Abstract
Purpose
Pseudoprogression mimicking recurrent glioblastoma remains a diagnostic challenge that may adversely confound or delay appropriate treatment or clinical trial enrollment. We sought to build a radiomic classifier to predict pseudoprogression in patients with primary isocitrate dehydrogenase wild type glioblastoma.
Methods and Materials
We retrospectively examined a training cohort of 74 patients with isocitrate dehydrogenase wild type glioblastomas with brain magnetic resonance imaging including dynamic contrast enhanced T1 perfusion before resection of an enhancing lesion indeterminate for recurrent tumor or pseudoprogression. A recursive feature elimination random forest classifier was built using nested cross-validation without and with O6-methylguanine–DNA methyltransferase status to predict pseudoprogression.
Results
A classifier constructed with cross-validation on the training cohort achieved an area under the receiver operating curve of 81% for predicting pseudoprogression. This was further improved to 89% with the addition of O6-methylguanine–DNA methyltransferase status into the classifier.
Conclusions
Our results suggest that radiomic analysis of contrast T1-weighted images and magnetic resonance imaging perfusion images can assist the prompt diagnosis of pseudoprogression. Validation on external and independent data sets is necessary to verify these advanced analyses, which can be performed on routinely acquired clinical images and may help inform clinical treatment decisions.
Introduction
Glioblastoma is the most common primary brain cancer and is rapidly fatal with a median survival of 12 to 18 months and 5-year survival of 9.8%.1,2 Standard-of-care treatment consists of maximal safe surgical resection followed by radiation therapy with concomitant and adjuvant temozolomide.2 Recent advances in the field include improved understanding of the molecular drivers of disease and the prognostic significance of isocitrate dehydrogenase (IDH) mutations and O6-methylguanine–DNA methyltransferase (MGMT) promoter methylation status.3 Parallel advances in neuroimaging include the development and adoption of standardized brain tumor imaging protocols and standardized response criteria to improve outcome homogeneity across clinical trials.4, 5, 6 These clinical trials often disappoint7; one vexing challenge is the difficulty of distinguishing between the overlapping manifestations of recurrent tumor and pseudoprogression, as both may manifest with new and/or increasing enhancing lesions on magnetic resonance imaging (MRI). Pseudoprogression, a transient early form of treatment-related change, occurs in 25% to 30% of glioblastomas.5 Pseudoprogression is associated with MGMT promoter status, is often asymptomatic, and has spontaneous stabilization or resolution.5,8 The uncertainty in diagnosis has prompted recommendations for additional follow-up scans and exclusion from clinical trials for the first 3 months after completing chemotherapy and radiation therapy in the Modified Criteria for Radiographic Response Assessment in Glioblastoma Clinical Trials.5 Some patients will undergo repeat surgery out of concern for recurrent tumor, albeit pseudoprogression is self-limited by definition and does not require resection. Advanced imaging techniques such as MRI perfusion, diffusion, spectroscopy, and positron emission tomography (PET) are helpful,9, 10, 11, 12, 13 but their acquisition, analysis, interpretation, and availability remain highly variable, making better imaging techniques and analyses necessary for the prompt and accurate diagnosis of pseudoprogression.
Discovery of a more specific noninvasive imaging biomarker would help patients avoid unnecessary surgery and treatment delays, expedite triage for clinical trials, and inform physician-patient counseling and treatment decisions. Radiomics is the high-throughput quantitative analysis of digital medical images, and machine learning techniques provide robust tools to this end. We hypothesize that sophisticated radiomic modeling of contrast T1-weighted images and perfusion images incorporating both structural and biologic data will enable the accurate distinction of pseudoprogression from recurrent tumor in patients with glioblastoma. Because of the innate biological and imaging differences between IDH wild type glioblastomas and IDH mutant “glioblastomas” (with only the former likely to remain in an upcoming revision to the World Health Organization classification), this study aimed to develop a machine learning model using standard radiomic features computed from perfusion MRI to predict histopathologically confirmed pseudoprogression in a homogenous cohort of IDH wild type primary glioblastomas.
Methods and Materials
Patient cohort
From a single National Cancer Institute Designated Comprehensive Cancer Center, we queried institutional and departmental databases for adult patients ≥18 years who had primary IDH wild type glioblastoma treated with standard of care maximal resection, radiation therapy and temozolomide, and adjuvant temozolomide. All patients had brain MRI with perfusion ≤6 weeks before repeat resection for worsening (new and/or increasing size) enhancing lesion in the radiation field indeterminate for recurrent tumor or pseudoprogression, with the repeat resection occurring <1 year after completing radiation therapy. Chart reviews were performed by CNS-specializing radiation oncologists (EW and ATW, with 3 and 4 years of experience, respectively). This study was approved by the local Institutional Review Board and performed under a Waiver of Informed Consent following Health Insurance Portability and Accountability Act regulations under retrospective protocol #16-552. The patient cohort and mutation testing details are provided in Supplemental Methods (in the Supplemental Materials).
Lesion diagnosis at repeat surgery
An experienced board-certified neuropathologist (TAB, 7 years of experience) reviewed the histopathology specimens of all patients and determined lesion outcomes in a binary manner: if any tumor was present, the lesion was classified as “tumor.” If no tumor was present, the lesion was classified as “non-tumor” (pseudoprogression).
