Abstract
The novel coronavirus disease (COVID‐19) outbreak that emerged at the end of 2019 has now swept the world for more than 2 years, causing immeasurable damage to the lives and economies of the world. It has drawn so much attention to discovering how the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) originated and entered the human body. The current argument revolves around two contradictory theories: a scenario of laboratory spillover events and human contact with zoonotic diseases. Here, we reviewed the transmission, pathogenesis, possible hosts, as well as the genome and protein structure of SARS‐CoV‐2, which play key roles in the COVID‐19 pandemic. We believe the coronavirus was originally transmitted to human by animals rather than by a laboratory leak. However, there still needs more investigations to determine the source of the pandemic. Understanding how COVID‐19 emerged is vital to developing global strategies for mitigating future outbreaks.
Keywords: genome similarities, laboratory origin, protein structure evolution, SARS‐CoV‐2, zoonotic origin
1. INTRODUCTION
The most widespread coronavirus to date was discovered in Wuhan, China in November 2019, and the disease it causes is named coronavirus disease (COVID‐19). The disease is a class of epidemics with human‐to‐human transmission caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) (Kumar et al., 2021; Li et al., 2021; Rosenthal et al., 2022). The high infectivity and pathogenicity of the virus was discovered at the beginning of the epidemic, and it has been trending worldwide. The World Health Organization (WHO) subsequently declared the disease a global pandemic. Because the public awareness on the virus prevention was initially bewildered, SARS‐CoV‐2 had a great opportunity to spread and mutate around the globe (Davis et al., 2021; Hu et al., 2020; Kumar et al., 2021). Globally, there was considerable uncertainty regarding when SARS‐CoV‐2 was introduced and began spreading locally (Davis et al., 2021).
In this global pandemic of SARS‐CoV‐2, five mainly mutated strains have emerged, namely alpha, beta, gamma, delta and omicron (Dhar et al., 2021; Gu et al., 2022; Hart et al., 2022; Ozer et al., 2022; Viana et al., 2022; Vohringer et al., 2021). In July 2020, the first discovered spike protein (S protein) mutation D614G swept the world. Two months later, the variant alpha strain was discovered in the United Kingdom (Davies et al., 2021; Hart et al., 2022). While beta variant was discovered in December 2020 in South Africa (Viana et al., 2022), gamma variant in January 2021 in Brazil, delta variant in the United Kingdom in March 2021 (Dhar et al., 2021; Vohringer et al., 2021), and omicron variant in November 2021 in the Botswana were discovered (Gu et al., 2022). By 10 July 2022, 551 million cases had been confirmed in more than 194 countries, with more than 6 million fatalities and the daily infections continue to rise rapidly (Figure 1).
FIGURE 1.
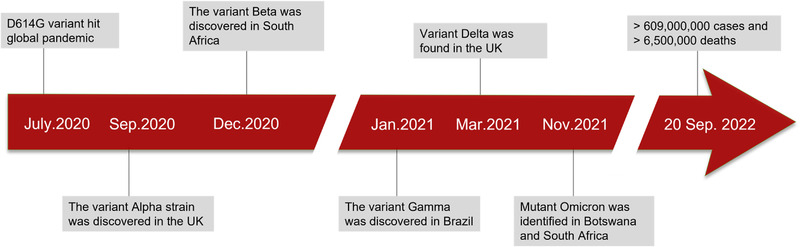
Timeline of the emergence of SARS‐CoV‐2 mutants. D614G is the first determined variant that is prevalent in the world. In the past 2 years of the pandemic, approximately 551 million cases have been confirmed to date, with over 6 million deaths worldwide.
Some pathogenic mechanisms of COVID‐19 have been demonstrated. SARS‐CoV‐2 combines with angiotensin‐converting enzyme‐2 (ACE2), a cellular receptor, to facilitate its access into the target cell. ACE2 receptors are extensively expressed in oral epithelial cells. The mean expression levels of ACE2 in the tongue are higher compared to other oral tissues, and are also more abundant in the minor salivary glands than in the lungs (Ferrer et al., 2021; Wang et al., 2022). These evidences strongly suggest that the oral cavity, especially the saliva, may pose a significant risk of transmission of SARS‐CoV‐2 (Wang et al., 2022). In the process of human resistance to the virus, it was found that patients' body reaction in the process of virus invasion has the following phases: asymptomatic stage, incursion and infection of the upper respiratory tract, involvement of the lower airways, development of acute respiratory distress syndrome (ARDS), and virus transmission and clinical manifestations (Parasher, 2021). In the air transmission path, respiratory aerosols carrying SARS‐CoV‐2 attach to the nasal epithelial cells of the upper respiratory tract (Parasher, 2021). It is possible to generate an immune response through this stage, but its strength is limited. Although the individuals have a low viral load, they are highly contagious. The next step is the immigration of virus from the nasal epithelium to the upper respiratory tract via respiration. The disease presents with symptoms of weariness, dry cough, fever, and body aches due to the upper respiratory tract involvement. A more intense immune response is evident at this stage than at the previous stages (Wang et al., 2020). About one in five infected patients will progress to the ARDS stage with severe symptoms (Kikkenborg Berg et al., 2022). The virus infects type 2 alveolar epithelial cells via ACE2, and begins to replicate to produce more viral nucleocapsids. Sustained damage from massive inflammatory cells and viral replication leads to loss of type 1 and 2 pneumocytes with diffuse alveolar damage, culminating in acute respiratory distress syndrome (Bain & MacDonald, 2022; Borges et al., 2022). SARS‐CoV‐2 can cause some common symptoms including fever, fatigue, cough, anosmia, hypogeusia, headaches and myalgia, just like the symptoms of classic coronaviruses infection (D'Onofrio et al., 2022; Parasher, 2021). Following the above primary stage of the disease, the lower respiratory tract will be attacked, which can progress to severe respiratory disease, resulting in respiratory failure, multi‐organ failure, and even loss of life (Barizien et al., 2021; D'Onofrio et al., 2022).
Since the advent of SARS‐CoV‐2, two competing ideas have emerged: one claimed that the pathogen escaped from a laboratory, while the other believed that the virus arose from a zoonotic outbreak. Here, we reviewed the evidence of current scientific research that may be helpful in clarifying the origin of SARS‐CoV‐2.
2. CARDING THE CLUES OF SARS‐COV‐2 ORIGIN
2.1. The development history of coronavirus
Coronavirus (CoV) has long been regarded as a virus with a high hazard of spreading. SARS‐CoV‐2, the ninth human‐infecting coronavirus on record, and the seventh coronavirus identified in the past 20 years, has become an unprecedented epidemic (Lednicky et al., 2021; Vlasova et al., 2022). Most human virus strains are zoonotic in origin, including all previous human coronavirus strains (Holmes et al., 2021). Animal CoVs, such as bovine coronavirus (BCoV), porcine transmissible gastroenteritis virus (TGEV), infectious bronchitis virus (IBV), and feline infectious peritonitis virus (FIPV), have been known since the late 1930s (Saif, 2004). Recent studies have linked the evolution of human CoVs (HCoVs) to a high level of urbanization and poultry breeding, which facilitates the exchange of species and simplify crossing of species barriers, as well as genomic reorganization of these viruses (Jones et al., 2013). Bats are a large‐capacity ‘reservoir’ of CoVs, virus sampling by the China Ecological Health Alliance alone found around 400 new coronaviruses strains (Olival, Weekley, & Daszak, 2015; Xie et al., 2018). In addition, bats' unique immune system enables them to deal with several viruses more effectively than human beings (Kirtipal, Bharadwaj, & Kang, 2020). Furthermore, the presence of a variety of zoonotic alphaCoV and betaCoV has been detected in Western Europe's bat populations (Wacharapluesadee et al., 2015). Although definitive spread of coronaviruses from bats to humans has not been fully identified, direct contact with intermediate hosts to humans is widely considered the most likely mode of spread (Hart et al., 2022; Kirtipal et al., 2020; Z. Zhu, Meng, & Meng, 2020).
The first cases of SARS‐CoV appeared in Foshan, Guangdong province, China in November 2002 (Ge, Hu, & Shi, 2015). It eventually became a SARS epidemic, affecting 28 countries around the world, with 8096 cases and 774 deaths (Ge et al., 2015). After a series of investigations, it was found that the SARS‐CoV virus first existed in masked civet cats and raccoon dogs. Antibodies to the virus in a badger were later found at a China's Shenzhen live animal market, and were suspected to be the source of the human infection (Drexler, Corman, & Drosten, 2014; Guan et al., 2003; Song et al., 2019). Additionally, Chinese rhinolophid bats were also found to harbour genetically differentiated CoVs related to SARS‐CoV (Rhinolophus spp.), suggesting that these animals are hosts for this novel HCoV (Figure 2) (Lau et al., 2005; Li et al., 2005).
FIGURE 2.

Schematic diagram of the transmission process of three HCoVs. Humans acquired SARS‐CoV and MERS‐CoV from bats through civet cats and dromedary camels, respectively. It is unclear how SARS‐CoV‐2 spread to humans.
Likewise, in Jeddah, Saudi Arabia, human infection with MERS‐CoV was first recorded in June 2012 (Ge et al., 2015). In November 2019, a total of 2494 MERS‐CoV cases were detected in 27 countries, with 858 fatalities (WHO, 2020). Beginning, bats were thought to be MERS‐CoV hosts. However, researchers found a significant prevalence of MERS‐CoV‐neutralizing antibodies in camels from Oman and the Canary Islands (Kirtipal et al., 2020; Reusken et al., 2013). Serological studies suggest that dromedary camels were carrying MERS‐CoV‐like viruses in East Africa, North Africa and the Middle East as early as 1983. In addition, in Saudi Arabia, dromedary camels have been found to have multiple viral genetic lineages (Sabir et al., 2016), involving crossing various barriers and causing outbreaks in humans. Collectively, these evidences strongly suggest that dromedary camels are an important reservoir for MERS‐CoV; viruses originating from bats have been predicted to enter dromedary camels more than 30 years ago, based on laboratory evidence (Figure 2) (Müller et al., 2014).
Recent SARS‐CoV‐2 development gives prominence to the potential for deadly viruses hidden in wild zoonotic reservoirs and the threat of zoonotic spillovers (Malik et al., 2020). With the help of sequencing technology, bats were again assessed as a natural host for SARS‐CoV‐2, because it shares 96% of its genome with two SARS‐like CoVs from bats, bat‐SLCoVZX45 and bat‐SL‐CoVZX2 (Xu et al., 2020; Zhou et al., 2020). Comparatively, SARS‐CoV‐2 has a lower similarity genome to SARS‐CoV (∼79%) or MERS‐CoV (∼50%). Until later, pangolins were considered a possible intermediate host for SARS‐CoV‐2 because it shared 99% genetic similarity with the CoV in pangolins (Yi, Lagniton, Ye, Li, & Xu, 2020). However, the 1% difference between the two genome sequences is considered significantly different. Therefore, the conclusive specific evidence is still under investigation (Yi et al., 2020).
Looking back, we see clear similarities between SARS‐CoV‐2 and SARS‐CoV. Both occurrences of the SARS‐CoVs have been linked to markets selling live wild animals and related species, especially civet cats and pangolin animal traders who worked in 2003 but were not diagnosed with SARS, while had high levels of SARS‐CoV immunoglobulin G (IgG) found in their serum (13% overall, over 50% of traders specializing in civet cats). Further serological surveys found that Yunnan residents living near the bat caves were 3% positive for SARS‐related coronavirus (SARSr‐CoV), indicating that spillover events may have occurred in zoonotic CoVs. This often occurs when bats are important hubs. As to the epidemic of coronaviruses, Yunnan may be considered the most possible location of a spillover event because of the high potentiality of human contact to bat. However, the places of the first reported cases, either SARS‐CoV (Foshan, Guangdong Province) or SARS‐CoV‐2 (Wuhan, Hubei Province), are far from Yunnan in terms of geographic disparity. This highlights the difficulty of determining the exact route by which the virus emerged. In addition, enough attention needs to be paid to sampling in areas other than Yunnan (Holmes et al., 2021).
According to epidemiological studies, the Huanan Market in Wuhan was the early and main epicentre of SARS‐CoV‐2 infection. Two of the three earliest recorded cases of COVID‐19 were directly related to the sale of wildlife at the market, and 28% of all cases reported in December 2019 were directly related to the market (WHO, 2021). During December 2019, about 55% of the cases had contact with other markets in Wuhan, and these cases were more concentrated in the fifteen days of early December (WHO, 2021). Early cases were predominantly located near the Huanan Market, according to an examination of the locations, which provided the basis for the identification of the outbreak site. These regions were also the first to experience excess deaths from pneumonia in January 2020, a measure less sensitive to reporting bias (Holmes et al., 2021). There may be cases in the early stages of the epidemic that do not appear to have a direct link to the market, possibly due to higher occult transmission rates and undiscovered secondary transmissions, and a similar situation was shown in early SARS‐CoV cases (Xu et al., 2004). However, whether the first infection case in the Huanan Market is the real first case of SARS‐CoV‐2 infection in the world depends on extensive investigations in other regions and countries.
2.2. Discussion on laboratory‐derived SARS‐COV‐2
In general, the storage and handling of biological products in biological laboratories follow strict management measures, and personnel involved in the experiments have also undergone professional training, so it is difficult to cause laboratory infection. If the relevant experiments carried out in the laboratory do not meet the corresponding laboratory safety level, the researchers engaged in irregular operations, and the waste disposal is improper, the risk of laboratory leakage will be increased. Although rare, laboratory accidents do occur, with bat coronaviruses being used in different laboratories around the world. There has indeed been precedent for events leading to sporadic infections and short chains of transmission in past laboratory studies (Senior, 2003). With the exception of Marburg virus, all known laboratory escape viruses are easily identifiable viruses that can infect humans. Those escape events were associated with persistently high titre cultures (Geddes, 2006; Lim et al., 2004; Senior, 2003). There is only one documented instance of epidemic or pandemic resulting from a wide‐ranging vaccine test: the A/H1N1 influenza pandemic in 1977 (Rozo & Gronvall, 2015). Early in the COVID‐19 pandemic, the Wuhan Institute of Virology (WIV) became the target of widespread criticism of the laboratory origin hypothesis. However, there is no indication that the Wuhan Institute of Virology had studied viruses similar to SARS‐CoV‐2 or artificially modified coronaviruses before the COVID‐19 pandemic (Holmes et al., 2021).
Prior to December 2019, no virus highly consistent to SARS‐CoV‐2 had been recorded by any laboratory, and no genome was available to provide the SARS‐CoV‐2 genome. A known laboratory outbreak has been confirmed to be associated with the index case's workplace, household contacts, and the laboratory of origin (Geddes, 2006; Lim et al., 2004; Ristanović, Kokoškov, Crozier, Kuhn, & Gligić, 2020; Senior, 2003). The Wuhan‐based Institute of Biology, all three laboratories working on CoV diagnostics, isolation of CoV and vaccine development are managed well and have high biosafety standards (BSL3 or 4) and the researchers have health surveillance program. Study shows that SARS‐CoV‐2 was not tested positive in workers despite extensive epidemiological tracing of early cases (WHO, 2021). At the time of testing in March 2020, the laboratory was reported to have followed proper biosafety protocols during coronavirus research.
