Abstract
The purpose of this study was to examine the genetic structure of the typical commensal Streptococcus mitis biovar 1 in its natural habitat in the human oral cavity and pharynx and to investigate the role that selected microbial properties and host, spatial, and temporal factors play in determining the structure of the bacterial population. Consecutive samples were collected from buccal and pharyngeal mucosal surfaces of two infants, their four parents, and two elderly individuals over a period of approximately 1 year. A total of 751 isolates identified as S. mitis biovar 1 were typed by restriction endonuclease analysis (REA) and representative clones were typed by multilocus enzyme electrophoresis (MLEE). The genetic diversity of the S. mitis biovar 1 isolates collected from single infant hosts over a period of 9 to 10 months was found to be between 0.69 and 0.76, which is considerably higher than that previously observed for intestinal populations of Escherichia coli. The study provides evidence of the existence of both transient and persistent clones in adult individuals. In the two infants, however, none of 42 demonstrated clones were detected on more than a single occasion. Statistical calculations showed that the ability to persist was not distributed at random in the S. mitis biovar 1 population. However, neither immunoglobulin A1 protease activity nor the ability to bind α-amylase from saliva was a preferential characteristic of persistent genotypes. In contrast to current concepts of climax ecosystems, the species niche in the habitat appears to be maintained predominantly by a succession of clones rather than by stable strains. Several lines of evidence suggest that the major origin of “new” clones is the many other habitats in the respiratory tract that are occupied by this species.
Population genetic analyses have provided detailed insight into the genetic diversity and molecular epidemiology of an array of pathogenic bacteria. Such studies have not only demonstrated various degrees of diversity within species with direct relevance to pathogenic potential but have also disclosed basic differences in the genetic population structure of species. While some species evolve slowly and, to a certain extent, predictably along distinct phylogenetic lineages, other species undergo frequent changes due to recombination, which may result in dramatic fluctuations in the severity and abundance of infections with which they are associated.
Even at the global level, most pathogenic species that have been examined consist of a limited number of clones, of which a few are often responsible for the majority of cases of disease (44). In contrast, genetic typing of bacteria that are commensals or opportunistic pathogens have revealed considerably more diversity, sometimes even within a single host (1, 2, 7, 11, 13, 15, 22, 23, 37, 49), although typing methods such as restriction endonuclease analysis (REA), ribotyping, or other highly sensitive DNA-based typing methods may miss the clonal relationships of types. However, only rarely are the causes and ecological significance of this diversity considered.
Pioneering longitudinal studies of fecal populations of Escherichia coli reported by Caugant and coworkers (11, 12) revealed a highly dynamic pattern with a mixture of transient and persistent clones. E. coli differs from the majority of commensals of human mucosal surfaces in having two habitats: a primary habitat in the lower intestine of warm-blooded animals and a secondary habitat in the environment external to the host (water, soil, and contaminated food) (41). In addition, E. coli populations in intestinal contents do not necessarily reflect only populations colonizing the gut mucosa. Both of these factors may have significant implications for the population biology and dynamics of this species.
Streptococcus mitis biovar 1 is a typical representative of the commensal microbiota of the respiratory tract. It colonizes several surfaces in the oral cavity and pharynx after birth and is believed to remain a numerically important member of those ecosystems throughout life (8, 15, 16, 22, 34, 45). Like a few other Streptococcus species, it participates in the initial colonization of tooth enamel (32) and may be implicated in nursing bottle and root surface caries (5, 51). S. mitis is also recognized as an increasingly important cause of bacteremia in patients with hematologic illnesses (3). Some but not all strains of S. mitis biovar 1 are characterized by immunoglobulin A1 (IgA1) protease production and the ability to bind salivary α-amylase, both of which have been suggested to confer ecological advantages (26, 42). Two previous studies have demonstrated significant genetic diversity in the populations of S. mitis biovar 1 on mucosal surfaces in the oral cavity and pharynx (15, 22). Combined, these properties make S. mitis biovar 1 an attractive model for studies of bacterial population dynamics in defined ecological habitats of the human body.
The purpose of this study was to examine the genetic structure of S. mitis biovar 1 in its natural habitat in the human oral cavity and pharynx and to investigate the role that selected microbial properties (IgA1 protease production and the ability to bind salivary α-amylase) and host, spatial, and temporal factors play in determining the structure of the bacterial population.
MATERIALS AND METHODS
Study population.
Eight individuals—two infants and six adults—participated in the study. Six of these were members of two families, each consisting of a mother, father, and infant. At the start of the sampling period, the infants in families 1 and 2 were 10 and 5 months of age, respectively. The age of the parents was between 23 and 27 years. The remaining individuals included in the study were two unrelated men aged 67 and 68 years, respectively, at the start of the study. All subjects had their full natural dentition. There was no history of antibiotic treatment at least 6 months prior to or during the study, and all subjects were healthy at the times of sampling. Within 261-day (family 1) and 313-day (family 2) periods, samples of oral and pharyngeal bacteria were collected four times from each individual (Fig. 1). At each of these four sampling occasions, samples from all family members were collected within 30 min. Dental plaque samples from buccal surfaces of upper molars were collected 5 years after the initial sampling from the parents of family 2. The four sampling occasions involving the two elderly men spanned a total of 358 (elderly 1) and 374 (elderly 2) days, respectively.
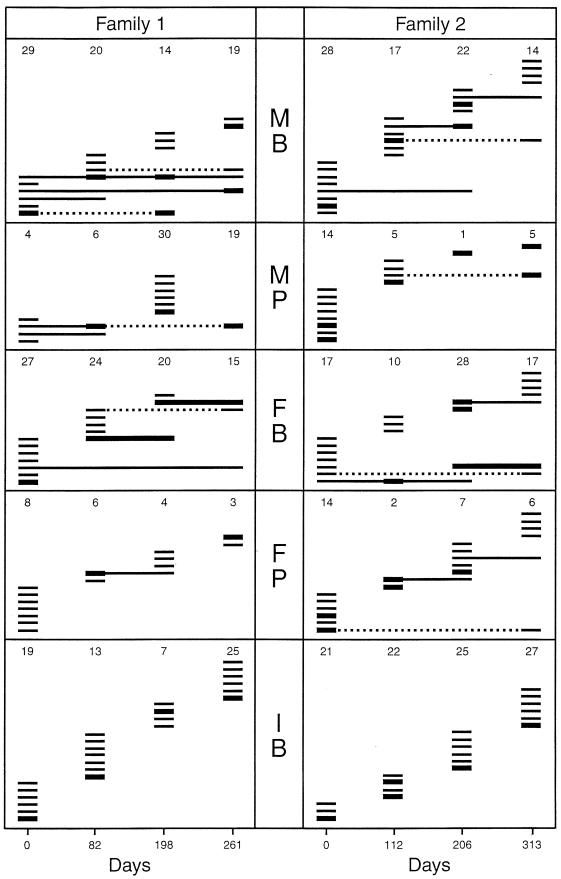
FIG. 1.
