Abstract
With the continuation of the coronavirus disease 2019 pandemic and the emergence of new severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) variants, the control of the spread of the virus remains urgent. Various animals, including cats, ferrets, hamsters, nonhuman primates, minks, tree shrews, fruit bats, and rabbits, are susceptible to SARS‐CoV‐2 infection naturally or experimentally. Therefore, to avoid animals from becoming mixing vessels of the virus, vaccination of animals should be considered. In the present study, we report the establishment of an efficient and stable system using Newcastle disease virus (NDV) as a vector to express SARS‐CoV‐2 spike protein/subunit for the rapid generation of vaccines against SARS‐CoV‐2 in animals. Our data showed that the S and S1 protein was sufficiently expressed in rNDV‐S and rNDV‐S1‐infected cells, respectively. The S protein was incorporated into and displayed on the surface of rNDV‐S viral particles. Intramuscular immunization with rNDV‐S was found to induce the highest level of binding and neutralizing antibodies, as well as strong S‐specific T‐cell response in mice. Intranasal immunization with rNDV‐S1 provoked a robust T‐cell response but barely any detectable antibodies. Overall, the NDV‐vectored vaccine candidates were able to induce profound humoral and cellular immunity, which will provide a good system for developing vaccines targeting both T‐cell and antibody responses.
Keywords: humoral immunity, Newcastle disease virus vector, SARS‐CoV‐2 vaccine, severe acute respiratory syndrome coronavirus‐2, T‐cell immunity
1. INTRODUCTION
The transmission of viruses between different species has caused great loss to public health and the global economy in the past several decades. According to data from the United States Agency for International Development, nearly 75% of emerging or re‐emerging infectious diseases in the last century have originated from animals, such as human immunodeficiency virus, Ebola virus, H5N1 avian influenza virus, and H1N1 swine influenza virus 1 ; All pathogenic human coronaviruses (hCoVs) have originated from animals. 2 , 3 , 4 Since the coronavirus disease 2019 (COVID‐19) pandemic, scientists have considered the impact of the virus on pets, livestock, and wild animals. The causative agent of COVID‐19 is severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), a positive‐strand RNA virus, containing an unusually large genome (29.8 kb) that encodes four structural proteins: the membrane (M), envelope (E), spike (S), and nucleocapsid phosphoprotein (N), and 16 nonstructural proteins (nsp1‐16). 5 Recent studies have shown that various animals can be infected with SARS‐CoV‐2. Animals infected under experimental conditions include cats, ferrets, hamsters, nonhuman primates, minks, tree shrews, fruit bats, and rabbits, indicating that these species are susceptible to SARS‐CoV‐2 infection. 6 Many natural infections have occurred in animals. SARS‐CoV‐2 RNA has been detected in dogs in the United States and Hong Kong. 7 , 8 Some studies have demonstrated that SARS‐CoV‐2 infection in dogs and cats was detected in households with SARS‐CoV‐2‐infected human(s), which suggests that the transmission may have occurred from humans to pets. 9 , 10 , 11 In addition, SARS‐CoV‐2 has been detected in tigers and lions in a zoo in New York. 12 , 13 SARS‐COV‐2 antibodies have been detected in the sera of cats and dogs. 14 , 15 , 16 Mink farms in the Netherlands have reported human‐to‐mink and mink‐to‐human transmission, which poses a high risk of cross‐species transmission of SARS‐CoV‐2. 2 , 6 Besides, there is growing evidence that SARS‐CoV‐2 could be transmitted by direct contact, droplet spread and aerosols, therefore, more caution should be paid to the mitigation of SARS‐CoV‐2 among animals and the countermeasures against it. 17
The discussion on wildlife infection may lead to disputes regarding evacuation. Pet infection may lead to animal‐to‐animal or animal‐to‐human transmission, increasing the possibility of cross‐species transmission of the virus and virus mutations. Although the intermediate host of SARS‐CoV‐2 is still unclear, it was believed to have evolved in bats and then spread to certain other animals, including human beings. 18 Furthermore, based on the rapid mutations of the virus, once a certain animal becomes a mixing vessel, the resulting infection may become uncontrollable and unpredictable. In view of the prolonged COVID‐19 pandemic, the range of susceptible animals may continue to expand, coupled with the fact that animals themselves can be infected with specific coronaviruses, which increases the possibility of animals serving as mixing vessels to promote viral mutation. Globally, apart from clinical trials on COVID‐19 vaccines for minks and cats in Russia, research and development on SARS‐CoV‐2 vaccines for animals is rare. There is an urgent need to develop vaccine candidates for animals, as they are also susceptible in a considerable proportion. Therefore, to prevent animals from becoming mixing vessels for the virus, it is important to develop animal‐oriented vaccines against SARS‐CoV‐2.
Newcastle disease virus (NDV) is a fast‐replicating virus prevalent in all avian species. It causes severe contagious disease in chickens and is a natural host range‐restricted virus in other species, in which NDV infection does not cause any disease symptoms. 19 NDV strains vary widely in virulence. Naturally occurring low‐virulence NDV strains, such as LaSota and B1, are widely used as live attenuated vaccines in the poultry industry and other animal vaccines; thus, they represent ideal vector candidates for the development of animal vaccines against SARS‐CoV‐2. 19 In the present study, we aimed to develop universal COVID‐19 vaccines for animals, including livestock, pets, and wild animals, by using the NDV vector to avoid potential competition for human vaccine resources and to prevent animals from becoming virus‐mixing vessels.