MRI scans
MRI scans including contrast-enhanced axial T1-weighted images and whole brain axial dynamic contrast-enhanced (DCE) T1 perfusion images were acquired at 3T (65%) and 1.5T using 14 different MRI scanners consisting of 5 different scanner types (Signa Excite, Signa HDxt, Signa PET/MR, Discover 750w, Optima 450w; GE Healthcare, Chicago, IL). MRI including MRI perfusion was performed as part of the standard of care for glioblastoma patients every 2 months. Three hundred radiomic features were computed with 100 features from each contrast T1-weighted image as well as Ktrans and plasma volume (VP) maps. Details of MRI acquisition and segmentation, image preprocessing, and radiomic feature extraction using the open-source Computational Environment for Radiological Research (CERR) software14 are provided in Supplemental Methods. Ktrans is the volume transfer coefficient quantifying contrast leakiness from the intravascular compartment into the extravascular extracellular space – and is affected by blood flow, capillary wall surface area, capillary permeability and blood-brain barrier disruption.15 VP is a measure of blood volume, analogous to cerebral blood volume calculated from dynamic susceptibility contrast T2* perfusion images, and is correlated with histopathologic and angiographic vascularity and thereby neoangiogenesis. Ktrans and VP have been proposed as useful techniques to determine recurrent glioblastoma from pseudoprogression.10,11,16
Radiomic feature extraction
Three hundred radiomic features from T1-weighted MR images and Ktrans and VP maps were computed using the open-source CERR software.14 CERR extracts several radiomic features including first order histogram features and second order features using gray level correlation matrix, gray level size zone, gray level run length matrix, shape-based metrics, and edge descriptors including Sobel and directional Gabor edge filters, all of which are Image Biomarker Standard Initiative (IBSI) compliant.17 The default settings were used for extracting features.
Machine learning classification for distinguishing tumor versus pseudoprogression
Feature selection
Feature preselection was based on maximum relevance and minimum redundancy (MRMR),18 whereby the top 80% of features (corresponding to 0.8 threshold) correlated with response (pseudoprogression vs tumor) were selected. For robust feature selection resilient to sample variability, we used an ensemble MRMR approach to combine multiple sets of MRMR feature selections. Ensemble MRMR feature selections were performed using a computationally fast ensemble method in the mRMRe package available in R version 3.3.0 (The R Foundation).18 For our ensemble MRMR model, we used 10 ensembles and a subset of 50 cases with a fixed causality measure of −0.2 to select the features. This step reduced the number of features from 300 to 70. A subset of 50 random cases was used in lieu of the whole set to prevent favorable biasing of the feature preselection step and to reduce the chance of producing an overoptimistic classification model.
Recursive feature elimination random forest (RFE-RF) classifier without and with MGMT status
Classifiers were constructed using recursive feature elimination random forests (RFE-RF) with 250 decision trees to classify patients by response (residual tumor vs pseudoprogression). The classifier was constructed by first eliminating redundant features through the feature preselection step. A radiomic classifier was trained using repeated (n = 5) 3-fold nested cross-validation with the preselected 70 features from the feature selection step to ensure reasonable generalization to unseen data. Training was performed by splitting the data into 3 distinct folds, where in each fold, models were constructed from two-thirds of the data, with the remaining one-third data held out for testing the model. Nested cross-validation was performed by optimizing for the hyperparameter; specifically, the number of features used in constructing the classifier6,10,15,19,20 was optimized within the individual training folds. The results were reported for the held-out testing set in each cross-validation fold. The hyperparameter optimization was done within each fold, which prevented data used in the held-out testing to ever be used in training in that fold.
The RFE-RF classifier then combined both an explicit recursive feature elimination-based feature selection and implicit feature selection through the random forest model to obtain a robust classifier, despite having an initially large set of radiomic features,19,20 to reduce the chance of overfitting when using many features in a classifier.21 The explicit feature selection was performed by the recursive feature elimination whereby the number of relevant set of features were identified. The random forest classifier performed implicit feature selection by ranking the relevance of the individual features with respect to classification by computing a Gini importance score,20 which measured the overall probability of misclassification when using a given feature in the individual trees of the RF classifier.21 Features with Gini feature importance of > 0 were considered relevant to the classification. All features were scaled using z score normalization centered using the feature mean and variance for the individual features. Near zero-variance predictors or features with one unique value (zero variance) and those features with few unique values relative to the number of samples (default percentage of unique values less than 10%) and large ratio of frequency of the most common value (default frequency ration of 95/5) were automatically identified and removed from being used in the classifier construction. The classifiers were implemented using the Caret package in the R version 4.0.2. The set of radiomic features used in the classifiers are listed in the Table E1.
In addition to the radiomic classifier, we also evaluated the performance of a classifier combining MGMT methylation status with radiomic features. Researchers have previously demonstrated that combining genomic information such as the MGMT methylation with radiomics improved the performance of outcome classification in a treatment-naïve glioblastoma cohort.20 This combined classifier treated the binary MGMT status as one hot encoded feature (positive and negative classifications were treated as separate features) to increase its chance of inclusion a priori. One hot encoding is a frequently used method in machine learning to handle categorical data, where different categories of the data are treated as separate features (eg, MGMT positive as one feature and MGMT negative as another feature). We evaluated the utility of performing feature preselection using MRMR by constructing the radiomics-only classifier using all of the 300 radiomic features. The same model settings as used for radiomics-only and radiomics + MGMT classifier was used.