Clonal composition of the S. mitis biovar 1 population on buccal (B) and pharyngeal (P) mucosae of members of two families examined over 261 and 313 days, respectively. Each family consisted of an infant (I), the mother (M), and the father (F). The figures for each observation represent the total number of S. mitis biovar 1 isolates examined, and each line represents a distinct clonal type. Bold lines indicate clones representing more than 25% of the total number of isolates. Broken lines indicate periods during which redetected clones are missing.
Bacteriological sampling.
Samples were collected with cotton swabs, and in the case of adults after thorough rinsing of the oral cavity with sterile saline. From the adults, separate samples were obtained from the buccal mucosae and from the posterior wall of the oropharynx; whereas in the infants, only the buccal mucosae were sampled. The samples were immediately transferred to 3 ml of cold brain heart infusion broth (Difco Laboratories, Detroit, Mich.) and were vigorously shaken with a Vortex mixer to disperse the bacteria. At the initial sampling occasion, which has previously been described in detail (22), serial dilutions of the suspensions were plated on a medium selective for streptococci, consisting of Todd-Hewitt agar (Difco) supplemented with 75 mg of trypan blue, 0.8 mg of crystal violet, and 1 mg of tellurite per liter of medium (16). The plates were incubated for 3 days under anaerobic conditions. An agar plate with 50 to 300 colonies was selected for each sample, and 50 colonies, including all colonies in a section of the agar plate, were subcultured to purity on Todd-Hewitt agar. On the three last sampling occasions, mitis salivarius agar was used as a cultivation medium. Experiments had shown that using mitis salivarius agar, which contains 5% sucrose, it was possible to exclude colonies of many of the non-S. mitis streptococci (i.e., extracellular polysaccharide producing) on the basis of the morphology and consistency of their colonies. Consequently, the number of initial isolates could be reduced to 30, representing the remaining colonial morphologies located within a certain section of the agar plate. This procedure was used for all samples collected from the two elderly men. Streptococcal isolates were identified and tested for IgA1 protease activity by methods and principles described previously (24). The ability of all isolates to bind salivary α-amylase was examined using the method described by Kilian and Nyvad (25).
REA.
Genomic DNA was extracted and purified as previously described (22, 40). Restriction endonuclease digestion with HaeIII (Boehringer GmbH, Mannheim, Germany), and electrophoresis was performed as described (22). Gels were stained with ethidium bromide, and the band patterns were compared visually. Isolates were considered to represent different genotypes when more than one band differed in the HaeIII REA profile. In cases where a HaeIII-defined genotype was reisolated from a subsequent sample from the same site, single isolates of the genotype from each sample were compared after digestion with EcoRI and TaqI. First and second isolates were allocated to different genotypes if more than one band differed in profiles obtained with any of these enzymes.
MLEE.
Bacterial lysates for multilocus enzyme electrophoresis (MLEE) analysis were prepared and stored as described by Helmig et al. (21). The lysates were electrophoresed in starch gels and selectively stained for activity of each of nine metabolic enzymes as described by Selander et al. (43). The enzymes assayed were as follows: adenylate kinase, esterase, glutamate dehydrogenase 2, glucose-6-phosphate dehydrogenase, hexokinase, leucine amino peptidase, nucleoside phosphorylase, 6-phosphogluconate dehydrogenase, and phosphoglucomutase. Buffer system A (Tris-citrate, pH 8.0) was used for all enzymes. Genetic diversity at each of the examined gene loci was calculated as h = (1 − Σ xi2) (n/[n −1]), where xi is the frequency of the ith allele and n is the number of electrophoretic types (ETs) or isolates (43). Each combination of electrophoretic mobility of the enzymes defined an ET. Genetic diversity between ETs was expressed as the proportion of enzymes at which dissimilar electrophoretic mobility was observed. Genetic diversity at individual enzyme loci was calculated using the ETDIV program, version 2.2, developed by T. S. Whittam, Department of Biology, Institute of Molecular Evolutionary Genetics, Pennsylvania State University, University Park, Pa. (www.http://foodsate.msu.edu/whittam/#programs). Genetic similarity between pairs of isolates from the two infants was calculated manually as the proportion of enzymes at which identical eletrophoretic mobility occurred.
Statistical evaluations.
Comparisons of proportional differences between sites or individuals were performed using the Mann-Whitney U test, and comparisons of the persistence of clones were performed using Fisher's exact test. The best-fit graph of the relationship between the number of isolates examined and the number of genotypes detected in a particular sample was determined with the program Fig.P for Microsoft, version 2 (Biosoft, Ferguson, Mo.). The index of association (IA), which compares the observed variance in the number of allelic mismatches between isolates with that expected for a population in linkage equilibrium, was calculated with the ETLINK program of T. S. Whittam, based on principles described by Smith et al. (46), according to which IA does not differ significantly from zero for a population in linkage equilibrium.
RESULTS
Identification and characterization of isolates.
A total of 1,319 streptococcal isolates were recovered from the six subjects belonging to two families, 614 of which were identified as S. mitis biovar 1. Among these 614 isolates, 155 and 321 were from buccal mucosae of the two infants and four parents, respectively, and 138 were isolated from the pharyngeal mucosae of the four parents. The median number of isolates per sample identified as S. mitis biovar 1 was 16 (range, 1 to 30). All isolates allocated to S. mitis biovar 1 were negative in tests for extracellular polysaccharide production, hydrolysis of arginine and esculin, and fermentation of amygdalin and mannitol. The majority (87.4%) failed to ferment raffinose and melibiose. Six percent of the S. mitis biovar 1 isolates were unusual relative to the description by Kilian et al. (24) in fermenting sorbitol or inulin. Among all the S. mitis isolates, 52.3% had IgA1 protease activity, 27.6% bound α-amylase, and 9.9% demonstrated both of these traits.
