Abstract
The northern Gulf of Mexico (nGOM) hypoxic zone is a shallow water environment where methane, a potent greenhouse gas, fluxes from sediments to bottom water and remains trapped due to summertime stratification. When the water column is destratified, an active planktonic methanotrophic community could mitigate the efflux of methane, which accumulates to high concentrations, to the atmosphere. To investigate the possibility of such a biofilter in the nGOM hypoxic zone we performed metagenome assembly, and metagenomic and metatranscriptomic read mapping. Methane monooxygenase (pmoA) was an abundant transcript, yet few canonical methanotrophs have been reported in this environment, suggesting a role for non-canonical methanotrophs. To determine the identity of these methanotrophs, we reconstructed six novel metagenome-assembled genomes (MAGs) in the Planctomycetota, Verrucomicrobiota and one putative Latescibacterota, each with at least one pmoA gene copy. Based on ribosomal protein phylogeny, closely related microbes (mostly from Tara Oceans) and isolate genomes were selected and co-analyzed with the nGOM MAGs. Gene annotation and read mapping suggested that there is a large, diverse and unrecognized community of active aerobic methanotrophs in the nGOM hypoxic zone and in the global ocean that could mitigate methane flux to the atmosphere.
Keywords: hypoxic zone, metagenome assembled genome (MAG), metagenomics, metatranscriptomics, methane, methanotrophs
Multi-omics approaches and analyses were used to identify and characterize novel, uncultured, non-canonical marine methanotrophs, including the pathways by which they carry out methane oxidation and their activity.
Introduction
Methane, a potent greenhouse gas, at 150% of pre-industrial levels and rising (Saunois et al. 2016), has reached the highest level in the last 800 000 years (IPCC 2013). Emissions from marine environments are an important source of atmospheric methane, with the coastal and open ocean accounting for 1%–13% of natural emissions (Saunois et al. 2016). Methane produced by biological processes in sediments or in the water column can escape to the atmosphere (Reeburgh 2007, IPCC 2013, Kirschke et al. 2013, Saunois et al. 2020, Rosentreter et al. 2021). Conversely, microbial oxidation (aerobic and anaerobic) can capture methane and reduce atmospheric efflux. Methanotrophs account for 5% of the global methane sink (Saunois et al. 2020). While this takes place primarily in soils and marine sediments, methane oxidation also occurs in the marine water column (Reeburgh 2007, IPCC 2013, Kirschke et al. 2013, Saunois et al. 2020, Rosentreter et al. 2021).
In the Gulf of Mexico (GOM), methane enters the water column primarily from natural seeps, but also from drilling operations and accidents, such as the Deepwater Horizon oil spill in 2010, and from the flux of methane from anoxic sediments. When methane fluxes from sediments to the stratified water column in the northern GOM (nGOM) hypoxic zone, also called the “dead zone”, it remains trapped in bottom water due to density stratification during the summer (Rogener et al. 2021). Importantly, methane concentrations are high relative to concentrations expected from equilibrium with atmospheric concentrations in this coastal dead zone (Rogener et al. 2018), with concentrations ranging from 5 to 641 nM (Rogener et al. 2021). Overturning of the shallow, hypoxic bottom water in the nGOM dead zone can result in the flux of trapped gases (e.g. methane, nitrous oxide) to the atmosphere (Walker et al. 2010, Rogener et al. 2021). Thus, understanding the fate of this bottom water methane is important (Knief 2015) and requires determining the function of methanotrophs that could act as a biofilter in the water column.
Methanotrophs use methane as a carbon and energy source and are a subgroup of methylotrophs, microbes that degrade single-carbon compounds (Anthony 1982). Methanotrophs thrive at oxic-anoxic interfaces (Hanson and Hanson 1996, Knief 2015), but exist across a range of environments, reflecting their physiological diversity. For example, Kalyuzhnaya et al. (2019) suggested methanotrophic bacteria are ubiquitous in the environment as both planktonic microbes and as symbionts of several organisms, including mussels, snails, sponges and tubeworms (reviewed in Dubilier et al. 2008).
Canonical aerobic methanotrophs, described as early as 1906 (Söhngen 1906), are categorized based on morphological characteristics, such as membrane type, physiology, methane oxidation pathway, methane monooxygenase sequence similarity and 16S rRNA phylogeny (Hanson and Hanson 1996, Kalyuzhnaya et al. 2019). In the marine environment, canonical aerobic marine methanotrophs are mainly affiliated with Proteobacteria, and more specifically the Gammaproteobacteria and Alphaproteobacteria (Hanson and Hanson 1996). More recently, methylotrophy is viewed as a modular genetic system encompassing an indeterminate number of gene combinations that enable transformation of single-carbon compounds into biomass and/or other metabolically useful compounds (Chistoserdova 2011).
The first step in aerobic methane oxidation (MOx) is the conversion of methane to methanol via methane monooxygenase (particulate or soluble, pMMO/sMMO) (Hanson and Hanson 1996). Virtually all methanotrophs code for pMMO (Murrell et al. 2000); however, only a few methanotrophs encode sMMO, which is utilized in copper-deficient environments (Semrau et al. 2010). The ubiquity of pMMO in the marine environment makes the alpha subunit of particulate methane monooxygenase (pmoA) a robust functional marker gene to identify methanotrophs (Mcdonald and Murrell 1997). Crespo-Medina et al. (2014) used this approach to identify a diversity of methanotrophs that responded to the input of methane during the Deepwater Horizon oil spill in the Gulf of Mexico.
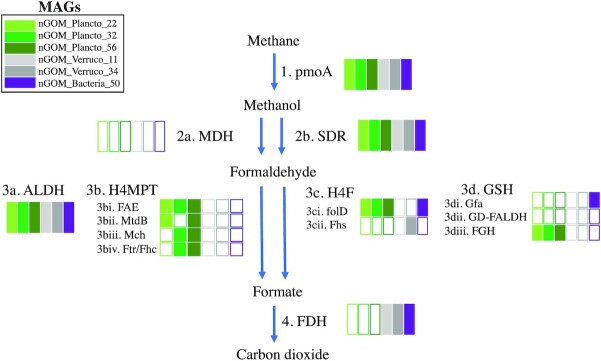
Fewer studies have looked beyond pmoA and evaluated the complete four-step pathway that is utilized by aerobic, planktonic methanotrophs. Methane oxidation proceeds via its enzymatic conversion to methanol (discussed above) followed by transformation of methanol to formaldehyde, which is mediated by methanol dehydrogenase (MDH) or a short-chain alcohol dehydrogenase (SDR). Formaldehyde is a central intermediate for methanotrophs, assimilated or oxidized to formate through multiple pathways, and formate is then assimilated via the serine pathway or is oxidized to carbon dioxide (CO2) via formate dehydrogenase (FDH) (Hanson and Hanson 1996, Vorholt 2002, Chistoserdova et al. 2009) (Fig. 1). In this study, we assessed the complete pathways for methane oxidation in multiple novel genomes. We would also note that some methanotrophs, such as Candidatus Methylomirabilis oxyfera, and Proteobacteria and Verrucomicrobiaota, can assimilate CO2, utilizing the serine or Calvin-Benson-Bassham cycles (Khadem et al. 2011, Rasigraf et al. 2014), which were also evaluated in our study.
Figure 1.
Schematic of the aerobic methane oxidation pathway. The colored boxes, which are color coded for taxonomy, correspond to the presence (colored boxes) or absence (empty boxes) of genes coding for enzymes necessary for these steps in the nGOM MAGs. Blue arrows indicate oxidation steps. The abbreviations shown in the figure are as follows: pmoA/AMO: particulate methane monooxygenase/ammonia monooxygenase; MDH: methanol dehydrogenase; SDR: short-chain alcohol dehydrogenase; ALDH: aldehyde dehydrogenase; H4MPT: tetrahydromethanopterin pathway; FAE: formaldehyde-activating enzyme; MtdB: methylene tetrahydromethanopterin dehydrogenase; Mch: methenyl-tetrahydromethanopterin cyclohydrolase; Ftr/Fhc: formylmethanofuran-tetrahydromethanopterin formyltransferase; H4F: tetrahydrofolate pathway; folD: tetrahydrofolate dehydrogenase/cyclohydrolase; Fhs: formate–tetrahydrofolate ligase; GSH: glutathione-dependent pathway; Gfa: glutathione-dependent formaldehyde activating enzyme; GD-FALDH: glutathione-dependent formaldehyde dehydrogenase; FGH: Formyl-glutathione hydrolase; FDH: formate dehydrogenase.
Despite the high methane concentrations in the nGOM dead zone, microbial ecology studies using iTag sequencing of 16S rRNA gene amplicons have reported low to undetectable abundances of canonical methanotrophs in this environment (Gillies et al. 2015, Campbell et al. 2019). Observations of high methane concentrations and oxidation rates in the nGOM dead zone (Kelley 2003, Rogener et al. 2018, 2021), coupled with the lack of canonical methanotrophs, suggested a role for an active planktonic methanotrophic community. Here, we sought to determine what planktonic microorganisms could carry out methane oxidation (MOx) through assembly of metagenomes and metabolic reconstruction. Further, to determine which microbes were actively mediating MOx, metatranscriptomic reads were mapped to our metagenome-assembled genomes (MAGs). Finally, complete to near complete genomes of close relatives of those nGOM microbes were identified and included in this analysis to evaluate active MOx in the 2013 nGOM dead zone and more broadly in the marine environment. Inclusion of these close relatives was done for two reasons. First, the nGOM MAGs assembled and presented herein were not complete, which presents challenges when trying to determine the full genetic repertoire encoded in these uncultured microbes. For example, co-analysis of relatives with complete to near-complete genomes provided information on genes that may have been missing due to an incomplete genome assembly in the nGOM MAGs. Second, the additional microbes were sampled from the global ocean, which expands our understanding of microbes mediating MOx outside of the nGOM.
Materials and Methods
Sample location and collection
Samples were collected at the oxygen minimum zone (OMZ) on the R/V Pelican in the dead zone along transects running perpendicular to the hypoxic zone starting from the mouth of the Mississippi River and ending near the Louisiana-Texas border in 2013 (Gillies et al. 2015) (see Supp. Fig. 1). The depth of water sample collection ranged from 6 to 35 meters below sea level, with an average sampling depth of 16 m, with up to 10 L collected at each location (Gillies et al. 2015). Oxygen concentrations were determined with a CTD oxygen sensor and calibrated using the Winkler method (Gillies et al. 2015). For our study, the samples O_D1, H_D2, H_D3, O_E2 and H_E4 (O means oxic, H means hypoxic) were selected for “omics” sequencing (Supp. Fig. 1).
