Hazard ratios, 95% confidence intervals, and P values are derived from Cox regression multivariate analysis adjusted for age, menopausal status, histological grade, tumor size, stage, breast surgery, region, nodal status, and chemotherapy regimen. Pinteraction indicates results of tests of heterogeneity for biomarker-defined subgroups in relation to treatment arm. Results were adjusted for multiple testing using the Benjamini–Hochberg method (BH). IHC basal phenotype is defined as triple-negative breast cancer with any staining for CK5/6+ or EGFR, whereas IHC non-basal phenotype is defined as triple-negative breast cancer with negative staining for both CK5/6 and EGFR. Abbreviations: DRFS, distant recurrence-free survival; OS, overall survival.
To determine the biological characteristics of cases classified as non-basal by PAM50, we performed a differential expression analysis to identify which genes in the codeset most significantly distinguished non-basal from basal TNBC (adjusted BH P value < 0.05; Supplementary Table S4). We focused on genes other than the ones used for PAM50 subtyping, to identify the biological processes characteristic of the PAM50 non-basal subtype. This analysis revealed the enrichment of PAM50 non-basal tumors for the capecitabine activation gene CES1, and for genes expressed by mast cells (TPSAB1 and CPA3), extracellular matrix and angiogenesis, while showing lower expression of genes involved in immune response (Supplementary Table S4).
RNA biomarkers for capecitabine benefit: hypothesis testing
In a prespecified multivariate analysis of the four signatures representing metagenes for cytotoxic, mast, endothelial cells, and the single gene PDL2 as continuous variables (Table 2), mast cell metagene was associated with significantly lower DRFS on the observation arm (HRobservation, 1.35; 95% CI, 1.12–1.62; P = 0.002, adjusted P value = 0.006). However, these findings were not significant by the interaction test (Pinteraction = 0.35; Table 2). When exploring whether findings could be related to PAM50 subtype, a trend toward a predictive association was observed among PAM50 non-basal tumors (HRobservation, 2.70; 95% CI, 0.99–7.35; P = 0.01, Pinteraction = 0.08; Table 2). A secondary analysis for the OS endpoint revealed that within PAM50 non-basal tumors, a continuous increase in mast cells expression was associated with poor survival on the observation arm when compared with capecitabine (HRobservation, 2.79; 95% CI, 1.27–6.12, P = 0.004; Table 2). However, results were not significant by the interaction test (Pinteraction = 0.22). Findings were similar for the DFS endpoint (Supplementary Table S5).
Table 2.
Multivariate survival analysis and interaction tests for the four (meta)genes included in the prespecified hypotheses testing their association with DRFS and OS.
| Multivariate analysis for the primary endpoint DRFS for selected biologically important genes and metagenes | |||||
|---|---|---|---|---|---|
| Gene/metagene | HRcapecitabine (95% CI) P value | Adjusted BH (capecitabine) | HRobservation (95% CI) P value | Adjusted BH (observation) | P interaction |
| Mast cells score | 1.23 (1.01–1.49) 0.04 | 0.15 | 1.35 (1.12–1.62) 0.002 | 0.006 | 0.35 |
| Endothelial score | 1.41 (0.94–2.11) 0.09 | 0.19 | 1.2 (0.85–1.7) 0.31 | 0.41 | 0.45 |
| PDL2 score | 0.94 (0.69–1.28) 0.69 | 0.87 | 1.35 (1.12–1.62) 0.16 | 0.65 | 0.80 |
| Cytotoxic cells score | 0.86 (0.7–1.07) 0.18 | 0.25 | 1.2 (0.85–1.7) 0.63 | 0.63 | 0.42 |
| Mast cells score in PAM50 non-basal | 0.7 (0.24–2.1) 0.53 | — | 2.7 (0.99–7.35) 0.01 | — | 0.08 |
| Mast cells score in PAM50 basal | 1.33 (1.08–1.64) 0.009 | — | 1.25 (1.01–1.54) 0.04 | — | 0.96 |
| Multivariate analysis for the secondary endpoint OS for selected biologically important genes and metagenes | |||||
| Mast cells score | 1.14 (0.9–1.43) 0.28 | 0.34 | 1.26 (1.01–1.57) 0.04 | 0.17 | 0.22 |
| Endothelial score | 1.26 (0.78–2.04) 0.34 | 0.34 | 1.13 (0.74–1.74) 0.57 | 0.69 | 0.70 |
| PDL2 score | 0.71 (0.49–1.03) 0.07 | 0.27 | 0.85 (0.59–1.21) 0.37 | 0.69 | 0.54 |
| Cytotoxic cells score | 0.83 (0.65–1.07) 0.16 | 0.32 | 0.95 (0.76–1.2) 0.69 | 0.69 | 0.51 |
| Mast cells score in PAM50 non-basala | 1.38 (0.75–2.54) 0.29 | — | 2.79 (1.27–6.12) 0.004 | — | 0.15 |
| Mast cells score in PAM50 basal | 0.98 (0.76–1.26) 0.87 | — | 1.1 (0.86–1.41) 0.44 | — | 0.50 |
Abbreviations: DRFS, distant recurrence-free survival; OS, overall survival.
aResults were not adjusted for multivariate analysis due to a very low number of events.
When assessing the predictive capacity of the continuous expression of the 38 individual genes previously linked to capecitabine benefit in the TNBC subset of the FinXX trial, after adjustment for multiplicity none of these genes were significantly associated with capecitabine DRFS benefit (Supplementary Table S6). Similar findings were observed for OS and DFS (Supplementary Tables S7 and S8).