Finally, we benchmarked the performance of the classifier against a simple model constructed using known clinical predictors, MGMT, age, and gender using a generalized linear regression model trained using 3-fold cross-validation using the Caret package. Results produced on the held-out data in each fold was used to compute accuracy.
Statistical analysis
Nonparametric Wilcoxon signed-rank tests were performed to determine the association of radiomic measures with lesion diagnosis (tumor recurrence vs pseudoprogression). Significance was set to P < .05.
Results
Patients and lesion diagnosis
Between August 2011 and October 2016, a total of 505 patients were newly diagnosed with glioblastoma and received radiation therapy and chemotherapy. Only 24 patients (4.8%) had IDH mutant glioblastomas. Of the 108 patients (21.4%) who underwent repeat brain surgery, a total of 74 patients met all inclusion criteria and each patient had only one lesion. The median age was 58 years (range, 27-77) and slightly more than two-thirds were male (n = 52, 70.3%). In addition to IDH wild type status, most patients with available molecular sequencing data demonstrated canonical molecular features of glioblastoma with alterations in either EGFR or TERT (n = 51/74, 68.9%).
All patients received partial brain radiation therapy using biologically similar doses at 6000 cGy (n = 66), 5940 cGy (n = 6), or 5400 cGy (n = 2) with concomitant and adjuvant temozolomide. MRI scans were acquired a median of 6.5 months after the end of radiation therapy (range, 0.2-11.9) and a median of 2 days before repeat surgery (range, 1-38).
Histopathology after repeat surgery revealed more patients with recurrent tumors (n = 57, 77%) than with pseudoprogression (n = 17, 23%). The time from the end of radiation therapy to repeat surgery was similar between patients with recurrent tumors (median time, 5.1 months; range, 0.2-12.0), and patients with pseudoprogression (median time, 5.0 months; range, 0.9-12.0) (P = .51). Representative cases are shown in Fig. 1. In tumors with known MGMT promoter status (n = 69, 93.2%), there were more unmethylated (n = 60, 87%) than methylated (n = 9, 13%) tumors. Methylated MGMT promoter was more common in pseudoprogression (n = 7/17, 41%) than in recurrent tumors (n = 2/57, 3.5%; P < .001). There were 71 deaths (95%). Overall survival from the MRI before repeat surgery was 8.7 months (range, 0.7-76.5) for all patients and was shorter for recurrent tumors at 7.7 months (range, 0.7-51.3) than for pseudoprogression at 10.8 months (range, 3.3-76.5), although this was not statistically significant (P = .16).
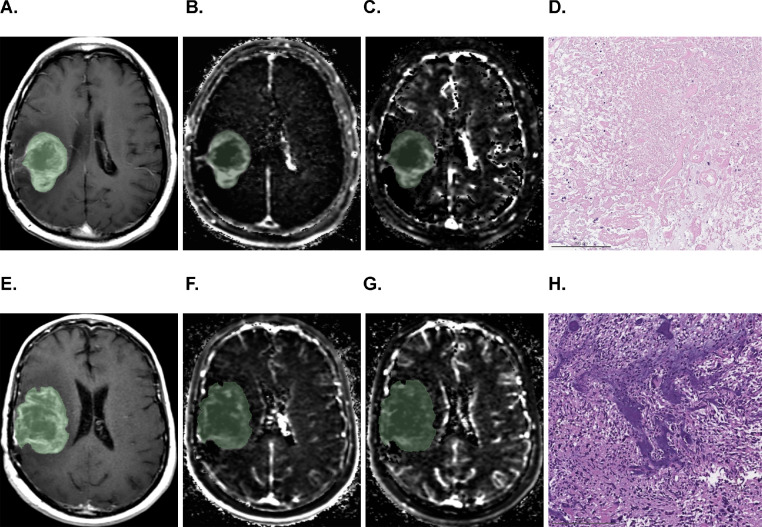
Figure 1.
Axial contrast-enhanced T1-weighted (A, E), Ktrans (B, F), and plasma volume (C, G) images with overlaid segmentation in 2 patients (top and bottom rows). Both lesions presented with ill-defined cystic or necrotic heterogeneously enhancing lesions with peripherally increased Ktrans and plasma volume. Corresponding histopathologic sections (H&E, 10X) showed almost entirely devitalized, necrotic tissue (D) in a patient with pseudoprogression who survived for another 29.2 months, and viable-appearing glioblastoma (H) in a patient who had recurrent tumor and survived for only 3.2 months.
RFE-RF classifier combining radiomics plus MGMT methylation to differentiate recurrent tumor versus pseudoprogression
From a total of 300 radiomic features,14 the MRMR procedure18 selected 70 radiomic features as being relevant for predicting pseudoprogression using a predetermined threshold of 0.8 (range, 0-1).