2. MATERIALS AND METHODS
2.1. Gene segment selection and recombinant plasmid construction
Receptor‐binding domain (RBD) is an important immunogen that induces the production of neutralizing antibodies. The S protein contains a large number of B‐cell and T‐cell epitopes; thus, 3819 nucleotides of the S gene segment, encoding a full‐length of SARS‐CoV‐2 S (1273 aa), consisting of a signal peptide (amino acids 1–13) located at the N‐terminus, the S1 subunit (14–685 residues), and the S2 subunit (686–1273 residues) was selected. The S1 subunit contains an N‐terminal domain (14–305 residues) and an RBD (319–541 residues); and the S2 subunit consists of the fusion peptide (FP) (788–806 residues), heptapeptide repeat sequence 1 (HR1) (912–984 residues), HR2 (1163–1213 residues), TM domain (1213–1237 residues), and cytoplasm domain (1237–1273 residues) comprise the S2 subunit (Figure 1B). 20 S1 contains the RBD domain, maybe a smaller gene unit that confers full immunogenicity, thus was constructed for comparison. NDV, as a vaccine vector, often uses the independent transcription unit (ITU) to introduce exogenous genes. The exogenous gene consists of a gene start (GS), Kozak sequence, exogenous gene ORF, and gene end (GE) components. Using primers containing GS and GE sequences, the sequence of S protein gene codons optimized for SARS‐CoV‐2 (GenBank: MN908947.3) from pCDNA3.1‐SARS‐CoV‐2‐S (Synbio Tech) was amplified, and inserted into the pNDV‐LaSota plasmid containing the LaSota full‐length reverse genome sequence via in‐fusion cloning method, and pNDV‐S and pNDV‐S1 were constructed. These gene fragments, S and S1, designed using different primers and expressed by ITU, were inserted between the P and M genes in the NDV genome, respectively (Figure 1A,B).
Figure 1.
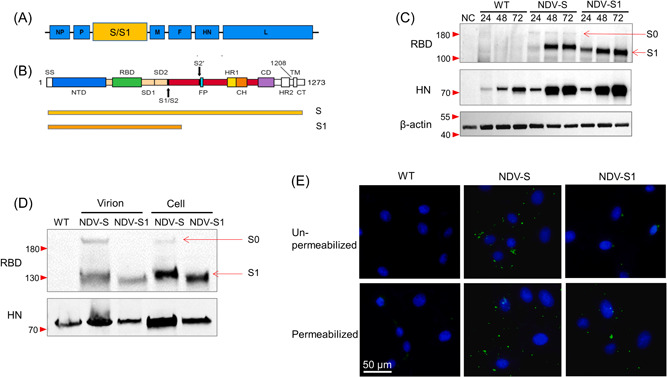
Construction and expression validation of rNDV‐S and rNDV‐S1. (A) The insertion site of the S gene into the whole genome of NDV. (B) Selection of full‐length S and S1 subunit for construction. (C) Western blot analysis of RBD expression in DF‐1 cells infected with NDV control (WT), rNDV‐S (NDV‐S), or rNDV‐S1 (NDV‐S1) with a multiplicity of infection of 0.1 for 24, 48, and 72 h. The expression of the HN protein was evaluated as an internal control. The corresponding protein ladder adjacent to the target band was (were) labeled with a red triangle(s) on the left. HN, hemagglutinin‐neuraminidase, internal control; NC, negative control; NDV, Newcastle disease virus; RBD, ribosome‐binding domain; S, spike; WT, wild type, the NDV control; β‐actin loading control. S0: the uncleaved form of S full‐length protein. (D) Immunofluorescence staining of RBD in virus‐bound Vero 81 cells. Vero 81 cells were incubated with rNDV‐S and rNDV‐S1 at 4°C for 2 h. The fixed cells were permeabilized or not (un‐permeabilized) with 0.1% Triton. Immunofluorescence staining was performed to detect viral RBD (green) and cell nuclei were counterstained (blue). (E) Western blot analysis of RBD expression in purified virions. HN protein was evaluated as an internal control.
2.2. Rescue of the recombinant viruses
The recombinant viruses were rescued via cotransfection of recombinant plasmids pNDV‐S and pNDV‐S1 expressing the exogenous gene S/S1 and helper plasmids expressing NDV NP, P, and L proteins into MVA/T7 virus‐infected Baby hamster kidney BHK‐21 (CCL‐10; ATCC) cells. 21 By infecting cells with MVA/T7 to provide T7 polymerases, three helper plasmids encoding NP, P, and L proteins under the control of T7 promoters were maintained to help cells produce infectious rNDVs. After 72 h from transfection, the cells were freeze‐thawed twice and harvested. The rescued viruses were amplified by inoculating 300 µl of the transfected cell lysate into the allantoic cavity of 10‐day‐old SPF chicken embryos and incubating the embryos for 4 days, then the allantoic fluid (AF) was harvested and the rescued virus was detected by hemagglutination (HA) assay. 22 HA‐positive AF was filtered through a 0.22 µm Nalgene Syringe Filter and amplified in chicken embryos two more times, and the purified recombinant viruses were harvested and named rNDV‐S and rNDV‐S1.
2.3. Western blot (WB) analysis
The expression of S (RBD) proteins was confirmed via WB analysis. The DF‐1 cells (CRL‐12203; ATCC), chicken embryo fibroblast cell line, were infected with the recombinant viruses rNDV‐S and rNDV‐S1 at the indicated time points. Cells were lysed and the protein concentration was measured using a BCA kit (Thermo Scientific™). Fifteen micrograms of total protein were loaded and separated using a 10% polyacrylamide gel. After trans‐blotting, the membrane was incubated with an in‐house‐developed monoclonal antibody, 4D11, for 1 h and then with a horseradish peroxidase‐conjugated goat antimouse immunoglobulin G (IgG) for 1 h. The blot was visualized using an enhanced chemiluminescence kit (Thermo Fisher Scientific).
2.4. Characterization of the recombinant viruses
To evaluate the biological properties of the recombinant virus rNDV‐S and rNDV‐S1, the pathogenicity of the recombinant viruses was examined by conducting mean death time (MDT) in 9‐day‐old SPF chicken embryos and intracerebral pathogenicity index (ICPI) in 1‐day‐old chickens and the virus titration was measured by the standard haemagglutinating activity (HA) test in a 96‐well microplate, and the 50% egg infective dose (EID50) assay in 9‐day‐old SPF chicken embryos according to standard procedures. 22
2.5. Virus attachment and immunofluorescence (IF) staining
Vero 81 cells (CCL81.4; ATCC) were inoculated with rNDV‐S and rNDV‐S1 at a multiplicity of infection of 20 and incubated at 4°C for 1 h. The cells were fixed with 4% paraformaldehyde (PF), followed by permeabilization using 0.1% Triton X‐100. The cells were incubated with mouse anti‐RBD antibody (4D11) at 37°C for 1 h, followed by incubation for 1 h with AF488‐conjugated goat anti‐mouse IgG secondary antibodies. The expression of RBD was analyzed and images were taken under an inverted fluorescence microscope (Nikon Eclipse TI‐U).
