Abstract
The present study aimed to determine whether current commercial immunoassays are adequate for detecting anti‐Omicron antibodies. We analyzed the anti‐SARS‐CoV‐2 antibody response of 23 unvaccinated individuals 1–2 months after an Omicron infection. All blood samples were tested with a live virus neutralization assay using a clinical Omicron BA.1 strain and four commercial SARS‐CoV‐2 immunoassays. We assessed three anti‐Spike immunoassays (SARS‐CoV‐2 IgG II Quant [Abbott S], Wantaï anti‐SARS‐CoV‐2 antibody ELISA [Wantaï], Elecsys Anti‐SARS‐CoV‐2 S assay [Roche]) and one anti‐Nucleocapsid immunoassay (Abbott SARS‐CoV‐2 IgG assay [Abbott N]). Omicron neutralizing antibodies were detected in all samples with the live virus neutralization assay. The detection rate of the Abbott S, Wantai, Roche, and Abbott N immunoassays were 65.2%, 69.6%, 86.9%, and 91.3%, respectively. The sensitivities of Abbott S and Wantai immunoassays were significantly lower than that of the live virus neutralization assay (p = 0.004, p = 0.009; Fisher's exact test). Antibody concentrations obtained with anti‐S immunoassays were correlated with Omicron neutralizing antibody concentrations. These data provide clinical evidence of the loss of performance of some commercial immunoassays to detect antibodies elicited by Omicron infections. It highlights the need to optimize these assays by adapting antigens to the circulating SARS‐CoV‐2 strains.
Keywords: assay sensitivity, binding antibodies, COVID‐19, immunoassay, neutralizing antibodies, Omicron, SARS‐CoV‐2
1. INTRODUCTION
SARS‐CoV‐2 antibody immunoassays are widely used to evaluate the spread of the virus 1 and to measure the immunity provided by vaccination or infection. 2 , 3 Several commercial assays are now available. Some detect antibodies that interact with the nucleocapsid (N); these are present only in infected individuals. Others target antibodies directed against the spike (S) protein; these are found in both vaccinated and infected individuals. These assays have been developed at the beginning of the pandemic using antigens based on ancestral SARS‐CoV‐2 strains. SARS‐CoV‐2 pandemic has been since dominated by the emergence of new variants, the latest being the Omicron variant characterized by numerous mutations in the gene coding the S protein. Therefore, Omicron‐specific antibodies could bind less efficiently to the ancestral SARS‐CoV‐2 antigens. While many commercial assays have been evaluated, 4 , 5 data regarding their capacity to detect antibodies induced by Omicron SARS‐CoV‐2 infections are lacking.
2. METHODS
2.1. Participants and sample collection
We analyzed the SARS‐CoV‐2 antibody response of 23 unvaccinated individuals who experienced mild or asymptomatic Omicron infection. Serological samples were taken between February 22 and March 18, 2022, 1–2 months after Omicron infection. Individual's symptoms at the time of infection, comorbidities, and history of previous SARS‐CoV‐2 infection were collected before blood sampling.
We used a control panel including 31 samples from nonhospitalized immunocompetent healthcare workers infected with an ancestral D614G strain before vaccination. 6 These control samples were collected 1–3 months postinfection. There was no significant difference in age and sex between individuals from the control panel and the study group.
