Abstract
SARS‐CoV‐2 viral infection led to the devastating COVID‐19 pandemic, where illness stemmed from interactions between virions and recipient host cells resulting in multi‐layered pathological consequences. The role of the infection portal is now understood to be the cellular angiotensin converting enzyme‐2 (ACE2) receptor, which binds to viral spike (S) protein initiating virion internalisation process. Since SARS‐CoV‐2 virions bear some resemblance to endogenously produced small extracellular vesicles (sEVs) we reasoned that EVs engineered to express S protein (viral mimics) may interfere with viral infection. Here, we report generation of HEK293T cells producing sEVs enriched for transmembrane S‐protein tagged with green fluorescent protein (S/GFP). Strikingly, S protein drove the GFP tag to the membrane of sEVs, while GFP alone was not efficiently included in the sEV cargo. High‐throughput quantitative proteomics revealed that S/GFP sEVs contained over 1000 proteins including canonical components of the exosomal pathway such as ALIX, syntenin‐1, and tetraspanins (CD81, CD9), but depleted for calnexin and cytochrome c. We found that 84 sEV proteins were significantly altered by the presence of S/GFP. S protein expressing EVs efficiently adhered to target cells in an ACE2‐dependent manner, but they were poorly internalised. Importantly, prolonged administration of S/GFP EV to K18‐hACE2 mice provided a significant protection against SARS‐CoV‐2 infection. Thus, the generation of sEV containing S protein can be considered as a novel therapeutic approach in reducing the transmission of SARS‐CoV‐2.
Keywords: COVID‐19, exosomes, mimics, proteomics, SARS‐CoV‐2, SEC, spike, ultracentrifuge, vaccine
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) caused the coronavirus disease of 2019 (COVID‐19) pandemic resulting, at the time of this writing, in over 400 millions of cases and 5.8 millions of deaths worldwide (http://www.who.int/publications/m/item/weekly‐operational‐update‐on‐covid‐19‐15‐february‐2022) and counting. SARS‐CoV‐2 is a spherical enveloped virus with a typical size range of 80–120 nm in diameter and a virion surrounded by a lipid bilayer with embedded spike (S) protein trimers (Ke et al., 2020). SARS‐CoV‐2 S protein plays a role in virus entry into airway epithelial cells and other targets via the specific recognition of the angiotensin converting enzyme‐2 (ACE2) receptor in recipient cells (Lan et al., 2020). As this process is critical for the disease pathogenesis, S protein is considered to be the main therapeutic target of interventions (vaccines) aiming at preventing viral infection (Heinz & Stiasny, 2021).
Viral particles bear some resemblance to, or their formation may utilise (Gould et al., 2003), extracellular vesicles (EV) naturally generated by various cells, released to their microenvironment and taken up by their counterparts locally and systemically (Nolte‐’t Hoen et al., 2016). Like viruses, EVs are nano‐sized spherical structures surrounded by a lipid bilayer, which encases a diverse repertoire of molecular cargo, including proteins, nucleic acids (RNA and DNA), and metabolites (Choi et al., 2013). EVs are heterogeneous due to the existence of diverse pathways of EV biogenesis, cargo loading, subcellular origin, and cellular states they represent (van Niel et al., 2018). The best characterised EV subtypes include exosomes derived from multivesicular endosomes (MVBs) and ectosomes, commonly known as microvesicles (MVs), shed directly from the plasma membrane (Choi et al., 2019).
Pathways of EV biogenesis and uptake intersect with viral life cycle at several points. For example, host cells infected by certain viruses may release EVs carrying the viral and host components that could modulate the immune response (Schorey et al., 2015). Moreover, recent report revealed that mRNA‐based vaccines against the SARS‐CoV‐2 virus infection devised by Pfizer and Moderna (Baden et al., 2021; Teo, 2021) can generate circulating EVs carrying the SARS‐CoV‐2 S protein and promoting the generation of anti‐S neutralising antibodies in vaccinated healthy individuals (Bansal et al., 2021). Other reports suggest a negative effect of S protein‐carrying EVs that may serve as decoys for the anti‐S neutralising antibodies reducing their effectiveness in blocking viral entry (Troyer et al., 2021). These observations suggest that S protein‐carrying EVs may play important roles in SARS‐CoV‐2 virus‐related pathogenesis and response to neutralising antibodies. This also indicates that due to their resemblance of viruses, modified EVs could be utilised as a novel therapeutic approach against pulmonary viral infections.
In the present study, we explored the impact of S protein expression on the characteristics of EVs produced by human cells. Thus, we generated HEK293T cells expressing the SARS‐CoV‐2 S protein coupled with green‐fluorescent protein (S/GFP). We isolated small EVs with high purity and observed appreciable degree of S‐protein incorporation into the EV membrane. High‐throughput, label‐free quantitative proteomics revealed the content of approximately 1000 proteins in S‐protein containing EVs carrying canonical markers such as ALIX, syntenin‐1, and tetraspanins (CD81, CD9), but depleted for calnexin and cytochrome c. We found 84 proteins that were significantly affected by S/GFP expression with some underrepresentation of the endosomal sorting complexes required for transport (ESCRT). S protein expressing EVs adhered to target cells in ACE2‐dependent manner. Prolonged administration of S/GFP EV to K18‐hACE2 mice provided a significant protection against lethal dose of SARS‐CoV‐2 infection.
2. MATERIAL AND METHODS
2.1. Animals
Six‐ to 8‐week old B6.Cg‐Tg(K18‐ACE2)2Prlmn/J mice were purchased from Jackson Laboratories. Animal studies were conducted in accordance with the guidelines of, and approved by, the Animal Research Ethics Board of McGill University and Facility Animal Care Committee (FACC project ID: 5860). Animals were housed under SPF conditions with ad libitum access to food and water and were randomly assigned to experimental groups. Experiments were performed using age‐ and sex‐matched animals.
2.2. Cell culture and establishment of S/GFP expressing HEK293T cells
HEK293T cells were grown in Dulbecco's modified essential medium (DMEM; Wisent, Canada) supplemented with 10% heat‐inactivated fetal bovine serum (FBS) (Wisent) and 1% penicillin‐streptomycin (Gibco‐Life Technologies, Grand Island, NY) at 37°C in 5% CO2. The cloning of COVID‐19 proteins was performed using the Gateway technology (Invitrogen, Carlsbad, CA). Briefly, 150 ng of Entry vector containing codon‐optimised viral SARS‐CoV‐2 S protein cDNA (Kim et al., 2020) was incubated with 150 ng of the Destination vector pLVU‐GFP (Addgene, Watertown, MA) (Krupka et al., 2010), overnight at 25°C with 2 μl of LR clonase II (Invitrogen). Then 3 μl of the clonase reaction volume was used to transform One Shot Stl3 Escherichia coli competent cells (Invitrogen) as per company protocol. From each transformation, 12 colonies were checked for successful sequence integration by PCR (using primers flanking the destination vector insertion site) and subsequently sequenced to verify the correct expression of the S protein. This PCR amplification entailed an initial denaturation at 95°C 10 min, followed by 35 cycles of 30 s at 95°C, 30 s at 62°C, and 4 min at 72°C. PCR products were analysed on a 0.8% agarose gel. Finally, six positive colonies were further checked by PCR to confirm the correct orientation of the insert using one primer corresponding to the S protein coding sequence, and one primer corresponding to the vector. This PCR reaction involved an initial denaturation for 10 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at 62°C and 30 s at 72°C. PCR products were analysed on a 2% agarose gel.
2.3. pLVU‐GFP and pLVU‐S/GFP lentiviral vector transduction
After DNA isolation from single positive colony by midiprep (Qiagen, Valencia, CA), the lentiviral vector DNA was incubated with the packaging vector DNA and used to transform HEK293T cells. The lentiviral particles were collected at 48‐h and used to transduce HEK293T cells. Transformed cells were monitored for GFP presence and media was replaced with media containing Blasticidin (5 μg/ml) every 48‐h to obtain stable clones.
