Abstract
Chalcone is an interesting scaffold found in the structure of many naturally occurring molecules. Medicinal chemists are commonly interested in designing new chalcone‐based structures because of having the α, β‐unsaturated ketone functional group, which allows these compounds to participate in Michael's reaction and create strong covalent bonds at the active sites of the targets. Some studies have identified several natural chalcone‐based compounds with the ability to inhibit the severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus proteases. A few years after the advent of the coronavirus disease 2019 pandemic and the publication of many findings in this regard, there is some evidence that suggests chalcone scaffolding has great potential for use in the design and development of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) inhibitors. Artificial placement of this scaffold in the structure of optimized anti‐SARS‐CoV‐2 compounds can potentially provide irreversible inhibition of the viral cysteine proteases 3‐chymotrypsin‐like protease and papain‐like protease by creating Michael interaction. Despite having remarkable capabilities, the use of chalcone scaffold in drug design and discovery of SARS‐CoV‐2 inhibitors seems to have been largely neglected. This review addresses issues that could lead to further consideration of chalcone scaffolding in the structure of SARS‐CoV‐2 protease inhibitors in the future.
Keywords: chalcone, COVID‐19, cysteine proteases, Michael reaction, SARS‐CoV‐2
1. INTRODUCTION
Modern viral genetic studies immediately after the advent of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) revealed that the genome of this virus has approximately 80% similarity with severe acute respiratory syndrome coronavirus (SARS‐CoV) and about 50% similarity with Middle East respiratory syndrome coronavirus (MERS‐CoV) (Lu et al., 2020). Identifying the high genetic similarity of these viruses made it possible to use past scientific findings to purposefully design new studies. Accordingly, the inhibitory effects of the identified natural‐based/synthetic structures with an inhibitory effect against the SARS‐CoV were investigated against SARS‐CoV‐2 and it was found that most of them are active.
Natural compounds are always one of the most important resources for finding bioactive structures that are readily available to us. Natural resources easily provide us with complex structures that are sometimes very difficult and costly to produce in the laboratory (Corona et al., 2022; DeCorte, 2016; Harvey, Edrada‐Ebel, & Quinn, 2015; Valipour, 2022c). Since the advent of the SARS‐CoV‐2, many natural‐based bioactive molecules have been introduced with extensive capabilities to inhibit this virus (Derosa, Maffioli, D'Angelo, & Di Pierro, 2021; El‐Missiry, Fekri, Kesar, & Othman, 2021; Gour, Manhas, Bag, Gorain, & Nandi, 2021; Haq et al., 2020; Sisakht, Mahmoodzadeh, & Darabian, 2021; Yang et al., 2021; Zahedipour et al., 2020; Zalpoor, Bakhtiyari, Liaghat, Nabi‐Afjadi, & Ganjalikhani‐Hakemi, 2022). We have recently reviewed the therapeutic potential of natural compounds chelerythrine, emetine, and papaverine to fight the SARS‐CoV‐2 from different aspects (Valipour, 2022b; Valipour, Irannejad, & Emami, 2022a, 2022b; Valipour, Zarghi, Ebrahimzadeh, & Irannejad, 2021). Continuing our efforts in this field, we have reached a structural platform called “Chalcone,” which is of interest to us from different angles for future in silico, in vitro, and in vivo studies. This study tries to highlight the potential and importance of this scaffold for use in the structure of novel SARS‐CoV‐2 inhibitors for future research. It is obvious that an accurate conclusion regarding the potential of chalcone‐based compounds to inhibit SARS‐CoV‐2 is possible after conducting adequate in vitro and in vivo research.
Chalcone structure with the IUPAC name (international union of pure and applied chemistry name) of “1,3‐diphenylprop‐2‐en‐1‐one” is a simple scaffold that is found in abundance in the structure of diverse natural‐based compounds. Structurally, Chalcones can exist as E and Z isomers, but the E isomer is more thermodynamically stable (Figure 1). As shown, chalcones are made by connecting two phenyl rings with a three‐carbon α‐β unsaturated ketone bridge. Typically, the phenyl ring attached to the carbonyl group is referred to as the A ring and the farther phenyl ring as the B ring. Due to the development of diverse synthetic methods for the production of chalcones derivatives (such as Claisen−Schmidt condensation, Friedel−Crafts acylation, Photo‐Fries rearrangement and various cross‐coupling methods including the Suzuki reaction, Heck reaction, Julia−Kocienski reaction, and Wittig's reaction), this scaffold is of great interest to medicinal/organic chemists for the design and production of new compounds (Zhuang et al., 2017). One of the important chemical characteristics of chalcones that play a key role in the exertion of diverse biological activities is having α, β‐unsaturated ketone functional group. This structural part can act as Michael acceptors in chemical interactions in vitro and in vitro. In these reactions, the carbon of the beta region is attacked by various nucleophiles and establishes a covalent bond with the attacking nucleophile (from this place) and finally, a stable secondary structure is formed. Chemically, the more electron‐withdrawing substituents on ring A, the more active the beta carbon will be and the tendency of chalcone compounds to participate in the Michael reaction will be further.
FIGURE 1.
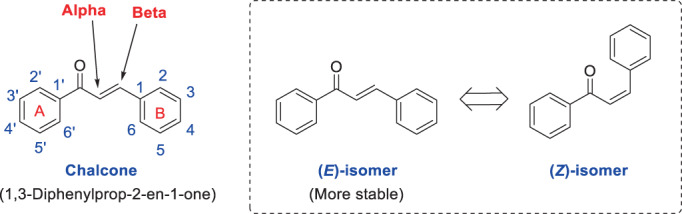
Chemical structure of chalcone and determination of its different chemical positions, along with structure of E/Z isomers
Alteration and bio‐isosteric replacement of chalcone phenyl rings (rings A and B) by aromatic nuclei usually do not cause a problem in the biological effects, but changing the α, β‐unsaturated ketone motif can lead to a fundamental change in the activity of these compounds. Since this component (α, β‐unsaturated ketone) can potentially cause irreversible inhibition of SARS‐CoV‐2 cysteine proteases, this study suggests that chalcone‐based structures can be considered in the future discovery and development of anti‐coronavirus disease 2019 (COVID‐19) agents.
2. ANTIVIRAL ACTIVITY OF CHALCONE‐BASED COMPOUNDS
Chalcone derivatives show a wide range of biological activities such as anti‐cancer, anti‐inflammatory, antibiotic, antioxidant, antileishmanial, antimalarial, antitubercular, antidiabetic, etc. (Gaonkar & Vignesh, 2017; Kar Mahapatra, Asati, & Bharti, 2019; Singh, Anand, & Kumar, 2014). Some chalcone‐based structures, such as metochalcone (as a choleretic drug) and sofalcone (as an antiulcer and mucoprotective drug), have even been approved for clinical use. Previous studies have also reported excellent antiviral activity of chalcones against a wide range of viruses, including different types of influenza virus (Dao et al., 2011; Malbari et al., 2018; Nguyen et al., 2010; Park et al., 2011), herpes simplex viruses (HSVs) (Phrutivorapongkul et al., 2003), hepatitis C virus (HCV) (Adianti et al., 2014; Mateeva et al., 2017), human immunodeficiency virus (HIV) (Cole, Hossain, Cole, & Phanstiel IV, 2016; Hameed et al., 2016; Sharma et al., 2011; Wu, Wang, Yi, & Lee, 2003), Zika virus (ZIKV), tobacco mosaic virus (Guo et al., 2019), cucumber mosaic virus (Gan, Wang, Hu, & Song, 2017), etc. (Figure 2a). Mechanistic evaluations in these studies show that chalcones can exert their direct antiviral effects in different ways. A recent review by Elkhalifa et al. summarizes well the results of mechanistic studies of chalcone‐based compounds against various viruses (Elkhalifa, Al‐Hashimi, Al Moustafa, & Khalil, 2021).
FIGURE 2.