Microbial sampling and DNA and RNA extraction
Samples for microbial analysis were collected by filtering up to 10 L of seawater through 2.7-μm pre-filters and then onto 0.22-μm Sterivex filters, which were preserved in RNAlater and immediately frozen. Details regarding DNA and RNA extraction, sequencing and analysis can be found in Gillies et al. (2015) and Thrash et al. (2017). Briefly, DNA and RNA were extracted directly off of the frozen, RNAlater-preserved filters by placing half of a Sterivex filter in a Lysing matrix E glass/zirconia/silica beads tube (MP Biomedicals, Santa Ana, CA, USA) following the protocol in Gillies et al. (2015) that combines phenol: chloroform: isoamyalcohol (25 : 24 : 1) and bead beating. Genomic DNA and RNA were purified using a QIAGEN (Valencia, CA, USA) AllPrep DNA/RNA Kit. DNA was quantified using a Qubit2.0 Fluorometer (Life Technologies, Grand Island, NY, USA). RNA quality was analyzed using an Agilent TapeStation with an RNA integrity number (RIN) (16S/23S rRNA gene ratio) to assess degradation (scale of 1 to 10, 10 being undegraded RNA). RNA with RIN scores of ≥8 was chosen for metatranscriptomic sequencing. Prior to sequencing, rRNA was subtracted from total RNA using a Ribo-Zero kit (Illumina) and mRNA was reverse transcribed to cDNA, as described in Mason et al. (2012).
16S rRNA gene sequence data
16S rRNA gene data from the 2013 and 2014 nGOM dead zone presented in Gillies et al. (2015) and Campbell et al. (2019) were used to determine the abundances of canonical methanotrophs (Tavormina et al. 2008). In these datasets, the relative abundances of Methylococcales, specifically Methylobacter, Methylococcus and Methylomicrobium in the Gammaproteobacteria, and Methylosinus and Methylocystis in the Alphaproteobacteria, were determined.
Metagenome and metatranscriptome sequencing and analyses
Metagenomes and metatranscriptomes were sequenced separately using six samples per lane with the Illumina HiSeq 2000, to produce 100 bp, paired-end reads (Thrash et al. 2017). Co-assembly of the metagenomes is described in Thrash et al. (2017). Briefly, reads were quality-filtered, pooled and assembled using IDBA-UD (Peng et al. 2012), binning was performed using emergent self-organizing maps (ESOM) of contigs ≥5 kb, bin quality control was done with CheckM (Parks et al. 2015) and annotation was carried out with the Integrated Microbial Genomes (IMG) (Markowitz et al. 2014), which resulted in 70 MAGs published previously (Thrash et al. 2017, 2018), plus an additional seven MAGs that were assembled but not published. An in silico search for pmoA annotations in all MAGs was carried out using the IMG annotation pipeline v. 5.0.0 (https://img.jgi.doe.gov/docs/pipelineV5/) and searched for functions and genes within these annotations, with six nGOM MAGs identified as having this gene. Of these six MAGs, Thrash et al. (2017) presented an analysis of only the unclassified Bacteria, putatively identified as Latescibacterota (bin 50) analyzed here, and while its pmoA gene was annotated and reported, this finding was not discussed. Taxonomic classification of the nGOM MAGs was assigned using GTDB-Tk (v. 1.0.2) (Parks et al. 2018, Chaumeil et al. 2019).
Five unassembled metatranscriptomes and five unassembled metagenomes from Thrash et al. (2017) were mapped to the six nGOM MAGs that had at least one pmoA copy using Bowtie2 (Langmead and Salzberg 2012). Prior to mapping, ribosomal RNA was subtracted from the metatranscriptome reads using riboPicker v. 0.4.3 with the default settings (Schmieder et al. 2012). On average, 8% of the metatranscriptomic reads were ribosomal RNA that were subsequently removed in silico with riboPicker.
Using the GTDB-tk ribosomal protein phylogenetic tree, close relatives of the nGOM MAGs were identified using phylogeny with monophyly and branch length as decision-making criteria for inclusion (see Supp. Fig. 2). All of these close relatives encoded pmoA. Four additional Proteobacteria MAGs were included here because: (1) they represent a canonical methanotroph clade; (2) they encode pmoA, as the nGOM MAGs do; and (3) they were also sampled from the GOM (deepwater asphalt seeps) (Rubin-Blum et al. 2019). All together, we assessed six nGOM MAGs, 28 non-nGOM MAGs and four canonical southern GOM MAGs that were included in all subsequent analyses (Table 1). A second phylogenetic tree (Fig. 2C) was constructed with the 38 genomes analyzed herein using the FastTree (v. 2) (Price et al. 2010) implementation in Anvi'o (v. 6.2) (Eren et al. 2015) with all 71 single-copy core genes (ribosomal and non-ribosomal) in the default bacterial collection and visualized using iTOL (v. 6) (Letunic and Bork 2021).
Table 1.
Microbial taxonomy, study and sample information for MAGs and genomes from cultivated representatives. The nGOM MAGs are shown in bold text.
| Genome | Taxonomy | Reference | Sample location | GenBank accession* |
|---|---|---|---|---|
| Methylococcales symb Hymedesmia L1 | Proteobacteria;Gammaproteobacteria;Methylococcales;Methylomonadaceae | Rubin-Blum et al. 2019 | GOM (southern) | GCA_003666335.1 |
| Methylococcales symb Hymedesmia N1 | Proteobacteria;Gammaproteobacteria;Methylococcales;Methylomonadaceae | Rubin-Blum et al. 2019 | GOM (southern) | GCA_003666385.1 |
| Methylococcales symb Iophon M1 | Proteobacteria;Gammaproteobacteria;Methylococcales;Methylomonadaceae | Rubin-Blum et al. 2019 | GOM (southern) | GCA_003666325.1 |
| Methylococcales symb Iophon M2 | Proteobacteria;Gammaproteobacteria;Methylococcales;Methylomonadaceae | Rubin-Blum et al. 2019 | GOM (southern) | GCA_003666345.1 |
| nGOM Plancto 22 | Planctomycetota;Planctomycetes;Planctomycetales;Planctomycetaceae | this study | GOM (northern) | 2693429805 (IMG ID) |
| nGOM Plancto 32 | Planctomycetota;Planctomycetes;Planctomycetales;Planctomycetaceae | this study | GOM (northern) | 2651870066 (IMG ID) |
| nGOM Plancto 56 | Planctomycetota;Planctomycetes;Planctomycetales;Planctomycetaceae | this study | GOM (northern) | 2651870065 (IMG ID) |
| Fuerstia marisgermanicae NH11*** | Planctomycetota;Planctomycetes;Planctomycetales;Planctomycetaceae | Kohn et al. 2016 | Wadden Sea Germany; crab shell | GCA_001983935.1 |
| Planctomyces sp SHPL14*** | Planctomycetota;Planctomycetes;Planctomycetales;Planctomycetaceae | NA^ | sugar beet rhizosphere | GCA_001610835.1 |
| Planctomycetaceae bacterium Baikal G1 2R | Planctomycetota;Planctomycetes;Pirellulales;UBA1268 | Cabello-Yeves et al. 2018 | Lake Baikal (Siberia, Russia) | GCA_002737405.1 |
| Planctomycetaceae bacterium NAT223 | Planctomycetota;Planctomycetes;Pirellulales;Pirellulaceae | Tully et al. 2018 | **Eastern Tropical North Pacific (TARA_137) | GCA_002705165.1 |
| Planctomycetaceae bacterium SAT2750 | Planctomycetota;Planctomycetes;Pirellulales;Pirellulaceae | Tully et al. 2018 | **Equatorial Tropical Pacific (TARA_128) | GCA_002708345.1 |
| Planctomycetaceae bacterium SP166 | Planctomycetota;Planctomycetes;Pirellulales;Pirellulaceae | Tully et al. 2018 | **Equatorial Tropical Pacific (TARA_128) | GCA_002709045.1 |
| Planctomycetaceae bacterium SP4 | Planctomycetota;Planctomycetes;Pirellulales;Pirellulaceae | Tully et al. 2018 | **coast of Chile (TARA_093) | GCA_002717145.1 |
| Planctomycetaceae bacterium UBA2671 | Planctomycetota;Planctomycetes;Planctomycetales;Planctomycetaceae | Parks et al. 2017 | NA (marine) | GCA_002359185.1 |
| Planctomycetaceae bacterium UBA2972 | Planctomycetota;Planctomycetes;Pirellulales;Pirellulaceae | Parks et al. 2017 | NA (marine) | GCA_002348315.1 |
| Planctomycetaceae bacterium UBA4655 | Planctomycetota;Planctomycetes;Pirellulales;UBA1268 | Parks et al. 2017 | NA (freshwater) | GCA_002405515.1 |
| Rhodopirellula sp NAT14 | Planctomycetota;Planctomycetes;Pirellulales;Pirellulaceae | Tully et al. 2018 | **Arabian Sea (TARA_036) | GCA_002698965.1 |
| Rhodopirellula sp NAT69 | Planctomycetota;Planctomycetes;Pirellulales;Pirellulaceae | Tully et al. 2018 | **Arabian Sea (TARA_036) | GCA_002701385.1 |
| Rhodopirellula sp SAT12 | Planctomycetota;Planctomycetes;Pirellulales;Pirellulaceae | Tully et al. 2018 | **Arabian Sea (TARA_036) | GCA_002714565.1 |