2.4. Isolation of EVs
Cells were plated in 12‐well plates for 24‐h at a concentration of 100,000 cells/ml in culture media containing EV‐depleted FBS (recovered from the supernatant after ultracentrifugation of FBS at 150,000 g for 18‐h at 4°C with 0.2 μm filtration). Conditioned medium was collected and centrifuged once at 400 g for 10 min. The resulting supernatant was concentrated up to a 100 μl volume using Amicon Ultra‐0.5 Centrifugal Filter Unit (EMD Millipore, Billerica, MA) with 100,000 NMWL molecular cut off. EV isolation was conducted by size exclusion chromatography (SEC) as described previously (Choi et al., 2019). Briefly, 100 μl of concentrated medium was loaded into qEVsingle SEC column followed by addition of 900 μl of phosphate buffered saline (PBS). Flow‐through was removed and EV‐enriched fractions, as determined by NTA (below), were collected following addition of 600 μl of PBS. For 3,3′‐Dioctadecyloxacarbocyanine Perchlorate (DiO) labelling of EVs, 100 μl of concentrated medium was incubated with 50 μM of DiO for 2‐h at RT. To remove unlabelled DiO dye, EVs were further isolated using qEVsingle SEC column as described above and in our previous study (Choi et al., 2019).
2.5. Nanoparticle tracking analysis (NTA)
The concentration and size of EVs were obtained through nanoparticle tracking analysis (NTA) using NanoSight NS500 instrument containing 532 nm laser (NanoSight Ltd., UK). Three recordings of 30‐s at 37°C were captured with camera level 15 and processed with detection threshold of 5 and blur size on auto using NTA software (version 3.0)
2.6. Nano‐flow cytometry
Nano‐flow cytometry was performed using CytoFLEX system (Beckman Coulter, Pasadena, CA) equipped with three lasers (405, 488, and 640 nm wavelength). The 405 nm violet laser was selected for side scatter (SSC) analysis with 1800 of manual threshold setting in the violetSSC‐height (violetSSC‐H) channel and gain set at 100 of violetSSC‐H signal in the acquisition mode. Samples were loaded and run with slow flow rate (10 μl/min) for 1 min until the event/s rate became stable, and then a 20 s acquisition run was saved. Data were acquired and analysed using CytExpert 2.0 software (Beckman Coulter) with events/s and events/ml outputs. The GFP positive region was gated on the basis of control EVs with no GFP content.
2.7. Confocal microscopy
A total 10,000 of Calu‐3 cells, which express high levels of ACE2 (Hoffmann et al., 2020), were seeded in μ‐Slide 8‐Well ibiTreat chambered coverslips (ibidi, Germany) and cultured for 24‐h before addition of S/GFP EVs or DiO‐labelled EVs at 5 × 109 particles/ml for 18‐h. For immunofluorescence, cells were fixed with 4% paraformaldehyde and permeabilised with 0.1% tween‐20 in PBS. The cells were then blocked with 1% of BSA and stained with fluorophore conjugated anti‐ACE2 and anti‐GFP antibodies. Images were collected using LSM780 confocal microscope (Carl Zeiss, Thornwood, NY) with the 63×/1.40 objective. For the calculation of Pearson correlation coefficient to address the colocalisation between proteins, correlation diagram was visualised in Fiji using colocalisation finder plugin. Pearson correlation coefficients greater than 0.49 represent strong colocalisation (Zinchuk et al., 2013).
2.8. Isolation of EVs for the proteomics
EV preparation was conducted as described earlier (Choi et al., 2019). Briefly, each cell population (control, GFP, S/GFP HEK293T cells) was seeded into two 150 mm plates at 4 × 106 cells per 22 ml of EV‐depleted culture media. After 3‐day culture, a total of 44 ml of conditioned media was collected and cell debris was pre‐cleared by centrifugation at 400 g for 10 min. The supernatant was concentrated using Amicon Ultra‐15 Centrifugal Filter Unit (EMD Millipore) with 100,000 NMWL molecular cut off to achieve a volume of 500 μl. A total of 500 μl of the concentrate was loaded into qEVoriginal SEC column and resolved. After loading the sample, 2.5 ml of PBS was added and flow‐through removed. EV‐enriched fractions (as determined by NTA ‐ above) were collected after adding 1.5 ml of PBS. EV‐enriched fraction was mixed with 7 ml of 1.2 M KCl in PBS and incubated for 30 min at 4°C (Choi et al., 2021). EVs were recovered by ultracentrifugation at 110,000 g for 1 h and resuspended in 100 μl of Radioimmunoprecipitation assay buffer (RIPA) buffer containing: 50 mM Tris‐HCl, 1% NP‐40, 0.25% Na‐ deoxycholate, 100 mM NaCl, 1 mM EDTA, pH 7.5) with protease inhibitor cocktail (Roche, Mannheim, Germany). The concentration of EV proteins was quantified using the micro BCA assay (Pierce Biotechnology, Rockford, IL).
2.9. Mass spectrometry
EV samples (control, GFP, S/GFP) were purified independently at three different times with parallel isolation on different days, along with corresponding cell lysates (also collected using RIPA buffer). For mass spectrometry, the same amounts of protein (10 μg) were desalted with SDS–PAGE, loaded onto the stacking gel followed by staining and de‐staining. The in‐gel trypsin digestion was carried out under reducing conditions afforded by DTT, and alkylation was achieved using iodoacetic acid as previously described (Shevchenko et al., 2006). The lyophilised peptides were resolubilised in 0.1% aqueous formic acid/2% acetonitrile, the peptides were loaded onto a Thermo Acclaim Pepmap (75 μm inner diameter × 2 cm, C18, 3 μm particle size, 100 Å pore size) (Thermo Fisher Scientific, San Jose, CA) pre‐column and the onto an Acclaim Pepmap Easyspray (75 μm inner diameter × 15 cm, C18, 2 μm particle size, 100 Å pore size) (Thermo Fisher Scientific) analytical column. Separation was achieved using a Dionex Ultimate 3000 uHPLC at 220 nl/min with a gradient of 2%–35% organic solvents (0.1% formic acid in acetonitrile) over 3 h. Peptides were analysed using an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific) operating at 120,000 resolution (FWHM in MS1, 15,000 for MS/MS) with higher‐energy collisional dissociation of all peptides with a charge of 2+ or greater. The raw data were converted into *.mgf format (Mascot generic format) by MS‐ Convert (ProteinWizard, version 3.0.6150), and searched using Mascot 2.5.1 against SwissProt (http://www.uniprot.org) human protein database (release 2020_09, 20316 entries) with SARS‐CoV‐2 S protein (www.uniprot.org/uniprot/P0DTC2) and with GFP sequence. The tolerance was 5 ppm monoisotopic for precursor ions and 0.1 Da for fragment ions. The permission of two potential missed cleavages was selected for trypsin digestion. The following modifications were used: fixed modification for the carbamidomethylation of cysteine (58 Da) and variable modification for the oxidation of methionine (16 Da) and the deamidation of asparagine and glutamine (1 Da). The database search results were further analysed by Scaffold Q software (proteomesoftware.com/products/scaffold/) (Proteome Sciences, Portland) (Protein threshold > 0.99%, Peptide threshold > 0.99%). We quantified the relative protein abundance by normalized weighted total spectra and calculated the p value by Student's t test using Scaffold Q+ software. Proteins with less than 0.05 of p value were considered as significantly changed.
2.10. Protein interaction network analyses
Protein–protein interactions were deducted from BioGRID database (version 4.4.206; http://www.thebiogrid.org/). The network for identified EV proteins was constructed using Cytoscape (http://www.cytoscape.org/).
2.11. Therapeutic effect of EV for SARS‐CoV‐2 infection in K18‐hACE2 transgenic mice
Six‐ to 8‐week old female C57BL/6J and B6.Cg‐Tg(K18‐ACE2)2Prlmn/J mice were purchased from Jackson Laboratories or bred at the RI‐MUHC, Montreal, QC, Canada. K18hACE2 mice received EVs (control, GFP, S/GFP) intravenously (i.v.) at day –14 with a concentration of 20 μg/100 μl and at day 0, intranasally (i.n.) with a concentration of 16 μg/40 μl. After 5‐h (day 0), mice were infected with SARS‐CoV‐2 via i.n. route with the lethal dose (1 × 104 TCID50). SARS‐CoV‐2/SB2 was kindly provided by Dr. Samira Mubareka, University of Toronto, ON, Canada. All animal studies were conducted in accordance with the guidelines of, and approved by, the Animal Research Ethics Board of McGill University and FACC (project ID: 5860). Mice were housed under SPF conditions with ad libitum access to food and water.