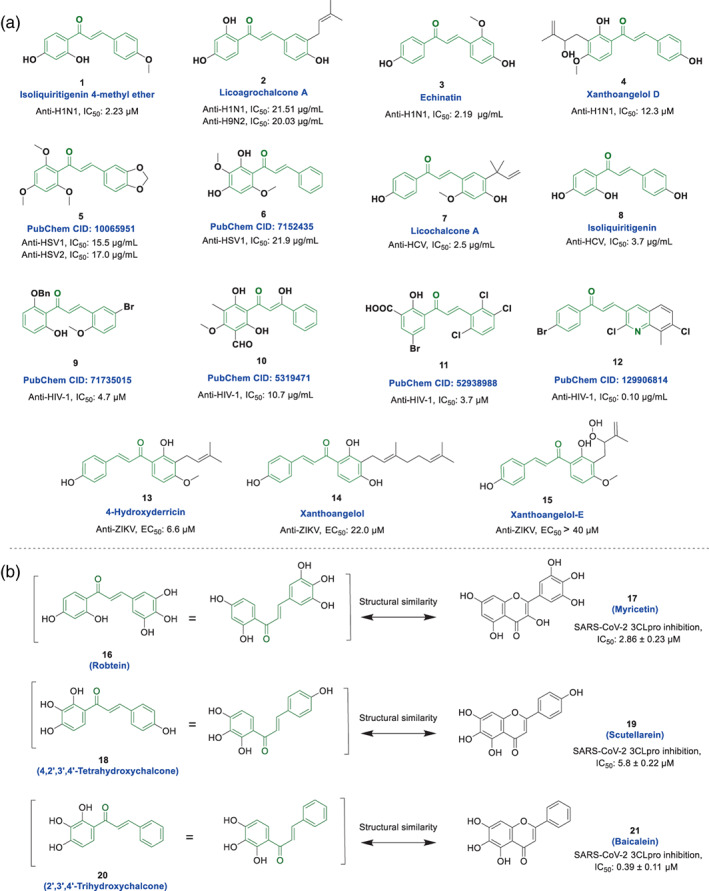
(a) Chemical structure of some chalcone‐based compounds with anti‐influenza (compounds 1–4) (Dao et al., 2011; Malbari et al., 2018; Nguyen et al., 2010; Park et al., 2011), anti‐HSV (compounds 5 and 6) (Brandão et al., 2010; Phrutivorapongkul et al., 2003), anti‐HCV (compounds 7 and 8) (Adianti et al., 2014), anti‐HIV (compounds 9–12) (Cole et al., 2016; Hameed et al., 2016; Sharma et al., 2011; Wu et al., 2003), and anti‐ZIKV activities (compounds 13–15) (Mottin et al., 2022); (b) Structural similarity of some chalcone‐based compounds (16, 18, and 20) and corresponding anti‐SARS‐CoV/SARS‐CoV‐2 flavonoids (compounds 17, 19, and 21) (Liu et al., 2021)
Chemically, chalcones are very similar to flavonoids and are actually their open and flexible analogs, which are made by connecting two aromatic rings with a three‐carbon α‐β unsaturated ketone bridge. Due to the similarity in structure and size, if these compounds have the same substitutions on rings A and B, they can potentially have similar physicochemical properties and ADME‐T parameters (absorption, distribution, metabolism, excretion, and toxicity), which is important in evaluations of pharmacokinetics properties and drug‐likeness of these compounds during drug discovery processes. Despite these similarities, chalcones and flavonoids have some important differences that are noteworthy. The most important difference is probably related to the possibility of chalcones participating in Michael interactions and forming covalent bonds with nucleophiles, which flavonoids lack this feature. Another important difference between these compounds is related to the different structural flexibility. Chalcones are clearly more flexible due to having an aliphatic linker in the middle section of the scaffold and creating E and Z structural isoforms, but flavonoids are more rigid structures. In general, this difference in flexibility cannot be considered an advantage or a disadvantage, because the structure–activity relationships in medicinal chemistry studies show that each of these characteristics can be useful or harmful in different conditions in optimizing the activity of structures. In general, chalcones can have more advantages for creating further interaction in the active site of targets due to the possibility of participating in Michael's reactions and also having more flexibility, but on the other hand, these features make chalcones have more possibility of unwanted interaction with other targets, which can be considered a defect.
Although the antiviral effects of flavonoids have been well studied against coronaviruses especially SARS‐CoV and SARS‐CoV‐2 (Bardelčíková, Miroššay, Šoltýs, & Mojžiš, 2022; Jo, Kim, Kim, Kim, & Shin, 2020; Liskova et al., 2021; Ngwa et al., 2020; Russo, Moccia, Spagnuolo, Tedesco, & Russo, 2020; Solnier & Fladerer, 2020), the potential of chalcone‐based compounds to inhibit these viruses have received less attention. Due to the significant structural similarity (as mentioned above), further investigation of the anti‐SARS‐CoV‐2 activity of chalcone structures (such as compounds 16, 18, and 20) corresponding to potent anti‐SARS‐CoV‐2 flavonoids such as myristin (17), scotlarine (19), baikaline (21), etc. seems necessary (Figure 2b). The greater flexibility along with having the same functional groups and pharmacophores, as well as the ability to participate in Michael's reaction in the active site of SARS‐CoV‐2 cysteine proteases 3‐chymotrypsin‐like protease (3CLpro) and papain‐like protease (PLpro), could potentially enable these compounds to have more effective interactions against SARS‐CoV‐2 targets.
Although polyphenolic compounds (shown in Figure 2b) could potentially have inhibitory effects against SARS‐CoV‐2, their clinical use may be of concern. Based on the structure‐toxicity relationships (STRs) findings, compounds containing catechol moieties (in which two hydroxyl groups are adjacent to each other on the phenyl ring) could be toxic by forming stable complexes in the presence of heavy metals such as iron or copper (Glaser & Holzgrabe, 2016; Schweigert, Zehnder, & Eggen, 2001). However, in drug discovery and development processes, toxicity issues must also be seriously considered.
3. INHIBITORY EFFECTS OF CHALCONES AGAINST CORONAVIRUSES
As stated in recent studies, enzymes 3CLpro and PLpro are the key agents for the replication and pathogenesis of SARS‐CoV‐2 and are highly regarded as critical targets for finding the treatment of COVID‐19 (Gonzalez et al., 2022). In previous studies, some natural‐based chalcones have been identified with a significant ability to inhibit coronaviruses 3CLpro and PLpro enzymes. Given that numerous natural‐based and synthetic derivatives of chalcones have been identified so far, it makes sense to search further to find more active derivatives against SARS‐CoV‐2. Previous studies have shown that chalcone‐based structures have remarkable effects in inhibiting SARS and MERS‐CoV infections. For example, in a study by Park et al., the inhibitory activities of some chalcone derivatives isolated from Angelica keiskei against important SARS‐CoV enzymes 3CLpro and PLpro were investigated using cell‐free/based assays. Among tested compounds, Xanthoangelol B (22), Xanthoangelol D (23), Xanthoangelol E (24), and Xanthoangelol F (25) showed better inhibitory activities than others (Figure 3). Mechanistic evaluations of this study suggested that the tested chalcones inhibit the 3CLpro enzyme through competitive inhibition, while PLpro inhibition is non‐competitive (Park et al., 2016).
FIGURE 3.
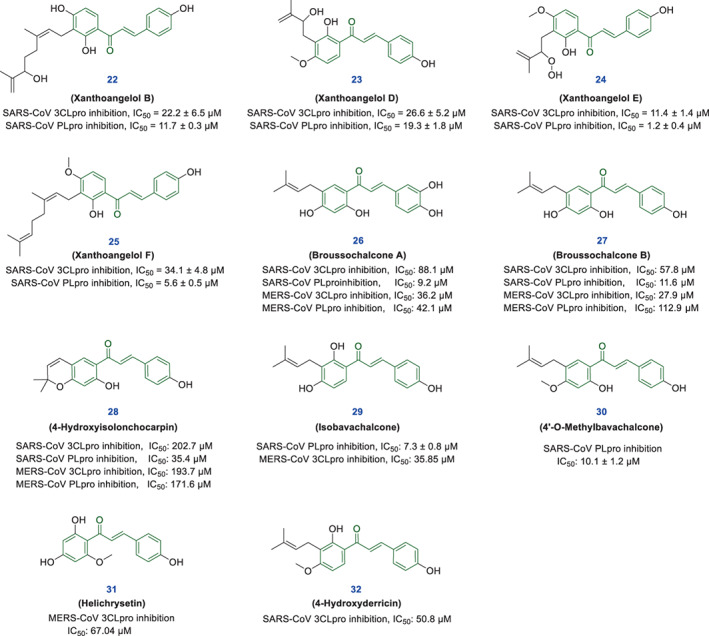
Chemical structures of some chalcone‐based compounds 22–32 with anti‐SARS‐CoV and anti‐MERS‐CoV activity
Park and co‐workers also performed a study to evaluate the inhibitory effects of some polyphenolic compounds extracted from Broussonetia papyrifera against 3CLpro and PLpro enzymes of SARS and MERS CoVs, in the presence of isoliquiritigenin, kaempferol, quercetin, and quercetin‐β‐galactoside as positive controls. Among the evaluated compounds, three alkaloids Broussochalcone A (26), Broussochalcone B (27) and 4‐hydroxyisolonchocarpin (28) have chalcone skeletons that showed stronger inhibitory effects compared to standards. Also, the results showed that these chalcone‐derivatives showed significantly more inhibitory effects against PLpro enzyme compared to 3CLpro enzyme with IC50 ranging between 9.2 to 35.4 μM, and can suppress SARS‐CoV more effectively than MERS‐CoV (Figure 3) (Park et al., 2017).