Genotypes of S. mitis biovar 1.
REA of genomic DNA digested with HaeIII revealed several genotypes of S. mitis biovar 1 in each sample (Fig. 1). With the exception of the pharyngeal sample from mother 1 at day 261, in which all 19 S. mitis biovar 1 isolates were identical (see below), and one pharyngeal sample from mother 2 (day 206), from which only 1 isolate was identified as S. mitis biovar 1, all samples contained from 2 to 8 genotypes (mean, 5.0) of this taxon. Reexamination with EcoRI and TaqI of DNA from isolates allocated to the same genotype upon analysis with HaeIII rarely revealed additional diversity. Only on one occasion did isolates from one sample (pharynx of mother 1, day 261), which showed consistent identity by HaeIII and EcoRI digestion, show minor diversity in the TaqI profile. We consider these isolates to be closely related, and they were consequently allocated to a single genotype (Fig. 1). Isolates allocated to the same genotype were always identical with respect to IgA1 protease activity and α-amylase binding. No significant differences were noted between the number of genotypes recovered from buccal and pharyngeal surfaces of adults (P = 0.06; two-tailed Mann-Whitney U test) and from buccal surfaces of infants and adults, respectively (P = 0.98). Combined, a total of 54 genotypes were detected among 134 pharyngeal isolates of S. mitis biovar 1 (1:2.5) compared to 107 genotypes among 480 buccal isolates (1:4.5).
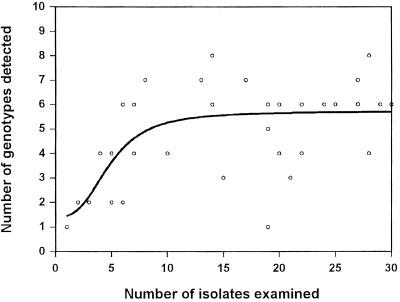
The relationship between the number of S. mitis biovar 1 isolates examined from each sample and the number of genotypes detected among these isolates is demonstrated in Fig. 2. The direct association between the number of isolates examined and the number of genotypes detected in the beginning of the graph suggests that the number of genotypes present in samples is likely to have been underestimated when less than 10 to 15 S. mitis biovar 1 isolates were examined. However, the data suggest that the method revealed close to actual clonal diversity when more than 20 isolates were examined.
FIG. 2.
Best-fit graph showing the relationship between the total number of S. mitis biovar 1 isolates examined and the number of distinct clones detected in individual samples from buccal and pharyngeal mucosae of two infants and their parents.
Figure 1 summarizes the dynamics observed in the population of S. mitis biovar 1 at each site in the members of the two families. In the two infants, from which a median number of 22 (range, 7 to 27) S. mitis biovar 1 isolates were analyzed per sample, no single genotype was detected on more than one occasion, corresponding to less than 112 days of persistence (Fig. 1). Contrasting with this, a total of 23.3% (24 of 103) of the genotypes detected in adults were found to persist over a period corresponding to 2, 3, or 4 sampling occasions (Fig. 1). Some genotypes reappeared after being undetected at intervening examinations. Unless otherwise stated (see below), the periods of persistence summarized in Table 1 were determined on the assumption that the genotypes were present throughout the whole period although occasionally in proportions below detectable levels.
TABLE 1.
Persistence of S. mitis biovar 1 clones on buccal and pharyngeal surfaces of two infants and four adults
| Subject group, mucosa(e) | No. of clones | % Clones persisting over 2- to 3-month intervals
|
Total no. of clones redetected | ||
|---|---|---|---|---|---|
| No. of periods
| |||||
| 2 | 3 | 4 | |||
| Infants, buccal | 42 | 0 (30)a | 0 (20) | 0 (9) | 0 (30) |
| Adults, buccal | 65 | 9.1 (55) | 18.2 (44) | 14.3 (28) | 30.9 (55) |
| Adults, pharyngeal | 57 | 10.4 (48) | 3.0 (33) | 4.0 (25) | 14.6 (48) |
| Adults, buccal and pharyngeal | 122 | 9.7 (103) | 11.7 (77) | 9.4 (53) | 23.3 (103) |
Numbers in parentheses indicate n, the number of clones for which persistence over the indicated number of occasions could be examined, given the time scale of the study.
In 17 of 40 samples, a single genotype accounted for 50% or more of the isolates. Figure 1 indicates genotypes constituting more than 25% of the S. mitis biovar 1 isolates. As shown, at only three sites did a certain genotype account for more than 25% of the S. mitis biovar 1 population on more than one occasion, indicating constant fluctuations in the proportions of individual genotypes.
Proportions of S. mitis biovar 1 isolates with IgA1 protease activity and proportions binding α-amylase also varied widely over time and with no consistent pattern in the four sequential samples collected from the two infants (buccal mucosae) and their four parents (buccal and pharyngeal mucosae) (data not shown).
Characterization of persisting genotypes.
Theoretically, the frequent turnover of genotypes might be a random phenomenon in that the probability of elimination over time is equal for all genotypes. Alternatively, some genotypes might possess properties that make them more likely to persist than others. In relation to this question, it was calculated for buccal isolates from the four parents that, among genotypes present on any one occasion, 46.5% (33 of 71) were present also on the subsequent occasion. In comparison, among genotypes present on two successive occasions, 69.6% (16/23) were also present on the subsequent occasion. The latter probability is significantly higher than the former (P = 0.04; Fisher's exact test, one tailed). By similar analysis of the data for pharyngeal clones, the corresponding P value was 0.05. When the same type of calculations was done under the assumption that a temporary failure of detection reflected a true absence of clones, the consequences of dual versus single detection on previous occasions were even more significant (P < 0.02). These results indicate that prolonged persistence was not a random phenomenon among S. mitis biovar 1 genotypes.
Hypothetically, the prolonged persistence of certain genotypes might be explained by potential ecological advantages conferred by properties such as IgA1 protease activity or the ability to bind salivary α-amylase. However, neither in the case of IgA1 protease nor in the case of α-amylase binding did the proportion of genotypes with these phenotypes differ significantly between genotypes detected only once and genotypes detected on two, three, or four occasions (Table 2). No other variable biochemical character used for isolate identification was preferentially associated with persistent clones.
TABLE 2.