3. RESULTS
3.1. Expression of SARS‐CoV‐2 spike‐GFP fusion protein in EV‐producing cells
We reasoned that introduction of S protein into human cells may result in its incorporation into cellular membranes and subsequent release as EVs endowed with the ability to interact with SARS‐CoV‐2 target cells. To explore this possibility, HEK293T cells that are well characterised for robust EV biogenesis (Lázaro‐Ibáñez et al., 2021) were transduced with lentivirus harbouring SARS‐CoV‐2 S/GFP sequence. GFP was attached to the short internal domain (33 amino acids) of S protein and its location in EVs was thereby directed to the intra‐vesicular side of the membrane, so as to prevent GFP from affecting S protein functionality on the external surfaces of EVs. Stable cell lines were established by selection in blasticidin containing media and then S/GFP positive cells were further sorted by FACS. The expression of S/GFP fusion protein in HEK293T cells was validated by Western blotting whereby the expected 170 kDa molecular weight band was visualised using anti‐GFP antibody (Figure 1a). To isolate the EVs, HEK293T cells were cultured for 3 days and the resulting conditioned media was processed using a validated protocol based on centrifugation and SEC (Choi et al., 2019). NTA analysis of this material revealed that overexpression of S/GFP protein or GFP proteins did not visibly affect the concentration or size distribution of HEK293T‐derived EVs (Figure 1b–d). Thus, introduction of viral S protein did not induce, or markedly change EV release by these cells.
FIGURE 1.
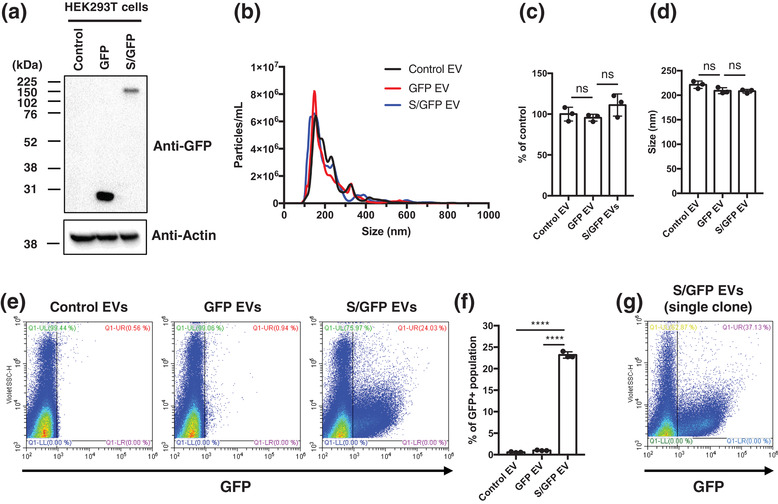
Characterisation of SARS‐CoV‐2 S/GFP EVs derived from HEK293T cells. (a) Western blotting shows the expression of free GFP and S/GFP in the transduced HEK293T cells. S/GFP is composed of viral S protein linked to GFP at its C‐terminus resulting in about 170 kDa molecular weight. (b) NTA shows the size distribution of isolated EVs with average size of 221.4, 209.1, and 208.3 nm in control, GFP, and S/GFP HEK293T cell‐derived EV, respectively. (c and d) Concentration and size of the released EVs were not significantly affected by the overexpression of free GFP and S/GFP protein expression. (e and f) Nano‐flow cytometry represents the single EV distribution of GFP‐positive EVs. GFP‐positive region was gated on EVs derived from control HEK293T cells (control EV). EVs derived from HEK239 GFP cells (GFP EV) rarely contained GFP‐positive EVs, but S/GFP EVs showed a significantly higher levels of GFP‐expressing EVs with about 24% positivity, which may represent selective sorting of viral S protein into EV. (g) Clonal cells obtained by single colony selection based on the increased cellular S/GFP expression showed the increased GFP‐positive EV about 37.1%
3.2. SARS‐CoV‐2 S‐protein drives the expression of GFP into the EV membrane
In order to determine whether S protein is exported as cargo of EVs, and exposed on their surfaces, and to what extent, we further explored their characteristics using nano flow cytometry at a single vesicle resolution (Figure 1e,f). This analysis revealed that approximately 24% of all EVs released from S/GFP‐expressing HEK293T cells were positive for GFP, while only a negligible number of GFP‐positive EVs were detected in the supernatant of cells transduced with GFP alone (<1%), in spite of a somewhat higher overall cellular levels of this protein (Figure 1a). This observation suggests that fusion of GFP with S protein selectively drives this fluorescent protein into the EV compartment and therefore S protein may have the ability to interact with machinery involved in small EV biogenesis. While the propensity of S/GFP protein to associate with EVs is of interest from both a biological and therapeutic perspective (e.g., as viral mimic or protein loading device) only a fraction of EVs carried S/GFP, possibly due to the heterogeneity of their producing cells or mechanis. To increase the EV content of S/GFP protein, we subjected S/GFP transduced HEK293T cells to random cloning followed by nano‐flow cytometry screening for high S/GFP producing clones. This enabled us to identify a clonal cell line with an increased output of S/GFP‐carrying EVs bringing it from 24.0% to 37.1% (Figure 1g). This cell line has been used throughout our subsequent studies.
3.3. Quantitative proteomic analysis of S‐protein carrying EVs
Since proteins strongly associated with the vesicle cargo may act as hubs for EV‐forming protein–protein interactions (Luck et al., 2020) we asked whether S protein impacts the proteome of EVs released from HEK293T cells. For this purpose, we modified the EV isolation protocol to include a combination of SEC, high‐salt washing, and ultracentrifugation to ensure the purity and correct representation of true EV integral proteins (Figure 2a), as we described earlier (Choi et al., 2019, 2021). The EV‐enriched fraction of SEC was washed with KCl to remove the outer membrane associated proteins (Choi et al., 2021; Zhao et al., 2004) and EVs were re‐isolated by ultracentrifugation at 110,000 g for 1 h and lysed. As expected, our isolated EVs were highly enriched for canonical EV proteins such as CD81, and TSG101, but depleted for mitochondrial cytochrome c (Figure 2b). Approximately, 15 μg of EV proteins were isolated from control, GFP, and spike/GFP expressing HEK293T cells (three biological replicated) with yield comparable among the groups and consistent with EV numbers as determined by NTA (Figure 2c).
FIGURE 2.

Quantitative proteomics of isolated EVs. (a) Diagram shows the schematic workflow of the purification of EVs. (b) Western blotting shows the enrichment of canonical EV marker proteins including CD81 and TSG101, but depletion of non‐EV marker cytochrome c in the EVs. A total 5 μg of S/GFP cell and EV proteins are loaded. (c) The protein amounts of each EV isolation were determined by micro BCA assay. Released EV amounts are not significantly affected by expression of GFP or S/GFP protein. (d) Numbers of identified proteins by proteomics are represented (Protein threshold > 0.99%, Peptide threshold > 0.99% in Scaffold software). Cellular proteomes from control, GFP, and S/GFP HEK293T were analysed once in mass spectrometry. EV proteomes of control, GFP, and S/GFP were analysed using three independent biological replicates. (e and f) Abundance of each protein was calculated by label‐free quantitation based on the weighted total spectra. Canonical EV marker proteins including ALIX, SDCBP, CD81, TSG101, CD9, and CD63 and depleted EV maker proteins such as calnexin (CANX) and cytochrome c (CYCS) are indicated. (g) Venn diagram shows the overlapped control EV proteins with top 100 identified EV proteins in Vesiclepedia database (Kalra et al., 2012). Proteins identified in our control EV proteome and not listed in Vesiclepedia include 9 histone proteins (HIST1H4A, HIST1H4B, HIST1H4I, HIST4H4, HIST1H4K, HIST1H4H, HIST1H4J, HIST1H4F, HIST1H4D)
A total of 10 μg of each EV protein sample were processed for in‐gel digestion, and the resulting tryptic peptides were analysed by mass spectrometry (three biological replicates per each data point). As a result, a total of 1258, 1395, and 1251 proteins were identified in control, GFP, and S/GFP EVs, respectively (Protein threshold > 0.99%, Peptide threshold > 0.99% in Scaffold software). Cellular proteomes from control, GFP, and S/GFP HEK293T cells were also resolved by mass spectrometry (Figure 2d). The abundance of each protein was calculated by label‐free quantification based on weighted total spectra (Table S1).