| Rubinisphaera brasiliensis DSM5305 | Planctomycetota;Planctomycetes;Planctomycetales;Planctomycetaceae | Scheuner, C. et al. 2014 | Water from salt pit, Lagoa Vermelha, Brazil | GCA_000165715.2 |
| Gemmatimonadetes bacterium EAC654 | Latescibacterota;UBA2968;UBA8231;UBA8231 | Tully et al. 2018 | **Eastern Tropical North Pacific (TARA_137) | GCA_002693325.1 |
| Gemmatimonadetes bacterium NP81 | Latescibacterota;UBA2968;UBA8231;UBA8231 | Tully et al. 2018 | southern coast of Africa (TARA_065) | GCA_002726335.1 |
| Gemmatimonadetes bacterium NP952 | Latescibacterota;UBA2968;UBA8231;UBA8231 | Tully et al. 2018 | Red Sea (TARA_032) | GCA_002725915.1 |
| Gemmatimonadetes bacterium RS821 | Latescibacterota;UBA2968;UBA8231;UBA8231 | Tully et al. 2018 | southern coast of Africa (TARA_065) | GCA_002727195.1 |
| Gemmatimonadetes bacterium RS822 | Latescibacterota;UBA2968;UBA8231;UBA8231 | Tully et al. 2018 | southern coast of Africa (TARA_065) | GCA_002724215.1 |
| Gemmatimonadetes bacterium SAT128 | Latescibacterota;UBA2968;UBA2968;GCA-2 709 665 | Tully et al. 2018 | **Equatorial Tropical Pacific (TARA_128) | GCA_002714465.1 |
| Gemmatimonadetes bacterium SAT162 | Latescibacterota;UBA2968;UBA2968;UBA2968 | Tully et al. 2018 | **Equatorial Tropical Pacific (TARA_128) | GCA_002712305.1 |
| Gemmatimonadetes bacterium SP138 | Latescibacterota;UBA2968;UBA2968;UBA2968 | Tully et al. 2018 | **Equatorial Tropical Pacific (TARA_128) | GCA_002709665.1 |
| Latescibacteria bacterium UBA6620 | Latescibacterota;UBA2968;UBA2968;UBA2968 | Parks et al. 2017 | NA (marine) | GCA_002433075.1 |
| nGOM Bacteria 50 | unclassified;unclassified;unclassified;unclassified | Thrash et al. 2017 | GOM (northern) | 2693429804 (IMG ID) |
| nGOM Verruco 11 | Verrucomicrobiota;Verrucomicrobiae;Verrucomicrobiales;unclassified | this study | GOM (northern) | 2651870083 (IMG ID) |
| nGOM Verruco 34 | Verrucomicrobiota;Verrucomicrobiae;Verrucomicrobiales;unclassified | this study | GOM (northern) | 2651870086 (IMG ID) |
| Pedosphaera sp ARS72 | Verrucomicrobiota;Verrucomicrobiae;Pedosphaerales;AAA164-E04 | Tully et al. 2018 | **Arabian Sea (TARA_036) | GCA_002686885.1 |
| Pedosphaera sp MED719 | Verrucomicrobiota;Verrucomicrobiae;Pedosphaerales;AAA164-E04 | Tully et al. 2018 | Mediterranean Sea (TARA_018) | GCA_002690655.1 |
| Verrucomicrobia bacterium SCGCAAA164E04 | Verrucomicrobiota;Verrucomicrobiae;Pedosphaerales;AAA164-E04 | Stepanauskas et al. 2013 | Gulf of Maine | GCA_000383715.1 |
| Verrucomicrobia bacterium UBA2970 | Verrucomicrobiota;Verrucomicrobiae;Pedosphaerales;AAA164-E04 | Parks et al. 2017 | NA (marine) | GCA_002348345.1 |
| Verrucomicrobiales bacterium SP5 | Verrucomicrobiota;Verrucomicrobiae;Pedosphaerales;AAA164-E04 | Tully et al. 2018 | **Arabian Sea (TARA_036) | GCA_002715965.1 |
*Unless otherwise indicated
** site where annual mean oxygen <2 ml/l (Pesant et al. 2015)
*** Cultured microbe
^submitter is Victor de Jager (v.dejager@nioo.knaw.nl)
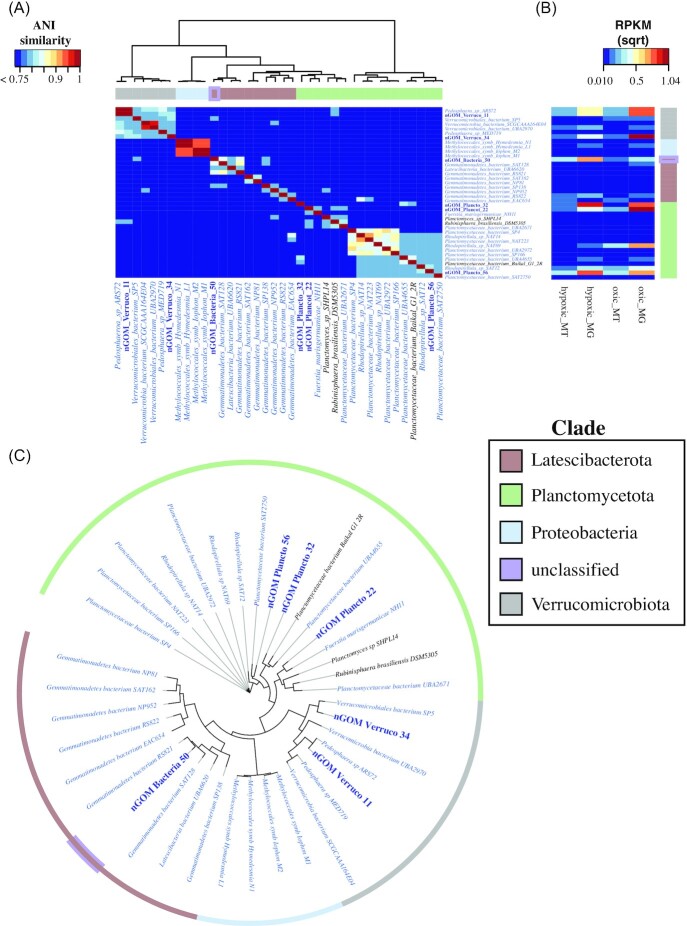
Figure 2.
ANI heatmap (A), square root RPKM abundance (B) and phylogenetic tree (C) of the 38 genomes. Genome names are color-coded blue and bold (nGOM MAGs), black (non-marine) and blue (marine). Only ANI above 75% is shown in 2A. The clade color code for nGOM Bacteria 50 is brown with a purple border to indicate that is putatively identified as Latescibacteria, but is herein classified as unknown.
Average nucleotide identity (ANI) was determined using Sourmash with -containment -ani -ksize 31 (Brown and Irber 2016). All genomes were analyzed using Anvi’o (v. 6.2) (Eren et al. 2015). Genes were annotated with functions by using the “anvi-run-pfams” and “anvi-run-ncbi-cogs” with the Pfam (Bateman et al. 2004) and COGs (Tatusov et al. 2000) databases. Outside of Anvi'o, the Pfam and COGs annotations were verified by reviewing annotations using blastx with DIAMOND (v. 0.9.30) (Buchfink et al. 2014) and NCBI's non-redundant RefSeq protein dataset (accessed from NCBI on 03/10/2020) (Tatusova et al. 2016). Functional annotation using COGs and Pfams databases, or blastx with the non-redundant RefSeq protein database, were confirmed if two of three database annotations agreed. These annotations were then used to identify the gene and transcript sequences comprising the different modules of the methane oxidation pathways. Using Bowtie2, metatranscriptome and metagenome reads were mapped to the annotated genes in each genome to determine abundance and expression by calculating reads per kilobase per million (RPKM) values following Thrash et al. (2017). RPKM values were also calculated to determine genome abundances and activity by mapping metagenome and metatranscriptome reads to each genome. To classify functions that were part of the full pangenome core, “anvi-get-enriched-functions-per-pan-group” was used in Anvi'o to identify individual clade cores using the ribosomal protein tree phylogeny (see above). This command was also used to determine what functions were statistically enriched in each clade by using a generalized linear model with logit linkage function.
Results
Chemistry
Of the five samples from the nGOM hypoxic zone selected for metagenomic and metatranscriptomic sequencing (Supp. Fig. 1) that were originally reported on in Thrash et al. (2017), oxygen concentrations were reported in Gillies et al. (2015) and additionally in Thrash et al. (2017) for the five metagenomic and metatranscriptomic samples. Of the five samples selected, two were oxic, O_D1 and O_E2 (4.12 and 2.64 mg L-1 dissolved oxygen (O2), respectively), and three were hypoxic, H_D3, H_D2 and H_E4 (0.4, 0.33 and 0.31 mg L-1 O2, respectively). Sampling at the same time and at directly adjacent sites, Rogener et al. (2021) found that methane concentrations were negatively correlated with O2 concentrations and reported MOx rates as high as 192 nmol L−1d−1 and depth-integrated methane oxidation rates (Fmox) of 0.2 µmol m−2d−1 in 2013, which was lower than Fmox in the other two years they analyzed (2.4 in 2015 and 322 in 2016). The estimated atmospheric flux of methane was also lowest in 2013 compared with the other two years (27 vs. 97 and 278 µmol m−2d−1) (Rogener et al. 2021).
Paucity of canonical methanotrophs in a legacy 16S rRNA gene sequence dataset
Reanalysis of the 16S rRNA gene amplicon sequence data from the 2013 dead zone that was presented in Gillies et al. (2015) and the 2014 dead zone reported on in Campbell et al. (2019) revealed that the relative proportions of canonical methanotrophs (Tavormina et al. 2008) were very low. Specifically, Methylococcales (Gammaproteobacteria) comprised an average of 0.07% of the population in the oxic samples and an average of 0.09% in hypoxic samples. Other canonical methanotrophs, such as Methylobacter, Methylococcus and Methylomicrobium in the Gammaproteobacteria, and Methylosinus and Methylocystis in the Alphaproteobacteria, were either not observed or were below 0.004% relative abundance in samples collected at any site in either year.
Taxonomy, phylogenetic relatedness, abundance and activity
Six nGOM MAGs were co-assembled from the 2013 dead zone metagenome data (Thrash et al. 2017) with average genome completeness of 58% and 2.2% contamination (Table 2). Five of these microbes were classified as Planctomycetota (three MAGs) and Verrucomicrobiota (two MAGs). The remaining MAG was putatively classified as Latescibacterota (formerly WS3) by ribosomal protein phylogeny, but as PAUCF/SAUL by 16S rRNA genes and amplicon data (Thrash et al. 2017), so is herein designated as unclassified. These nGOM MAGs all encoded genes for PmoA, part of the pMMO enzyme necessary for methane oxidation to methanol, as well as a full or partial MOx pathway (Fig. 1). In addition to these six nGOM MAGS, relatives chosen using the GTDB phylogenetic tree monophyly and branch length as criteria for inclusion, resulted in 28 non-nGOM genomes being co-analyzed, with an additional four genomes included that represented canonical methanotrophs. Of these 32 additional genomes that were co-analyzed here, 92% were sampled from the marine environment (Table 1, Fig. 2C) and all encoded PmoA and a partial to full MOx pathway (Fig. 3).
Table 2.
Statistics for MAGs and genomes from cultivated representatives. The nGOM MAGs are shown in bold text.
