Abstract
Humans have significant individual variations in odor perception, derived from their experience or sometimes from differences in the olfactory receptor (OR) gene repertoire. In several cases, the genetic variation of a single OR affects the perception of its cognate odor ligand. Musks are widely used for fragrance and are known to demonstrate specific anosmia. It, however, remains to be elucidated whether the OR polymorphism contributes to individual variations in musk odor perception. Previous studies reported that responses of the human musk receptor OR5AN1 to a variety of musks in vitro correlated well with perceptual sensitivity to those odors in humans and that the mouse ortholog, Olfr1440 (MOR215-1), plays a critical role in muscone perception. Here, we took advantage of genetic variation in OR5AN1 to examine how changes in receptor sensitivity are associated with human musk perception. We investigated the functional differences between OR5AN1 variants in an in vitro assay and measured both perceived intensity and detection threshold in human subjects with different OR5AN1 genotypes. Human subjects homozygous for the more sensitive L289F allele had a lower detection threshold for muscone and found macrocyclic musks to be more intense than subjects homozygous for the reference allele. These results demonstrate that the genetic variation in OR5AN1 contributes to perceptual differences for some musks. In addition, we found that the more functional variant of OR5A1, a receptor involved in β-ionone perception, is associated with the less functional variant of OR5AN1, suggesting that the perceived intensities of macrocyclic musks and β-ionone are inversely correlated.
Keywords: genotype, musk, olfactory receptor, perception
Introduction
Despite decades of research exploring the relationship between molecular structure and odor character, there are a number of odorant pairs where molecular similarity and perceptual similarity are uncorrelated (Sell 2014). Musks are particularly puzzling, as molecules from at least 5 distinct structural classes (macrocyclic, polycyclic, nitro, steroid-based, and alicyclic) evoke the “musk” character, but small deviations within each structural class, including enantiomers, can ablate the character (Rossiter 1996; Eh 2004). How does the olfactory system encode the “musk” percept?
One possibility is that the convergence of structures to a common odor character occurs at the receptor level. For example, all structures that evoke the “musk” percept may activate 1 receptor or a subset of receptors. Previous studies have shown that 1 human olfactory receptor (OR), OR5AN1, responds to macrocyclic and nitro musks, but not polycyclic or alicyclic musks, in cell-based assays (McClintock et al. 2014; Shirasu et al. 2014; Sato-Akuhara et al. 2016). OR5AN1 is also more sensitive to nitro musks than to muscone, consistent with human sensitivity. In addition, mice lacking the mouse ortholog, Olfr1440, cannot detect low concentrations of muscone (Sato-Akuhara et al. 2016). These results suggest that this OR plays a crucial role in the encoding of some musky odor characters in animals.
It has been shown that functional variation in a single OR can affect the olfactory perception of its ligands (Keller et al. 2007; Menashe et al. 2007; McRae et al. 2012, 2013; Jaeger et al. 2013; Mainland et al. 2014; Trimmer et al. 2019). OR5AN1 is genetically variable across individuals and thus, we set out to determine if this genetic variation also influences the perception of musks including detection thresholds, intensity, and pleasantness. We performed both in vitro and behavioral assays on various musk odorants among subjects with different genotypes of OR5AN1, demonstrating that the genetic variation of OR5AN1 accounts for differences in musk perception. Moreover, we found that a receptor associated with β-ionone perception, OR5A1 (Jaeger et al. 2013), was in linkage disequilibrium with OR5AN1, showing that the individual perception of musk and β-ionone are correlated.
Materials and methods
Reagents
Odorants and solvents used for in vitro assays and psychophysical tests were purchased from MP BioMedical, Wako, TCI, or Sigma (Supplementary Table 1). Ethylene brassylate, exaltolide, galaxolide, and tonalide were kindly provided by Takasago International Corporation, as described previously (Shirasu et al. 2014; Sato-Akuhara et al. 2016). Propylene glycol (ADEKA), used for the detection threshold tests, was kindly provided by T. Hasegawa Corporation. The odorants used for the luciferase assay were prepared as 100 mM stock solutions in dimethyl sulfoxide (Wako).
Cloning ORs and mutagenesis
All ORs used in the luciferase assays were amplified from human genomic DNA (Promega). The cloned OR sequences of humans are identical to those in GenBank (http://www.ncbi.nlm.nih.gov/genbank/). Single nucleotide polymorphisms (SNPs) in OR5AN1 were identified from the 1000 Genomes Phase 3. We generated OR5AN1 and OR5A1 variants by inserting each nucleotide into reference sequences or L289F sequence using a Prime STAR Mutagenesis Basal Kit (TaKaRa).
A polynucleotide encoding the first 20 aa of human rhodopsin (Rho-tag) was included as a tag in N-terminal of a pME18S vector kindly provided by Dr. Kazuo Maruyama for expression in HEK293T cells.
Cell culture and luciferase assay
HEK293T cells were grown in a 37 °C incubator containing 5% CO2. The luciferase reporter gene assay was performed using Dual-Glo Luciferase Assay System (Promega) as described previously (Zhuang and Matsunami 2008; Shirasu et al. 2014; Sato-Akuhara et al. 2016). Luminescence was measured using Centro LB960 plate reader (Berthold Technologies). Relative luciferase activity, L, was calculated as luminescence of firefly luciferase divided by luminescence of Renilla luciferase in a given well. Fold increase was calculated by the following formula: , where Ln represents the relative luciferase activity in a given well and Lno odor represents the relative luciferase activity in a well not stimulated by any odorants. The response (%) is normalized to the maximum values for each odorant of the reference sequence. Each assay was done in triplicate. Mean values of 3 screening triplicates are defined as n = 1.