As expected, the canonical EV marker proteins including ALIX, syntenin‐1, CD81, TSG101, CD9, and CD63 showed a higher abundance in control EV proteome than in the corresponding cellular proteomes, while calnexin (CANX) and cytochrome c (CYCS) were expectedly depleted in control EVs (Figure 2e,f). Notably, our control EV proteome included 91 proteins frequently identified in Vesiclepedia database (Kalra et al., 2012) (Figure 2g), which did not report nine histone proteins (HIST1H4A, HIST1H4B, HIST1H4I, HIST4H4, HIST1H4K, HIST1H4H, HIST1H4J, HIST1H4F, HIST1H4D) that we have identified.
3.4. Impact of S protein expression on the EV proteome
The strong propensity of S/GPF fusion protein to be incorporated into the EV membranes prompted us to examine the corresponding EV proteome and compare it to that of EVs released from control GFP expressing and uninfected cells. We first asked whether S protein may have an impact on canonical EV marker proteins as defined by international consensus (Lotvall et al., 2014). We reasoned that if this was the case such a change may suggest an impact of S protein on one or more of core mechanisms responsible for EV biogenesis (van Niel et al., 2018). To this end, we evaluated several marker proteins for their enrichment in EVs relative to corresponding cells (Figure 3a). As expected, such enrichment was observed in proteins related to endosome (ALIX, syntenin‐1), tetraspanins (CD9, CD63, CD81), membrane‐binding proteins (MFGE8, annexins), and proteins often associated with EV surfaces (albumin and extracellular matrix proteins). In keeping with this pattern, we observed low abundance, or absence in the EV proteome of proteins that normally tend to be excluded from the EV cargo such as CANX (calnexin), CYCS (cytochrome c), histones (H4C1, H2BU1), and HSP90B1, also known as Grp94 (Lotvall et al., 2014). Collectively, our EV isolations and proteomic analyses confidently identified the expected marker proteins, but there was no consistent difference in their expression between EVs carrying S protein or GFP alone.
FIGURE 3.

Enrichment of canonical EV marker proteins in EV proteomes. (a) Heat map shows the enrichment of EV marker proteins related in endosome (ALIX, syntenin‐1), tetraspanins (CD9, CD63, CD81), membrane‐binding proteins (MFGE8, annexins), and EV‐associated proteins (albumin and extracellular matrix proteins), which are canonical EV proteins suggested by ISEV guidelines (Lotvall et al., 2014). (b) Identified peptides in S/GFP protein are highlighted in yellow. Amino acids with red letter are glycosylated peptide on asparagine (Watanabe et al., 2020). Bold letters with underline are the sequence of GFP
Mass spectrometry analysis also reinforced our observation as to the enrichment of S/GFP protein in the EV cargo, which was not the case for free GFP (Figure 3a). Interestingly, when we mapped the identifiable peptides of the S/GFP protein (Figure 3b) we were able to assign 722 out of 1538 amino acids comprising the S/GFP protein (47% sequence coverage). The unidentified tryptic peptides are known to be glycosylated on asparagine (Watanabe et al., 2020), an observation that suggests that mass spectrometry could be applied to find the ‘naked’ peptide sequences associated with foreign viral proteins.
To glean more insights as to the possible impact of the viral S protein on the EV composition we compared the relative abundance of proteins detected in S/GFP EVs and those of control EVs (GFP alone and non‐infected). A total of 84 differentially expressed EV proteins were identified of which 35 (2.8% of 1251 total S/GFP EV proteins) (Table S2) and 49 (3.3% of 1472 total other EV proteins) (Table S2) were significantly upregulated in either S/GFP EVs or control EVs, relative to their respective counterparts (Figure 4a). It is noteworthy that the numbers of EV‐associated proteins that are affected by the introduction of the exogenous S protein (S/GFP) into HEK293T cells are considerably lower than is the case for cellular transformation following the expression of exogenous oncogenic EGFRvIII (Choi et al., 2018) or HRAS (Choi et al., 2021) proteins, either of which impacted more than 20% of EV proteins.
FIGURE 4.

Quantitative profiles of EV proteins affected by S/GFP expression. (a) Relative abundances of EV proteins were calculated based on weighted spectral count by the Scaffold program. Volcano plot shows the significantly changed proteins with more than 2‐fold upregulation in S/GFP EV proteomes comparing with other (control and GFP) EV proteomes. (b) 27 proteins with more than average three quantitative value of proteins from total 39 upregulated proteins in the S/GFP EV proteome are represented. Functionally related proteins are grouped into categories designated in bold. (c) Interaction partners of S protein in EV proteomes are visualised by Cytoscape (Shannon et al., 2003) using BioGRID database (Oughtred et al., 2019). Inferred interactions between proteins with more than 2‐fold upregulation in S/GFP EV are indicated with red edge
Interestingly, among 35 proteins upregulated in the S/GFP EV proteome, the main functional groups include: post‐translational modification (PTM), endoplasmic reticulum (ER), protein folding, and mitochondria‐related proteins (Figure 4b). This may suggest that S protein translation and PTM could be involved in its sorting into EVs. The protein–protein interaction network linking S protein to other elements of the S/GFP EV proteome also points to a functional connection with ER‐ and glycosylation‐related pathways (Figure 4c). In this regard, VDAC1 and VDAC2 are enriched in S/GFP EVs by a factor of 4.1‐ and 3.3‐fold, respectively, and these proteins are also connected with ER/glycosylation‐related processes. Curiously, in severely SARS‐CoV‐2‐infected patients, a higher expression of VDAC1 is observed in T‐cells, resulting in mitochondrial dysfunction and apoptosis (Thompson et al., 2021). Whether S protein interactions, or EV biogenesis play a role in these effects remains unknown.
3.5. Sorting of S/GFP protein into an EV subpopulation thatshed from the plasma membrane
Among 49 proteins down‐regulated in S/GFP carrying EVs (Table S3), ubiquitin‐ and vesicle trafficking‐related proteins were especially noticeable (Figure 5a). This included BAG5 (BAG family molecular chaperone regulator 5), NEDD4L (E3 ubiquitin‐protein ligase NEDD4‐like), STUB1 (E3 ubiquitin‐protein ligase CHIP), and STAMBP (STAM‐binding protein), all of which play a role in ubiquitin‐dependent cellular trafficking of proteins. In addition, we observed down‐regulation of VAMP3 (Vesicle‐associated membrane protein 3), which is a member of SNAP Receptor (SNARE) family of proteins involved in vesicular transport from the late endosomes to the trans‐Golgi network (Hu et al., 2007). We also observed somewhat reduced representation in S/GFP EVs of CHMP4A (Charged multivesicular body protein 4a), which is an ESCRT‐III component involved in protein sorting into exosome compartment (Baietti et al., 2012). A network analysis of primary interaction partners of BAG5, NEDD4L, VAMP3, and CHMP4A showed the down‐regulated functional module of ESCRTs and vesicle trafficking (Figure 5b). Based on these observations, we postulated that exosome biogenesis is unlikely to be the main pathway resulting in formation of S/GFP‐carrying small EVs.
FIGURE 5.

Linkages of S/GFP carrying EV to subcellular regions of vesiculation. (a) Heat map shows selected proteins related to EV biogenesis from the pool of down‐regulated 49 proteins in S/GFP EVs. (b) Interaction partners of BAG5, NEDD4L, STUB1, and STAMBP in EV proteomes as visualised by Cytoscape (Shannon et al., 2003) using BioGRID database (Oughtred et al., 2019). ESCRT and vesicle trafficking functional modules are visualised by dashed box. (c and d) Confocal images show notable differences of subcellular location of cellular S/GFP protein, mostly associated with plasma membrane, versus intracellular localisation of ALIX and syntenin‐1. (e and f) Diagrams show the correlation of subcellular localisation of S/GFP protein versus ALIX or syntenin‐1. Proteins in UL (upper left), UR (upper right), and LR (lower right) are indicated with white colour and their Pearson correlation coefficient is indicated in the lower part of the image. Pearson correlation coefficients greater than 0.49 represent strong colocalisation (Zinchuk et al., 2013)
To explore this notion further, we analysed the subcellular localisation of S/GFP protein relative to the representative exosomal marker proteins, ALIX and syntenin‐1, which are among the main proteins regulating exosome release (Baietti et al., 2012) (Figure 5c,d). Confocal images show that most of the S/GFP protein signal localises in the plasma membrane rather than intracellular compartments, where ALIX and syntenin‐1 can be readily visualised. Correlation diagrams were also generated, and they reinforce the notion that S/GFP protein is not strongly colocalised with ALIX and syntenin‐1, with less than 0.49 value of the Pearson correlation coefficient (Figure 5e,f). These results suggest that S/GFP is mainly directed to the plasma membrane, from where it is likely released as small non‐exosomal EVs (ectosomes) (van Niel et al., 2018).