In another effort, Kim et al. also conducted a study for assessment of inhibitory activity of phenolic phytochemicals bavachinin, neobavaisoflavone, isobavachalcone, 4′‐O‐methylbavachalcone, psoralidin, and corylifol A driven from Psoralea corylifolia seeds against SARS‐CoV PLpro. Results of evaluations showed that chalcone‐based compounds isobavachalcone (29) and 4′‐O‐methylbavachalcone (30) have remarkable inhibitory activities against SARS‐CoV PLpro in a dose‐dependent manner by the IC50 = 7.3 μM and 10.1 μM, respectively (Figure 3) (Kim et al., 2014).
In a recent study, Jo et al. examined the anti‐MERS‐CoV 3CLpro activity of a flavonoid library. Of the six compounds with the best activity, two compounds called isobavachalcone with IC50 = 35.85 μM, and helichrysetin (31) with IC50 = 67.04 μM had chalcone structures. Computational evaluations suggested that S1 and S2 subsites of the MERS‐CoV 3CLpro active site are further involved in interactions with tested chalcones and flavonoid structures. These researchers stated that chalcones and flavonols are the preferred scaffolds for binding to the MERS‐CoV 3CLpro catalytic site (Jo, Kim, Kim, Shin, & Kim, 2019).
In the post‐COVID‐19 era, there are few in silico studies in which chalcones are suggested as potential therapeutic agents for the treatment of COVID‐19 (Alsafi, Hughes, & Said, 2020; Duran et al., 2021; Mathpal, Joshi, Sharma, Pande, & Chandra, 2022). In a recent in vitro study, the antiviral activity of a new ciprofloxacin‐chalcone hybrid structure (compound 33) was assessed against SARS‐CoV‐2 3CLpro in Vero cells by Alaaeldin et al. (2022; Figure 4). Results of evaluations showed that this compound significantly inhibited viral load replication (EC50 = 3.93 nM), and the plaque formation ability of the virus was inhibited to 86.8%. Enzymatic evaluations also showed that this compound exhibited a significant inhibitory effect against 3CLpro (IC50 = 0.6 ± 0.05 μM) in vitro in a dose‐dependent manner. In the evaluations, the desired ciprofloxacin‐chalcone hybrid compound showed stronger inhibitory activity against SARS‐CoV‐2 replication as well as 3CLpro inhibition compared to ciprofloxacin as the standard positive control. Since crystallographic evaluations have not been done in this study to determine how this compound binds in the active site of the 3CLpro, it is not possible to conclude with certainty about the role of the chalcone part in increasing the inhibitory activity. Interestingly, as a result of the hybridization of ciprofloxacin with chalcone, an N,2‐diarylacetamide structural core is created in the middle part of the molecule 33, which structures similar to this core have recently been recognized as special structural fragments that create very strong interactions in the active site of the SARS‐CoV‐2 3CLpro (see co‐crystal ligands 34–37 attached to the 3CLpro active site with the protein data bank (PDB) codes 5RGV, 5RGZ, 5R84, and 5RH0).
FIGURE 4.
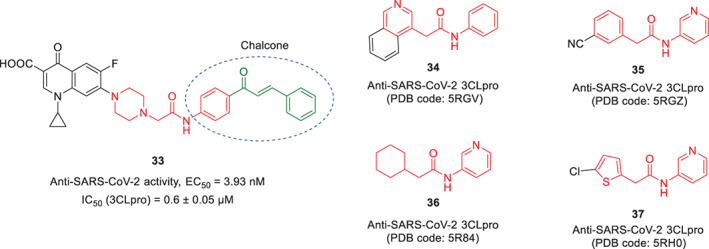
Chemical structure of a newly synthesized ciprofloxacin‐chalcone hybrid structure 33, and some small molecules 34–37 having N,2‐diarylacetamide structure with significant anti‐SARS‐CoV‐2 3CLpro activity (Alaaeldin, Mustafa, Abuo‐Rahma, & Fathy, 2022)
4. BENEFITS OF USING CYSTEINE REACTIVE WARHEADS IN THE STRUCTURE OF SARS‐COV‐2 INHIBITORS
There are many approved drugs and hit/lead compounds whose structural design is based on reaction with cysteine residues in the active sites of specific targets and covalent inhibition of these targets. As can be seen in the structure of the approved drugs afatinib, ibrutinib, osimertinib, and zanubrutinib (Figure 5a), the participant component in Michael's reaction in these structures is the α, β‐unsaturated part attached to an amide group. It seems that the nitrogen atom of the amide group here acts electronically as a balancer of activity, modulating/reducing the reactivity of molecules in vivo, which could potentially have a significant effect on reducing unwanted interactions and side effects. For this purpose and to prevent unwanted side reactions, the rings of piperidine and piperazine can be tested and used instead of the A‐ring in chalcone structures.
FIGURE 5.
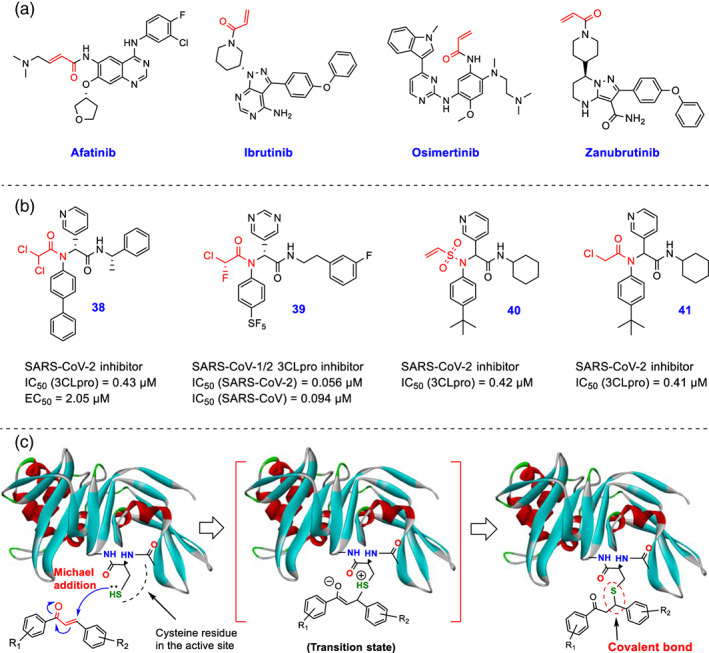
(a) The chemical structure of the FDA‐approved drugs afatanib, ibrutinib, osimertinib, and zanubrutinib containing α, β‐unsaturated carbonyl moieties, are previously designed to inhibit their targets (containing a unique cysteine residue in a specific protein of the active site) by irreversible hetero‐Michael addition reaction; (b) Chemical structures of novel developed covalent SARS‐CoV‐2 3CLpro inhibitors 38–41 containing different cysteine reactive warheads (Ma, Xia, et al., 2021; Stille et al., 2022; Yamane et al., 2022); (c) Schematic representation of the interaction of chalcone‐based derivatives containing α, β‐unsaturated ketone functional group at the active site of SARS‐CoV‐2 PLpro/3CLpro cysteine proteases through Michael's reaction and constructing a covalent carbon_sulfur (C–S) bond (images were prepared using Discovery Studio Visualizer v4.5)
In SARS‐CoV‐2 replication processes, viral proteases 3CLpro and PLpro have a critical responsibility for the production of non‐structural proteins by processing crude polyproteins translated by viral RNA. Because of these vital roles, these enzymes are considered highly important targets for the drug development of anti‐COVID‐19 agents (Cho et al., 2022; Ghahremanpour et al., 2020; Shcherbakov et al., 2021). In the designs made to make effective molecules, the possibility of covalent and non‐covalent inhibition of these enzymes has been considered. Among these studies, some useful research has been carried out recently for the development of covalent inhibitors of the SARS‐CoV‐2 proteases. Unlike most of the past studies in which the α, β‐unsaturated carbonyl groups were used in the structure of the molecules to inhibit the enzyme covalently, in recent studies, different structural warheads such as acrylamide, dichloroacetamide, 2‐chloroacrylamide, dibromoacetamide, ethyl‐4‐amino‐4‐oxobut‐2‐enoate, tribromoacetamide, 2‐bromo‐2,2‐dichloroacetamide, 4‐(dimethylamino)but‐2‐enamide, 2‐chloro‐2,2‐dibromoacetamide, etc. have been used in order to block the active site of the cysteine proteases.
