Abstract
The probiotic attributes and genomic profiles of amylase-producing Lactobacillus strains from rice-based fermented foods of Meghalaya in the North-Eastern India were evaluated in the study. A preliminary screening of 17 lactic acid bacteria strains was performed based on their starch hydrolysis and glucoamylase activities. Out of 17 strains, 5 strains (L. fermentum KGL4, L. rhamnosus RNS4, L. fermentum WTS4, L. fermentum KGL2, and L. rhamnosus KGL3A) were selected for further characterization of different probiotic attributes. Whole-genome sequencing of two of the best strains was carried out using a shotgun sequencing platform based on their rich probiotic attributes. The EPS production was in the range of 2.89–3.92 mg/mL. KGL2 (41.5%) and KGL3A (41%) showed the highest antioxidant activity. The highest antibiotic susceptibility was exhibited by all the five Lactobacillus strains against ampicillin, ranging from 24.66 to 27.33 mm. The lactobacilli isolates used in the study could survive the simulated gastric/intestinal juices. Genomic characterization of KGL4 and KGL3A illustrated their possible adherence to the intestinal wall, specialized metabolic patterns, and possible role in boosting host immunity.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13197-022-05633-8.
Keywords: Probiotics, Amylase, Lactobacillus, Whole genome, Fermented foods
Introduction
Amylases catalyze the conversion of starch's α-D-1,4-glycosidic bonds to short oligosaccharides, which are in high demand in the food and dairy sectors. Besides that, microbial-based amylase enzymes are advantageous due to their ease of manipulation and robustness as compared to other sources, as well as their cost-effectiveness in bulk processing. The majority of conventional fermented foods are manufactured from cereals and are readily available in most areas. Since cereals are high in amylose and amylopectin-based starch, amylolytic bacterial cultures are more common in fermented cereals (Ghosh et al. 2015). Lactic acid bacteria (LAB) with amylolytic activity, on the other hand, has an advantage due to its health benefits and social acceptance as a food ingredient (Gobbetti et al. 2005). Since starch is a less expensive source of saccharification than sucrose in the food industry, amylolytic LAB is advantageous and effective in the fermented food industry. Traditional fermented foods are consumed by a group or province as part of an ancient practice and provide considerable health benefits when ingested. Furthermore, as a part of traditional fermented foods, the process and recipe have been around for a long time, making it a reliable source of native microorganisms (Marco and Golomb 2016). These microorganisms could be employed as starter cultures with high nutritional value, health advantages, and functional probiotic properties. As a result, food industries are looking for starter cultures to use in the fermentation process to achieve the necessary consistency, texture, nutritional content, and additional health benefits with natural safety (Leroy and Vuyst 2004). Probiotics are live bacteria that can withstand the rigors of the intestinal tract and promote the host's health. Before a microbial strain is considered for use as a probiotic, it must pass safety tests. However, each strain should be evaluated for several factors, including its potential to thrive in the gastrointestinal system, and substantially modulate the gut microbiota composition imbalance by increasing the bacterial population (Azad et al. 2018). In the gut, prebiotics combined with probiotics provide an additional health benefit and also supply the feed for the gut bacteria for their growth and nourishment. Polysaccharides and starches have previously been thought to have prebiotic qualities (Lovegrove et al. 2017). The Northeastern Indian States are home to a wide variety of ethnic tribal cultures and traditions that have been well preserved over ancient times, so has been their lifestyle and eating habits. Rice is a major component of the diet in this region, and people prepare a variety of rice-based fermented foods as well as rice-based beverages (Mishra et al. 2021a). As a result, the study aimed was to isolate amylase-producing LAB from rice-based traditional fermented foods and beverages common in Meghalaya's tribal area in the North-Eastern part of India, characterize their probiotic potential, and then sequence the whole genomes of LAB isolates with high probiotic attributes.
Material and methods
Isolation and biochemical analysis of lactic cultures
Samples of traditional starter culture, i.e., wanti (50 g) and traditional fermented rice beverage were collected (100 mL) from the three hills (Khasi, Jaintia, and Garo) of Meghalaya state. The isolates were obtained from the rice-based fermented foods and beverages following the methods of Mishra et al. (2021b). These isolates were further screened based on the classical characteristics of Lactobacillus sp. as shown in Table S1 and S4. The isolates were further classified using biochemical methods. The API 50 CHL kit for the identification of the genus Lactobacillus and related genera (bioMérieux, France) were used as per the manufacturer’s instructions for various carbohydrate utilization patterns.
Physiological and molecular analysis
Tolerance of the isolates against 6.5% NaCl was determined adapting the methods from Mulaw et al. (2019). The catalase test was also conducted by dripping two drops of hydrogen peroxide (3%) on 24 h-old cultures on a glass slide (Mulaw et al. 2019). The PCR amplification techniques were adopted from Mishra et al. (2021b). Molecular characterization of isolates was done by PCR using primers, 27F (5′ TACGGYTACCTTGGACGACTT 3′) and 1492R (5′ AGAGTTTGATCAMTGGCTCAG 3′) (Eurofins Scientific India Pvt. Ltd., Bengaluru, India).
Total amylase activity (starch hydrolysis) and glucoamylase activity
The bacterial strains positive for amylase activity were confirmed by the method adapted from Okolo et al. (1996). The glucoamylase activity of the LAB isolates was measured spectrophotometrically at 305 nm using the method of McCleary et al. (1991), using p-nitrophenyl-glucopyronoside as the substrate.
in vitro assessment of safety attributes
Lactobacilli cultures were screened for hemolytic activity by streaking on Sheep Blood Agar Plate (MP1301 HiMedia, India) (Guerra et al. 2018). The LAB isolates were evaluated to degrade mucin in vitro, in order to assess their potential pathogenicity and local toxicity following the methodology of Zhou et al. (2001). For the DNAse activity, the isolates were streaked on the DNase Test Agar Base (M482, HiMedia, India), and the plates were incubated at 37 °C for 48 h, followed by the addition of 1 M HCl solution to the plates (Fonseca et al. 2020).
Probiotic potential of selected isolates (in-vitro)
pH tolerance
pH 2 and 3 with time intervals of 0, 1, and 3 h, were examined in MRS broth solution by adjusting with a hydrochloric acid solution. The survival of Lactobacillus isolates was calculated in terms of log CFU/mL (Hati et al. 2018).
Bile salt tolerance and bile salt hydrolase activity
Bile tolerance and hydrolase activity was studied according to the method suggested by Singhal et al. (2019).
Cholesterol assimilation potential
The method of Gilliland and Walker (1990) adopted by Das et al. (2020) was followed for determining the cholesterol assimilation potential of the lactic isolates. Fifty µg/mL of cholesterol was aseptically added into 10 mL of MRS broth base containing 0.2% sodium thioglycollate and 0.3% sodium taurocholate. To this broth media tubes, 24 h active test strain of LAB was inoculated at the rate of 2%. The tubes were incubated anaerobically under reduced O2 conditions in a gas pack jar at 37 °C up to 24 h. Thereafter, the content of the tubes was centrifuged at 10,000 rpm for 10 min at 4 °C (Eppendorf Centrifuge, US). Supernatant broth obtained thus was treated as sample and 0.5 mL of the same was transferred into a clean test tube.