3.6. Interaction of S protein expressing EVs with cells susceptible to SARS‐CoV‐2 infection
Since EVs expressing S protein could act as mimics of SARS‐CoV‐2, we interrogated their interactions with Calu‐3 cells. This non‐small cell lung cancer cell line is often used as a model of airway epithelium and is known to express ACE2 receptor, which serves as a portal for viral infection (Hoffmann et al., 2020). To this end, S/GFP EVs were incubated with Calu‐3 cells for 18‐h and their fate was monitored by GFP‐ and immunofluorescence and visualised by confocal microscopy (Figure 6a). This imaging confirmed the presence of a largely membrane‐associated ACE2 signal in Calu‐3 cells and revealed extensive contacts between S/GFP‐containing EVs and cellular surfaces. While S/GFP EVs accumulated at the plasma membrane of Calu‐3 cells in a manner reminiscent of SARS‐CoV‐2 virions (Jackson et al., 2022), they were not efficiently internalised for the duration of the experiment (18‐h). However, lipophilic dye DiO‐labelled HEK293T EVs readily accumulated in perinuclear region in Calu‐3 cells suggesting a functional EV internalisation pathway in these cells (Figure 6b). Interestingly, S protein expression appears to divert EVs into a different mode of surface interactions with recipient cells. Overall, these observations may suggest that S/GFP‐EVs could potentially occupy ACE2 receptors and act as decoys interfering with initial phases of SARS‐CoV‐2 infection.
FIGURE 6.

Cellular interactions in vitro and therapeutic activity in vivo of S/GFP EVs. (a and b) Confocal images show contacts of S/GFP‐containing EVs with cellular surfaces of Calu‐3 cells which endogenously express high levels of ACE2. In contrast, DiO‐labelled HEK293T EVs accumulate in perinuclear region in the cells indicating internalisation. (c) Experimental model of EVs administration and i.n infection with SARS‐CoV‐2 in K18‐hACE‐2 mice (n = 13–16 mice per group). EV concentration was approximately 1.43 × 109 particles/μg. (d and e) Morbidity and mortality of EVs treated mice after i.n. infection with SARS‐CoV‐2 as measured by body weight (d), and overall survival (e) at ethical endpoint.
3.7. Therapeutic activity of S/GFP EVs resulting in decreased lethality of SARS‐CoV‐2 infection in mice
To evaluate the therapeutic effect of S/GFP EVs on the SARS‐CoV‐2 infection in an animal model, control, GFP, and S/GFP were administered intravenously (i.v.) into K18‐hACE2 mice. These transgenic mice express human ACE2 receptor and are susceptible to SARS‐CoV‐2 infection (Winkler et al., 2020). After 14 days of initial systemic exposure, EVs were locally administered via the intranasal (i.n.) route and 5‐h later the mice were infected via i.n. route with lethal doses of SARS‐CoV‐2 (Figure 6c). Because of the intranasal route of infection in this model i.n. administration of EVs was chosen to enable a direct contact of S/GFP EVs with the luminal side of the plasma membrane of ACE2‐expressing lung epithelial cells prior to viral exposure. Based on our aforementioned in vitro observations we reasoned that this application would be more efficient at blocking the binding of SARS‐CoV‐2 virions to host cells than systemic delivery of S/GFP EVs. The number of total EVs in i.n. inoculum (16 μg) corresponds to approximately 2.3 × 1010 particles, according to NTA counts, which is about 2.3 million‐fold higher than the number of viral particles administered to these animals (1 × 104 particles).
We found that K18‐hACE2 mice treated with S/GFP EV had markedly reduced morbidity (early body weight loss) after infection with a lethal dose of SARS‐CoV‐2 (Figure 6c). The apparent transiency of the weight loss, or rebound in surviving control mice is consistent with reported data (Falach et al., 2021; Li et al., 2021; Sun et al., 2021) and may represent a selection of more resilient animals in the group. Importantly, the overall survival of mice treated with S/GFP EVs was significantly increased to 65% compared to 20% in control mice, which received only GFP‐containing EVs (Figure 6d). Therefore, treatment with EVs containing S‐protein can significantly enhance host defences against SARS‐CoV‐2 infection.
4. DISCUSSION
In this study, we documented changes in properties, ACE‐2‐dependent cellular uptake and biological activity of EVs engineered to carry surface associated viral S protein. Our observations indicate that S protein is preferentially incorporated into membranes of eukaryotic cells (HEK293T), from which it is released on the surface of small ectosome‐like EVs. Incorporation of S protein into membranes of EVs elicits limited changes in their proteome with upregulation of proteins involved in posttranslational modifications, endoplasmic reticulum, protein folding, and mitochondria. Moreover, S protein carrying EVs bind to surfaces of ACE‐2 expressing recipient cells, but unlike the SARS‐CoV‐2 virus, they do not undergo rapid internalisation. Importantly, S protein decorated EVs attenuate the severity of SARS‐CoV‐2 infection in mice.
Our analysis suggests that EV‐based viral mimics could possess infection‐attenuating properties in vivo by interfering with interactions between the pathogen and susceptible host cells. In this regard, it is crucial to understand whether insertion of a viral protein, such as S protein of SARS‐CoV‐2, may adversely alter the properties of eukaryotic donor cells and their EVs. In order to address this question efficient EV isolation techniques and thorough analysis of their composition and cargo are crucial (Lázaro‐Ibáñez et al., 2021). To this end we applied a combination of SEC and ultracentrifugation for the isolation of EVs based on their size and density to achieve a high purity preparation (Guan et al., 2020; Wei et al., 2020). This purity was further enhanced by 1 M KCl washing, to remove peripheral membrane proteins by disruption of ionic interaction among molecules (Zhao et al., 2004). In our hands this protocol enabled us a more rapid EV isolation and at higher purity (within 3 h) than density gradient ultracentrifugation (Jeppesen et al., 2019).
Quantitative proteomics revealed 84 EV‐associated proteins, the content of which was altered in the presence of S/GFP protein (Figure 4a). This is a relatively modest change in the proteome, suggesting that S protein could be introduced into EVs at high levels and without major uncontrolled rearrangements in the overall EV composition. Indeed, this observation implies that in spite of its high levels in the EV cargo S protein does not recruit many host proteins into the vesiculation pathway. This is consistent with the fact that the S protein, despite having a length of 1271 amino acids, contains only a very short (33 amino acid) cytoplasmic domain. This would be expected to reduce the potential for S protein to orchestrate protein–protein interaction‐based sorting into EVs, which is one of the driving forces of EV protein assembly (Lotvall et al., 2014; Watanabe et al., 2020). Neither does the expression of S protein appear to perturb the regulatory mechanisms responsible for vesiculation of EV donor cells. This is at variance with EV production resulting from introduction of overtly transforming/signalling EV‐associated proteins, such as oncogenic EGFRvIII in glioma cells (Choi et al., 2018) or mutant RAS in intestinal epithelial cells (Choi et al., 2021), both of which impact hundreds of EV‐associated proteins.
Several upregulated proteins and potential interaction partners of S protein within S/GFP EV proteome appear to form a functional module within the EV protein network (Figure 4). This grouping includes calnexin (CANX), protein disulfide isomerases (PDIA3, PDIA4), and Dolichyl‐diphosphooligosaccharide‐protein glycosyltransferases (RPN1, RPN2), which are components of oligosaccharyltransferase (OST) complex. These results may suggest that PTM, especially glycosylation in the ER could play a role in S protein sorting into the membrane EV compartments. This is of interest as N‐linked glycosylation of S protein with the involvement of the OST complex components (STT3A) was found to influence the interactions with the ACE2 receptor and COVID‐19 disease severity (Huang et al., 2021). Moreover, a therapeutic strategy to target OST showed the decreased infectivity of SARS‐CoV‐2 due to a defect in N‐linked glycosylation of S protein (Huang et al., 2021). Whether similar mechanisms may explain packaging of S protein into membrane‐derived EVs remains to be studied in more detail.
