Abstract
Astrocytes, a type of glia, are abundant and morphologically complex cells. Here, we report astrocyte molecular profiles, diversity, and morphology across the mouse CNS. We identified shared and region-specific astrocytic genes and functions, and explored the cellular origins of their regional diversity. We identified gene networks correlated with astrocyte morphology, several of which unexpectedly contained Alzheimer’s disease (AD) risk genes. CRISPR/Cas9-mediated reduction of candidate genes reduced astrocyte morphological complexity and resulted in cognitive deficits. The same genes were downregulated in human AD, in an AD mouse model that displayed reduced astrocyte morphology, and in other human brain disorders. We thus provide comprehensive molecular data on astrocyte diversity and mechanisms across the CNS, and on the molecular basis of astrocyte morphology in health and disease.
One-Sentence Summary:
We report the molecular basis of astrocyte diversity and of their complex morphology in health and in CNS disorders.
In order to explore how the brain performs its functions, it is critical to understand its diverse cell populations in detail, as articulated by the BRAIN Initiative (1, 2). Furthermore, it is important to explore both neuronal and non-neuronal cells such as glia (1-3).
Astrocytes, the most populous glial cells, were documented at the same time as neurons, but for a long time their role was relegated to a type of glue that held the brain together (4). With recent technical advances (5), these fascinating cells are emerging as essential contributors of brain physiology, animal behavior, and disease (6, 7). Astrocytes are ubiquitous, morphologically complex bushy cells that make extensive contacts with other brain cells. They serve diverse roles, including ion and neurotransmitter homeostasis, synapse formation/removal, synaptic modulation, and contributions to neurovascular coupling and the blood brain barrier. Astrocytes are widely implicated in neurological and psychiatric diseases (7, 8).
Unlike neurons, which are extremely diverse within neural circuits, astrocytes have been viewed as homogeneous. However, recent gene expression and functional studies employing different methods in the context of health and brain tumors show that astrocytes are separable between several CNS regions (9-18). This has led to the realization that astrocytes may be diverse, an attribute that together with their highly complex morphology may allow them to mediate their multiple roles in different parts of the CNS. However, there has been no broad molecular assessment of astrocyte diversity, similarity or of astrocyte morphology across the CNS of any species.
Results
Strategy
We studied astrocytes from 13 regions of the adult mouse CNS (Fig. 1A; olfactory bulb, OB; motor cortex, MCX; somatosensory cortex, SCX; visual cortex, VCX; hippocampus, HIP; striatum, STR; thalamus, TH; hypothalamus, HY; cerebellum, CB; midbrain, MB; hindbrain, HB; ventral spinal cord, VSC; dorsal spinal cord, DSC). Firstly, we assessed astrocyte-neuron density as well as astrocyte marker expression in different CNS regions. Next, we assessed molecular profiles and signaling pathways using astrocyte-specific RNA sequencing (RNA-seq), bulk tissue RNA-seq, and single-cell RNA-seq (scRNA-seq). Additionally, we documented the morphology of astrocytes and used weighted gene co-expression network analyses (WGCNA) to identify gene networks correlated with morphological complexity. We also tested the consequences of reducing the expression of genes predicted to affect astrocyte morphology using CRISPR/Cas9. Then, we related molecular signatures of astrocyte morphology to gene expression and astrocyte morphology changes in an AD mouse model and to human CNS disorders, including AD.
Fig. 1. Astrocyte density and astrocyte-to-neuron ratios across mouse CNS.
(A) Experimental design for investigating astrocytes in 13 CNS regions. (B) Illustration and representative images of whole-brain astrocyte imaging by FAST (A: anterior, L: lateral, V: vertical). The brains were from Aldh1l1-Cre/ERT2 x Ai14 (Aldh1l1 tdTomato) mice. (C) Representative coronal sections of Aldh1l1-Cre/ERT2 x Ai14 (Aldh1l1 tdTomato) mouse brains imaged by FAST. Magnified images show tdTomato+ astrocytes in the motor cortex (MCX), hippocampus (HIP), and striatum (STR), the other 10 regions are shown in fig. S1. The color images show exemplar Sox9 and Aldh1l1 driven tdTomato images (see fig. S2). (D) Astrocyte density measurements by FAST in 13 CNS regions (N = 3 mice for spinal cord; N = 6 mice for brain). (E) Plot shows the astrocyte-to-neuron ratio in 13 CNS regions (N = 8 mice). (F,G) Plot of astrocyte and neuron numbers in 13 CNS regions. (H) Correlation between astrocyte and neuron numbers. Data are shown as mean ± SEM.
Cellular and anatomical evaluation of astrocytes
We mapped astrocyte density across 13 CNS regions by using whole-brain imaging by block-face serial microscopy tomography (FAST) (19) in mice expressing tdTomato in astrocytes (20) (from Aldh1l1 locus; Fig. 1B). We observed tdTomato+ astrocytes throughout the brain (Movie S1) and spinal cord with densities that varied between regions, but that overlapped with Sox9 (21) by 95 ± 2% (Fig. 1C; fig. S1, S2). While Aldh1l1 and Sox9 are reliable markers of astrocytes (20, 21), no marker labels all astrocytes and 100% overlap was not expected. We quantified tdTomato+ astrocyte density and found significant, but modest 2-fold differences between areas (Fig. 1C,D; N = 3-6 mice; all regions in fig. S1). Next, we labelled neurons and most astrocytes by immunohistochemistry (IHC) for S100β and glial fibrillary acidic protein (GFAP) in mice expressing GCaMP6f in astrocytes (fig. S3). The astrocyte-neuron ratios varied almost 10-fold, being highest in ventral spinal cord and lowest in cerebellum, where our evaluations included Bergmann glia (Fig. 1E; N = 8 mice). These differences were not driven by major variation in astrocytes, but by differences in neurons (Fig. 1F,G; N = 8 mice). There was no one-to-one scaling between neuron and astrocyte density across regions, although the two parameters were correlated (r = 0.495, p = 0.118; Fig. 1H). These data show that the density of astrocytes per region did not scale simply with neuron density, but that astrocytes tiled the CNS with modest variation, likely reflecting homeostatic functions. Large variation of astrocyte-neuron ratios between CNS regions was driven by neurons. Because astrocytes form part of the neurovascular unit (NVU), we also mapped astrocyte-endothelial cell density and ratios across CNS regions (fig. S4A-D). We found that endothelial cell and astrocyte density were strongly correlated (fig. S4E), reflecting critical NVU biology.
We performed IHC for GFAP, S100β, and GCaMP6f (from the Aldh1ll locus) to determine if these markers were equivalent or dissimilar between CNS regions (20). There was ~500-fold variation between CNS regions for these established markers (fig. S5; N = 8 mice; Movie S2). Although the density of astrocytes varied little (Fig. 1), quantitative IHC experiments (fig. S5) provided evidence for diversity between CNS regions.
Shared CNS-wide astrocyte genes and pathways
To evaluate astrocyte gene expression, we performed astrocyte-specific RNA-seq (9). Whole tissue (input) and astrocyte RNA (IP) was extracted from 13 regions (Fig. 2A; N = 3-5 mice). Multidimensional scaling showed separation between input, IP, and CNS areas (Fig. 2B). IP fractions were enriched with astrocyte markers (fig. S6A).