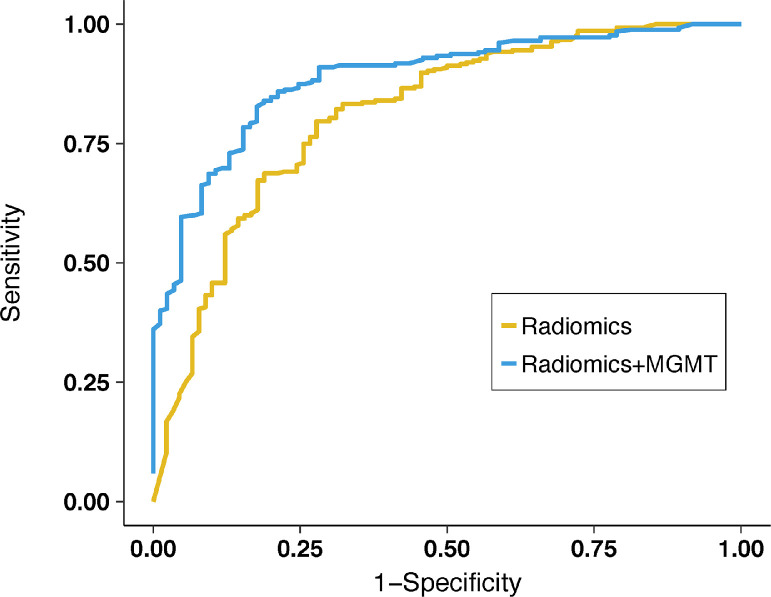
Both the radiomics-only classifier and the combined radiomics plus MGMT classifier achieved an excellent area under the receiver operating characteristic (AUROC): 0.81 (95% confidence interval [CI], 0.76-0.87) and 0.89 (95% CI, 0.85-0.93), respectively. The combined classifier had a higher accuracy than the radiomics-only classifier (P = .03). The other performance metrics including specificity, sensitivity, positive predictive value, and negative predictive value of the 2 classifiers are presented in Table 1. The ROC curve of the 2 classifiers is illustrated in Fig. 2.
Table 1.
Performance metrics for the machine learning classifier
| Method | AUROC (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | PPV | NPV |
|---|---|---|---|---|---|
| Radiomics, all features | 0.79 (0.74-0.85) | 0.60 (0.49-0.70) | 0.84 (0.79-0.88) | 0.54 (0.43-0.64) | 0.87 (0.83-0.91) |
| Radiomics | 0.81 (0.76-0.87) | 0.67 (0.56-0.76) | 0.83 (0.78-0.87) | 0.57 (0.47-0.66) | 0.88 (0.84-0.92) |
| Radiomics + MGMT | 0.89 (0.85-0.93) | 0.72 (0.61-0.81) | 0.90 (0.86-0.94) | 0.71 (0.60-0.80) | 0.91 (0.86-0.94) |
| MGMT + age + sex* | 0.60 (0.53-0.66) | 0.67 (0.56-0.77) | 0.27 (0.22-0.33) | 0.23 (0.18-0.29) | 0.71 (0.61-0.80) |
Abbreviations: AUROC = area under the receiver operating characteristic; CI = confidence interval; MGMT = O6-methylguanine–DNA methyltransferase; NPV = negative predictive value; PPV = positive predictive value.
Logistic regression classifier was used for clinical variables and fit using 3-fold cross-validation.
The positive class is that of pseudoprogression. Radiomic analysis was used to predict histopathologically confirmed pseudoprogression in glioblastoma patients. Results on testing sets used in the validation folds not used in the training are shown for all methods.
Figure 2.
Receiver operating characteristic curves show improved performance in discriminating pseudoprogression from recurrent tumor for the radiomics + O6-methylguanine–DNA methyltransferase (MGMT) (blue) classifier than for the radiomics only (yellow) classifier.
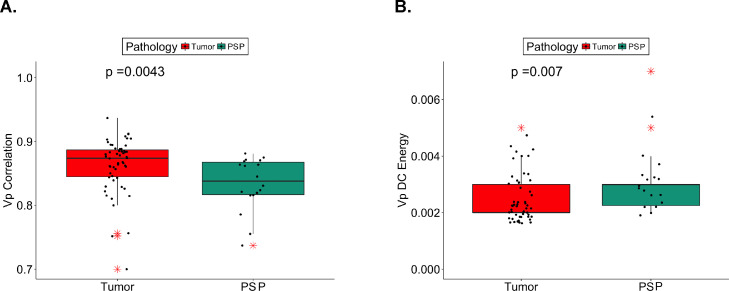
The radiomics-only classifier ranked 9 features as relevant (Gini importance > 0) for predicting pseudoprogression, while the combined radiomics + MGMT classifier ranked 19 features as relevant. Of these, 4 features were commonly identified by the 2 classifiers, namely, VP dependence count energy (VPDCE), VP correlation (VPC), contrast T1-weighted Gabor (0°, 1.414) correlation (cT1GC), and VP Gabor (135°, 1.414). VPDCE quantifies the heterogeneity in the distribution of the texture measures with respect to a voxel neighborhood computed from a VP map. VP correlation quantifies how much the pixels are correlated with respect to their neighborhood pixels when computed from a VP image. The cT1GC feature quantifies the correlation of the Gabor edge image computed from the T1-weighted contrast image. The Gabor edges are directionally sensitive edges that characterize the edges in specific orientations. The VP Gabor quantifies the Gabor edge of the VP image. From these 4 features, recurrent tumor was significantly correlated with VPC and inversely correlated with VPDCE. The VPDCE had a tight distribution of values with a median of 0.002 and an interquartile range (IQR) of 0.002 to 0.003 for the whole data set. The VPC also had a tight distribution of values with a median of 0.866 and an IQR of 0.830 to 0.884 for the whole data set. The median and IQR values for all relevant features are summarized in Table E1. The box plots for the VPC and VPDCE are shown in Fig. 3.