To determine that a receptor responds to an odorant, we adopted the criteria as described previously (Mainland et al. 2015). We defined an odorant as an agonist if the 95% confidence intervals of the top and bottom parameters did not overlap, the SD of the fitted log EC50 was less than 1 log unit, and the extra sum-of-squares F-test confirmed that the odorant activated the receptor significantly more than the control (empty vector, Rho-pME18S). For ethylene brassylate and exaltolide, it is suggested that cells are damaged by high concentrations of odor stimuli, or that the maximum response is not measured as a result of the OR being too responsive to odors and depletion of downstream G protein or adenylate cyclase. In such cases, the correct EC50 cannot be obtained from the experimental values. Therefore, for these odors, the estimated EC50 was calculated using the average of the Hill coefficients obtained from odors that showed an ideal sigmoidal response for each SNP. To validate the difference in efficacy among the receptor haplotypes, we performed 2-tailed paired t-test at the maximum response of each allele. For EC50, we defined the difference between the OR5AN1 reference and 289F variants if the 95% confidence intervals of the EC50 did not overlap. Data were analyzed using Microsoft Excel and GraphPad Prism.
Psychophysical testing and genotyping in a New York cohort
Collection of psychophysical data was previously reported (Li et al. 2022) and approved by the Rockefeller University’s Institutional Review Board. Five hundred and thirty subjects rated the intensity and pleasantness of 90 stimuli, 16 of which are reported here (Table 1). The high and low odorant dilutions were intensity matched to 1/1,000 and 1/10,000 dilutions of 1-butanol, respectively. Odors were rated on a 100-point scale. To control for scale usage, odor ratings were ranked within each subject, such that the odorant with the highest rated intensity or pleasantness was ranked as 90, and the odor with the lowest rated intensity or pleasantness was ranked as 1.
Table 1.
Musk odorants and dilutions for intensity/pleasantness test.
| Musk odorant | High | Low | Note | Solvent |
|---|---|---|---|---|
| Ambretone | 1/10 | 1/10,000 | Paraffin oil | |
| Cyclopentadecanone | Neat | 1/250 | Solid diluted to 1 M in paraffin oil | Paraffin oil |
| Ethylene brassylate | 1/100 | 1/500 | Paraffin oil | |
| Exaltolide | 1/5 | 1/150 | Solid diluted to 1 M in paraffin oil | Paraffin oil |
| Galaxolide | 1/10 | 1/1,000 | Paraffin oil | |
| Muscone | Neat | 1/10,000 | Paraffin oil | |
| Musk ketone | 1/2 | 1/20,000 | Solid diluted to 1 M in paraffin oil | Paraffin oil |
| Tonalide | 1/25 | 1/1,000,000 | Solid diluted to 1 M in diethyl phthalate | Diethyl phthalate |
Genomic DNA was extracted from saliva samples using the Oragene Discover 2 mL kit and sequenced as previously described (Li et al. 2022). OR5AN1 was amplified using primers up- (5ʹ-CCAGTGCTTGTTGGAAGAAA) and downstream (5ʹ-GGGTTAGGGAGATTCTGATGG) of OR5AN1’s open reading frame and sequenced (ABI 3730XL) at the University of Pennsylvania DNA Sequencing Facility.
The effect of genotype on the perceived intensity or pleasantness of each musk was examined using a Kruskal–Wallis test, and differences between individual alleles were determined using a Wilcoxon rank-sum test with Bonferroni correction.
Psychophysical testing and genotyping in a Japanese cohort
All procedures were approved by the ethics committee of the University of Tokyo and carried out in accordance with their guidelines. Written consent was obtained from all subjects before tests or collecting samples. Pregnant women, smokers, and subjects with general anosmia, as indicated by a score below 8 out of 12 on the Open Essence (Wako) odor identification test, were excluded from the analysis.
We determined odor detection thresholds for 54 subjects between the ages of 19 and 47 (mean = 27.3 ± 8.33, 24 females) from the University of Tokyo. We used the single staircase method as described previously (Doty et al. 1995; Keller et al. 2007; Doty and Laing 2015) with binary dilutions of muscone from 1:1,024 (dilution 10) to 1:134,217,728 (dilution 27), starting at 1:8,388,608 (dilution 23). Participants smelled 1 mL of odor in propylene glycol in 125-mm long glass test tubes with a 30- to 60-s interstimulus interval. Individual thresholds were the averages of 2 experiments from different visits separated by at least 1 day. We excluded subjects who did not converge at the highest dilution of muscone.
Fifty-four subjects between the ages of 19 and 55 (mean = 26.0 ± 9.01, 20 females) rated the intensity of β-ionone, 3 dilutions of n-butanol, and solvent (propylene glycol) using a 7-point scale (Keller et al. 2007; Trimmer et al. 2019). The order of sample presentation was randomized for each subject, and a 1-min interval was enforced between samples. Subjects were presented with 6,000 ppm of β-ionone, a detectable concentration for all OR5A1 genotypes (Jaeger et al. 2013), and 3 dilutions of n-butanol (1/10,000, 1/30,000, and 1/50,000) to standardize scale usage. Because subjects on average rated the 1/30,000 dilution as “neither weak not strong” (mean = 4.02 ± 1.52), we calculated the relative intensity value by subtracting the intensity rating of 1/30,000 dilution of n-butanol from that of β-ionone.