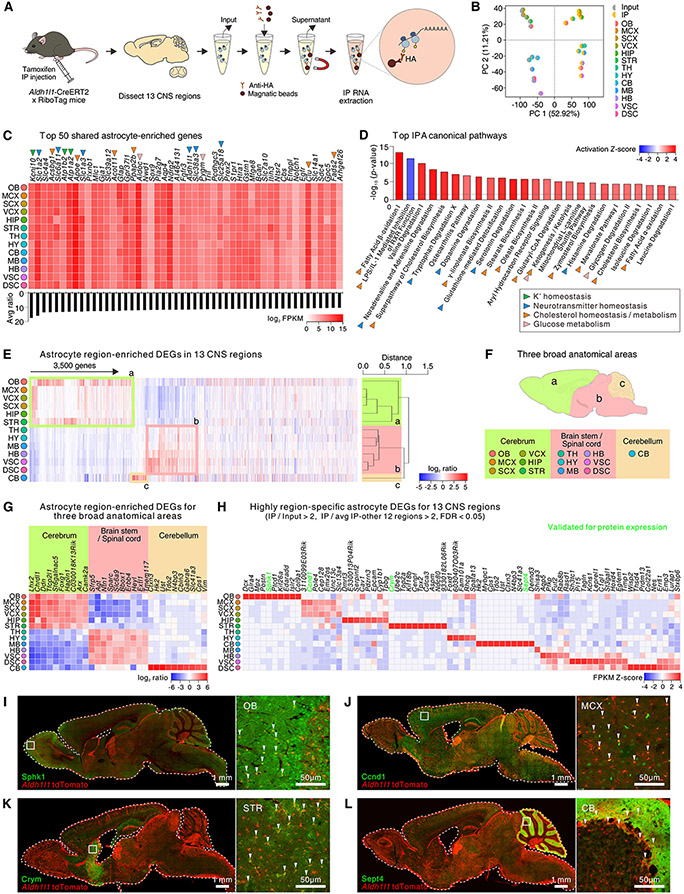
Fig. 2: Shared and unique astrocyte CNS molecular signatures and mechanisms.
(A) Approach for astrocyte-specific RNA-seq. (B) Multi-dimensional scaling plot for input and immunoprecipitated (IP) (N = 4-5 mice) RNA-seq data. (C) Heat map shows the log2 FPKM values of the top 50 most enriched astrocyte genes shared in 13 regions. The bar graph shows the average ratio of IP versus input. Colored arrowheads indicate functional class. (D) The bar graph depicts the top 25 Ingenuity Pathway Analysis (IPA) canonical pathways for the 825 shared astrocyte-enriched genes with a threshold of p value < 0.05. The colored arrowheads indicate functional class. (E) Heat map showing 3,500 astrocyte region-enriched DEGs in 13 CNS regions with a cluster dendrogram. Astrocyte region-specific DEGs are defined by comparing IP in one CNS region with the average of IP samples from the other 12 regions with a threshold of log2 ratio > 1, FDR < 0.05, and FPKM > 1. From the dendrogram, astrocytes classified into 3 broad groups. Genes defining OB, MCX, SCX, HIP, and STR (cerebrum) are outlined in green (a). Genes highly expressed within and defining TH, HY, MB, HB, VSC, and DSC (brain stem or spinal cord) are outlined in pink (b). Genes highly expressed within and defining CB (cerebellum) are outlined in orange (c) and include Bergmann glia genes. (F) Schematic of anatomical grouping of astrocytes in 13 CNS regions based on RNA-seq. (G) Heat map depicts the log2 ratio of IP vs the averaged IP from the other 12 regions of the top 10 astrocyte-specific DEGs for anatomically relevant CNS regions. (H) The heat map shows the top region specific astrocyte genes from RNA-seq. (I-L) Representative (N = 4 mice) IHC images for Sphk (I), Ccnd1 (J), Crym (K), and Sept4 (L), which reproduced regional expression in OB, MCX, striatum and cerebellum, respectively.
We identified 4,314 astrocyte enriched genes (relative to input; log2 ratio > 1; FDR < 0.05, FPKM > 1) and of these 825 (~20%) were common across the 13 regions and termed “shared” (fig. S6B-D). 939 genes (~22%) were “unique” to a particular region and the remaining 2,588 genes (~60%) were “partially shared” in 2-12 regions (fig. S6B-D). To explore the pathways and signaling mechanisms shared by astrocytes across all regions, we assessed the molecules represented by the 825 genes. Around half were related to enzymatic and transporter activity or transcriptional regulation (fig. S6E). Fig. 2C shows the top 50 astrocyte enriched genes shared across 13 regions in order of enrichment, with a color scale representing abundance, and with annotations illustrating the major signaling mechanisms. Recalling IHC (Fig. 1; fig. S2), Aldh1l1 and Sox9 were abundant across regions (Fig. 1C) and known astrocyte genes (e.g. Kcnj10, Slc1a2, Apoe) were among the top 50 highly expressed (Fig. 2C). Unbiased analyses for the 825-shared astrocyte-enriched genes showed that the top pathways were related to neurotransmitter homeostasis, cholesterol biosynthesis, and glucose metabolism (Fig. 2D).
We performed several analyses for astrocytes across 13 regions (fig. S7-8). For K+ homeostasis, the most highly expressed genes were Kcnj10, Kcnj16, and Atp1a2, encoding K+ channels and Na+/K+ ATPase (fig. S7A). Astrocytes highly expressed genes related to neurotransmitter transport and metabolism, but they lacked or expressed at very low levels genes related to Ca2+-dependent vesicular release (fig. S7B-E). Shared functional properties of astrocytes may be cell-autonomously regulated because >50% of all upstream regulators were categorized as transcriptional regulators and ligand-dependent nuclear receptors (fig. S8A,B). The top 20 genes across 13 regions reflecting functional classes of molecules such as enzymes, transporters, kinases, receptors, peptidases, and ion channels are provided in fig. S8C-J. For example GPCR genes Gpr37l1, S1pr1, Ntsr2, Enrb, Smo, Adora2b, Olfr287, Gpr146, Agtrap, Fzd1, Fzd9, and Npr2 were expressed in astrocytes across the CNS. These evaluations of shared genes and pathways represent core features of CNS astrocytes (Fig. 2C,D; Excel file S1). Little is known about the physiology of many astrocyte genes within the top 50 (Fig. 2C).
Region-specific astrocyte genes and pathways
We analyzed astrocyte DEGs in each CNS area relative to the other 12 regions and identified 3,500 region-enriched DEGs (Fig. 2E). Hierarchical clustering of these region-enriched DEGs revealed a trend consistent with anatomical proximity (Fig. 2F). Thus, astrocytes from the cerebrum (OB, MSC, SCX, VCX, HIP and STR) were clustered distinctly from broad regions of the brain stem and spinal cord (TH, HY, MB, VSC, and DSC), and cerebellum (Fig. 2E,F). The number of region-enriched genes varied (fig. S10A), but regional differences were mainly driven by upregulated genes (fig. S10A). Overlap of the top 100 astrocyte region-enriched DEGs is illustrated in fig. S10B for the cerebrum, brain stem/spinal cord, and cerebellum. Clustering for the bulk tissue-specific DEGs, with or without astrocyte-enriched genes for the 13 CNS regions, showed a similar pattern as astrocyte DEGs (fig. S9C-E), suggesting that the regional identity of astrocytes reflects the region-specific microenvironment (reflected in bulk RNA-seq) as well as astrocyte lineage-related gene expression (reflected in IP RNA-seq data).