| Genome | Completeness | Contamination | Contig count | Scaffold count | Genome size | % GC |
|---|---|---|---|---|---|---|
| Methylococcales symb Hymedesmia L1 | 94.96% | 0.29% | 86 | 84 | 2 010 086 | 37.83 |
| Methylococcales symb Hymedesmia N1 | 96.45% | 0.31% | 64 | 63 | 2 219 015 | 37.76 |
| Methylococcales symb Iophon M1 | 95.59% | 0.47% | 123 | 123 | 2 092 041 | 37.73 |
| Methylococcales symb Iophon M2 | 94.55% | 0.46% | 128 | 126 | 2 006 182 | 37.67 |
| nGOM Plancto 22 | 46.41% | 2.22% | 3330 | 688 | 3 841 030 | 55.52 |
| nGOM Plancto 32 | 52.07% | 7.02% | 2123 | 362 | 2 492 421 | 62.50 |
| nGOM Plancto 56 | 83.84% | 2.56% | 3836 | 322 | 4 807 217 | 51.78 |
| Fuerstia marisgermanicae NH11** | 96.60% | 1.11% | 1 | 1 | 8 920 478 | 55.90 |
| Planctomyces sp SHPL14** | 98.89% | 2.22% | 1 | 1 | 8 442 773 | 65.56 |
| Planctomycetaceae bacterium Baikal G1 2R | 93.04% | 13.22% | 98 | 98 | 3 383 196 | 57.49 |
| Planctomycetaceae bacterium NAT223 | 67.86% | 2.60% | 157 | 157 | 2 943 028 | 48.09 |
| Planctomycetaceae bacterium SAT2750 | 56.03% | 0.00% | 102 | 102 | 1 910 127 | 58.19 |
| Planctomycetaceae bacterium SP166 | 92.87% | 0.00% | 69 | 69 | 4 535 358 | 46.53 |
| Planctomycetaceae bacterium SP4 | 70.54% | 0.00% | 115 | 115 | 4 424 693 | 47.85 |
| Planctomycetaceae bacterium UBA2671 | 95.56% | 0.00% | 330 | 127 | 6 652 450 | 48.68 |
| Planctomycetaceae bacterium UBA2972 | 95.87% | 0.11% | 401 | 253 | 4 362 043 | 49.07 |
| Planctomycetaceae bacterium UBA4655 | 90.08% | 0.00% | 626 | 320 | 3 844 742 | 68.04 |
| Rhodopirellula sp NAT14 | 89.32% | 2.30% | 192 | 192 | 5 424 119 | 46.52 |
| Rhodopirellula sp NAT69 | 97.58% | 0.00% | 100 | 100 | 4 890 980 | 48.08 |
| Rhodopirellula sp SAT12 | 94.64% | 1.18% | 119 | 119 | 4 537 602 | 48.43 |
| Rubinisphaera brasiliensis DSM5305 | 95.56% | 2.22% | 1 | 1 | 6 006 602 | 56.45 |
| Gemmatimonadetes bacterium EAC654 | 89.50% | 7.14% | 166 | 166 | 3 573 524 | 41.14 |
| Gemmatimonadetes bacterium NP81 | 83.62% | 4.92% | 279 | 279 | 5 970 290 | 63.93 |
| Gemmatimonadetes bacterium NP952 | 85.61% | 5.01% | 293 | 293 | 3 810 359 | 62.74 |
| Gemmatimonadetes bacterium RS821 | 86.81% | 1.10% | 38 | 38 | 5 422 662 | 59.09 |
| Gemmatimonadetes bacterium RS822 | 93.59% | 3.30% | 266 | 266 | 5 850 589 | 62.44 |
| Gemmatimonadetes bacterium SAT128 | 97.74% | 8.24% | 78 | 78 | 5 214 681 | 56.54 |
| Gemmatimonadetes bacterium SAT162 | 90.11% | 2.75% | 146 | 146 | 4 765 829 | 60.21 |
| Gemmatimonadetes bacterium SP138 | 94.44% | 4.40% | 84 | 84 | 5 266 773 | 52.96 |
| Latescibacteria bacterium UBA6620 | 95.60% | 1.10% | 151 | 61 | 5 642 168 | 59.49 |
| nGOM Bacteria 50 | 84.80% | 1.40% | 5484 | 455 | 5 346 994 | 57.52 |
| nGOM Verruco 11 | 51.72% | 0% | 1636 | 327 | 1 757 369 | 53.66 |
| nGOM Verruco 34 | 27.77% | 0% | 1128 | 222 | 1 175 995 | 58.54 |
| Pedosphaera sp ARS72 | 97.41% | 7.37% | 140 | 140 | 4 267 059 | 53.30 |
| Pedosphaera sp MED719 | 86.49% | 2.27% | 35 | 35 | 4 083 318 | 47.78 |
| Verrucomicrobia bacterium SCGCAAA164E04 | 71.90% | 2.36% | 222 | 218 | 3 949 105 | 47.70 |
| Verrucomicrobia bacterium UBA2970 | 86.61% | 2.70% | 779 | 500 | 4 780 578 | 57.17 |
| Verrucomicrobiales bacterium SP5 | 95.27% | 2.36% | 101 | 101 | 6 081 546 | 56.46 |
** Cultured microbe
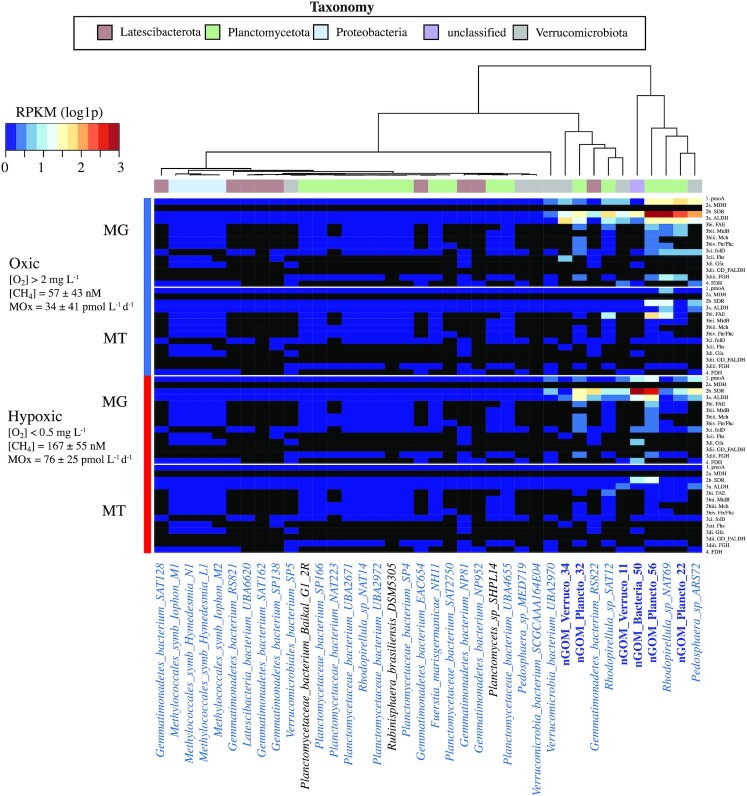
Figure 3.
Heatmap of abundance (metagenome, MG) and expression (metatranscriptome, MT) of genes involved in aerobic marine methane oxidation. Genome names at the bottom are color-coded blue and bold (nGOM MAGs), black (non-marine) and blue (marine). Genes not encoded in a particular genome are shown as a black box in the figure. The FDH of nGOM Verruco 34 was annotated only in IMG and not confirmed by COGs and Pfam annotations and is therefore shown as not annotated in this figure.
The ribosomal protein tree phylogeny revealed the most similar microbes to nGOM MAGs were largely from globally distributed TARA Oceans samples (Tully et al. 2018), many of which were low oxygen marine environments (Table 1). For example, the nGOM Planctomycetota MAGs were most similar to Planctomycetales and Pirellulales obtained from sites with annual mean oxygen less than 2.66 mg L−1 (Pesant et al. 2015) (Table 1) (see Fig. 2C and Supp. Fig. 2, Table 1 for full taxonomy, Table 2 for genome statistics). Similarly, the nGOM Verrucomicrobiota MAGs were most similar to members of the Pedosphaerales (see Fig. 2C and Supp. Fig. 2, Table 1 for full taxonomy, Table 2 for genome statistics) sampled from TARA Oceans samples (Tully et al. 2018), two of which were collected from Arabian Sea sites with annual mean oxygen less than 2.66 mg L−1 (Pesant et al. 2015). The nGOM Bacteria 50 was most similar to Latescibacterota in the UBA2968 and UBA8231 orders (see Fig. 2C and Supp. Fig. 2, Table 1 for full taxonomy, Table 2 for genome statistics) primarily from TARA Oceans samples in the Eastern Tropical North Pacific, the coast of southern Africa and the Red Sea (Tully et al. 2018).
In addition to analyzing ribosomal protein tree phylogenetic relationships, ANI analysis was carried out, revealing that nGOM MAGs were more similar to non-nGOM microbes that were sampled from the global ocean than they were to one another (Fig. 2A and C). This is best exemplified by nGOM Verruco 11 and 34 with an ANI of 79%, while nGOM Verruco 11 had an ANI of 98% with Pedosphaera sp ARS72(Fig. 2A and C), suggesting that the latter pair could be the same species (Goris et al. 2007, Richter and Rosselló-Móra 2009, Kim et al. 2014). Similarly, the nGOM Planctomycetota MAGs were <75% similar to one another, but up to 80% similar to non-nGOM Planctomycetaceae, such as nGOM Verruco 34 and bacterium UBA4655 (Fig. 2A and C). The nGOM Bacteria 50 is the only representative of the unclassified, putative Latescibacterota in the nGOM data, thus its ANI was highest with non-GOM genomes, such as that of Gemmatimonadetes bacterium RS821(ANI = 82%) and Gemmatimonadetes bacterium SAT128(ANI = 81%; Fig. 2A).
To determine genome abundance and activity, DNA and RNA reads were recruited to each of the 38 genomes. On average, the nGOM MAGs recruited the greatest number of DNA and RNA reads of the 38 genomes (Fig. 2B), with DNA RPKM values ranging from an average of 0.1 to 1.07. For example, nGOM Plancto 22 and 56 had DNA RPKM values up to 1.50, while the nGOM Verruco 34 maximum was 2.09 and nGOM Bacteria 50 was up to 1.15 (Fig. 2B). There were some exceptions to this, with the non-nGOM Rhodopirellula sp NAT69having DNA RPKM values of up to 1.23. No other genomes had DNA RPKM values >1.0. As a point of comparison with Thrash et al. (2017), MAGs from the nGOM, our nGOM MAGs, averaged 0.46 RPKM, while those in Thrash et al. (2017) ranged in abundance from 13.9 RPKM for Euryarchaeoata to similar abundances to our MAGs, particularly the Candidate Phyla. The four canonical methanotroph Proteobacteria were classified as Methylococcales (see Table 1 for full taxonomy, Table 2 for genome statistics) and are common methane-oxidizing symbionts of various marine organisms (Rubin-Blum et al. 2019). These MAGs are included here to represent canonical methanotrophs, not because of close phylogenetic relationships with the nGOM MAGs (Fig. 2C). While these genomes recruited DNA reads from each sample, RPKM values were lower than the nGOM MAGs and other microbes discussed previously.
The RNA read recruitment pattern was generally the same as that for DNA, with the nGOM MAGs presented herein recruiting the greatest number of RNA reads of the microbes analyzed here, with RPKM values up to 0.34 (Fig. 2B). However, three non-GOM Planctomycetota and Verrucomicrobiota from TARA Oceans samples (Tully et al. 2018), collected from Arabian Sea sites with an annual mean oxygen of less than 2.66 mg L−1 (Pesant et al. 2015), had similar RNA RPKM values as the nGOM MAGS, with RPKM values up to 0.30. The nGOM Bacteria 50 recruited RNA reads from each sample, with a maximum RPKM of 0.19 (Fig. 2B). Thus, nGOM Bacteria 50 was active, and for sample H_E4, recruited the greatest number of RNA reads of any of the 38 microbes analyzed herein, which agrees with Thrash et al. (2017), who reported this as one of the most active microbes in the 2013 dead zone analysis based on its high relative DNA to RNA recruitment rank and cytochrome c oxidase expression. All four canonical methanotrophs recruited low levels of RNA reads, relative to the other 34 genomes, particularly the nGOM MAGs.
The pangenome and clade core functions
The microbes analyzed herein are united in their functional potential to oxidize methane, which is a novel metabolism for many of these taxa, and none are recognized as marine methanotrophs. Therefore, we sought to determine their pangenome core functions. These methanotroph core functions were defined as having the same annotation in a minimum of two out of the three results obtained using COGs and Pfams databases, or blastx with the non-redundant RefSeq protein database from NCBI, as well as being found in all 38 genomes. We also defined clade-specific core functions the same way.