Saliva samples were collected from all subjects using Oragene DNA (DNA Genotek), and genomic DNA was prepared with prepIT L2P (DNA Genotek). OR5AN1 was amplified from genomic DNA using Tks Gflex DNA polymerase (TaKaRa) with primers upstream (5ʹ-AGAGTGAAACATTTGTCCATTTCCAGTGCTTGTTG-3ʹ) and downstream (5ʹ-CCTGATGTCTGTGGCATTGAGTCATCTGTTGGTC-3ʹ) and sequenced. For OR5A1, primers upstream (5ʹ-TGCTAATACCACCTATAATGTGGACTGTCATTACT-3ʹ) and downstream (5ʹ-CCTTGCCTGAGGCAGAGGTGGAGGTGAAG-3ʹ) were used. Because the frequency of OR5AN1 and OR5A1 reference sequences is lower than those of OR5AN1 L289F (66.1%) and OR5A1 D183N (62.9%), the number of L/L and D/D subjects was relatively small in our Japanese cohort (8.3KJPNv2; Tohoku Medical Megabank Organization [ToMMo] [jMORP]).
Microsoft Excel and R 3.6.1 were used for data analysis. The thresholds and intensities of the different genotypes were compared with linear regression.
Linkage disequilibrium analysis
For the linkage disequilibrium analysis of the human OR SNPs, haplotypes of 2,504 human genomes from the 1000 Genome Projects Phase 3 were used. R 3.6.1 and its package “LDheatmap” were used for data analysis (Shin et al. 2006). Nonsynonymous SNPs were used for analysis if those allele frequencies were 1% or higher, including a frameshift mutation (rs77048571).
Results
OR5AN1 genetic variation affects receptor responses to musk odorants
We investigated the frequency of genetic variation in OR5AN1 in 1000 Genomes data and found that the top 5 most frequent haplotypes had 4 distinct SNPs (Abecasis et al. 2010; Fig. 1A). Two variants have high allele frequencies in the population: the reference sequence (31.7%) and a variant with a single amino acid change at residue 289 (L289F) (66.1%). Three additional variants G3R/L289F, T247K, and G162A have low allele frequencies (0.66%, 0.56%, and 0.23%, respectively). We next investigated functional differences among these haplotypes in a heterologous HEK293T cell-based assay. Fluorescence activated cell sorting (FACS) analysis did not show a significant difference in cell surface expression levels across haplotypes (Supplementary Fig. 1) (Student’s t-test, P > 0.56). In the luciferase reporter gene assay, 3 haplotypes including the reference and L289F responded to macrocyclic musks (muscone, cyclopentadecanone, ambretone, exaltolide, and ethylene brassylate) and a nitro musk (musk ketone) (Fig. 1B–G, Supplementary Table 2), consistent with our previous report (Sato-Akuhara et al. 2016). Notably, the L289F variant responded to macrocyclic musks with greater efficacy than that of the reference variant (paired t-test, P < 0.05, at the maximum response of each odorant) (Fig. 1B–F, Supplementary Table 2). On the other hand, no difference was found for musk ketone efficacy between these 2 alleles (paired t-test, P = 0.77) (Fig. 1G, Supplementary Table 2). There was no difference in sensitivity (EC50) for 5 macrocyclic musk odorants among the 3 haplotypes (Fig. 1B–F, Supplementary Table 2), while only for musk ketone, there was a little difference in sensitivity between reference and L289F alleles (Fig. 1G, Supplementary Table 2A). The other 2 variants, G162A and T247K, did not respond to macrocyclic musks except EB to G162A, but showed a weak response to musk ketone (Supplementary Fig. 2, Supplementary Table 2). No OR5AN1 variants responded to polycyclic musks (galaxolide and tonalide) in either a luciferase assay or a GloSensor cAMP assay (Fig. 1H and I, Supplementary Fig. 2, Supplementary Table 2).
Fig. 1.
OR5AN1 variants had different responses to 6 musk odorants. (A) OR5AN1 snake plot with amino acid changes indicated. (B–G) Dose–response curves to musk odorants of empty vector (pME18S) (cross) and OR5AN1 responsive variants; reference sequence (ref, square), L289F (triangle), G3R, L289F (inverted triangle) in HEK293T cells using luciferase reporter gene assay. The EC50 values (µM) to muscone were 11.45 for reference sequence, 9.192 for L289F, 12.81 for G3R/L289F, to cyclopentadecanone 19.96 for reference sequence, 14.12 for L289F, 15.87 for G3R/L289F, to ambretone 4.304 for reference sequence, 3.390 for L289F, 5.500 for G3R/L289F, to ethylene brassylate 52.81 for reference sequence, 40.91 for L289F, 35.67 for G3R/L289F, to exaltolide 67.68 for reference sequence, 58.41 for L289F, 66.90 for G3R/L289F, to musk ketone 0.1860 for reference sequence, 0.09705 for L289F, and 0.1400 for G3R/L289F. The asterisks indicate significant differences at the maximum response of each odorant between reference sequence and L289F using 2-tailed paired t-test (*P < 0.05). (H and I) Responses to polycyclic musks (PCMs), galaxolide (H) and tonalide (I) of 3 OR5AN1 haplotypes. 30 µM muscone (Mus. 30 µM) was used as the positive control. The Y-axis showed fold increase (see Materials and methods). Error bars indicate SEM (n = 3).