Because astrocytes from 13 CNS regions clustered into three broad anatomical classes, we identified the top 10 marker genes from the cerebrum, brain stem/spinal cord and cerebellum (Fig 2G). Furthermore, we identified region-specific astrocyte marker genes for each of the 13 CNS areas (Fig. 2H) and classified the top 20 based on their functions (fig. S10). Some of the genes we identified as regionally enriched within astrocytes (e.g. Chrdl1, Crym) were known (9, 14, 22), but many are new. We validated expression of genes for which reliable antibodies were available: IHC for proteins encoded by Sphk1, Ccnd1, Crym, and Sept4 confirmed regional location at the protein level (Fig. 2I-L; N = 4 mice). Additional analyses for the astrocyte region-enriched genes across the 13 CNS regions revealed differences and similarities in transcription factors that may partly explain the inter-regional differences of astrocytes (fig. S11A). GO analysis for the region-enriched DEGs identified putative astrocyte region-specific functions (fig. S11B). We also assessed genes related to astrocyte functions such as phagocytosis, synapses, reactivity (fig. S12), and Ca2+ signaling (fig. S13), noting differences between regions for transmembrane Ca2+ flux pathways and GPCR signaling (fig. S13E-F) that foreshadow functional studies.
Bioinformatic evaluation of astrocyte diversity
To explore how differences between astrocytes from different CNS regions may arise, we performed scRNA-seq for the cortex, hippocampus, and striatum, which we chose because they shared the same anatomical area (Fig. 2E,F) and are widely studied in the field. We asked several questions. Firstly, do subclusters of astrocytes exist within these regions? Next, do different combinations of these subclusters contribute to the regional nature of astrocytes? Moreover, are regional differences between astrocytes related to a metric of the local tissue in which they reside?
We analyzed scRNA-seq data of 89,553 cells from the cortex, hippocampus, and striatum, (Fig. 3A, B; fig. S14A; N = 13 mice). Astrocytes accounted for ~15% (fig. S14B) in accord with expectations (4). The percentage should be treated cautiously as it results from tissue dissociation, but the % of Sox9 and tdTomato positive astrocytes relative to DAPI positive cells (e.g. from fig. S2) in cortex, hippocampus, and striatum was 10 ± 1, 11 ± 1 and 13 ± 1% (N = 3). Further analysis showed that these CNS regions shared seven astrocyte subclusters (AST1 to AST7; Fig. 3C) representative of the cortex, hippocampus, and striatum (Fig. 3D). Genes known to be enriched in cortical (Chrdl1), hippocampal (Gfap), and striatal (Crym) astrocytes showed high expression within their cognate region (Fig. 3E). By analyzing the subclusters we identified the top 100 genes enriched within each (Fig. 3F). Furthermore, the seven subclusters represented different proportions of astrocytes per brain region, implying that regional diversity may be cast partly from variable representation of the subclusters (Fig. 3G). For example, subcluster AST2 was dominant in the cortex, subcluster AST7 was dominant in the hippocampus, and subclusters AST5 and AST6 were dominant in the striatum (Fig. 3G). Upstream analysis for the seven subclusters revealed differences and similarities in the top 5 upstream transcriptional regulators (fig. S14C). We also found upstream regulators related to external cues such as neurotransmitters (e.g. GABA), growth factors (e.g. EGF), and cytokines (e.g. IL9; fig. S14D), implying that astrocyte regional differences were related to tissue-dependent profiles within the local microenvironment. We assessed functional features of the astrocyte subclusters (fig. S14E) and performed candidate gene analyses (fig. S15). Genes related to glucose homeostasis were upregulated in AST2, AST4, AST5, and AST6 (fig. S15A). Related to K+ homeostasis, Kcnj10 was upregulated in AST2 while Atp1a2 and Atp1b2 were upregulated in AST1 (fig. S15B). Subcluster-specific enrichment was observed for genes related to cholesterol homeostasis, neurotransmitter homeostasis, dopamine metabolism, synapse function, and phagocytosis (fig. S15), indicating that combinations of subclusters between CNS regions imparted separable functional features to astrocytes in different CNS regions.
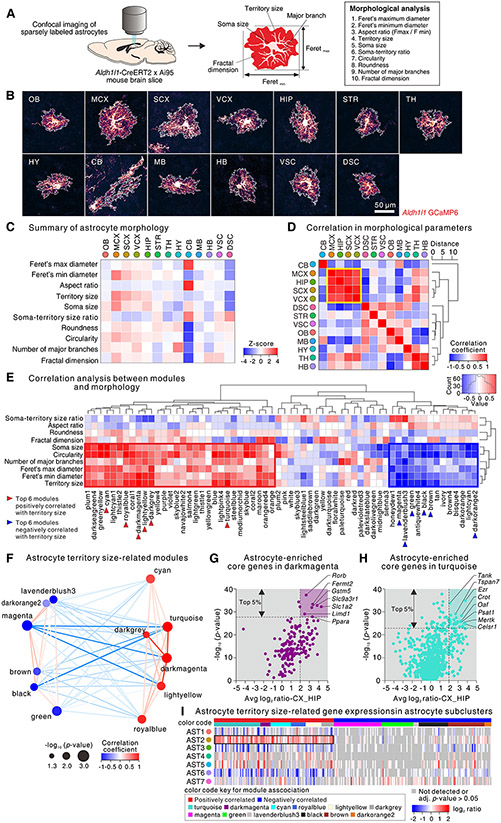
Fig. 3: Origins of astrocyte diversity from regional and scRNA-seq data.
(A) Experimental procedure for scRNA-seq. Single cells were dissociated from the cortex, hippocampus, and striatum of wild type mice (N = 4-7 mice). Data of striatal cells were partly from our recent study (34) and combined with new cortical, hippocampal, and striatal single cell data. (B) Uniform manifold approximation and projection (UMAP) plot of 89,553 brain cells from 3 CNS regions grouped by expression similarity identified 26 major cell populations (10 major cell types). (C) UMAP plot of cluster analysis for 8,898 astrocytes (AST1 and AST2 in panel B) from 3 CNS regions. The cell numbers of astrocyte subclusters are shown as AST1 to AST7. (D) UMAP plot of the astrocyte subclusters to illustrate the three different brain regions examined. (E) UMAP plots from panel C showing expression of Chrdl1, Gfap, and Crym. (F). The heat map shows the top 100 genes that were upregulated in each astrocyte subcluster relative to the others. Example genes are identified. (G) Proportional bar graph showing the percentage of each astrocyte subcluster representation in the 3 CNS regions from scRNA-seq data. (H) Correlations between astrocyte-region specific RNA-seq data (left), bulk RNA-seq data (middle), and bulk RNA-seq minus astrocyte region specific RNA-seq (right) that were gathered as part of Fig 2. The correlation plots show that astrocytes grouped into three broad anatomical areas (a, b, and c).
The existence of several extracellular species in the top 10 upstream regulators for the astrocyte subclusters (e.g. GABA, IL4, EGF, TNF, IL5; fig. S14D) suggested regional astrocyte diversity may reflect the tissue microenvironment, which we explored using astrocyte specific and bulk RNA-seq data (Fig. 2). In Fig. 2 astrocyte RNA-seq data for the 13 CNS regions could be hierarchically clustered into three broad anatomical regions. This trend was also observed when we compared the % shared genes between all 13 regions in a pairwise manner for astrocyte RNA-seq and bulk RNA-seq data (Fig. 3H). Next, we compared the % of shared genes between the astrocyte region-specific RNA-seq data with the bulk RNA-seq data (minus astrocyte enriched genes) and found a similar trend (Fig. 3H). Because the astrocyte-specific RNA-seq data were correlated with the equivalent bulk RNA-seq data per region, these data provide evidence that region specific features were shaped, at least in part, by the local tissue environment (Fig. 3H) as well as by variable representation of the subclusters in a region-specific manner (Fig. 3G). This recalls the finding that many postnatal cortical astrocytes arise from local proliferation, and are likely tuned to their environment (23). Our analyses provide the rationale and data to explore such relationships when specific genetic tools become available.