RNA biomarkers for capecitabine benefit: exploratory
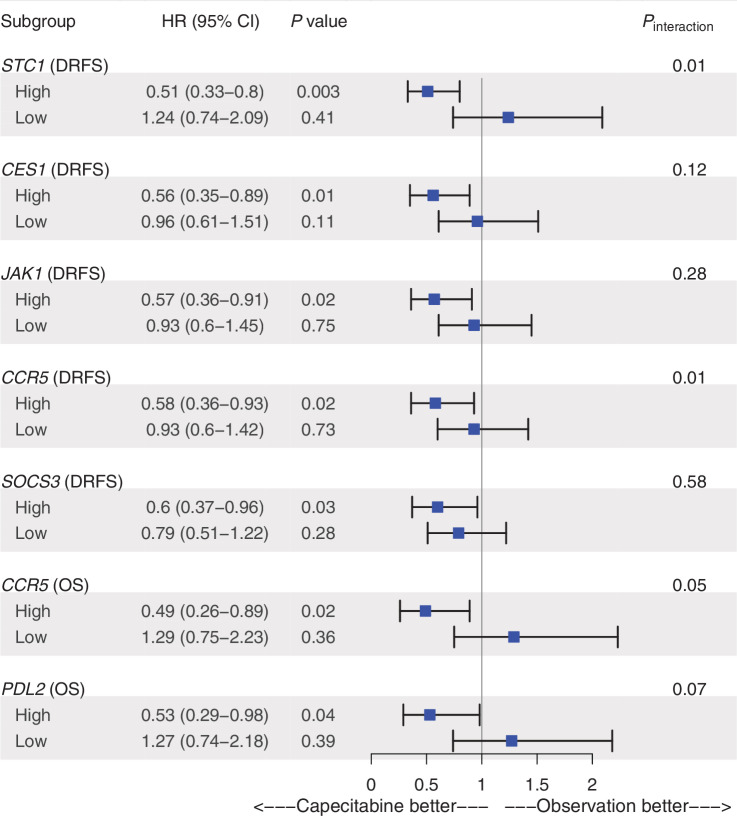
Analysis for the predictive capacity of selected genes and metagenes tested in the primary and secondary hypotheses was further performed using categorical classifications as “high” versus “low” based on the median cutoff point. Tumors above the median for genes involved in angiogenesis (STC1), capecitabine metabolism (CES1), JAK1/STAT3 signaling (JAK1, SOCS3), and immune response (CCR5) were found to be significantly associated with favorable DRFS rates on the capecitabine arm (HRcapecitabine ranged between 0.51 and 0.60; P = 0 < 0.05; Fig. 4). Among them, only high STC1 showed a significant interaction test for DRFS on capecitabine (HRcapecitabine = 0.51; 95% CI, 0.33–0.8; Pinteraction = 0.01; Fig. 4; Supplementary Fig. S3). These results were further observed when assessing the DFS endpoint (Supplementary Fig. S4) and were significant specifically within PAM50 non-basal tumors (Supplementary Table S9). High expression of genes involved in immune response (CCR5, PDL2) was found to be associated with improved OS on the capecitabine arm (Fig. 4), particularly within the PAM50 non-basal subtype (Supplementary Table S9). Results of the multivariate analysis and interaction tests for the remaining genes and metagenes are displayed in Supplementary Fig. S5.
Figure 4.
Forest plot of the categorical expression scores of selected genes and metagenes tested in the exploratory analysis that their high expression was found to be significantly associated with a higher survival on the capecitabine arm over observation. Expression status was derived from the median gene expression scores. Hazard ratios, 95% confidence intervals, and P values are derived from Cox regression analyses adjusted for age, menopausal status, histological grade, tumor size, stage, breast surgery, region, nodal status, chemotherapy regimen, and phenotype by IHC. Results for the remaining selected genes and metagenes are presented in Supplementary Figs. S3 and S5.
Assessment of the continuous expression of metagenes for CD8 T cells and exhausted CD8 cells did not reveal a significant association with capecitabine benefit (Supplementary Data S1).
Prognostic analysis of RNA biomarkers in TNBC
Finally, we performed an exploratory prognostic analysis of DRFS in association with continuous increases in the scores of the selected genes and metagenes tested in the primary and secondary hypotheses. Increases in the expression of the mast cell metagene and the angiogenesis biomarker ANGPT1 were found to be significantly associated with shorter DRFS, DFS, and OS (Supplementary Data S2). In contrast, increases in the expression of the immune biomarkers GZMH, NKG7 and KLRK1 were associated with longer DRFS, OS, and DFS (Supplementary Data S2). Increases in the continuous scores for PDL1 and IDO1 were associated with longer OS and DFS, whereas the endothelial metagene was associated with shorter OS (Supplementary Data S2). When exploring the prognostic capacity of the continuous expression of these RNA biomarkers within the PAM50 non-basal tumors, increases in the continuous scores for the mast cell and endothelial metagenes were found to be associated with shorter DRFS and OS (Supplementary Table S10). Increase in the expression of the angiogenesis biomarker ANGPT1 was associated with shorter DRFS, whereas the immune biomarker KLRK1 was associated with longer DRFS.
Discussion
The current study presents a prespecified correlative analysis using high-quality materials from the GEICAM/CIBOMA trial assessing the predictive capacity of intrinsic PAM50 subtype and RNA biomarkers for adjuvant capecitabine benefit in TNBC. Hypotheses generated from previous analyses of capecitabine trials were formally tested, and our results confirmed the independent predictive value of PAM50 non-basal status to identify patients with early-stage TNBC who gain the greatest survival benefit from capecitabine.
The predictive capacity of the non-basal subtype in TNBCs was previously observed on the GEICAM/CIBOMA trial using an IHC assay; however, the magnitude of capecitabine's benefit was even more strongly predicted using the multigene RNA definition. Non-basal subtype was the most significant predictor for capecitabine benefit when adjusted for multiple testing assessing metagenes and 38 individual genes previously identified to predict capecitabine benefit in TNBC in the FinXX trial. These results demonstrate the improved predictive information that can be obtained from more detailed, quantitative multigene expression subtyping assays that reflect the underlying biology more reliably than IHC results derived from the addition of two protein biomarkers (CK5/6 or EGFR). In addition, IHC methods are only semiquantitative, less reproducible, and influenced by several preanalytic and analytic factors that make them hard to standardize, factors that favor integrating RNA-based biomarkers in clinical practice. Although the secondary analysis for OS showed that IHC non-basal significantly predicted capecitabine benefit, results were not significant when adjusted for multiplicity testing. These findings highlight the additive value that can be achieved from PAM50 data over IHC to inform future clinical trial designs assessing the predictive capacity of non-basal RNA TNBC subtypes for capecitabine benefit.