Figure 3.
Pseudoprogression (PSP) demonstrated lower plasma volume correlation (VP Correlation) (A) and higher plasma volume dependence count energy (VP DC Energy) (B) than did recurrent tumor. Absolute values of VP Correlation and VP DC Energy for the 2 groups tumor versus PSP are plotted. The horizontal line represents median, the box indicates interquartile range, and outliers are represented with red asterisks.
Utility of feature preselection
Analysis of the radiomics-only classifier constructed using all the radiomics features produced an AUROC of 0.79 (95% CI, 0.74-0.85), a specificity of 0.84 (95% CI, 0.79-0.88), and sensitivity of 0.60 (95% CI, 0.49-0.70). This AUROC was slightly worse than that of the radiomics-only classifier using feature preselection.
Classification accuracy of known clinical predictors
The linear regression model constructed using MGMT, age, and gender produced a low AUROC of 0.60 (95% CI, 0.53-0.66), a specificity of 0.27 (95% CI, 0.22-0.33), and a sensitivity of 0.67 (95% CI, 0.56-0.77). This result indicates that including radiomics features with clinical predictors improves the accuracy of predicting pseudoprogression.
Discussion
We developed a standard radiomics-based machine learning classifier to predict pseudoprogression. This model was further improved in cross-validation accuracy when standard radiomic features were combined with MGMT features. These analyses demonstrate the power of machine learning to extract useful, discriminative information from standard-of-care contrast T1-weighted images and Ktrans and plasma volume MRI perfusion images. The strength of our analyses includes the use of routine contrast T1-weighted and DCE perfusion images, which are part of the institutional standard of care for glioma imaging; use of open-source CERR software for radiomic feature extraction; and use of accepted dual machine learning approaches after preselection by ensemble MRMR and RFE-RF classifiers. Our analysis also showed that including radiomics features with MGMT produced a much higher accuracy than a simple model constructed with known clinical predictors. Researchers have previously demonstrated that combining genomic information such as the MGMT methylation with radiomics improved the performance of outcome classification in a treatment-naïve glioblastoma cohort.20
Pseudoprogression represents the paramount diagnostic challenge during the treatment of primary glioblastoma, where not all new or increasing enhancing lesions represent recurrent tumors and require treatment. Nearly three-fourths of all patients with pseudoprogression are asymptomatic 21; in reverse, this suggests that about one-quarter are symptomatic and may have findings that mimic the clinical signs and symptoms of tumor progression. The distinction is critical as there is no standard of care treatment for recurrent glioblastoma; instead, current guidelines recommend enrollment in a clinical trial. Pseudoprogression has been correlated with improved survival in part due to its association with methylated MGMT promoter status.19,22,23 Recent studies and meta-analyses have corroborated the utility of advanced techniques such as MRI perfusion, MRI spectroscopy, and FDG and novel radiotracer PET scans to assist in determining pseudoprogression,9,10, 11, 12, 13,23,24 although persistent heterogeneity in technique temper their application. Developing a semiautomated radiomics approach would help streamline data analysis and may facilitate data interpretation. Prompt, accurate diagnosis of pseudoprogression would reduce follow-up imaging and enable appropriate triage of patients who might otherwise undergo repeat resection or clinical trial enrollment for experimental antitumor therapies rather than supportive measures.
We constructed a combined radiomics plus MGMT classifier that outperformed a radiomics only classifier. This combined classifier leveraged the known MGMT status available for most (93.2%) of our patients and is novel compared with prior radiomics studies of pseudoprogression. The association between methylated MGMT promoter status and pseudoprogression has been well described, as has the correlation with improved survival.8,22 Advanced MRI parameters may perform better in methylated tumors than in unmethylated tumors.25,26 We found that recurrent tumor was significantly correlated with VPC and inversely correlated with VPDCE. The radiomics results derived by automated high-throughput feature extractions are usually visually imperceptible and lack any biological correlation, unlike the early semantic features manually assigned by radiologists. Although efforts are underway to only use specific radiomic features with demonstrable biologic correlations,27 expert consensus has advocated the testing and validation of radiomics in clinical trials as exploratory or even primary or secondary endpoints with biological correlation suggested but not mandatory.28
A recent multicenter study examined radiomic analysis of DCE T1 perfusion and dynamic susceptibility contrast T2* perfusion images in pseudoprogression in IDH wild type as well as IDH mutant (9.2%) and IDH unknown (69.4%) glioblastomas.29 In our study, we elected to examine only IDH wild type glioblastomas because their biological behavior, course, and prognosis are known to be different from IDH mutant glioblastoma.3 These innate tumor differences may manifest with intrinsic imaging differences; a small series by Juratli et al30 suggested that pseudoprogression is distinctly uncommon in secondary glioblastoma. Therefore, our study represents the largest and most comprehensive radiomic evaluation of pseudoprogression specifically in IDH wild type primary glioblastomas. Radiomic imaging analysis with standard-of-care MR images including noncontrast T1, T2, and fluid attenuated inversion recovery (FLAIR) images may provide additional information to distinguish pseudoprogression. A recent study by Ismail et al31 also used IDH wild type tumors and showed that shape features computed from within the tumor habitat extracted from T2-weighted, FLAIR, and T1-weighted MRI were predictive of pseudoprogression. A similar multi-institutional study using deep learning and standard radiomic features computed from T1-weighted, FLAIR, T2-weighted, and apparent diffusion coefficient images in patients with IDH wild type tumors using cross-validation showed similar accuracy as our method.32 By contrast, we used standard radiomic features computed exclusively on contrast T1-weighted and DCE perfusion images. We believe that the application of open-source CERR tools broadens the appeal and application of our radiomic approach over proprietary in-house developed software approaches. CERR is compliant with the IBSI, an independent international collaboration providing consensus-based guidelines for the extraction and translation of acquired images into high-throughput image biomarkers.