Proportions of S. mitis biovar 1 isolates with IgA1 protease or α-amylase-binding activity related to period of persistence on buccal and pharyngeal mucosae of four adult individualsa
| No. of periods | % Activity
|
|
|---|---|---|
| IgA1 protease | α-amylase binding | |
| 1 | 58.2 (79)b | 20.1 (72) |
| 2 | 40.0 (10) | 40.0 (10) |
| 3 | 55.6 (9) | 22.2 (9) |
| 4 | 60.0 (5) | 20.0 (5) |
| Total | 55.7 (122) | 25.9 (116) |
The percentage of clones with activity among clones detected on one to four successive sampling occasions at 2- to 3-month intervals is shown.
Numbers in parentheses indicates n, the number of clones for which persistence over the indicated number of occasions could be examined given the time scale of the study.
S. mitis biovar 1 in elderly individuals.
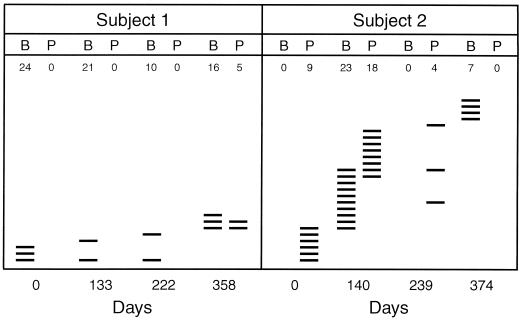
To examine if the stability of the S. mitis biovar 1 population continues to increase with age, we monitored this taxon in two elderly men over a period of approximately 1 year. A total of 137 isolates of S. mitis biovar 1 were examined. While S. mitis biovar 1was a constant component of the buccal and pharyngeal microbiotas in the infants and their parents, this was not the case in the samples from the two elderly men, although the same number of streptococcal isolates was examined. In subject 1, S. mitis biovar 1 was not detected in three of four pharyngeal samples, although it was detected in all buccal samples. In subject 2, it was undetected in two buccal samples and in one of four pharyngeal samples (Fig. 3).
FIG. 3.
Clonal composition of the S. mitis biovar 1 population on buccal (B) and pharyngeal (P) mucosae of two elderly men examined over 358 and 374 days, respectively. The figure for each sample represents the number of strains recovered and examined; each line represents distinct clones.
The total number of genotypes detected among 137 isolates from the two individuals was 8 of 76 (number of genotypes per number of S. mitis biovar 1 isolates) and 26 of 61, respectively. All isolates from each of the two elderly individuals were compared with regard to genotype. Figure 3 summarizes the results of these analyses. A few genotypes were detected in both buccal and pharyngeal samples on the same occasion, but only one and three genotypes, respectively, were detected in the two individuals on more than one of the sampling occasions.
Genetic diversity and relationships revealed by MLEE.
Theoretically, the large number and frequent turnover of S. mitis biovar 1 genotypes detected in each individual may be explained by (i) frequent acquisition of “new” clones from external sources and subsequent loss, (ii) frequent emergence of recombinant forms as a result of horizontal gene transfer within the population of streptococci inhabiting each individual, or (iii) a combination of the two. To address this question, a total of 13 and 19 isolates from the two infants were examined by MLEE to determine the genetic relationships within and between each of the two populations. The 13 and 19 isolates were chosen to represent the individual REA types recovered from the two infants during the observation period.
Examination of the 32 REA types by MLEE analysis of nine enzymes revealed from 2 to 13 alleles (mean, 6.78) of the corresponding gene loci. The number of alleles detected at each locus and the corresponding genetic diversity at each locus (h value) are shown in Table 3 for each of the two populations of isolates and for the two populations combined. These figures demonstrate that the degree of genetic diversity within each of the two populations was as high as that observed within the combined population. Analysis of the MLEE data by the ETDIV program revealed the maximum number of ETs, i.e., 32, supporting the conclusion based on REA that each of the 32 isolates was genetically distinct.
TABLE 3.
Genetic diversity observed in nine structural gene loci by MLEE analysis of 19 and 13 isolates representing different REA types of S. mitis biovar 1 isolates from two individuals, compared to the diversity in the two collections combined
| Locusa | Combined material
|
Infant 1 (n = 19)
|
Infant 2 (n = 13)
|
|||
|---|---|---|---|---|---|---|
| No. of alleles | h | No. of alleles | h | No. of alleles | h | |
| GD2 | 2 | 0.514 | 2 | 0.515 | 2 | 0.462 |
| EST | 11 | 0.849 | 10 | 0.901 | 6 | 0.718 |
| ADK | 6 | 0.708 | 6 | 0.825 | 4 | 0.423 |
| G6P | 8 | 0.853 | 8 | 0.877 | 4 | 0.782 |
| LAP | 7 | 0.819 | 6 | 0.778 | 7 | 0.872 |
| 6PG | 4 | 0.659 | 4 | 0.573 | 4 | 0.769 |
| NSP | 13 | 0.913 | 11 | 0.924 | 8 | 0.897 |
| PGM | 5 | 0.615 | 5 | 0.690 | 4 | 0.526 |
| HEX | 5 | 0.750 | 5 | 0.719 | 4 | 0.731 |
| Mean | 6.8 | 0.742 | 6.3 | 0.756 | 4.8 | 0.687 |
Abbreviations: GD2, glutamate dehydrogenase 2; EST, esterase; ADK, adenylate kinase; G6P, glucose-6-phosphate dehydrogenase; LAP, leucine amino peptidase; 6PG, 6-phosphogluconate dehydrogenase; NSP, nucleoside phosphorylase; PGM, phosphoglucomutase; HEX, hexokinase.
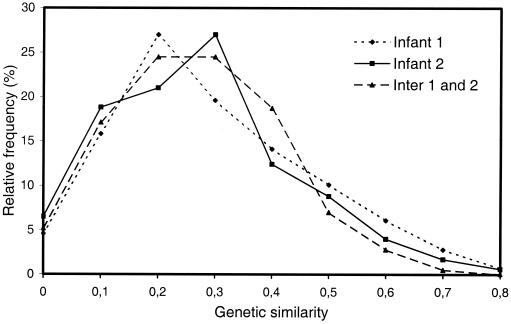
The MLEE data were further analyzed in an attempt to obtain information on the origin of the new genotypes observed at each examination of the two infants. The degree of genetic similarity between all possible pairs of isolates of each of the two populations and of the two populations combined was calculated manually and expressed as relative frequencies of the different degrees of genetic similarities (Fig. 4). These analyses revealed no more mutual similarity of isolates from a single infant than among combined isolates from the two infants. Furthermore, no single allele was unique to isolates from a single individual.