2.6. Virus purification and WB analysis
RBD expression on viral particles was further characterized using WB and purified virions. The viral suspension was propagated in Vero 81 cells. After 24 h of infection, the culture medium was collected and clarified via centrifugation at 3200g for 20 min. After filtration using a 0.22 µm filter, the virus suspension was loaded onto a discontinuous gradient consisting of 10%, 15%, 20%, and 25% iodixanol diluted from OptiPrep™ Density Gradient Medium (Sigma‐Aldrich). After ultra‐centrifugation at 100 000g for 2 h, the opalescent band was collected, the virions were lysed in radioimmunoprecipitation acid (RIPA) buffer, and WB analysis was performed using mouse anti‐RBD (4D11) monoclonal antibodies.
2.7. Detection of anti‐RBD IgG production via enzyme‐linked immunosorbent assay (ELISA)
C57/BL6 mice were immunized twice at a 2‐week interval using recombinant viruses rNDV‐S and rNDV‐S1. Fourteen days post the prime and 8 days post‐boost immunization, blood was drawn from the tail veins of the mice, and serum was collected. The production of IgG against RBD in the mouse serum was detected using a commercial RBD assay kit (Darui). All animal procedures were approved by the Ethics Commission of the First Affiliated Hospital of Guangzhou Medical University (No. 2021‐011).
2.8. Angiotensin‐converting enzyme‐2 (ACE2) RBD competition assay
Microtiter plates in 96‐well were coated overnight with 100 μl of purified RBD recombinant protein (1 μg/ml) (YP_009724390.1, SinoBiological, Cat# 40592‐V31H) in phosphate‐buffered saline (PBS) pH 7.4 at 4°C, followed by blocking with PBS containing 3% (wt/vol) bovine serum albumin) at 37°C for 2 h. Mice sera were 1:10 with dilution buffer. Biotinylated human ACE2 (hACE2‐biotin) was diluted to 0.1 μg/ml. Diluted serum was mixed with hACE2‐biotin in an assay buffer at a 1:1 volume ratio. These sample series were transferred to the RBD‐coated microtiter plate and incubated for 1 h at room temperature. Duplicated wells were performed for each sample. Finally, the wells were incubated with streptavidin conjugated with horseradish peroxidase (Invitrogen, Cat# S911) for 1 h at room temperature. Chemiluminescent development was performed with SuperSignal™ ELISA Pico substrate solution (Thermo Scientific, Cat# 37069) for 10 min at room temperature. Chemiluminescence was measured at 425 nm using a chemiluminescence reader (Biotek, Synergy Neo2). The relative light units (RLUs) of each sample were divided by the mean of the ACE2 normalization control. The normalized values were converted into percent and subtracted from 100 resulting in the percentage of inhibition. The inhibition rate >25% was determined as a positive read for neutralizing antibody (nAb) by using positive and negative controls (Data not shown).
2.9. Focus reduction neutralization test (FRNT)
Fifty microliters of serum samples were fourfold serially diluted, starting from fivefold dilution, and were then mixed with 50 μl of SARS‐CoV‐2 containing 100 focus‐forming units for 1 h at 37°C. Mixtures were transferred to 96‐well plates, previously seeded with Vero E6 cells (C1008, ECACC, China), and incubated at 37°C for 1 h. Inoculums were then removed, and 100 μl of overlay (MEM containing 1.2% carboxymethylcellulose) was added. After 24 h, the overlay was removed and the cells were fixed with 4% PF solution for 30 min, followed by permeabilization with 0.1% Triton X‐100 in PBS. The cells were then incubated with rabbit anti‐SARS‐CoV‐N IgG (Sino Biological, Cat# 40143‐R001) for 1 h and then HRP‐conjugated goat antirabbit IgG (H + L) antibody (Jackson ImmunoResearch, Cat# 111‐035‐144) for 1 h. Finally, the reactions were developed with KPL TrueBlue Peroxidase substrates (Seracare Life Sciences Inc.). The numbers of SARS‐CoV‐2 foci were calculated using an EliSpot reader (Cellular Technology Ltd.). FRNT50, expressed by IC50 of virus neutralization was calculated using the 4‐parameter logistic model.
2.10. Detection of neutralizing antibodies with pseudovirus‐based assays
Neutralizing antibodies were measured by reduction of luciferase gene expression assays, as described previously. 23 Briefly, the mice sera, starting from fivefold dilution, were threefold‐serially diluted and incubated with an equal volume of pseudovirus for 1 h. Afterward, freshly trypsinized Huh‐7 cells (0403, Japanese Collection of Research Bioresources [JCRB]) were added to each well. Following 24 h of incubation at 37°C, the luminescence was measured. The 50% inhibitory dilution (ID50) was defined as the serum dilution at which the RLUs were reduced by 50% compared with the virus control wells (virus + wells) after subtraction of the background RLUs in the control groups with cells only. The ID50 values were calculated with nonlinear regression, that is, log (inhibitor) versus response (four parameters), using GraphPad Prism 8 (GraphPad Software).
2.11. Detection of cellular response in mouse lungs and spleens via fluorescence‐activated cell sorting (FACS) analysis
Twenty‐eight days after boost immunization, the mice were euthanized, the lungs and spleens were collected, and single cells were isolated via homogenization. Cells were stimulated with an S protein peptide pool overnight. After intracellular cytokine staining (ICS) staining using a panel consisting of anti‐CD3 PE‐Cy7, anti‐CD4 APC‐Cy7, anti‐CD8 PerCP Cy5.5, anti‐IFN‐γ FITC, and anti‐TNF‐α APC (Table 1), the T cell response based on the expression of interferon‐gamma (IFN‐γ) and tumor necrosis factor α (TNF‐α) was determined using BD FACS Aria III (BD Biosciences). These data were analyzed using FlowJo software (Ttree; FlowJo).
Table 1.