Genetic variation in OR5AN1 correlates with perceived intensity of some musk odorants
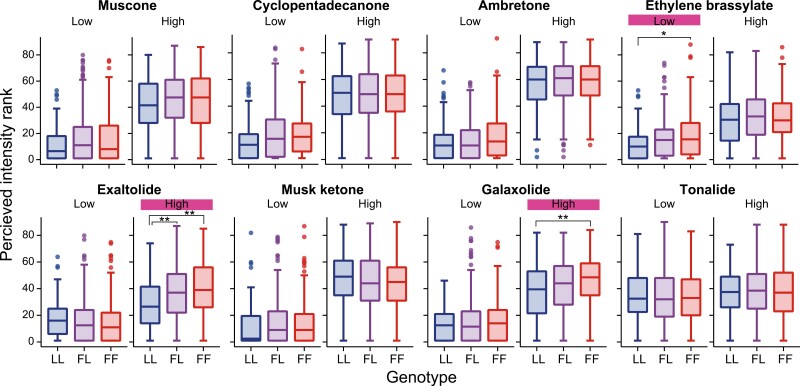
Next, we investigated whether functional variation in OR5AN1 affects the perception of musk odorants. We asked 530 participants to rate the intensity and pleasantness of 2 different dilutions (high and low dilution) of 8 different musk odorants (Table 1). Ten OR5AN1 diplotypes were found in our subject pool (Supplementary Fig. 3). The most common haplotype combinations found in our subject pool were LL (homozygous reference (289L) sequences) (12.83% of 530), LF (L289F and reference sequences) (45.09% of 530), and FF (homozygous L289F sequences) (38.68% of 530). Other amino acid changes (T247K, G162A, G223C, and G3R/L289F) were found in combination with L289F and the reference variant at low frequencies. We next examined the perceived intensity of musk odorants in subjects with the 3 most common genotype combinations: LL, FL, and FF (Fig. 2).
Fig. 2.
The association of OR5AN1 common genotypes and musk odorant intensity. LL, FL, and FF indicate the OR5AN1 genotypes of subjects, subjects with homozygous reference sequences (n = 68, 12.83% of 530), subjects with L289F and reference sequences (n = 239, 45.09% of 530), and homozygous L289F (n = 205, 38.68% of 530), respectively. Odorants and their dilutions are showed above each graph. The Y-axis shows the perceived odor rank of odor intensities. The odorant with the highest rated intensity was ranked as 90, and the odor with the lowest rated intensity was ranked as 1. The height of the box indicates the first and third quartiles, the line indicates the median rank, and the whiskers extend to 1.5*IQR (Tukey’s style). The pink labels indicate cases wherein the association between genotype and phenotype is significant (Kruskal–Wallis test, P < 0.05). The asterisk indicates a significant difference between the 2 genotypes using the Wilcoxon rank-sum test with Bonferroni correction (*P < 0.05, **P < 0.01).
Significant differences in intensities among the 3 genotypes were observed for the 3 musks: ethylene brassylate (low dilution) (P = 0.024, Kruskal–Wallis test; P = 0.030, Wilcoxon rank-sum test with Bonferroni correction [LL vs. FF]), exaltolide (high dilution) (P = 0.00007 overall; P = 0.00004 LL vs. FF), and galaxolide (high dilution) (P = 0.0046 overall; P = 0.0050 LL vs. FF). The FF subjects rated these 3 musks as more intense than the LL subjects, while the FL subjects rated their intensity between the homozygous groups. For ethylene brassylate and exaltolide, OR5AN1 L289F responded with between 6- and 3-fold greater efficacy than that of the reference in an in vitro assay (paired t-test, P = 0.04, P = 0.02, respectively) (Fig. 1E and F, Supplementary Table 2), whereas galaxolide did not activate OR5AN1 in vitro (Sato-Akuhara et al. 2016; Fig. 1H, Supplementary Table 2). The intensity of the other macrocyclic musks did not vary significantly with genetic variation, but there were some trends (ambretone [Low]: P = 0.076, the Kruskal–Wallis test; cyclopentadecanone [Low]: P = 0.060; muscone [Low]: P = 0.17). These macrocyclic musks showed a 1.3- to 2-fold difference in efficacy in the in vitro assay (Fig. 1B–D, Supplementary Table 2). For musk ketone, a nitro musk, there was no difference in either the in vitro assay (paired t-test, P = 0.77) or the perceived intensity (Fig. 1G). Taken together, the cell culture assay and genetic association data strongly agree for 3 musks (ethylene brassylate, exaltolide, musk ketone, and tonalide), somewhat agree for 3 musks (muscone, ambretone, and cyclopentadecanone), and do not agree at all for galaxolide.
Detection thresholds for muscone using binary steps
We observed functional differences in the response of OR5AN1 to muscone in vitro, but there was no significant association between genetic variation in OR5AN1 and perceived intensity. We next examined the association of genetic variation with the detection threshold of muscone using a staircase method with a greater range of odor dilutions (18 steps). (Fig. 3). We examined 54 subjects consisting of 11 LL, 21 FL, and 22 FF. The FF participants were more sensitive to muscone than the LL participants (mean of binary dilution 15.72 in FF and 13.53 in LL), and the sensitivity of FL subjects was intermediate between these 2 groups (mean of binary dilution 14.31) (P = 0.0027, linear regression, adjusted R-squared: 0.14). These results demonstrate that the more functional 289F allele is correlated with a higher sensitivity to muscone (lower detection threshold).
Fig. 3.

OR5AN1 L289F variants associate with the detection threshold for muscone. LL, FL, and FF indicate the OR5AN1 genotypes of subjects, subjects with homozygous reference sequences (n = 11), subjects with L289F and reference sequences (n = 21), and homozygous L289F (n = 22), respectively. The Y-axis shows the detection thresholds of the subjects at the binary dilution of muscone. The upper dots indicate that the subjects can detect muscone more sensitively. The height of the box plot indicates the first and third quartiles, the line indicates the median rank, and the upper and lower whiskers indicate the upper and lower limits, respectively. The asterisk indicates a significant difference among the 3 genotypes (P = 0.0027, adjusted R-square: 0.14, linear regression).