FIG. 4.
Distribution of genetic similarities revealed by MLEE analysis of nine enzymes in pair-wise comparisons of ETs detected in each of two infants and in the combined collection (Inter 1 and 2).
Finally, evaluation of the MLEE data for evidence of linkage disequilibrium in the population of the 32 ETs using the ETLINK program revealed an IA value of 0.985 ± 0.242 (mean ± standard error), which is significantly different from zero and indicates linkage disequlibrium. The IA values calculated for the ETs from each of the two infants were 1.658 ± 0.377 and 1.490 ± 0.315, respectively. Both of these values indicate populations in linkage disequilibrium.
S. mitis biovar 1 in dental plaque samples.
To test if clones persisting on mucosal membranes of the parents were permanent members of the dental plaque microflora, we collected samples from the buccal surfaces of upper molars in one of the parent pairs 5 years after the initial sampling. A total of 29 and 22 isolates of S. mitis biovar 1 were recovered from the two individuals, and each of these were compared with representatives of each of the previously detected persisting clones by REA with HaeIII. In no case was an identical clone redetected after 5 years.
DISCUSSION
This study confirms the ubiquitous nature of S. mitis biovar 1 in the upper respiratory tracts of children and young adults, but the observations suggest that it may be a less regular member of the microbiota in the elderly population. The results furthermore confirm that the S. mitis biovar 1 population, even in well-defined, narrow, and homogeneous habitats like the buccal and oropharyngeal mucosae, consists of a multitude of distinct genotypes (15, 22). Degrees of clonal diversity within single individuals approaching that of S. mitis biovar 1 have been demonstrated for Eikenella corrodens, Actinomyces naeslundii, and Haemophilus parainfluenzae (4, 13, 23).
The additional dimension of this study is that it elucidates the temporal factor. Several methodological considerations are important for studies of population dynamics, i.e., the sensitivity of the typing method employed and the size and representativeness of the sample. REA of whole genomic DNA, as employed in this study, is a very sensitive typing method surpassed only by DNA sequencing. In contrast to our previous cross-sectional study (22), this longitudinal study allowed for deviation in one REA band in isolates allocated to the same genotype. Single-band differences between two isolates observed by REA may result from transmissible DNA elements whereas deviations in two or more bands are likely to reflect differences in genomic DNA. Therefore, by deliberately changing the scoring of REA profiles, we minimized the risk that occasionally acquired DNA elements would prevent clones from being recognized on a later occasion. That isolates allocated to a common REA type were always identical with respect to IgA1 protease and α-amylase binding activity and that isolates allocated to different REA types also were distinct by MLEE analysis support the validity of the genotyping by REA. The correlation between typing results based on REA and MLEE furthermore excludes the possibility that differences in REA patterns were due to genome rearrangements or inserts, as has been observed in sequential isolates of Pseudomonas aeruginosa and Helicobacter pylori from single individuals (31, 38).
With the significant degree of clonal diversity detected in single individuals, the number of isolates examined per sample becomes an important factor. The analysis of the relationship between the number of isolates examined and the number of genotypes detected (Fig. 2) clearly suggests that we underestimated the degree of diversity at sites from which less than 15 S. mitis biovar 1 isolates were examined, as a result of low proportions of this taxon in the local streptococcal flora. Conversely, the data indicate that our examinations revealed close to actual diversity in the majority of samples and that examination of additional isolates, in these cases, likely would not have resulted in extra diversity.
While mucosal bacterial pathogens usually are eliminated by the immune system within a limited period of time, it has been generally assumed that commensal bacteria exist in a state of equilibrium with the host (17). However, this longitudinal study spanning approximately 1 year clearly demonstrates a succession of genotypes rather than equilibrium in the S. mitis flora both on the buccal and pharyngeal mucosae. Of the 42 genotypes detected in the two infants, none were isolated on more than one occasion. However, in the four parents of these infants, one to three clones of the heterogeneous population persisted throughout the entire observation period, although they fluctuated markedly in abundance (Fig. 1). The difference between infants and their parents prompted us to include two elderly individuals to test the hypothesis that indigenous streptococcal flora becomes increasingly stable with age. Surprisingly, S. mitis biovar 1 was often undetectable in samples from the elderly men, either on the buccal or on the pharyngeal mucosae, and there was no evidence of stability additional to that observed in the four young adults (Fig. 3).
Succession of strains of E. coli, lactobacilli, and bifidobacteria has been previously demonstrated in gastrointestinal contents of humans, pigs, chicken, and feral house mice (2, 11, 18, 48, 52). Caugant and coworkers (11) have demonstrated patterns in the fecal E. coli population of a single individual very similar to that observed for S. mitis biovar 1 in the young adults with a combination of transient and resident clones. However, the direct applicability of those findings to this study is not clear, because the bacteria in feces are likely to reflect a multitude of habitats not restricted to mucosal surfaces. Several lines of evidence support our assumption that even genotypes of S. mitis biovar 1 that were detected only on a single occasion were indeed established on the mucosal membranes and, although transient, remained part of the microbiota for some weeks but for less than 3 months. Firstly, in the adults, samples were collected directly from the mucosal surfaces immediately following rinsing with saline, which is likely to eliminate nonattached bacteria. Secondly, single genotypes often predominated both in adults and in infants. Finally, the infant study reported by Fitzsimmons et al. (15), although not designed to address this question, did reveal cases in which identical genotypes were observed at 2- to 4-week intervals.
One of the intriguing questions raised by our results is the origin of the constantly changing genotypes. The current dogma in microbial ecology is that climax communities of indigenous bacteria are highly stable in the absence of ecological perturbation and resist exogenous colonization (6, 17). However, the monolayers of bacteria that colonize continuously desquamating surface epithelia of mucosal membranes may be different in this respect from true biofilms, such as dental plaque. While the few dental plaque bacteria that have been examined show relative clonal stability over time (10, 39), studies of pharyngeal populations of noncapsulated H. influenzae and Moraxella catarrhalis in children have demonstrated frequent clonal replacement (27, 47, 50).