Astrocyte morphology-related gene networks
A key feature of astrocytes is their morphological complexity. To exploit transcriptomic datasets, we documented the 2-dimensional (2D) morphological features of astrocytes from 13 CNS regions and related these to RNA-seq data using WGCNA. To quantify the morphology of astrocytes we expressed GCaMP6f sparsely in astrocytes, obtained diffraction-limited 2D images from flattened confocal volumes using IHC (Fig. 4A,B; S16), and subjected them to identical evaluations (Fig. 4A, fig. S16). For example, astrocytes in MCX displayed the largest territory size and showed a relatively round shape, whereas astrocytes in CB (Bergmann glia) showed the most elongated shape (fig. S16B-I). We found no differences in the number of major branches per astrocyte from the 13 CNS regions (average: 4.0-5.6; fig. S16J). The fractal dimension (Df), a parameter reflecting morphological complexity, revealed significant differences between astrocytes in 13 CNS regions (fig. S16K-L). Astrocytes in each region showed distinct morphological features, summarized with a z-score (Fig. 4C). By assessing the correlation coefficient for the morphological z-score of astrocytes in 13 CNS regions, we found strong concordance among astrocytes from four closely related forebrain regions (Fig. 4D), recalling RNA-seq (Fig. 2).
Fig. 4: Astrocyte morphology-related gene networks.
(A) Procedure for morphological analysis of astrocytes (an artificial red LUT was used). (B) Representative images of single astrocytes from 13 CNS regions. (C) A heat map of aggregate Z-score of the astrocyte morphological parameters across 13 CNS regions (raw data in fig. S16). (D) A heat map showing the correlation coefficient along with a cluster dendrogram of morphological parameter Z-scores of astrocytes in 13 CNS regions. Four regions (MCX, SCX, VCX, and HIP) that are the most correlated are outlined in yellow. (E) Heat map showing correlation coefficients along with cluster dendrograms among 62 module eigengenes (MEs) in WGCNA as well as the morphological parameters of astrocytes from 13 CNS regions. A histogram in the color scale represents the coefficient values. The WGCNA modules highly correlated with morphological parameters, including for territory size, are outlined in red (positive correlation) or blue (negative correlation). (F) Multidimensional scaling plot of MEs significantly correlated with astrocyte territory size (p value < 0.05). Size of the node indicates p value, color of the node indicates correlation coefficient with astrocyte territory size, and color of the line indicates correlation coefficient between two MEs. (G, H) Scatter plots showing log2 ratio (IP vs input) for the average value from MCX, SCX, VCX, and HIP and -log10 (p value) for correlation of genes within darkmagenta (G) and turquoise (H) modules. Several genes are named and the top 5% are emphasized in the graphs with background coloring. (I) Heat map shows expression of the top 5% morphology-related genes within astrocyte subclusters from 3 CNS regions identified in Fig. 3.
We performed WGCNA for astrocyte RNA-seq data of the 13 CNS regions and identified 62 module eigengenes (MEs; fig. S17). We compared morphological parameters and the 62 modules for astrocytes from the 13 CNS regions and found significant correlations between the two (Fig. 4E). We identified several modules with high correlation to astrocyte morphological parameters (red box in Fig. 4E). Of these, we focused on two modules, “darkmagenta” and “turquoise” (Fig. 4F), because they displayed the highest correlation with other modules (cyan, lightyellow), astrocyte territory size, and with Feret’s min and max (Fig. 4E,F). We analyzed genes within these modules using stringent criteria of > 4-fold enrichment within astrocytes for both hippocampus and cortex (MCX, SCX, VCX), as well as by focusing on the top 5% of genes displaying the highest significance (Fig. 4G,H). Using these criteria, within the darkmagenta module we identified genes Rorb, Fermt2, Gstm5, Slc9a3r1, Slc1a2, Limdl1 and Ppara. Within the turquoise module, we identified genes Tank, Tspan7, Ezr, Crot, Oaf, Psat1, Mertk, and Celsr1. We also assessed morphology-related genes (top 5%) in astrocyte subclusters from scRNA-seq (Fig. 4I). Most of the genes positively correlated with astrocyte territory size were highly expressed in subcluster AST2 and depleted in subcluster AST6. Because astrocytes in the cortex (MCX, SCX, and VCX) displayed larger territory size than the striatum (Fig. 4B,C, S16) and because subcluster AST2 was dominant in the cortex while subcluster AST6 was dominant in the striatum (Fig. 3G), our data suggest that distinct astrocyte subclusters contribute morphological features of astrocytes.
Evaluation of astrocyte morphology-related genes with CRISPR/Cas9
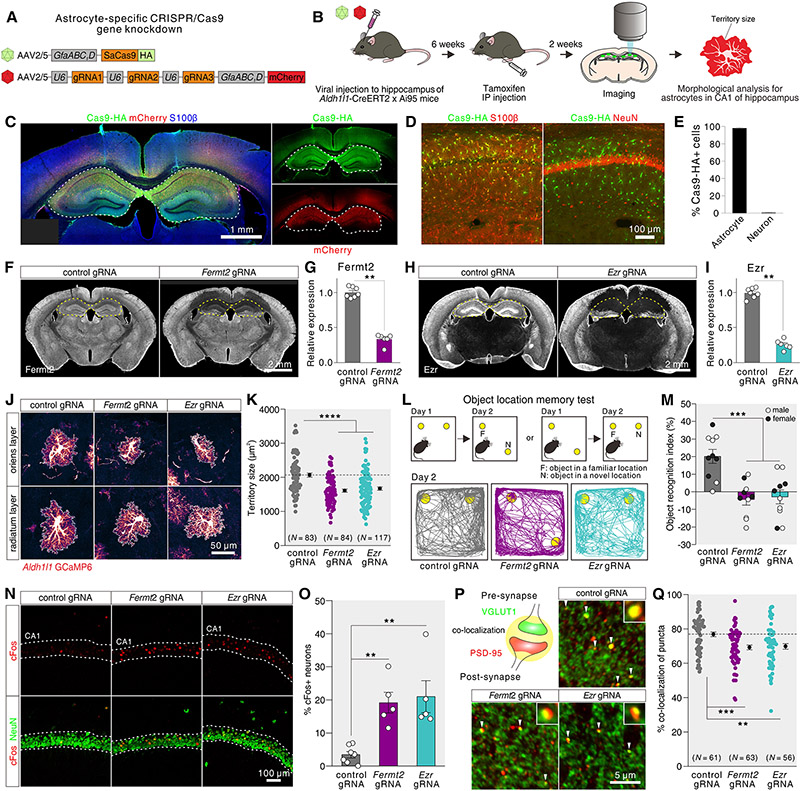
From analyses of astrocyte territory size-related modules (Fig. 4), our attention was drawn to the proteins encoded by Fermt2 and Ezr, i.e. FERMT2 (FERM Domain Containing Kindlin 2) and ezrin, because they are known putative actin interacting proteins that may contribute to astrocyte morphology. To test this possibility, we used astrocyte-specific CRISPR/Cas9-based gene knockdown by expressing Staphylococcus aureus Cas9 (SaCas9) in astrocytes together with gRNAs to reduce expression of candidate genes in astrocyte reporter mice to image astrocyte morphology (Aldh1l1-Cre/ERT2 x Ai95; Fig. 5A-C; fig. S18). We focused on Fermt2 and Ezr, achieving cell-specific SaCas9 expression within astrocytes (Fig. 5D, E N = 3 mice) and 72% and 75% reduction in Fermt2 and Ezr expression relative to controls (Fig. 5F-I; S18A,B; N = 6-7 mice). Having established significant reduction of Fermt2 and Ezr, we assessed the morphology of sparsely labeled hippocampal CA1 astrocytes (Fig. 5J,K; S18C). We found that astrocytes with reduced Fermt2 or Ezr showed significantly reduced territory sizes and Feret’s dimensions by ~20% (22% for Fermt2, 19% for Ezr; Fig. 5J,K; S18D-E) showing that they are involved in maintenance of astrocyte morphology, supporting the predictions from WGCNA (Fig. 4). The observed ~20% reductions in astrocyte morphology (p < 0.0001) are consistent with single gene evaluations (24-28) and suggest that multiple mechanisms contributed to astrocyte morphology, reflected within WGCNA modules.