Extraction: To the above sample, 3 mL of 95% ethanol followed by 2 mL of 50% KOH were added to the tubes and the contents were mixed thoroughly on a cyclomixer. Thereafter the tubes were heated for 10 min in a water bath maintained at 60 °C and cooled subsequently. Further, 5 mL of hexane was added, and the tubes were mixed thoroughly. Then, 3 mL of distilled water was added, and the mixing was repeated. To permit phase separation, the tubes were allowed to stand for 15 min at room temperature. Thereafter, 2.5 mL hexane layer was transferred into clean test tubes. The hexane was evaporated from the tubes by heating them at 60 °C using hot water bath for an overnight period.
Estimation: In this method, 4 mL of OPA reagent (50 mg OPA per liter of glacial acetic acid) was added in above dried extracts and the tubes were allowed to stand at room temperature for 10 min. Then, 2 mL of concentrated sulphuric acid was added slowly from the side of the test tube and the contents mixed thoroughly on cyclomixer. The tubes were allowed to stand at room temperature for further 10 min. Then, the test absorbance was read against blank at 550 nm wavelength on Systronic PC based double beam Spectrophotometer, 2206. Results were recorded in terms of percentage reduction in cholesterol in the test supernatant broth as compared to that in the uninoculated blank supernatant broth. A % reduction in cholesterol in the test supernatant broth when compared to the uninoculated blank supernatant broth was reported.
where, C0: OD550 of MRS broth supernatant containing culture, C1: OD550 of MRS broth supernatant without any culture.
Antibiotic susceptibility
The antibiotic zone scale was used to measure the diameter (mm) of the zone of inhibition around the antibiotic discs, and the results were expressed in terms of resistance or susceptibility by comparing the results to the interpretative zone diameters provided by the Performance Standards for Antimicrobial Disk Susceptibility tests (Mishra et al. 2021b).
Antibacterial activity
The Agar well diffusion method was used to analyze the antibacterial behavior of the lactobacilli isolates towards the test organisms (Schillinger 1989).
Microbial adhesion to hydrocarbon and cellular auto-aggregation
The method of Das et al. (2020) was used to evaluate bacterial adhesion to n-hexadecane and the cellular aggregation efficacy of the isolates respectively.
Exopolysaccharide (EPS) production
For estimating EPS production, MRS broth was infused with 5% sucrose (w/v) as the carbon source, and lactobacilli isolates were added at the rate of 2%, followed by a 24 h of incubation at 37 °C following the method prescribed by Kimmel and Roberts (1998).
Antioxidant activity
The antioxidative assessment was based on the ability of LAB isolates to scavenge the 2, 2'- azino-bis ethylbenzthiazoline-6-sulfonic acid (ABTS) radical cation (Emad and Sanaa 2012).
Resistance to simulated gastric and intestinal fluids
Viable cell counts (log CFU/mL) of Lactobacillus isolates in gastric fluid (pepsin) and in intestinal fluid (pancreatin) at different incubation hours was analyzed following the method of Vidhyasagar and Jeevaratnam (2013).
Whole genome sequencing
NextSeq500 shotgun library preparation and quantity/quality check on agilent 4200 tape station
The query genomes were sequenced on the IlluminaNextSeq® 500 shotgun sequencing platform. The TruSeq Nano DNA Library Prep kit was employed to develop paired end (PE) sequencing libraries from QC passed genomic DNA samples. According to the manufacturer's specifications, the PCR enriched libraries were evaluated on an Agilent Technologies 4200 Tape Station system utilizing high sensitivity D1000 Screen tape.
Cluster generation and sequencing and genome annotation
The PE illumina libraries were loaded onto NextSeq500 for cluster creation and sequencing after getting the Qubit concentration for the libraries and the mean peak sizes from the Agilent Tape Station profile. The respective reference genomic assemblies of Lactobacillus fermentum [Assembly ID: ASM1014v1] and Lactobacillus rhamnosus [Assembly ID: ASM401097v1] were obtained from NCBI and their annotations were utilized. Further, the subsystem category distributions in reference and query genomes were compared using the SEED viewer and RAST server (Overbeek et al. 2005; Aziz et al. 2008). To assay the evidenced genome reduction in LAB, the number of pseudogenes and insertion sequences were estimated through the DFAST server and ISfinder (Tanizawa et al. 2018; Siguier et al. 2006). The role of the query LABs in host immunization against bacteriophages was assessed using the PHAST tool (Zhou et al. 2011).
Statistical analysis
The results were provided as mean ± standard deviations (M ± SD). Using IBM SPSS Statistical software (Ver. 19), two-way analysis of variance (ANOVA) was used to establish significance, and Tukey's HSD test was performed to evaluate post-hoc comparisons. A 0.05 p-value was used as the significance criteria.
Results
Isolation and biochemical characterization of LAB
Initially, 17 LAB isolates were screened based on Gram reaction, growth in 6.5% NaCl, and catalase activity (Table S1). The sugar fermentation profiles of the LAB isolates using API 50 CHL kits are depicted in Table S2. The morphological, physiological, and biochemical tests were carried out for the initial characterization of lactic acid bacteria isolated from the samples. The reactions were interpreted by comparing the turbidity that indicated their growth against growth controls, and the lactic strains were identified by referring to the API manual and also by analyzing the results online at apiweb™ (https://apiweb.biomerieux.com).
Bacterial identification
Bacterial species were confirmed through 16 s rRNA gene sequencing. An 800 to 1400 bp fragment of the retrieved sequences was identified using the Basic Local Alignment Search Tool (BLAST) in the National Center for Biotechnology Information (NCBI) nucleotide database and sequence similarity with closed sequences in the database was analyzed to identify each strain at the species level. All of the 17 isolates were identified based on 16 s sequences and were submitted to GenBank to obtain their specific accession numbers (Table S3).
Starch hydrolysis and glucoamylase activity
Initially, 17 lactic strains were analyzed for starch hydrolysis and glucoamylase activities, out of which 5 lactobacilli strains, viz. KGL4, RNS4, WTS4, KGL2, and KGL3A, showed maximum enzyme activity (Fig. 1a, Figure S1a). L. fermentum KGL4 exhibited the highest starch hydrolysis activity (1.44 µM/min), followed by L. rhamnosus KGL3A (1.17 µM/min) and L. fermentum WTS4 (1.09 µM/min). The obtained results of glucoamylase activity are given in Fig. 1b. Three isolates, KGL4 (0.094 µM/min), RNS4 (0.092 µM/min), and WTS4 (0.05 µM/min) had shown higher glucoamylase activity compared to others. However, the obtained activity was much less, but it represents approx. 5 to 10% of total amylase activity.
Fig. 1.