Much of the diversity of the clonal composition of intestinal E. coli populations is thought to be due to continuous acquisition and extinction of clones (11). In contrast to E. coli, which in addition to its primary habitat (the lower intestine of warm-blooded animals) has a secondary habitat in the environment external to the host (water, soil, and contaminated food) (41), no other habitat than the human upper respiratory tract is known for S. mitis. If the S. mitis biovar 1 clones detected in this study had been acquired close to the time of sampling, one would also expect them to be detectable in family members in close contact or, in the case of the infants, in other infants and caretakers in the nursery that they attended. Previous studies have demonstrated that the source of strains of S. mutans and certain other oral bacterial species in infancy and childhood is usually the mother (9, 30, 35). However, our previous cross-sectional study of one of the families included in this study revealed very limited sharing of genotypes among the three individuals (22). We have no data that allow us to elucidate the significance of contacts in nurseries.
The steadily changing genotypes of S. mitis biovar 1 observed in this study might, theoretically, represent local fluctuations in the proportions of individual clones in a very large population residing in the two habitats examined. That examination of more than 20 isolates was unlikely to have significantly increased the number of detectable genotypes (Fig. 2) argues against this explanation. However, the oral cavity and pharynx consist of a large variety of habitats that offer dramatically different ecological conditions for the bacteria, e.g., biofilms on the different surfaces of teeth above and below the gingival margin; a number of relatively smooth mucosal surfaces on the cheek and vestibulum and in the pharynx; and the rough surfaces of the dorsum of the tongue and the tonsils. While some bacteria like S. mutans, Porphyromonas gingivalis, and Actinobacillus actinomycetemcomitans are restricted to dental plaques, S. mitis and several other species appear to be able to colonize all, or the majority, of these habitats (16, 28). Our previous cross-sectional examination of one of the families included in this study revealed completely different clonal compositions of the S. mitis biovar 1 populations on buccal and pharyngeal mucosae, in spite of the fact that these two surfaces are among the most similar. Likewise, sampling from buccal mucosal surfaces in different parts of the mouth of individuals, although showing the same predominant clones, revealed differences in less-abundant clones (22). It is therefore conceivable that transmission between different habitats play a major role in the emergence and disappearance of clones at a particular site.
A final hypothetical model is that part of the observed emergence and disappearance of genotypes is a result of mutations and in situ recombination between members of the population of S. mitis biovar 1 and possibly other closely related species present in the respiratory tract. Although accumulation of mutations is an important contributor to long-term genetic diversification in bacteria, it is unlikely to explain the significant differences observed by MLEE analysis between the frequently emerging S. mitis clones (Fig. 4). Recombination in vitro has been previously shown both within and between related species of oral streptococci (20, 29, 36), and there is evidence of horizontal transfer of genes encoding altered penicillin-binding proteins between S. mitis, S. oralis, and S. pneumoniae (14, 19). However, the frequency of these events in vivo is not known. In the mentioned cases, even rare recombinatorial events become detectable because they result in antibiotic resistance for which there is a strong selection pressure.
To test the hypothesis that all or part of the genotypes of S. mitis biovar 1 detected on oral mucosa had emerged as a result of recombination, we compared by MLEE analysis the pairwise genetic relatedness of isolates from each of the two infants to isolates of the two populations combined. The lack of closer genetic relationships of isolates within each individual (Fig. 4) and the absence of alleles unique to either of the two populations do not support the in situ recombination theory. It is striking that the genetic diversity observed within each infant was as extensive as or approached that of the combined populations (Table 3). Theoretically, this situation could be a result of extensive in situ recombination. However, the calculated IA, which compares the observed variance in the allelic mismatches between isolates with that expected for a population in linkage equilibrium, was statistically significantly different from zero (IA = 0.985 ± 0.242) and rules out the possibility that maximal genetic diversity had developed by recombination within each subpopulation as has been observed in local populations of gonococci (33). The small sample sizes do not allow us to exclude that a minor part of the detected genotypes were indeed results of in situ recombination; however, there is no support for the hypothesis that recombination in situ plays a major role in the emergence and disappearance of clones. This conclusion is also supported by the finding of persisting clones in the adults but not in the infants. The same conclusion was reached after analyses of intestinal populations of E. coli (11).
It remains to be explained why some genotypes in adults but not in infants persist over long periods of time. Statistical calculations showed that this ability was not distributed at random in the S. mitis biovar 1 population. However, neither IgA1 protease activity nor the ability to bind α-amylase from saliva was a preferential characteristic of persisting genotypes (Table 2). Both of these factors have been hypothesized to promote bacterial colonization either directly or by evasion of secretory IgA1-mediated immunity (26, 42). It is possible that such resident genotypes have a different habitat from which they are constantly seeded to the buccal and pharyngeal mucosae in adults. A likely habitat, which was missing in the infants during the initial part of the study, might be tooth surfaces. However, our previous observation that the genotypes persisting on buccal and pharyngeal mucosae were distinct (22) does not support this hypothesis. Furthermore, we were unable to redetect the persisting genotypes on the tooth surfaces of two of the parents 5 years after the initial samples were collected. Further genetic and physiological studies are required to determine how such resident clones differ from transient clones.
In conclusion, this study demonstrates that the S. mitis biovar 1 population on mucosal membranes of the upper respiratory tract is in a state of constant change, although partial stability develops in adults. The majority of strains appear to be no more stable than pathogenic bacteria that transiently colonize mucosal membranes. Thus, in contrast to current concepts of climax ecosystems, the species niche in the habitat appears to be maintained by a succession of clones rather than by stable strains. We conclude that the major origin of new clones is in some of the many other habitats in the respiratory tract occupied by this species. The ecological pressure driving this constant replacement of clones is yet unknown.
ACKNOWLEDGMENTS
This study was supported by the Danish Medical Research Council, by the Research Foundation of the University of Aarhus, and by a Ph.D. stipend to J.H. from the Faculty of Health Sciences, University of Aarhus.
We thank Flemming Scheutz for statistical help.