The analyses reported in this work were performed with a single discovery cohort without a separate validation set by using cross-validation training due to sample size limitations as well as typical data imbalance in the prevalence of pseudoprogression and tumor recurrence. Cross-validation analysis to limit overfitting and small sample size has also been previously used for predicting pseudoprogression previously using machine learning methods.32 Prior work by Jang et al33 used hold-out testing and training sets with a more balanced class prevalence, but 10-fold cross-validation analysis was required in a subsequent study using a larger data set34 owing to large differences in imaging features. Cross-validation has also been used in other prior works because of large class imbalance for predicting pseudoprogression35,36 as well as cancer.37 In addition to the aforementioned limitation of the lack of validation set, this retrospective study included patients with histopathology that predated the 2016 World Health Organization classification.3 The incorporation of Response Assessment in Neuro-Oncology (RANO) criteria into our algorithm was not feasible from a practical standpoint. Nevertheless, all tumors were IDH wild type glioblastomas, most (74%) by PCR or next generation sequencing, with the remaining (26%) by negative IDH R132H immunohistochemistry and considered very likely to represent “primary” IDH wild type glioblastoma.38 Third, there is considerable variability in the literature in the definition of “pseudoprogression.” We temporally defined pseudoprogression as occurring within the first year after completing radiation therapy and concomitant chemotherapy. Although usually occurring <3 to 6 months after radiation therapy, we have seen pseudoprogression and other treatment-related changes develop and evolve over many months and often up to 1 year. For the purposes of our study, we defined pseudoprogression as the complete absence of recurrent tumor at repeat resection. This is an accepted albeit nonconsensus definition, as other studies have included small amounts of recurrent tumor in the setting of overwhelming necrosis.10,12,29 Commonly, IDH wild type glioblastomas are heterogeneous lesions with coexisting radiation necrosis and tumor cells, and these remaining tumor cells may demonstrate a low proliferative index. Because this pathologic presentation may also represent clinical pseudoprogression, the proportion of tumor cells to background necrosis may hold additional relevance to predict outcomes. The use of complete absence of recurrent tumor derives from our experience with salvage resections for irradiated brain metastases, which has suggested no difference in recurrence when stratifying by the proportion of viable tumor versus treatment related changes. A recent position paper by the RANO working group acknowledges the absence of a radiographic and/or histologic gold standard for the diagnosis of pseudoprogression, and although such guidelines are in development, there is at present considerable outstanding work to establish standardized robust, objective quantitative or semiquantitative criteria.39 Fourth, there is inherent bias in requiring repeat surgery, as the surgery itself may be diagnostic and therapeutic in probable recurrent tumors and therefore usually avoided in patients with probable pseudoprogression. This bias is further exacerbated by the fact that nearly three-fourths of all patients with pseudoprogression are asymptomatic.21
DCE perfusion is incorporated into our routine brain tumor scans, although the clinical implementation relies on visual analysis only, not quantitative. Despite diametric differences between visual analysis and multidimensional radiomic analysis, both used the same input (ie, Ktrans and VP maps), and there is the potential for further selection bias in directing high perfusion lesions likely to represent tumor to surgery. We do not believe that how the patients were selected for surgery has any material influence on the performance of our radiomic model, although it is possible for worse (or better) performance in a differently selected cohort.
Although our data was acquired across many different scanners at 3T and 1.5T, suggesting potential repeatability despite technical variations, all scanners were manufactured by a single manufacturer and further work is necessary to determine the potential effect of differences between various scanner manufacturers. As repeatability is a significant concern for MRI-based radiomics, we should note that this is just one source of variability that could limit the generalizability of our model.
Conclusion
In summary, we constructed a radiomic model using structural and biological MR images to predict pseudoprogression in primary glioblastomas, and then improved the model by adding MGMT data into a radiomics plus MGMT model. Without disruption or changes to the clinical MRI scans, these models may be executed by trained data scientists and/or technologists in an imaging laboratory and may provide useful adjunct data to assist radiologist and clinician reading of MRI scans to inform treatment decisions and improve patient outcomes. Independent validation of the developed machine learning models on multi-institutional data sets is first necessary to establish potential for clinical translation.