2.2. SARS‐CoV‐2 omicron detection
After RNA extraction from nasopharyngeal swab samples (MGI Easy Nucleic Acid Extraction kit), amplification was performed with the Thermofisher TaqPath RT‐PCR assay (ThermoFisher) running on a QuantStudio 5 Real‐Time PCR System. 7 Infection with the omicron variant was established by performing SARS‐CoV‐2 sequencing on each sample. As previously described, SARS‐CoV‐2 RNA was sequenced by long‐read sequencing (Pacific Biotechnology, USA). 8
2.3. Immunoassays and neutralization assay
We compared the sensitivities of four commercial immunoassays to that of a live virus‐neutralization assay based on Vero cells (ATCC CCL'−81TH) and a clinical SARS‐CoV‐2 Omicron BA.1 strain (EPI‐ISL_10316329). 9 , 10 One immunoassay, the Abbott SARS‐CoV‐2 IgG assay (Abbott N) was designed to detect anti‐N antibodies, the three others detected either anti‐S IgG antibodies (SARS‐CoV‐2 IgG II Quant [Abbott S]) or anti‐S total antibodies (Wantaï anti‐SARS‐CoV‐2 antibody ELISA [Wantaï], and Elecsys Anti‐SARS‐CoV‐2 S assay [Roche]). The limit of detection of the three anti‐Spike immunoassays according to manufacturer insert are 7.1, 1, and 0.8 BAU/ml for Abbott S, Wantai, and Roche respectively.
2.4. Statistical analysis
Statistical analyses were performed using GraphPad Prism 7 (GraphPad Software, Inc.). Assay sensitivities were compared using Fisher's exact test. Kruskal–Wallis test was used to compare antibody concentrations. The relationship between immunoassay results and neutralizing antibody titers was assessed using Spearman's rank correlation. A p‐value < 0.05 is considered statistically significant.
3. RESULTS
The time between Omicron infection and blood collection from the 23 subjects (16 males, 69.6%, median age: 35 years, range: 22–61) was 42 days (range: 32–66) (Table 1). All the individuals were infected with Omicron BA.1. They were all immunocompetent, with no risk factors for severe disease. Most (19, 82.6%) experienced minor symptoms but none required hospitalization for their SARS‐CoV‐2 infection. Among the 23 individuals, 5 (21.7%) had had a previous SARS‐CoV‐2 infection (median time between the two infections: 282 days; range: 198–499).
Table 1.
Patient characteristics and detection of anti‐SARS‐CoV‐2 antibodies
| Patient | Age | Sex | Symptoms at the time of Omicron BA.1 infection | Days post‐Omicron BA.1 infection | Previous infection | Time between Omicron BA.1 and non‐Omicron infection (days) | Neutralizing antibody titer | Abbott S anti‐S (BAU/ml) | Wantaï anti‐S (BAU/ml) | Roche anti‐S (BAU/ml) | Abbott N anti‐N (S/CO) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 22 | M | Mild | 50 | ‐ | 128 | 2737.5 | 15000 | 7000 | 10.2 | |
| 2 | 48 | M | Mild | 37 | Sep‐2020 | 499 | 8 | 196.7 | 3774 | 1647 | 7.3 |
| 3 | 39 | M | Mild | 38 | ‐ | 8 | <7.1 | 1.1 | 2.8 | 0.8 | |
| 4 | 33 | M | Asymptomatic | 48 | ‐ | 32 | 47.1 | 87.9 | 105 | 6 | |
| 5 | 50 | F | Mild | 45 | ‐ | 4 | <7.1 | 0.6 | 7.7 | 6 | |