Fig. 5: Evaluation of astrocyte morphology-related genes with CRISPR/Cas9.
(A) AAVs used for astrocyte-specific CRISPR-Cas9 gene knockdown. (B) Schematic of morphology analysis of hippocampal astrocytes using astrocyte-specific CRISPR/Cas9 gene knockdowns. (C) Exemplar images of brain sections immunostained for SaCas9-HA (green) and mCherry (red). (D) Images of hippocampal astrocytes in 2-3 month wild type mice injected with AAV for astrocyte-specific SaCas9 immunostained for SaCas9-HA (green), S100β (red, left) or NeuN (red, right). (E) Percentage of SaCas9-HA+ astrocytes or neurons in the hippocampus of wild type mice injected with AAV for SaCas9 (N = 3). (F) Exemplar images of sections immunostained with Fermt2 antibody. Brains were microinjected with AAV for astrocyte-specific SaCas9 and with AAV for control gRNAs or AAV for Fermt2 gRNAs in the hippocampus. Loss of Fermt2 immunostaining was observed in the hippocampus. (G) Knocking-down efficiency for Fermt2 protein in the hippocampus of Aldh1l1-Cre/ERT2 x Ai95 mice injected with astrocyte-specific AAV for SaCas9 and AAV for Fermt2 gRNAs (N = 6-7). (H-I) As in F-G, but for Ezr (N = 6-7). (J, K) Images and data for astrocytes in the hippocampal CA1 layers of Aldh1l1-Cre/ERT2 x Ai95 mice injected with AAV for astrocyte-selective SaCas9 and AAV for control gRNAs, Fermt2 gRNAs, or Ezr gRNAs (83-117 astrocytes from N = 3 mice per group). (L, M) Object location memory test to assess control, Fermt2, and Ezr knockdown mice. Schematic, representative traces and average data are shown (N = 10 mice; males and females shown). The differences were significant for both sexes (p = 0.015 for males, p = 0.0076 for females) and for pooled data (p < 0.0001) using ANOVA; sex differences were not our focus and so average data are shown. (N, O) Representative images and average data for cFos positive neurons (NeuN) in the CA1 region of the hippocampus from control, Fermt2, and Ezr knockdown mice (N = 5-7 mice). (P, Q) Representative images and average data for VGLUT1 and PSD-95 co-localization in the CA1 region of the hippocampus from control, Fermt2, and Ezr knockdown mice (25-35 images from N = 5-7 mice per group). The graph shows the % of PSD95 puncta co-localized with VGLUT1. In panel Q, the points represent the average co-localization between VGLUT1 and PSD-95 puncta for each image (see fig. S19 for additional analyses). Data are shown as mean ± SEM.
We also performed experiments using the object location memory test as a measure of cognition in the Fermt2 and Ezr knockdown mice and noted significant impairments relative to controls (Fig. 5L,M; N = 10 mice). At a cellular level, these impairments were accompanied by elevated activity-dependent immediate early gene, cFos, expression in neurons of the CA1 pyramidal cell layer (Fig. 5N, O; N = 5-7 mice) and reduced co-localization between pre- and post-synapse markers PSD-95 and VGLUT1 (Fig. 5P, Q), which was accompanied by reduced immunostaining for these molecules (fig. S19). Thus, these data show that reducing expression of genes predicted from our WGCNA modules to contribute to astrocyte morphology, led to reduced astrocyte morphology, and to cognitive, cellular, and synaptic deficits (Fig. 5).
Astrocyte morphology-related genes in AD
We identified several known AD risk genes (e.g. Apoe, Clu, Fermt2) among the WGCNA module genes that were highly correlated with astrocyte territory size (Fig. 4F-H). These were enriched in astrocytes from the cortex and hippocampus, areas affected in AD (fig. S20A). To explore a possible association between morphology related genes and AD, we performed scRNA-seq on 10-11 month APP/PS1 AD model mice (29) relative to controls in order to identify astrocyte DEGs. We sequenced 45,391 cortical cells, and found 640 astrocyte DEGs (Fig. 6A,B). Eleven DEGs were upregulated relative to controls and 629 were downregulated (p-value < 0.05, log2 fold change > 0.1 or < −0.1; Fig. 6B). From the volcano plots, several of the downregulated genes in AD were related to those correlated with astrocyte morphology, implying that astrocyte territory size may be reduced in AD. We also assessed genes related to astrocyte reactivity and found no significant trend in scRNA-seq of APP/PS1 mice or snRNA-seq of human AD (Fig. S20B), recalling recent work (30).
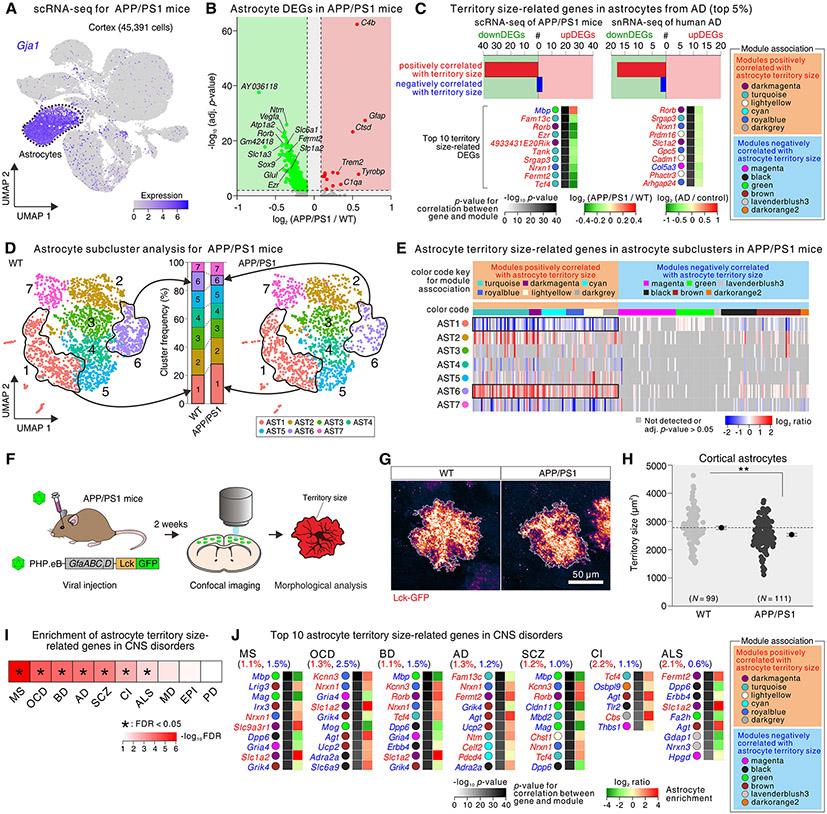
Fig. 6: Astrocyte morphology-related genes in an AD mouse model, human AD, and for other CNS disorders.