Investigations into the WIV indicate that WIV is not engaged in any research related to SARS‐CoV‐2. Although WIV possesses a large collection of SARSr‐CoVs samples form bats (Latinne et al., 2020) and has cultured three of them (WIV1, WIV16 and Rs4874), but these three viral lineages are more related to SARS‐CoV (Ge et al., 2013; Ristanović et al., 2020; Yang et al., 2015). Additionally, the RaTG13 virus from WIV was never further experimentally manipulated and exists only as a nucleotide sequence (Cohen, 2020). There is no open report or study showing that WIV used other ideas, involving the development of new reverse genetics systems for SARSr‐CoV transmission based on bats sequence data. Researches that obtain predicted function will use the genomic backbone of SARSr‐CoV, or at least viruses formerly identified by sequencing (Holmes et al., 2021). However, Research on recombinant coronaviruses in WIV previously used a genetic framework unrelated to SARS‐CoV‐2 (B. Hu et al., 2017), which produced no evidence of laboratory‐derived markers. In any case of lab accident, the escaped virus must have left clues in the scientific establishment before a pandemic, but support for this idea has not yet been found, and no sequences have been identified that could serve as precursors.
Assuming that the escape event occurred in a laboratory, the accidental infection of SARSr‐CoV could occur during the continuous transmission among animals commonly used in laboratories. Nevertheless, some early isolates of SARS‐CoV‐2 failed to infect wild‐type mice (Wan, Shang, Graham, Baric, & Li, 2020). Human ACE2 (hACE2) transgenic mice can be used for studying infections and testing vaccines in vivo. However, the mice usually develop atypical disease when exposed to SARS‐CoV‐2 (Bao et al., 2020; Hassan et al., 2020; Israelow et al., 2020; Rathnasinghe et al., 2020; Sun et al., 2020). As a result of serial passage in Vero E6 cells of three viruses isolated from faeces, the SARS‐CoV‐2‐specific furin cleavage site consistently disappeared (Davidson et al., 2020; Klimstra et al., 2020; Liu, Zheng, et al., 2020; Ogando et al., 2020; Sasaki et al., 2021; Wong et al., 2021; Zhu et al., 2021). Therefore, these techniques are highly unlikely to isolate SARS‐CoV‐2 progenitors with intact furin cleavage sites. These studies suggest that it is less likely that the virus has increased affinity for humans through serial passage in susceptible animals (Holmes et al., 2021). However, it cannot be ignored that there are some non‐common animals in the laboratory, such as hamsters, dogs, monkeys, etc. Their ACE2 receptors have been shown to be able to bind to the S protein of SARS‐CoV‐2 (primary) with moderate to high strength and are susceptible to infection (Piplani, Singh, Winkler, & Petrovsky, 2021). This adds loophole to laboratory animal models for studying SARS‐CoV‐2 passaging. Of note, the susceptibility of non‐human species to SARS‐CoV‐2 varies widely. Piplani, Singh, Winkler, and Petrovsky (2021) tested the affinity of ACE2 to the spike protein in more than 10 animals and humans. Human ACE2 has the strongest affinity for S protein, followed by pangolin, and mouse is the weakest, which is consistent with its experimental observation (Piplani et al., 2021). This finding is surprising because typical zoonotic viruses exhibit high affinity for the original host and low affinity for the new host. As the virus gradually adapts to the new host, the affinity will gradually increase. This indicates the possible existence of an intermediate host whose ACE2 structure is very similar to that of humans, but this intermediate host is still very mysterious.
Among the variants of SARS‐CoV‐2, D614G was the first strong mutant to emerge. G614 greatly enhanced the transmissibility of the virus, rapid replacing of the D614 mutant was the dominant strain within months. Using lung, liver and intestinal cell lines, Daniloski et al. (2021) separately demonstrated the robust infectious capacity of G614 compared to D614. Although D614G produced strong infectivity, it was a mutation event in the non‐RBD region of S protein. Only two months later, the B.1.1.7 (also known as Alpha) variant carrying a mutation marked by N501Y rose rapidly. The strong infectivity of 501Y lineage depends on the deletion of the two amino acids Δ69/Δ70. The 501Y lineage Δ69/Δ70 deletion is about 60% different from 501N without deletion (Leung, Shum, Leung, Lam, & Wu, 2021). The advantage of B.1.1.7 was short‐lived. Within a few months of the appearance of Alpha, two other important mutations occurred in the RBD region of SARS‐CoV‐2, resulting in the beta strains (N501Y, E484K, K417N). The increase of E484K and K417N enables Beta to acquire the ability to avoid binding to neutralizing antibodies, that is, the ability to escape from immunity, thus quickly becoming a new dominant strain. In the newest Omicron lineage, SARS‐CoV‐2 tends to infect the upper respiratory tract rather than the lung, and with a high vaccination rate, thus its pathogenicity appears to be milder (Fan et al., 2022). Such changes are conducive to the coexistence of viruses and humans, which could be considered the result of highly adaptive evolution of viruses to human hosts.
In the evolution of the virus for more than 2 years, it is not difficult to see that SARS‐CoV‐2 has been undergoing adaptive changes with the human host, and the cycle of this change is often within a few months. Its increasing infectivity and immune evasion capabilities underscore the virus' excellent adaptability. If we backward reasoning this law, we should also have found some lineages with similar evolutionary characteristics to SARS‐CoV‐2‐Wuhan, but this assumption has not been proven so far. The failure to find an evolutionarily similar ancestor of SARS‐CoV‐2 is undoubtedly a major shock to the theory of the origin of zoonosis.
2.3. Likelihood of origin in other regions of the world
When it comes to the origin of this epidemic, Wuhan, China, must be the first place that comes to mind (Lytras, Xia, Hughes, Jiang, & Robertson, 2021; Singh & Yi, 2021). China was the first country to announce the outbreak, so the world's attention naturally turned to the country. Nonetheless, the place of first sharing must be the true birthplace? This is unlikely to be the case.
A retrospective survey found that sewage samples collected in Barcelona, Spain, on 12 March 2019, were positive for SARS‐CoV‐2 RNA, but other samples collected between January 2018 and December 2019 were negative. This indicates that at least as early as March 2019, SARS‐CoV‐2 may appear in other areas of the world (Chavarria‐Miró et al., 2021).
In France, Carrat et al. (2021) identified 353 participants who tested positive for anti‐SARS‐CoV‐2 IgG in the samples of 9144 adults with routine serum collection in a French general population (Carrat et al., 2021). Subsequently, the samples were further analysed using ELISA‐S and SN positive tests to screen 13 participants who were both ELISA‐S and SN positive. Through an epidemiological survey of participants (Carrat et al., 2021), it was found that some of them had suffered from unknown respiratory diseases in October 2019, and the others had no history of close contact and travel. Through comprehensive analysis, it can be deduced that the latent transmission of SARS‐CoV‐2 in Europe was much earlier than the time of reporting (Carrat et al., 2021; WHO, 2021), and those infected with no travel history may have acquired the infection through local transmission. Sewage testing by RT‐PCR for SARS‐CoV‐2 conducted in Brazil on 27 November 2019 was positive, much earlier than the first case reported in the Americas. In the United States, a serological survey of 7389 archived donated blood samples collected from 9 states over the period 13 December 2019 to 17 January 2020 detected 106 positive specimens (Basavaraju et al., 2021). The report also made many explanations for the possibility of ‘false positives’: on the one hand, ‘true positives’ can only be collected from individuals with positive molecular diagnostic tests or from pairs of acute convalescent sera with rising titres (Basavaraju et al., 2021); on the other hand, samples from individuals are not representative for all states. Furthermore, only a few positive cases have a history of outbound travel within 28 days (Basavaraju et al., 2021). This shows that SARS‐CoV‐2 may have existed locally before, and it is difficult to determine whether it was introduced from abroad. After all, it cannot be determined that Asia is the earliest origin of the virus according to the existing evidence; it is possible that some early modes of transmission, such as blood product transmission, are unknown.
It is also worth mentioning that, in the light of the US Centers for Disease Control and Prevention (CDC), there was an outbreak of ‘e‐cigarette pneumonia’ in the United States from mid‐2019 to the end of the year. As of April 2019, there were sporadic reports, followed by a sharp uptick in infections from June to September, with a peak in September and then a decline thereafter (CDC, 2021b). The CDC stopped collecting and updating ‘vaping pneumonia’ data on 25 February 2020. So far, there have been 2807 hospitalizations and 68 deaths, with a mortality rate of about 2.4% (CDC, 2021b). Sales of electronic cigarettes in the United States increased from 2014 to 2020, increasing by 122.2% during this period (i.e., from 7.7 million to 17.1 million every 4 weeks) (CDC, 2021a). Such a huge consumption of e‐cigarettes suddenly broke out in ‘e‐cigarette pneumonia’ in June 2019, and magically disappeared in February 2020, which is confusing.
In Italy, Apolone et al. (2021) collected blood samples from 959 participants in a prospective lung cancer screening trial from September 2019 to February 2020. To determine the probability of early infection in Italy, using proprietary SARS‐CoV‐2‐neutralizing antibodies from plasma sample banks, they looked into various epidemiological characteristics of exposure to SARS‐CoV‐2 in 959 asymptomatic individuals, including temporal, spatial, and distribution across different populations. With SARS‐CoV‐2 receptor binding protein specific ELISA, 111 (11.6%) samples were identified as positive. Of these, 4 samples collected in October, 1 sample collected in November, and 1 sample in February were also positive in qualitative microneutralization tests (Apolone et al., 2021). Overall, the 111 positive samples all showed SARS‐CoV‐2 RBD‐specific antibodies (IgM, IgG or both), and further analysis can assess the time of arrival and early prevalence of the virus. Analysis of anti‐SARS‐CoV‐2 antibody reactions in COVID‐19 patients revealed that almost 100% of patients seroconverted to antiviral immunoglobulin G (IgG) or immunoglobulin M (IgM) within 13 days of symptom onset.
SARS‐CoV‐2 virus infections most commonly result in the development of antibodies against the virus, except in immunocompromised patients. IgM is the first antibody detected in the body after infection, which can be produced within three days of infection and provide the first line of humoral immune defence. High‐affinity antibody IgG starts later than IgM, but it is the main force in long‐term immunity (Racine & Winslow, 2009). Data from Hou et al. (2020) showed that IgM was produced within 1 week of symptom onset in COVID‐19 patients, peaked within 2−3 weeks, and then decreased. IgG appeared later than IgM, but remained high for 2 months. Hence, detectable IgM and IgG antibodies can be used to assess serological routines during the disease progression, since the presence of IgM antibodies indicates recent pathogen invasion, whereas the presence of IgG antibodies indicates an earlier exposure to the virus (Hou et al., 2020).
Among the 162 samples collected in Italy in September 2019, three samples were positive for IgG antibodies and 20 were positive for IgM antibodies, and most of the positive samples were collected in early September. The small number of samples suggests that there may be a large number of positives undetected, and with the available evidence, the arrival of SARS‐CoV‐2 in Italy can be retrospected back to July or even earlier. However, given the speed of transmission, the number of hospitalizations and ICU treatment of patients, the initial arrival of the virus in Italy may manifest milder symptoms in patients, which may be related to different ethnic groups (Apolone et al., 2021). Then wave after wave of virus peaks appeared, speculating that the virus has achieved adaptive evolution in different regions. It was only between November and December 2019 that many general practitioners (GPs) began to report patients with atypical bilateral bronchitis and severe respiratory symptoms, mostly the elderly and infirm (Apolone et al., 2021). As the new coronavirus was unknown at the time, this phenomenon was attributed to invasive seasonal influenza. Furthermore, phylogenetic analysis of SARS‐CoV‐2 genomes from three Lombard patients who were involved in the first COVID‐19 outbreak revealed that these strains share the common origin that can be traced back to weeks before the first reported case of COVID‐19 pneumonia in China (Apolone et al., 2021).
Subsequently, the Italian Ministry of Health conducted a large‐scale SARS‐CoV‐2 seroprevalence study in a representative sample of 64,660 people from 25 May to 15 July in 2020. The result suggests an overall prevalence of 2.5%, with a peak of 7.5% in Lombardy, and 24% in the province of Bergamo in particular (Apolone et al., 2021). The Lombardy region has the highest rate of positive detections. After SARS‐CoV‐2 being confirmed, Lombardy was also the region worst hit by the pandemic in Italy. This seems to contradict the Wuhan Huanan Seafood Market‐related hypothesis of zoonotic origin. Apparently, there is no such raw poultry market in Lombardy to create direct bird‐human contact.
La Rosa et al. (2021) analysed 40 composite influent wastewater samples taken from 5 wastewater treatment plants between October 2019 and February 2020, and identified 15 positive samples using techniques such as RT‐PCR, which can be traced back to Milan and Turin on 18 December 2019. A child who was suspected of having measles had SARS‐CoV‐2 antibodies detected by PCR in early December in the same area (Amendola et al., 2021). Environmental studies have also shown that the virus may exist earlier in Italy.
3. EVIDENCE FROM THE GENOME AND PROTEIN STRUCTURE EVOLUTION OF SARS‐CoV‐2
3.1. Genome comparison of SARS‐CoV‐2 with other coronaviruses
The process of evolution is continuous, and genes are the imprints of this process. Whenever a new virus appears, scientists can always try to trace its source through genetic clues. The tracking methods of CoVs ancestors are based on a series of nucleotides sequence analysis in the coronavirus genome data set. For example, dinucleotide composition analysis, relative synonymous codon usage analysis (also known as RSCU), and maximum likelihood analysis (Kumar et al., 2021). Kumar et al. (2021) study suggests that direct transmission of SARS‐CoV‐2 from bats to humans without an intermediate host is unlikely. A SARS‐CoV‐2‐like coronavirus found in a dead Malayan pangolin shares 91.02% homology with SARS‐CoV‐2 in an analysis of coronavirus sequences in species worldwide over the past 2 years (Zhang, Wu, & Zhang, 2020); the nucleotide sequences of SARS‐CoV‐2‐related coronaviruses from two Rhinolophus shameli bats (RshSTT200 and RshSTT182), which were collected in Cambodia in 2010, shared 92.6% identity with the SARS‐CoV‐2 genome (Delaune et al., 2021); a bat‐derived coronavirus (the RmYN02 strain) shared 93.3% similarity with SARS‐CoV‐2 (Zhou et al., 2020). Based on these studies, it is obvious that only little difference of the coronaviruses genomes was found between regional species, but the similarity to the SRAS‐2 genome is relatively small. This phenomenon may indicate that viruses circulate frequently among different species in different regions, and the evolution of viruses is inseparable from this circulation. Further research indicated that a Horseshoe bat virus strain found in China's Yunnan Province has a genome similarity of 96.2% with SARS‐CoV‐2 (Zhou et al., 2020). Coincidentally, the later discovered that BANAL‐52 strain is 96.8% similar to SARS‐CoV‐2 (Tyshkovskiy & Panchin, 2021). These two strains are the most similar to SARS‐CoV‐2 reported to date, but the evidence to extrapolate from them is still very limited.