| 6 | 40 | F | Asymptomatic | 40 | ‐ | 4 | 11.2 | 0.2 | 1.4 | 2.6 | |
| 7 | 29 | F | Mild | 66 | ‐ | 8 | 55.4 | 6.0 | 44.6 | 4.5 | |
| 8 | 35 | M | Mild | 42 | Apr‐2021 | 282 | 32 | 1190.8 | 14184 | 7649 | 10.2 |
| 9 | 24 | M | Mild | 46 | ‐ | 8 | 21.8 | 2.8 | 27.4 | 3.8 | |
| 10 | 23 | M | Mild | 43 | Jul‐2021 | 198 | 8 | 242.2 | 4958 | 1618 | 3.6 |
| 11 | 28 | M | Mild | 45 | ‐ | 4 | <7.1 | 0.03 | <0.4 | 1.3 | |
| 12 | 50 | M | Mild | 42 | ‐ | 4 | <7.1 | 0.2 | 1.6 | 1.6 | |
| 13 | 28 | M | Mild | 41 | ‐ | 32 | 487.9 | 11947 | 5001 | 8.4 | |
| 14 | 22 | F | Mild | 37 | Nov‐2020 | 454 | 32 | 1315.4 | 13505 | 6045 | 8.4 |
| 15 | 61 | M | Mild | 43 | ‐ | 4 | <7.1 | 0.05 | <0.4 | 2.9 | |
| 16 | 35 | F | Mild | 42 | ‐ | 8 | <7.1 | 0.5 | 1.7 | 3.9 | |
| 17 | 48 | M | Asymptomatic | 45 | ‐ | 16 | 21.5 | 2.4 | 9.4 | 7.3 | |
| 18 | 48 | M | Mild | 41 | ‐ | 4 | 8.9 | 1.1 | 3.3 | 2.2 | |
| 19 | 45 | M | Mild | 43 | ‐ | 4 | <7.1 | 1.7 | 5.2 | 2.5 | |
| 20 | 40 | F | Mild | 40 | May‐2021 | 250 | 32 | 455.3 | 5063 | 1893 | 3.9 |
| 21 | 35 | F | Mild | 48 | ‐ | 8 | 12.5 | 3.3 | 11.1 | 3.2 | |
| 22 | 41 | M | Asymptomatic | 39 | ‐ | 4 | <7.1 | 0.02 | <0.4 | 2.0 | |
| 23 | 30 | M | Mild | 32 | ‐ | 16 | 54 | 65.1 | 21.8 | 4.9 | |
| Sensitivity (%) | 100 | 65.2 | 69.6 | 86.9 | 91.3 | ||||||
Note: M, male; F, female; anti‐S, anti‐spike; anti‐N, anti‐nucleocapsid; S/CO, sample to cut‐off, according to the manufacturer's instructions positive result if S/CO ≥1.4.
While Omicron neutralizing antibodies were detected in all the subjects, the detection rates varied for commercial immunoassays: Abbott S: 65.2%, Wantai: 69.6%, Roche: 86.9%, and Abbott N: 91.3% (Table 1). The Abbott S and Wantai assays were significantly less sensitive than the live virus‐neutralization assay (p = 0.004, p = 0.009; Fisher's exact test). The sensitivities of the Roche, Abbott N and neutralization assays did not differ significantly. For the 31 samples of control panel, the detection rate was 96.8%, 100%, 100%, and 87.1% for Abbott S, Wantaï, Roche, and Abbott N, respectively.
The median neutralizing antibody (Nab) titer was 8 [IQR:4–32] (Figure 1A), while the median antibody concentrations, as BAU/ml, were: Abbott S: 55.4 [IQR:21.5–487.9], Wantai: 76.5 [IQR:2.5–10226], and Roche: 24.6 [IQR:3.8–1832], (p = 0.79; Kruskal–Wallis test) (Figure 1A). Spearman's rank correlation test indicated that the three anti‐S immunoassays results were positively and significantly correlated with the Nab titers (Figure 1B–D). Anti‐N results (Abbott N) and Nab titers were also correlated (r = 0.65, p = 0.009). Moreover, the antibody concentrations obtained with the three anti‐S immunoassays correlated very well with each other with r‐values of 0.96 (Abbott S/Wantai), 0.96 (Abbott S/Roche), and 0.98 (Wantai/Roche). Anti‐N results (Abbott N) were also correlated with anti‐S concentrations obtained with Abbott S (r = 0.73, p = 0.002), Wantaï (r = 0.80, p < 0.001) and Roche (r = 0.83, p < 0.001).
Figure 1.