(A) UMAP plot (45,391 cells) from the cortex of wild type and APP/PS1 mice grouped by expression similarity identified 10 cell populations. Astrocytes were identified by the expression of Gja1. (B) Volcano plot showing DEGs within astrocytes of APP/PS1 vs wild type mice (adj. p-value < 0.05, log2 fold change > 0.10 or < −0.10); several genes named. (C) The upper graphs show the number of territory size-related genes from the up and downregulated DEGs in astrocytes that were altered in scRNA-seq data from APP/PS1 mice and in human AD snRNA-seq data. The heat maps show the top 10 territory size-related genes that displayed high log10 p values in the WGCNA analyses (from the top 5% from WGCNA). The WGCNA modules that the genes belong to are on the right. (D) UMAP plot of cluster analysis (3,808 astrocytes) from wild type (WT) and APP/PS1 mice in relation to the bar graph showing cluster frequency of astrocyte subclusters for wild type (WT) and APP/PS1 mice (N = 9-10 mice). (E) Heat map shows expression of the morphology-related genes (top 5% from Fig. 4) within astrocyte subclusters in APP/PS1 mice. Genes positively correlated with astrocyte morphology were enriched in cluster 6, which was reduced in APP/PS1 mice. In contrast, genes positively correlated with morphology were depleted in cluster 1, which was increased in APP/PS1 mice. (F) Procedure for morphology analysis of cortical astrocytes in APP/PS1 mice. (G) Images of single astrocytes from the cortex labeled with Lck-GFP in wild type (WT) and APP/PS1. (H) Analysis of Lck-GFP labeled astrocyte territory size in the cortex of WT and APP/PS1 mice (N = 99-111 astrocytes from 3 mice per group). (I) Hypergeometric heat map shows enrichment of astrocyte territory related genes within those associated with CNS disorders. * indicates significance with a FDR < 0.05. (J) Top 10 astrocyte territory size-related genes that overlapped with genes associated with AD, amyotrophic lateral sclerosis (ALS), cerebral infarction (CI), multiple sclerosis (MS), schizophrenia (SCZ), bipolar disorder (BD), and obsessive-compulsive disorder (OCD). Data are shown as mean ± SEM.
We determined how many of the astrocyte territory size-related genes (from the top 5%, Fig. 4F-H) were also significantly up or downregulated DEGs in APP/PS1 mice (adjusted p < 0.05). We found that 40 genes that were positively correlated with astrocyte territory size were downregulated, and 5 genes that were negatively correlated with astrocyte territory size were up regulated in APP/PS1 mice, and this was also seen in human AD snRNA-seq data (Fig. 6C) (31). To further explore these associations, we performed subcluster analysis for astrocytes in APP/PS1 mice and found that wild type and transgenic APP/PS1 mice shared seven astrocyte subclusters in distinct proportions, whereby subcluster AST1 was significantly increased and subcluster AST6 was significantly decreased in APP/PS1 mice compared with wild type mice (Fig. 6D; fig. S20D). We assessed expression of the top 5% morphology-related genes in astrocyte subclusters from APP/PS1 mice. Most of the genes positively correlated with astrocyte territory size were highly expressed in subcluster AST6 and depleted in subcluster AST1 (Fig. 6E), indicating that astrocyte territory size may be reduced in the context of AD. To test this, we injected AAV PHP.eB:GfaABC1D Lck-GFP (32) into the retro-orbital sinus of 9 month wild type and APP/PS1 mice and measured territory sizes of sparsely labeled astrocytes at ~10 months age (Fig. 6F). Astrocyte territory sizes were significantly reduced in the cortex of APP/PS1 mice relative to controls (Fig. 6G,H; fig. S20E), which was in accord with our predictions (Fig. 5), and notable because astrocytes were assessed irrespective of subcluster identity. Furthermore, because physiological functions of astrocytes are implicated in pathophysiology of AD (7, 33), we performed candidate gene analyses for the astrocyte subclusters and found that genes related to cholesterol homeostasis, K+ homeostasis, neurotransmitter homeostasis, and synapse function were enriched in subcluster AST6 and depleted in subcluster AST1 (fig. S20F-I). Note that scRNA-seq data lack spatial information, which could be evaluated by mapping of astrocyte types in relation to plaque markers in AD model mice and in human tissue. Our data on disease-specific astrocyte subclusters (AST1 and AST6) suggest these as potential therapeutic targets for AD.
Astrocyte morphology-related genes in other CNS disorders
Our analysis of astrocytes in wild type mice identified genes associated with astrocyte territory sizes (Fig. 4), which we tested with CRISPR/Cas9 mediated knockdown (Fig. 5). Furthermore, these genes were downregulated in human AD and in a mouse model of AD that displayed reduced astrocyte territory sizes (Fig. 6). To further explore such associations we evaluated astrocytes from Huntington’s disease (HD), because they displayed reduced territory sizes (27, 28, 34, 35). In accord with our AD findings, we found downregulation of genes positively correlated with astrocyte territory sizes, and upregulation of genes negatively correlated with astrocyte territories in a HD mouse model and postmortem human HD tissue (Fig. S21) (36). To explore the association between astrocyte territory-related genes and other common CNS disorders, we identified disease-associated genes from Phenopedia (37) for AD, Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), HD, cerebral infarction (CI), multiple sclerosis (MS), epilepsy (Epi), schizophrenia (SCZ), major depression (MD), bipolar disorder (BD), and obsessive-compulsive disorder (OCD) and performed hypergeometric analyses. Significant enrichment of astrocyte territory related genes was found for MS, OCD, BD, AD, SCZ, CI, and ALS (Fig. 6I). Fig. 6J shows the top 10 astrocyte territory size-related genes that overlapped with the genes associated with these CNS disorders, indicating astrocyte morphological changes may be a common underappreciated feature of diverse CNS disorders.
Discussion
There is renewed interest in understanding astrocyte biology in health and disease, necessitating exploration of astrocytes with high-dimensional molecular data in different regions (9-18). By comparing astrocytes across 13 CNS regions, we found ~850 genes that were shared, with functions related to metabolism, cholesterol biosynthesis, and neurotransmitter uptake and biosynthesis. These represent the core functions of astrocytes, i.e. to maintain tissue homeostasis and this realization is important because many of these pathways are disrupted in brain disorders. However, from the top genes shared between astrocytes, little is known about approximately a third, implying that fully understanding core functions of astrocytes across CNS areas is an important experimental goal. For example, K+ homeostasis is a core function of astrocytes and both Kir4.1 and ATP1A2 contribute (38). Our data show that both Kcnj10 and Atp1a2 are highly expressed in astrocytes across the CNS. Further studies are needed to explore which dominates under specific settings. Across all regions, we found little evidence for the molecular machinery supporting astrocyte Ca2+-dependent vesicular glutamate release. We found abundant evidence that cholesterol metabolism and fatty acid β-oxidation were core functions of astrocytes.
Our data provided strong evidence to indicate that astrocytes have molecular features and functions specific to CNS regions. Our data suggested that these region-specific functions arise from the local tissue environment and by variable representation of subclusters AST1 to AST7 in a region-specific manner. Once specific genetic tools become available, one could disentangle these two contributions to astrocyte diversity. Our data also provide molecules that are highly enriched within astrocytes from specific brain regions. For example, Chrdl1 is highly enriched in cortical astrocytes and regulates synaptic function (22). Crym, encoding μ-crystallin, was unique to striatal astrocytes, but its functional relevance is unexplored. Region-specific genes suggest it may be possible to target astrocytes in a brain region-specific manner to interrogate neural circuits and for therapeutic strategies.