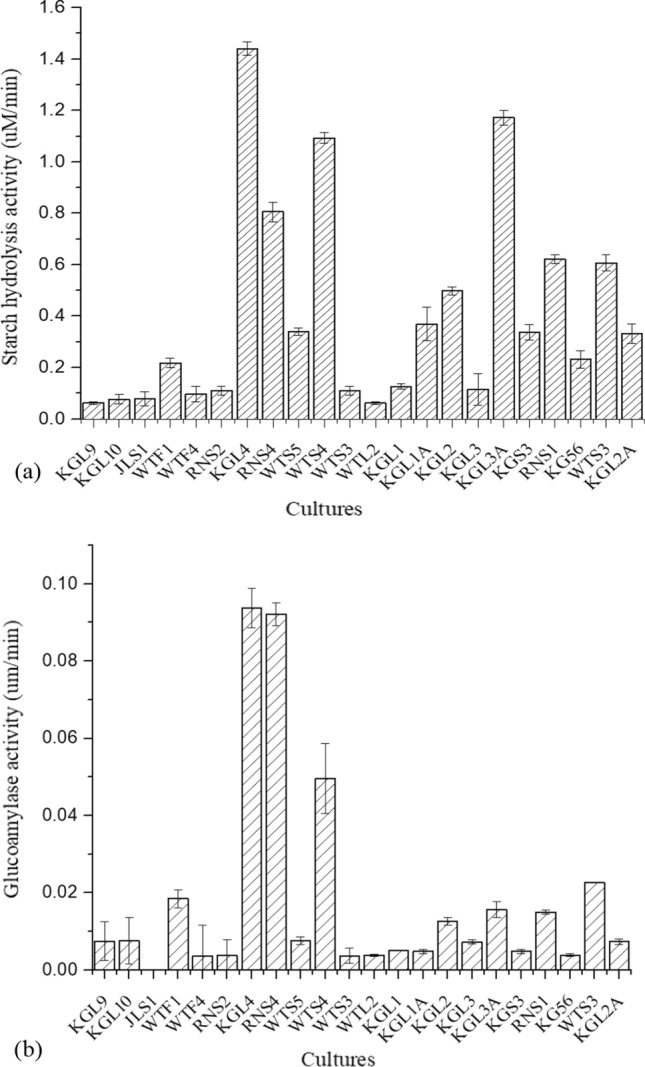
a Starch hydrolysis and b Glucoamylase activity of the Lactobacillus isolates. Each bar represents the average of three independent experiments, error bars are standard deviations. (P ≤ 0.05, n = 3, means ± SD)
in vitro probiotic attributes of the LAB isolates
Based on significantly higher starch hydrolysis and glucoamylase activities, five Lactobacillus cultures, viz. KGL2, KGL4, WTS4, RNS4, and KGL3A, were further selected to study their probiotic potential. The summary of morphological, biochemical, and primary safety characteristics of these five isolates are depicted in Table S4.
Safety assessment tests (in vitro)
Haemolysis was not shown by any of the five Lactobacillus isolates after investigating α-hemolysis (gray-green halo) and β-hemolysis (transparent halo). All the five lactobacilli strains also showed negative DNase activity. Based on the fact that no visible discolored halos around the colonies were observed, it was concluded that selected Lactobacillus strains cannot degrade mucin (Table S4).
pH tolerance
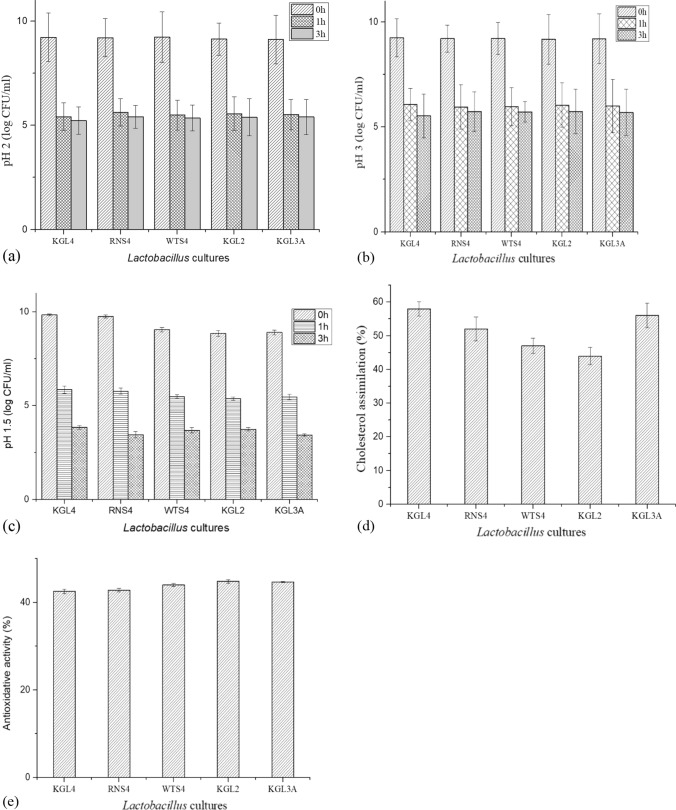
Five Lactobacillus isolates were evaluated for pH tolerance (pH 1.5, 2 and 3) for 0, 1, and 3 h. The three pH levels were tolerated by most isolates. RNS4 showed the best tolerance to pH 2 with 5.6 log CFU/mL at 1 h and 5.4 log CFU/mL at 3 h incubation (Fig. 2a). With 6.5 log CFU/mL at 1 h and 5.5 log CFU/mL at 3 h, KGL4 had the best resistance to pH 3 and at pH 1.5 (3.85 log CFU/mL after 3 h incubation) among the isolates (Fig. 2b, 2c). As the incubation duration and acidic conditions were increased, the viability of bacterial strains dropped. Between isolates, there was no discernible variation in the number of viable cells.
Fig. 2.
Probiotic attributes of Lactobacillus isolates. a Tolerance to pH 2, b pH 3 and c pH 1.5; d Cholesterol assimilation; e Antioxidative activity. Each bar represents the average of three independent experiments. Error bars are standard deviations. (P < 0.05), n = 3, means ± SD)
Bile salt tolerance and bile salt hydrolase activity
The survival of selected isolates in bile salt was investigated, and the findings are shown in Table 1. KGL4 (72.98%) had the highest tolerance to the bile salts, followed by KGL2 (65.50%), and KGL3A (59.61%). RNS4 had the lowest tolerance (52.02%) to bile salts. All of the LAB isolates, however, demonstrated greater than 50% resistance to the 0.5% bile salt, which contains sodium taurocholate and glycocholate. Additionally, bile salt hydrolase activity was investigated, which resulted in a white hallo of precipitation (Figure S1b).
Table 1.
Probiotic attributes of the Lactobacillus isolates
| Lactobacillus isolates | Bile tolerance (%) | Cell surface hydrophobicity (%) | Cellular auto-aggregation (%) | EPS production (mg/mL) |
|---|---|---|---|---|
| KGL4 | 72.98 ± 1.11c | 68.30 ± 0.78e | 70.57 ± 0.68 g | 3.67 ± 0.73i |
| RNS4 | 52.02 ± 1.46a | 65.80 ± 0.89e | 73.68 ± 1.09 g | 3.92 ± 0.34i |
| WTS4 | 55.26 ± 0.83a | 47.44 ± 0.64c | 62.55 ± 1.01f | 2.89 ± 0.12 h |
| KGL2 | 65.50 ± 1.04b | 56.32 ± 1.57d | 59.11 ± 1.15f | 3.72 ± 0.35i |
| KGL3A | 59.61 ± 0.88b | 66.03 ± 1.60e | 68.43 ± 0.76 g | 3.85 ± 0.24i |
Values are represented in mean ± SD with three independent determinations (n = 3) from each sample. Values bearing different superscripts in each cell differ significantly from each other as evident by Tukey’s test at P ≤ 0.05
Cholesterol assimilation
The LAB isolates in the study showed cholesterol assimilation rates ranging from 44 to 58% significantly (P ≤ 0.05) (Fig. 2d). However, the KGL4 strain assimilated the most cholesterol (58%) followed by the KGL3A strain (56%). RNS4, WTS4, and KGL2 showed 52%, 47%, and 44% cholesterol assimilation, respectively.