Correlation between galaxolide perception and the OR5AN1 gene cluster
Although OR5AN1 did not respond to galaxolide in vitro (Sato-Akuhara et al. 2016; Fig. 1H, Supplementary Table 2, Supplementary Fig. 2), the genetic variation in OR5AN1 was significantly correlated with differences in the perceived intensity of galaxolide (Fig. 2). One potential explanation for this observation is that genetic variation in OR5AN1 is in linkage disequilibrium with variation in other nearby odorant receptors, one of which may be the causal receptor. The genetic association of SNPs in OR4D6, located in the same cluster as OR5AN1, has been previously shown to be significantly associated with the perceived intensity of galaxolide (Trimmer et al. 2019; Li et al. 2022). Thus, we analyzed linkage disequilibrium at this locus using 1000 Genomes data (Fig. 4). We observe pairs of SNPs with high linkage disequilibrium in different OR genes (namely OR5A2 and OR4D6, OR5A1 and OR4D6). We investigated the responses of these ORs to galaxolide in in vitro, but no OR responded to galaxolide (Supplementary Fig. 4, consistent with previous reports (Trimmer et al. 2019; Li et al. 2022). Thus, it remains unknown which OR is causal for the galaxolide association.
Fig. 4.
Linkage disequilibrium of OR5AN1 cluster. Linkage disequilibrium (r2 metric) heatmap for the genomic region containing OR5AN1 on Chromosome11_13 associated with galaxolide perception (Trimmer et al. 2019). The OR gene name indicates above each SNP. Red asterisks and circle emphasize the SNPs, OR5AN1 L289F (rs7941190) and OR5A1 D183N (rs6591536). LD (r2) was calculated from human OR haplotypes data in 1000 Genomes Phase 3 and visualized as a heatmap using “LDheatmap” R package (Shin et al. 2006).
Perceptual correlation in musk and β-ionone
We found that L289F in OR5AN1 (rs7941190) is in linkage disequilibrium with D183N in OR5A1 (rs6591536) (r2 = 0.4975) (Fig. 4). OR5A1 has previously been reported as an important receptor for β-ionone perception, with its genotype almost completely determining detection and perceived intensity, which in turn affects odor experience and food selection (Jaeger et al. 2013; McRae et al. 2013). The D183N haplotype of OR5A1 is less sensitive to β-ionone and the individuals with 2 copies of this allele have higher detection thresholds for β-ionone. From the 1000 Genomes dataset, approximately 80% of the people homozygous for L289F in OR5AN1 are also homozygous for D183N in OR5A1 (880/1,136), and 84% of the people homozygous for D183N in OR5A1 are also homozygous for L289F in OR5AN1 (880/1,047). Likewise, approximately 70% of the people homozygous for the OR5AN1 reference (L) allele are also homozygous for the OR5A1 reference (D) allele (183/273).
Given how commonly genetic variation in OR5AN1 and OR5A1 are found together, we hypothesized that musk-sensitive individuals are less sensitive to β-ionone and that musk-insensitive individuals are more sensitive to β-ionone. We first examined the OR5A1 genotype in subjects that were phenotyped for muscone odor detection threshold (Supplementary Fig. 5). These 54 subjects consisted of DD (homozygous for the reference allele) (n = 8), DV (heterozygous reference and I52V alleles) (n = 1), DN (heterozygous reference and D183N alleles) (n = 21), NN (homozygous for D183N allele) (n = 20), and NV (heterozygous for D183N and I52V) (n = 3). I52V is a minor haplotype of OR5A1 found only in the Asian population (allele frequency: 3.59% in 1000 Genomes, 7.09% in ToMMo 8.3KJPN). OR5A1 D183N is correlated with lower muscone detection thresholds (higher sensitivity) (P = 0.00085, adjusted R-squared: 0.20, linear regression), despite the fact that OR5A1 does not respond to musks (Supplementary Fig. 6, Supplementary Table 2). Next, we analyzed how the perceived intensity of β-ionone correlated with both OR5A1 and OR5AN1 genotype. The subjects rated the intensity of a 6,000 ppm dilution of β-ionone on a 7-point scale, which was then compared with the rating of a control odor (n-butanol) for each subject. The perceived intensity of β-ionone was significantly higher for the subjects with OR5A1 DD than those with D183N (DN and NN) (Fig. 5A) (P = 0.0043, adjusted R-squared: 0.15, linear regression [DD, DN, and NN]), generally consistent with previous results that the D allele to be dominant (Jaeger et al. 2013). In addition, OR5AN1 FF individuals rated the perceived intensity of β-ionone to be lower than LL individuals (P = 0.00018, adjusted R-squared: 0.22, linear regression), which is the opposite of what we observe for muscone threshold (Figs. 3 and 5B). These results indicate that the detection threshold and perceived intensity of muscone are inversely correlated with those of β-ionone. We also note that the follow-up experiment for some participants in Fig. 3 showed that people with low sensitivity to muscone (LL-DD: n = 4, FL-DN: n = 2) can be predicted to be highly sensitive to β-ionone with fairly good certainty (data not shown). However, regardless of genotypes, muscone high sensitivity type may not be necessarily predicted to be low sensitive to β-ionone at the level of sensory evaluation.
Fig. 5.
OR5AN1 variants associate with the intensities for β-ionone. The data are the same for A and B. The Y-axis shows the relative intensity values for β-ionone to the standard odorant, n-butanol. The upper dots (>0) indicate that the subjects perceived the odor more strongly than n-butanol. The height of the box plot indicates the first and third quartiles, the line indicates the median rank, and the upper and lower whiskers indicate the upper and lower limits. (A) DD, DN, NN, VN, and DV indicate the OR5A1 genotypes; subjects with homozygous reference sequences (n = 6), subjects with heterozygous of D183N and reference sequences (n = 23), and homozygous D183N (n = 17), subjects with heterozygous of I52V and D183N (n = 7), and subjects with heterozygous of reference sequence and I52V (n = 1), respectively. The asterisk indicates a significant difference among the 3 genotypes; DD, DN, and NN (**P = 0.0043, adjusted R-square: 0.15, linear regression). (B) LL, FL, and FF indicate the OR5AN1 genotypes, subjects with homozygous reference sequences (n = 7), subjects with heterozygous of L289F and reference sequences (n = 28), and homozygous L289F (n = 19), respectively. The asterisk indicates a significant difference among genotypes (**P = 0.00018, adjusted R-square: 0.22, linear regression).