In a recent study by Ma et al., some novel covalent SARS‐CoV‐2 3CLpro inhibitors containing various cysteine reactive warheads were designed and tested in order to inhibit the active site of the enzyme (Ma, Xia, et al., 2021). Among tested compounds in this study, compound 38 (Figure 5b) containing a dichloroacetamide moiety represented promising antiviral activity (EC50 = 2.05 μM) and selective enzymatic inhibition against 3CLpro (IC50 = 0.43 μM) without inhibition of the host cysteine proteases cathepsins (B, K, and L) calpain I, and caspase‐3. Importantly, the co‐crystal structure of SARS‐CoV‐2 3CLpro with compound 38 (PDB code: 7RN1) shows that this compound forms a critical covalent bond with the catalytic Cys145. In another recent study by Yamane et al., a new potent irreversible inhibitor of SARS‐CoV‐2 3CLpro with the IC50 of 0.056 μM (Figure 5b), compound 39 was developed that has chlorofluoroacetamide moiety as a warhead for the covalent blockage of the thiol group of Cys145 in 3CLpro active site (Yamane et al., 2022). Stille et al. also recently introduced two optimized SARS‐CoV‐2 3CLpro covalent inhibitors compound 40 (IC50 = 0.42 μM, PDB code: 7MLG) and compound 41 (IC50 = 0.41 μM, PDB code: 7MLF), in which vinyl sulfonamide and 2‐chloroacetamide moieties are used respectively to create covalent interaction in the active site of 3CLpro (Figure 5b) (Stille et al., 2022).
As stated above, cysteine moieties are present as important interacting residues in the active site of SARS‐CoV‐2 PLpro/3CLpro enzymes, so chalcones containing α, β‐unsaturated carbonyl functional groups can potentially form covalent carbon_sulfur (C–S) bonds with the sulfhydryl group of these residues (Figure 5c). Because covalent bond formation is irreversible, a fatal blow to the virus will occur if this interaction occurs (Stille et al., 2022). However, it should not be overlooked that there are infinite proteins (such as a variety of enzymes and receptors) in the in vivo environment to which irreversible covalent binding of chalcones with them may cause side effects (Srinivasan, Johnson, Lad, & Xing, 2009; Zhou, Chan, Duan, Huang, & Chen, 2005). The more reactive a molecule is, the more likely it is that adverse reactions will occur. Chemically, the reactivity of chalcones can be adjusted by placing electron withdrawing/donating groups on the rings. For example, placing highly electron‐withdrawing substitutions on A‐ring can significantly increase the reactivity of chalcones to participate in Michael's reactions (Dinkova‐Kostova, Abeygunawardana, & Talalay, 1998; Srinivasan et al., 2009). Of course, the presence of α, β‐unsaturated carbonyl group in the structure of a molecule does not mean that this compound necessarily establishes a covalent interaction. Since molecules can form many interactions such as hydrogen bonding, van der Waals, etc. in the active sites with amino acid side chains and even with amide backbone (carbonyl group and –NH moieties), so performing the Michael reaction requires proper spatial orientation of the molecule and availability of the α, β‐unsaturated portion for the sulfhydryl group of the cysteine residues.
Sequencing the SARS‐CoV‐2 RNA genome shortly after its emergence is one of the valuable steps to better fight COVID‐19. Discovering the high genetic similarity between SARS‐CoV and SARS‐CoV‐2 allows medicinal chemists to a faster and more targeted design of structures to find effective anti‐COVID‐19 agents. Based on the previous structure–activity relationship findings, several new studies were designed immediately, the results of which showed that most SARS‐CoV inhibitors are also effective against SARS‐CoV‐2. As highlighted in some recent publications, SARS‐CoV‐2 PLpro is a promising antiviral drug target for several reasons, including its role in virus replication and modulation of host innate immunity during viral infection (Tan, Hu, Jadhav, Tan, & Wang, 2022). Since the discovery of the first selective SARS‐CoV PLpro inhibitors by Gosh and colleagues in 2008 (Ratia et al., 2008), major advances in PLpro targeting have been made, which have been reviewed in several studies. As a result of high throughput screenings (HTSs) and lead optimizations in different studies, some highly potent structures with good profiles of activity have been obtained, which can be considered important steps to achieving the anti‐COVID‐19 drugs (Ma, Sacco, et al., 2021; Ma, Hu, Wang, Choza, & Wang, 2022; Osipiuk et al., 2021; Shan et al., 2021; Shen et al., 2021; Welker et al., 2021). Among the compounds reported so far, some compounds have been identified with promising and selective effects in inhibiting SARS‐CoV/SARS‐CoV‐2 PLpro proteases (known as the first generation of SARS‐CoV/SARS‐CoV‐2 PLpro inhibitors; Valipour, 2022a) whose central scaffold is N‐benzylpiperidine‐4‐carboxamide (N‐BPC). For this skeleton, some structural correlations with chalcone and chalcone‐amide scaffolds can be imagined (Figure 6). All three scaffolds have the same size and consist of two rings (A and B) connected by an aliphatic three‐atom linker. As stated above, chalcone‐amides are obtained by isosteric replacement of the carbon atom in the α position of the chalcones with a nitrogen atom. Recently, I introduced the chalcone‐amide scaffold as a unique backbone for the design and synthesis of selective SARS‐CoV‐2 PLpro inhibitors along with providing detailed structure–activity relationships (Valipour, 2022a). Although the absence of the amide functional group in the structure of chalcones makes the possibility of hydrogen interaction of these compounds disappear in this part of the molecule, the presence of α, β unsaturated ketone functional group makes these compounds have a high potential for creating covalent bonds with nucleophiles, especially cysteine residues (other mentioned scaffolds chalcone‐amide and N‐BPC do not have this capability). However, although chalcones are capable of reacting with nucleophiles as recipients in the Michael reaction, it is important that the interaction of these structures in biological environments is through not only Michael's reaction and the formation of covalent bonds with nucleophiles.
FIGURE 6.
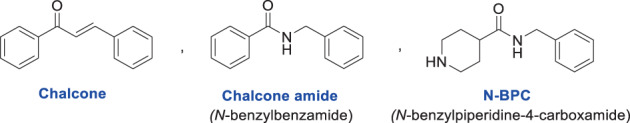
Structural similarity between scaffolding chalcone, chalcone‐amide, and N‐benzylpiperidine‐4‐carboxamide (N‐BPC)
HTS is an efficient approach to identifying specific bioactive compounds, which has been useful and widely used for the identification and development of SARS‐CoV/SARS‐CoV‐2 inhibitors. The success and efficiency of this approach depend a lot on the capacity and diversity of screened chemical libraries, as well as the convenience and cost‐effectiveness of the performed HTS methods. For the first time, Ghosh and colleagues identified a hit compound bearing an N‐BPC backbone with a moderate inhibitory effect against SARS‐CoV PLpro (IC50 = 59.2 μM) by performing an extensive fluorescence‐based HTS on a diverse structural library with 50,080 compounds (Figure 7, HTS 1, compound 42) (Ratia et al., 2008). Structural optimization of this lead structure led to the achievement of two highly potent optical isomers (compound 43 and 44) with sub‐micromolar IC50 values in which the N‐BPC backbone is available for structural modifications without any spatial disturbance (Ghosh et al., 2010). This hit compound was also used in other studies as the initial lead structure for the design and development of the subsequent SARS‐CoV PLpro inhibitors (Báez‐Santos et al., 2014). Shan and colleagues also recently performed an HTS on a library with 35,360 bioactive compounds, which identified some active structures bearing the N‐BPC backbone as the best candidates for developing SARS‐CoV‐2 PLpro inhibitors (Figure 7, HTS 2) (Shan et al., 2021). As a result of structural optimizing the lead compounds identified in this study, three highly active compounds were produced in which there are bulk amide/ureide substitutions in their meta position of the B ring (compounds 47–49).
FIGURE 7.
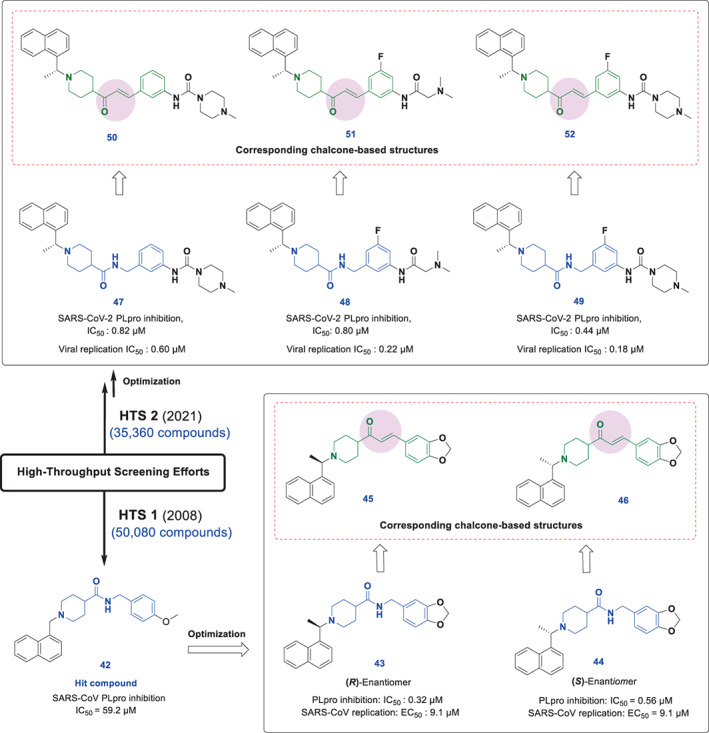
Chemical structure of compounds 42–44 and 47–49 containing N‐BPC core as the important selective SARS‐CoV PLpro inhibitors identified by two independent HTS, along with corresponding chalcone‐based structures 45–46 and 50–52 having the ability to establish covalent bonds in the active site of the targets by participating in Michael's reaction (The chalcone‐based structures drawn in this figure have never been synthesized anywhere).