3.2. Plot the origin of SARS‐CoV‐2 based on proteins
SARS‐CoV‐2 is a kind of Sarbecovirus belonging to the Betacoronavirus genus of the coronavirus family (Wu et al., 2020). Compared with other typical human coronaviruses, SARS‐CoV‐2 has a single stranded RNA genome which encodes 4 structural proteins including spike (S), membrane (M), envelope (E) and nucleocapsid (N) (Kim et al., 2020); 16 non‐structural proteins (nsp1‐16); and 9 other cofactors (3a, 3b, 6, 7a, 7b, 8b, 9a, 9b and orf10) (Figure 3) (Al‐Qaaneh et al., 2021). Two open reading frames (ORFs) 1a and 1b at the 5' terminal of SARS‐CoV‐2 genome encode the polyproteins pp1a and pp1ab. Pp1a and pp1ab are hydrolysed into 16 non‐structural proteins (nsp1‐16) by two proteases (nsp3 and nsp5) encoded by the viral genome itself (Al‐Qaaneh et al., 2021). Nsp3 was named Papain‐like protease (PLpro) and nsp5 was referred to as 3‐chymotrypsin‐like main protease (3CL pro or Mpro), respectively (Klemm et al., 2020; Shin et al., 2020). These two proteases are shared by coronaviruses (Tan, Fung, Shen, Huang, & Liu, 2018). The 16 non‐structural proteins are involved in the replication, transcription, translation, and modification of viral genomes (Al‐Qaaneh et al., 2021). As indispensable proteins that regulate the coronavirus life cycle, the sequences of nsp3, nsp5 and nsp12 as well as N protein are highly conserved and therefore are also used for potential targets of multiple antiviral drugs (Anand et al., 2021). Nsp12 (RNA‐dependent‐RNA‐polymerase, RdRp) acts as a catalytic subunit of the replication transcription complex (RTC) (Bertolin et al., 2021; Chen et al., 2020; de et al., 2021). RdRp gene is a common gene necessary for the replication of all RNA viruses and is considered as a stable genetic marker that can be used for evolutionary analysis of RNA viruses worldwide because of its high conservation and universality (Holmes & Duchêne, 2019; Wolf et al., 2018, 2019).
FIGURE 3.
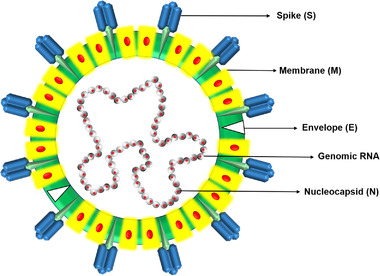
Schematic diagram of the typical structure of coronavirus (80–120 nm), showing different structural proteins of the virus, i.e., S, M, E and genomic RNA encapsulated within the granule by the N protein.
3.2.1. S protein
S protein, a type I transmembrane protein, can polymerize on the envelope surface of the virus to form a homotrimer (Papa et al., 2021). Two S proteins from the bat coronavirus, BANAL‐20‐52 and RaTG13, show 95% and 93.1% nucleotide sequence similarities with SARS‐CoV‐2, separately (Zhou et al., 2020). RaTG13 S protein has 98% identities at the protein sequence level (Wrapp et al., 2020), of which the RBD region has 89.3% amino acid sequence homology with SARS‐CoV‐2 (Boni et al., 2020; Wu et al., 2020; Zhou et al., 2020). The pangolin‐CoV‐2019 S protein sequence shares 97.5% homology with SARS‐CoV‐2, higher than that of BANAL‐20‐52 (Figure 4) (Zhou et al., 2020; Z. Zhu et al., 2020).
FIGURE 4.
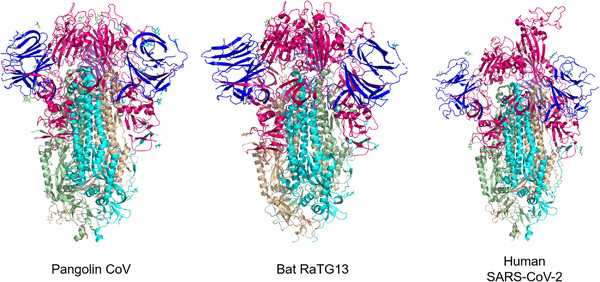
Schematic diagrams of the spike proteins from three coronaviruses: pangolin coronavirus (PDB ID:7BBH, left), bat RaTG13 coronavirus (PDB ID:6ZGF, middle) and human SARS‐CoV‐2 Spike D614G variant (PDB ID:7EAZ, right). The N‐terminus is shown in blue, the middle sequence is pink and the C‐terminus is coloured by cyan, wheat, and light‐orange. The diagrams were based on Cryo‐EM data and visualized using PymoL software.
3.2.2. Furin cleavage site
There are two subunits in the S protein: S1 and S2. S1 mediates virus binding to host cell surface receptors, whereas S2 mediates cell fusion (Papa et al., 2021). The two subunits can be cleaved by proteases in host cells at the furin cleavage site (FCS), but furin protease is not required for the cleavage, as other proteases in cells may be able to achieve this (Papa et al., 2021). This cleavage site was first found in Beta‐CoVs of lineage B with 12 nucleotide insertions at the S1/S2 junction encoding four residues (681PRRA684), forming a multi‐base furin site (NSPRRAR↓SV) (Chan & Zhan, 2022), which is the main feature of SARS‐CoV‐2 (Segreto & Deigin, 2021). Nevertheless, this also makes it possible for someone to speculate that it is artificially inserted (Liu et al., 2021). Hence, Liu et al. (2021) argued that SARS‐CoV‐2 was an artificial virus because of the furin cleavage site in the early stages of the epidemic. However, FCS designed in the laboratory is known to be produced preferentially through substitution rather than insertion, and it is based on the typical R‐X‐[R/K]‐R furin motif (Klenk & Garten, 1994; Yamada & Liu, 2009). In 2006, Nunberg's group first inserted a synthetic furin recognition site (RRSRR) into the S protein at the putative R667 S1/S2 cleavage site to investigate whether the proteolytic cleavage could benefit the fusion activity of SARS‐CoV (Follis, York, & Nunberg, 2006). Clearly, the PRRA insertion in SARS‐CoV‐2 does not coincide with the canonical furin motif. Besides, the RRAR amino acid sequence at S1/S2 cleavage sites in feline coronaviruses (FCoV) cannot be cleaved by furin protease (Deng, Xing, & He, 2022; Holmes et al., 2021). There is no reason for scientists to create such a cleavage site with no successful precedent. Yet, this is not enough to discredit the artificial origin of this insertion site.
The emergence of furin cleavage sites is common in the process of natural evolution. A total of 86 different furin cleavage sites have been identified in 249 coronavirus species worldwide, distributed in 24 animal species in 28 countries since 1954 (Liu et al., 2021). Four of the seven coronaviruses known to infect humans carry furin cleavage sites. These include HCoV‐OC43, HCoV‐HKU1, MERS‐CoV and SARS‐CoV‐2 (Liu et al., 2021). It was discovered in the United States in 1967 that Furin cleavage site (KNRRSRR↓AI) appears in HCoV‐OC43 (beta‐CoV) (Lau et al., 2011; McIntosh, Becker, & Chanock, 1967; Su et al., 2016), and furin cleavage site (SSRRKRR↓SI) of HCoV‐HKU1 was first discovered in China in 2004 (Woo et al., 2005). MERS‐CoV with a furin cleavage site (LTPRSVR↓SV) first identified in Saudi Arabia in 2012, originated in bats, can infect humans and camels (Coutard et al., 2020; Memish et al., 2013; Wong, Li, Lau, & Woo, 2019). In addition, a furin site (TKRRSRR↓AI) was also detected in human intestinal coronaviruses (HECV‐4408, beta‐CoV) first in Germany in 1988 (Zhang, Herbst, Kousoulas, & Storz, 1994). Furthermore, other viruses, such as the H5 and H7 influenza viruses, have also been reported possessing a new multi‐base cleavage site that naturally arises by insertion (Decha et al., 2008; Garushyants, Rogozin, & Koonin, 2021; Taubenberger et al., 2005; Tyshkovskiy & Panchin, 2021).
Kumar et al. (2021) reported six arginine‐coding codons with the highest frequency (AGA, AGG, CGA, CGC, CGG and CGT) in common chiroptera‐hosted CoVs (Kumar et al., 2021). CGG is the least frequently used among the six codons, while CGT and AGA are the most usual arginine‐coding codons. Most preferred codons encoding for arginine have host‐specific preferences, such as human‐adapted CoVs (either CGT or AGA and CGT), SARS‐CoV (AGA and CGT), MERS‐CoV (CGT), Pangolin‐CoVs (either AGA or AGA and CGT), BtCoV‐ RaTG13 (AGA), and SARS‐CoV‐ 2‐Wuhan‐Hu‐1 (AGA) (Kumar et al., 2021). Among them, AGA and CGT are the most frequently occurring codons among the above‐mentioned six codons. Even with host differences, CGG is still the most hardly seen one in codons of all CoVs species.
Focusing on each CoV's furin‐cleavage site, avian, bovine, human (229E, HKU1, NL3 and OC43), pangolins and bat frequently carry either CGT or CGT and AGA. Rarely, in the murine coronavirus (MHV‐A59) furin cleavage site, arginine is encoded by three different synonymous codons: CGC, AGG and CGA. More interestingly, SARS‐CoV‐2 possesses one highest frequency codon (CGT) and two least frequent and consecutive codons CGG in the furin cleavage site (Kumar et al., 2021).
Studies have shown that the furin cleavage site has played an important role in the past decades of coronavirus infection in humans. The precedent of multiple independent occurrences and insertions of such multi‐base sites suggests that it is not uncommon for such events to occur (Tyshkovskiy & Panchin, 2021). In addition, the frequent interchange of furin cleavage site motifs among alpha, beta and gamma coronavirus genera indicates the relatively high recombination frequency of this site (Liu et al., 2021). It can therefore be assumed that coronavirus nucleotide fragments can exchange naturally to produce this particular furin cleavage site, but further research is needed to verify this possibility.
3.2.3. RBD and ACE2
SARS‐CoV‐2 relies on the receptor binding domain (RBD) of the S1 subunit and heparan sulphate on host cell surfaces to enter cells (Clausen et al., 2020). Some sarbecoviruses are unable to use hACE2 owing to the short RBD (Letko & Munster, 2020). The RBD of SARS‐CoV‐2 consists of two subdomains, the core, and external subdomains (Li, Li, Farzan, & Harrison, 2005; Lu et al., 2013). The core subdomain is responsible for the forming of S trimer (Yuan et al., 2017), while the external one contains two exposed loops that bound to ACE2 on the surface of the host cells (Letko & Munster, 2020). Investigating the relationship of other known coronaviruses in the RBD sequence may help to trace the virus origin trends (Liu, Xiao, et al., 2020). At the nucleotide (93.6%) and amino acid (97.4%) levels in the RBD sequences, the coronavirus found in horseshoe bats from Laos is the most similar one to SARS‐CoV‐2, followed by RaTG13 (85.5% and 89.2%, respectively) and MP789 (86.6% and 96.9%, respectively) (Temmam et al., 2021). The RBD region of viruses found in Laos differs from SARS‐CoV‐2 with only by one or two residues and binds hACE2 as efficiently as the earlier isolated SARS‐CoV‐2 strains in Wuhan, then relies on hACE2 to enter human cells. Besides, viruses found in Laos can be inhibited by antibodies that neutralize SARS‐CoV‐2 (Temmam et al., 2021). RaTG13 infection in humans, however, has never been documented. RaTG13 cannot interact effectively with human ACE2 receptors (Wrobel et al., 2020), suggesting that the bat virus does not appear to have the capacity to infect humans directly. However, due to a single amino acid change in RaTG13 (T403R), the S protein can now use human ACE2 for viral invasion (Zech et al., 2021).
The RBD sequence of SARS‐CoV‐2 has a higher affinity for ACE2 on host cells compared to SARS‐CoV (Segreto & Deigin, 2021). Xu et al. (2020) demonstrated that, relatively to SARS‐CoV, loss of hydrogen bond interactions by changing R426 to N426 in the S protein increased the binding free energy of the SARS‐CoV‐2 S protein by 28 kcal/mol, which indicates that the S protein of SARS‐CoV‐2 has a stronger binding affinity with human ACE2. It has been shown that six amino acids in the RBD are essential for binding to ACE2 and for determining the host range of SARS‐COV‐like viruses. These six amino acids are L455, F486, Q493, S494, N501 and Y505 in SARS‐CoV‐2 (Wan et al., 2020). While SARS‐CoV only has one identical residue of these six amino acid residues (Andersen, Rambaut, Lipkin, Holmes, & Garry, 2020). Surprisingly, the Pangolin‐CoV mentioned above possesses the same six key RBD residues as SARS‐CoV‐2 in the RBD, showing closer homology with SARS‐CoV‐2 (Figure 4) (Tao Zhang, Qunfu Wu, & Zhigang Zhang, 2020). This SARS‐CoV‐2 S protein was probably selected for its affinity for human ACE2 by natural selection (Andersen et al., 2020). It is reported that SARS‐CoV‐2 is mutating rapidly, various variants have been reported worldwide (Das & Roy, 2021). Notably, the most crucial mutations have occurred in the S protein. The D614G mutation was the first adaptive mutation detected among wild SARS‐COV‐2 isolates since early 2020 (Bhattacharya, Chatterjee, Sharma, Agoramoorthy, & Chakraborty, 2021; Gobeil et al., 2021; Korber et al., 2020; Volz et al., 2021). These single mutations, such as N501Y, S477N, N439K and Y453F, significantly increased the affinity of RBD to hACE2 to enhance the pathogenicity and transmissibility (Amanat et al., 2021; Barton et al., 2021; Han et al., 2021). Therefore, it can be deduced that the emergence of SARS‐CoV‐2 is simply the result of natural adaptation.