REFERENCES
- 1.Barsotti O, Morrier J J, Decoret D, Benay G, Rocca J P. An investigation into the use of restriction endonuclease analysis for the study of transmission of Actinomyces. J Clin Periodontol. 1993;20:436–442. doi: 10.1111/j.1600-051x.1993.tb00385.x. [DOI] [PubMed] [Google Scholar]
- 2.Bateup J M, Dobbinson S, McConnell M A, Munro K, Tannock G W. Molecular analysis of the composition of Lactobacillus populations inhabiting the stomach and caecum of pigs. Microb Ecol Health Dis. 1998;10:95–102. [Google Scholar]
- 3.Bochud P Y, Calandra T, Francioli P. Bacteremia due to viridans streptococi in neutropenic patients: a review. Am J Med. 1994;97:256–264. doi: 10.1016/0002-9343(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 4.Bowden G H W. Oral biofilm: an archive of past events? In: Newman H N, Wilson M, editors. Dental plaque revisited. Oral biofilms in health and disease. Cardiff, United Kingdom: Bioline; 1999. pp. 211–235. [Google Scholar]
- 5.Bowden G H W, Ekstrand J, McNaughton B, Challacombe S J. The association of selected bacteria with the lesions of root surface caries. Oral Microbiol Immunol. 1990;5:346–351. doi: 10.1111/j.1399-302x.1990.tb00439.x. [DOI] [PubMed] [Google Scholar]
- 6.Bowden G H W, Hamilton I R. Survival of oral bacteria. Crit Rev Oral Biol Med. 1998;9:54–85. doi: 10.1177/10454411980090010401. [DOI] [PubMed] [Google Scholar]
- 7.Bowden G H W, Nolette N, Ryding H, Cleghorn B M. The diversity and distribution of the predominant ribotypes of Actinomyces naeslundii genospecies 1 and 2 in samples from enamel and from healthy and carious root surfaces of teeth. J Dent Res. 1999;78:1800–1809. doi: 10.1177/00220345990780120601. [DOI] [PubMed] [Google Scholar]
- 8.Carlsson J. Presence of various types of non-haemolytic streptococci in dental plaque and in other sites of the oral cavity in man. Odontol Revy. 1967;18:55–74. [PubMed] [Google Scholar]
- 9.Carlsson J, Gothefors L. Transmission of Lactobacillus jensenii and Lactobacillus acidophilus from mother to child at the time of delivery. J Clin Microbiol. 1975;1:124–128. doi: 10.1128/jcm.1.2.124-128.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caufield P W, Walker T M. Genetic diversity within Streptococcus mutans evident from chromosomal DNA restriction fragment polymorphism. J Clin Microbiol. 1989;27:274–278. doi: 10.1128/jcm.27.2.274-278.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caugant D A, Levin B R, Selander R K. Genetic diversity and temporal variation in the E. coli population of a human host. Genetics. 1981;98:467–490. doi: 10.1093/genetics/98.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caugant D A, Levin B R, Selander R K. Distribution of multilocus genotypes of Escherichia coli within and between host families. J Hyg (London) 1984;92:377–384. doi: 10.1017/s0022172400064597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C, Ashimoto A. Clonal diversity of oral Eikenella corrodens within individual subjects by arbitrarily primed PCR. J Clin Microbiol. 1996;34:1837–1839. doi: 10.1128/jcm.34.7.1837-1839.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowson C G, Coffey T J, Kell C, Whiley R A. Evolution of penicillin resistance in Streptococcus pneumoniae; the role of Streptococcus mitis in the formation of a low affinity PBP2B in S. pneumoniae. Mol Microbiol. 1993;9:635–643. doi: 10.1111/j.1365-2958.1993.tb01723.x. [DOI] [PubMed] [Google Scholar]
- 15.Fitzsimmons S, Evans M, Pearce C, Sheridan M J, Wientzen R, Bowden G, Cole M F. Clonal diversity of Streptococcus mitis biovar 1 isolates from the oral cavity of human neonates. Clin Diagn Lab Immunol. 1996;3:517–522. doi: 10.1128/cdli.3.5.517-522.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frandsen E V G, Pedrazzoli V, Kilian M. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol Immunol. 1991;6:129–133. doi: 10.1111/j.1399-302x.1991.tb00466.x. [DOI] [PubMed] [Google Scholar]
- 17.Freter R. Mechanisms that control the microflora in the large intestine. In: Hentges D J, editor. Human intestinal microflora in health and disease. London, United Kingdom: Academic Press; 1983. pp. 33–54. [Google Scholar]
- 18.Gordon D M. The genetic structure of Escherichia coli populations in feral house mice. Microbiology. 1997;143:2039–2046. doi: 10.1099/00221287-143-6-2039. [DOI] [PubMed] [Google Scholar]
- 19.Hakenbeck R, König A, Kern I, van der Linden M, Keck W, Billot-Klein D, Legrand R, Schoot B, Gutmann L. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level β-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J Bacteriol. 1998;180:1831–1840. doi: 10.1128/jb.180.7.1831-1840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartley D L, Jones K R, Tobian J A, LeBlanc D J, Macrina F L. Disseminated tetracycline resistance in oral streptococci: implication of a conjugative transposon. Infect Immun. 1984;45:13–17. doi: 10.1128/iai.45.1.13-17.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helmig R, Uldbjerg N, Boris J, Kilian M. Clonal analysis of Streptococcus agalactiae isolated from infants with neonatal sepsis or meningitis and their mothers and from healthy pregnant women. J Infect Dis. 1993;168:904–909. doi: 10.1093/infdis/168.4.904. [DOI] [PubMed] [Google Scholar]
- 22.Hohwy J, Kilian M. Clonal diversity of the Streptococcus mitis biovar 1 population in the human oral cavity and pharynx. Oral Microbiol Immunol. 1995;10:19–25. doi: 10.1111/j.1399-302x.1995.tb00113.x. [DOI] [PubMed] [Google Scholar]
- 23.Kerr G R D, Forbes K J, Williams A, Pennington T H. An analysis of the diversity of Haemophilus parainfluenzae in the adult human respiratory tract by genomic DNA fingerprinting. Epidemiol Infect. 1993;111:89–98. doi: 10.1017/s0950268800056715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilian M, Mikkelsen L, Henrichsen J. Taxonomic study of viridans streptococci: description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis (White and Niven 1946), Streptococcus oralis (Bridge and Sneath 1982), and Streptococcus mitis (Andrewes and Horder 1906) Int J Syst Bacteriol. 1989;39:471–484. [Google Scholar]