Discussion
Musks are commercially important compounds in the fragrance industry, but many individuals are unable to smell a subset of musk compounds (Whissell-Buechy and Amoore 1973; Amoore et al. 1977; Gilbert and Kemp 1996; Wysocki 2009). These specific anosmias may help to identify how ORs underly olfactory perception, much as color blindness helped to identify the receptor mechanisms underlying color vision (Nathans et al. 1986). We previously reported that OR5AN1 responded to macrocyclic and nitro musks, and not to polycyclic and alicyclic musks (Shirasu et al. 2014; Sato-Akuhara et al. 2016). In this study, we investigated whether variations in the human musk receptor OR5AN1 accounts for musk anosmia. As a result, we showed that 3 OR5AN1 haplotypes responded to macrocyclic and nitro musks, while 2 minor alleles did not respond to macrocyclic musks (Fig. 1, Supplementary Fig. 2, Supplementary Table 2). Notably, the L289F allele was more effective than the other alleles, and individuals with 1 or 2 copies of the L289F allele found certain macrocyclic musks (exaltolide and ethylene brassylate) to be more intense and are more sensitive to muscone than individuals with reference alleles (Figs. 2 and 3, Supplementary Table 2, Supplementary Fig. 5).
Leu289 in OR5AN1 is located at the first “x” of the highly conserved motif, NPxxY in transmembrane 7 (Trzaskowski et al. 2012). The mutation in this motif is considered to indirectly affect the coupling with the G protein (Rovati et al. 2007). Indeed, for the β2 adrenergic receptor, also a class A G-protein-coupled receptor, a mutation in NPxxY resulted in the stabilization of the inactive conformation and the reduction of the agonist response (Ragnarsson et al. 2019). Given that there was no difference in the surface expression level among OR5AN1 haplotypes (Supplementary Fig. 1), the 289th amino acid of OR5AN1 may be involved in the conformational change from the inactive state to the active state, resulting in an effect on G protein signals and thus the haplotypes exhibit a difference in receptor activity.
OR5AN1 variation affected odor intensity but not pleasantness (Supplementary Fig. 3). In the intensity/pleasantness ratings, OR5AN1 genetic variation was significantly associated with the perceived intensity of exaltolide [high] (P = 0.000072, Kruskal–Wallis test), ethylene brassylate [low] (P = 0.024), and galaxolide [high] (P = 0.0046). Subjects with L289F allele(s) rated these odors as more intense (Fig. 2). Genetic variation in OR5AN1 was not significantly associated with the intensity of the other macrocyclic musks (muscone, cyclopentadecanone, and ambretone), although there were trends toward significance. These intensity perception results are consistent with the efficacy (maximum value) differences in the cell culture assay (Figs. 1 and 2).
In the muscone detection threshold test, there was a significant difference among the OR5AN1 genotypes (P = 0.0027, linear regression). Individuals with the L289F haplotype detected muscone with higher sensitivity (Fig. 3), although no significant difference was observed in perceptional intensity (Fig. 2). Similarly, some previous studies have demonstrated cases where an OR’s genetic variation was correlated with its ligand’s detection threshold, but not suprathreshold intensity/pleasantness ratings (Menashe et al. 2007; McRae et al. 2012; Trimmer et al. 2019). One possible explanation for this is that the effects occurred at only one of the 2 tested dilutions. Indeed, genetic variation in an OR may be perceptually relevant only at certain concentrations (Trimmer et al. 2019). For muscone, we set up a range of binary steps. According to the threshold result, the 2 tested dilutions for intensity/pleasantness may be too high to observe a difference among the genotypes (Table 1).
It is reported that 7.2%–9% of Caucasians have specific anosmia for exaltolide and 6% of people have specific anosmia for muscone (Whissell-Buechy and Amoore 1973; Amoore et al. 1977; Wysocki 2009). In heterologous assays, 2 minor alleles, G162A and T247K showed a loss of ability to respond to OR5AN1 ligands (Supplementary Fig. 1, Supplementary Table 2). Individuals with these null mutants are predicted to be insensitive to some musk odors; however, these alleles are too rare in the population to allow us to test this hypothesis (1 KK and 0 AA) (Supplementary Fig. 3). Furthermore, the reported percentage of musk anosmia is not consistent with allele frequencies of null mutants (G162A: 0.31%, T247K: 0.31%). This contradictory observation may be due to 2 possibilities: (i) musk odor perception may not be fully explained by only OR5AN1 variation, or (ii) because the reported estimated percentages, methods used, and the definition of specific anosmia varied from study to study, they may not be specific anosmia but include specific hyposmia to musk (Klyuchnikova and Voznessenskaya 2019).
Galaxolide, one of the most commonly used musk compounds for cosmetic detergents, did not activate any of the tested OR5AN1 variants (Sato-Akuhara et al. 2016; Fig. 1H and I, Supplementary Fig. 2). None of the ORs in the OR5AN1 cluster responded in the luciferase reporter gene assay (Supplemental Fig. 4; Sato-Akuhara et al. 2016; Trimmer et al. 2019; Li et al. 2022), the GloSensor cAMP assay (Supplementary Fig. 2B), or a Ca2+ imaging assay (data not shown). Nevertheless, the OR5AN1 L289F mutant was significantly associated with galaxolide perception (P = 0.0046, the Kruskal–Wallis test; P = 0.0050, Wilcoxon rank-sum test with Bonferroni correction [LL vs. FF]) (Fig. 2). OR5A2 and OR4D6, both in the same genetic locus as OR5AN1, were shown to be associated with galaxolide perception (Huysseune et al. 2018; Trimmer et al. 2019; Li et al. 2022), but did not respond to any musk in our in vitro assays (Supplemental Fig. 4, partly data not shown). The inability to identify a galaxolide receptor may be due to its cytotoxicity at concentrations above 30 µM in the luciferase assay (Fig. 1). It should be of note that musk odorants are generally cytotoxic for heterologous cells at high concentrations (Sato-Akuhara et al. 2016).