The interaction of RBD‐ACE2 is a critical factor that determines the host range of coronaviruses (Liu, Xiao, et al., 2020). SARS‐CoV‐2 may also recognize ACE2 in a variety of animal species (except mice and rats). Pigs, ferrets, cats and non‐human primates contain a larger number of residues that favour the interaction of SARS‐CoV‐2 with ACE2 and therefore serve as animal models or intermediate hosts for the SARS‐CoV‐2 (Wan et al., 2020). Similarly, Liu, Xiao, et al. (2020) suggest that turtles (Chrysemys picta bellii, Chelonia mydas and Pelodiscus sinensis) may be potential intermediate hosts for SARS‐CoV‐2 transmission to humans other than pangolins and snakes based on the interaction between the key amino acids in RBD with ACE2. However, it has been found in ex vivo tests that, unlike the early isolated WHU01, variants beta and gamma are able to bind and use mouse ACE2, suggesting that these variants may have obtained the ability to infect mice through evolution (Yao et al., 2021). A study reported that the beta variant was able to infect common BALB/c laboratory mice and caused pulmonary changes in contrast to WT SARS‐CoV‐2 (Kant et al., 2021). Indeed, a single amino acid change within the RBD (Q498H, Q498Y, N501Y) could be sufficient to allow SARS‐CoV‐2 to leverage mouse ACE2 (Dinnon et al., 2020; Gu et al., 2020; Kuiper et al., 2022; Wang et al., 2020). There is still no evidence that mice and rats have the capacity to act as immediate hosts of the initially isolated WT SRAS‐CoV‐2. The ability of SARS‐CoV‐2 to cross the barrier of murine species may be obtained through simple mutations. It is also possible for SARS‐CoV‐2 to acquire the ability to infect humans and spread between humans by some simple mutation.
Moreover, bats are also important reservoir species for a great range of coronaviruses (Cui, Li, & Shi, 2019). In an experimental study, ACE2 from 25 species of bats were reported to support SARS‐CoV‐2 entry, and the infection rates of several bats were comparable to those of humans ACE2 (Yan et al., 2021).
3.2.4. Nsp5/M pro
SARS‐CoV‐2 Mpro is a 33.8 kDa protein with 306 amino acids containing a highly conserved substrate‐binding domain (Hsu et al., 2005; Muramatsu et al., 2016). The Mpro of SARS‐CoV‐2 forms a homodimer identical to the biological state of SARS‐CoV and shares 96% amino acid sequence with SARS‐CoV, about 12−13 amino acids are different in the 306 residues (Macchiagodena, Pagliai, & Procacci, 2020). Mpro from SARS‐CoV‐2 shows a slightly increased proteolytic activity than that of SARS‐CoV (Zhang et al., 2020; Zou et al., 2020). Compared to SARS‐CoV, the substitution T285A is the only difference in the dimer interface (Prates et al., 2021). In addition, the sequence in the active site pocket of Mpro is nearly indistinguishable to SARS‐CoV except for Ser46 and Val86 (Macchiagodena et al., 2020). This substitution is thought to play a role in enhancing the efficiency of nsp5 by improving hydrophobic packing within monomers (Prates et al., 2021).
3.2.5. Nsp12/RdRp
There are three subunits in the SARS‐COV‐2 RdRp complex: a nsp12 core component, a heterodimeric nsp7‐nsp8 homodimer, and an additional nsp8 subunit (nsp8‐2). The complex is in charge of the replication and transcription of genomic RNA (Biswas & Mudi, 2020; Gao et al., 2020). Zhou et al. (2020) has found the high degree of homology of BatCoV RaTG13 to SARS‐CoV‐2 in the RdRp region (Zhou et al., 2020). Molecular phylogenetic analysis of the RdRp sequence of SARS‐CoV‐2 generally shows that RaTG13, Pangolin‐CoV and SARS‐CoV‐2 is part of the same group (Zhang et al., 2020; Zhang et al., 2020). Moreover, Kasibhatla et al. (2020) pointed out that, in addition to Bat‐SARS‐isolate (RaTG13), the SARS‐CoV‐2 cluster has also branched off from Pangolin‐CoVs and evolved as an autonomous isomorphic cluster. All the three clusters share the same ancestor. However, SARS‐CoV detaches form another unique cluster. SARS‐Bat‐CoVs and Bat‐CoVs were separated into four clusters based on different host species. Independent or mixed evolution within different hosts may have given Bat‐CoVs the potential in zoonotic spillover effects (Kasibhatla et al., 2020).
4. CURRENT LIMITATIONS IN UNDERSTANDING THE ORIGIN Of SARS‐CoV‐2
Pertaining to the origin of SARS‐CoV‐2, this review has made a certain summary based on existing research and findings. Currently, there are hypotheses about zoonotic spillovers, as well as debates over the laboratory leak hypothesis. Since there are differing opinions on this matter, it has not reached a point of resolution yet. Judging from the investigation on the origin of the virus in the past 2 years, it seems that some limitations have emerged, which may be one of the reasons for the delay in determining the origin.
4.1. Suspensive place of origin
Initially, reports of the virus appeared in Wuhan, and the world's eyes were riveted on the city. It seems reasonable in conventional thinking to find the cause of the problem where it was appearing, but in the case of virus spread, it has huge uncertainties. Equal attention should be given to all advanced biological laboratories worldwide. It is imperative that all areas with suspected epidemics and all laboratories involved in CoVs research prior to the outbreak of SARS‐CoV‐2 should undergo an intensive investigation of epidemiology, strain collections and research records.
Several recent studies on the Huanan Seafood Market (HSM) have emerged. George et al. (2022) showed SARS‐CoV‐2 test results of 1380 samples (including 923 environmental samples and 457 animal samples) collected from the environment and market animals at the HSM in early 2020. As a result of RT‐qPCR for the test of SARS‐CoV‐2, 73 environmental samples were positive for the virus, and three live viruses were also isolated. The virus from the market shares 99.980% to 99.993% nucleotide identity with the human isolate HCoV/Wuhan/IVDC‐HB‐01. Nevertheless, all the 457 samples of animals were negative for SARS‐CoV‐2 nucleic acids, suggesting that animals carrying SARS‐CoV‐2 are rare on the market. Further analysis of environmental samples showed the possibility that sewage contamination in the market acts as a mediator in clustered cases, or where infected people contaminate sewage (Gao et al., 2022). Additional results showed an association of cases with different products, including aquatic products, cold chain products, seafood, poultry, livestock, wildlife products and vegetables. The results suggest that SARS‐CoV‐2 may have circulated for a short period in the market in December 2019, especially in the western part of the market, resulting in the widespread distribution of the virus in the market, possibly due to crowded buyers and polluted environment. At this stage, since the market attracts many visitors daily, it may have acted as an amplifier, resulting in many initially identified clusters of infections early in the outbreak.
The above studies are also in line with the WHO's investigation report in Wuhan, which included four possible routes of transmission of the virus in its March report, but did not provide definitive evidence as to which route is the most possible. Zoonotic introductions or spillovers of SARS‐CoV‐2 from wild animals are direct routes to transmission. Additionally, there are three indirect routes of infection: consumption of contaminated food or infected animal products, including frozen food imported from other countries that sold at the Wuhan market; handling infected farm animals; from laboratories that study viruses in animals. However, in this preliminary study, no definitive conclusion can be made about the origin of SARS‐CoV‐2.
Most of the available data showed that the earliest reports about the outbreak were concentrated in November and December 2019 (Wilensky, 2021). Determining an epidemic outbreak requires a certain infection base and precise medical analysis. In addition, the official reports are always well thought out and coordinated, so there will be a little bit of a delay in timing. Therefore, it can be inferred that the epidemic has spread quietly in Wuhan before November. As mentioned earlier, the incubation period of SARS‐CoV‐2 is five days to 2 weeks, so we can roughly speculate that the virus spread in Wuhan in mid‐October.
4.2. Inadequate global information sharing
Although COVID‐19 has been widely developed so far, the traceability progress is not optimistic. In addition to the complexity of the coronavirus itself, the other reason is the lack of harmonious international cooperation, which led to delayed traceability work (Holmes et al., 2021; Wilensky, 2021; Zhu et al., 2020). Human beings share the same destiny, as well as the same weal and woe. The borders of countries do not separate epidemics from others. Every pandemic will cause immeasurable harm to human beings. The only way to stop the next epidemic is working together to trace its origin. A global expert group should be established as soon as possible to carry out joint traceability studies in all the countries and regions suspected to be the source of the epidemic. Time is running out for tracing the origin. If the only window period is missed, the task of tracing the origin gradually becomes an insurmountable challenge. Even though science and politics are often influenced by each other. The traceability of the epidemic is expected to advance only through concerted efforts of the international community, and the truth will emerge with the cooperation of all countries.
5. CONCLUSION
Despite the fact that animal host of SARS‐CoV2 has not yet been identified, zoonotic origin is supported by a substantial body of research. Even though laboratory accidents cannot entirely be ruled out, it can be deduced from the current research that the emergence of SARS‐CoV‐2 likely stemmed from natural adaptation. It is imperative to conduct comprehensive investigations on the source of COVID‐19 by tight international collaborations. Otherwise, the world remains vulnerable to future pandemic of serious infectious diseases.
CONFLICT OF INTEREST
The authors declare that the work was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AUTHOR CONTRIBUTIONS
J.M.C and D.P.W conceived the article structure. Y.J.H and M.Y.W draw the figures. Y.J.H and Y.L.W co‐wrote the manuscript. J.M.C, D.P.W, L.Z and J.Y.S provided help in article revisions. All authors read and edited the manuscript.
ETHICAL STATEMENT
The authors confirm that ethics approval is not applicable.
Hao, Y.‐J. , Wang, Y.‐L. , Wang, M.‐Y. , Zhou, L. , Shi, J.‐Y. , Cao, J.‐M. , & Wang, D.‐P. (2022). The origins of COVID‐19 pandemic: A brief overview. Transboundary and Emerging Diseases, 00, 1–17. 10.1111/tbed.14732
Ying‐Jian Hao and Yu‐Lan Wang contributed equally to this work.
Contributor Information
Ji‐Min Cao, Email: caojimin@sxmu.edu.cn.
De‐Ping Wang, Email: wangdeping@sxmu.edu.cn.
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study. cd_value_code=text
REFERENCES
- Al‐Qaaneh, A. M. , Alshammari, T. , Aldahhan, R. , Aldossary, H. , Alkhalifah, Z. A. , & Borgio, J. F. (2021). Genome composition and genetic characterization of SARS‐CoV‐2. Saudi Journal of Biological Sciences, 28(3), 1978–1989. 10.1016/j.sjbs.2020.12.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat, F. , Thapa, M. , Lei, T. , Ahmed, S. M. S. , Adelsberg, D. C. , Carreno, J. M. , Strohmeier, S. , Schmitz, A. J. , Zafar, S. , Zhou, J. Q. , Rijnink, W. , Alshammary, H. , Borcherding, N. , Reiche, A. G. , Srivastava, K. , Sordillo, E. M. , van Bakel, H. , Personalized Virology, I. , Turner, J. S. , & Krammer, F. (2021). SARS‐CoV‐2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell, 184(15), 3936–3948. e3910. 10.1016/j.cell.2021.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola, A. , Bianchi, S. , Gori, M. , Colzani, D. , Canuti, M. , Borghi, E. , Raviglione, M. C. , Zuccotti, G. V. , & Tanzi, E. (2021). Evidence of SARS‐CoV‐2 RNA in an oropharyngeal swab specimen, Milan, Italy, early December 2019. Emerging Infectious Diseases, 27(2), 648–650. 10.3201/eid2702.204632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand, N. M. , Liya, D. H. , Pradhan, A. K. , Tayal, N. , Bansal, A. , Donakonda, S. , & Jainarayanan, A. K. (2021). A comprehensive SARS‐CoV‐2 genomic analysis identifies potential targets for drug repurposing. PLoS One, 16(3), e0248553. 10.1371/journal.pone.0248553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, K. G. , Rambaut, A. , Lipkin, W. I. , Holmes, E. C. , & Garry, R. F. (2020). The proximal origin of SARS‐CoV‐2. Nature Medicine, 26(4), 450–452. 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apolone, G. , Montomoli, E. , Manenti, A. , Boeri, M. , Sabia, F. , Hyseni, I. , Mazzini, L. , Martinuzzi, D. , Cantone, L. , Milanese, G. , Sestini, S. , Suatoni, P. , Marchiano, A. , Bollati, V. , Sozzi, G. , & Pastorino, U. (2021). Unexpected detection of SARS‐CoV‐2 antibodies in the prepandemic period in Italy. Tumori, 107(5), 446–451. 10.1177/0300891620974755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain, C. C. , & MacDonald, A. S. (2022). The impact of the lung environment on macrophage development, activation and function: Diversity in the face of adversity. Mucosal Immunology, 15(2), 223–234 . 10.1038/s41385-021-00480-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, L. , Deng, W. , Huang, B. , Gao, H. , Liu, J. , Ren, L. , Wei, Q. , Yu, P. , Xu, Y. , Qi, F. , Qu, Y. , Li, F. , Lv, Q. , Wang, W. , Xue, J. , Gong, S. , Liu, M. , Wang, G. , Wang, S. , … Qin, C. (2020). The pathogenicity of SARS‐CoV‐2 in hACE2 transgenic mice. Nature, 583(7818), 830–833. 10.1038/s41586-020-2312-y [DOI] [PubMed] [Google Scholar]