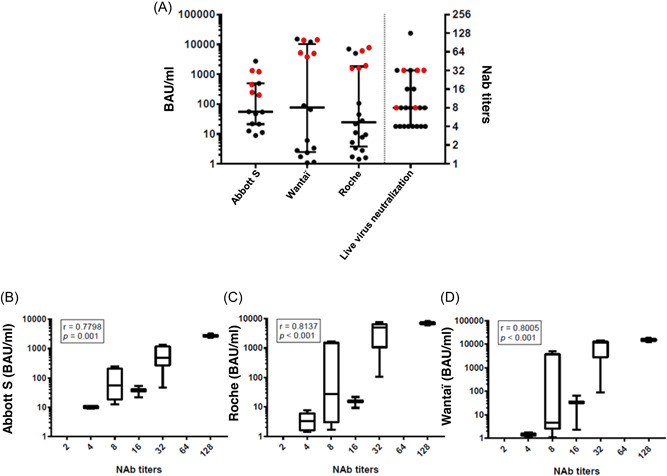
Immunoassays results and neutralizing antibody titers. (A) Distribution of anti‐S SARS‐CoV‐2 antibody concentrations expressed in BAU/ml according to the immunoassay used (left side). Distribution of anti‐SARS‐CoV‐2 neutralizing antibody (NAb) titers using Omicron BA.1 strain (right side): previously infected individuals (red symbols) and ancestral strain‐naïve individuals (black symbol). Data are represented as median plus interquartile range. (B–D) Correlation between Abbott S/Roche/Wantaï antibody concentrations and the NAb titers for all positive results. Data are represented as medians (midlines) plus interquartile ranges (IQR) (top and bottom box edges). Whiskers represent the upper and lower values. Spearman's rank coefficients (r) and their p‐value are indicated.
4. DISCUSSION
Among more than 90 serologic assays authorized by the FDA EUA, only four immunoassays were evaluated in this study. The Wantaï microplate assay was one of the first and most performant available test. 11 The Abbott S, Abbott N, and Roche assays were chosen given the frequent use of these automated multiparametric systems in French laboratories.
The sensitivities we found for commercial serological assays are much lower than those previously reported in pre‐Omicron era. Using these assays, the detection rate after infection with an ancestral D614G strain (control panel) ranged from 96.8% to 100% for anti‐S antibodies; it was 87.1% for anti‐N antibodies. A systematic review and meta‐analysis of SARS‐CoV‐2 serological assay performance found that the overall sensitivity of commercial assays was 91% (IC95:85–94) after 15 days of infection regardless of the technique used. 5 The pre‐Omicron sensitivities of the Abbott S and Wantaï assays were 100%, 9 while those of the Roche and Abbott N assays were 97.9% and 96%, respectively, for patients infected for 15 days. 12 , 13 This indicates that these assays are less sensitive for detecting anti‐Omicron antibodies than for detecting antibodies induced by ancestral strains.
Our results indicate that the decrease in sensitivity varies from one assay to another and might depend on the antigen used and the limit of detection of the assay. Indeed, the better sensitivity rate was found for the anti‐N assay (Abbott N) that might be less impacted by Omicron mutations and for the Roche assay characterized by the lowest limit of detection among anti‐S assays. Anti‐N assays seem to be better than anti‐S assays for the serodiagnosis of previous SARS‐CoV‐2 Omicron infections. They can also differentiate between natural and vaccine‐derived seroreactivity. Nevertheless, decrease in sensitivity due to anti‐N waning over time has been described 14 and only anti‐S assays are adequate for assessing the antibody response after vaccination with currently available vaccines. Although these commercial immunoassays failed to detect some Omicron SARS‐CoV‐2 antibodies, the Ab concentrations in positive samples quantified by anti‐S immunoassays were still correlated with the NAb concentrations. All the specimen missed by these commercial immunoassays had relatively low NAb titers. Moreover, a weaker humoral response to the Omicron variant compared to previous variants has been recently reported. 15 , 16
To conclude, the use of unvaccinated Omicron‐infected patient's serum enabled us to demonstrate that while Omicron neutralizing antibodies were detected in all serum in viral culture, some antibodies were not detected by the commercial kits. Despite the small sample size, this manuscript presents the first evidence that several commercial immunoassays lack sensitivity for detecting antibodies elicited by Omicron infections. It highlights the importance to reevaluate serological assays with emerging variants, the need to optimize these assays with revised antigens and raises the question of antigenic tests sensitivity.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENT
The English text was edited by Dr Owen Parkes.