S/GFP protein is enriched in small EVs that resemble exosomes. However, S/GFP is also strongly associated with the plasma membrane and EVs whose protein composition are inconsistent with ESCRT‐dependent pathways of exosome biogenesis. For example, among 49 proteins down‐regulated in S/GFP EVs (vs. controls), BAG5 (BAG family molecular chaperone regulator 5), NEDD4L (E3 ubiquitin‐protein ligase NEDD4‐like), STUB1 (E3 ubiquitin‐protein ligase CHIP), and STAMBP (STAM‐binding protein) are closely related to protein ubiquitination. Since this process is also closely linked to exosome biogenesis, especially the assembly of ESCRT complexes prior to membrane invagination and intraluminal vesicle formation within MVBs, downregulation of these proteins may suggest the involvement of a different mechanism of S/GFP EV biogenesis (Pisitkun et al., 2004) . Similarly downregulation of CHMP4A (Charged multivesicular body protein 4a), an ESCRT‐III component, suggests an involvement of exosome‐unrelated pathway (Baietti et al., 2012), as does downregulation of VAMP3 (Vesicle‐associated membrane protein 3), a member of SNARE proteins involved in vesicular transport, which is a part of exosome release machinery (van Niel et al., 2018). Moreover, confocal images indicate a sharp separation of subcellular S/GFP distribution (mainly plasma membrane) and that of other exosomal proteins such as ALIX and syntenin‐1 (throughout the cytoplasm).
It is puzzling and presently unclear why S/GFP EVs are ostensibly retained on the plasma membrane of Calu‐3 cells and do not become internalised through one of the canonical EV uptake mechanisms, such as endocytosis or macropinocytosis (Choi et al., 2021; Mulcahy et al., 2014). These pathways are clearly operative in Calu‐3 cells, as documented by robust internalisation of fluorescent EVs that do not expose S protein. It could be speculated that protracted docking of S/GFP EVs at the plasma membrane and mediated by ACE2 may signify the distinction between viral entry pathways (aborted in this case) and canonical EV uptake mechanisms.
Nonetheless, in a mouse model of COVID‐19 the administration of S/FP EVs attenuated the severity of the disease and improved the overall survival of infected animals. This effect is reminiscent of those achieved with neutralising antibodies (Weinreich et al., 2021), ACE2 mimics (ACE2‐expressing EVs) (Cocozza et al., 2020), or other agents. Especially, ACE2 mimics could play a role as decoy receptors competing for SARS‐CoV‐2 binding against ACE2 expressed by susceptible host cells, thereby exerting protective effects during viral infection (El‐Shennawy et al., 2022; Kim et al., 2022; Zhang et al., 2021). In this regard, El‐Shennawy et al. explored the consequences of pre‐incubation of viral particles with 130 μg of ACE2 EV mimics before i.n administration of SARS‐CoV‐2 in the K18‐hACE2 mouse model. Although in this model the effective blockage of viral infection has been achieved (El‐Shennawy et al., 2022), pre‐incubation of viral particles with ACE2 mimics could be impractical in view of clinical applications. Kim et al. injected 200 μg of ACE2 mimics via i.p. route every 6‐days (total 1200 μg), but their protective effect was very limited in the K18‐hACE2 mouse model of COVID‐19. In this case only individual mice gained survival advantage in groups administered ACE2 mimics, compared to controls (Kim et al., 2022).
In this context our approach is based on a different principle. Rather than using EVs to bind the virus prior to contact with target cells we sought to shield the viral portal sites on these cells, namely by targeting ACE2 receptors with an intent to limit viral access in vivo. We used a total of 36 μg of S/GFP EVs, which is considerably below the amount of therapeutic EVs reported in prior studies, and we applied two administration cycles of EVs via i.n. route. It could be argued that introducing the viral mimics into the airways could improve their effects compared to other approaches (El‐Shennawy et al., 2022; Kim et al., 2022).
While promising, our survival data ought to be viewed with caution. For example, interaction of S protein on EVs with the host ACE2 receptor may potentially drive the downregulation of host ACE2, resulting in prolonged activation of angiotensin II triggering pulmonary inflammation and thrombosis (Shirbhate et al., 2021; Verdecchia et al., 2020). This should be addressed in future studies. Also, Troyer et al. reported that EVs carrying S protein were able to inhibit the action of neutralising antibodies against the coronavirus suggesting a negative impact on disease outcome. This is an important consideration in the context of S protein containing EV decoys and one pertaining to the importance of timing and route of administration during therapy with EV mimics. In our experiments S/GFP EV administration occurred mainly locally (in the airways) and prior to virus infection, and thus their mode of action is likely different than that described by Troyer et al. Indeed, we observed a remarkable improvement in survival of infected mice when S/GFP EVs were pre‐inoculated. It could be argued that the optimal use of different EV mimics should be tailored to the stage of SARS‐CoV‐2 infection with different approaches being used prior to, or during a full‐blown disease.
While in the current study we have not been able to show whether the protection of S/GFP EVs is mediated via the interference with the viral internalisation into the cells, generation of neutralising antibodies, or both, the capacity of EV‐containing viral proteins to significantly reduce the severity of the disease provides a novel avenue for developing therapy against pulmonary viral infections. Considering the rapid mutation of SARS‐CoV‐2 and the variants of concern, which are highly resistant to current vaccines, alternative approaches, such as EV‐containing S‐protein, may become useful in efforts to overcome some of the limitation of the current vaccines.
CONFLICT OF INTEREST
The authors report no conflicts of interest.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
This work was supported by the Soonchunhyang University Research Fund to DC and McGill Interdisciplinary Initiative in Infection and Immunity (MI4) Emergency Research grant to DC, MD and JR. D‐K.K. and F.P.R. were supported bya Canadian Institutes for Health Research Foundation Grant and by the Thistledown Foundation. FRQS provided infrastructure support.
Choi, D. , Khan, N. , Montermini, L. , Tawil, N. , Meehan, B. , Kim, D.‐K. , Roth, F. P. , Divangahi, M. , & Rak, J. (2022). Quantitative proteomics and biological activity of extracellular vesicles engineered to express SARS‐CoV‐2 spike protein. Journal of Extracellular Biology, 1, e58. 10.1002/jex2.58
Dongsic Choi, Nargis Khan, and Laura Montermini equally contributed to this work.
Contributor Information
Dongsic Choi, Email: dongsicchoi@gmail.com.
Janusz Rak, Email: janusz.rak@mcgill.ca.