List of FACS antibodies used
| Antigen | Color | Clone | Supplier |
|---|---|---|---|
| CD3 | PE‐Cy7 | 145‐2C11 | BD Bioscience |
| CD4 | APC‐Cy7 | GK1.5 | BD Bioscience |
| CD8 | PerCP‐Cy5.5 | 53‐6.7 | BioLegend |
| TNF‐α | APC | MP6‐XT22 | BioLegend |
| IFN‐γ | FITC | XMG1.2 | BioLegend |
| CD44 | PE | IM7 | BioLegend |
| CD62L | PE‐Cy7 | MEL‐14 | BioLegend |
Abbreviation: FACS, fluorescence‐activated cell sorting.
3. RESULTS
3.1. SARS‐CoV‐2 spike (S) protein was displayed on the surface of recombinant NDV‐S (rNDV‐S)
Two recombinant viruses expressing the S and S1 proteins were rescued using a highly efficient reverse genetic system. The total length of recombinant viruses rNDV‐S and rNDV‐S1 is 19 242 and 17 442 bps, respectively, and all are divisible by 6 abiding by the “Rule of Six.” 24 To evaluate biological properties of the parental and the rescued viruses rNDV‐S and rNDV‐S1 on viral pathogenicity and growth ability, the viruses were examined in vitro and in vivo by testing the virus titration (EID50 and HA test), MDT, and ICPI. As shown in Table 2, the recombinant viruses showed similar attenuated characteristics in SPF embryonated eggs and 1‐day‐old chickens with a longer MDT (>150 h) and low ICPI (0.0) compared to the parental strain rLaSota. The results of the EID50 and HA test showed that the titer of rNDV‐S was slightly lower but was comparable to that of rNDV‐S1 and rLaSota.
Table 2.
Biological assessments of the recombinant viruses
| Viruses | MDT | ICPI | HA | EID50 |
|---|---|---|---|---|
| rLaSota | 130 h | 0.15 | 210 | 3.16 × 109 |
| rNDV‐S | >150 h | 0.0 | 29 | 3.98 × 108 |
| rNDV‐S1 | >150 h | 0.0 | 210 | 5.62 × 108 |
Abbreviations: EID50, the 50% egg infectious dose assay in embryonated eggs; HA, hemagglutination titer expressed in Log2; ICPI, intracerebral pathogenicity index assay in day‐old chickens; MDT, mean death time assay in embryonated eggs.
To verify the expression of S and S1 proteins in the recombinant viruses, DF‐1 cells were infected with NDV control, rNDV‐S, and rNDV‐S1, respectively. WB was performed using the anti‐RBD monoclonal antibody 4D11 (in‐house developed). RBD was considerably expressed in both rNDV‐S‐ and rNDV‐S1‐infected cells and the NDV hemagglutinin‐neuraminidase protein was detected in all infected cells as an infection control (Figure 1C). To confirm whether the exogenous S protein was integrated into the recombinant viral particles, WB was performed to analyze S/S1 protein expression in the purified virus. The WB results clearly showed that RBD protein was detected on both rNDV‐S and rNDV‐S1 viruses. S0, the uncleaved form of S protein, was also detected in rNDV‐S, indicating that both S0 and S1 forms of protein were present in the expression system, similar to the protein expression format in the infected cells (Figure 1C). In the expression system, the full length of the S gene is inserted between the P and M genes, the transmembrane domain theoretically leads the S protein to be assembled and displayed on the surface of the virions. To confirm if the S protein was displayed on the surface of virions, the recombinant virus was absorbed onto Vero 81 cells at 4°C for 1 h, followed by fixation and IF staining. Based on the results, RBD, representing S protein, was detected on the virion‐attached cells, whether permeabilized or not, suggesting that S protein was likely displayed on the surface of rNDV‐S (Figure 1D). RBD, representing the S1 protein, was only detected in permeabilized rNDV‐S1‐attached cells, suggesting that the S1 protein was integrated into but not displayed on the virions.
3.2. rNDV‐S induced profound antibody production and strong T‐cell immunity
To assess the immunogenicity of the recombinant viruses, mice were vaccinated and the anti‐RBD antibodies and virus‐specific T‐cell responses were evaluated. Specifically, C57/BL6 mice were inoculated with live rNDV‐S and rNDV‐S1 intramuscularly (IM) or intranasally (IN), because NDV rarely replicates in the muscles, and LaSota strain infection in the lungs of mice is restricted and causes no symptoms. The production of IgG against RBD in mice was measured using an RBD ELISA, and the levels of neutralizing antibodies were measured using an in‐house developed ACE2 competition assay procedure. Fourteen days post prime immunization, only mice injected with rNDV‐S via IM were found to produce relatively higher levels of RBD‐specific IgG (Figure 2A). Fourteen days post‐boost immunization, mice that received low (median tissue culture infectious dose [TCID50], 106) and high (TCID50, 107) doses of rNDV‐S via IN showed clear antibody responses, though the antibody levels were still lower than those in mice injected via IM. High‐dose‐immunized mice also had relatively higher antibody levels. Notably, mice immunized via IN with rNDV‐S1 did not produce anti‐RBD antibodies (Figure 2B). Furthermore, ACE2 competitive ELISA analysis showed that only mice in the IM rNDV‐S group produced considerable levels of neutralizing antibodies (Figure 2C). This was consistent with neutralizing antibodies determined by authentic virus neutralization assays (Figure 2D). To assess if the vaccine candidates show cross‐protection against different SARS‐CoV‐2 variants of concern (VOCs), we performed pseudovirus‐based neutralizing assays using VSV expressing S of SARS‐CoV‐2 prototype Wuhan‐hu‐1, delta (B.1.617.2) variant, and Omicron (BA.4/5, the S of BA.4 and BA.5 have the identical aa sequence) variant. Results showed that immunization via IM efficiently induced a high amount of neutralizing antibodies against the WT virus and B.1.617.2 variant, however, these were escaped by a more recent variant BA.4/5 (Figure 2E). Taken together, rNDV‐S administered via IM induced the strongest humoral binding and neutralizing antibodies.
Figure 2.