Our finding that tumors displaying a non-basal molecular subtype benefit from adjuvant capecitabine is consistent with recent findings from the ECOG-ACRIN EA1131 trial (30) of early-stage patients with TNBC with residual disease after neoadjuvant standard chemotherapy. A pre-planned analysis in that trial according to PAM50 basal versus non-basal subtype showed that non-basal patients appeared to display superior invasive DFS when treated with capecitabine than with a platinum agent, whereas no significant differences between the two arms were observed for patients with the PAM50 basal subtype. However, biomarkers that characterize the non-basal molecular subtype were not proposed in this trial (30). In our study, the non-basal molecular subtype was found to represent a group of TNBC particularly enriched for mast cells, extracellular matrix, and angiogenesis (8, 9), suggesting that biomarkers involved in these pathways may contribute to the survival benefit obtained from the addition of capecitabine to standard adjuvant chemotherapy.
High expression of mast cell metagene could contribute to the survival benefit from capecitabine among non-basal because mast cells play a role as regulators of immune response and angiogenic processes (31). Mast cells have been shown to augment the activity of myeloid-derived suppressor cells (MDSC), which are known to inhibit T-cell activation through several mechanisms—including the secretion of immune suppressive enzymes and of cytokines such as indoleamine 2,3-dioxygenase (IDO), arginase, and IL10 (32); through expression of T-cell exhaustion biomarkers such as PDL1; and by inducing regulatory T-cell expansion (33). The active metabolite of capecitabine, 5-FU, is known to specifically deplete MDSCs, relieving their inhibitory effect on cytotoxic T cells (34) and thereby unleashing a stronger antitumor immune response when capecitabine is added to standard chemotherapy.
Mast cells have also been shown to play a role in inducing tumor angiogenesis through mechanisms, including the secretion of proangiogenic factors such as VEGF, bFGF, TGF-beta, TNF-alpha, and IL8 (31, 35, 36). In addition, mast cells release proteases (e.g., tryptase and chymase) and heparin-binding growth factors that promote the release of proangiogenic factors essential for neovascularization during tumor progression (31, 35, 37). These factors further modulate the tumor microenvironment, activating pathways involved in epithelial-to-mesenchymal transition (37–39). 5-FU–based drugs, including capecitabine, have been reported to induce thrombospondin-1 expression that has anti-angiogenic effects (40). Furthermore, the enzyme thymidine phosphorylase, responsible for activating capecitabine in tumor tissue, has been shown to facilitate the formation of a proangiogenic microenvironment (13). These findings support that the TNBC subset with a non-basal RNA profile enriched for angiogenesis, a feature further enhanced by mast cells, would benefit the most from the capecitabine's anti-angiogenic activity.
To date, capecitabine's anti-angiogenic effect has been best demonstrated in preclinical models using metronomic chemotherapy schedules (40–42), findings that informed the design of the recent SYSUCC-001 phase III clinical trial (20). This trial tested the addition of 1-year metronomic capecitabine therapy (650 mg/m2 twice daily) in patients with TNBC otherwise treated with standard chemotherapy and reported a significant improvement in 5-year DFS in the capecitabine arm compared with observation (20). The administration of metronomic capecitabine has less toxicity and appears to represent an effective and safer regimen, with improved quality of life, when compared with conventional capecitabine protocols (20). However, considering the inconsistent findings observed across recent trials evaluating various dosages of adjuvant capecitabine in TNBC (3, 15–21), the results of SYSUCC-001 highlight the importance of identifying the subset of patients deriving the most benefit from capecitabine irrespective of regimen.
Metronomic capecitabine is known to exert antitumor activity at least in part through selective inhibition of endothelial cell migration, induction of TSP-1, and downregulating proangiogenic factors such as VEGF, all contributing to angiogenic dormancy that prevents tumor neovascularization, proliferation, recurrence, and metastasis (41, 43). Although the antiangiogenic properties of capecitabine in its metronomic schedule could be applicable to lower risk tumors with low proliferation rates (43), this analysis suggests that mast cells and other angiogenesis-related genes could be biomarkers for sensitivity to the anti-angiogenic effects of capecitabine in its conventional higher dose that targets the rapidly proliferating breast cancers in the high-risk patients enrolled in the GEICAM/CIBOMA trial.
Mast cells have been proposed to play a critical role in inducing the “angiogenic switch,” an early hallmark in malignant transformation reported to occur before the emergence of an actively invasive tumor phenotype (35, 43). In line with our data, mast cell expression has been shown to be most predominant in non-basal subtypes of TNBC (44). In the context of the well-established heterogeneity of TNBC, including at least 4 main subtypes (basal-like immune activated, basal-like immune suppressed, mesenchymal, and luminal androgen receptor), mast cells have been reported to be most enriched in the mesenchymal subtype of TNBC (6, 7), and specifically to be most characteristic of the subset defined as mesenchymal-stem-like by a recent refined 5-subtype TNBC classification (8) that approximately accounts for 15% of TNBC overall close to the fraction of PAM50 non-basal cases (16%) identified in our study (6–8). Compared with mesenchymal TNBC, mesenchymal-stem-like TNBCs are more likely to display a non-basal PAM50 profile and to be highly enriched for angiogenesis signatures (8, 9), supporting that they might be the TNBC subtype most likely to benefit from chemotherapies possessing anti-angiogenic properties (such as capecitabine). Although anti-angiogenic therapies have previously failed to show a significant benefit in otherwise unselected populations of early-stage patients with TNBC (45), they might be a good option in mesenchymal-stem-like tumors that warrant further investigation. Thus, mast cells, along with the biomarkers involved in angiogenesis such as STC1 and JAK1/STAT3 signaling we found to be enriched in non-basal TNBCs, might be marking these mesenchymal-stem-like tumors. Importantly, mesenchymal subtypes have to date lacked a particular strategy for subtype-specific therapy and our findings suggest that capecitabine might be a good option for this enigmatic subgroup of TNBC.