- Barizien, N. , Le Guen, M. , Russel, S. , Touche, P. , Huang, F. , & Vallee, A. (2021). Clinical characterization of dysautonomia in long COVID‐19 patients. Scientific Reports, 11(1), 14042. 10.1038/s41598-021-93546-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, M. I. , MacGowan, S. A. , Kutuzov, M. A. , Dushek, O. , Barton, G. J. , & van der Merwe, P. A. (2021). Effects of common mutations in the SARS‐CoV‐2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. ELIFE, 10, 10.7554/eLife.70658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavaraju, S. V. , Patton, M. E. , Grimm, K. , Rasheed, M. A. U. , Lester, S. , Mills, L. , Stumpf, M. , Freeman, B. , Tamin, A. , Harcourt, J. , Schiffer, J. , Semenova, V. , Li, H. , Alston, B. , Ategbole, M. , Bolcen, S. , Boulay, D. , Browning, P. , Cronin, L. , … Stramer, S. L. (2021). Serologic testing of US Blood donations to identify severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2)‐reactive antibodies: December 2019‐January 2020. Clinical Infectious Diseases, 72(12), e1004–e1009. 10.1093/cid/ciaa1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolin, A. P. , Weissmann, F. , Zeng, J. , Posse, V. , Milligan, J. C. , Canal, B. , Ulferts, R. , Wu, M. , Drury, L. S. , Howell, M. , Beale, R. , & Diffley, J. F. X. (2021). Identifying SARS‐CoV‐2 antiviral compounds by screening for small molecule inhibitors of nsp12/7/8 RNA‐dependent RNA polymerase. Biochemical Journal, 478(13), 2425–2443. 10.1042/BCJ20210200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya, M. , Chatterjee, S. , Sharma, A. R. , Agoramoorthy, G. , & Chakraborty, C. (2021). D614G mutation and SARS‐CoV‐2: Impact on S‐protein structure, function, infectivity, and immunity. Applied Microbiology and Biotechnology, 105(24), 9035–9045. 10.1007/s00253-021-11676-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, S. K. , & Mudi, S. R. (2020). Spike protein D614G and RdRp P323L: The SARS‐CoV‐2 mutations associated with severity of COVID‐19. Genomics Inform, 18(4), e44. 10.5808/GI.2020.18.4.e44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni, M. F. , Lemey, P. , Jiang, X. , Lam, T. T. , Perry, B. W. , Castoe, T. A. , Rambaut, A. , & Robertson, D. L. (2020). Evolutionary origins of the SARS‐CoV‐2 sarbecovirus lineage responsible for the COVID‐19 pandemic. Nature Microbiology, 5(11), 1408–1417. 10.1038/s41564-020-0771-4 [DOI] [PubMed] [Google Scholar]
- Borges, V. , Isidro, J. , Trovão, N. S. , Duarte, S. , Cortes‐Martins, H. , Martiniano, H. , Gordo, I. , Leite, R. , Vieira, L. , Lira, A. J. S. , Sousa Fernandes, A. M. , Estrada, A. , Nunes, A. , Rodrigues, A. , Caldas, A. , Constança, A. , Henriques, A. M. , Matos, A. M. , Oliveira, A. , … Gomes, J. P. (2022). SARS‐CoV‐2 introductions and early dynamics of the epidemic in Portugal. Communications Medicine, 2(1). 10.1038/s43856-022-00072-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrat, F. , Figoni, J. , Henny, J. , Desenclos, J. C. , Kab, S. , de Lamballerie, X. , & Zins, M. (2021). Evidence of early circulation of SARS‐CoV‐2 in France: Findings from the population‐based “CONSTANCES” cohort. European Journal of Epidemiology, 36(2), 219–222. 10.1007/s10654-020-00716-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . (2021a). Economic trends in tobacco. Retrieved from https://www.cdc.gov/tobacco/data_statistics/fact_sheets/economics/econ_facts/
- CDC . (2021b). Outbreak of lung injury associated with the use of E‐cigarette, or vaping, products. Retrieved from https://www.cdc.gov/tobacco/basic_information/e‐cigarettes/severe‐lung‐disease.html
- Chan, Y. A. , & Zhan, S. H. (2022). The emergence of the spike furin cleavage site in SARS‐CoV‐2. Molecular Biology and Evolution, 39(1). 10.1093/molbev/msab327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarria‐Miró, G. , Anfruns‐Estrada, E. , Martínez‐Velázquez, A. , Vázquez‐Portero, M. , Guix, S. , Paraira, M. , Galofré, B. , Sánchez, G. , Pintó, R. M. , & Bosch, A. (2021). Time evolution of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) in wastewater during the first pandemic wave of COVID‐19 in the metropolitan area of Barcelona, Spain. Applied and Environmental Microbiology, 87(7). 10.1128/aem.02750-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Malone, B. , Llewellyn, E. , Grasso, M. , Shelton, P. M. M. , Olinares, P. D. B. , Maruthi, K. , Eng, E. , Vatandaslar, H. , Chait, B. T. , Kapoor, T. , Darst, S. A. , & Campbell, E. A. (2020). Structural basis for helicase‐polymerase coupling in the SARS‐CoV‐2 replication‐transcription complex. bioRxiv. 10.1101/2020.07.08.194084 [DOI] [PMC free article] [PubMed]
- Clausen, T. M. , Sandoval, D. R. , Spliid, C. B. , Pihl, J. , Perrett, H. R. , Painter, C. D. , Narayanan, A. , Majowicz, S. A. , Kwong, E. M. , McVicar, R. N. , Thacker, B. E. , Glass, C. A. , Yang, Z. , Torres, J. L. , Golden, G. J. , Bartels, P. L. , Porell, R. N. , Garretson, A. F. , Laubach, L. , … Esko, J. D. (2020). SARS‐CoV‐2 infection depends on cellular heparan sulfate and ACE2. Cell, 183(4), 1043–1057. e1015. 10.1016/j.cell.2020.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J. (2020). Wuhan coronavirus hunter Shi Zhengli speaks out. Science, 369(6503), 487–488. 10.1126/science.369.6503.487 [DOI] [PubMed] [Google Scholar]
- Coutard, B. , Valle, C. , de Lamballerie, X. , Canard, B. , Seidah, N. G. , & Decroly, E. (2020). The spike glycoprotein of the new coronavirus 2019‐nCoV contains a furin‐like cleavage site absent in CoV of the same clade. Antiviral Research, 176, 104742. 10.1016/j.antiviral.2020.104742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, J. , Li, F. , & Shi, Z. L. (2019). Origin and evolution of pathogenic coronaviruses. Nature Reviews Microbiology, 17(3), 181–192. 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Onofrio, V. , Keulen, L. , Vandendriessche, A. , Dubois, J. , Cartuyvels, R. , Vanden Abeele, M. E. , Fraussen, J. , Vandormael, P. , Somers, V. , Achten, R. , Dendooven, A. , Driessen, A. , Augsburg, L. , Hellings, N. , Lammens, M. , Vanrusselt, J. , & Cox, J. (2022). Studying the clinical, radiological, histological, microbiological, and immunological evolution during the different COVID‐19 disease stages using minimal invasive autopsy. Scientific Reports, 12(1), 1360. 10.1038/s41598-022-05186-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski, Z. , Jordan, T. X. , Ilmain, J. K. , Guo, X. , Bhabha, G. , tenOever, B. R. , & Sanjana, N. E. (2021). The Spike D614G mutation increases SARS‐CoV‐2 infection of multiple human cell types. ELIFE, 10, 10.7554/eLife.65365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, J. K. , & Roy, S. (2021). A study on non‐synonymous mutational patterns in structural proteins of SARS‐CoV‐2. Genome, 64(7), 665–678. 10.1139/gen-2020-0157 [DOI] [PubMed] [Google Scholar]
- Davidson, A. D. , Williamson, M. K. , Lewis, S. , Shoemark, D. , Carroll, M. W. , Heesom, K. J. , Zambon, M. , Ellis, J. , Lewis, P. A. , Hiscox, J. A. , & Matthews, D. A. (2020). Characterisation of the transcriptome and proteome of SARS‐CoV‐2 reveals a cell passage induced in‐frame deletion of the furin‐like cleavage site from the spike glycoprotein. Genome Medicine, 12(1), 68. 10.1186/s13073-020-00763-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, N. G. , Jarvis, C. I. , Group, C. C.‐W. , Edmunds, W. J. , Jewell, N. P. , Diaz‐Ordaz, K. , & Keogh, R. H. (2021). Increased mortality in community‐tested cases of SARS‐CoV‐2 lineage B.1.1.7. Nature, 593(7858), 270–274. 10.1038/s41586-021-03426-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, J. T. , Chinazzi, M. , Perra, N. , Mu, K. , Pastore, Y. P. A. , Ajelli, M. , Dean, N. E. , Gioannini, C. , Litvinova, M. , Merler, S. , Rossi, L. , Sun, K. , Xiong, X. , Longini, I. M., Jr. , Halloran, M. E. , Viboud, C. , & Vespignani, A. (2021). Cryptic transmission of SARS‐CoV‐2 and the first COVID‐19 wave. Nature, 600(7887), 127–132. 10.1038/s41586-021-04130-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- de, O. A. J. , Pinheiro, S. , Zamora, W. J. , Alves, C. N. , Lameira, J. , & Lima, A. H. (2021). Structural, energetic and lipophilic analysis of SARS‐CoV‐2 non‐structural protein 9 (NSP9). Scientific Reports, 11(1), 23003. 10.1038/s41598-021-02366-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decha, P. , Rungrotmongkol, T. , Intharathep, P. , Malaisree, M. , Aruksakunwong, O. , Laohpongspaisan, C. , Parasuk, V. , Sompornpisut, P. , Pianwanit, S. , Kokpol, S. , & Hannongbua, S. (2008). Source of high pathogenicity of an avian influenza virus H5N1: Why H5 is better cleaved by furin. Biophysical Journal, 95(1), 128–134. 10.1529/biophysj.107.127456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaune, D. , Hul, V. , Karlsson, E. A. , Hassanin, A. , Ou, T. P. , Baidaliuk, A. , Gambaro, F. , Prot, M. , Tu, V. T. , Chea, S. , Keatts, L. , Mazet, J. , Johnson, C. K. , Buchy, P. , Dussart, P. , Goldstein, T. , Simon‐Loriere, E. , & Duong, V. (2021). A novel SARS‐CoV‐2 related coronavirus in bats from Cambodia. Nature Communications, 12(1), 6563. 10.1038/s41467-021-26809-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, S. , Xing, K. , & He, X. (2022). Mutation signatures inform the natural host of SARS‐CoV‐2. National Science Review, 9(2), nwab220. 10.1093/nsr/nwab220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar, M. S. , Marwal, R. , Vs, R. , Ponnusamy, K. , Jolly, B. , Bhoyar, R. C. , Sardana, V. , Naushin, S. , Rophina, M. , Mellan, T. A. , Mishra, S. , Whittaker, C. , Fatihi, S. , Datta, M. , Singh, P. , Sharma, U. , Ujjainiya, R. , Bhatheja, N. , Divakar, M. K. , … Cherian, S. S. (2021). Genomic characterization and epidemiology of an emerging SARS‐CoV‐2 variant in Delhi, India. Science, 374(6570), 995–999. 10.1126/science.abj9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon, K. H., 3rd , Leist, S. R. , Schäfer, A. , Edwards, C. E. , Martinez, D. R. , Montgomery, S. A. , West, A. , Yount, B. L., Jr. , Hou, Y. J. , Adams, L. E. , Gully, K. L. , Brown, A. J. , Huang, E. , Bryant, M. D. , Choong, I. C. , Glenn, J. S. , Gralinski, L. E. , Sheahan, T. P. , & Baric, R. S. (2020). A mouse‐adapted model of SARS‐CoV‐2 to test COVID‐19 countermeasures. Nature, 586(7830), 560–566. 10.1038/s41586-020-2708-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler, J. F. , Corman, V. M. , & Drosten, C. (2014). Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Research, 101, 45–56. 10.1016/j.antiviral.2013.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Y. , Li, X. , Zhang, L. , Wan, S. , Zhang, L. , & Zhou, F. (2022). SARS‐CoV‐2 Omicron variant: Recent progress and future perspectives. Signal Signal Transduction and Targeted Therapy, 7(1), 141. 10.1038/s41392-022-00997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer, M. D. , Barrueco, A. S. , Martinez‐Beneyto, Y. , Mateos‐Moreno, M. V. , Ausina‐Marquez, V. , Garcia‐Vazquez, E. , Puche‐Torres, M. , Giner, M. J. F. , Gonzalez, A. C. , Coello, J. M. S. , Rueda, I. A. , Auba, J. M. V. , Espanol, C. C. , Velasco, A. L. , Abad, D. S. , Garcia‐Esteban, S. , Artacho, A. , Lopez‐Labrador, X. , & Mira, A. (2021). Clinical evaluation of antiseptic mouth rinses to reduce salivary load of SARS‐CoV‐2. Scientific Reports, 11(1), 24392. 10.1038/s41598-021-03461-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follis, K. E. , York, J. , & Nunberg, J. H. (2006). Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell‐cell fusion but does not affect virion entry. Virology, 350(2), 358–369. 10.1016/j.virol.2006.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y. , Yan, L. , Huang, Y. , Liu, F. , Zhao, Y. , Cao, L. , Wang, T. , Sun, Q. , Ming, Z. , Zhang, L. , Ge, J. , Zheng, L. , Zhang, Y. , Wang, H. , Zhu, Y. , Zhu, C. , Hu, T. , Hua, T. , Zhang, B. , & Rao, Z. (2020). Structure of the RNA‐dependent RNA polymerase from COVID‐19 virus. Science, 368(6492), 779–782. 10.1126/science.abb7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garushyants, S. K. , Rogozin, I. B. , & Koonin, E. V. (2021). Insertions in SARS‐CoV‐2 genome caused by template switch and duplications give rise to new variants that merit monitoring. bioRxiv. 10.1101/2021.04.23.441209 [DOI] [PMC free article] [PubMed]
- Ge, X.‐Y. , Hu, B. , Wang, L.‐F. , & Shi, Z.‐L. (2015). Bat origin of human coronaviruses. Virol J, 12, 221. 10.1186/s12985-015-0422-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, X. Y. , Li, J. L. , Yang, X. L. , Chmura, A. A. , Zhu, G. , Epstein, J. H. , Mazet, J. K. , Hu, B. , Zhang, W. , Peng, C. , Zhang, Y. J. , Luo, C. M. , Tan, B. , Wang, N. , Zhu, Y. , Crameri, G. , Zhang, S. Y. , Wang, L. F. , Daszak, P. , & Shi, Z. L. (2013). Isolation and characterization of a bat SARS‐like coronavirus that uses the ACE2 receptor. Nature, 503(7477), 535–538. 10.1038/nature12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes, A. M. (2006). The history of smallpox. Clinics in Dermatology, 24(3), 152–157. 10.1016/j.clindermatol.2005.11.009 [DOI] [PubMed] [Google Scholar]
- Gao, G. , Liu, W. , Liu, P. , Lei, W. , Jia, Z. , He, X. , Liu, L.‐1L. , Shi, W. , Tan, Y. , Zou, S. , Zhao, X. , Wong, G. , Wang, J. , Wang, F. , Wang, G. , Qin, K. , Gao, R. , Zhang, J. , Li, M. , … Wu, G. (2022). Surveillance of SARS‐CoV‐2 in the environment and animal samples of the Huanan Seafood Market. Nature, 10.21203/rs.3.rs‐1370392/v1.