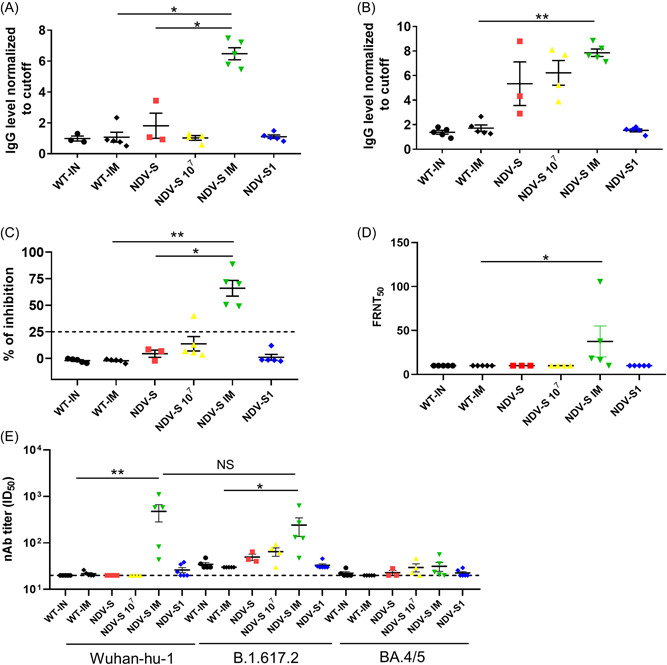
Comparison of antibody levels and T‐cell response between different routes and doses of immunization in NDV‐S, and NDV‐S1‐vaccinated mice: 6‐week‐old female C57/BL6 mice were vaccinated via intranasal inoculation of 106 TCID50 of NDV control (WT‐IN) or recombinant rNDV‐S (NDV‐S) and rNDV‐S1 (NDV‐S1), or 107 TCID50 of rNDV‐S (NDV‐S 107), or via intramuscular inoculation of 106 TCID50 of NDV control (WT‐IM) and rNDV‐S (NDV‐S IM) twice with a 14‐day interval. (A, B) Anti‐RBD antibody levels were determined using ELISA at (A) 14 days postprime and (B) 14 days postboost immunization. (C) Neutralizing antibodies in the sera of mice at 14 days postboost immunization were detected using an ACE2 competitive inhibition assay. (D) Detection of neutralizing antibodies with authentic virus‐based FRNT50 assays at 14 days postboosting. FRNT, focus reduction neutralization test. (E) Neutralizing antibodies detected with pseudovirus‐based neutralizing assays at 14 days postboosting. B.1.617.2, Delta variant; BA4/5 (S of BA.4 and BA.5 have the identical aa sequence), Omicron variant; ID50, 50% inhibitory dilution; Wuhan‐hu‐1, pseudovirus expressing SARS‐CoV‐2 S prototype. The data are presented as the mean ± SE. The asterisks (* and **) show the significance difference: *p < 0.05, and **p < 0.01 by Student's t test. ELISA, enzyme‐linked immunosorbent assay.
Fourteen days postboost immunization, ICS was performed using antibodies against CD3, CD4, CD8, TNF‐α, and IFN‐γ (Table 1). Robust T‐cell response was observed in the lungs of mice that were IN inoculated with low‐dose rNDV‐S and rNDV‐S1, with proportions of IFN‐γ+ TNF‐α+ CD8+ T cell ranging from 0.96%–7.99% to 0.1%–16.19%, respectively. Considerable T‐cell responses were observed in the splenocytes of mice immunized either via IM or IN with either high‐ or low‐dose recombinant viruses, with a slight reverse correlation to the antibody levels (Figure 3A–C).
Figure 3.
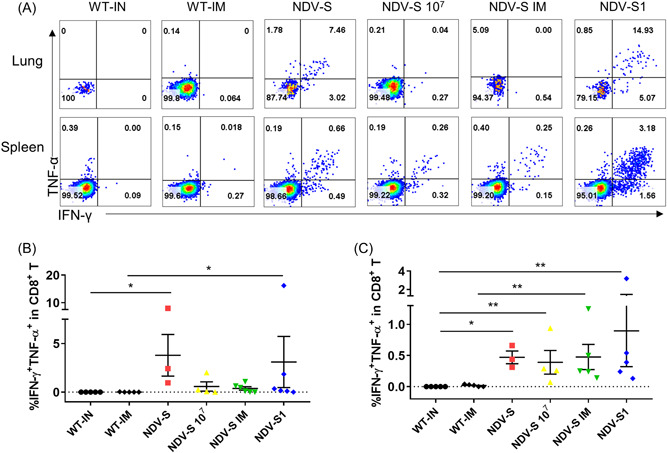
T‐cell response detection in the lungs and splenocytes of mice immunized with rNDV‐S and rNDV‐S1 with different administration routes. Grouping has been detailed previously in Figure 2. (A) Representative FACS plots of S‐specific CD8+ T cell in the lungs and spleens of vaccinated mice. Single cells were separated from the lungs and spleens of mice 14 days postboost immunization. After stimulation using an S peptide pool overnight, the cells were stained and analyzed using FACS. (B, C) Dot plot comparison of frequencies of S‐specific CD8+ T cell present in the (B) lungs and (C) spleens of vaccinated mice. FACS, fluorescence‐activated cell sorting; TCID50, median tissue culture infectious dose. The data are presented as the mean ± SE. The asterisks show the significance difference: *p < 0.05, and **p < 0.01 by Student's t test.
3.3. Inactivated rNDV‐S stimulated antibody production comparable to that of the live virus but did not induce a T‐cell response
To access if inactivated virus exerted similar immunogenicity, NDV control (WT) and rNDV‐S were inactivated with 0.05% of PF solution for a week. The inactivation was confirmed by inoculating Vero 81 cells with the highest amount of virus that did not cause any infection (Figure 4A). Subsequently, 6‐week‐old C57/BL6 mice were administered via IM with 106 TCID50 of live rNDV‐S or an equivalent amount of inactivated rNDV‐S. Fourteen days after boost immunization, inactivated rNDV‐S induced antibody production that was comparable to that induced by the live compartment. Thirty days postboost immunization, the mice were euthanized and single splenocytes and lung cells were isolated. After overnight stimulation with the SARS‐CoV‐2 S peptide pool, the cells were FACS‐stained with antibodies to CD3, CD8, TNF‐α, IFN‐γ, CD44, and CD62L (Table 1) and analyzed using FACS BD AriaIII. Both live and inactivated viruses induced production of a sufficient level of antibodies, with a slight decrease in antibody levels against the inactivated virus (Figure 4B,C). Moderate levels of T‐cell response were still detected in mice 30 days postboost immunization in splenocytes, but not in the lung cells (Figure 4D,E). Based on the expression of CD62L and CD44, most virus‐specific T cells in the spleen were found to be still present in the effector memory phase (Figure 4F). These cells have a high probability of differentiating into central memory T cells and may persist for a long period. The T‐cell response induced by the recombinant virus vaccine needs to be characterized in depth.