Our findings might be of clinical value to guide therapeutic choices of adjuvant therapies in the setting of early-stage TNBC with residual disease. The CREATE-X trial, which recruited predominantly Japanese and Korean women, showed that capecitabine improved survival in early-stage women with residual TNBC after neoadjuvant therapy (3), and based on our findings we suggest that this trial was positive in part because a relatively high fraction of the population enrolled in CREATE-X is likely to be of molecular non-basal subtypes known to be relatively more prevalent among Asian populations, specifically with the luminal androgen receptor biology (46, 47), and/or because the eligibility criteria in CREATE-X were limited to patients with TNBC with residual disease after neoadjuvant therapy that are more likely to be of non-basal like subtypes with poor chemotherapy responses, especially those profiled as mesenchymal-stem-like, as these are known to less likely experience pathologic complete response (pCR) when compared with tumors classified as basal-like. In contrast, patients with basal-like immune–activated profile might be better candidates for the immunotherapy of pembrolizumab, based on recent results from KEYNOTE-522 (10). Our study could have a direct clinical implication as the role of adjuvant capecitabine in early-stage TNBC, in the context of the recent approval of the immunotherapy of pembrolizumab (KEYNOTE-522 trial) in the same adjuvant setting raises many questions regarding the best treatment approach for early-stage TNBC (48). Of note, the approval of pembrolizumab in KEYNOTE-522 was granted for all comers with TNBC as the pCR rates and event-free survival reported on the arm, including pembrolizumab, were consistent across both PDL1-positive and negative subgroups as defined by the SP142 assay (10, 11). However, the finding that pembrolizumab benefit was also observed among PDL1-negative tumors and the approval of pembrolizumab irrespective of TNBC subtype suggests that predicting which TNBC tumors will benefit most from immune checkpoint blockade will require a more reliable classifier than PDL1 to inform the selection of patients with TNBC who achieve the greatest benefit from immunotherapy versus other patients with TNBC who are unlikely to benefit from immunotherapy and could be candidates for the option of capecitabine approved in the same setting. In light of recent exploratory analyses in metastatic TNBC (49, 50), it might be of interest to test whether the RNA basal-like immune–activated profile defines the TNBC subset that benefits most from immunotherapy in KEYNOTE-522 whereas non-basal tumors, especially those with the mesenchymal RNA profile, may benefit more from capecitabine (48).
Our study could have another direct clinical implication in the context of the current recommendation for the use of adjuvant olaparib in germline BRCA-mutated TNBC. Although the original GEICAM/CIBOMA trial did not assess BRCA1 germline status, and none of the GEICAM/CIBOMA patients were considered for adjuvant olaparib as the trial was designed before its FDA approval in 2022, BRCA1 germline mutations are known to be found in approximately 15% of TNBC (51) and these women could be good candidates for adjuvant olaparib rather than capecitabine or pembrolizumab in the same adjuvant setting. Furthermore, whether the addition of olaparib in the adjuvant setting of TNBC should be considered in combination with the current use of pembrolizumab also requires further investigation (48).
Strengths of our study include use of high-quality clinical trial materials with adherence to REMARK guidelines (25) following a formal design testing prespecified primary and secondary hypotheses (26). The specific findings validating the predictive capacity of non-basal TNBC phenotype using the large number of TNBC cases enrolled in GEICAM/CIBOMA support a relatively high level of evidence for the clinical use of this biomarker to select patients most likely to benefit from adjuvant capecitabine in other clinical trials. However, these findings still require a confirmation in a second, similar prospective–retrospective clinical trial series to reach level 1B evidence (26). Moreover, our findings could be further extended to inform prospective study designs for patients with TNBC, specifically those classified as mesenchymal-stem-like, with residual disease after neoadjuvant therapy who may most benefit from adjuvant capecitabine.
Our study has some limitations. Although the classification of the tested metagenes was derived from an RNA quantitative assay, compared with IHC, RNA-based assays are not easily accessible in routine clinical settings and do not provide information about spatial context within a complex tumor microenvironment that includes carcinoma cells, immune subsets, and extracellular matrix compartments where the localization of mast cells could potentially have different predictive value. Thus, integrating our findings with methods able to detail in situ morphologic characteristics could further define the phenotype that approximates a relevant TNBC subgroup of otherwise poor prognosis patients who will benefit most from adjuvant capecitabine. Second, although the 164-gene custom RNA panel run in this study was prespecified to validate the predictive capacity of genes and metagenes found to be significant in the correlative analysis of FinXX TNBC cases using the 658 available FFPE patient specimens from the GEICAM/CIBOMA trial, there might still be other biomarkers that could predict adjuvant capecitabine benefit; for example, by more directly highlighting mesenchymal-stem-like TNBCs. In addition, the evaluation of TNBC molecular subtypes based on a more detailed multigene expression assay could provide a higher predictive value in comparison with the subset of genes and metagenes included in our codeset or than IHC biomarkers that are less reproducible due to analytic variability issues such as those described previously for the luminal androgen receptor subtype (52–54). Thus, future trials should incorporate molecular TNBC subtype classification as strata or inclusion criteria in their prospective design. Third, our study included 105 PAM50 non-basal patients in total with 26 events. Among these, 20 events were observed in the 0–5 years period, whereas only 6 occurred after >5 years group. Thus, the small number of late events identified decreased the power to observe significant findings when assessing the interaction between PAM50 non-basal subtype and capecitabine benefit beyond 5 years. In addition, our cohort was underpowered to assess any biologically important estrogen-related genes and their interaction with treatment benefit beyond 5 years. Thus, whether late recurrences beyond 5 years and interaction with capecitabine within the PAM50 non-basal subgroup could be related to luminal biology or the luminal androgen receptor TNBC subtype rather than the mesenchymal-stem-like subtype requires further investigation in future studies powered for analyses of late events. Finally, although several adjuvant capecitabine regimens have been evaluated in clinical trials, regimen-specific predictive biomarkers are still lacking. Thus, it would be valuable to apply our findings to other adjuvant capecitabine clinical trials, particularly those exploring metronomic capecitabine.