The full pangenome core contained 26 functions that included mostly housekeeping genes, ABC transporters and aromatic amino acid synthesis enzymes (Supp. Table 1). However, three core functions were worth noting: (1) NAD-dependent aldehyde dehydrogenase (ALDH), which is a non-specific enzyme in numerous pathways such as C1 and alkane degradation, including the formaldehyde oxidation step of the methane oxidation pathway (Patel et al. 1979, Anthony 1982); (2) NAD(P)-dependent dehydrogenase, SDR family protein, which can be utilized in a variety of transformations, including the second step of methane oxidation (methanol conversion to formaldehyde) (Arfman et al. 1997, Guo et al. 2019); and (3) pmoA (Supp. Table 1).
In the individual clades, 114 core functions were determined for the Planctomycetota, which included housekeeping genes, such as those for tRNA synthesis, outer membrane proteins and iron-containing alcohol dehydrogenases (Supp. Table 2). Two functions (tRNA-modifying enzymes and nucleotide sugar biosynthesis) were statistically enriched in the Planctomycetota (Supp. Table 2). In the verrucomicrobial clade, 89 functions were determined to be core (Supp. Table 3), most of which were housekeeping genes, including ribosomal proteins and phosphorylation enzymes (Supp. Table 3). Neither the Planctomycetota nor the Verrucomicrobiota encoded functions unique to either group as they were observed in other phylogenetic clades. The unclassified, putative Latescibacterota nGOM Bacteria 50, encoded 878 functions, one of which, relating to cell membrane biogenesis (LrgB-like family), was unique, not found in any other microbe in this study (Supp. Table 4). As noted, this microorganism was not definitively part of Latescibacterota, therefore core functions across this clade were not determined. The Proteobacteria clade core was comprised of 708 functions, 90 of which were statistically enriched in this group and 65 functions were found only in this clade, including rubredoxin, cobalamin synthesis, nitrate reductase delta subunit and several uncharacterized conserved proteins (Supp. Table 5).
Methane oxidation pathway
Methane to methanol
The conversion of methane to methanol is mediated by methane monooxygenase (Fig. 1); therefore, pmoA (or sMMO) is often used as a marker gene for methanotrophs. Pfam and blastx annotations revealed that 34 genomes encoded pmoA, while the remaining four Methylococcales gene annotations were less clear, with blastx suggesting PmoA/AmoA were encoded while Pfam annotations revealed PmoC/AmoC were encoded in these genomes. Despite the discrepancy in the Methylococcales gene annotations based on the database searched, for the sake of clarity we will refer to the genes in this group as pmoA. Thus, all 38 microbes in this study encoded pmoA (Fig. 3 and Supp. Table 1), but none had the complete operon (pmoA, pmoB and pmoC), with the genes surrounding pmoA being annotated as hypothetical proteins. None of the 38 genomes coded for any part of the sMMO operon.
To determine pmoA abundance and expression, DNA and RNA reads were mapped to the 38 genomes. The Planctomycetota had the highest DNA RPKM for pmoA genes (2.2–8.1) of the pmoA in our study (Fig. 3). Verrucomicrobiota clade members pmoA also recruited DNA reads, but had lower DNA RPKM values (a maximum of 3.9) than the Planctomycetota (Fig. 3). The remaining Latescibacterota pmoA recruited few DNA reads (RPKM < 0.6), while Proteobacteria pmoA did not recruit any DNA reads (Fig. 3). Eleven genomes encoded the most highly expressed pmoA genes (the RNA RPKM range was 0.042–0.58) of all 38 microbes. These 11 genomes represented all clades, including the Proteobacteria, and three nGOM MAGs (nGOM Plancto 56, nGOM Bacteria 50 and nGOM Verruco 11) (Fig. 3).
Methanol to formaldehyde
No genes encoding MDH, which mediates the conversion of methanol to formaldehyde (Fig. 1), were identified in any of the genomes, including the canonical proteobacterial methanotrophs. However, every microbe in this study encoded NAD(P)-dependent dehydrogenase, in the SDR family (Fig. 3). SDR genes are present in a variety of organisms and are capable of oxidizing primary and secondary alcohols, including methanol, albeit with low affinity (Brändén et al. 1975, Arfman et al. 1997, Guo et al. 2019).
Of the 38 genomes, the nGOM Bacteria 50 SDR gene had the highest RPKM values compared with all other methane oxidation genes in this study. For this MAG, RPKM ranged from an average of 19.1 in hypoxic sites to an average of 2.98 in oxic sites, with a maximum DNA RPKM of 43.9 from site H_E4 (Fig. 3). Unlike DNA read recruitment, the nGOM Bacteria 50 SDR gene only recruited RNA reads from hypoxic samples (the RPKM range was 1.08–2.83) (Fig. 3). Some Planctomycetota SDR genes had similar DNA RPKM values to that of nGOM Bacteria 50. For example, the nGOM Plancto 56 SDR genes recruited DNA reads from oxic (RPKM max was 31.6) and hypoxic (RPKM max was 19.7) samples (Fig. 3). Planctomycetota SDR gene RNA read recruitment RPKM values were the highest of all 38 microbes, with oxic sample RPKM of up to 5.12 and hypoxic sample RPKM of up to 5.3 (Fig. 3). In the Verrucomicrobiota, the non-nGOM Pedosphaera sp ARS72SDR recruited the largest number of reads in this clade with average DNA RPKM values of 6.47 in oxic samples and 3.58 in hypoxic samples and the largest number of RNA reads, with an RPKM average of 1.11 in oxic sites and 0.23 RNA RPKM in hypoxic sites (Fig. 3). The remaining genomes recruited a lower number of DNA and RNA reads to SDR genes compared with those microbes discussed above (Fig. 3).
Formaldehyde to formate
Methanotrophs can employ multiple pathways for formaldehyde detoxification, assimilation and oxidation (Vorholt 2002, Chistoserdova et al. 2009) (Fig. 1). The most straightforward conversion of formaldehyde to formate is facilitated by formaldehyde dehydrogenase (FALDH), however, no FALDH genes were identified in COGs or Pfams annotations in any of the 38 microbes analyzed. Instead, a non-specific ALDH, which can be used to oxidize formaldehyde to formate (Patel et al. 1979, Anthony 1982) (Fig. 1), was identified as a core gene in all 38 genomes (discussed in detail below) (Fig. 3). Additional pathways to convert formaldehyde to formate include the tetrahydromethanopterin (H4MPT), the tetrahydrofolate (H4F) and the glutathione-dependent (GSH) pathway (Fig. 1), and are presented below. The H4MPT pathway has four steps: formaldehyde activating enzyme (FAE), tetrahydromethanopterin dehydrogenase (MtdB), tetrahydromethanopterin cyclohydrolase (Mch) and formylmethanofuran-tetrahydromethanopterin formyltransferase (Ftr/Fhc) (Fig. 1). The tetrahydrofolate (H4F) pathway is comprised of two main enzymes: the bifunctional methenyltetrahydrofolate cyclohydrolase/methylenetetrahydrofolate dehydrogenase (folD) and formate—tetrahydrofolate ligase (also called formyltetrahydrofolate synthetase; Fhs) (Fig. 1). The GSH includes glutathione-dependent formaldehyde activating enzyme (Gfa), glutathione-dependent formaldehyde dehydrogenase (GD-FALDH) and formyl-glutathione hydrolase (FGH) (Fig. 1).
ALDH pathway
The ALDH gene was expressed primarily in hypoxic sites, by all clades, except for Proteobacteria and Latescibacterota, whose ALDH genes did not recruit any DNA or RNA reads (Fig. 3). Planctomycetota ALDH genes recruited the most DNA reads compared with other clades in this study (Fig. 3). Specifically, Planctomycetota recruited the greatest number of DNA and RNA reads to a maximum of 8.19 DNA RPKM and 2.35 RNA RPKM (Fig. 3). The three nGOM Planctomycetota and Verrucomicrobiota ALDH genes recruited DNA and RNA reads, however, RPKM values were typically lower than those of the non-nGOM Planctomycetota (Fig. 3). The ALDH genes encoded in the remaining genomes recruited the fewest DNA and RNA reads. Proteobacterial ALDH genes not recruiting any DNA or RNA reads (Fig. 3).
Tetrahydromethanopterin (H4MPT) pathway
Planctomycetota and Proteobacteria clades encoded the full or partial H4MPT pathway (Fig. 3), but members of the other clades evaluated did not encode any part of the H4MPT pathway in their genomes (Fig. 3). Rhodopirellula sp NAT69 and Rhodopirellula sp SAT12 genes that are part of the H4MPT pathway recruited the greatest number of DNA and RNA reads (the RPKM maxima were 5.3 and 3.51, respectively), relative to the other microbes that encoded this full or partial pathway (Fig. 3). The nGOM Plancto 56, the only nGOM microbe that encoded a complete H4MPT pathway, had minimal DNA and RNA read recruitment to the genes in this pathway (RPKM < 1; (Fig. 3)), which is of the same order of magnitude as other Planctomycetota in this group (Fig. 3). While this full pathway was encoded in the Proteobacteria genomes analyzed herein, genes in this pathway did not recruit any DNA or RNA reads (Fig. 3).
Tetrahydrofolate (H4F) pathway
Of the 38 genomes, analyzed only three non-nGOM Latescibacterota encoded a complete H4F pathway (Fig. 3), with some members of the remaining clades encoding partial pathways. For the complete pathway, only the Gemmatimonadetes bacterium RS822 genes in the H4F pathway recruited DNA reads, and this was from all sites except one (average oxic RPKM value of 0.04 and average hypoxic RPKM value of 0.135), but recruited RNA reads only from a hypoxic sample (RPKM was 0.03; Fig. 3). The remaining microbes encoded either a partial H4F pathway, or no genes in this pathway (Fig. 3). For those that encoded a partial pathway, six non-nGOM Planctomycetota H4F pathway genes recruited the greatest number of DNA reads compared with the other microbes that encoded this pathway (RPKM maximum value was 1.6), but no RNA reads were recruited (Fig. 3). The nGOM Bacteria 50 partial H4F pathway genes recruited a low number of DNA reads but had the highest RNA RPKM values for this pathway compared with other microbes, with average DNA RPKM of 0.63 and average RNA RPKM of 0.12 from hypoxic sites compared with oxic sites (average DNA RPKM was 0.13, average RNA RPKM was 0) (Fig. 3). Members of the other clades H4F pathway genes recruited a low number, or no DNA or RNA reads (Fig. 3).
Glutathione-dependent (GSH) pathway
Non-nGOM Verrucomicrobiales bacterium SP5 and Gemmatimonadetes bacterium NP81 genomes represented the only microbes analyzed herein that encoded two parts of the GSH pathway; however, no DNA or RNA reads were recruited (Fig. 3). Representatives of the Planctomycetota, Verrucomicrobiota, Latescibacterota and the Proteobacteria encoded only one part of the GSH pathway (Figs. 1 and 3). The Planctomycetota Rhodopirellula sp NAT69 partial GSH pathway had the highest RPKM value for DNA read recruitment (RPKM maximum was 2.53) compared with the other microbes that encode this partial or full pathway (Fig. 3). The genes in this partial pathway of the remaining microbes recruited a low number or no DNA reads (Fig. 3). The only genes in the GSH pathway that were represented in the metatranscriptomic data were Rhodopirellula sp SAT12with RNA RPKM ranging from 0.053 to 0.54 (Fig. 3). None of 38 microbial genomes analyzed here encoded GD-FALDH.