Genes for OR5AN1 and OR5A1 are located in the same genomic cluster on chromosome 11, and their SNPs L289F and D183N are in high linkage disequilibrium. Cell-based assays show that OR5A1 responds to β-ionone and OR5AN1 responds to a variety of musks, but due to genetic correlations, variation in both genes can predict perception of both odors. Similarly, an individual’s detection threshold for muscone can be predicted by their detection threshold for β-ionone. Genetic linkage occurs throughout the human OR subgenome, and the diversity of human phenotypes is smaller than it would be if the variation in the perception of each odor was independent of the perception of other odors.
OR genes constitute the largest multigene family in mammals, and duplications and deletions of OR genes during evolution have been extremely frequent (Niimura and Nei 2003, 2007). In nonhuman primate species, OR5AN1 orthologous genes have the less functional leucine residue at position 289 (Sato-Akuhara et al. 2016), suggesting that a higher functional allele emerged in humans. Muscone is a male pheromone in musk deer that humans have used for medicine for >2,000 years. Indeed, some reports have been suggested to have physiological effects on humans (Kato et al. 2004; Fukui et al. 2007). Taken together, these findings suggest that L289F might be a mutation i.e. advantageous for human survival.
In summary, OR5AN1 is a crucial macrocyclic and nitro musk receptor in humans, and genetic variation appears to cause variable responsiveness to macrocyclic musks in in vitro assay. Significant differences were observed among OR5AN1 genotypes in musk odor perception; genetic variation of OR5AN1 affects the perceived intensity of exaltolide and ethylene brassylate, and the detection threshold for muscone. Moreover, we found that the sensitive variant of OR5AN1, L289F, is genetically linked to the insensitive variant of OR5A1, D183N, providing a case wherein the detection threshold of 1 odorant could predict the detection threshold of another odorant.
Supplementary Material
Acknowledgments
We thank Takasago International Corporation for providing highly pure musk odorants, T. Hasegawa Corporation for providing solvents, and all participants in this study.
Contributor Information
Narumi Sato-Akuhara, Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Casey Trimmer, Monell Chemical Senses Center, Philadelphia, PA 19104, United States.
Andreas Keller, Laboratory of Neurogenetics and Behavior, The Rockefeller University, New York, NY, United States.
Yoshihito Niimura, Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan; Department of Veterinary Sciences, Faculty of Agriculture, University of Miyazaki, 1-1, Gakuen Kibanadai Nishi, Miyazaki, Miyazaki 889-2192, Japan.
Mika Shirasu, Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Joel D Mainland, Monell Chemical Senses Center, Philadelphia, PA 19104, United States; Department of Neuroscience, University of Pennsylvania School of Medicine, Philadelphia, PA, United States.
Kazushige Touhara, Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Funding
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI grant number JP16J09029 to NS-A, 18K06359 to YN, 18K14651 and 22K06418 to MS, by Japan Society for Bioscience, Biotechnology, and Agrochemistry (JSBBA) Grant for Women Scientists in Challenging Research to MS, and by Exploratory Research for Advanced Technology (ERATO) Touhara Chemosensory Signal Project (grant number 501100009024; JPMJER1202) and the Japan Science and Technology Agency (JST)-Mirai Program (JPMJMI17DC and JPMJMI19D1) to KT and by the National Institutes of Health grants R01 DC013339 to JM; T32 DC000014 and F32 DC014202 to CT, by the National Center for Advancing Translational Sciences (NCATS, National Institutes of Health (NIH)) Clinical and Translational Science Award (CTSA) program grant # UL1 TR000043 to AK..
Authors’ contributions
NS-A, AK, MS, JM, and KT designed research; NS-A, CT, YN, and MS performed research, NS-A, CT, YN, MS, JM, and KT analyzed data, NS-A, CT, MS, JM, and KT wrote the paper.
Conflict of interest
None declared.
Data availability
The data supporting the results of this article will be shared on reasonable request to the corresponding author.