REFERENCES
- Baden, L. R. , El Sahly, H. M. , Essink, B. , Kotloff, K. , Frey, S. , Novak, R. , Diemert, D. , Spector, S. A. , Rouphael, N. , Creech, C. B. , McGettigan, J. , Khetan, S. , Segall, N. , Solis, J. , Brosz, A. , Fierro, C. , Schwartz, H. , Neuzil, K. , Corey, L. , … Group, C. S. (2021). Efficacy and safety of the mRNA‐1273 SARS‐CoV‐2 vaccine. New England Journal of Medicine, 384(5), 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti, M. F. , Zhang, Z. , Mortier, E. , Melchior, A. , Degeest, G. , Geeraerts, A. , Ivarsson, Y. , Depoortere, F. , Coomans, C. , Vermeiren, E. , Zimmermann, P. , & David, G. (2012). Syndecan‐syntenin‐ALIX regulates the biogenesis of exosomes. Nature Cell Biology, 14(7), 677–685. [DOI] [PubMed] [Google Scholar]
- Bansal, S. , Perincheri, S. , Fleming, T. , Poulson, C. , Tiffany, B. , Bremner, R. M. , Mohanakumar, T. , & Edge, C. (2021). Circulating exosomes with COVID spike protein are induced by BNT162b2 (Pfizer‐BioNTech) vaccination prior to development of antibodies: A novel mechanism for immune activation by mRNA vaccines. Journal of Immunology, 207(10), 2405–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, D. , Montermini, L. , Jeong, H. , Sharma, S. , Meehan, B. , & Rak, J. (2019). Mapping subpopulations of cancer cell‐derived extracellular vesicles and particles by nano‐flow cytometry. ACS Nano, 13(9), 10499–10511. [DOI] [PubMed] [Google Scholar]
- Choi, D. , Montermini, L. , Kim, D. K. , Meehan, B. , Roth, F. P. , & Rak, J. (2018). The impact of oncogenic EGFRvIII on the proteome of extracellular vesicles released from glioblastoma cells. Molecular & Cellular Proteomics, 17(10), 1948–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, D. , Montermini, L. , Meehan, B. , Lazaris, A. , Metrakos, P. , & Rak, J. (2021). Oncogenic RAS drives the CRAF‐dependent extracellular vesicle uptake mechanism coupled with metastasis. Journal of Extracellular Vesicles, 10(8), e12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, D. , Spinelli, C. , Montermini, L. , & Rak, J. (2019). Oncogenic Regulation of Extracellular Vesicle Proteome and Heterogeneity. Proteomics, 19(1‐2), e1800169. [DOI] [PubMed] [Google Scholar]
- Choi, D. S. , Kim, D. K. , Kim, Y. K. , & Gho, Y. S. (2013). Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics, 13(10‐11), 1554–1571. [DOI] [PubMed] [Google Scholar]
- Cocozza, F. , Névo, N. , Piovesana, E. , Lahaye, X. , Buchrieser, J. , Schwartz, O. , Manel, N. , Tkach, M. , Théry, C. , & Martin‐Jaular, L. (2020). Extracellular vesicles containing ACE2 efficiently prevent infection by SARS‐CoV‐2 Spike protein‐containing virus. Journal of Extracellular Vesicles, 10(2), e12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Shennawy, L. , Hoffmann, A. D. , Dashzeveg, N. K. , McAndrews, K. M. , Mehl, P. J. , Cornish, D. , Yu, Z. , Tokars, V. L. , Nicolaescu, V. , Tomatsidou, A. , Mao, C. , Felicelli, C. J. , Tsai, C. F. , Ostiguin, C. , Jia, Y. , Li, L. , Furlong, K. , Wysocki, J. , Luo, X. , … Liu, H. (2022). Circulating ACE2‐expressing extracellular vesicles block broad strains of SARS‐CoV‐2. Nature Communication, 13(1), 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falach, R. , Bar‐On, L. , Lazar, S. , Kadar, T. , Mazor, O. , Aftalion, M. , Gur, D. , Evgy, Y. , Shifman, O. , Aminov, T. , Israeli, O. , Cohen‐Gihon, I. , Zaide, G. , Gutman, H. , Vagima, Y. , Makdasi, E. , Stein, D. , Rosenfeld, R. , Alcalay, R. , … Sabo, T. (2021). Mice with induced pulmonary morbidities display severe lung inflammation and mortality following exposure to SARS‐CoV‐2. JCI Insight, 6(12), e145916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, S. J. , Booth, A. M. , & Hildreth, J. E. (2003). The Trojan exosome hypothesis. Proceedings of the National Academy of Sciences of the United States of America, 100(19), 10592–10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, S. , Yu, H. , Yan, G. , Gao, M. , Sun, W. , & Zhang, X. (2020). Characterization of urinary exosomes purified with size exclusion chromatography and ultracentrifugation. Journal of Proteome Research, 19(6), 2217–2225. [DOI] [PubMed] [Google Scholar]
- Heinz, F. X. , & Stiasny, K. (2021). Distinguishing features of current COVID‐19 vaccines: Knowns and unknowns of antigen presentation and modes of action. NPJ Vaccines, 6(1), 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M. , Kleine‐Weber, H. , Schroeder, S. , Krüger, N. , Herrler, T. , Erichsen, S. , Schiergens, T. S. , Herrler, G. , Wu, N. H. , Nitsche, A. , Müller, M. A. , Drosten, C. , & Pöhlmann, S. (2020). SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, C. , Hardee, D. , & Minnear, F. (2007). Membrane fusion by VAMP3 and plasma membrane t‐SNAREs. Experimental Cell Research, 313(15), 3198–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. C. , Lai, Y. J. , Liao, C. C. , Yang, W. F. , Huang, K. B. , Lee, I. J. , Chou, W. C. , Wang, S. H. , Wang, L. H. , Hsu, J. M. , Sun, C. P. , Kuo, C. T. , Wang, J. , Hsiao, T. C. , Yang, P. J. , Lee, T. A. , Huang, W. , Li, F. A. , Shen, C. Y. , … Li, C. W. (2021). Targeting conserved N‐glycosylation blocks SARS‐CoV‐2 variant infection in vitro. EBioMedicine, 74, 103712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. J. , Zhao, H. , Huang, X. , Deng, Y. Q. , Li, X. F. , Ye, Q. , Li, R. T. , Xu, Y. P. , Cao, T. S. , & Qin, C. F. (2021). Identification of oligosaccharyltransferase as a host target for inhibition of SARS‐CoV‐2 and its variants. Cell Discovery, 7(1), 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, C. B. , Farzan, M. , Chen, B. , & Choe, H. (2022). Mechanisms of SARS‐CoV‐2 entry into cells. Nature Reviews Molecular Cell Biology, 23(1), 3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Fenix, A. M. , Franklin, J. L. , Higginbotham, J. N. , Zhang, Q. , Zimmerman, L. J. , Liebler, D. C. , Ping, J. , Liu, Q. , Evans, R. , Fissell, W. H. , Patton, J. G. , Rome, L. H. , Burnette, D. T. , & Coffey, R. J. (2019). Reassessment of exosome composition. Cell, 177(2), 428–445.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra, H. , Simpson, R. J. , Ji, H. , Aikawa, E. , Altevogt, P. , Askenase, P. , Bond, V. C. , Borras, F. E. , Breakefield, X. , Budnik, V. , Buzas, E. , Camussi, G. , Clayton, A. , Cocucci, E. , Falcon‐Perez, J. M. , Gabrielsson, S. , Gho, Y. S. , Gupta, D. , Harsha, H. C. , … Mathivanan, S. (2012). Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biology, 10(12), e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, Z. , Oton, J. , Qu, K. , Cortese, M. , Zila, V. , McKeane, L. , Nakane, T. , Zivanov, J. , Neufeldt, C. J. , Cerikan, B. , Lu, J. M. , Peukes, J. , Xiong, X. , Krausslich, H. G. , Scheres, S. H. W. , Bartenschlager, R. , & Briggs, J. A. G. (2020). Structures and distributions of SARS‐CoV‐2 spike proteins on intact virions. Nature, 588(7838), 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. K. , Knapp, J. J. , Kuang, D. , Chawla, A. , Cassonnet, P. , Lee, H. , Sheykhkarimli, D. , Samavarchi‐Tehrani, P. , Abdouni, H. , Rayhan, A. , Li, R. , Pogoutse, O. , Coyaud, E. , van der Werf, S. , Demeret, C. , Gingras, A. C. , Taipale, M. , Raught, B. , Jacob, Y. , & Roth, F. P. (2020). A comprehensive, flexible collection of SARS‐CoV‐2 coding regions. G3 (Bethesda), 10(9), 3399–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. K. , Cho, J. , Kim, E. , Kim, J. , Yang, J. S. , Kim, K. C. , Lee, J. Y. , Shin, Y. , Palomera, L. F. , Park, J. , Baek, S. H. , Bae, H. G. , Cho, Y. , Han, J. , Sul, J. H. , Lee, J. , Park, J. H. , Cho, Y. W. , Lee, W. , & Jo, D. G. (2022). Engineered small extracellular vesicles displaying ACE2 variants on the surface protect against SARS‐CoV‐2 infection. Journal of Extracellular Vesicles, 11(1), e12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupka, N. , Strappe, P. , Gotz, J. , & Ittner, L. M. (2010). Gateway‐compatible lentiviral transfer vectors for ubiquitin promoter driven expression of fluorescent fusion proteins. Plasmid, 63(3), 155–160. [DOI] [PubMed] [Google Scholar]