Figure 4.
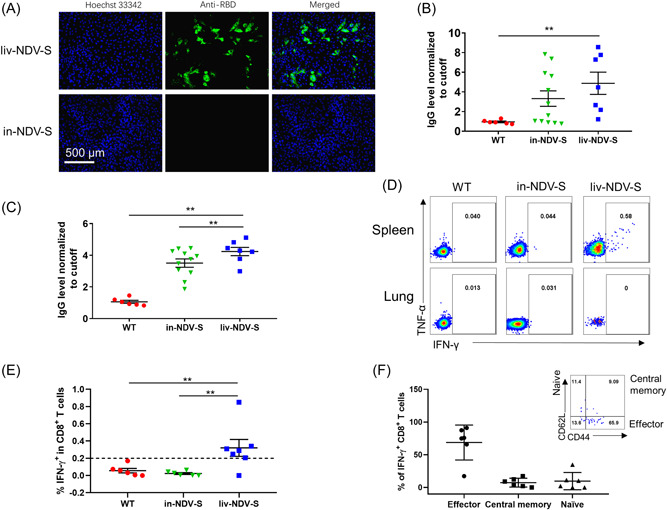
Comparison of antibody levels and T‐cell response post intramuscular immunization between live and inactivated viruses: recombinant NDV‐S virus was inactivated via treatment with 0.05% PF solution for 1 week. Six‐week‐old female C57/BL6 mice were inoculated with 106 TCID50 of NDV control (WT), inactivated rNDV‐S (in‐NDV‐S) or live rNDV‐S (liv‐NDV‐S) virus twice with 14‐day intervals. Sera were collected 14 days postprime and postboost immunization, respectively. (A) Immunofluorescence staining of RBD in Vero 81 cells infected with live or inactivated rNDV‐S. (B, C) Anti‐RBD antibodies present in sera of mice (B) 14 days post‐prime and (C) 14 days postboost immunization. (D) Representative FACS plots showing the expression of TNF‐α and IFN‐γ response to S peptide pool stimulation. (E) Proportions of IFN‐γ+CD8+ T cell in mouse spleens immunized with WT, inactivated rNDV‐S, or live rNDV‐S. (F) The phenotypes of S‐specific CD8+ T cell in the spleen of live NDV‐S‐immunized mice. FACS, fluorescence‐activated cell sorting; IFN‐γ, interferon‐gamma; NDV, Newcastle disease virus; S, spike; TCID50, median tissue culture infectious dose; TNF‐α, tumor necrosis factor‐α; WT, wild type. The data are presented as the mean ± SE. The asterisks (*and **) show the significance difference: *p < 0.05, and **p < 0.01 by Student's t test.
4. DISCUSSION
The vaccination of susceptible animal species, such as cats, minks, and great apes, is essential for public health, and the successful elimination of SARS‐CoV‐2 will only be possible by controlling transmission in all susceptible animal species. Eliminating the spread of the virus between animals will not only help prevent the re‐emergence of viruses in humans but also prevent the emergence of novel variants such as the SARS‐CoV‐2 mink variant at the human‐animal interface, facilitating the successful prevention and control of the pandemic.
NDV‐based SARS‐CoV‐2 vaccine has been reported since 2021 and the immunogenicity and effectiveness have been tested in mice, rats, and pigs, and some candidates have been already in phase 1/2 trial. 25 , 26 , 27 , 28 , 29 These reports have shown the induction of strong humoral immunity by NDV‐based vaccine candidates, and the report by Sun et al. 27 has shown the protection of mice by challenge experiments. However, only two of the studies have reported cellular response results, in which none or weak T‐cell immunity was induced by the candidate vaccine. 25 , 27 In the present study, we showed a strong S‐specific T‐cell response which will add valuable information to the development of SARS‐CoV‐2 vaccines that provoke both humoral and cellular immunity. Here we report the establishment of an efficient and stable system using NDV as a vector to express the SARS‐CoV‐2 S protein for the rapid generation of vaccines against SARS‐CoV‐2 in animals. The S and S1 proteins were sufficiently expressed in rNDV‐S‐ and rNDV‐S1‐infected cells. The S protein was integrated and displayed on the surface of viral particles (Figure 1C,D). IM immunization with live rNDV‐S was found to induce both humoral and cellular immunity in mice. IN immunization with live virus induced a higher S‐specific T‐cell response in the lungs than that induced by IM injection (Figure 3A‐C). Overall, using the NDV vector vaccination system, both robust humoral response and cellular immunity were induced in mice. The virus‐specific T‐cell response to an overnight stimulation using the S peptide pool, which is represented by the dual expression of IFN‐γ and TNF‐α in CD4+ or CD8+ T cells, accounted for up to 15% of the lung T cell and up to 3.2% of the splenocytes of the rNDV‐S/S1‐immunized mice and were considerably higher than those reported in similar studies using mRNA vaccine, adenovirus‐vectored vaccine, and replicon RNA vaccines as candidates. 30 , 31 , 32
Although the protection of T‐cell immunity in SARS‐CoV‐2 infection remains less studied, several studies have demonstrated the important roles of T‐cell memory in the persistence of protective antibodies. With the coordination of humoral and cellular immunity, antibodies, especially neutralizing antibodies, can be maintained for a long period according to several studies. 33 , 34 Memory B cells require T follicular helper cells to transmit signals to facilitate preferential selection after encountering the same antigens, which subsequently leads to antibody maturation and persistence. 35 In our previous study, we also demonstrated that SARS‐CoV‐2‐specific CD4+ T cells, including virus‐specific T helper 1, circulating T follicular helper 1, and circulating T follicular helper 17 cells, is associated with a relatively slower decay of neutralizing antibodies. 36 The coordination of memory B and T cells in the maintenance of antibodies, especially protective antibodies, should be investigated in depth in future studies. The rNDV‐S will provide a good model to study the role of B and T cell interactions in antibody persistence.