Formate to carbon dioxide
FDH catalyzes the conversion of formate directly to carbon dioxide (Anthony 1991, Dijkhuizen et al. 1992, Hanson and Hanson 1996) (Fig. 1). All Proterobacteria and most Latescibacterota encoded FDH, as did the two nGOM Verrucomicrobiota MAGs, and the unclassified nGOM Bacteria 50, but none of the Planctomycetota encoded FDH (Fig. 3). The nGOM Bacteria 50 FDH gene recruited DNA reads from each site (average oxic RPKM was 0.16 and average hypoxic RPKM was 0.98) and was the only one out of all 38 microbes whose FDH genes recruited any RNA reads (site H_E4 only with an RPKM of 0.21; Fig. 3). The nGOM Verruco 11 and Pedosphaera sp ARS72 FDH genes recruited DNA reads from all sites (RPKM range was 0.06–0.39) (Fig. 3). The remaining Verrucomicrobiota that encoded FDH recruited a low number of DNA reads, while no DNA or RNA reads were recruited to this gene encoded in Latescibacterota or Proteobacteria (Fig. 3).
CO2 fixation
A key gene in autotrophic carbon dioxide fixation is ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) and some more recently discovered methanotrophs classified as Verrucomicrobiota and NC10 have been shown to encode this gene (Khadem et al. 2011, Rasigraf et al. 2014). For example, the analysis by Thrash et al. (2017) identified this functional annotation in nGOM Bacteria 50 using IMG. According to annotations by both COGs and Pfams in this study, this gene was encoded by nGOM Bacteria 50 and some of the Latescibacterota (Gemmatimonadetes bacterium RS821, Gemmatimonadetes bacterium RS822, Gemmatimonadetes bacterium SAT128 and Latescibacteria bacterium UBA6620), but these genes recruited few (RPKM < 0.8) if any DNA reads and no RNA reads in any sample.
Discussion
The average methane concentrations and oxidation rates in the 2013 dead zone (Rogener et al. 2021) were well above background levels of <4 nM (Joye et al. 2011) and 0.05 nmol L−1d−1 (Crespo-Medina et al. 2014) observed in the offshore GOM. Specifically, Rogener et al. (2021) reported depth-integrated methane oxidation rates (Fmox) of 0.2 µmols m−2d−1 in 2013. They also reported that methane concentrations in the nGOM were negatively correlated with O2 concentrations, which have been previously described (Abril and Iversen 2002, Kelley 2003, Mau et al. 2013, Osudar et al. 2015, Steinle et al. 2017, Rogener et al. 2021). Gillies et al. (2015) and Campbell et al. (2019) datasets revealed that relative proportions of canonical methanotrophs were very low. Rogener et al. (2021) also made measurements in the 2015 dead zone and reported Fmox of 2.4 µmols m−2d−1. In parallel, 2015 dead zone samples, Campbell and Mason (unpublished) used iTag sequencing to evaluate the microbial community in the 2015 dead zone and found Methylococcales, which contain canonical marine methanotrophs, reached a maximum abundance of 1% of the community, averaging 0.09% relative abundance. Methylosinus was even less abundant, averaging 0.003% in relative abundance. Given that the abundance of canonical methanotrophs was very low and invariant during 2013–2015, but Fmox was significantly higher in 2015, the data suggest that unknown, non-canonical methanotrophs are likely carrying out methane oxidation in this shallow water, methane-rich environment.
The microbes we presented herein with genomes encoding partial to complete pathways for methane oxidation are united in that they represent non-canonical aerobic, largely marine methanotrophs, which are not known to mediate methane oxidation. The presence of these novel methanotrophs in the nGOM dead zone begins to reconcile the high methane oxidation rates that have been reported in this ecosystem (Rogener et al. 2021) with the low to undetectable levels of canonical methanotrophs (Gillies et al. 2015, Campbell et al. 2019 and Campbell and Mason unpublished data). The level of DNA and RNA read recruitment to these genomes and genes was highest for nGOM MAGs, which is not unexpected. The lower read recruitment to non-nGOM microbes suggested that while these microbes were not abundant or highly active in the nGOM dead zone at the time we sampled, they encode a previously unrecognized capacity to oxidize methane in a diversity of marine environments, in which they may be both abundant and actively consuming methane.
These 38 methanotrophs, both from the nGOM and outside of the GOM, were further united in that beyond pmoA they all have the metabolic capacity for methanol and formaldehyde oxidation (SDR and ALDH). Additional modes of formaldehyde oxidation (H4MPT, H4F, GSH) were also detected, but these pathways were incomplete in most genomes, if they were encoded at all. Further, all clades except Planctomycetota, contained at least some members encoding FDH. As expected, the canonical methanotrophs in the Proteobacteria encoded complete methane oxidation pathways. In addition, the non-canonical methanotrophs, including the unclassified nGOM MAG, as well as most Latescibacterota and Verrucomicrobiota in this study, encoded complete methane oxidation pathways, while all Planctomycetota encoded partial methane oxidation pathways. The partial to complete methane oxidation pathways encoded in these non-canonical planktonic methanotrophs reflect the phylogenetic diversity of methanotrophs, as well as the potential role in mitigating methane efflux from the ocean.
All the Planctomycetota genomes encoded partial MOx pathways, with their pmoA genes recruiting more RNA reads in oxic sites than hypoxic. The Planctomycetota nGOM and global microbes analyzed herein encoded the full or partial H4MPT pathway and partial H4F pathway (Figs. 1 and 3), which is consistent with previous reports for members of this phylogenetic clade (Chistoserdova et al. 2004, Woebken et al. 2007, Fuerst and Sagulenko 2011). The nGOM Planctomycetota MAGs and the majority of global Planctomycetota genomes encoded a partial GSH pathway (FGH) (Figs. 1 and 3), as has been previously reported (Woebken et al. 2007). Not all Planctomycetota encode FDH (Kim et al. 2016), and none of the nGOM or global Planctomycetota in this study did. While the three Planctomycetota from non-marine environments (Planctomyces sp SHPL14, Planctomycetaceae bacterium Baikal G1 2R and Rubinisphaera brasiliensis DSM5305) encoded enough modules to complete the methane oxidation pathway to formate (because none encode FDH), they do not appear to be actively carrying out methane oxidation in this environment (Fig. 3).
The nGOM MAGs and two microbes from the Arabian Sea (Tully et al. 2018) recruited the most DNA and RNA reads to their genomes compared with other Planctomycetota (Fig. 3). Both ecosystems experience hydrocarbon contamination and low oxygen concentrations (temporary in the case of nGOM, permanent in the Arabian Sea), and it appears these microbes encode similar modules of the methane oxidation pathway (Fig. 3). Members of this clade contain genes involved in C1 transfer pathways (H4MPT, H4F) and although the potential for methylotrophy has been suggested (Buckley et al. 2006), thus far they have only been hypothesized to be key ancestral players in the evolution of the global methane cycle (Glöckner et al. 2003, Bauer et al. 2004, Chistoserdova et al. 2004, Kalyuzhnaya et al. 2004, Chistoserdova et al. 2005, Woebken et al. 2007). This diverse clade is found in numerous environments and is one of the most abundant groups in OMZs (Wright et al. 2012), where they are primary mediators of nitrogen loss through anaerobic oxidation of ammonium (Kuypers et al. 2003, 2005, Schmid et al. 2007, Woebken et al. 2008, Galán et al. 2009, Lam et al. 2016). Our study results suggest a new function for Planctomycetota in OMZs: methane oxidation, which expands their role in biogeochemical cycles and supports the previously hypothesized role of this clade in global methane cycles.
Verrucomicrobiota are also common in OMZs (Wright et al. 2012), but are not known to carry out methane oxidation in the marine environment. The Verrucomicrobiota we analyzed were actively expressing pmoA in low oxygen environments, which suggested they may be like the thermoacidophilic verrucomicrobial methanotrophs that oxidize methane, but in acidic, geothermal environments (Dunfield et al. 2007; Pol et al. 2007; Islam et al. 2008). These methanotrophic Verrucomicrobiota do not employ the same MOx pathway modules that are observed in canonical (Proteobacteria) methanotrophs. For example, they have been shown to code for XoxF-MDH, which converts methanol directly to formate (Keltjens et al. 2014), thereby lacking common formaldehyde oxidation modules (Dunfield et al. 2007). XoxF-MDH was not annotated in the Verrucomicrobiota genomes analyzed here (only SDR), nor was a more common, complete formaldehyde oxidation pathway (H4MPT, H4F, GSH).
Acidophilic verrucomicrobial methanotrophs have been shown to encode the complete H4F pathway (Picone et al. 2021, Schmitz et al. 2021), but none in this study did, only a partial one if at all. Thus, the formaldehyde oxidation step remains unclear in the Verrucomicrobiota analyzed herein. The last step in methane oxidation encoded by FDH was annotated in five of the seven Verrucomicrobiota, which is consistent with acidophilic terrestrial methanotrophs (Dunfield et al. 2007, Picone et al. 2021, Schmitz et al. 2021). Pedosphaera sp ARS72, which is from a site in the Arabian Sea where annual mean oxygen is <2.66 mg L−1 (Pesant et al. 2015), and nGOM Verruco 11, both encode the same modules of the methane oxidation pathway (except H4F) (Fig. 3). These microorganisms are from areas that experience hydrocarbon contamination and low oxygen concentrations and were some of the most abundant (Fig. 2) and active microbes in this study (Fig. 3). Thus, the Verrucomicrobiota presented herein are like their terrestrial, acidophilic relatives in that they can carry out methane oxidation, but do so in a basic, aquatic marine environment where they may act as an important methane biofilter in the water column using novel modules within the overall methane oxidation pathway (Dunfield et al. 2007).
Of the nine Latescibacterota in this study, eight were originally classified as Gemmatimonadetes. Members of the Gemmatimonadetes (now classified as either Gemmatimonadota or Latescibacterota) are also abundant in OMZs, although typically less so than Planctomycetota (Wright et al. 2012), and while not recognized as aerobic marine methanotrophs, some genomes have been shown to encode MDH, classifying them as methylotrophs (Butterfield et al. 2016). Members of Latescibacterota (previously WS3 and candidate phylum Lastescibacteria) were first discovered in a hydrocarbon-contaminated aquifer and have since been found in marine sediments, hydrothermal vents, soil and other hydrocarbon-contaminated environments (Farag et al. 2017). Recently, members of this clade were shown to encode for fermentation (García-Lozano et al. 2019) and anaerobic hydrocarbon degradation (Dombrowski et al. 2017), still none have been reported to encode the capacity for aerobic methane oxidation. The Latescibacterota in this study all have pmoA, SDR and ALDH, although most of those genes recruited minimal DNA and RNA reads. These microbes did not encode MDH or any part of the H4MPT pathway, which contrasts with previous research (Butterfield et al. 2016); however, Butterfield et al. (2016) did not find that their microbes encoded pmoA. Most of the Latescibacterota in this study encoded a partial H4F pathway, but few encoded the GSH pathway. Despite the presence of pmoA and other modules in MOx, Latescibacterota did not appear to be active in MOx in the nGOM. However, the Latescibacterota presented here represent the first aerobic methane oxidizing members of this clade in the marine environment. It is possible that at other timepoints in the nGOM, or in other environments with different methane or oxygen concentrations, this group of bacteria could become active and serve as a methane biofilter.