Acknowledgments
The authors are grateful for the expert editorial advice of Joanne Chin, MFA, and Alyssa Duck, PhD. They are also thankful for the generous support from the Departments of Radiology and Neurosurgery and the Brain Tumor Center at Memorial Sloan Kettering Cancer Center.
Footnotes
Sources of support: This project was funded in part by the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748, Radiologic Society of North America Research Medical Student Grant (Dr McKenney), and MSK Department of Radiology Developmental Project Fund (Dr Young).
Disclosures: Dr Young has engaged in consulting for Agios, ICON plc, NordicNeuroLab, and Puma, and received research funding from Agios. Dr Moss has engaged in consulting for AstraZeneca in the previous year and is performing research funded in part by GT Medical Technologies. All other authors have no disclosures to declare.
Data sharing statement: Research data are stored in an institutional repository and will be shared upon request to the corresponding author.
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.adro.2022.100916.
Appendix. Supplementary materials
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Louis DN, Perry A, Wesseling P, et al. The 2016 World Health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 4.Ellingson BM, Bendszus M, Boxerman J, et al. Consensus recommendations for a standardized Brain Tumor Imaging Protocol in clinical trials. Neuro Oncol. 2015;17:1188–1198. doi: 10.1093/neuonc/nov095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellingson BM, Wen PY, Cloughesy TF. Modified criteria for radiographic response assessment in glioblastoma clinical trials. Neurotherapeutics. 2017;14:307–320. doi: 10.1007/s13311-016-0507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen PY, Chang SM, Van den Bent MJ, Vogelbaum MA, Macdonald DR, Lee EQ. Response assessment in neuro-oncology clinical trials. J Clin Oncol. 2017;35:2439–2449. doi: 10.1200/JCO.2017.72.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanderbeek AM, Rahman R, Ge Fell, et al. The clinical trials landscape for glioblastoma: Is it adequate to develop new treatments? Neuro Oncol. 2018;5:1034–1043. doi: 10.1093/neuonc/noy027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandes AA, Franceschi E, Tosoni A, et al. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J Clin Oncol. 2008;26:2192–2197. doi: 10.1200/JCO.2007.14.8163. [DOI] [PubMed] [Google Scholar]
- 9.Prager AJ, Martinez N, Beal K, et al. Diffusion and perfusion MRI to differentiate treatment-related changes including pseudoprogression from recurrent tumors in high-grade gliomas with histopathologic evidence. AJNR Am J Neuroradiol. 2015;36:877–885. doi: 10.3174/ajnr.A4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas AA, Arevalo-Perez J, Kaley T, et al. Dynamic contrast enhanced T1 MRI perfusion differentiates pseudoprogression from recurrent glioblastoma. J Neurooncol. 2015;125:183–190. doi: 10.1007/s11060-015-1893-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatzoglou V, Yang TJ, Omuro A, et al. A prospective trial of dynamic contrast-enhanced MRI perfusion and fluorine-18 FDG PET-CT in differentiating brain tumor progression from radiation injury after cranial irradiation. Neuro Oncol. 2016;18:873–880. doi: 10.1093/neuonc/nov301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel P, Baradaran H, Delgado D, et al. MR perfusion-weighted imaging in the evaluation of high-grade gliomas after treatment: A systematic review and meta-analysis. Neuro Oncol. 2017;19:118–127. doi: 10.1093/neuonc/now148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Dijken BRJ, van Laar PJ, Holtman GA, van der Hoorn A. Diagnostic accuracy of magnetic resonance imaging techniques for treatment response evaluation in patients with high-grade glioma, a systematic review and meta-analysis. Eur Radiol. 2017;27:4129–4144. doi: 10.1007/s00330-017-4789-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Apte AP, Iyer A, Crispin-Ortuzar M, et al. Technical note: Extension of CERR for computational radiomics: A comprehensive MATLAB platform for reproducible radiomics research [e-pub ahead of print]. Med Phys. doi: 10.1002/mp.13046, accessed March 15, 2022. [DOI] [PMC free article] [PubMed]
- 15.Lin X, Lee M, Buck O, et al. Diagnostic accuracy of T1-weighted dynamic contrast-enhanced-MRI and DWI-ADC for differentiation of glioblastoma and primary CNS lymphoma. AJNR Am J Neuroradiol. 2017;38:485–491. doi: 10.3174/ajnr.A5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu LS, Baxter LC, Smith KA, et al. Relative cerebral blood volume values to differentiate high-grade glioma recurrence from posttreatment radiation effect: Direct correlation between image-guided tissue histopathology and localized dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging measurements. AJNR Am J Neuroradiol. 2009;30:552–558. doi: 10.3174/ajnr.A1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zwanenburg A, Vallières M, Adbalah MA, et al. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiol. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng H, Long F, Ding C. Feature selection based on mutual information: Criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell. 2005;27:1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 19.Bani-Sadr A, Berner L, Barritault M, et al. Combined analysis of MGMT methylation and dynamic-susceptibility-contrast MRI for the distinction between early and pseudo-progression in glioblastoma patients. Rev Neurol (Paris) 2019;175:534–543. doi: 10.1016/j.neurol.2019.01.400. [DOI] [PubMed] [Google Scholar]