- 25.Kilian M, Nyvad B. Ability to bind salivary α-amylase discriminates certain viridans group streptococcal species. J Clin Microbiol. 1990;28:2576–2577. doi: 10.1128/jcm.28.11.2576-2577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kilian M, Reinholdt J, Lomholt H, Poulsen K, Frandsen E V G. Biological significance of IgA1 proteases in bacteriological colonization and pathogenesis: critical evaluation of experimental evidence. APMIS. 1996;104:321–338. doi: 10.1111/j.1699-0463.1996.tb00724.x. [DOI] [PubMed] [Google Scholar]
- 27.Klingman K L, Pye A, Murphy T F, Hill S. Dynamics of respiratory tract colonization by Branhamella catarrhalis in bronchiectasis. Am J Respir Crit Care Med. 1995;152:1072–1078. doi: 10.1164/ajrccm.152.3.7663786. [DOI] [PubMed] [Google Scholar]
- 28.Könönen E, Asikanen S, Saarela M, Karjalainen J, Jousimies-Somer H. The oral gram-negative anaerobic microflora in young children: longitudinal changes from edentulous to dentate mouth. Oral Microbiol Immunol. 1994;9:136–141. doi: 10.1111/j.1399-302x.1994.tb00049.x. [DOI] [PubMed] [Google Scholar]
- 29.Kuramitsu H K, Trapa V. Genetic exchange between oral streptococci during mixed growth. J Gen Microbiol. 1984;130:2497–2500. doi: 10.1099/00221287-130-10-2497. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Caufield P W. The fidelity of initial acquisition of mutans streptococci by infants from their mothers. J Dent Res. 1995;74:681–685. doi: 10.1177/00220345950740020901. [DOI] [PubMed] [Google Scholar]
- 31.Marshall D G, Dundon W G, Beesley S M, Smyth C J. Helicobacter pylori—a conundrum of genetic diversity. Microbiology. 1998;144:2925–2939. doi: 10.1099/00221287-144-11-2925. [DOI] [PubMed] [Google Scholar]
- 32.Nyvad B, Kilian M. Comparison of the initial streptococcal microflora on dental enamel in caries active and in caries inactive individuals. Caries Res. 1990;24:267–272. doi: 10.1159/000261281. [DOI] [PubMed] [Google Scholar]
- 33.O'Rourke M, Stevens E. Genetic structure of Neisseria gonorrhoeae populations: a non-clonal pathogen. J Gen Microbiol. 1993;139:2603–2611. doi: 10.1099/00221287-139-11-2603. [DOI] [PubMed] [Google Scholar]
- 34.Pearce C, Bowden G H, Evans M, Fitzsimmons S P, Johnson J, Sheridan M J, Wientzen R, Cole M F. Identification of pioneer viridans streptococci in the oral cavity of human neonates. J Med Microbiol. 1995;42:67–72. doi: 10.1099/00222615-42-1-67. [DOI] [PubMed] [Google Scholar]
- 35.Petit M D A, van Steenbergen T J M, Scholte L M H, van der Velden U, de Graff J J. Epidemiology and transmission of Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans among children and their family members. A report of 4 surveys. J Clin Periodontol. 1993;20:641–650. doi: 10.1111/j.1600-051x.1993.tb00709.x. [DOI] [PubMed] [Google Scholar]
- 36.Poulsen K, Reinholdt J, Jespersgaard C, Boye K, Brown T A, Hauge M, Kilian M. A comprehensive genetic study of streptococcal immunoglobulin A1 proteases: evidence for recombination within and between species. Infect Immun. 1998;66:181–190. doi: 10.1128/iai.66.1.181-190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poulsen K, Theilade E, Lally E T, Demuth D R, Kilian M. Population structure of Actinobacillus actinomycetemcomitans: a framework for studies of disease-associated properties. Microbiology. 1994;140:2049–2060. doi: 10.1099/13500872-140-8-2049. [DOI] [PubMed] [Google Scholar]
- 38.Römling U, Geipel J, Tümmler B. Gradient of genomic diversity in the Pseudomonas aeruginosa chromosome. Mol Microbiol. 1995;17:323–332. doi: 10.1111/j.1365-2958.1995.mmi_17020323.x. [DOI] [PubMed] [Google Scholar]
- 39.Saarela M H, Dogan B, Alaluusua S, Asikainen S. Persistence of oral colonization by the same Actinobacillus actinomycetemcomitans strain(s) J Periodontol. 1999;70:504–509. doi: 10.1902/jop.1999.70.5.504. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Savageau R K. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am Nat. 1983;122:732–744. [Google Scholar]
- 42.Scanapieco F A, Torres G, Levine M J. Salivary α-amylase: role in dental plaque and caries formation. Crit Rev Oral Biol Med. 1993;4:301–307. doi: 10.1177/10454411930040030701. [DOI] [PubMed] [Google Scholar]
- 43.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selander R K, Musser J M. Population genetics of bacterial pathogenesis. In: Iglewski B H, Clark V L, editors. Molecular basis of bacterial pathogenesis. New York, N.Y: Academic Press, Inc.; 1990. pp. 11–36. [Google Scholar]
- 45.Smith D J, Andersen J M, King W F, van Houte J, Taubman M A. Oral colonization of infants. Oral Microbiol Immunol. 1993;8:1–4. doi: 10.1111/j.1399-302x.1993.tb00535.x. [DOI] [PubMed] [Google Scholar]
- 46.Smith J M, Smith N H, O'Rourke M, Spratt B S. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spinola S M, Peacock J, Denny F W, Smith D L, Cannon J G. Epidemiology of colonization by nontypable Haemophilus influenzae in children. J Infect Dis. 1986;154:100–109. doi: 10.1093/infdis/154.1.100. [DOI] [PubMed] [Google Scholar]
- 48.Tannock G W, Fuller R, Pedersen K. Lactobacillus succession in the piglet digestive tract demonstrated by plasmid profiling. Appl Environ Microbiol. 1990;56:1310–1316. doi: 10.1128/aem.56.5.1310-1316.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thurnheer T, Guggenheim B, Gruica B, Gmür R. Infinite serovar and ribotype heterogeneity among oral Fusobacterium nucleatum strains? Anaerobe. 1999;5:79–92. [Google Scholar]
- 50.Trottier S, Stenberg K, Svanborg-Edén C. Turnover of nontypable Haemophilus influenzae in the nasopharynxes of healthy children. J Clin Microbiol. 1989;27:2175–2179. doi: 10.1128/jcm.27.10.2175-2179.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Houte J, Lopman J, Kent R. The predominant cultivable flora of sound and carious root surfaces. J Dent Res. 1994;73:1727–1734. doi: 10.1177/00220345940730110801. [DOI] [PubMed] [Google Scholar]
- 52.Whittam T S. Clonal dynamics of Escherichia coli in its natural habitat. Antonie Leeuwenhoek. 1989;55:23–32. doi: 10.1007/BF02309616. [DOI] [PubMed] [Google Scholar]