Antibiotic susceptibility
Vancomycin, kanamycin, norfloxacin, and nalidixic acid resistance were found in all of the LAB isolates used in the study. Other than LAB isolates viz. KGL4 and RNS4, the rest of the isolates (WTS4, KGL2, and KGL3A) showed intermediate susceptibility towards streptomycin. The highest susceptibility was shown by all the five Lactobacillus isolates against ampicillin, ranging from 24.66 to 27.33 mm, and against erythromycin (21.66 to 22.66 mm) (Table 2). The intermediate sensitivity of the five Lactobacillus isolates was shown mainly towards methicillin, oxacillin, gentamycin, and tetracycline.
Table 2.
Antibiotic susceptibility of the Lactobacillus isolates
| Antibiotics (conc. in mcg/disk) | Lactobacillus isolates | ||||
|---|---|---|---|---|---|
| KGL4 | RNS4 | WTS4 | KGL2 | KGL3A | |
| ZOI (in mm) | |||||
| K30 | ND | ND | ND | ND | ND |
| MET15 | 15.66 ± 0.57b | 11.66 ± 0.57f | 17.00 ± 1.00 l | 19.33 ± 1.15q | 18.33 ± 1.15u |
| OX1 | 18.66 ± 0.57c | 12.66 ± 0.57f | 15.66 ± 0.57 k | 19.33 ± 1.15q | 19.33 ± 0.57u |
| VA30 | ND | ND | ND | ND | ND |
| RIF5 | 17.66 ± 0.57c | 20.33 ± 0.50 h | 19.00 ± 1.00 m | 18.00 ± 1.00q | 21.33 ± 0.57v |
| E15 | 22.33 ± 0.57d | 22.00 ± 1.00i | 22.66 ± 0.57n | 22.66 ± 0.57r | 21.66 ± 0.57v |
| TE30 | 19.33 ± 0.57c | 18.66 ± 1.15 g | 18.33 ± 0.57 m | 16.66 ± 0.57p | 20.33 ± 0.57v |
| NX10 | ND | ND | ND | ND | ND |
| S10 | ND | ND | 15.66 ± 0.57 k | 15.33 ± 0.57p | 15.66 ± 0.57t |
| AMP10 | 25.00 ± 1.00e | 24.66 ± 0.57j | 27.33 ± 0.57o | 26.00 ± 1.00 s | 24.66 ± 0.57w |
| GEN10 | 12.66 ± 0.57a | 11.66 ± 0.57f | 15.66 ± 0.57 k | 16.00 ± 1.00p | 14.33 ± 0.57t |
| NA30 | ND | ND | ND | ND | ND |
K30 kanamycin (SD017); MET10 methicillin (SD136); OX1 oxacillin (SD088); VA30 vancomycin (SD045); RIF5 rifampicin (SD030); E15 Erythromycin (SD013); TE30 tetracycline (SD037); NX10 norfloxacin (SD057); S10: streptomycin (SD031), AMP10 ampicillin (SD002); GEN10 gentamycin (SD016); NA30 nalidixic acid (SD021)
mm = diameter of zone of inhibition around the antibiotic discs; ZOI zone of inhibition; ND Not Detected; R Resistant (0 to 15 mm), I Intermediate (15.01 to 20.00), S Sensitive (more than 20 mm). Values are represented in mean ± SD with three independent determinations (n = 3) from each sample. Values bearing different superscripts in each cell differ significantly from each other as evident by Tukey’s test at P ≤ 0.05
Antimicrobial activity
The agar well diffusion method was employed to study the antimicrobial activity of Lactobacillus isolates against eight test organisms, as shown in Fig. 3. The zones of inhibition of the indicator organisms were analyzed by measuring the zone of inhibition that ranged from 10 to 15 mm in diameter (borer diameter-15 mm). The highest antibacterial activity was witnessed against E. coli ATCC 25,922 (15 mm), S. aureus MTCC 114 (14.6 mm), B. cereus ATCC 11,778 (14.4 mm) and E. faecalis ATCC 29,212 (14 mm) by the KGL4 strain. The strain, KGL3A, also showed potent antibacterial activity against a few test organisms, viz. B. cereus ATCC 11,778 (12 mm), S. dysenteriae NCDC 107 (12 mm), S. aureus MTCC 114 (13 mm), E. coli ATCC 25,922 (14 mm), E. faecalis ATCC 29,212 (12.6 mm), S. typhi NCDC 6017 (12.6 mm), and M. luteus NCDC 174 (13.3 mm). None of the Lactobacillus isolates showed antagonistic action against L. monocytogenes MTCC 657.
Fig. 3.
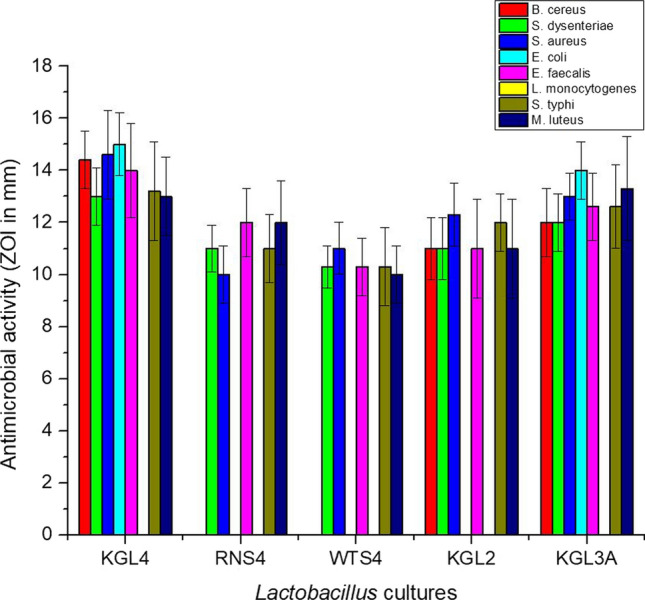
Antimicrobial activity of Lactobacillus isolates against major indicator organisms, viz. B. cereus (ATCC 11,778), S. dysenteriae (NCDC 107), S. aureus (MTCC 114), E. coli (ATCC 25,922), E. faecalis (ATCC 29,212), L. monocytogenes (MTCC 657), S. typhi (NCDC 6017), and M. luteus (NCDC 174). Each bar represents the average of three independent experiments. Error bars are standard deviations. (P < 0.05, n = 3, means ± SD)
Microbial adhesion to hydrocarbons
The adhesion of bacterial isolates to the hydrocarbon (n-hexadecane) was evaluated as the fraction partitioned to the hydrocarbon phase. The fraction partition co-efficient and the cell surface hydrophobicity capability percentage were accordingly calculated. The majority of the bacteria adhered effectively to the hydrocarbon, justifying its adhesion efficacy by absorbance at 600 nm. Among the lactobacilli isolates, KGL4 (68.30%), RNS4 (65.80%), and KGL3A (66.03%) had the highest cell surface hydrophobicity (Table 1). The isolate viz. KGL2 (56.32%) followed byWTS4 (47.44%) had the least hydrophobicity, respectively.
Cellular auto-aggregation
One of the essential phenomena for bacterial adhesion and colonization of the gut is cellular auto aggregation in combination with surface hydrophobicity. As a result, all of the LAB isolates were put to the test to see whether they could auto-aggregate. Auto-aggregation was good in all of the isolates, ranging from 59.11% to 73.68% (Table 1). The maximum auto-aggregation behavior was seen in RNS4 (73.68%), KGL4 (70.57%), and KGL3A (68.43%). KGL2 had the least amount of aggregation (59.11%).