In conclusion, we present data from a prespecified correlative analysis of the phase III GEICAM/CIBOMA clinical trial, reporting that by RNA analysis non-basal subtype identifies those early-stage patients with TNBC who are most likely to benefit from adjuvant capecitabine.
Supplementary Material
Supplementary Table S1 shows the gene list for the NanoString custom nCounter codeset included in the study. Supplementary Table S2 shows patient characteristics in the GEICAM/CIBOMA translational study cohort versus the original trial. Supplementary Table S3 shows the distribution of PAM50 intrinsic subtypes in the GEICAM/CIBOMA translational study cohort. Supplementary Table S4 shows the differential gene expression analysis performed on the 164 genes included in the custom codeset comparing PAM50 non-basal vs. basal-like subtype and their biological processes. Supplementary Table S5 shows multivariate survival analysis and interaction tests for the four genes and metagenes included in the prespecified hypotheses testing their association with DFS. Supplementary Table S6 shows multivariate survival analysis and interaction tests for the continuous expression of the 38 individual genes included in the prespecified hypotheses testing their association with DRFS. Supplementary Table S7 shows multivariate survival analysis and interaction tests for the continuous expression of the 38 individual genes included in the prespecified hypotheses testing their association with OS. Supplementary Table S8 shows multivariate survival analysis and interaction tests for the continuous expression of the 38 individual genes included in the prespecified hypotheses testing their association with DFS. Supplementary Table S9 shows multivariate survival analysis and interaction tests restricted to the PAM50 non-basal subgroup. Supplementary Table S10 shows Exploratory analysis for the prognostic capacity of the continuous expression of genes and metagenes restricted to the PAM50 non-basal subgroup.
Supplementary Data S1 shows exploratory analysis for the predictive capacity of the continuous expression of additional genes and metagenes included in the codeset. Supplementary Data S2 shows exploratory analysis for the prognostic capacity of the continuous expression of the selected genes and metagenes
Supplementary Figure S1 shows the expression of the 50 genes in the PAM50 signature, obtained from the 164-gene codeset.
Supplementary Figure S2 shows a forest plot comparing basal vs. non-basal subtype as defined by PAM50 or by IHC, and their associations with disease free survival on capecitabine vs. observation arm.
Supplementary Figure S3 shows a forest plot of the exploratory categorical analysis of selected (meta)gene expression and their association with distant recurrence free survival on the capecitabine arm vs. observation.
Supplementary Figure S4 shows a forest plot of the exploratory categorical analysis of selected (meta)gene expression and their association with disease free survival on the capecitabine arm vs. observation.
Supplementary Figure S5 shows a forest plot of the exploratory categorical analysis of selected (meta)gene expression and their association with overall survival on the capecitabine arm vs. observation.
Acknowledgments
We thank the women in Spain and Latin America who contributed their specimens to the GEICAM/CIBOMA clinical trial. We thank the following for their contributions: Drs. R. Caballero and D. Fernández García for their input in study conception and design, data interpretation, article editing and for overall project coordination and management; Drs. P. López Serra and N. Martín for assistance in sample preparation and management. Research reported in this publication was supported by funds from the Canadian Cancer Society (grant number 705463; to T.O. Nielsen) and from GEICAM (Spanish Breast Cancer Group). K. Asleh was supported by the Vanier Canada Graduate Scholarship—Canadian Institutes of Health Research. Publication costs were supported by GEICAM (Spanish Breast Cancer Group).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 297
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
A. Lluch reports personal fees from Roche, AstraZeneca, Novartis, Pfizer, and Eisai during the conduct of the study. C. Barrios reports personal fees from Boehringer-Ingelheim, GSK, Novartis, Pfizer, Roche/Genentech, Eisai, Bayer, MSD, AstraZeneca, Zodiac, Lilly, Sanofi, and Daiichi during the conduct of the study as well as grants from AbbVie, Nektar, Pfizer, Polyphor, Amgen, Daiichi Sankyo, Sanofi, Exelixis, Regeneron, Novartis, Henlius, Shanghai, GSK, Janssen, OBI Pharma, Lilly, Seagen, Checkpoint Therapeutics, Roche, BMS, MSD, Merck Serono, AstraZeneca, Novocure, Aveo Oncology, Takeda, TRIO, PharmaMar, Celgene, Myovant, PPD, Syneos Health, Docs, Labcorp, ICON, IQVIA, Parexel, Nuvisan, PSI, and Medpace outside the submitted work. J. Bines reports personal fees from Roche, AstraZeneca, AbbVie, Daiichi Sankyo, GenomicHealth, Libbs, Lilly, Pfizer, and Sanofi outside the submitted work. Á. Guerrero-Zotano reports grants, personal fees, and nonfinancial support from Pfizer; personal fees and nonfinancial support from Novartis and AstraZeneca; and personal fees from Exact Science, Lilly, and Pierre Fabre outside the submitted work. J.Á. García-Sáenz reports personal fees from Lilly, Novartis, Roche, Daiichi Sankyo, and Seagen; personal fees and nonfinancial support from AstraZeneca; and grants from Tecnofarma, Asofarma, and Gilead outside the submitted work. J.M. Cejalvo reports other support from Pfizer and Novartis outside the submitted work. F. Ayala reports personal fees from Pfizer, Seagen, Roche, Eisai, Pierre Fabre, AstraZeneca, Novartis, Sanofi, Lilly, MSD, and Daichi Sankyo and grants from BMS, Roche, and Celgene outside the submitted work. F. Rojo reports personal fees from Roche, AstraZeneca, BMS, MSD, Novartis, Lilly, GSK, Pfizer, and Pierre Fabre outside the submitted work. T.O. Nielsen reports grants from Canadian Cancer Society during the conduct of the study as well as personal fees from Bioclassifier outside the submitted work; in addition, T.O. Nielsen reports a patent for Prosigna issued, licensed, and with royalties paid from Veracyte. M. Martin reports grants, personal fees, and nonfinancial support from Roche and Lilly; grants from Puma; personal fees from Pfizer, Novartis, AstraZeneca, and Seagen; and personal fees and nonfinancial support from Daiichi-Sankyo outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