Migueres M, Chapuy‐Regaud S, Miédougé M, et al. Current immunoassays and detection of antibodies elicited by Omicron SARS‐CoV‐2 infection. J Med Virol. 2022;95:e28200. 10.1002/jmv.28200
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Cohen C, Kleynhans J, von Gottberg A, et al. SARS‐CoV‐2 incidence, transmission, and reinfection in a rural and an urban setting: results of the PHIRST‐C cohort study, South Africa, 2020‐21. Lancet Infect Dis. 2022;22(6):821‐834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dimeglio C, Herin F, Da‐Silva I, et al. Post‐vaccination severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) antibody kinetics and protection duration. Clin Infect Dis. 2022;75(1):e924‐e925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vacharathit V, Aiewsakun P, Manopwisedjaroen S, et al. CoronaVac induces lower neutralising activity against variants of concern than natural infection. Lancet Infect Dis. 2021;21(10):1352‐1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deshpande PS, Abraham IE, Pitamberwale A, Dhote RH. Review of clinical performance of serology based commercial diagnostic assays for detection of severe acute respiratory syndrome coronavirus 2 antibodies. Viral Immunol. 2022;35(2):82‐111. [DOI] [PubMed] [Google Scholar]
- 5. Macedo ACL, Prestes GDS, Colonetti T, et al. A systematic review and meta‐analysis of the accuracy of SARS‐COV‐2 IGM and IGG tests in individuals with COVID‐19. J Clin Virol. 2022;148:105121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dimeglio C, Herin F, Miedougé M, et al. Screening for SARS‐CoV‐2 antibodies among healthcare workers in a university hospital in southern France. J Infect. 2021;82(1):e29‐e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Migueres M, Lhomme S, Trémeaux P, et al. Evaluation of two RT‐PCR screening assays for identifying SARS‐CoV‐2 variants. J Clin Virol. 2021;143:104969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Migueres M, Dimeglio C, Mansuy JM, et al. The influence of the nasopharyngeal viral load on the spread of the Omicron BA.2 variant. Clin Infect Dis. 2022;ciac563. [DOI] [PubMed] [Google Scholar]
- 9. Chapuy‐Regaud S, Miédougé M, Abravanel F, et al. Evaluation of three quantitative anti‐SARS‐CoV‐2 antibody immunoassays. Microbiol Spectr. 2021;9(3):e0137621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dimeglio C, Herin F, Da‐Silva I, et al. Decreased efficiency of neutralizing antibodies from previously infected or vaccinated individuals against the B.1.617.2 (delta) SARS‐CoV‐2 variant. Microbiol Spectr. 2022;10(4):e0270621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS‐CoV‐2 in patients with novel coronavirus disease 2019. Clin Infect Dis. 2020;71(16):2027‐2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Manalac J, Yee J, Calayag K, et al. Evaluation of Abbott anti‐SARS‐CoV‐2 CMIA IgG and Euroimmun ELISA IgG/IgA assays in a clinical lab. Clin Chim Acta. 2020;510:687‐690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Riester E, Findeisen P, Hegel JK, et al. Performance evaluation of the Roche Elecsys Anti‐SARS‐CoV‐2 S immunoassay. J Virol Methods. 2021;297:114271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Montesinos I, Dahma H, Wolff F, et al. Neutralizing antibody responses following natural SARS‐CoV‐2 infection: dynamics and correlation with commercial serologic tests. J Clin Virol. 2021;144:104988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He C, He X, Yang J, et al. Spike protein of SARS‐CoV‐2 Omicron (B.1.1.529) variant have a reduced ability to induce the immune response. Signal Transduct Target Ther. 2022;7(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Servellita V, Syed AM, Morris MK, et al. Neutralizing immunity in vaccine breakthrough infections from the SARS‐CoV‐2 Omicron and Oelta variants. Cell. 2022;185(9):1539‐1548.e1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