- Gobeil, S. M. C. , Janowska, K. , McDowell, S. , Mansouri, K. , Parks, R. , Manne, K. , Stalls, V. , Kopp, M. F. , Henderson, R. , Edwards, R. J. , Haynes, B. F. , & Acharya, P. (2021). D614G mutation alters SARS‐CoV‐2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Reports, 34(2). 10.1016/j.celrep.2020.108630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, H. , Chen, Q. , Yang, G. , He, L. , Fan, H. , Deng, Y. Q. , Wang, Y. , Teng, Y. , Zhao, Z. , Cui, Y. , Li, Y. , Li, X. F. , Li, J. , Zhang, N. N. , Yang, X. , Chen, S. , Guo, Y. , Zhao, G. , Wang, X. , … Zhou, Y. (2020). Adaptation of SARS‐CoV‐2 in BALB/c mice for testing vaccine efficacy. Science, 369(6511), 1603–1607. 10.1126/science.abc4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, H. , Xie, R. , Adam, D. C. , Tsui, J. L. H. , Chu, D. K. , Chang, L. D. J. , Cheuk, S. S. Y. , Gurung, S. , Krishnan, P. , Ng, D. Y. M. , Liu, G. Y. Z. , Wan, C. K. C. , Cheng, S. S. M. , Edwards, K. M. , Leung, K. S. M. , Wu, J. T. , Tsang, D. N. C. , Leung, G. M. , Cowling, B. J , … Poon, L. L. M. (2022). Genomic epidemiology of SARS‐CoV‐2 under an elimination strategy in Hong Kong. Nature Communications, 13(1). 10.1038/s41467-022-28420-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Y. , Zheng, B. J. , He, Y. Q. , Liu, X. L. , Zhuang, Z. X. , Cheung, C. L. , Luo, S. W. , Li, P. H. , Zhang, L. J. , Guan, Y. J. , Butt, K. M. , Wong, K. L. , Chan, K. W. , Lim, W. , Shortridge, K. F. , Yuen, K. Y. , Peiris, J. S. , & Poon, L. L. (2003). Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science, 302(5643), 276–278. 10.1126/science.1087139 [DOI] [PubMed] [Google Scholar]
- Han, P. , Su, C. , Zhang, Y. , Bai, C. , Zheng, A. , Qiao, C. , Wang, Q. , Niu, S. , Chen, Q. , Zhang, Y. , Li, W. , Liao, H. , Li, J. , Zhang, Z. , Cho, H. , Yang, M. , Rong, X. , Hu, Y. , Huang, N. , … Qi, J. (2021). Molecular insights into receptor binding of recent emerging SARS‐CoV‐2 variants. Nature Communications, 12(1), 6103. 10.1038/s41467-021-26401-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, W. S. , Miller, E. , Andrews, N. J. , Waight, P. , Maini, P. K. , Funk, S. , & Thompson, R. N. (2022). Generation time of the alpha and delta SARS‐CoV‐2 variants: An epidemiological analysis. The Lancet Infectious Diseases, 22, P603–610. 10.1016/s1473-3099(22)00001-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan, A. O. , Case, J. B. , Winkler, E. S. , Thackray, L. B. , Kafai, N. M. , Bailey, A. L. , McCune, B. T. , Fox, J. M. , Chen, R. E. , Alsoussi, W. B. , Turner, J. S. , Schmitz, A. J. , Lei, T. , Shrihari, S. , Keeler, S. P. , Fremont, D. H. , Greco, S. , McCray, P. B., Jr. , Perlman, S. , … Diamond, M. S. (2020). A SARS‐CoV‐2 infection model in mice demonstrates protection by neutralizing antibodies. Cell, 182(3), 744–753. e744. 10.1016/j.cell.2020.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, E. C. , & Duchêne, S. (2019). Can sequence phylogenies safely infer the origin of the global virome? mBio, 10(2). 10.1128/mBio.00289-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, E. C. , Goldstein, S. A. , Rasmussen, A. L. , Robertson, D. L. , Crits‐Christoph, A. , Wertheim, J. O. , Anthony, S. J. , Barclay, W. S. , Boni, M. F. , Doherty, P. C. , Farrar, J. , Geoghegan, J. L. , Jiang, X. , Leibowitz, J. L. , Neil, S. J. D. , Skern, T. , Weiss, S. R. , Worobey, M. , Andersen, K. G. , … Rambaut, A. (2021). The origins of SARS‐CoV‐2: A critical review. Cell, 184(19), 4848–4856. 10.1016/j.cell.2021.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, H. , Wang, T. , Zhang, B. , Luo, Y. , Mao, L. , Wang, F. , Wu, S. , & Sun, Z. (2020). Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin Transl Immunology, 9(5), e01136. 10.1002/cti2.1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, M. F. , Kuo, C. J. , Chang, K. T. , Chang, H. C. , Chou, C. C. , Ko, T. P. , Shr, H. L. , Chang, G. G. , Wang, A. H. , & Liang, P. H. (2005). Mechanism of the maturation process of SARS‐CoV 3CL protease. Journal of Biological Chemistry, 280(35), 31257–31266. 10.1074/jbc.M502577200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , Guo, H. , Zhou, P. , & Shi, Z.‐L. (2020). Characteristics of SARS‐CoV‐2 and COVID‐19. Nature Reviews Microbiology, 19(3), 141–154. 10.1038/s41579-020-00459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , Zeng, L. P. , Yang, X. L. , Ge, X. Y. , Zhang, W. , Li, B. , Xie, J. Z. , Shen, X. R. , Zhang, Y. Z. , Wang, N. , Luo, D. S. , Zheng, X. S. , Wang, M. N. , Daszak, P. , Wang, L. F. , Cui, J. , & Shi, Z. L. (2017). Discovery of a rich gene pool of bat SARS‐related coronaviruses provides new insights into the origin of SARS coronavirus. PLOS Pathogens, 13(11), e1006698. 10.1371/journal.ppat.1006698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow, B. , Song, E. , Mao, T. , Lu, P. , Meir, A. , Liu, F. , Alfajaro, M. M. , Wei, J. , Dong, H. , Homer, R. J. , Ring, A. , Wilen, C. B. , & Iwasaki, A. (2020). Mouse model of SARS‐CoV‐2 reveals inflammatory role of type I interferon signaling. Journal of Experimental Medicine, 217(12). 10.1084/jem.20201241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, B. A. , Grace, D. , Kock, R. , Alonso, S. , Rushton, J. , Said, M. Y. , McKeever, D. , Mutua, F. , Young, J. , McDermott, J. , & Pfeiffer, D. U. (2013). Zoonosis emergence linked to agricultural intensification and environmental change. Proceedings of the National Academy of Sciences of the United States of America, 110(21), 8399–8404. 10.1073/pnas.1208059110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant, R. , Kareinen, L. , Smura, T. , Freitag, T. L. , Jha, S. K. , Alitalo, K. , Meri, S. , Sironen, T. , Saksela, K. , Strandin, T. , Kipar, A. , & Vapalahti, O. (2021). Common laboratory mice are susceptible to infection with the SARS‐CoV‐2 Beta variant. Viruses, 13(11). 10.3390/v13112263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasibhatla, S. M. , Kinikar, M. , Limaye, S. , Kale, M. M. , & Kulkarni‐Kale, U. (2020). Understanding evolution of SARS‐CoV‐2: A perspective from analysis of genetic diversity of RdRp gene. Journal of Medical Virology, 92(10), 1932–1937. 10.1002/jmv.25909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkenborg Berg, S. , Dam Nielsen, S. , Nygaard, U. , Bundgaard, H. , Palm, P. , Rotvig, C. , & Vinggaard Christensen, A. (2022). Long COVID symptoms in SARS‐CoV‐2‐positive adolescents and matched controls (LongCOVIDKidsDK): A national, cross‐sectional study. The Lancet Child & Adolescent Health, 6(4), 240–248. 10.1016/s2352-4642(22)00004-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Lee, J. Y. , Yang, J. S. , Kim, J. W. , Kim, V. N. , & Chang, H. (2020). The architecture of SARS‐CoV‐2 transcriptome. Cell, 181(4), 914–921. e910. 10.1016/j.cell.2020.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirtipal, N. , Bharadwaj, S. , & Kang, S. G. (2020). From SARS to SARS‐CoV‐2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infection, Genetics and Evolution, 85, 104502. 10.1016/j.meegid.2020.104502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm, T. , Ebert, G. , Calleja, D. J. , Allison, C. C. , Richardson, L. W. , Bernardini, J. P. , Lu, B. G. , Kuchel, N. W. , Grohmann, C. , Shibata, Y. , Gan, Z. Y. , Cooney, J. P. , Doerflinger, M. , Au, A. E. , Blackmore, T. R. , van der Heden van Noort, G. J. , Geurink, P. P. , Ovaa, H. , Newman, J. , … Komander, D. (2020). Mechanism and inhibition of the papain‐like protease, PLpro, of SARS‐CoV‐2. The EMBO Journal, 39(18), e106275. doi: 10.15252/embj.2020106275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk, H. D. , & Garten, W. (1994). Host cell proteases controlling virus pathogenicity. Trends in Microbiology, 2(2), 39–43. 10.1016/0966-842x(94)90123-6 [DOI] [PubMed] [Google Scholar]
- Klimstra, W. B. , Tilston‐Lunel, N. L. , Nambulli, S. , Boslett, J. , McMillen, C. M. , Gilliland, T. , Dunn, M. D. , Sun, C. , Wheeler, S. E. , Wells, A. , Hartman, A. L. , McElroy, A. K. , Reed, D. S. , Rennick, L. J. , & Duprex, W. P. (2020). SARS‐CoV‐2 growth, furin‐cleavage‐site adaptation and neutralization using serum from acutely infected hospitalized COVID‐19 patients. Journal of General Virology, 101(11), 1156–1169. 10.1099/jgv.0.001481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber, B. , Fischer, W. M. , Gnanakaran, S. , Yoon, H. , Theiler, J. , Abfalterer, W. , Hengartner, N. , Giorgi, E. E. , Bhattacharya, T. , Foley, B. , Hastie, K. M. , Parker, M. D. , Partridge, D. G. , Evans, C. M. , Freeman, T. M. , de Silva, T. I. , McDanal, C. , Perez, L. G. , Tang, H. , … Montefiori, D. C. (2020). Tracking changes in SARS‐CoV‐2 spike: Evidence that D614G increases infectivity of the COVID‐19 virus. Cell, 182(4), 812–827.e819. 10.1016/j.cell.2020.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper, M. J. , Wilson, L. O. W. , Mangalaganesh, S. , Lee, C. , Reti, D. , & Vasan, S. S. (2022). “But Mouse, You Are Not Alone”: On some severe acute respiratory syndrome coronavirus 2 variants infecting mice. ILAR Journal, 62(1–2), 48–59. 10.1093/ilar/ilab031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, N. , Kaushik, R. , Tennakoon, C. , Uversky, V. N. , Mishra, A. , Sood, R. , Srivastava, P. , Tripathi, M. , Zhang, K. Y. J. , & Bhatia, S. (2021). Evolutionary signatures governing the codon usage bias in coronaviruses and their implications for viruses infecting various bat species. Viruses, 13(9). 10.3390/v13091847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Tao, Q. , Weaver, S. , Sanderford, M. , Caraballo‐Ortiz, M. A. , Sharma, S. , Pond, S. L. K. , & Miura, S. (2021). An evolutionary portrait of the progenitor SARS‐CoV‐2 and its dominant offshoots in COVID‐19 pandemic. Molecular Biology and Evolution, 38(8), 3046–3059. 10.1093/molbev/msab118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa, G. , Mancini, P. , Bonanno Ferraro, G. , Veneri, C. , Iaconelli, M. , Bonadonna, L. , Lucentini, L. , & Suffredini, E. (2021). SARS‐CoV‐2 has been circulating in northern Italy since December 2019: Evidence from environmental monitoring. Science of the Total Environment, 750, 141711. 10.1016/j.scitotenv.2020.141711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latinne, A. , Hu, B. , Olival, K. J. , Zhu, G. , Zhang, L. , Li, H. , Chmura, A. A. , Field, H. E. , Zambrana‐Torrelio, C. , Epstein, J. H. , Li, B. , Zhang, W. , Wang, L. F. , Shi, Z. L. , & Daszak, P. (2020). Origin and cross‐species transmission of bat coronaviruses in China. Nature Communications, 11(1), 4235. 10.1038/s41467-020-17687-3 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lau, S. K. , Lee, P. , Tsang, A. K. , Yip, C. C. , Tse, H. , Lee, R. A. , So, L. Y. , Lau, Y. L. , Chan, K. H. , Woo, P. C. , & Yuen, K. Y. (2011). Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. Journal of Virology, 85(21), 11325–11337. 10.1128/jvi.05512-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, S. K. P. , Woo, P. C. Y. , Li, K. S. M. , Huang, Y. , Tsoi, H.‐W. , Wong, B. H. L. , Wong, S. S. Y. , Leung, S.‐Y. , Chan, K.‐H. , & Yuen, K.‐Y. (2005). Severe acute respiratory syndrome coronavirus‐like virus in Chinese horseshoe bats. Proceedings of the National Academy of Sciences of the United States of America, 102(39), 14040–14045. 10.1073/pnas.0506735102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky, J. A. , Tagliamonte, M. S. , White, S. K. , Elbadry, M. A. , Alam, M. M. , Stephenson, C. J. , Bonny, T. S. , Loeb, J. C. , Telisma, T. , Chavannes, S. , Ostrov, D. A. , Mavian, C. , De Rochars, V. M. B. , Salemi, M. , & Morris, J. G. (2021). Emergence of porcine delta‐coronavirus pathogenic infections among children in Haiti through independent zoonoses and convergent evolution. medRxiv. 10.1101/2021.03.19.21253391 [DOI]
- Letko, M. , & Munster, V. (2020). Functional assessment of cell entry and receptor usage for lineage B β‐coronaviruses, including 2019‐nCoV. bioRxiv. 10.1101/2020.01.22.915660 [DOI] [PMC free article] [PubMed]
- Leung, K. , Shum, M. H. , Leung, G. M. , Lam, T. T. , & Wu, J. T. (2021). Early transmissibility assessment of the N501Y mutant strains of SARS‐CoV‐2 in the United Kingdom, October to November 2020. Eurosurveillance, 26(1). 10.2807/1560-7917.Es.2020.26.1.2002106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Li, W. , Farzan, M. , & Harrison, S. C. (2005). Structure of SARS coronavirus spike receptor‐binding domain complexed with receptor. Science, 309(5742), 1864–1868. 10.1126/science.1116480 [DOI] [PubMed] [Google Scholar]
- Li, J. , Lai, S. , Gao, G. F. , & Shi, W. (2021). The emergence, genomic diversity and global spread of SARS‐CoV‐2. Nature, 600(7889), 408–418. 10.1038/s41586-021-04188-6 [DOI] [PubMed] [Google Scholar]
- Li, W. , Shi, Z. , Yu, M. , Ren, W. , Smith, C. , Epstein, J. H. , Wang, H. , Crameri, G. , Hu, Z. , Zhang, H. , Zhang, J. , McEachern, J. , Field, H. , Daszak, P. , Eaton, B. T. , Zhang, S. , & Wang, L. F. (2005). Bats are natural reservoirs of SARS‐like coronaviruses. Science, 310(5748), 676–679. 10.1126/science.1118391 [DOI] [PubMed] [Google Scholar]
- Lim, P. L. , Kurup, A. , Gopalakrishna, G. , Chan, K. P. , Wong, C. W. , Ng, L. C. , Se‐Thoe, S. Y. , Oon, L. , Bai, X. , Stanton, L. W. , Ruan, Y. , Miller, L. D. , Vega, V. B. , James, L. , Ooi, P. L. , Kai, C. S. , Olsen, S. J. , Ang, B. , & Leo, Y. S. (2004). Laboratory‐acquired severe acute respiratory syndrome. The New England Journal of Medicine, 350(17), 1740–1745. 10.1056/NEJMoa032565 [DOI] [PubMed] [Google Scholar]
- Liu, X. , Wu, Q. , & Zhang, Z. (2021). Global diversification and distribution of coronaviruses with Furin cleavage sites. Frontiers in Microbiology, 12, 649314. 10.3389/fmicb.2021.649314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Xiao, X. , Wei, X. , Li, J. , Yang, J. , Tan, H. , Zhu, J. , Zhang, Q. , Wu, J. , & Liu, L. (2020). Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS‐CoV‐2. Journal of Medical Virology, 92(6), 595–601. 10.1002/jmv.25726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Zheng, H. , Lin, H. , Li, M. , Yuan, R. , Peng, J. , Xiong, Q. , Sun, J. , Li, B. , Wu, J. , Yi, L. , Peng, X. , Zhang, H. , Zhang, W. , Hulswit, R. J. G. , Loman, N. , Rambaut, A. , Ke, C. , Bowden, T. A. , … Lu, J. (2020). Identification of common deletions in the spike protein of severe acute respiratory syndrome coronavirus 2. Journal of Virology, 94(17). 10.1128/jvi.00790-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, G. , Hu, Y. , Wang, Q. , Qi, J. , Gao, F. , Li, Y. , Zhang, Y. , Zhang, W. , Yuan, Y. , Bao, J. , Zhang, B. , Shi, Y. , Yan, J. , & Gao, G. F. (2013). Molecular basis of binding between novel human coronavirus MERS‐CoV and its receptor CD26. Nature, 500(7461), 227–231. 10.1038/nature12328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytras, S. , Xia, W. , Hughes, J. , Jiang, X. , & Robertson, D. L. (2021). The animal origin of SARS‐CoV‐2. Science, 373(6558), 968–970. 10.1126/science.abh0117 [DOI] [PubMed] [Google Scholar]