K. Asleh: Conceptualization, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. A. Lluch: Investigation, methodology, writing–review and editing. A. Goytain: Investigation, methodology, writing–review and editing. C. Barrios: Investigation, methodology, writing–review and editing. X.Q. Wang: Methodology, writing–review and editing. L. Torrecillas: Resources, methodology, writing–review and editing. D. Gao: Formal analysis, methodology, writing–review and editing. M. Ruiz-Borrego: Resources, methodology, writing–review and editing. S. Leung: Investigation, methodology, writing–review and editing. J. Bines: Resources, methodology, writing–review and editing. Á. Guerrero-Zotano: Resources, methodology, writing–review and editing. J.Á García-Sáenz: Resources, methodology, writing–review and editing. J.M. Cejalvo: Resources, methodology, writing–review and editing. J. Herranz: Formal analysis, investigation, writing–review and editing. R. Torres: Resources, methodology, writing–review and editing. J. de la Haba-Rodriguez: Resources, methodology, writing–review and editing. F. Ayala: Resources, methodology, writing–review and editing. H. Gómez: Resources, methodology, writing–review and editing. F. Rojo: Investigation, methodology, writing–review and editing. T.O. Nielsen: Conceptualization, supervision, funding acquisition, writing–review and editing. M. Martin: Conceptualization, supervision, funding acquisition, writing–review and editing.
References
- 1. Waks AG, Winer EP. Breast cancer treatment: a review. JAMA 2019;321:288–300. [DOI] [PubMed] [Google Scholar]
- 2. Cardoso F, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rubio IT, et al. Early breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up†. Ann Oncol 2019;30:1194–220. [DOI] [PubMed] [Google Scholar]
- 3. Masuda N, Lee SJ, Ohtani S, Im YH, Lee ES, Yokota I, et al. Adjuvant capecitabine for breast cancer after preoperative chemotherapy. N Engl J Med 2017;376:2147–59. [DOI] [PubMed] [Google Scholar]
- 4. Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121:2750–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist 2010;15:39–48. [DOI] [PubMed] [Google Scholar]
- 6. Jiang YZ, Ma D, Suo C, Shi J, Xue M, Hu X, et al. Genomic and transcriptomic landscape of triple-negative breast cancers: subtypes and treatment strategies. Cancer Cell 2019;35:428–40. [DOI] [PubMed] [Google Scholar]
- 7. Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res 2015;21:1688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bareche Y, Venet D, Ignatiadis M, Aftimos P, Piccart M, Rothe F, et al. Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann Oncol 2018;29:895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bareche Y, Buisseret L, Gruosso T, Girard E, Venet D, Dupont F, et al. Unraveling triple-negative breast cancer tumor microenvironment heterogeneity: towards an optimized treatment approach. J Natl Cancer Inst 2020;112:708–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmid P, Cortes J, Pusztai L, McArthur H, Kümmel S, Bergh J, et al. Pembrolizumab for early triple-negative breast cancer. N Engl J Med 2020;382:810–21. [DOI] [PubMed] [Google Scholar]
- 11. Schmid P, Cortes J, Dent R, Pusztai L, McArthur H, Kümmel S, et al. Event-free survival with pembrolizumab in early triple-negative breast cancer. N Engl J Med 2022;386:556–67. [DOI] [PubMed] [Google Scholar]
- 12. Tutt ANJ, Garber JE, Kaufman B, Viale G, Fumagalli D, Rastogi P, et al. Adjuvant olaparib for patients with BRCA1- or BRCA2-mutated breast cancer. N Engl J Med 2021;384:2394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Toi M, Atiqur Rahman M, Bando H, Chow LW. Thymidine phosphorylase (platelet-derived endothelial-cell growth factor) in cancer biology and treatment. Lancet Oncol 2005;6:158–66. [DOI] [PubMed] [Google Scholar]
- 14. Blum JL. The role of capecitabine, an oral, enzymatically activated fluoropyrimidine, in the treatment of metastatic breast cancer. Oncologist 2001;6:56–64. [DOI] [PubMed] [Google Scholar]
- 15. O'Shaughnessy J, Koeppen H, Xiao Y, Lackner MR, Paul D, Stokoe C, et al. Patients with slowly proliferative early breast cancer have low five-year recurrence rates in a phase III adjuvant trial of capecitabine. Clin Cancer Res 2015;21:4305–11. [DOI] [PubMed] [Google Scholar]
- 16. Martín M, Ruiz Simón A, Ruiz Borrego M, Ribelles N, Rodríguez-Lescure Á, Muñoz-Mateu M, et al. Epirubicin plus cyclophosphamide followed by docetaxel versus epirubicin plus docetaxel followed by capecitabine as adjuvant therapy for node-positive early breast cancer: results from the GEICAM/2003–10 study. J Clin Oncol 2015;33:3788–95. [DOI] [PubMed] [Google Scholar]
- 17. Joensuu H, Kellokumpu-Lehtinen PL, Huovinen R, Jukkola-Vuorinen A, Tanner M, Kokko R, et al. Adjuvant capecitabine, docetaxel, cyclophosphamide, and epirubicin for early breast cancer: final analysis of the randomized FinXX trial. J Clin Oncol 2012;30:11–8. [DOI] [PubMed] [Google Scholar]