A cardinal feature of astrocytes is their bushy morphology. We mined our data to explore the molecular underpinning of morphological differences between astrocytes from different CNS regions: several previously proposed molecules were detected, including Ntrk2 (39), Hepacam, Gja1 (24), Gjb6 (40), Nlgn1-3 (25), β-integrin signaling genes (41), and Mertk, Chrdl1, Cyp4f15 (14, 42). We then identified gene networks related to astrocyte morphology across 13 CNS regions by assessing the correlation of 10 morphological parameters to modules from astrocyte RNA-seq. To test the hypothesis that the astrocyte morphology-related genes contributed to morphology, we developed astrocyte-specific CRISPR/Cas9-based gene knockdown to reduce expression of core genes in the hippocampus where astrocytes showed highest morphological complexity. We found significant reduction of hippocampal astrocyte territory sizes following reduction of Fermt2 and Ezr proteins. We also observed concomitant changes in a cognitive task, cFos expression in neurons, and co-localization of pre- and post-synapse markers. This suggested that astrocyte morphological changes that accompany disease states have causal effects on neural circuit and synaptic function, thus contributing to disease phenotypes. Our molecular studies portend physiological studies to explore these relationships systematically for AD across age and sex.
Unexpectedly, we identified several known AD risk genes (Apoe, Clu, Fermt2) as significant astrocyte morphology-related genes. This is notable, because astrocyte morphology changes in neurological and psychiatric diseases, including AD (43-46). Moreover, disease-associated astrocytes displayed higher Gfap expression in an AD mouse model (47). By exploring associations between astrocyte morphology-related genes and AD, we found that positively correlated morphology-related genes were predominantly downregulated. A similar trend was also observed for human AD (31) and for mouse and human HD (28, 34, 35). Because our findings predicted that astrocyte territory size might be reduced in the context of AD, we assessed astrocyte territory sizes in APP/PS1 mice and found that astrocyte territory sizes were indeed significantly reduced in the cortex of APP/PS1 mice. In subcluster analysis for astrocytes, we identified seven astrocyte subclusters in distinct proportions. Subcluster AST1 was significantly increased and subcluster AST6 was significantly decreased in APP/PS1 mice. The increased subcluster AST1 population was characterized by higher Gfap, and reduced homeostatic and morphology-related genes in APP/PS1 mice. On the other hand, subcluster AST6 that was decreased was characterized by lower Gfap, and by upregulated homeostatic and morphology-related genes. These data indicate dynamic alterations of astrocytes in response to early pathological states in an amyloid AD model, reflecting morphology and cellular composition.
We provide comprehensive molecular data permitting new physiological and hypothesis-driven experiments for astrocytic genes and pathways that are core features across the CNS and for those unique to specific regions. Our findings identify the molecular basis of complex astrocyte morphology and indicate that reduced astrocyte morphological complexity, and the attendant loss of homeostatic and metabolic tissue support, is a common feature of CNS disorders that is potentially amenable to therapeutic manipulation.
Methods summary
All animal experiments were conducted in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals and were approved by the Chancellor’s Animal Research Committee at the University of California, Los Angeles (UCLA). Mice were housed in the vivarium managed by the Division of Laboratory Animal Medicine at UCLA with a 12 hr light/dark cycle and ad libitum access to food and water. All animals were healthy with no obvious behavioral phenotype at the start of the experiments other than those expected, were not involved in previous studies, and were sacrificed during the light cycle. Mice were assigned randomly to the experiments based on the genotype. Wild-type C57BL/6NTac mice were maintained in an in-house breeding colony or purchased from Taconic Biosciences. Ai95 (B6;129S-Gt(ROSA)26Sortm95.1(CAG-GCaMP6f)Hze/J, JAX Stock #028865) or Ai14 (B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J, JAX Stock #007908) mice were acquired from JAX and crossed with Aldh1l1-Cre/ERT2 BAC mice (B6N.FVB-Tg(Aldh1l1-Cre/ERT2)1Khakh/J, JAX Stock #029655) fully backcrossed to C57BL/6NTac background in an in-house breeding colony. RiboTag mice (B6N.129-Rpl22tm1.1Psam/J, JAX Stock # 011029) were acquired from JAX and crossed with Aldh1l1-Cre/ERT2 mice. APP/PS1dE9 mice (B6;C3-Tg(APPswe,PSEN1dE9)85Dbo/Mmjax, JAX MMRRC Stock #034829-JAX) were acquired from JAX MMRRC and maintained in an in-house breeding colony. (APP/PS1 mice carry human transgenes for both APP bearing the Swedish mutation and PSEN1 the deletion mutation of exon 9, both under the control of the mouse prion promoter).
All statistical tests were performed in GraphPad Prism 9. Power analyses were performed in Graphpad StatMate 2.0. The graphs were created in OriginPro 2018b and assembled in CorelDraw 2018. Data are presented as mean ± SEM along with the individual data points. Of note, in some of the graphs, the error bars representing the SEM were smaller than the symbols representing the mean. OriginPro 2018b was used to determine whether the data were normally distributed or not. Parametric tests were used for normally distributed datasets while non-parametric tests were applied to data not normally distributed or data with a small sample size. For comparisons of two groups, paired and unpaired two-tailed Student’s t-tests or two-tailed Mann-Whitney tests were used. For comparisons of more than three groups, one-way ANOVA tests followed by Tukey’s or Kruskal-Wallis tests were used. In the figures, p values were stated by asterisk(s): *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. When p values were greater than 0.05, it was stated as non-significant (n.s.). The number of replicates (n values), exact p values, sample numbers and details of statistical analyses in each case are stated in Table S4 for every experiment.
Supplementary Material
Acknowledgments
Thanks to F. Gao and the UCLA Neuroscience Genomics Core for assistance with RNA-seq. The sequencing core was supported partly by the Genetics, Genomics and Informatics Core of the Semel Institute of Neuroscience. Thanks to M. Gangwani for AAV delivery (Fig. 6F) and discussions, S. Cheong for help counting astrocytes (Fig 1D), and D. Geschwind for discussions. Thanks to A. Huang and L. Wu for comments and N. Okabe for IHC guidance.
Funding:
This work, BSK, JS, FE, and XY were supported by the National Institutes of Health (R01DA047444, R35NS111583, R01AG0759655), an Allen Distinguished Investigator Award, a Paul G. Allen Frontiers Group advised grant of the Paul G. Allen Family Foundation, and the Ressler Family Foundation (BSK). JSS was supported by the National Science Foundation Graduate Research Fellowship Program (NSF-GRFP; DGE-2034835), by the UCLA Eugene V. Cota-Robles Fellowship. AK and HH were supported by JSPS KAKENHI JP21K19335 (HH), JP20H00492 (HH), JP20H03391 (AK); MEXT KAKENHI JP18H05416 (HH); AMED JP21dm0207117 (HH); grants from the Takeda Science Foundation, Japan (AK, HH). VG was supported by a NIH Pioneer Award (DP1OD025535), the Vallee Foundation, the CZI Neurodegeneration Challenge Network, and the Beckman Institute for CLARITY, Optogenetics and Vector Engineering Research (CLOVER) for technology development and dissemination.
Footnotes
Competing interests: the authors declare no competing interests related to this manuscript.