The x‐ray crystal structure of compound 43 in the active site of SARS‐CoV PLpro has been previously released under PDB code 3MJ5. As shown in Figure 8, this protein is made of 316 amino acids, eight of which are cysteine (residues Cys112, Cys149, Cys190, Cys193, Cys225, Cys227, Cys261, and Cys271). The most important interacting residues with various parts of compound 43 in the active site of the PLpro enzyme are Leu163, Gly164, Asp165, Pro248, Pro249, Tyr265, Tyr269, and Tyr274, which are highlighted in Figure 8 and Table 1. Examining the location of cysteine residues (highlighted in blue in Figure 8) shows that most of them are located far from the active site, and only two residues Cys112 and Cys271 are present in the active site of PLpro. As mentioned above, the amide functional group of compound 43 is readily available for modification and replacement with an α, β unsaturated ketone part. Of course, it is important to note that this part is involved in creating an important hydrogen bond interaction (derived from the interaction of –NH– with the Tyr269 residue), and this important interaction is also lost when this part is removed. Attention to the amino acid sequence of the mentioned PLpro enzyme shows that there is a cysteine residue (Cys271) near Tyr269, which can potentially trigger Michael's reaction if the conditions are met. Interestingly, if compound 45 is placed in the active site of PLpro (instead of compound 43), the carbon in the β position of the unsaturated part is located at a suitable distance within reach of the sulfur atom of Cys271. As shown in Figure 9, the distance between β‐carbon in compound 45 and the carbon atom carrying the side chain of Cys271 is less than 7 Å. In addition, this side chain is quite flexible and can be directed towards molecule 45, so its sulfur atom can be placed much closer to the carbon in the β position. However, if compound 45 interacts at the active site of the PLpro, it is not clear how the structure directs in the active site. Using the insights obtained from the crystallographic evaluations, the role of the naphthyl moiety (which is an important lipophilic scaffold in medicinal chemistry; Valipour, Naderi, et al., 2021) in the orientation of compound 45 in a similar pattern to that of compound 43 in the active site of the PLpro is critical. Accordingly, the naphthyl moiety is most likely located inside a lipophilic pocket containing Pro248 and Pro249 residues (see the interactions represented in the crystal structure of PLpro bound to some potent N‐BPC‐based inhibitors with the PDB codes of 4OVZ, 4OW0, 3MJ5, 3E9S, and 7E35). Although the orientation of compound 45 can also be predicted using computational methods, more accurate conclusions can be obtained through crystallographic studies. In general, the presence of α, β‐unsaturated carbonyl part of chalcone‐based compounds greatly increases the possibility of Michael's interaction and irreversible inhibition of the viral cysteine proteases.
FIGURE 8.
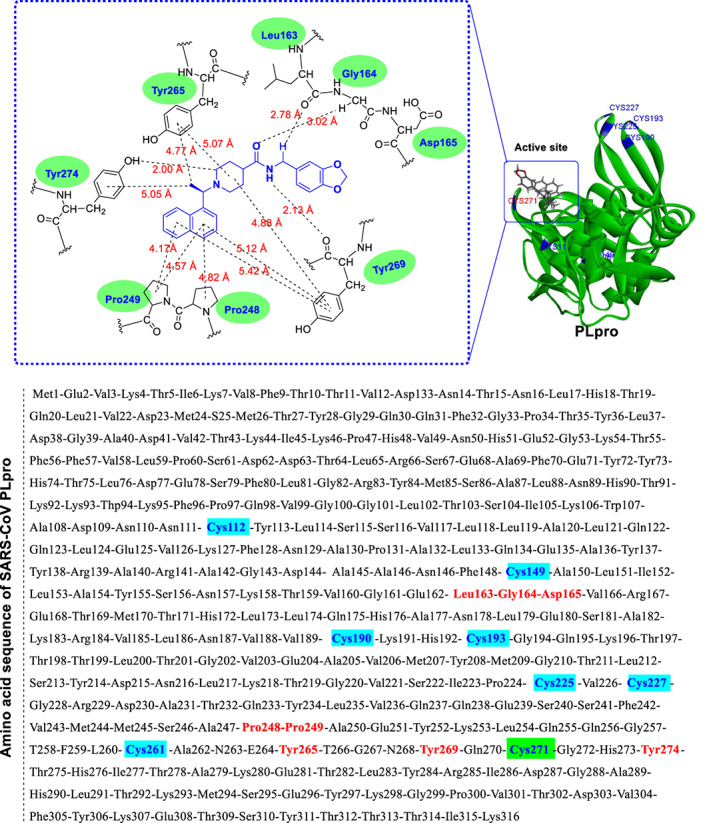
Schematic representation of binding of compound 43 to the SARS‐CoV PLpro active site (PDB code: 3MJ5). As highlighted in red, the most important interacting sequences in the active site of the PLpro are Leu163, Gly164, Asp165, Pro248, Pro249, Tyr265, Tyr269, and Tyr274. In total, this enzyme has eight cysteine residues Cys112, Cys149, Cys190, Cys193, Cys225, Cys227, Cys261, and Cys271, two of which (Cys112 and Cys271) are located in the active site (graphic images were prepared and analyzed by Discovery Studio Visualizer v4.5).
TABLE 1.
The most important interactions of the compound 43 (PDB code: 3MJ5) in the active site of SARS‐CoV PLpro (interactions and distances were analyzed by Discovery Studio Visualizer v4.5)
| Interaction | Residues | Interacting structural parts (ligand) | Distance (Å) |
|---|---|---|---|
| Carbon | Leu163 (carbonyl group) | H (methylene group) | 2.78 |
| Carbon | Gly164 (methylene group) | Carbonyl group (amide) | 2.74 |
| Carbon | Asp165 (carboxylic acid group) | H (piperidine ring) | 1.69 |
| Pi‐alkyl | Pro248 (pyrrolidine ring) | First ring of the naphthyl moiety | 4.82 |
| Pi‐alkyl | Pro249 (pyrrolidine ring) | First ring of the naphthyl moiety | 4.57 |
| Pi‐alkyl | Pro249 (pyrrolidine ring) | Second ring of the naphthyl moiety | 4.17 |
| Pi‐alkyl | Tyr265 (phenyl group) | Piperidine ring | 5.07 |
| Pi‐alkyl | Tyr265 (phenyl group) | Chiral methyl group | 4.77 |
| Conventional H‐bond | Tyr269 (carbonyl group) | NH (amide) | 2.13 |
| Pi‐pi T‐shaped | Tyr269 (phenyl group) | First ring of the naphthyl moiety | 5.12 |
| Pi‐pi T‐shaped | Tyr269 (phenyl group) | Second ring of the naphthyl moiety | 5.42 |
| Pi‐alkyl | Tyr269 (phenyl group) | Piperidine ring | 4.88 |
| Pi‐alkyl | Tyr274 (phenyl group) | Chiral methyl group | 5.05 |
| Carbon | Tyr274 (phenyl group) | H (Piperidine ring) | 2.00 |
FIGURE 9.