- 20.Tixier F, Um H, Bermudez D, et al. Preoperative MRI-radiomics features improve prediction of survival in glioblastoma patients over MGMT methylation status alone. Oncotarget. 2019;10:660–672. doi: 10.18632/oncotarget.26578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brandsma D, Stalper L, Taal W, et al. Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas. Lancet Oncol. 2008;9:453–461. doi: 10.1016/S1470-2045(08)70125-6. [DOI] [PubMed] [Google Scholar]
- 22.Gahramanov S, Varallyaya C, Tyson RM, et al. Diagnosis of pseudoprogression using MRI perfusion in patients with glioblastoma multiforme may predict improved survival. CNS Oncol. 2014;3:389–400. doi: 10.2217/cns.14.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Li J, Cheng G, et al. IDH mutation and MGMT promoter methylation are associated with the pseudoprogression and improved prognosis of glioblastoma multiforme patients who have undergone concurrent and adjuvant temozolomide-based chemoradiotherapy. Clin Neurol Neurosurg. 2016;151:31–36. doi: 10.1016/j.clineuro.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Young R, Souza França de PD, Pirovano G, et al. Preclinical and first-in-human-brain-cancer applications of [(18)F]poly (ADP-ribose) polymerase inhibitor PET/MR. Neurooncol Adv. 2020;2:vdaa119. doi: 10.1093/noajnl/vdaa119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong DS, Kim ST, Lim DH, et al. Diagnostic dilemma of pseudoprogression in the treatment of newly diagnosed glioblastomas: The role of assessing relative cerebral blood flow volume and oxygen-6-methylguanine-DNA methyltransferase promoter methylation status. AJNR Am J Neuroradiol. 2011;32:382–387. doi: 10.3174/ajnr.A2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaracz J, Gattner K, Jaracz K, Górna K. Unexplained painful physical symptoms in patients with major depressive disorder: Prevalence, pathophysiology and management. CNS Drugs. 2016;30:293–304. doi: 10.1007/s40263-016-0328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomaszewski MR, Gillies RJ. The biological meaning of radiomic features. Radiol. 2021;298:505–516. doi: 10.1148/radiol.2021202553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fournier L, Costaridou L, Bidaut L, et al. Incorporating radiomics into clinical trials: Expert consensus endorsed by the European Society of Radiology on considerations for data-driven compared to biologically driven quantitative biomarkers. Eur Radiol. 2021;31:6001–6012. doi: 10.1007/s00330-020-07598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elshafeey N, Kotrotsou A, Hassan A, et al. Multicenter study demonstrates radiomic features derived from magnetic resonance perfusion images identify pseudoprogression in glioblastoma. Nat Commun. 2019;10:3170. doi: 10.1038/s41467-019-11007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juratli TA, Engellandt K, Lautenschlaeger T, et al. Is there pseudoprogression in secondary glioblastomas? Int J Radiat Oncol Biol Phys. 2013;87:1094–1099. doi: 10.1016/j.ijrobp.2013.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ismail M, Hill V, Statsevych V, et al. Shape features of the lesion habitat to differentiate brain tumor progression from pseudoprogression on routine multiparametric MRI: A multisite study. AJNR Am J Neuroradiol. 2018;39:2187–2193. doi: 10.3174/ajnr.A5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akbari H, Rathore S, Bakas S, et al. Histopathology-validated machine learning radiographic biomarker for noninvasive discrimination between true progression and pseudo-progression in glioblastoma. Cancer. 2020;126:2625–2636. doi: 10.1002/cncr.32790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jang BS, Jeon SH, Kim IH, Kim IA. Prediction of pseudoprogression versus progression using machine learning algorithm in glioblastoma. Sci Rep. 2018;8:12516. doi: 10.1038/s41598-018-31007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jang BS, Park AJ, Jeon SH, et al. Machine learning model to predict pseudoprogression versus progression in glioblastoma using MRI: A multi-institutional study (KROG 18-07) Cancers (Basel) 2020:12. doi: 10.3390/cancers12092706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vézina M, Plaa GL. Methyl isobutyl ketone metabolites and potentiation of the cholestasis induced in rats by a manganese-bilirubin combination or manganese alone. Toxicol Appl Pharmacol. 1988;92:419–427. doi: 10.1016/0041-008x(88)90181-0. [DOI] [PubMed] [Google Scholar]
- 36.Lee J, Wang N, Turk S, et al. Discriminating pseudoprogression and true progression in diffuse infiltrating glioma using multi-parametric MRI data through deep learning. Sci Rep. 2020;10:20331. doi: 10.1038/s41598-020-77389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fehr D, Veeraraghavan H, Wibmer A, et al. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci U S A. 2015;112:E6265–E6273. doi: 10.1073/pnas.1505935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brat DJ, Aladape K, Colman H, et al. cIMPACT-NOW update 3: recommended diagnostic criteria for Diffuse astrocytic glioma, IDH-wild type, with molecular features of glioblastoma, WHO grade IV. Acta Neuropathol. 2018;136:805–810. doi: 10.1007/s00401-018-1913-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haider AS, Bent MVD, Wen PY, et al. Toward a standard pathological and molecular characterization of recurrent glioma in adults: A response assessment in neuro-oncology effort. Neuro Oncol. 2020;22:450–456. doi: 10.1093/neuonc/noz233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.