- 18. Joensuu H, Kellokumpu-Lehtinen PL, Huovinen R, Jukkola-Vuorinen A, Tanner M, Kokko R, et al. Adjuvant capecitabine in combination with docetaxel, epirubicin, and cyclophosphamide for early breast cancer: the randomized clinical FinXX trial. JAMA Oncol 2017;3:793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li J, Yu K, Pang D, Wang C, Jiang J, Yang S, et al. Adjuvant capecitabine with docetaxel and cyclophosphamide plus epirubicin for triple-negative breast cancer (CBCSG010): an open-label, randomized, multicenter, phase III trial. J Clin Oncol 2020;38:1774–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X, Wang SS, Huang H, Cai L, Zhao L, Peng RJ, et al. Effect of capecitabine maintenance therapy using lower dosage and higher frequency vs. observation on disease-free survival among patients with early-stage triple-negative breast cancer who had received standard treatment: the SYSUCC-001 randomized clinical trial. JAMA 2021;325:50–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lluch A, Barrios CH, Torrecillas L, Ruiz-Borrego M, Bines J, Segalla J, et al. Phase III trial of adjuvant capecitabine after standard neo-/adjuvant chemotherapy in patients with early triple-negative breast cancer (GEICAM/2003–11_CIBOMA/2004–01). J Clin Oncol 2020;38:203–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van Mackelenbergh MT, Seither F, Möbus V, O'Shaughnessy J, Martin M, Joensuu H, et al. Effects of capecitabine as part of neo-/adjuvant chemotherapy—a meta-analysis of individual breast cancer patient data from 13 randomised trials, including 15,993 patients. Eur J Cancer 2022;166:185–201. [DOI] [PubMed] [Google Scholar]
- 23. Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 2004;10:5367–74. [DOI] [PubMed] [Google Scholar]
- 24. Asleh K, Brauer HA, Sullivan A, Lauttia S, Lindman H, Nielsen TO, et al. Predictive biomarkers for adjuvant capecitabine benefit in early-stage triple-negative breast cancer in the FinXX clinical trial. Clin Cancer Res 2020;26:2603–14. [DOI] [PubMed] [Google Scholar]
- 25. Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. PLoS Med 2012;9:e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst 2009;101:1446–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nielsen T, Wallden B, Schaper C, Ferree S, Liu S, Gao D, et al. Analytical validation of the PAM50-based prosigna breast cancer prognostic gene signature assay and nCounter analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer 2014;14:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Danaher P, Warren S, Dennis L, D'Amico L, White A, Disis ML, et al. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer 2017;5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Picornell AC, Echavarria I, Alvarez E, López-Tarruella S, Jerez Y, Hoadley K, et al. Breast cancer PAM50 signature: correlation and concordance between RNA-Seq and digital multiplexed gene expression technologies in a triple-negative breast cancer series. BMC Genomics 2019;20:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mayer IA, Zhao F, Arteaga CL, Symmans WF, Park BH, Burnette BL, et al. Randomized phase III postoperative trial of platinum-based chemotherapy versus capecitabine in patients with residual triple-negative breast cancer following neoadjuvant chemotherapy: ECOG-ACRIN EA1131. J Clin Oncol 2021;39:2539–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krystel-Whittemore M, Dileepan KN, Wood JG. Mast cell: a multi-functional master cell. Front Immunol 2015;6:620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu J, Du W, Yan F, Wang Y, Li H, Cao S, et al. Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J Immunol 2013;190:3783–97. [DOI] [PubMed] [Google Scholar]
- 33. Weber R, Fleming V, Hu X, Nagibin V, Groth C, Altevogt P, et al. Myeloid-derived suppressor cells hinder the anticancer activity of immune checkpoint inhibitors. Front Immunol 2018;9:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 2009;9:162–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Norrby K. Mast cells and angiogenesis. APMIS 2002;110:355–71. [DOI] [PubMed] [Google Scholar]
- 36. Cimpean AM, Tamma R, Ruggieri S, Nico B, Toma A, Ribatti D. Mast cells in breast cancer angiogenesis. Crit Rev Oncol Hematol 2017;115:23–6. [DOI] [PubMed] [Google Scholar]
- 37. de Souza DA, Toso VD, Campos MR, Lara VS, Oliver C, Jamur MC. Expression of mast cell proteases correlates with mast cell maturation and angiogenesis during tumor progression. PLoS ONE 2012;7:e40790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xiang M, Gu Y, Zhao F, Lu H, Chen S, Yin L. Mast cell tryptase promotes breast cancer migration and invasion. Oncol Rep 2010;23:615–9. [DOI] [PubMed] [Google Scholar]
- 39. Tekpli X, Lien T, Røssevold AH, Nebdal D, Borgen E, Ohnstad HO, et al. An independent poor-prognosis subtype of breast cancer defined by a distinct tumor immune microenvironment. Nat Commun 2019;10:5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ooyama A, Oka T, Zhao HY, Yamamoto M, Akiyama S, Fukushima M. Anti-angiogenic effect of 5-Fluorouracil-based drugs against human colon cancer xenografts. Cancer Lett 2008;267:26–36. [DOI] [PubMed] [Google Scholar]
- 41. Pasquier E, Kavallaris M, André N. Metronomic chemotherapy: new rationale for new directions. Nat Rev Clin Oncol 2010;7:455–65. [DOI] [PubMed] [Google Scholar]
- 42. Kerbel RS, Kamen BA. The anti-angiogenic basis of metronomic chemotherapy. Nat Rev Cancer 2004;4:423–36. [DOI] [PubMed] [Google Scholar]
- 43. Natale G, Bocci G. Does metronomic chemotherapy induce tumor angiogenic dormancy? A review of available preclinical and clinical data. Cancer Lett 2018;432:28–37. [DOI] [PubMed] [Google Scholar]
- 44. Majorini MT, Cancila V, Rigoni A, Botti L, Dugo M, Triulzi T, et al. Infiltrating mast cell-mediated stimulation of estrogen receptor activity in breast cancer cells promotes the luminal phenotype. Cancer Res 2020;80:2311–24. [DOI] [PubMed] [Google Scholar]