- Macchiagodena, M. , Pagliai, M. , & Procacci, P. (2020). Identification of potential binders of the main protease 3CL(pro) of the COVID‐19 via structure‐based ligand design and molecular modeling. Chemical Physics Letters, 750, 137489. 10.1016/j.cplett.2020.137489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, Y. S. , Sircar, S. , Bhat, S. , Sharun, K. , Dhama, K. , Dadar, M. , Tiwari, R. , & Chaicumpa, W. (2020). Emerging novel coronavirus (2019‐nCoV)‐current scenario, evolutionary perspective based on genome analysis and recent developments. Veterinary Quarterly, 40(1), 68–76. 10.1080/01652176.2020.1727993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh, K. , Becker, W. B. , & Chanock, R. M. (1967). Growth in suckling‐mouse brain of “IBV‐like” viruses from patients with upper respiratory tract disease. Proceedings of the National Academy of Sciences of the United States of America, 58(6), 2268–2273. 10.1073/pnas.58.6.2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish, Z. A. , Mishra, N. , Olival, K. J. , Fagbo, S. F. , Kapoor, V. , Epstein, J. H. , Alhakeem, R. , Durosinloun, A. , Al Asmari, M. , Islam, A. , Kapoor, A. , Briese, T. , Daszak, P. , Al Rabeeah, A. A. , & Lipkin, W. I. (2013). Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerging Infectious Diseases, 19(11), 1819–1823. 10.3201/eid1911.131172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, M. A. , Corman, V. M. , Jores, J. , Meyer, B. , Younan, M. , Liljander, A. , Bosch, B. J. , Lattwein, E. , Hilali, M. , Musa, B. E. , Bornstein, S. , & Drosten, C. (2014). MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983–1997. Emerging Infectious Diseases, 20(12), 2093–2095. 10.3201/eid2012.141026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu, T. , Takemoto, C. , Kim, Y. T. , Wang, H. , Nishii, W. , Terada, T. , Shirouzu, M. , & Yokoyama, S. (2016). SARS‐CoV 3CL protease cleaves its C‐terminal autoprocessing site by novel subsite cooperativity. Proceedings of the National Academy of Sciences of the United States of America, 113(46), 12997–13002. 10.1073/pnas.1601327113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando, N. S. , Dalebout, T. J. , Zevenhoven‐Dobbe, J. C. , Limpens, R. , van der Meer, Y. , Caly, L. , Druce, J. , de Vries, J. J. C. , Kikkert, M. , Bárcena, M. , Sidorov, I. , & Snijder, E. J. (2020). SARS‐coronavirus‐2 replication in Vero E6 cells: Replication kinetics, rapid adaptation and cytopathology. Journal of General Virology, 101(9), 925–940. 10.1099/jgv.0.001453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival, K. J. , Weekley, C. C. , & Daszak, P. (2015). Are bats really “special” as viral reservoirs? What we know and need to know. In L.‐F. Wang, & C. Cowled (Eds.), Bats and viruses (pp. 281–294). Wiley.
- Ozer, E. A. , Simons, L. M. , Adewumi, O. M. , Fowotade, A. A. , Omoruyi, E. C. , Adeniji, J. A. , Olayinka, O. A. , Dean, T. J. , Zayas, J. , Bhimalli, P. P. , Ash, M. K. , Maiga, A. I. , Somboro, A. M. , Maiga, M. , Godzik, A. , Schneider, J. R. , Mamede, J. I. , Taiwo, B. O. , Hultquist, J. F. , & Lorenzo‐Redondo, R. (2022). Multiple expansions of globally uncommon SARS‐CoV‐2 lineages in Nigeria. Nature Communications, 13(1), 688. 10.1038/s41467-022-28317-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa, G. , Mallery, D. L. , Albecka, A. , Welch, L. G. , Cattin‐Ortola, J. , Luptak, J. , Paul, D. , McMahon, H. T. , Goodfellow, I. G. , Carter, A. , Munro, S. , & James, L. C. (2021). Furin cleavage of SARS‐CoV‐2 Spike promotes but is not essential for infection and cell‐cell fusion. PLOS Pathogens, 17(1), e1009246. 10.1371/journal.ppat.1009246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasher, A. (2021). COVID‐19: Current understanding of its pathophysiology, clinical presentation and treatment. Postgraduate Medical Journal, 97(1147), 312–320. 10.1136/postgradmedj-2020-138577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piplani, S. , Singh, P. K. , Winkler, D. A. , & Petrovsky, N. (2021). In silico comparison of SARS‐CoV‐2 spike protein‐ACE2 binding affinities across species and implications for virus origin. Scientific Reports, 11(1), 13063. 10.1038/s41598-021-92388-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prates, E. T. , Garvin, M. R. , Pavicic, M. , Jones, P. , Shah, M. , Demerdash, O. , Amos, B. K. , Geiger, A. , & Jacobson, D. (2021). Potential pathogenicity determinants identified from structural proteomics of SARS‐CoV and SARS‐CoV‐2. Molecular Biology and Evolution, 38(2), 702–715. 10.1093/molbev/msaa231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine, R. , & Winslow, G. M. (2009). IgM in microbial infections: Taken for granted? Immunology Letters, 125(2), 79–85. 10.1016/j.imlet.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnasinghe, R. , Strohmeier, S. , Amanat, F. , Gillespie, V. L. , Krammer, F. , García‐Sastre, A. , Coughlan, L. , Schotsaert, M. , & Uccellini, M. B. (2020). Comparison of transgenic and adenovirus hACE2 mouse models for SARS‐CoV‐2 infection. Emerging Microbes & Infections, 9(1), 2433–2445. 10.1080/22221751.2020.1838955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken, C. B. , Haagmans, B. L. , Müller, M. A. , Gutierrez, C. , Godeke, G. J. , Meyer, B. , Muth, D. , Raj, V. S. , Smits‐De Vries, L. , Corman, V. M. , Drexler, J. F. , Smits, S. L. , El Tahir, Y. E. , De Sousa, R. , van Beek, J. , Nowotny, N. , van Maanen, K. , Hidalgo‐Hermoso, E. , Bosch, B. J. , & Koopmans, M. P. (2013). Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: A comparative serological study. The Lancet Infectious Diseases, 13(10), 859–866. 10.1016/s1473-3099(13)70164-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristanović, E. S. , Kokoškov, N. S. , Crozier, I. , Kuhn, J. H. , & Gligić, A. S. (2020). A forgotten episode of Marburg virus disease: Belgrade, Yugoslavia, 1967. Microbiology and Molecular Biology Reviews, 84(2). 10.1128/mmbr.00095-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal, S. H. , Gerasimova, A. , Ruiz‐Vega, R. , Livingston, K. , Kagan, R. M. , Liu, Y. , Anderson, B. , Owen, R. , Bernstein, L. , Smolgovsky, A. , Xu, D. , Chen, R. , Grupe, A. , Tanpaiboon, P. , & Lacbawan, F. (2022). Development and validation of a high throughput SARS‐CoV‐2 whole genome sequencing workflow in a clinical laboratory. Scientific Reports, 12(1). 10.1038/s41598-022-06091-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozo, M. , & Gronvall, G. K. (2015). The reemergent 1977 H1N1 strain and the gain‐of‐function debate. mBio, 6(4). 10.1128/mBio.01013-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabir, J. S. , Lam, T. T. , Ahmed, M. M. , Li, L. , Shen, Y. , Abo‐Aba, S. E. , Qureshi, M. I. , Abu‐Zeid, M. , Zhang, Y. , Khiyami, M. A. , Alharbi, N. S. , Hajrah, N. H. , Sabir, M. J. , Mutwakil, M. H. , Kabli, S. A. , Alsulaimany, F. A. , Obaid, A. Y. , Zhou, B. , Smith, D. K. , … Guan, Y. (2016). Co‐circulation of three camel coronavirus species and recombination of MERS‐CoVs in Saudi Arabia. Science, 351(6268), 81–84. 10.1126/science.aac8608 [DOI] [PubMed] [Google Scholar]
- Saif, L. J. (2004). Animal coronaviruses: What can they teach us about the severe acute respiratory syndrome? Revue Scientifique et Technique, 23(2), 643–660. doi: 10.20506/rst.23.2.1513 [DOI] [PubMed] [Google Scholar]
- Sasaki, M. , Uemura, K. , Sato, A. , Toba, S. , Sanaki, T. , Maenaka, K. , Hall, W. W. , Orba, Y. , & Sawa, H. (2021). SARS‐CoV‐2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2‐deficient cells. PLOS Pathogens, 17(1), e1009233. 10.1371/journal.ppat.1009233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segreto, R. , & Deigin, Y. (2021). The genetic structure of SARS‐CoV‐2 does not rule out a laboratory origin: SARS‐COV‐2 chimeric structure and furin cleavage site might be the result of genetic manipulation. Bioessays, 43(3), e2000240. 10.1002/bies.202000240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior, K. (2003). Recent Singapore SARS case a laboratory accident. The Lancet Infectious Diseases, 3(11), 679. 10.1016/s1473-3099(03)00815-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, D. , Mukherjee, R. , Grewe, D. , Bojkova, D. , Baek, K. , Bhattacharya, A. , Schulz, L. , Widera, M. , Mehdipour, A. R. , Tascher, G. , Geurink, P. P. , Wilhelm, A. , van der Heden van Noort, G. J. , Ovaa, H. , Muller, S. , Knobeloch, K. P. , Rajalingam, K. , Schulman, B. A. , Cinatl, J. , … Dikic, I. (2020). Papain‐like protease regulates SARS‐CoV‐2 viral spread and innate immunity. Nature, 587(7835), 657–662. 10.1038/s41586-020-2601-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, D. , & Yi, S. V. (2021). On the origin and evolution of SARS‐CoV‐2. Experimental & Molecular Medicine, 53(4), 537–547. 10.1038/s12276-021-00604-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Z. , Xu, Y. , Bao, L. , Zhang, L. , Yu, P. , Qu, Y. , Zhu, H. , Zhao, W. , Han, Y. , & Qin, C. (2019). From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses, 11(1). 10.3390/v11010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, S. , Wong, G. , Shi, W. , Liu, J. , Lai, A. C. K. , Zhou, J. , Liu, W. , Bi, Y. , & Gao, G. F. (2016). Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends in Microbiology, 24(6), 490–502. 10.1016/j.tim.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S. H. , Chen, Q. , Gu, H. J. , Yang, G. , Wang, Y. X. , Huang, X. Y. , Liu, S. S. , Zhang, N. N. , Li, X. F. , Xiong, R. , Guo, Y. , Deng, Y. Q. , Huang, W. J. , Liu, Q. , Liu, Q. M. , Shen, Y. L. , Zhou, Y. , Yang, X. , Zhao, T. Y. , & Wang, Y. C. (2020). A Mouse model of SARS‐CoV‐2 infection and pathogenesis. Cell Host & Microbe, 28(1), 124–133. e124. 10.1016/j.chom.2020.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Y. W. , Fung, T. S. , Shen, H. , Huang, M. , & Liu, D. X. (2018). Coronavirus infectious bronchitis virus non‐structural proteins 8 and 12 form stable complex independent of the non‐translated regions of viral RNA and other viral proteins. Virology, 513, 75–84. 10.1016/j.virol.2017.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger, J. K. , Reid, A. H. , Lourens, R. M. , Wang, R. , Jin, G. , & Fanning, T. G. (2005). Characterization of the 1918 influenza virus polymerase genes. Nature, 437(7060), 889–893. 10.1038/nature04230 [DOI] [PubMed] [Google Scholar]
- Temmam, S. , Vongphayloth, K. , Salazar, E. B. , Munier, S. , Bonomi, M. , Régnault, B. , Douangboubpha, B. , Karami, Y. , Chretien, D. , Sanamxay, D. , Xayaphet, V. , Paphaphanh, P. , Lacoste, V. , Somlor, S. , Lakeomany, K. , Phommavanh, N. , Pérot, P. , Donati, F. , Bigot, T. , … Eloit, M. (2021). Coronaviruses with a SARS‐CoV‐2‐like receptorbinding domain allowing ACE2‐mediated entry into human cells isolated from bats of Indochinese peninsula. Nature, 604(7905), 330–336. doi: 10.21203/rs.3.rs-871965/v1 [DOI]
- Tyshkovskiy, A. , & Panchin, A. Y. (2021). There is still no evidence of SARS‐CoV‐2 laboratory origin: Response to Segreto and Deigin (10.1002/bies.202100137). Bioessays, 43(12), e2100194. 10.1002/bies.202100194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana, R. , Moyo, S. , Amoako, D. G. , Tegally, H. , Scheepers, C. , Althaus, C. L. , Anyaneji, U. J. , Bester, P. A. , Boni, M. F. , Chand, M. , Choga, W. T. , Colquhoun, R. , Davids, M. , Deforche, K. , Doolabh, D. , du Plessis, L. , Engelbrecht, S. , Everatt, J. , Giandhari, J. , … de Oliveira, T. (2022). Rapid epidemic expansion of the SARS‐CoV‐2 Omicron variant in southern Africa. Nature, 603(7902), 679–686. 10.1038/s41586-022-04411-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova, A. N. , Diaz, A. , Damtie, D. , Xiu, L. , Toh, T. H. , Lee, J. S. , Saif, L. J. , & Gray, G. C. (2022). Novel canine coronavirus isolated from a hospitalized patient with pneumonia in East Malaysia. Clinical Infectious Diseases, 74(3), 446–454. 10.1093/cid/ciab456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vohringer, H. S. , Sanderson, T. , Sinnott, M. , De Maio, N. , Nguyen, T. , Goater, R. , Schwach, F. , Harrison, I. , Hellewell, J. , Ariani, C. V. , Goncalves, S. , Jackson, D. K. , Johnston, I. , Jung, A. W. , Saint, C. , Sillitoe, J. , Suciu, M. , Goldman, N. , Panovska‐Griffiths, J. , … Gerstung, M. (2021). Genomic reconstruction of the SARS‐CoV‐2 epidemic in England. Nature, 600(7889), 506–511. 10.1038/s41586-021-04069-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz, E. , Hill, V. , McCrone, J. T. , Price, A. , Jorgensen, D. , O'Toole, Á. , Southgate, J. , Johnson, R. , Jackson, B. , Nascimento, F. F. , Rey, S. M. , Nicholls, S. M. , Colquhoun, R. M. , da Silva Filipe, A. , Shepherd, J. , Pascall, D. J. , Shah, R. , Jesudason, N. , Li, K. , … Connor, T. R. (2021). Evaluating the effects of SARS‐CoV‐2 spike mutation D614G on transmissibility and pathogenicity. Cell, 184(1), 64–75.e11. 10.1016/j.cell.2020.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee, S. , Duengkae, P. , Rodpan, A. , Kaewpom, T. , Maneeorn, P. , Kanchanasaka, B. , Yingsakmongkon, S. , Sittidetboripat, N. , Chareesaen, C. , Khlangsap, N. , Pidthong, A. , Leadprathom, K. , Ghai, S. , Epstein, J. H. , Daszak, P. , Olival, K. J. , Blair, P. J. , Callahan, M. V. , & Hemachudha, T. (2015). Diversity of coronavirus in bats from Eastern Thailand. Virology Journal, 12, 57. 10.1186/s12985-015-0289-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y. , Shang, J. , Graham, R. , Baric, R. S. , & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade‐long structural studies of SARS coronavirus. Journal of Virology, 94(7). 10.1128/jvi.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Shuai, L. , Wang, C. , Liu, R. , He, X. , Zhang, X. , Sun, Z. , Shan, D. , Ge, J. , Wang, X. , Hua, R. , Zhong, G. , Wen, Z. , & Bu, Z. (2020). Mouse‐adapted SARS‐CoV‐2 replicates efficiently in the upper and lower respiratory tract of BALB/c and C57BL/6J mice. Protein Cell, 11(10), 776–782. 10.1007/s13238-020-00767-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Xu, Y. , Gao, R. , Lu, R. , Han, K. , Wu, G. , & Tan, W. (2020). Detection of SARS‐CoV‐2 in different types of clinical specimens. JAMA, 323(18), 1843–1844. 10.1001/jama.2020.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Lv, J. , Yu, P. , Qu, Y. , Zhou, Y. , Zhou, L. , Zhu, Q. , Li, S. , Song, J. , Deng, W. , Gao, R. , Liu, Y. , Liu, J. , Tong, W. M. , Qin, C. , & Huang, B. (2022). SARS‐CoV‐2 treatment effects induced by ACE2‐expressing microparticles are explained by the oxidized cholesterol‐increased endosomal pH of alveolar macrophages. Cellular & Molecular Immunology, 19(2), 210–221. 10.1038/s41423-021-00813-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . (2021). WHO‐convened global study of origins of SARS‐CoV‐2. China Part (World Health Organization).