EPS production
EPS production potential was checked for the five LAB isolates used in the study (Table 2). The EPS-producing efficacy of the four Lactobacillus isolates was at par with one another, viz. RNS4 (3.92 mg/mL), KGL3A (3.85 mg/mL), KGL2 (3.72 mg/mL) and KGL4 (3.67%). WTS4 depicted the least EPS production with 2.89 mg/mL.
Antioxidant activity
The antioxidant capacity of selected LAB isolates ranged from 38 to 42% in terms of free radical scavenging ability. KGL2 and KGL3A had the highest antioxidant activity of all the isolates, at 41.5% and 41% respectively (Fig. 2e). However, no significant differences were discovered between isolates.
Resistance to simulated gastric and intestinal fluids
The isolates KGL4 and KGL3A showed the highest tolerance to gastric fluids with pepsin supplementation at pH 2 (5.32 log CFU/mL, 5.06 log CFU/mL) and pH 3 (5.76 log CFU/mL, 5.60 log CFU/mL) conditions after 4 h incubation (Table 3). Similarly, on exposure to simulated intestinal fluid viz. pancreatin, the two Lactobacillus isolates displayed the highest resistance with the maximum cell count after 4 h incubation, viz. KGL4 (6.65 log CFU/mL) and KGL3A (6.42 log CFU/mL) at 5% (P ≤ 0.05) level of significance (Table 4). Therefore, it can be claimed from the above findings that Lactobacillus isolates have been adapted to grow significantly in both acidic and neutral conditions.
Table 3.
Viable cell counts (log CFU/mL) of Lactobacillus isolates in gastric fluid (pepsin) at different incubation hours
| Isolates | Viable cell counts (log CFU/ml) at different incubation hours | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 h | 2 h | 4 h | 0 h | 2 h | 4 h | 0 h | 2 h | 4 h | |
| Gastric fluid (pH 2) | Gastric fluid (pH 3) | Control (pH 7) | |||||||
| KGL4 | 8.15 ± 0.05b | 6.55 ± 0.03f | 5.32 ± 0.04 g | 8.23 ± 0.05B | 7.33 ± 0.08E | 5.76 ± 0.18H | 8.30 ± 0.06I | 8.86 ± 0.04II,III | 9.10 ± 0.18III,IV |
| RNS4 | 7.12 ± 0.06a | 5.36 ± 0.07d | 3.10 ± 0.15e | 8.04 ± 0.09A | 6.40 ± 0.11C | 4.26 ± 0.07F | 8.15 ± 0.09I | 8.53 ± 0.08II | 8.86 ± 0.12II,III |
| WTS4 | 7.33 ± 0.05a | 5.65 ± 0.12d | 3.36 ± 0.06e | 7.90 ± 0.13A | 6.06 ± 0.07C | 4.13 ± 0.16F | 8.18 ± 0.17I | 8.72 ± 0.20II | 9.01 ± 0.06II,III |
| KGL2 | 8.05 ± 0.10b | 6.10 ± 0.10e | 4.77 ± 0.02f | 8.10 ± 0.18A,B | 7.03 ± 0.08D | 5.04 ± 0.13G | 8.26 ± 0.08I | 8.63 ± 0.08II | 8.98 ± 0.09II,III |
| KGL3A | 8.54 ± 0.15c | 6.90 ± 0.08f | 5.06 ± 0.13 g | 8.40 ± 0.04B | 7.82 ± 0.06E | 5.60 ± 0.08H | 8.43 ± 0.10I | 8.94 ± 0.05II,III | 9.20 ± 0.14IV |
Values are represented in mean ± SD with three independent determinations (n = 3) from each sample. Values bearing different superscripts in each cell differ significantly from each other as evident by Tukey’s test at P ≤ 0.05
Table 4.
Viable cell counts (log CFU/ml) of Lactobacillus isolates in intestinal fluid (pancreatin) at different incubation hours
| Isolates | 0 h | 2 h | 4 h | 0 h | 2 h | 4 h |
|---|---|---|---|---|---|---|
| Control (pH 7) | Intestinal fluid (pH 8) | |||||
| KGL4 | 8.35 ± 0.11a | 8.88 ± 0.10b,c | 9.15 ± 0.15d | 8.35 ± 0.06B,C | 7.30 ± 0.05A | 6.65 ± 0.11D |
| RNS4 | 8.18 ± 0.22a | 8.58 ± 0.20b | 8.92 ± 0.25b,c | 8.10 ± 0.11B | 7.04 ± 0.16E | 5.80 ± 0.16F |
| WTS4 | 8.20 ± 0.06a | 8.76 ± 0.15b | 9.07 ± 0.18b,c | 7.65 ± 0.01A | 6.64 ± 0.18D | 5.26 ± 0.20F |
| KGL2 | 8.31 ± 0.11a | 8.61 ± 0.14b | 8.89 ± 0.07b,c | 7.77 ± 0.09A | 6.50 ± 0.20D | 5.11 ± 0.09F |
| KGL3A | 8.48 ± 0.19a | 8.90 ± 0.07b,c | 9.30 ± 0.15d | 8.48 ± 0.20C | 7.56 ± 0.12A | 6.42 ± 0.10D |
Values are represented in mean ± SD with three independent determinations (n = 3) from each sample. Values bearing different superscripts in each cell differ significantly from each other as evident by Tukey’s test at P ≤ 0.05
Whole genome sequencing of L. fermentum KGL4
Based on the above in vitro probiotic attributes, L. fermentum KGL4 (Figure S2a) and L. rhamnosus KGL3A (Figure S2b) were further chosen for sequencing of their respective genomes. The paired-end DNA libraries of the L. fermentum KGL4 genome yielded 2,838,148 paired-end reads accounting for 837,584,859 bases. Mapping of the high-quality reads with the representative reference genome of L. fermentum IFO 3956 (firmicutes) (Assembly accession no. ASM1014v1) covers 85.05% of the genome, totaling a consensus sequence length of 1,733,183 bp. A total of 1674 genes were identified, comprising of 1600 protein-coding sequences (CDS), out of which 1434 were functionally assigned and the rest 166 were hypothetical. Further, 57 tRNA, 15 rRNA (5S, 16S, 23S:: 5, 5, 5) and one each of tmRNA (transfer-messenger RNA) and SRPsRNA (Signal Recognition Particle RNA small type) were recognized. The GC content of the KGL4 strain (53.9%) was observed to be relatively close to that of the reference genome (51.5%), but the reference genome documented a comparatively higher number of genes (2140) and genome size (2,098,685 bp) (Figure S2a) (Tanizawa et al. 2018). One phage region (≈17.9 kb) was recognized with 9 CDSs that matched the phage protein database with 92.3% similarity, but complete prophage or phage attachment sites had not been identified. 33 insertion sequences (IS) from 5 IS families, namely IS3, ISL3, IS30, IS200/IS605, IS 256, were identified from known Lactic Acid Bacteria genomes at the IS Finder database with an expected value < 10−5 (Overbeek et al. 2005).