The initial goal of this study was to select from two recombinant NDV‐vectored viruses expressing full‐length S and S1 subunits, and two routes (IM and IN) of immunization, to develop a vaccine candidate with optimal immunogenicity. Although the expression of S1 protein was similar between the rNDV‐S‐ and rNDV‐S1‐infected cells and the recombinant virions (Figure 1C,D), IN immunization with rNDV‐S1 failed to induce barely any antibodies against RBD, while provoked almost highest amount of T cell response, suggesting that the display of antigens likely affects the recognition and presentation of antigens by antigen‐presenting cells, and exposure manner of antigens may be important for antibody production. This may be explained by antigen form and exposure manner that affect B‐cell recognition and activation. B cells recognize and respond to soluble and membrane‐associated antigens, of which the latter is more important for B‐cell activation in vivo. 37 The S1 protein has been shown to be incorporated into but not displayed on the virions (Figure 1D,E), making it less likely an ideal form of antigen to be recognized by B cells. Another possible reason may be that the minimal amount of antigens, either because the antigens were not exposed to rNDV‐S1 virions or the replication was restricted in the lungs of mice, was not able to induce humoral immunity. However, strong CD8+ T‐cell response was detected after rNDV‐S1 boost immunization, suggesting that a negligible amount of antigen may be sufficient to induce a T‐cell response. This is consistent with our recent finding that T‐cell response was detected in the COVID‐19 close contacts even though they were never positive for nucleic acid amplification tests (NAAT) or antibodies or any symptoms of infection. 38 At present, although many efforts have been made in vaccine development, especially for hexa‐pro S and adenovirus vector vaccines, to induce T‐cell responses in humans, the T‐cell response in clinical trials reported in a few recent studies is still not optimal. 39 , 40 , 41 Regarding the generation of strong T‐cell responses, the NDV vector system is expected to provide valuable resources to T‐cell‐orientated vaccine candidates.
In this study, IN vaccination with NDV‐S induced strong T‐cell immunity and IM immunization with live NDV‐S induced the production of a sufficient titer of antibodies, providing valuable information and a potential mix‐and‐match to the strategy of mixed boost vaccination in present human vaccinations. For instance, matching live adenovirus‐vectored vaccine with spray vaccination may help produce virus‐specific T‐cell immunity in the lungs of humans. In addition to helping to decide a strategy, live NDV‐S may have the potential for application in humans based on its safety as NDV viruses replicate in the cytoplasm alone and do not integrate into the host cell genome. Furthermore, NDV vector vaccines may reduce the cost of animal vaccine production. NDV vectors easily replicate in chicken embryos, can produce recombinant viruses with high titers, and the production cost is considerably reduced, which can meet the demand for vaccines for pets, such as cats and dogs, as well as economic animals.
AUTHOR CONTRIBUTIONS
Zhongfang Wang and Xiaoyun Yang designed the experiments, analyzed data, and wrote the paper. Lei He, Jiaying Zhong, Guichang Li, Zhengfang Lin, Hairong Wang, and Yuhao Zhang performed the experiments and analyzed the data. Peijing Zhao, and Chuhua Yang handled the animals.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
This study was supported by the State Key Basic Research Project (2019YFC0810900), Ministry of Science and Technology of the people Republic ofChina (2021YFC0864400), NSFC (81971485, 82271801), Guangdong Key Basic Research Project (2019B1515120068,2020B1111330001,2022B1111070002), Guangdong Province General Colleges and Universities Youth Innovative Talents Project (2019KQNCX120), and Emergency Key Program of Guangzhou Laboratory (NO. EKPG21‐30‐1).
He L, Zhong J, Li G, et al. Development of SARS‐CoV‐2 animal vaccines using a stable and efficient NDV expression system. J Med Virol. 2022;95:e28237. 10.1002/jmv.28237
Xiaoyun Yang and Zhongfang Wang contributed equally to this study.
DATA AVAILABILITY STATEMENT
The data sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Development)., U.U.S.A.f.I . Emerging Pandemic Threats Program. 20220: Available from https://www.usaid.gov/news-information/fact-sheets/emerging-pandemic-threats-program.
- 2. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Forni D, Cagliani R, Clerici M, Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25(1):35‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019‐nCoV) originating in China. Cell Host Microbe. 2020;27(3):325‐328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, et al. Transmission of SARS‐CoV‐2 on mink farms between humans and mink and back to humans. Science. 2021;371(6525):172‐177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sit THC, Brackman CJ, Ip SM, et al. Infection of dogs with SARS‐CoV‐2. Nature. 2020;586(7831):776‐778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Morais HA, Dos Santos AP, do Nascimento NC, et al. Natural infection by SARS‐CoV‐2 in companion animals: a review of case reports and current evidence of their role in the epidemiology of COVID‐19. Front Vet Sci. 2020;7:591216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neira V, Brito B, Agüero B, et al. A household case evidences shorter shedding of SARS‐CoV‐2 in naturally infected cats compared to their human owners. Emerg Microbes Infect. 2021;10(1):376‐383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stout AE, André NM, Jaimes JA, Millet JK, Whittaker GR. Coronaviruses in cats and other companion animals: where does SARS‐CoV‐2/COVID‐19 fit. Vet Microbiol. 2020;247:108777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Segalés J, Puig M, Rodon J, et al. Detection of SARS‐CoV‐2 in a cat owned by a COVID‐19‐affected patient in Spain. Proc Natl Acad Sci U S A. 2020;117(40):24790‐24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartlett SL, Diel DG, Wang L, et al. Sars‐Cov‐2 infection and longitudinal fecal screening in Malayan Tigers (Panthera Tigris Jacksoni), Amur Tigers (Panthera Tigris Altaica), and African Lions (Panthera Leo Krugeri) at the Bronx Zoo, New York, USA. J Zoo Wildl Med. 2021;51(4):733‐744. [DOI] [PubMed] [Google Scholar]