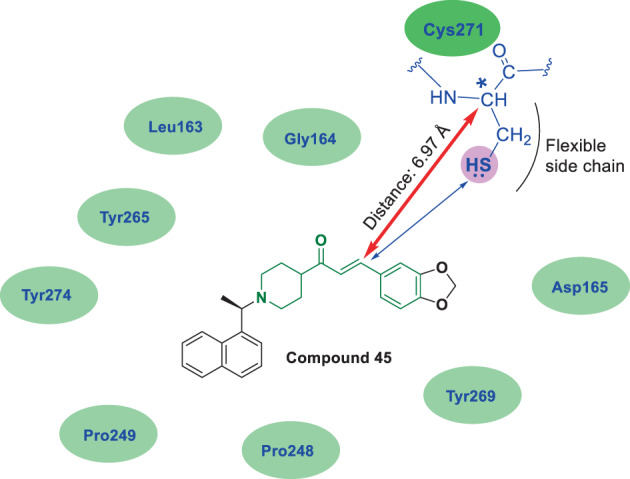
Hypothetical placement of compound 45 in the active site of SARS‐CoV PLpro enzyme (PDB code 3MJ5). As shown, if the placement of compound 45 in the active site of the PLpro is similar to compound 43 in terms of spatial orientation (which is very likely), the α, β unsaturated ketone functional group will be placed in a suitable orientation and distance to create Michael interaction with Cys271 (graphic image was prepared by Discovery Studio Visualizer v4.5., and distance was calculated by Molegro Molecular Viewer 6.5 software)
5. CONCLUSION
The targeted covalent modification which is commonly performed by an irreversible hetero‐Michael addition has expanded the druggable landscape by enhancing the ligand‐binding selectivity for proteins and increasing the binding affinities in targets with shallow binding sites (Jackson, Widen, Harki, & Brummond, 2017). Although covalent bonding and the formation of drug‐protein adducts can lead to drug toxicity, many approved drugs or their metabolites (such as acetaminophen, irinotecan, carbamazepine, ritonavir, clozapine, procainamide, hydralazine, cyclosporine A, halothane, tamoxifen, non‐steroidal anti‐inflammatory drugs, antibacterial sulfonamides, macrolide antibiotics, etc.) act via the construction of covalent bonds with protein targets in vivo (Zhou et al., 2005).
Given the potential for side effects from covalent inhibitors, irreversible inhibition of SARS‐CoV‐2 proteases is now considered a promising strategy for the treatment of COVID‐19 (Stille et al., 2022). In the structure of many important reported SARS‐CoV‐2 inhibitors, there are diverse electrophilic moieties with the ability to establish covalent bonds, such as α, β‐unsaturated carbonyl groups (Michael acceptors), epoxide, α‐ketoamide, aziridine, etc. Chalcones are one of the most well‐known Michael acceptors in chemical reactions. The interaction of α, β‐unsaturated carbonyl group of chalcones with the thiol (–SH) of cysteine residues protein targets can lead to the construction of covalent bonds. This activity of chalcones becomes more important when we know that key SARS‐CoV‐2 enzymes PLpro and 3CLpro are cysteine proteases with shallow binding sites. Interestingly, some previous studies have reported the significant inhibitory effects of chalcones (compounds 22–32) on both these enzymes, which may have occurred through covalent inhibition. As discussed in the text, chalcones are structurally similar to flavonoids (which are known as selective SARS‐CoV‐2 3CLpro inhibitors) and similar to compounds containing N‐BPC backbone (which are known as selective SARS‐CoV‐2 PLpro inhibitors). This evidence suggests that by modification of the chalcone‐based structures there is a good potential to achieve promising dual‐action anti‐SARS‐CoV‐2 compounds with inhibitory activity against both enzymes 3CLpro and PLpro.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
Valipour, M. (2022). Recruitment of chalcone's potential in drug discovery of anti‐SARS‐CoV‐2 agents. Phytotherapy Research, 36(12), 4477–4490. 10.1002/ptr.7651
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Adianti, M. , Aoki, C. , Komoto, M. , Deng, L. , Shoji, I. , Wahyuni, T. S. , … Hotta, H. (2014). Anti‐hepatitis C virus compounds obtained from Glycyrrhiza uralensis and other Glycyrrhiza species. Microbiology and Immunology, 58(3), 180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaaeldin, R. , Mustafa, M. , Abuo‐Rahma, G. E. D. A. , & Fathy, M. (2022). In vitro inhibition and molecular docking of a new ciprofloxacin‐chalcone against SARS‐CoV‐2 main protease. Fundamental & Clinical Pharmacology, 36(1), 160–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsafi, M. A. , Hughes, D. L. , & Said, M. A. (2020). First COVID‐19 molecular docking with a chalcone‐based compound: Synthesis, single‐crystal structure and Hirshfeld surface analysis study. Acta Crystallographica Section C: Structural Chemistry, 76(12), 1043–1050. [DOI] [PubMed] [Google Scholar]
- Báez‐Santos, Y. M. , Barraza, S. J. , Wilson, M. W. , Agius, M. P. , Mielech, A. M. , Davis, N. M. , … Mesecar, A. D. (2014). X‐ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain‐like proteases. Journal of Medicinal Chemistry, 57(6), 2393–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardelčíková, A. , Miroššay, A. , Šoltýs, J. , & Mojžiš, J. (2022). Therapeutic and prophylactic effect of flavonoids in post‐COVID‐19 therapy. Phytotherapy Research, 36(5), 2042–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão, G. C. , Kroon, E. G. , Duarte, M. G. R. , Braga, F. C. , de Souza Filho, J. D. , & de Oliveira, A. B. (2010). Antimicrobial, antiviral and cytotoxic activity of extracts and constituents from Polygonum spectabile Mart. Phytomedicine, 17(12), 926–929. [DOI] [PubMed] [Google Scholar]
- Cho, C. C. , Li, S. G. , Lalonde, T. J. , Yang, K. S. , Yu, G. , Qiao, Y. , … Ray Liu, W. (2022). Drug repurposing for the SARS‐CoV‐2 papain‐like protease. ChemMedChem, 17(1), e202100455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, A. L. , Hossain, S. , Cole, A. M. , & Phanstiel, O., IV . (2016). Synthesis and bioevaluation of substituted chalcones, coumaranones and other flavonoids as anti‐HIV agents. Bioorganic & Medicinal Chemistry, 24(12), 2768–2776. [DOI] [PubMed] [Google Scholar]
- Corona, A. , Wycisk, K. , Talarico, C. , Manelfi, C. , Milia, J. , Cannalire, R. , … Beccari, A. R. (2022). Natural compounds inhibit SARS‐CoV‐2 nsp13 unwinding and ATPase enzyme activities. ACS Pharmacology & Translational Science, 5(4), 226–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao, T. T. , Nguyen, P. H. , Lee, H. S. , Kim, E. , Park, J. , Lim, S. I. , & Oh, W. K. (2011). Chalcones as novel influenza A (H1N1) neuraminidase inhibitors from Glycyrrhiza inflata . Bioorganic & Medicinal Chemistry Letters, 21(1), 294–298. [DOI] [PubMed] [Google Scholar]
- DeCorte, B. L. (2016). Underexplored opportunities for natural products in drug discovery: Miniperspective. Journal of Medicinal Chemistry, 59(20), 9295–9304. [DOI] [PubMed] [Google Scholar]
- Derosa, G. , Maffioli, P. , D'Angelo, A. , & Di Pierro, F. (2021). A role for quercetin in coronavirus disease 2019 (COVID‐19). Phytotherapy Research, 35(3), 1230–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova‐Kostova, A. T. , Abeygunawardana, C. , & Talalay, P. (1998). Chemoprotective properties of phenylpropenoids, bis (benzylidene) cycloalkanones, and related Michael reaction acceptors: Correlation of potencies as phase 2 enzyme inducers and radical scavengers. Journal of Medicinal Chemistry, 41(26), 5287–5296. [DOI] [PubMed] [Google Scholar]
- Duran, N. , Polat, M. F. , Aktas, D. A. , Alagoz, M. A. , Ay, E. , Cimen, F. , … Algul, O. (2021). New chalcone derivatives as effective against SARS‐CoV‐2 agent. International Journal of Clinical Practice, 75(12), e14846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkhalifa, D. , Al‐Hashimi, I. , Al Moustafa, A.‐E. , & Khalil, A. (2021). A comprehensive review on the antiviral activities of chalcones. Journal of Drug Targeting, 29(4), 403–419. [DOI] [PubMed] [Google Scholar]
- El‐Missiry, M. A. , Fekri, A. , Kesar, L. A. , & Othman, A. I. (2021). Polyphenols are potential nutritional adjuvants for targeting COVID‐19. Phytotherapy Research, 35(6), 2879–2889. [DOI] [PubMed] [Google Scholar]
- Gan, X. , Wang, Y. , Hu, D. , & Song, B. (2017). Design, synthesis, and antiviral activity of novel chalcone derivatives containing a purine moiety. Chinese Journal of Chemistry, 35(5), 665–672. [Google Scholar]
- Gaonkar, S. L. , & Vignesh, U. (2017). Synthesis and pharmacological properties of chalcones: A review. Research on Chemical Intermediates, 43(11), 6043–6077. [Google Scholar]