References and notes
- 1.Bargmann C et al. , BRAIN 2025: a scientific vision. Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Working Group Report to the Advisory Committee to the Director, NIH, (2014). [Google Scholar]
- 2.Insel TR, Landis SC, Collins FS, Research priorities. The NIH BRAIN Initiative. Science (New York, N.Y.) 340, 687–688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barres BA, The mystery and magic of glia: a perspective on their roles in health and disease. Neuron 60, 430–440 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Allen NJ, Lyons DA, Glia as architects of central nervous system formation and function. Science (New York, N.Y.) 362, 181–185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu X, Nagai J, Khakh BS, Improved tools to study astrocytes. Nature reviews. Neuroscience 21, 121–138 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Nagai J et al. , Behaviorally consequential astrocytic regulation of neural circuits. Neuron 109, 576–596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee HG, Wheeler MA, Quintana FJ, Function and therapeutic value of astrocytes in neurological diseases. Nature reviews. Drug discovery, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelley KW, Nakao-Inoue H, Molofsky AV, Oldham MC, Variation among intact tissue samples reveals the core transcriptional features of human CNS cell classes. Nature neuroscience 21, 1171–1184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chai H et al. , Neural circuit-specialized astrocytes: transcriptomic, proteomic, morphological and functional evidence. Neuron 95, 531–549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akdemir ES et al. , Lunatic Fringe-GFP Marks Lamina-Specific Astrocytes That Regulate Sensory Processing. The Journal of neuroscience : the official journal of the Society for Neuroscience, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang AY et al. , Region-Specific Transcriptional Control of Astrocyte Function Oversees Local Circuit Activities. Neuron 106, 992–1008.e1009 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.John Lin CC et al. , Identification of diverse astrocyte populations and their malignant analogs. Nature neuroscience 20, 396–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lozzi B, Huang TW, Sardar D, Huang AY, Deneen B, Regionally Distinct Astrocytes Display Unique Transcription Factor Profiles in the Adult Brain. Frontiers in neuroscience 14, 61 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bayraktar OA et al. , Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map. Nature neuroscience 23, 500–509 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molofsky AV et al. , Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature 509, 189–194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Batiuk MY et al. , Identification of region-specific astrocyte subtypes at single cell resolution. Nature communications 11, 1220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boisvert MM, Erikson GA, Shokhirev MN, Allen NJ, The Aging Astrocyte Transcriptome from Multiple Regions of the Mouse Brain. Cell reports 22, 269–285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morel L et al. , Molecular and Functional Properties of Regional Astrocytes in the Adult Brain. The Journal of neuroscience : the official journal of the Society for Neuroscience 37, 8706–8717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seiriki K et al. , High-Speed and Scalable Whole-Brain Imaging in Rodents and Primates. Neuron 94, 1085–1100.e1086 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Srinivasan R et al. , New Transgenic Mouse Lines for Selectively Targeting Astrocytes and Studying Calcium Signals in Astrocyte Processes In Situ and In Vivo. Neuron 92, 1181–1195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun W et al. , SOX9 Is an Astrocyte-Specific Nuclear Marker in the Adult Brain Outside the Neurogenic Regions. The Journal of neuroscience : the official journal of the Society for Neuroscience 37, 4493–4507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanco-Suarez E, Liu TF, Kopelevich A, Allen NJ, Astrocyte-Secreted Chordin-like 1 Drives Synapse Maturation and Limits Plasticity by Increasing Synaptic GluA2 AMPA Receptors. Neuron 100, 1116–1132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ge WP, Miyawaki A, Gage FH, Jan YN, Jan LY, Local generation of glia is a major astrocyte source in postnatal cortex. Nature 484, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baldwin KT et al. , HepaCAM controls astrocyte self-organization and coupling. Neuron 109, 2427–2442 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stogsdill JA et al. , Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature 551, 192–197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou B et al. , Astroglial dysfunctions drive aberrant synaptogenesis and social behavioral deficits in mice with neonatal exposure to lengthy general anesthesia. PLoS biology 17, e3000086 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang R, Diaz-Castro B, Tong X, Looger LL, Khakh BS, Dysfunctional calcium and glutamate signaling in striatal astrocytes from Huntington's disease model mice. The Journal of neuroscience : the official journal of the Society for Neuroscience 36, 3453–3470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Octeau JC et al. , An Optical Neuron-Astrocyte Proximity Assay at Synaptic Distance Scales. Neuron 98, 49–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jankowsky JL et al. , Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Human molecular genetics 13, 159–170 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Jiwaji Z et al. , Reactive astrocytes acquire neuroprotective as well as deleterious signatures in response to Tau and Aß pathology. Nature communications 13, 135 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathys H et al. , Single-cell transcriptomic analysis of Alzheimer's disease. Nature 570, 332–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Challis RC et al. , Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nature protocols 14, 379–414 (2019). [DOI] [PubMed] [Google Scholar]
- 33.De Strooper B, Karran E, The Cellular Phase of Alzheimer's Disease. Cell 164, 603–615 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Yu X et al. , Context-Specific Striatal Astrocyte Molecular Responses Are Phenotypically Exploitable. Neuron 108, 1146–1162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benraiss A et al. , Cell-intrinsic glial pathology is conserved across human and murine models of Huntington's disease. Cell reports 36, 109308 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Lee H et al. , Cell Type-Specific Transcriptomics Reveals that Mutant Huntingtin Leads to Mitochondrial RNA Release and Neuronal Innate Immune Activation. Neuron 107, 891–908 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu W, Clyne M, Khoury MJ, Gwinn M, Phenopedia and Genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics (Oxford, England) 26, 145–146 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacAulay N, Molecular mechanisms of K(+) clearance and extracellular space shrinkage-Glia cells as the stars. Glia 68, 2192–2211 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Holt LM et al. , Astrocyte morphogenesis is dependent on BDNF signaling via astrocytic TrkB.T1. eLife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pannasch U et al. , Connexin 30 sets synaptic strength by controlling astroglial synapse invasion. Nature neuroscience 17, 549–558 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Cho S, Muthukumar AK, Stork T, Coutinho-Budd JC, Freeman MR, Focal adhesion molecules regulate astrocyte morphology and glutamate transporters to suppress seizure-like behavior. Proceedings of the National Academy of Sciences of the United States of America 115, 11316–11321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanjakornsiripan D et al. , Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nature communications 9, 1623 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arranz AM, De Strooper B, The role of astroglia in Alzheimer's disease: pathophysiology and clinical implications. The Lancet. Neurology 18, 406–414 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Pekny M et al. , Astrocytes: a central element in neurological diseases. Acta neuropathologica 131, 323–345 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Pekny M, Pekna M, Astrocyte reactivity and reactive astrogliosis: costs and benefits. Physiological reviews 94, 1077–1098 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Escartin C et al. , Reactive astrocyte nomenclature, definitions, and future directions. Nature neuroscience 24, 312–325 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Habib N et al. , Disease-associated astrocytes in Alzheimer's disease and aging. Nature neuroscience 23, 701–706 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seiriki K et al. , Whole-brain block-face serial microscopy tomography at subcellular resolution using FAST. Nature protocols 14, 1509–1529 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson WE, Li C, Rabinovic A, Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England) 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Law CW, Chen Y, Shi W, Smyth GK, voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biology 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langfelder P, Horvath S, WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu YE, Pan L, Zuo Y, Li X, Hong W, Detecting Activated Cell Populations Using Single-Cell RNA-Seq. Neuron 96, 313–329 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Belkina AC et al. , Automated optimized parameters for T-distributed stochastic neighbor embedding improve visualization and analysis of large datasets. Nature communications 10, 5415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shigetomi E et al. , Imaging calcium microdomains within entire astrocyte territories and endfeet with GCaMPs expressed using adeno-associated viruses. J Gen Physiol 141, 633–647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ran FA et al. , In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E, CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic acids research 42, W401–407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW, GenBank. Nucleic acids research 38, D46–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanjana NE, Shalem O, Zhang F, Improved vectors and genome-wide libraries for CRISPR screening. Nature methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chan KY et al. , Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nature neuroscience 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar SR et al. , Multiplexed Cre-dependent selection yields systemic AAVs for targeting distinct brain cell types. Nature methods 17, 541–550 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagai J et al. , Hyperactivity with Disrupted Attention by Activation of an Astrocyte Synaptogenic Cue. Cell 177, 1280–1292 e1220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.