Unlike the previously discussed taxa, the Proteobacteria contain canonical aerobic marine methanotrophs (Hanson and Hanson 1996). This clade has been reported as the most abundant group in global OMZs via 16S rRNA gene surveys (Wright et al. 2012). All four Proteobacteria in this study encoded pmoA, SDR and ALDH, but genomic and transcriptomic read representation was minimal. They also all encoded the full H4MPT pathway, partial H4F and GSH pathways and all encoded FDH. While these microbes represented canonical methanotrophs, like the other 34 genomes analyzed herein, they lacked MDH genes, thus their genomes encoded only some of typical modules to carry out MOx. These canonical methanotrophs were not abundant and or had low levels of activity in the nGOM hypoxic zone, where methane concentrations and oxidation rates are high, thus highlighting the importance of novel, non-canonical methanotrophs potentially acting as a biofilter.
Conclusion
Climate change-induced expansion of OMZs that are enriched in greenhouse gases suggests an active planktonic methanotrophic community is even more critical in mitigating greenhouse gas efflux from the water column to the atmosphere. Herein we show that canonical methanotrophs (Proteobacteria) appear to be less abundant and active in the 2013 nGOM dead zone than previously unrecognized methanotrophs belonging to Verrucomicrobiota, Planctomycetota and Latescibacterota. These non-canonical methanotrophs are globally distributed and may play a key role in oxidizing methane in the water column before it reaches the atmosphere. Further research should investigate the alternative modes of formaldehyde and formate oxidation by these uncultivated, non-canonical aerobic marine methanotrophs. Further, methanol oxidation via MDH was not encoded in any of 38 genomes, including the canonical methanotrophs. Thus, additional analyses are needed to better understand the metabolic capacities and potential to act as a methane biofilter by canonical and non-canonical methanotrophs, across an oxygen gradient, as well as to re-evaluate the marine microbial methane sink. Further, this study provides an illustration on why assessing taxonomy alone (e.g. 16S rRNA gene data) may obscure important metabolic processes that are revealed using multi-omics approaches. Finally, in the age of omics, our analyses are likely the first of many in which new, non-canonical methanotrophs are revealed.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the science and vessel crews of the R/V Pelican for their valuable shipboard and onshore support.
Contributor Information
Kathryn L Howe, Department of Earth, Ocean, and Atmospheric Science, Florida State University, 32306, Tallahassee, United States.
Kiley W Seitz, Department of Marine Science, Marine Science Institute, University of Texas at Austin, 78373, Port Aransas, United States.
Lauren G Campbell, Department of Earth, Ocean, and Atmospheric Science, Florida State University, 32306, Tallahassee, United States.
Brett J Baker, Department of Marine Science, Marine Science Institute, University of Texas at Austin, 78373, Port Aransas, United States; Department of Integrative Biology, University of Texas at Austin, 78712, Austin, United States.
J Cameron Thrash, Department of Biological Sciences, University of Southern California, 90089, Los Angeles, United States.
Nancy N Rabalais, Department of Oceanography and Coastal Sciences, Louisiana State University, 70803, Baton Rouge, United States; Louisiana Universities Marine Consortium, 70344, Chauvin, United States.
Mary-Kate Rogener, Department of Marine Sciences, University of Georgia, 30602, Athens, United States.
Samantha B Joye, Department of Marine Sciences, University of Georgia, 30602, Athens, United States.
Olivia U Mason, Department of Earth, Ocean, and Atmospheric Science, Florida State University, 32306, Tallahassee, United States.
Conflicts of interest statement
The authors declare that the research was conducted in the absence of any conflict of interest.
Funding
This work was supported by a grant from The Gulf of Mexico Research Initiative RFP-VI: Research Grants [OUM Consortium for Simulation of Oil-Microbial Interactions in the Ocean (CSOMIO)]. Data are publicly available through the Gulf of Mexico Research Initiative Information & Data Cooperative (GRIIDC) at https://data.gulfresearchinitiative.org DOI:10.7266/D7MJVFX0. This was also supported by Simons Early Career Investigator in Marine Microbial Ecology and Evolution Awards [BJB 687165] and [JCT 688424]. Vessel and logistical support was provided by the National Oceanic and Atmospheric Administration, Center for Sponsored Coastal Ocean Research, award numbers [NNR NA09NOS4780204] Louisiana Universities Marine Consortium [RET NA09NOS4780230] to Louisiana State University.
References
- Abril G, Iversen N. Methane dynamics in a shallow non-Tidal estuary (Randers Fjord, Denmark). Marine Ecology Progress Series. 2002;230:171–81. [Google Scholar]
- Anthony C. CHAPTER 4 - Assimilation of carbon by methylotrophs. In: Goldberg I, Rokem JS (eds). Biology of Methylotrophs, Butterworth-Heinemann, 1991, 79–109. 10.1016/B978-0-7506-9188-8.50011-5. [DOI] [PubMed] [Google Scholar]
- Anthony C. The Biochemistry of Methylotrophs. New York: Academic Press, 1982. [Google Scholar]
- Arfman N, Hektor HJ, Bystrykh LVet al. Properties of an NAD(H)-containing methanol dehydrogenase and its activator protein from Bacillus Methanolicus. Eur J Biochem. 1997;244:426–33. [DOI] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin Ret al. The Pfam protein Families Database. Nucleic Acids Res. 2004;32:138D–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M, Lombardot T, Teeling Het al. Archaea-like genes for C1-Transfer enzymes in planctomycetes: phylogenetic implications of their unexpected presence in this phylum. J Mol Evol. 2004;59:571–86. [DOI] [PubMed] [Google Scholar]
- Brändén C-I, Jürnvall H, Eklund Het al. In: Boyer PD (ed.), Alcohol dehydrogenases. The Enzymes. Vol. 11, Academic Press, 1975, 103–90. 10.1016/S1874-6047(08)60211-5. [DOI] [Google Scholar]
- Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014;12:59–60. [DOI] [PubMed] [Google Scholar]
- Buckley DH, Huangyutitham V, Nelson TAet al. Diversity of planctomycetes in soil in relation to soil history and environmental heterogeneity. 2006;72:4522–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield CN, Li Z, Andeer PFet al. Proteogenomic analyses indicate bacterial methylotrophy and archaeal heterotrophy are prevalent below the grass root zone. PeerJ. 2016;2016:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello-Yeves PJ, Zemskaya TI, Rosselli Ret al. Genomes of novel microbial lineages assembled from the sub-ice waters of Lake Baikal. Appl Environ Microbiol. 2018;84:e02132–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LG, Cameron Thrash J, Rabalais NNet al. Extent of the annual Gulf of Mexico hypoxic zone influences microbial community structure. 2019;14:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil PA, Mussig AJ, Hugenholtz Pet al. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy Database. Bioinformatics. 2019;36:1925–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L, Jenkins C, Kalyuzhnaya MGet al. The enigmatic planctomycetes may hold a key to the origins of methanogenesis and methylotrophy. Mol Biol Evol. 2004;21:1234–41. [DOI] [PubMed] [Google Scholar]
- Chistoserdova L, Kalyuzhnaya M, Lidstrom M. The expanding world of methylotrophic metabolism. Annu Rev Microbiol. 2009;63:477–99. 10.1146/annurev.micro.091208.073600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L, Kalyuzhnaya MG, Lidstrom ME. C 1 -Transfer modules: from genomics to ecology. ASM-NEWS. 2005;71:1–8. [Google Scholar]
- Chistoserdova L. Modularity of methylotrophy, revisited. Environ Microbiol. 2011;13:2603–22. [DOI] [PubMed] [Google Scholar]
- Crespo-Medina M, Meile CD, Hunter KSet al. The rise and fall of methanotrophy following a deepwater oil-Well blowout. Nat Geosci. 2014;7:423–7. 10.1038/NGEO2156. [DOI] [Google Scholar]
- Dijkhuizen L, R Levering P, de Vries GE. The physiology and biochemistry of aerobic methanol-utilizing gram-negative and gram-positive bacteria BT - Methane and methanol utilizers. Methane and methanol utilizers. In: Murrell CJ, Dalton H (eds). Boston, MA: Springer US, 1992, 149–81. 10.1007/978-1-4899-2338-7_5. [DOI] [Google Scholar]
- Dombrowski N, Seitz KW, Teske APet al. Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome. 2017;5:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubilier N, Bergin C, Lott C. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol. 2008;6:725–40. [DOI] [PubMed] [Google Scholar]
- Dunfield PF, Yuryev A, Senin Pet al. Methane oxidation by an extremely acidophilic bacterium of the phylum verrucomicrobia. Nature. 2007;450:879–82. [DOI] [PubMed] [Google Scholar]
- Eren AM, Esen ÖC, Quince Cet al. Anvi'o: an advanced Analysis and Visualization platform for ‘omics Data. PeerJ. 2015;3:e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag IF, Youssef NH, Elshahed MS. Global distribution patterns and pangenomic diversity of the candidate phylum ‘latescibacteria’ (WS3). Appl Environ Microbiol. 2017;83:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst JA, Sagulenko E. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol. 2011;9:403–13. 10.1038/nrmicro2578. [DOI] [PubMed] [Google Scholar]
- Galán A, Molina V, Thamdrup Boet al. Anammox bacteria and the Anaerobic oxidation of ammonium in the oxygen Minimum zone off Northern Chile. Deep-Sea Research Part II: Topical Studies in Oceanography. 2009;16:1021–31. [Google Scholar]
- García-Lozano M, Lira IOH-De, Huber DHet al. Spatial variations of bacterial communities of an Anaerobic lagoon-Type biodigester fed with dairy manure. Processes. 2019;7:408. 10.3390/pr7070408. [DOI] [Google Scholar]
- Gillies LELE, Cameron Thrash JC, DeRada Set al. Archaeal enrichment in the Hypoxic Zone in the Northern Gulf of Mexico. Environ Microbiol. 2015;17:3847–56. [DOI] [PubMed] [Google Scholar]
- Glöckner FO, Kube M, Bauer Met al. Complete genome sequence of the marine planctomycete pirellula sp. Strain 1. Proc Nat Acad Sci USA. 2003;100:8298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JAet al. DNA-DNA hybridization values and their relationship to whole-Genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. [DOI] [PubMed] [Google Scholar]
- Guo X, Feng Y, Wang Xet al. Characterization of the substrate scope of an alcohol dehydrogenase commonly used as methanol dehydrogenase. Bioorg Med Chem Lett. 2019;29:1446–9. [DOI] [PubMed] [Google Scholar]
- Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC . `Climate Change 2013 ’. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. 2013.