- Ghahremanpour, M. M. , Tirado‐Rives, J. , Deshmukh, M. , Ippolito, J. A. , Zhang, C.‐H. , Cabeza de Vaca, I. , … Jorgensen, W. L. (2020). Identification of 14 known drugs as inhibitors of the main protease of SARS‐CoV‐2. ACS Medicinal Chemistry Letters, 11(12), 2526–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, A. K. , Takayama, J. , Rao, K. V. , Ratia, K. , Chaudhuri, R. , Mulhearn, D. C. , … Johnson, M. E. (2010). Severe acute respiratory syndrome coronavirus papain‐like novel protease inhibitors: Design, synthesis, protein−ligand X‐ray structure and biological evaluation. Journal of Medicinal Chemistry, 53(13), 4968–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser, J. , & Holzgrabe, U. (2016). Focus on PAINS: False friends in the quest for selective anti‐protozoal lead structures from nature? MedChemComm, 7(2), 214–223. [Google Scholar]
- Gonzalez, B. L. , de Oliveira, N. C. , Ritter, M. R. , Tonin, F. S. , Melo, E. B. , Sanches, A. C. C. , … de Medeiros Araújo, D. C. (2022). The naturally‐derived alkaloids as a potential treatment for COVID‐19: A scoping review. Phytotherapy Research, 36(7), 2686–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gour, A. , Manhas, D. , Bag, S. , Gorain, B. , & Nandi, U. (2021). Flavonoids as potential phytotherapeutics to combat cytokine storm in SARS‐CoV‐2. Phytotherapy Research, 35(8), 4258–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, T. , Xia, R. , Chen, M. , He, J. , Su, S. , Liu, L. , … Xue, W. (2019). Biological activity evaluation and action mechanism of chalcone derivatives containing thiophene sulfonate. RSC Advances, 9(43), 24942–24950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed, A. , Abdullah, M. I. , Ahmed, E. , Sharif, A. , Irfan, A. , & Masood, S. (2016). Anti‐HIV cytotoxicity enzyme inhibition and molecular docking studies of quinoline based chalcones as potential non‐nucleoside reverse transcriptase inhibitors (NNRT). Bioorganic Chemistry, 65, 175–182. [DOI] [PubMed] [Google Scholar]
- Haq, F. U. , Roman, M. , Ahmad, K. , Rahman, S. U. , Shah, S. M. A. , Suleman, N. , … Ullah, W. (2020). Artemisia annua: Trials are needed for COVID‐19. Phytotherapy Research, 34(10), 2423–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, A. L. , Edrada‐Ebel, R. , & Quinn, R. J. (2015). The re‐emergence of natural products for drug discovery in the genomics era. Nature Reviews Drug Discovery, 14(2), 111–129. [DOI] [PubMed] [Google Scholar]
- Jackson, P. A. , Widen, J. C. , Harki, D. A. , & Brummond, K. M. (2017). Covalent modifiers: A chemical perspective on the reactivity of α, β‐unsaturated carbonyls with thiols via hetero‐Michael addition reactions. Journal of Medicinal Chemistry, 60(3), 839–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, S. , Kim, H. , Kim, S. , Shin, D. H. , & Kim, M. S. (2019). Characteristics of flavonoids as potent MERS‐CoV 3C‐like protease inhibitors. Chemical Biology & Drug Design, 94(6), 2023–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, S. , Kim, S. , Kim, D. Y. , Kim, M.‐S. , & Shin, D. H. (2020). Flavonoids with inhibitory activity against SARS‐CoV‐2 3CLpro. Journal of Enzyme Inhibition and Medicinal Chemistry, 35(1), 1539–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar Mahapatra, D. , Asati, V. , & Bharti, S. K. (2019). An updated patent review of therapeutic applications of chalcone derivatives (2014‐present). Expert Opinion on Therapeutic Patents, 29(5), 385–406. [DOI] [PubMed] [Google Scholar]
- Kim, D. W. , Seo, K. H. , Curtis‐Long, M. J. , Oh, K. Y. , Oh, J.‐W. , Cho, J. K. , … Park, K. H. (2014). Phenolic phytochemical displaying SARS‐CoV papain‐like protease inhibition from the seeds of Psoralea corylifolia . Journal of Enzyme Inhibition and Medicinal Chemistry, 29(1), 59–63. [DOI] [PubMed] [Google Scholar]
- Liskova, A. , Samec, M. , Koklesova, L. , Samuel, S. M. , Zhai, K. , Al‐Ishaq, R. K. , … Zarrabi, A. (2021). Flavonoids against the SARS‐CoV‐2 induced inflammatory storm. Biomedicine & Pharmacotherapy, 138, 111430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Ye, F. , Sun, Q. , Liang, H. , Li, C. , Li, S. , … Lai, L. (2021). Scutellaria baicalensis extract and baicalein inhibit replication of SARS‐CoV‐2 and its 3C‐like protease in vitro. Journal of Enzyme Inhibition and Medicinal Chemistry, 36(1), 497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Zhao, X. , Li, J. , Niu, P. , Yang, B. , Wu, H. , … Bi, Y. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C. , Hu, Y. , Wang, Y. , Choza, J. , & Wang, J. (2022). Drug‐repurposing screening identified tropifexor as a SARS‐CoV‐2 papain‐like protease inhibitor. ACS Infectious Diseases, 8(5), 1022–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C. , Sacco, M. D. , Xia, Z. , Lambrinidis, G. , Townsend, J. A. , Hu, Y. , … Gongora, M. (2021). Discovery of SARS‐CoV‐2 papain‐like protease inhibitors through a combination of high‐throughput screening and a FlipGFP‐based reporter assay. ACS Central Science, 7(7), 1245–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C. , Xia, Z. , Sacco, M. D. , Hu, Y. , Townsend, J. A. , Meng, X. , … Zhang, X. (2021). Discovery of di‐and trihaloacetamides as covalent SARS‐CoV‐2 main protease inhibitors with high target specificity. Journal of the American Chemical Society, 143(49), 20697–20709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malbari, K. , Gonsalves, H. , Chintakrindi, A. , Gohil, D. , Joshi, M. , Kothari, S. , … Kanyalkar, M. (2018). In search of effective H1N1 neuraminidase inhibitor by molecular docking, antiviral evaluation and membrane interaction studies using NMR. Acta Virologica, 62(2), 179–190. [DOI] [PubMed] [Google Scholar]
- Mateeva, N. , Eyunni, S. V. , Redda, K. K. , Ononuju, U. , Hansberry, T. D., II , Aikens, C. , & Nag, A. (2017). Functional evaluation of synthetic flavonoids and chalcones for potential antiviral and anticancer properties. Bioorganic & Medicinal Chemistry Letters, 27(11), 2350–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathpal, S. , Joshi, T. , Sharma, P. , Pande, V. , & Chandra, S. (2022). Assessment of activity of chalcone compounds as inhibitors of 3‐chymotrypsin like protease (3CLPro) of SARS‐CoV‐2: In silico study. Structural Chemistry, 33(5), 1815–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottin, M. , Caesar, L. K. , Brodsky, D. , Mesquita, N. C. , de Oliveira, K. Z. , Noske, G. D. , … Zorn, K. M. (2022). Chalcones from Angelica keiskei (ashitaba) inhibit key Zika virus replication proteins. Bioorganic Chemistry, 120, 105649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, P. H. , Na, M. , Dao, T. T. , Ndinteh, D. T. , Mbafor, J. T. , Park, J. , … Oh, W. K. (2010). New stilbenoid with inhibitory activity on viral neuraminidases from Erythrina addisoniae . Bioorganic & Medicinal Chemistry Letters, 20(22), 6430–6434. [DOI] [PubMed] [Google Scholar]
- Ngwa, W. , Kumar, R. , Thompson, D. , Lyerly, W. , Moore, R. , Reid, T.‐E. , … Toyang, N. (2020). Potential of flavonoid‐inspired phytomedicines against COVID‐19. Molecules, 25(11), 2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipiuk, J. , Azizi, S.‐A. , Dvorkin, S. , Endres, M. , Jedrzejczak, R. , Jones, K. A. , … Maki, S. L. (2021). Structure of papain‐like protease from SARS‐CoV‐2 and its complexes with non‐covalent inhibitors. Nature Communications, 12(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.‐Y. , Jeong, H. J. , Kim, Y. M. , Park, S.‐J. , Rho, M.‐C. , Park, K. H. , … Lee, W. S. (2011). Characteristic of alkylated chalcones from Angelica keiskei on influenza virus neuraminidase inhibition. Bioorganic & Medicinal Chemistry Letters, 21(18), 5602–5604. [DOI] [PubMed] [Google Scholar]
- Park, J.‐Y. , Ko, J.‐A. , Kim, D. W. , Kim, Y. M. , Kwon, H.‐J. , Jeong, H. J. , … Ryu, Y. B. (2016). Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS‐CoV. Journal of Enzyme Inhibition and Medicinal Chemistry, 31(1), 23–30. [DOI] [PubMed] [Google Scholar]