- 13. Murphy HL, Ly H. Understanding the prevalence of SARS‐CoV‐2 (COVID‐19) exposure in companion, captive, wild, and farmed animals. Virulence. 2021;12(1):2777‐2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith SL, Anderson ER, Cansado‐Utrilla C, et al. SARS‐CoV‐2 neutralising antibodies in dogs and cats in the United Kingdom. Curr Res Virol Sci. 2021;2 100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Udom K, Jairak W, Chamsai E, et al. Serological survey of antibodies against SARS‐CoV‐2 in dogs and cats, Thailand. Transbound Emerg Dis. 2022;69(4):2140‐2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barua S, Hoque M, Adekanmbi F, et al. Antibodies to SARS‐CoV‐2 in dogs and cats, USA. Emerg Microbes Infect. 2021;10(1):1669‐1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang S, Mao Y, Jones RM, et al. Aerosol transmission of SARS‐CoV‐2? Evidence, prevention and control. Environ Int. 2020;144:106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Choudhary OP, Priyanka ▫, Ali RK, Maulud SQ, Dhawan M, Mohammed TA. Will the next spillover pandemic be deadlier than the COVID‐19?: a wake‐up call. Int J Surg. 2022;97:106208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ganar K, Das M, Sinha S, Kumar S. Newcastle disease virus: current status and our understanding. Virus Res. 2014;184:71‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS‐CoV‐2 spike protein: potential antivirus drug development for COVID‐19. Acta Pharmacol Sin. 2020;41(9):1141‐1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Estevez C, King D, Seal B, Yu Q. Evaluation of Newcastle disease virus chimeras expressing the Hemagglutinin‐Neuraminidase protein of velogenic strains in the context of a mesogenic recombinant virus backbone. Virus Res. 2007;129(2):182‐190. [DOI] [PubMed] [Google Scholar]
- 22. Alexander DJ. Newcastle disease and other avian paramyxoviruses. Rev Sci Tech. 2000;19(2):443‐462. [DOI] [PubMed] [Google Scholar]
- 23. Li Q, Wu J, Nie J, et al. The impact of mutations in SARS‐CoV‐2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284‐1294.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kolakofsky D, Roux L, Garcin D, Ruigrok R. Paramyxovirus mRNA editing, the “rule of six” and error catastrophe: a hypothesis. J Gen Virol. 2005;86(Pt 7):1869‐1877. [DOI] [PubMed] [Google Scholar]
- 25. Pitisuttithum P, Luvira V, Lawpoolsri S, et al. Safety and immunogenicity of an inactivated recombinant Newcastle disease virus vaccine expressing SARS‐CoV‐2 spike: interim results of a randomised, placebo‐controlled, phase 1 trial. EClinicalMedicine. 2022;45:101323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lara‐Puente JH, Carreño JM, Sun W, et al. Safety and immunogenicity of a Newcastle disease virus vector‐based SARS‐CoV‐2 vaccine candidate, AVX/COVID‐12‐HEXAPRO (Patria), in pigs. mBio. 2021;12(5):e0190821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun W, Liu Y, Amanat F, et al. A Newcastle disease virus expressing a stabilized spike protein of SARS‐CoV‐2 induces protective immune responses. Nat Commun. 2021;12(1):6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tcheou J, Raskin A, Singh G, et al. Safety and immunogenicity analysis of a Newcastle disease virus (NDV‐HXP‐S) expressing the spike protein of SARS‐CoV‐2 in Sprague–Dawley rats. Front Immunol. 2021;12:791764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Duc Dang A, Dinh Vu T, Hai Vu H, et al. Safety and immunogenicity of an egg‐based inactivated Newcastle disease virus vaccine expressing SARS‐CoV‐2 spike: interim results of a randomized, placebo‐controlled, phase 1/2 trial in Vietnam. Vaccine. 2022;40(26):3621‐3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu S, Zhong G, Zhang J, et al. A single dose of an adenovirus‐vectored vaccine provides protection against SARS‐CoV‐2 challenge. Nat Commun. 2020;11(1):4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Erasmus JH, Khandhar AP, O'Connor MA, et al. An alphavirus‐derived replicon RNA vaccine induces SARS‐CoV‐2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci Transl Med. 2020;12(555):eabc9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Corbett KS, Edwards DK, Leist SR, et al. SARS‐CoV‐2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567‐571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seydoux E, Homad LJ, MacCamy AJ, et al. Analysis of a SARS‐CoV‐2‐Infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity. 2020;53(1):98‐105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boppana S, Qin K, Files JK, et al. SARS‐CoV‐2‐specific circulating T follicular helper cells correlate with neutralizing antibodies and increase during early convalescence. PLoS Pathog. 2021;17(7):e1009761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Law H, Venturi V, Kelleher A, Munier C. Tfh cells in health and immunity: potential targets for systems biology approaches to vaccination. Int J Mol Sci. 2020;21:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Z, Yang X, Mei X, et al. SARS‐CoV‐2‐specific CD4(+) T cells are associated with long‐term persistence of neutralizing antibodies. Signal Transduct Target Ther. 2022;7(1):132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Batista FD, Harwood NE. The who, how and where of antigen presentation to B cells. Nat Rev Immunol. 2009;9(1):15‐27. [DOI] [PubMed] [Google Scholar]
- 38. Wang Z, Yang X, Zhong J, et al. Exposure to SARS‐CoV‐2 generates T‐cell memory in the absence of a detectable viral infection. Nat Commun. 2021;12(1):1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lanini S, Capone S, Antinori A, et al. GRAd‐COV2, a gorilla adenovirus‐based candidate vaccine against COVID‐19, is safe and immunogenic in younger and older adults. Sci Transl Med. 2022;14(627):eabj1996. [DOI] [PubMed] [Google Scholar]
- 40. Pozzetto B, Legros V, Djebali S, et al. Immunogenicity and efficacy of heterologous ChAdOx1‐BNT162b2 vaccination. Nature. 2021;600(7890):701‐706. [DOI] [PubMed] [Google Scholar]
- 41. Woldemeskel BA, Garliss CC, Blankson JN. SARS‐CoV‐2 mRNA vaccines induce broad CD4+ T cell responses that recognize SARS‐CoV‐2 variants and HCoV‐NL63. J Clin Invest. 2021;131(10):e149335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.