- Wilensky, G. R. (2021). Seeking the origins of SARS‐CoV‐2—and more cooperative global responses to new viral threats. JAMA Health Forum, 2(9). 10.1001/jamahealthforum.2021.3547 [DOI] [PubMed] [Google Scholar]
- Wolf, Y. I. , Kazlauskas, D. , Iranzo, J. , Lucía‐Sanz, A. , Kuhn, J. H. , Krupovic, M. , Dolja, V. V. , & Koonin, E. V. (2018). Origins and evolution of the global RNA virome. mBio, 9(6). 10.1128/mBio.02329-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, Y. I. , Kazlauskas, D. , Iranzo, J. , Lucía‐Sanz, A. , Kuhn, J. H. , Krupovic, M. , Dolja, V. V. , & Koonin, E. V. (2019). Reply to Holmes and Duchêne, “Can Sequence Phylogenies Safely Infer the Origin of the Global Virome?”: Deep phylogenetic analysis of RNA viruses is highly challenging but not meaningless. mBio, 10(2). 10.1128/mBio.00542-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, A. C. P. , Li, X. , Lau, S. K. P. , & Woo, P. C. Y. (2019). Global epidemiology of bat coronaviruses. Viruses, 11(2). 10.3390/v11020174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, Y. C. , Lau, S. Y. , Wang To, K. K. , Mok, B. W. Y. , Li, X. , Wang, P. , Deng, S. , Woo, K. F. , Du, Z. , Li, C. , Zhou, J. , Chan, J. F. W. , Yuen, K. Y. , Chen, H. , & Chen, Z. (2021). Natural transmission of bat‐like severe acute respiratory syndrome coronavirus 2 without proline‐arginine‐arginine‐alanine variants in coronavirus disease 2019 patients. Clinical Infectious Diseases, 73(2), e437–e444. 10.1093/cid/ciaa953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, P. C. , Lau, S. K. , Chu, C. M. , Chan, K. H. , Tsoi, H. W. , Huang, Y. , Wong, B. H. , Poon, R. W. , Cai, J. J. , Luk, W. K. , Poon, L. L. , Wong, S. S. , Guan, Y. , Peiris, J. S. , & Yuen, K. Y. (2005). Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. Journal of Virology, 79(2), 884–895. 10.1128/jvi.79.2.884-895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp, D. , Wang, N. , Corbett, K. S. , Goldsmith, J. A. , Hsieh, C. L. , Abiona, O. , Graham, B. S. , & McLellan, J. S. (2020). Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science, 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel, A. G. , Benton, D. J. , Xu, P. , Roustan, C. , Martin, S. R. , Rosenthal, P. B. , Skehel, J. J. , & Gamblin, S. J. (2020). SARS‐CoV‐2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin‐cleavage effects. Nature Structural & Molecular Biology, 27(8), 763–767. 10.1038/s41594-020-0468-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, A. , Peng, Y. , Huang, B. , Ding, X. , Wang, X. , Niu, P. , Meng, J. , Zhu, Z. , Zhang, Z. , Wang, J. , Sheng, J. , Quan, L. , Xia, Z. , Tan, W. , Cheng, G. , & Jiang, T. (2020). Genome composition and divergence of the novel coronavirus (2019‐nCoV) originating in China. Cell Host & Microbe, 27(3), 325–328. 10.1016/j.chom.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. , Zhao, S. , Yu, B. , Chen, Y. M. , Wang, W. , Song, Z. G. , Hu, Y. , Tao, Z. W. , Tian, J. H. , Pei, Y. Y. , Yuan, M. L. , Zhang, Y. L. , Dai, F. H. , Liu, Y. , Wang, Q. M. , Zheng, J. J. , Xu, L. , Holmes, E. C. , & Zhang, Y. Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J. , Li, Y. , Shen, X. , Goh, G. , Zhu, Y. , Cui, J. , Wang, L. F. , Shi, Z. L. , & Zhou, P. (2018). Dampened STING‐dependent interferon activation in bats. Cell Host & Microbe, 23(3), 297–301.e294. 10.1016/j.chom.2018.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, R. H. , He, J. F. , Evans, M. R. , Peng, G. W. , Field, H. E. , Yu, D. W. , Lee, C. K. , Luo, H. M. , Lin, W. S. , Lin, P. , Li, L. H. , Liang, W. J. , Lin, J. Y. , & Schnur, A. (2004). Epidemiologic clues to SARS origin in China. Emerging Infectious Diseases, 10(6), 1030–1037. 10.3201/eid1006.030852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Chen, P. , Wang, J. , Feng, J. , Zhou, H. , Li, X. , Zhong, W. , & Hao, P. (2020). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China Life Sciences, 63(3), 457–460. 10.1007/s11427-020-1637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. , Shi, L. , Wang, Y. , Zhang, J. , Huang, L. , Zhang, C. , Liu, S. , Zhao, P. , Liu, H. , Zhu, L. , Tai, Y. , Bai, C. , Gao, T. , Song, J. , Xia, P. , Dong, J. , Zhao, J. , & Wang, F. S. (2020). Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. The Lancet Respiratory Medicine, 8(4), 420–422. 10.1016/s2213-2600(20)30076-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, Y. , & Liu, D. X. (2009). Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin‐dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. Journal of Virology, 83(17), 8744–8758. 10.1128/jvi.00613-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H. , Jiao, H. , Liu, Q. , Zhang, Z. , Xiong, Q. , Wang, B. J. , Wang, X. , Guo, M. , Wang, L. F. , Lan, K. , Chen, Y. , & Zhao, H. (2021). ACE2 receptor usage reveals variation in susceptibility to SARS‐CoV and SARS‐CoV‐2 infection among bat species. Nature Ecology and Evolution, 5(5), 600–608. 10.1038/s41559-021-01407-1 [DOI] [PubMed] [Google Scholar]
- Yang, X. L. , Hu, B. , Wang, B. , Wang, M. N. , Zhang, Q. , Zhang, W. , Wu, L. J. , Ge, X. Y. , Zhang, Y. Z. , Daszak, P. , Wang, L. F. , & Shi, Z. L. (2015). Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. Journal of Virology, 90(6), 3253–3256. 10.1128/jvi.02582-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, W. , Ma, D. , Wang, H. , Tang, X. , Du, C. , Pan, H. , Li, C. , Lin, H. , Farzan, M. , Zhao, J. , Li, Y. , & Zhong, G. (2021). Effect of SARS‐CoV‐2 spike mutations on animal ACE2 usage and in vitro neutralization sensitivity. bioRxiv. 10.1101/2021.01.27.428353 [DOI]
- Yi, Y. , Lagniton, P. N. P. , Ye, S. , Li, E. , & Xu, R.‐H. (2020). COVID‐19: what has been learned and to be learned about the novel coronavirus disease. International Journal of Biological Sciences, 16(10), 1753–1766. 10.7150/ijbs.45134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y. , Cao, D. , Zhang, Y. , Ma, J. , Qi, J. , Wang, Q. , Lu, G. , Wu, Y. , Yan, J. , Shi, Y. , Zhang, X. , & Gao, G. F. (2017). Cryo‐EM structures of MERS‐CoV and SARS‐CoV spike glycoproteins reveal the dynamic receptor binding domains. Nature Communications, 8, 15092. 10.1038/ncomms15092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech, F. , Schniertshauer, D. , Jung, C. , Herrmann, A. , Cordsmeier, A. , Xie, Q. , Nchioua, R. , Prelli Bozzo, C. , Volcic, M. , Koepke, L. , Muller, J. A. , Kruger, J. , Heller, S. , Stenger, S. , Hoffmann, M. , Pohlmann, S. , Kleger, A. , Jacob, T. , Conzelmann, K. K. , & Kirchhoff, F. (2021). Spike residue 403 affects binding of coronavirus spikes to human ACE2. Nature Communications, 12(1), 6855. 10.1038/s41467-021-27180-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Lin, D. , Kusov, Y. , Nian, Y. , Ma, Q. , Wang, J. , von Brunn, A. , Leyssen, P. , Lanko, K. , Neyts, J. , de Wilde, A. , Snijder, E. J. , Liu, H. , & Hilgenfeld, R. (2020). α‐Ketoamides as broad‐spectrum inhibitors of coronavirus and enterovirus replication: Structure‐based design, synthesis, and activity assessment. Journal of Medicinal Chemistry, 63(9), 4562–4578. 10.1021/acs.jmedchem.9b01828 [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Wu, Q. , & Zhang, Z. (2020). Pangolin homology associated with 2019‐nCoV. bioRxiv. 10.1101/2020.02.19.950253 [DOI]
- Zhang, T. , Wu, Q. , & Zhang, Z. (2020). Probable pangolin origin of SARS‐CoV‐2 associated with the COVID‐19 outbreak. Current Biology, 30(7), 1346–1351. e1342. 10.1016/j.cub.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. M. , Herbst, W. , Kousoulas, K. G. , & Storz, J. (1994). Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. Journal of Medical Virology, 44(2), 152–161. 10.1002/jmv.1890440207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , Chen, X. , Hu, T. , Li, J. , Song, H. , Liu, Y. , Wang, P. , Liu, D. , Yang, J. , Holmes, E. C. , Hughes, A. C. , Bi, Y. , & Shi, W. (2020). A novel bat coronavirus closely related to SARS‐CoV‐2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Current Biology, 30(11), 2196–2203. e2193. 10.1016/j.cub.2020.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P. , Yang, X. L. , Wang, X. G. , Hu, B. , Zhang, L. , Zhang, W. , Si, H. R. , Zhu, Y. , Li, B. , Huang, C. L. , Chen, H. D. , Chen, J. , Luo, Y. , Guo, H. , Jiang, R. D. , Liu, M. Q. , Chen, Y. , Shen, X. R. , Wang, X. , … Shi, Z. L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Feng, F. , Hu, G. , Wang, Y. , Yu, Y. , Zhu, Y. , Xu, W. , Cai, X. , Sun, Z. , Han, W. , Ye, R. , Qu, D. , Ding, Q. , Huang, X. , Chen, H. , Xu, W. , Xie, Y. , Cai, Q. , Yuan, Z. , & Zhang, R. (2021). A genome‐wide CRISPR screen identifies host factors that regulate SARS‐CoV‐2 entry. Nature Communications, 12(1), 961. 10.1038/s41467-021-21213-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z. , Meng, K. , & Meng, G. (2020). Genomic recombination events may reveal the evolution of coronavirus and the origin of SARS‐CoV‐2. Scientific Reports, 10(1), 21617. 10.1038/s41598-020-78703-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, L. , Ruan, F. , Huang, M. , Liang, L. , Huang, H. , Hong, Z. , Yu, J. , Kang, M. , Song, Y. , Xia, J. , Guo, Q. , Song, T. , He, J. , Yen, H. L. , Peiris, M. , & Wu, J. (2020). SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. The New England Journal of Medicine, 382(12), 1177–1179. 10.1056/NEJMc2001737 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study. cd_value_code=text