- Park, J.‐Y. , Yuk, H. J. , Ryu, H. W. , Lim, S. H. , Kim, K. S. , Park, K. H. , … Lee, W. S. (2017). Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. Journal of Enzyme Inhibition and Medicinal Chemistry, 32(1), 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phrutivorapongkul, A. , Lipipun, V. , Ruangrungsi, N. , Kirtikara, K. , Nishikawa, K. , Maruyama, S. , … Ishikawa, T. (2003). Studies on the chemical constituents of stem bark of Millettia leucantha: Isolation of new chalcones with cytotoxic, anti‐herpes simplex virus and anti‐inflammatory activities. Chemical and Pharmaceutical Bulletin, 51(2), 187–190. [DOI] [PubMed] [Google Scholar]
- Ratia, K. , Pegan, S. , Takayama, J. , Sleeman, K. , Coughlin, M. , Baliji, S. , … Baker, S. C. (2008). A noncovalent class of papain‐like protease/deubiquitinase inhibitors blocks SARS virus replication. Proceedings of the National Academy of Sciences, 105(42), 16119–16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, M. , Moccia, S. , Spagnuolo, C. , Tedesco, I. , & Russo, G. L. (2020). Roles of flavonoids against coronavirus infection. Chemico‐Biological Interactions, 328, 109211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweigert, N. , Zehnder, A. J. , & Eggen, R. I. (2001). Chemical properties of catechols and their molecular modes of toxic action in cells, from microorganisms to mammals: Minireview. Environmental Microbiology, 3(2), 81–91. [DOI] [PubMed] [Google Scholar]
- Shan, H. , Liu, J. , Shen, J. , Dai, J. , Xu, G. , Lu, K. , … Xiang, H. (2021). Development of potent and selective inhibitors targeting the papain‐like protease of SARS‐CoV‐2. Cell Chemical Biology, 28(6), 855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, H. , Patil, S. , Sanchez, T. W. , Neamati, N. , Schinazi, R. F. , & Buolamwini, J. K. (2011). Synthesis, biological evaluation and 3D‐QSAR studies of 3‐keto salicylic acid chalcones and related amides as novel HIV‐1 integrase inhibitors. Bioorganic & Medicinal Chemistry, 19(6), 2030–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakov, D. , Baev, D. , Kalinin, M. , Dalinger, A. , Chirkova, V. , Belenkaya, S. , … Sharlaeva, E. (2021). Design and evaluation of bispidine‐based SARS‐CoV‐2 main protease inhibitors. ACS Medicinal Chemistry Letters, 13(1), 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Z. , Ratia, K. , Cooper, L. , Kong, D. , Lee, H. , Kwon, Y. , … Sharlaeva, E. (2021). Design of SARS‐CoV‐2 PLpro inhibitors for COVID‐19 antiviral therapy leveraging binding cooperativity. Journal of Medicinal Chemistry, 65(4), 2940–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P. , Anand, A. , & Kumar, V. (2014). Recent developments in biological activities of chalcones: A mini review. European Journal of Medicinal Chemistry, 85, 758–777. [DOI] [PubMed] [Google Scholar]
- Sisakht, M. , Mahmoodzadeh, A. , & Darabian, M. (2021). Plant‐derived chemicals as potential inhibitors of SARS‐CoV‐2 main protease (6LU7), a virtual screening study. Phytotherapy Research, 35(6), 3262–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnier, J. , & Fladerer, J.‐P. (2020). Flavonoids: A complementary approach to conventional therapy of COVID‐19? Phytochemistry Reviews, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, B. , Johnson, T. E. , Lad, R. , & Xing, C. (2009). Structure−activity relationship studies of chalcone leading to 3‐hydroxy‐4, 3′, 4′, 5′‐tetramethoxychalcone and its analogues as potent nuclear factor κB inhibitors and their anticancer activities. Journal of Medicinal Chemistry, 52(22), 7228–7235. [DOI] [PubMed] [Google Scholar]
- Stille, J. K. , Tjutrins, J. , Wang, G. , Venegas, F. A. , Hennecker, C. , Rueda, A. M. , … Labarre, A. (2022). Design, synthesis and in vitro evaluation of novel SARS‐CoV‐2 3CLpro covalent inhibitors. European Journal of Medicinal Chemistry, 229, 114046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, H. , Hu, Y. , Jadhav, P. , Tan, B. , & Wang, J. (2022). Progress and challenges in targeting the SARS‐CoV‐2 papain‐like protease. Journal of Medicinal Chemistry, 65(11), 7561–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valipour, M. (2022a). Chalcone‐amide, a privileged backbone for the design and development of selective SARS‐CoV/SARS‐CoV‐2 papain‐like protease inhibitors. European Journal of Medicinal Chemistry, 240, 114572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valipour, M. (2022b). Different aspects of Emetine's capabilities as a highly potent SARS‐CoV‐2 inhibitor against COVID‐19. ACS Pharmacology & Translational Science, 5(6), 387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valipour, M. (2022c). Recent advances of antitumor shikonin/alkannin derivatives: A comprehensive overview focusing on structural classification, synthetic approaches, and mechanisms of action. European Journal of Medicinal Chemistry, 235, 114314. [DOI] [PubMed] [Google Scholar]
- Valipour, M. , Irannejad, H. , & Emami, S. (2022a). Application of emetine in SARS‐CoV‐2 treatment: Regulation of p38 MAPK signaling pathway for preventing emetine‐induced cardiac complications. Cell Cycle, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valipour, M. , Irannejad, H. , & Emami, S. (2022b). Papaverine, a promising therapeutic agent for the treatment of COVID‐19 patients with underlying cardiovascular diseases (CVDs). Drug Development Research, 83(6), 1246–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valipour, M. , Naderi, N. , Heidarli, E. , Shaki, F. , Motafeghi, F. , Amiri, F. T. , … Irannejad, H. (2021). Design, synthesis and biological evaluation of naphthalene‐derived (arylalkyl) azoles containing heterocyclic linkers as new anticonvulsants: A comprehensive in silico, in vitro, and in vivo study. European Journal of Pharmaceutical Sciences, 166, 105974. [DOI] [PubMed] [Google Scholar]
- Valipour, M. , Zarghi, A. , Ebrahimzadeh, M. A. , & Irannejad, H. (2021). Therapeutic potential of chelerythrine as a multi‐purpose adjuvant for the treatment of COVID‐19. Cell Cycle, 20(22), 2321–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker, A. , Kersten, C. , Müller, C. , Madhugiri, R. , Zimmer, C. , Müller, P. , … Sotriffer, C. (2021). Structure‐activity relationships of benzamides and isoindolines designed as SARS‐CoV protease inhibitors effective against SARS‐CoV‐2. ChemMedChem, 16(2), 340–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J.‐H. , Wang, X.‐H. , Yi, Y.‐H. , & Lee, K.‐H. (2003). Anti‐AIDS agents 54. A potent anti‐HIV chalcone and flavonoids from genus Desmos. Bioorganic & Medicinal Chemistry Letters, 13(10), 1813–1815. [DOI] [PubMed] [Google Scholar]
- Yamane, D. , Onitsuka, S. , Re, S. , Isogai, H. , Hamada, R. , Hiramoto, T. , … Ojida, A. (2022). Selective covalent targeting of SARS‐CoV‐2 main protease by enantiopure chlorofluoroacetamide. Chemical Science, 13(10), 3027–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. , Wei, J. , Huang, T. , Lei, L. , Shen, C. , Lai, J. , … Liu, Y. (2021). Resveratrol inhibits the replication of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) in cultured Vero cells. Phytotherapy Research, 35(3), 1127–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedipour, F. , Hosseini, S. A. , Sathyapalan, T. , Majeed, M. , Jamialahmadi, T. , Al‐Rasadi, K. , … Sahebkar, A. (2020). Potential effects of curcumin in the treatment of COVID‐19 infection. Phytotherapy Research, 34(11), 2911–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalpoor, H. , Bakhtiyari, M. , Liaghat, M. , Nabi‐Afjadi, M. , & Ganjalikhani‐Hakemi, M. (2022). Quercetin potential effects against SARS‐CoV‐2 infection and COVID‐19‐associated cancer progression by inhibiting mTOR and hypoxia‐inducible factor‐1α (HIF‐1α). Phytotherapy Research, 36(7), 2679–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, S. , Chan, E. , Duan, W. , Huang, M. , & Chen, Y.‐Z. (2005). Drug bioactivation covalent binding to target proteins and toxicity relevance. Drug Metabolism Reviews, 37(1), 41–213. [DOI] [PubMed] [Google Scholar]
- Zhuang, C. , Zhang, W. , Sheng, C. , Zhang, W. , Xing, C. , & Miao, Z. (2017). Chalcone: A privileged structure in medicinal chemistry. Chemical Reviews, 117(12), 7762–7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