Whole genome sequencing of L. rhamnosus KGL3A
The whole-genome sequencing of L. rhamnosus KGL3A reported 4,111,537 pair-end reads yielding 1,214,398,155 bases (Figure S2b). The high-quality reads of the KGL3A strain were mapped successfully onto the reference genome of L. rhamnosus (firmicutes) strain LR-B1 (Assembly accession no. ASM401097v1) covering 87.92% of the reference genome. The KGL3A genome included 2080 protein-coding sequences (CDS), 58 tRNA, 15 rRNA (5S, 16S, 23S:: 5, 5, 5) and one each of non-coding RNA (ncRNA) and transfer-messenger RNA (tmRNA), totaling 2596 CDS. The protein CDS tally was shared by 2080 functionally annotated proteins with 441 hypothetical ones (Tanizawa et al. 2018). Two phage-homologous regions (of size 57.7 Kb & 29.1 Kb) were predicted with 29 and 11 CDS matching the phage protein database and 54 insertion sequences (IS) were detected with an expected value < 10–6 from the BLAST search against the customized lactic acid bacteria IS finder database (Overbeek et al. 2005; Aziz et al. 2008).
Discussion
Amylolytic lactic fermentation can enhance the nutritional content of a rice-based diet by increasing the bioavailability of lysine and improving the digestibility of starch in young children (Gobbetti et al. 2005). Cereals have been used as a major ingredient in traditional fermented foods which are rich in starch (Nachi et al. 2018). Therefore, amylolytic microorganisms are commonly present in them. However, two major species of Lactobacillus were frequently reported in rice-based traditional fermented foods, viz. L. fermentum and L. plantarum (Nachi et al. 2018), which were in accordance with our results. Based on the previous literature, for the very first time, amylase-producing L. rhamnosus (KGL3A) has also been reported in this study.
One of the most important characteristics in in vitro testing for determining stomach acid resistance is pH tolerance. Because food stays in the stomach for at least 3 h (Nath et al. 2021), the in vitro experiment was designed with this time limit in consideration. The isolates used in the study survived pH 1.5, 2 and 3 conditions, having a high number of viable counts, indicating that they have the potential to be established as probiotics. However, Mulaw et al. (2019) found that incubation at low pH resulted in a significant drop in the survival rate of all LAB isolates. Low acidity, especially at pH 1.5 and pH 2, had a substantial impact on the viable counts of all lactic acid bacteria, according to the authors. Based on prior research, it's generally acknowledged that an isolate that can tolerate pH 3 for 3 h is a high-acid-resistant strain with promising probiotic capabilities. The ability of microorganisms in the gastric juice to survive low pH conditions is a critical parameter for judging their survival. Depending on the individual, food, and several other factors, the transit time might range from 1 to 3–4 h (Nath et al. 2021). Similarly, Mesquita et al. (2021) also reported the survival of L. paracasei in the fermented beverage after exposure to gastric juice and sequential exposure to gastric and pancreatic juice, i.e., 99.47% and 93.21% respectively, which supports our data. In order to survive in the small intestine, bile salt tolerance is frequently used as one of the probiotic attributes’ tests (Mulaw et al. 2019). Unlike the findings from our study, where more than 50% tolerance to bile salts was reported by the lactobacilli isolates, contrastingly, Handa (2012) found that all of the LAB isolates had limited resistance to bile salts, with a survival percentage of less than 50% after 24 h at 37 °C when exposed to 0.3% bile salts. BSH is an example of a phenomenon that works to reverse the negative effects of bile. Furthermore, the presence of BSH homologs in bile-containing environments strongly suggests that they play a role in bacterial colonization of the intestine. BSH is essential not only for bile tolerance and colonization of the gut but also plays a role in cholesterol absorption (Das et al. 2020). Pereira and Gibson (2002) recorded similar findings with their seven possible probiotic strains, ranging from 0.4 to 44% cholesterol assimilation activities. In a similar study, the cholesterol removal rates of 86 lactic acid bacteria isolates (screened from traditional fermented foods) ranged from 7.29 to 25.66%, and 18 isolates showed a cholesterol removal rate of more than 15% (Zhang et al. 2021). Cholesterol is a precursor to bile acid, so it's used to make fresh bile acid. Since the isolates used in our study showed good BSH activity (Figure S1b), they also had good cholesterol assimilation capacity. In another study reported by Shobharani and Halami, (2016), the cholesterol removal ability of three potential probiotics viz. B. flexus MCC 2458, B. flexus MCC 2427 and B. licheniformis MCC 2514, has been studied. All the three isolates were deprived of bile salt hydrolase enzyme, but they were still able to remove cholesterol by other mechanisms which were strain-dependent, thus facilitating cholesterol removal from the culture media.
A suitable probiotic candidate should not have or acquire any antibiotic resistance genes as a major trait. Notably, most were found to be vancomycin-resistant, which is consistent with prior investigations that have identified vancomycin resistance as an inherent characteristic of Lactobacillus, Leuconostoc, and Pediococcus. Many Lactobacillus strains, including L. fermentum, are employed in the food industry on a regular basis. Vancomycin resistance is chromosomally encoded, non-transferable, and non-inducible in this group of bacteria (Bazireh et al. 2020). Lactobacillus resistance to vancomycin, kanamycin, and norfloxacin is well reported in previous literature (Zhou et al. 2001). Lactobacilli are generally sensitive towards inhibitors for cell wall synthesis and stand resistant to various aminoglycosides (vancomycin, gentamycin kanamycin, streptomycin) due to the absence of cytochrome-mediated electron transport enabling antibiotic uptake. Moreover, LAB resistance to streptomycin and quinolones (Nofloxacin and Nalidixic acid) are due to alteration in the cell wall structure and permeability, or an efflux mechanism (Hummel et al. 2006). When compared to Gram-positive bacteria, it is clear that all of the lactobacilli strains had substantial antibacterial action against Gram-negative test organisms. These findings highlight the significance of the strains used in our research, as they are likely to have broad-spectrum antibiotic activity. Das et al. (2020) claimed that the antimicrobial effect of the lactic acid bacteria might be related to the lactic acid production or due to the reduction of pH, acetic acid, diacetyl, etc.
Hydrophobicity is considered as one of the important properties improving the first contact between bacteria and host cells. The findings are in concordance with Das et al. (2020) where L. rhamnosus K4E (71.57%) was reported to be more adherent to n-hexadecane followed by L. fermentum K16 (69.30%), L. plantarum RD7 (67.21%), and L. fermentum K7 (63.10%). The auto-aggregation test assesses isolates' ability to attach to the gut and exert antipathogenic effects. In contrast to our findings, Bazireh et al. (2020) found that auto-aggregation of Lactobacillus strains ranged from 2.23% to 33.43%, which was lower than the aggregation seen in our investigation. Cellular auto-aggregation aids in the aggregation of bacteria, which is accompanied by biofilm formation, which assists in adhesion to the gut wall, and pathogen eradication by selective competition. The results from the study are in concordance with Das et al. (2020), where the indigenous Lactobacillus isolates aggregated at a rate of 29.4–83.0%. Exopolysaccharides released from LAB strains, in combination with medium (carbon sources) and growth conditions play a critical role in improving texture, sensory quality, and total yield for formulations used in the food fermentation industry (Prete et al. 2021). Therefore, EPS production potential was determined for the five LAB isolates used in the study (Table 1). The study is in agreement with Das et al. (2020), where the Lactobacillus were isolated from traditional fermented foods and EPS production (L. rhamnosus K4E: 950 mg/L, L. plantarum RD7 (710 mg/L) was also reported. After 24 h, the Lactobacillus cultures used in the study were able to scavenge the ABTS+ and reduce the ferryl myoglobin radical, making them promising antioxidant suppressors.
