Abstract
The binding of platelets by bacteria is a proposed central mechanism in the pathogenesis of infective endocarditis. Platelet binding by Streptococcus mitis strain SF100 (an endocarditis isolate) was recently shown to be mediated in part by the surface proteins PblA and PblB. The genes encoding PblA and PblB are clustered with genes nearly identical to those of streptococcal phages r1t, 01205, and Dp-1, suggesting that pblA and pblB might reside within a prophage. To address this possibility, cultures of SF100 were exposed to either mitomycin C or UV light, both of which are known to induce the lytic cycle of many temperate phages. Both treatments caused a significant increase in the transcription of pblA. Treatment with mitomycin C or UV light also caused a substantial increase in the expression of PblA and PblB, as detected by Western blot analysis of proteins in the SF100 cell wall. By electron microscopy, phage particles were readily visible in the supernatants from induced cultures of SF100. The phage, designated SM1, had a double-stranded DNA genome of approximately 35 kb. Southern blot analysis of phage DNA indicated that pblA and pblB were contained within the SM1 genome. Furthermore, Western blot analysis of phage proteins revealed that both PblA and PblB were present in the phage particles. These findings indicate that PblA and PblB are encoded by a lysogenic bacteriophage, which could facilitate the dissemination of these potential virulence determinants to other bacterial pathogens.
The binding of microorganisms to human platelets may have a central role in the pathogenesis of infective endocarditis (23). The adherence of bacteria in the bloodstream to platelets on the damaged endocardial surface may be an important mechanism for the initial colonization of cardiac valves (8, 9, 16). The subsequent deposition of platelets onto this infected surface may also be mediated by direct binding of bacteria to platelets, leading to the formation of mature, macroscopic vegetations (7, 25). Recent studies further indicate that direct binding may contribute to one of the major complications of this disease, the formation of systemic emboli (24). Thus, the binding of microbes with platelets may be crucial for several steps in the pathogenesis of endocardial infection.
Although Streptococcus mitis is a leading cause of infective endocarditis, few potential virulence determinants of this organism have been identified. We have recently described two distinct genetic loci of S. mitis strain SF100 that contribute to platelet binding (2). The first locus encodes PblT, a probable transmembrane transporter. As yet, the mechanism by which PblT enhances binding to platelets has not been determined. The second locus encodes two cell wall-associated proteins, PblA and PblB, that also augment platelet binding. Both proteins must be expressed on the bacterial surface for normal levels of binding to occur. Although the precise interactions of PblA and PblB with the platelet membrane have not been defined entirely, data indicate that PblB may be a direct platelet adhesin, whereas PblA may affect the surface presentation of PblB (2).
Several features of PblA and PblB are somewhat atypical of surface proteins of gram-positive bacteria. PblA is predicted to have an unusually long (71-residue) amino-terminal signal peptide (in Bacillus subtilis, secretory signal peptides range from 19 to 44 residues, with an average length of 28 residues [26]). PblB has no apparent signal for transport mediated by the general secretory pathway. Neither protein has a signal for lipid anchoring [L(A/S)(A/G)C (26)] or a signal for covalent anchoring to the cell wall peptidoglycan (LPXTG motif [12, 19, 20]). However, PblA does have regular repeats of a tryptophan-rich pentapeptide motif, which may constitute a wall association domain similar to those of the choline-binding proteins of Streptococcus pneumoniae (31) and the lipoteichoic acid-binding proteins of Listeria monocytogenes (3).
PblA and PblB are also unusual because neither protein has strong similarity to known bacterial adhesins. Instead, these proteins resemble structural components of bacteriophages. The amino-terminal half of PblA is similar to a protein of unknown function encoded in the structural protein region of the Lactococcus lactis phage r1t (27). The carboxy-terminal half of PblB resembles the host attachment proteins of various temperate and lytic phages and is most similar to a tail fiber protein of the Streptococcus thermophilus phage 01205 (22).