References
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA; 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoore JE, Pelosi P, Forrester LJ.. Specific anosmias to 5α-androst-16-en-3-one and ω-pentadecalactone: the urinous and musky primary odors. Chem Senses. 1977;2(4):401–425. [Google Scholar]
- Doty RL, Laing DG.. Psychophysical measurement of human olfactory function. In: Doty RL (Ed.), Handbook of olfaction and gustation. Hoboken, NJ: John Wiley & Sons, Inc; 2015. p. 225–260. [Google Scholar]
- Doty RL, McKeown DA, Lee WW, Shaman P.. A study of the test-retest reliability of ten olfactory tests. Chem Senses. 1995;20(6):645–656. [DOI] [PubMed] [Google Scholar]
- Eh M. New alicyclic musks: the fourth generation of musk odorants. Chem Biodivers. 2004;1(12):1975–1984. [DOI] [PubMed] [Google Scholar]
- Fukui H, Komaki R, Okui M, Toyoshima K, Kuda K.. The effects of odor on cortisol and testosterone in healthy adults. Neuro Endocrinol Lett. 2007;28(4):433–437. [PubMed] [Google Scholar]
- Gilbert AN, Kemp SE.. Odor perception phenotypes: multiple, specific hyperosmias to musks. Chem Senses. 1996;21(4):411–416. [DOI] [PubMed] [Google Scholar]
- Huysseune S, Veithen A, Quesnel Y.. Olfactory receptor involved in the perception of musk fragrance and the use thereof. Patent WO/2019/110630. 2019/06/13. Belgium; 2018. [Google Scholar]
- Jaeger SR, McRae JF, Bava CM, Beresford MK, Hunter D, Jia Y, Chheang SL, Jin D, Peng M, Gamble JC, et al. A Mendelian trait for olfactory sensitivity affects odor experience and food selection. Curr Biol. 2013;23(16):1601–1605. [DOI] [PubMed] [Google Scholar]
- Kato Y, Tanaka H, Yamaoka M.. Study of the stimulation on the secretion of the female sex hormone by some perfumery raw material. Aroma Res. 2004;5:64–68. [Google Scholar]
- Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H.. Genetic variation in a human odorant receptor alters odour perception. Nature. 2007;449(7161):468–472. [DOI] [PubMed] [Google Scholar]
- Klyuchnikova MA, Voznessenskaya VV.. Specific anosmia in humans and animals: environmental and genetic influences. Ukr J Ecol. 2019;9(3):52–59. [Google Scholar]
- Li B, Kamarck ML, Peng Q, Lim F-L, Keller A, Smeets MAM, Mainland JD, Wang S.. From musk to body odor: decoding olfaction through genetic variation. PLoS Genet. 2022;18(2):e1009564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainland JD, Keller A, Li YR, Zhou T, Trimmer C, Snyder LL, Moberly AH, Adipietro KA, Liu WLL, Zhuang H, et al. The missense of smell: functional variability in the human odorant receptor repertoire. Nat Neurosci. 2014;17(1):114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainland JD, Li YR, Zhou T, Liu WL, Matsunami H.. PMC4412152; Human olfactory receptor responses to odorants. Sci Data. 2015;2:150002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock TS, Adipietro K, Titlow WB, Breheny P, Walz A, Mombaerts P, Matsunami H.. Pmc4236398; In vivo identification of eugenol-responsive and muscone-responsive mouse odorant receptors. J Neurosci. 2014;34(47):15669–15678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae JF, Jaeger SR, Bava CM, Beresford MK, Hunter D, Jia Y, Chheang SL, Jin D, Peng M, Gamble JC, et al. Identification of regions associated with variation in sensitivity to food-related odors in the human genome. Curr Biol. 2013;23(16):1596–1600. [DOI] [PubMed] [Google Scholar]
- McRae JF, Mainland JD, Jaeger SR, Adipietro KA, Matsunami H, Newcomb RD.. Pmc3408771; Genetic variation in the odorant receptor OR2J3 is associated with the ability to detect the “grassy” smelling odor, cis-3-hexen-1-ol. Chem Senses. 2012;37(7):585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menashe I, Abaffy T, Hasin Y, Goshen S, Yahalom V, Luetje CW, Lancet D.. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol. 2007;5(11):e284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J, Piantanida TP, Eddy RL, Shows TB, Hogness DS.. Molecular genetics of inherited variation in human color vision. Science. 1986;232(4747):203–210. [DOI] [PubMed] [Google Scholar]
- Niimura Y, Nei M.. Evolution of olfactory receptor genes in the human genome. Proc Natl Acad Sci USA. 2003;100(21):12235–12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M.. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS One. 2007;2(8):e708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragnarsson L, Andersson Å, Thomas WG, Lewis RJ.. Mutations in the NPxxY motif stabilize pharmacologically distinct conformational states of the α1B- and β2-adrenoceptors. Sci Signal. 2019;12(572):eaas9485. [DOI] [PubMed] [Google Scholar]
- Rossiter KJ. Structure–odor relationships. Chem Rev. 1996;96(8):3201–3240. [DOI] [PubMed] [Google Scholar]
- Rovati GE, Capra V, Neubig, RR.. The highly conserved DRY motif of class A G protein-coupled receptors: beyond the ground state. Mol Pharmacol. 2007;71(4):959–964. [DOI] [PubMed] [Google Scholar]
- Sato-Akuhara N, Horio N, Kato-Namba A, Yoshikawa K, Niimura Y, Ihara S, Shirasu M, Touhara K.. Ligand specificity and evolution of mammalian musk odor receptors: effect of single receptor deletion on odor detection. J Neurosci. 2016;36(16):4482–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sell CS. Chemistry and the sense of smell. Hoboken, NJ: John Wiley & Sons; 2014. ISBN 1118522966, 9781118522967. [Google Scholar]
- Shin J-H, Blay S, McNeney B, Graham J.. LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J Stat Softw. 2006;16(1):1–9. [Google Scholar]
- Shirasu M, Yoshikawa K, Takai Y, Nakashima A, Takeuchi H, Sakano H, Touhara K.. Olfactory receptor and neural pathway responsible for highly selective sensing of musk odors. Neuron. 2014;81(1):165–178. [DOI] [PubMed] [Google Scholar]
- Trimmer C, Keller A, Murphy NR, Snyder LL, Willer JR, Nagai MH, Katsanis N, Vosshall LB, Matsunami H, Mainland JD.. Genetic variation across the human olfactory receptor repertoire alters odor perception. Proc Natl Acad Sci USA. 2019;116(19):9475–9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A, Filipek S.. Action of molecular switches in GPCRs—theoretical and experimental studies. Curr Med Chem. 2012;19(8):1090–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whissell-Buechy D, Amoore JE.. Odour-blindness to musk: simple recessive inheritance. Nature. 1973;242(5395):271–273. [DOI] [PubMed] [Google Scholar]
- Wysocki CJ. Update on racial and gender differences in odor perception. Chem Senses. 2009;34(7):631–642. [Google Scholar]
- Zhuang H, Matsunami H.. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat Protoc. 2008;3(9):1402–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the results of this article will be shared on reasonable request to the corresponding author.