Mostly similar subsystem structure was also observed between L. fermentum KGL4 and the L. fermentum IFO 3956 over 24 subsystems, while the exclusive presence of subsystems like motility and chemotactic ability as well as secondary metabolism was noted in the KGL4 strain by comparatives of SEED & RAST servers (Overbeek et al. 2005; Aziz et al. 2008). The KGL4 strain might have advantages in gut colonization at the gastrointestinal mucus layer (Figure S2a) through its chemotactic ability, which may be involved in antibiotic and pigment production as secondary metabolites. This observation also found support from the notably higher tolerance to bile, cell adhesion to hydrocarbons, cellular auto aggregation, and survival of the KGL4 strain in the gastric and intestinal phases as elaborated above (Table 1, Table 3, Table 4). The L. fermentum KGL4 genome sequence was reported to be short of 214 functional genes in comparison to the reference genome of L. fermentum IFO 3956, comprising mostly of genes engaged in transposition, carbohydrate biosynthesis and metabolism, transcriptional regulation, and membrane transport-pointing toward the specialized carbohydrate utilization pattern of the KGL4 strain. The L. rhamnosus KGL3A genome shared a similar GC content (47.9%) with the reference genome (46.7%) and was found to be somewhat smaller in size with 2,506,971 base pairs compared to the reference genome of 3,007,503 bp (Figure S2b). Further, the KGL3A genome has reported 280 fewer functional genes in comparison to the reference genome, mostly comprised of membrane proteins, transcriptional regulators, transposable elements, and different types of enzymes like oxidoreductase, transferase, nuclease, and kinase involved in various metabolic pathways apart from hypothetical proteins. At the subsystem level, both the KGL3A and the reference genome documented similar categorization over 18 subsystems but differed in motility and chemotaxis; protein metabolism; carbohydrate utilization; biosynthesis and degradation of nucleosides and nucleotides; amino acids; fatty acids, lipids, and isoprenoids; and cofactors, vitamins, prosthetic groups, pigments; and in the expression of prophages, transposable elements (Overbeek et al. 2005; Aziz et al. 2008). Altogether the missing genes and the shrunk metabolic subsystem outline indicated a highly specialized metabolic pattern of L. rhamnosus KGL3A with a possible advantage in adherence to the intestinal epithelial cell through gained motility and chemotactic ability. The proposition was seconded by the report of substantial survival in gastric and intestinal juice; significant bile tolerance, microbial adhesion, and cellular aggregation as documented above (Table 1, Table 3, Table 4).
Conclusion
The five Lactobacillus isolates from traditional rice-based fermented foods and beverages showed promising probiotic characteristics in this study. These isolates were identified to have potent probiotic properties and were non-hemolytic in nature. Two LAB isolates, L. fermentum KGL4 and L. rhamnosus KGL3A were found to have potential in vitro probiotic qualities when compared to the others. Furthermore, the amylase activity of the isolates adds to their characteristics to be used along with natural cereals or starch-based fermentation processes, where isolates would enhance the bio-functionality of food products during the saccharification of starch. The genome of the strain KGL4 and KGL3A exhibited mostly similarity to the respective representative genomes. Both strains demonstrated genetic superiority in gut acclimatization and evidence of specialized metabolic patterns. Both strains have also depicted evidence of genomic reduction and the chances of boosting host immunity through the presence of incomplete prophages. The genomic annotations corroborated well with the experimental observations. Hence, the use of lactobacilli cultures from the study is recommended as starter cultures for functional food development followed by further clinical trials required to validate the health claims.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by the Department of Biotechnology (DBT), New Delhi, Government of India, under the DBT Twinning Project between NEHU, Meghalaya, India, and AAU, Anand, Gujarat, India (BT/PR15969/NER/95/39/2015).
Abbreviations
- LAB
Lactic acid bacteria
- API
Analytical profile index
- PCR
Polymerase chain reaction
- HCL
Hydrochloric acid
- CFU
Colony-forming units
- OPA
Ortho-phthalaldehyde
- MRS
De Man Rogosa and Sharpe
- EPS
Exopolysaccharide
- ABTS
2, 2′- Azino-bis ethylbenzthiazoline-6-sulfonic acid
- PE
Paired-end
- NCBI
National Center for Biotechnology Information
- RAST
Rapid annotation using subsystem technology
- PHAST
PHAge search tool
- ANOVA
Analysis of variance
- SPSS
Statistical package for social sciences
- HSD
Honestly significant difference
- BLAST
Basic local alignment search tool
- tmRNA
Transfer-messenger RNA
- SRPsRNA
Signal recognition particle RNA small type
- IS
Insertion sequences
- CDS
Coding sequences
- ncRNA
Non-coding RNA
- BSH
Bile salt hydrolase
Author contributions
Conceptualization: BKM, SH; Methodology: MP, SKN, SD; Formal analysis and investigation: MP. SD, SKN; Writing: SD, SKN, MP; Original draft preparation: MP, SD, SKN; Review and editing: SD, SH, BKM, SKN; Funding acquisition: BKM, SH; Resources: BKM, SH; Supervision: SH, BKM. All the authors viewed and approved the final version of the manuscript. SH, BKM, SD, and SKN. contributed equally to this study.
Funding
Not Applicable.
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Code availability
Not Applicable.
Declarations
Conflicts of interest
All the authors declare that there is no conflict of interest.
Ethical approval
Not Applicable.
Consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Birendra K. Mishra, Sujit Das, Suman K. Nandy, Subrota Hati have contributed equally to this work.