- Islam T, Jensen S, Reigstad LJet al. Methane oxidation at 55 C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. PNAS. 2008;105:300–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joye SB, Macdonald IR, Leifer Iet al. Magnitude and oxidation potential of hydrocarbon gases released from the BP oil well blowout. Nat Geosci. 2011;3:1–5. [Google Scholar]
- Kalyuzhnaya MG, Gomez OA, Murrell JC. The methane-Oxidizing bacteria (Methanotrophs). In: McGenity TJ (ed.). Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes, Cham, Switzerland: Springer International Publishing, 2019, 1–34. 10.1007/978-3-319-60053-6_10-1. [DOI] [Google Scholar]
- Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. Utility of environmental primers targeting ancient enzymes: methylotroph detection in Lake Washington. Microb Ecol. 2004;48:463–72. [DOI] [PubMed] [Google Scholar]
- Kelley C. Methane oxidation potential in the water column of two diverse coastal marine sites. Biogeochemistry. 2003;65:105–20. 10.1023/A:1026014008478. [DOI] [Google Scholar]
- Keltjens JT, Pol A, Reimann Jet al. PQQ-Dependent methanol dehydrogenases: rare-Earth elements make a difference. Appl Microbiol Biotechnol. 2014;98:6163–83. [DOI] [PubMed] [Google Scholar]
- Khadem AF, Pol A, Wieczorek Aet al. Autotrophic methanotrophy in Verrucomicrobia : methylacidiphilum fumariolicum SolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation ᰔ †. 2011;193:4438–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Brawley SH, Prochnik Set al. Genome analysis of planctomycetes inhabiting blades of the red alga Porphyra Umbilicalis. PLoS One. 2016;11:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Oh HS, Park SCet al. Towards a taxonomic coherence between average nucleotide identity and 16S RRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–51. [DOI] [PubMed] [Google Scholar]
- Kirschke S, Bousquet P, Ciais Pet al. Three decades of global methane sources and sinks. Nat Geosci. 2013;6:813–23. [Google Scholar]
- Knief C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on PmoA as molecular marker. Front Microbiol. 2015;6:1346. 10.3389/fmicb.2015.01346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn T, Heuer A, Jogler Met al. Fuerstia marisgermanicae gen. nov., sp. nov., an Unusual Member of the Phylum Planctomycetes from the German Wadden Sea. Front Microbiol. 2016;7:2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers MMM, Lavik G, Woebken Det al. Massive nitrogen loss from the Benguela upwelling system through Anaerobic ammonium oxidation. Proc Nat Acad Sci USA. 2005;102:6478–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers MMM, Silekers AO, Lavik Get al. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature. 2003;422:608–11. [DOI] [PubMed] [Google Scholar]
- Lam P, Strous M, Dumont MGet al. Methanotrophic community dynamics in a seasonally anoxic Methanotrophic community dynamics in a seasonally anoxic fjord: saanich Inlet, British Columbia. 2016;3:317–45. 10.3389/fmars.2016.00268. [DOI] [Google Scholar]
- Langmead B, Salzberg SL. Bowtie2. Nat Methods. 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree of Life (ITOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen IMA, Palaniappan Ket al. IMG 4 version of the Integrated Microbial Genomes Comparative Analysis system. Nucleic Acids Res. 2014;42:D560–7. 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason O, Hazen T, Borglin Set al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 2012;6:1715–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mau S, Blees J, Helmke Eet al. Vertical distribution of methane oxidation and methanotrophic response to elevated methane concentrations in stratified waters of the Arctic Fjord Storfjorden (Svalbard, Norway). Biogeosciences. 2013;10:6267–8. [Google Scholar]
- Mcdonald IR, Murrell JC. The particulate methane monooxygenase gene PmoA and its use as a functional gene probe for methanotrophs. 1997;156:156. [DOI] [PubMed] [Google Scholar]
- Murrell JC, Gilbert B, McDonald IR. Molecular biology and regulation of methane monooxygenase. Arch Microbiol. 2000;173:325–32. [DOI] [PubMed] [Google Scholar]
- Osudar R, Matoušů A, Alawi Met al. Environmental factors affecting methane distribution and bacterial methane oxidation in the German bight (North Sea). Estuarine Coastal Shelf Sci. 2015;160:10–21. [Google Scholar]
- Parks DH, Chuvochina M, Waite DWet al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996. [DOI] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CTet al. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Rinke C, Chuvochina Met al. Recovery of nearly 8,0000 metagenome-assembled genomes substantially expands the tree of life. Nature Microbiology. 2017;2:1533–42. 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- Patel R, Hou CT, Felix A. Microbial oxidation of methane and methanol: purification and properties of a heme-containing aldehyde dehydrogenase from Methylomonas Methylovora. Arch Microbiol. 1979;247:241–7. [DOI] [PubMed] [Google Scholar]
- Peng Yu, Leung HCM, Yiu SMet al. IDBA-UD: a de novo assembler for single-Cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–8. [DOI] [PubMed] [Google Scholar]
- Pesant S, Not F, Picheral Met al. Open science resources for the discovery and analysis of Tara Oceans data. no. Lmd: Sci. Data, 2015;1–16. 10.1038/sdata.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picone N, Blom P, Hogendoorn Cet al. Metagenome assembled genome of a novel verrucomicrobial methanotroph from Pantelleria Island. Front Microbiol. 2021;12:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A, Heijmans K, Harhangi Het al. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature. 2007;450:874–8. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2 – Approximately maximum-Likelihood trees for large alignments. PloS one. 2010;5:e9490. 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasigraf O, Kool DM, Jetten MSMet al. Autotrophic carbon dioxide fixation via the Calvin-Benson-Bassham cycle by the denitrifying methanotroph ‘ Candidatus Methylomirabilis Oxyfera’. AEM. 2014;80:2451–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeburgh WS. Oceanic methane biogeochemistry. Chem Rev. 2007;107:486–513. [DOI] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Nat Acad Sci USA. 2009;106:19126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogener MK, Bracco A, Hunter KSet al. Long-Term impact of the Deepwater Horizon oil well blowout on methane oxidation dynamics in the Northern Gulf of Mexico. Elementa. 2018;1:1–17.30345319 [Google Scholar]
- Rogener MK, Hunter KS, Rabalais NNet al. Pelagic denitrification and methane oxidation in oxygen-depleted waters of the Louisiana Shelf. Biogeochemistry. 2021;8:231–54. 10.1007/s10533-021-00778-8. [DOI] [Google Scholar]
- Rosentreter JA, Borges Av, Deemer BRet al. Half of global methane emissions come from highly variable aquatic ecosystem sources. Nat Geosci. 2021;14:225–30. [Google Scholar]
- Rubin-Blum M, Antony CP, Sayavedra Let al. Fueled by methane: deep-Sea sponges from asphalt seeps gain their nutrition from methane-Oxidizing symbionts. ISME J. 2019;13:1209–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunois M, Jackson RB, Bousquet Pet al. The growing role of methane in anthropogenic climate change. Environ Res Lett. 2016;11:120207. [Google Scholar]
- Saunois M, Stavert AR, Poulter Bet al. The global methane budget 2000–2017. Earth Syst Sci Data. 2020;1561–623.
- Scheuner C, Tindall BJ, Lu Met al. Complete genome sequence of Planctomyces brasiliensis type strain (DSM 5305T), phylogenomic analysis and reclassification of Planctomycetes including the descriptions of Gimesia gen. nov., Planctopirus gen. nov. and Rubinisphaera gen. nov. and emended descriptions of the order Planctomycetales and the family Planctomycetaceae. Stand in Genomic Sci. 2014;9:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MC, Risgaard-Petersen N, Van De Vossenberg Jet al. Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity. Environ Microbiol. 2007;9:1476–84. [DOI] [PubMed] [Google Scholar]
- Schmieder R, Lim YW, Edwards R. Identification and removal of ribosomal RNA sequences from metatranscriptomes. Bioinformatics. 2012;28:433–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RA, Peeters SH, Versantvoort Wet al. Verrucomicrobial methanotrophs: ecophysiology of metabolically versatile acidophiles. FEMS Microbiol Rev. 2021; 45:1–20. 10.1093/femsre/fuab007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrau JD, Dispirito AAet al. Methanotrophs and copper. FEMS Microbiol Rev. 2010;34:496–531. [DOI] [PubMed] [Google Scholar]
- Söhngen NL. Uber Bakterien, Welche Methan Ab Kohlenstoffnahrung and Energiequelle Gebrauchen. 1906;2:513–7. [Google Scholar]
- Steinle L, Maltby J, Treude Tet al. Effects of low oxygen concentrations on aerobic methane oxidation in seasonally hypoxic coastal waters. Biogeosciences. 2017;14:1631–45. [Google Scholar]
- Stepanauskas R, Huntemann M, Han Jet al. Verrucomicrobia bacterium SCGC AAA164-E04, whole genome shotgun sequencing project. DOE Joint Genome Institute, CA, USA. 2013. [Google Scholar]
- Tatusov RL, Galperin MY, Natale DAet al. The COG Database : a tool for genome-Scale analysis of protein functions and evolution. 2000;28:33–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T, Dicuccio M, Badretdin Aet al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina PL, Ussier W, Orphan VJ. Planktonic and sediment-Associated aerobic methanotrophs in two seep systems along the North American margin. Appl Environ Microbiol. 2008;74:3985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash JC, Baker BJ, Seitz KWet al. Metagenomic assembly and prokaryotic metagenome- Mexico ‘Dead Zone’. 2018;7:4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash JC, Seitz KW, Baker BJet al. Metabolic roles of uncultivated bacterioplankton lineages in the Northern Gulf of Mexico ‘Dead zone’. MBio. 2017;8:e01017–17. 10.1128/mBio. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus Brown C, Irber L. Sourmash: a library for MinHash sketching of DNA. J Open Source Softw. 2016;1:27. [Google Scholar]
- Tully BJ, Graham ED, Heidelberg JF. The reconstruction of 2631 draft metagenome-assembled genomes from the global oceans. Scientific Data. 2018;5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorholt JA. Cofactor-Dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch Microbiol. 2002;178:239–49. [DOI] [PubMed] [Google Scholar]
- Walker JT, Stow CA, Geron C. Nitrous oxide emissions from the Gulf of Mexico hypoxic zone. Environ Sci Technol. 2010;44:1617–23. [DOI] [PubMed] [Google Scholar]
- Woebken D, Lam P, Kuypers MMMet al. A microdiversity study of anammox bacteria reveals a novel candidatus scalindua phylotype in marine oxygen minimum zones. Environ Microbiol. 2008;10:3106–19. [DOI] [PubMed] [Google Scholar]
- Woebken D, Teeling H, Wecker Pet al. Fosmids of novel marine planctomycetes from the Namibian and Oregon coast upwelling systems and their cross-comparison with planctomycete genomes. ISME J. 2007;1:419–35. [DOI] [PubMed] [Google Scholar]
- Wright JJ, Konwar KM, Hallam SJ. Microbial ecology of expanding oxygen minimum zones. Nat Rev Microbiol. 2012;10:381–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.