- 45. Cameron D, Brown J, Dent R, Jackisch C, Mackey J, Pivot X, et al. Adjuvant bevacizumab-containing therapy in triple-negative breast cancer (BEATRICE): primary results of a randomised, phase 3 trial. Lancet Oncol 2013;14:933–42. [DOI] [PubMed] [Google Scholar]
- 46. Sweeney C, Bernard PS, Factor RE, Kwan ML, Habel LA, Quesenberry CP, et al. Intrinsic subtypes from PAM50 gene expression assay in a population-based breast cancer cohort: differences by age, race, and tumor characteristics. Cancer Epidemiol Biomarkers Prev 2014;23:714–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pan JW, Zabidi MMA, Ng PS, Meng MY, Hasan SN, Sandey B, et al. The molecular landscape of Asian breast cancers reveals clinically relevant population-specific differences. Nat Commun 2020;11:6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Korde LA, Somerfield MR, Hershman DL., Neoadjuvant chemotherapy EdT, and targeted therapy for breast cancer guideline expert panel. Use of immune checkpoint inhibitor pembrolizumab in the treatment of high-risk, early-stage triple-negative breast cancer: ASCO guideline rapid recommendation update. J Clin Oncol 2022;40:1696–98. [DOI] [PubMed] [Google Scholar]
- 49. Emens LA, Goldstein LD, Schmid P, Rugo HS, Adams S, Barrios CH, et al. The tumor microenvironment (TME) and atezolizumab + nab-paclitaxel (A+nP) activity in metastatic triple-negative breast cancer (mTNBC): IMpassion130. J Clin Oncol 2021;39:1006. [Google Scholar]
- 50. André F, Deurloo R, Qamra A, Cameron D, Gligorov J, Schneeweiss A, et al. Activity of atezolizumab (atezo) plus paclitaxel (pac) in metastatic triple-negative breast cancer (mTNBC) according to Burstein molecular subtype: analysis of the IMpassion131 trial. Cancer Res 2022;82:PD10-05. [Google Scholar]
- 51. Asleh K, Riaz N, Nielsen TO. Heterogeneity of triple-negative breast cancer: current advances in subtyping and treatment implications. J Exp Clin Cancer Res 2022;41:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rampurwala M, Wisinski KB, O'Regan R. Role of the androgen receptor in triple-negative breast cancer. Clin Adv Hematol Oncol 2016;14:186–93. [PMC free article] [PubMed] [Google Scholar]
- 53. Michmerhuizen AR, Spratt DE, Pierce LJ, Speers CW. ARe we there yet? Understanding androgen receptor signaling in breast cancer. NPJ Breast Cancer 2020;6:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Salvi S, Bonafè M, Bravaccini S. Androgen receptor in breast cancer: a wolf in sheep's clothing? A lesson from prostate cancer. Semin Cancer Biol 2020;60:132–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1 shows the gene list for the NanoString custom nCounter codeset included in the study. Supplementary Table S2 shows patient characteristics in the GEICAM/CIBOMA translational study cohort versus the original trial. Supplementary Table S3 shows the distribution of PAM50 intrinsic subtypes in the GEICAM/CIBOMA translational study cohort. Supplementary Table S4 shows the differential gene expression analysis performed on the 164 genes included in the custom codeset comparing PAM50 non-basal vs. basal-like subtype and their biological processes. Supplementary Table S5 shows multivariate survival analysis and interaction tests for the four genes and metagenes included in the prespecified hypotheses testing their association with DFS. Supplementary Table S6 shows multivariate survival analysis and interaction tests for the continuous expression of the 38 individual genes included in the prespecified hypotheses testing their association with DRFS. Supplementary Table S7 shows multivariate survival analysis and interaction tests for the continuous expression of the 38 individual genes included in the prespecified hypotheses testing their association with OS. Supplementary Table S8 shows multivariate survival analysis and interaction tests for the continuous expression of the 38 individual genes included in the prespecified hypotheses testing their association with DFS. Supplementary Table S9 shows multivariate survival analysis and interaction tests restricted to the PAM50 non-basal subgroup. Supplementary Table S10 shows Exploratory analysis for the prognostic capacity of the continuous expression of genes and metagenes restricted to the PAM50 non-basal subgroup.
Supplementary Data S1 shows exploratory analysis for the predictive capacity of the continuous expression of additional genes and metagenes included in the codeset. Supplementary Data S2 shows exploratory analysis for the prognostic capacity of the continuous expression of the selected genes and metagenes
Supplementary Figure S1 shows the expression of the 50 genes in the PAM50 signature, obtained from the 164-gene codeset.
Supplementary Figure S2 shows a forest plot comparing basal vs. non-basal subtype as defined by PAM50 or by IHC, and their associations with disease free survival on capecitabine vs. observation arm.
Supplementary Figure S3 shows a forest plot of the exploratory categorical analysis of selected (meta)gene expression and their association with distant recurrence free survival on the capecitabine arm vs. observation.
Supplementary Figure S4 shows a forest plot of the exploratory categorical analysis of selected (meta)gene expression and their association with disease free survival on the capecitabine arm vs. observation.
Supplementary Figure S5 shows a forest plot of the exploratory categorical analysis of selected (meta)gene expression and their association with overall survival on the capecitabine arm vs. observation.
Data Availability Statement
Clinical data for the patients included in this study are not publicly available per the GEICAM Spanish Breast Cancer Group policy to protect patient privacy. Any queries for data access used in this study should be directed to the corresponding author.