References
- Azad M, Kalam A, Sarker M, Li T, Yin J. Probiotic species in the modulation of gut microbiota: an overview. Biomed Res Int. 2018 doi: 10.1155/2018/9478630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F. The RAST server: rapid annotations using subsystems technology. BMC Genom. 2008;9(1):1–5. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazireh H, Shariati P, Azimzadeh Jamalkandi S, Ahmadi A, Boroumand MA. Isolation of novel probiotic Lactobacillus and enterococcus strains from human salivary and fecal sources. Front Microbiol. 2020;11:597946. doi: 10.3389/fmicb.2020.597946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Mishra BK, Hati S. Techno-functional characterization of Indigenous Lactobacillus isolates from the traditional fermented foods of Meghalaya, India. Curr Res Food Sci. 2020;3:9–18. doi: 10.1016/j.crfs.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emad AS, Sanaa MMS. Comparison of DPPH and ABTS assays for determining antioxidant potential of water and methanol extracts of Spirulina platensis. Indian J Geo Mar Sci. 2012;42:556–564. [Google Scholar]
- Fonseca HC, de Sousa MD, Ramos CL, Dias DR, Schwan RF. Probiotic properties of Lactobacilli and their ability to inhibit the adhesion of enteropathogenic bacteria to Caco-2 and HT-29 Cells. Probiotics Antimicrob Proteins. 2020;13:102–112. doi: 10.1007/s12602-020-09659-2. [DOI] [PubMed] [Google Scholar]
- Ghosh K, Ray M, Adak A, Dey P, Halder SK, Das A. Microbial, saccharifying and antioxidant properties of an Indian rice based fermented beverage. Food Chem. 2015;168:196–202. doi: 10.1016/j.foodchem.2014.07.042. [DOI] [PubMed] [Google Scholar]
- Gilliland SE, Walker DK. Factors to consider when selecting a culture of Lactobacillus acidophilus as a dietary adjunct to produce a hypocholesterolemic effect in humans. J Dairy Sci. 1990;73(4):905–911. doi: 10.3168/jds.S0022-0302(90)78747-4. [DOI] [PubMed] [Google Scholar]
- Gobbetti M, De Angelis M, Corsetti A, Di Cagno R. Biochemistry and physiology of sourdough lactic acid bacteria. Trends Food Sci Technol. 2005;16(1–3):57–69. doi: 10.1016/j.tifs.2004.02.013. [DOI] [Google Scholar]
- Guerra AF, Fernandes WJ, Geraldo LJ. Lactobacillus paracasei probiotic properties and survivability under stress-induced by processing and storage of ice cream bar or ice-lolly. Food Technol. 2018;48(9):1–9. doi: 10.1590/0103-8478cr20170601. [DOI] [Google Scholar]
- Handa S. Isolation of lactic acid bacteria and to study their potential as probiotics, Dissertation. Dr. Y. S: Parmar University of Horticulture and Forestry, Solan, Himachal Pradesh, India; 2012. [Google Scholar]
- Hati S, Patel N, Mandal S. Comparative growth behaviour and biofunctionality of lactic acid bacteria during fermentation of soy milk and bovine milk. Probiotics Antimicro Prot. 2018;10(2):277–283. doi: 10.1007/s12602-017-9279-5. [DOI] [PubMed] [Google Scholar]
- Hummel AS, Hertel C, Holzapfel WH, Franz CMAP. Antibiotic resistances of starter and probiotic strains of lactic acid bacteria. Appl Environ Microbiol. 2006;73:730–739. doi: 10.1128/aem.02105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel SA, Roberts RF. Development of a growth medium suitable for exopolysaccharide production by Lactobacillus delbrueckii ssp bulgaricus RR. Int J Food Microbiol. 1998;40(1–2):87–92. doi: 10.1016/S0168-1605(98)00023-3. [DOI] [PubMed] [Google Scholar]
- Leroy F, Vuyst LD. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Technol. 2004;15:67–78. doi: 10.1016/j.tifs.2003.09.004. [DOI] [Google Scholar]
- Lovegrove A, Edwards CH, De Noni I, Patel H, El SN, Grassby T, Zielke C, Ulmius M, Nilsson L, Butterworth PJ, Ellis PR, Shewry PR. Role of polysaccharides in food, digestion, and health. Crit Rev Food Sci Nutr. 2017;57(2):237–253. doi: 10.1080/10408398.2014.939263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco ML, Golomb BL. Fermented Foods, Lactobacillus, and Health. Microbe. 2016;11:349–354. doi: 10.1128/microbe.11.349.1. [DOI] [Google Scholar]
- McCleary BV, Bouhet F, Driguez H. Measurement of amyloglucosidase using P-nitrophenyl β-maltoside as substrate. Biotechnol Tech. 1991;5:255–258. doi: 10.1007/bf02438658. [DOI] [Google Scholar]
- Mesquita MC, dos Santos LE, Rodrigues de Alencar E, Botelho RB. Survival of Lactobacillus paracasei subsp paracasei LBC 81 in fermented beverage from chickpeas and coconut in a static in vitro digestion model. Fermentation. 2021;7(135):1–11. doi: 10.3390/fermentation7030135. [DOI] [Google Scholar]
- Mishra BK, Das S, Prajapati JB, Hati S. Bio-functional properties and storage study of ‘Chubitchi’-a fermented rice beverage of Garo hills Meghalaya. Indian J Tradit Know. 2021;20(2):498–511. [Google Scholar]
- Mishra BK, Hati S, Brahma J, Das S. Evaluation of probiotic potentials of yeast isolates from traditional fermented rice beverages of Meghalaya. India Rev Med Microbiol. 2021;32(1):28–38. doi: 10.1097/MRM.0000000000000230. [DOI] [Google Scholar]
- Mulaw G, Sisay Tessema T, Muleta D, Tesfaye A. In vitro evaluation of probiotic properties of lactic acid bacteria isolated from some traditionally fermented Ethiopian food products. Int J Microbiol. 2019 doi: 10.1155/2019/7179514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachi I, Fhoula I, Smida I, Ouzari H-I, Hassouna M. Microbiological analysis and assessment of biotechnological potential of lactic acid bacteria isolated from Tunisian flours. Ann Microbiol. 2018;69:29–40. doi: 10.1007/s13213-018-1365-8. [DOI] [Google Scholar]
- Nath S, Roy M, Sikidar J, Deb B, Sharma I, Guha A. Characterization and in-vitro screening of probiotic potential of novel Weissella confusa strain GCC_19R1 isolated from fermented sour rice. Curr Res Biotechnol. 2021;3:99–108. doi: 10.1016/j.crbiot.2021.04.001. [DOI] [Google Scholar]
- Okolo BN, Ezeogu LI, Ebisike CO. Raw starch digesting amylase from Thermoactinomyces thalpophilus F13. World J Microbiol Biotechnol. 1996;12:637–638. doi: 10.1007/bf00327728. [DOI] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira DIA, Gibson GR. Cholesterol Assimilation by Lactic Acid Bacteria and Bifidobacteria Isolated from the Human Gut. Appl Microbiol Biotechnol. 2002;68:4689–4693. doi: 10.1128/aem.68.9.4689-4693.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prete R, Alam MK, Perpetuini G, Perla C, Pittia P, Corsetti A. Lactic acid bacteria exopolysaccharides producers: a sustainable tool for functional foods. Foods. 2021;10(7):1653. doi: 10.3390/foods10071653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillinger U. Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol. 1989;55(8):1901–1906. doi: 10.1128/aem.55.8.1901-1906.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shobharani P, Halami PM. In vitro evaluation of the cholesterol-reducing ability of a potential probiotic Bacillus spp. Ann Microbiol. 2016;66(2):643–651. doi: 10.1007/s13213-015-1146-6. [DOI] [Google Scholar]
- Siguier P, Pérochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N, Maurya AK, Mohanty S, Kumar M, Virdi JS. Evaluation of bile salt hydrolases, cholesterol-lowering capabilities, and probiotic potential of Enterococcus faecium isolated from rhizosphere. Front Microbiol. 2019;10:1567. doi: 10.3389/fmicb.2019.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa Y, Fujisawa T, Nakamura Y. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics. 2018;34:1037–1039. doi: 10.1093/bioinformatics/btx713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidhyasagar V, Jeevaratnam K. Evaluation of Pediococcus pentosaceus strains isolated from Idly batter for probiotic properties in vitro. J Funct Foods. 2013;5(1):235–243. doi: 10.1016/j.jff.2012.10.012. [DOI] [Google Scholar]
- Zhang Q, Song X, Sun W, Wang C, Li C, He L, Wang X, Tao H, Zeng X. Evaluation and application of different cholesterol-lowering lactic acid bacteria as potential meat starters. J Food Protect. 2021;84(1):63–72. doi: 10.4315/JFP-20-225. [DOI] [PubMed] [Google Scholar]
- Zhou JS, Gopal PK, Gill HS. Potential probiotic lactic acid bacteria Lactobacillus rhamnosus (HN001), Lactobacillus acidophilus (HN017) and Bifidobacterium lactis (HN019) do not degrade gastric mucin in vitro. Int J Food Microbiol. 2001;63(1–2):81–90. doi: 10.1016/S0168-1605(00)00398-6. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:347–352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Not Applicable.