- Lan, J. , Ge, J. , Yu, J. , Shan, S. , Zhou, H. , Fan, S. , Zhang, Q. , Shi, X. , Wang, Q. , Zhang, L. , & Wang, X. (2020). Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature, 581(7807), 215–220. [DOI] [PubMed] [Google Scholar]
- Lázaro‐Ibáñez, E. , Faruqu, F. N. , Saleh, A. F. , Silva, A. M. , Tzu‐Wen Wang, J. , Rak, J. , Al‐Jamal, K. T. , & Dekker, N. (2021). Selection of fluorescent, bioluminescent, and radioactive tracers to accurately reflect extracellular vesicle biodistribution in vivo. ACS Nano, 15(2), 3212–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K. , Shen, Y. , Miller, M. A. , Stabenow, J. , Williams, R. W. , & Lu, L. (2021). Differing susceptibility of C57BL/6J and DBA/2J mice‐parents of the murine BXD family, to severe acute respiratory syndrome coronavirus infection. Cell & Bioscience, 11(1), 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotvall, J. , Hill, A. F. , Hochberg, F. , Buzas, E. I. , Di Vizio, D. , Gardiner, C. , Gho, Y. S. , Kurochkin, I. V. , Mathivanan, S. , Quesenberry, P. , Sahoo, S. , Tahara, H. , Wauben, M. H. , Witwer, K. W. , & Thery, C. (2014). Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. Journal of Extracellular Vesicles, 3, 26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck, K. , Kim, D. K. , Lambourne, L. , Spirohn, K. , Begg, B. E. , Bian, W. , Brignall, R. , Cafarelli, T. , Campos‐Laborie, F. J. , Charloteaux, B. , Choi, D. , Coté, A. G. , Daley, M. , Deimling, S. , Desbuleux, A. , Dricot, A. , Gebbia, M. , Hardy, M. F. , Kishore, N. , … Calderwood, M. A. (2020). A reference map of the human binary protein interactome. Nature, 580(7803), 402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy, L. A. , Pink, R. C. , & Carter, D. R. (2014). Routes and mechanisms of extracellular vesicle uptake. Journal of Extracellular Vesicles, 3, 10. 10.3402/jev.v3.24641 eCollection;%2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte‐’t Hoen, E. , Cremer, T. , Gallo, R. C. , & Margolis, L. B. (2016). Extracellular vesicles and viruses: Are they close relatives? Proceedings of the National Academy of Sciences of the United States of America, 113(33), 9155–9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oughtred, R. , Stark, C. , Breitkreutz, B. J. , Rust, J. , Boucher, L. , Chang, C. , Kolas, N. , O'Donnell, L. , Leung, G. , McAdam, R. , Zhang, F. , Dolma, S. , Willems, A. , Coulombe‐Huntington, J. , Chatr‐Aryamontri, A. , Dolinski, K. , & Tyers, M. (2019). The BioGRID interaction database: 2019 update. Nucleic Acids Research, 47(D1), D529–D541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisitkun, T. , Shen, R. F. , & Knepper, M. A. (2004). Identification and proteomic profiling of exosomes in human urine. Proceedings of the National Academy of Sciences of the United States of America, 101(36), 13368–13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorey, J. S. , Cheng, Y. , Singh, P. P. , & Smith, V. L. (2015). Exosomes and other extracellular vesicles in host‐pathogen interactions. EMBO Reports, 16(1), 24–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P. , Markiel, A. , Ozier, O. , Baliga, N. S. , Wang, J. T. , Ramage, D. , Amin, N. , Schwikowski, B. , & Ideker, T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13(11), 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko, A. , Tomas, H. , Havlis, J. , Olsen, J. V. , & Mann, M. (2006). In‐gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols, 1(6), 2856–2860. [DOI] [PubMed] [Google Scholar]
- Shirbhate, E. , Pandey, J. , Patel, V. K. , Kamal, M. , Jawaid, T. , Gorain, B. , Kesharwani, P. , & Rajak, H. (2021). Understanding the role of ACE‐2 receptor in pathogenesis of COVID‐19 disease: A potential approach for therapeutic intervention. Pharmacological Reports : PR, 73(6), 1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, C. P. , Jan, J. T. , Wang, I. H. , Ma, H. H. , Ko, H. Y. , Wu, P. Y. , Kuo, T. J. , Liao, H. N. , Lan, Y. H. , Sie, Z. L. , Chen, Y. H. , Ko, Y. A. , Liao, C. C. , Chen, L. Y. , Lee, I. J. , Tsung, S. I. , Lai, Y. J. , Chiang, M. T. , Liang, J. J. , … Tao, M. H. (2021). Rapid generation of mouse model for emerging infectious disease with the case of severe COVID‐19. PLoS Pathogens, 17(8), e1009758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo, S. P. (2021). Review of COVID‐19 mRNA vaccines: BNT162b2 and mRNA‐1273. Journal of Pharmacy Practice, 8971900211009650. [DOI] [PubMed] [Google Scholar]
- Thompson, E. A. , Cascino, K. , Ordonez, A. A. , Zhou, W. , Vaghasia, A. , Hamacher‐Brady, A. , Brady, N. R. , Sun, I. H. , Wang, R. , Rosenberg, A. Z. , Delannoy, M. , Rothman, R. , Fenstermacher, K. , Sauer, L. , Shaw‐Saliba, K. , Bloch, E. M. , Redd, A. D. , Tobian, A. A. R. , Horton, M. , … Powell, J. D. (2021). Metabolic programs define dysfunctional immune responses in severe COVID‐19 patients. Cell Reports, 34(11), 108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer, Z. , Alhusaini, N. , Tabler, C. O. , Sweet, T. , de Carvalho, K. I. L. , Schlatzer, D. M. , Carias, L. , King, C. L. , Matreyek, K. , & Tilton, J. C. (2021). Extracellular vesicles carry SARS‐CoV‐2 spike protein and serve as decoys for neutralizing antibodies. Journal of Extracellular Vesicles, 10(8), e12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel, G. , D'Angelo, G. , & Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology, 19(4), 213–228. [DOI] [PubMed] [Google Scholar]
- Verdecchia, P. , Cavallini, C. , Spanevello, A. , & Angeli, F. (2020). The pivotal link between ACE2 deficiency and SARS‐CoV‐2 infection. European Journal of Internal Medicine, 76, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y. , Allen, J. D. , Wrapp, D. , McLellan, J. S. , & Crispin, M. (2020). Site‐specific glycan analysis of the SARS‐CoV‐2 spike. Science (New York, N.Y.), 369(6501), 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, R. , Zhao, L. , Kong, G. , Liu, X. , Zhu, S. , Zhang, S. , & Min, L. (2020). Combination of size‐exclusion chromatography and ultracentrifugation improves the proteomic profiling of plasma‐derived small extracellular vesicles. Biological Procedures Online, 22, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich, D. M. , Sivapalasingam, S. , Norton, T. , Ali, S. , Gao, H. , Bhore, R. , Musser, B. J. , Soo, Y. , Rofail, D. , Im, J. , Perry, C. , Pan, C. , Hosain, R. , Mahmood, A. , Davis, J. D. , Turner, K. C. , Hooper, A. T. , Hamilton, J. D. , Baum, A. , … Yancopoulos, G. D. (2021). REGN‐COV2, a neutralizing antibody cocktail, in outpatients with COVID‐19. New England Journal of Medicine, 384(3), 238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, E. S. , Bailey, A. L. , Kafai, N. M. , Nair, S. , McCune, B. T. , Yu, J. , Fox, J. M. , Chen, R. E. , Earnest, J. T. , Keeler, S. P. , Ritter, J. H. , Kang, L. I. , Dort, S. , Robichaud, A. , Head, R. , Holtzman, M. J. , & Diamond, M. S. (2020). SARS‐CoV‐2 infection of human ACE2‐transgenic mice causes severe lung inflammation and impaired function. Nature Immunology, 21(11), 1327–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Jeppesen, D. K. , Higginbotham, J. N. , Franklin, J. L. , Crowe, J. E. Jr. , & Coffey, R. J. (2021). Angiotensin‐converting enzyme 2‐containing small extracellular vesicles and exomeres bind the severe acute respiratory syndrome coronavirus 2 spike protein. Gastroenterology, 160(3), 958–961.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Zhang, W. , Kho, Y. , & Zhao, Y. (2004). Proteomic analysis of integral plasma membrane proteins. Analytical Chemistry, 76(7), 1817–1823. [DOI] [PubMed] [Google Scholar]
- Zinchuk, V. , Wu, Y. , & Grossenbacher‐Zinchuk, O. (2013). Bridging the gap between qualitative and quantitative colocalization results in fluorescence microscopy studies. Science Reports, 3, 1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.