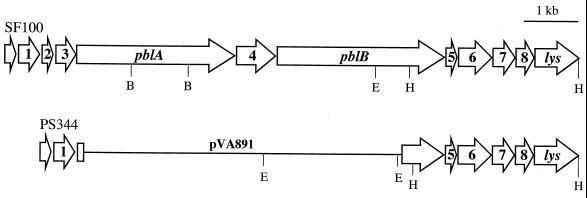
PblA and PblB thus appear to be remnants of bacteriophage components. In addition to the similarity of PblA and PblB to structural components of streptococcal phages, proteins encoded by open reading frames (ORFs) 1 to 3 upstream of pblA (Fig. 1) are 69 to 77% similar (47 to 61% identical) to proteins encoded by ORFs 37 to 39 of the L. lactis phage r1t (2). In aggregate, these data indicate that the pblAB locus might be a mosaic of streptococcal phage sequences.
FIG. 1.
Map of the pblAB locus in S. mitis strains SF100 and PS344. The pblAB locus is an apparent mosaic of streptococcal phage sequences. ORF1, ORF2, ORF3, and pblA are similar to sequences of the L. lactis phage r1t. pblB is similar to an S. thermophilus phage 01205 gene encoding a tail fiber protein. ORF4, ORF5, ORF6, and ORF8 are not similar to any sequences reported in GenBank. ORF7 and lys are similar to sequences of the S. pneumoniae phage Dp-1. In PS344, the pblAB locus was replaced with the integrative vector pVA891. EcoRI sites (E), as well as select BglII (B) and HindIII (H) sites, are indicated.
The high level of similarity of genes in the pblAB locus to phage genes, as well as their organization on the S. mitis chromosome, suggested that pblA and pblB might reside within a prophage. To address this possibility, we examined whether phage production could be induced from strain SF100. This report describes the isolation of a temperate bacteriophage, designated SM1, that carries pblA and pblB. Furthermore, these data indicate that PblA and PblB are incorporated into phage particles, as well as into the S. mitis cell wall.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
The bacterial strains and plasmids used in this study are listed in Table 1. S. mitis strain SF100 is a previously described isolate that binds human platelets in vitro (2). Strain PS344, a variant of SF100 with reduced platelet binding (2), contains a 6.6-kb chromosomal deletion in the pblAB locus (Fig. 1). S. mitis strains were grown in Todd-Hewitt broth (THB) (Difco Laboratories) or on sheep blood agar (Remel) at 37°C in a 5% CO2 environment. When indicated, chloramphenicol was added to the media at a concentration of 5 μg per ml. Escherichia coli strains were grown in Luria-Bertani broth (Fisher) containing 100 μg of ampicillin per ml or 15 μg of chloramphenicol per ml, when appropriate. Mitomycin C, sodium lauroylsarcosinate, and o-nitrophenyl β-d-galactoside were obtained from Sigma.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | Fr− m+ φ80dlacZΔM15 | Gibco BRL |
| S. mitis | ||
| SF100 | Platelet-binding parental strain | 2 |
| PS344 | SF100ΔORF2-pblB | 2 |
| PS291 | SF100 pblA::pEVP3 with lacZ in the forward orientation | This study |
| PS294 | SF100 pblA::pEVP3 with lacZ in the reverse orientation | This study |
| Plasmids | ||
| pBluescript | Apr | Stratagene |
| pEVP3 | cat lacZ ori (E. coli) | 4 |
| pSF100AZ | pEVP3 with a BglII fragment of pblA in the same orientation as lacZ | This study |
| pSF100ArevZ | pEVP3 with a BglII fragment of pblA oriented opposite to lacZ | This study |
Apr, ampicillin resistance.
DNA sequence analysis.
The sequence of a region of the SF100 chromosome downstream of pblB was obtained by inverse PCR. Chromosomal DNA was digested with HindIII and then treated with DNA ligase under dilute conditions to allow circular ligation of fragments. The circularized DNA fragments were used as a template for PCR, using primers reading outward from pblB (5′-CGCAGATACTACAACAGACC-3′ and 5′-TCCCCTCAATACAAACGAATG-3′). The resulting 3-kb PCR product was cloned in pBluescript (Stratagene) and sequenced by primer walking. DNA sequencing was performed by the Biomolecular Resource Center at the University of California, San Francisco, using the ABI Prism system (Applied Biosystems). The sequence was assembled, formatted, and translated, using Gene Construction Kit 2 software (Textco, Inc., West Lebanon, N.H.). Protein similarity searches were conducted, using BLAST programs, against sequences in the nonredundant databases available at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
Construction of the pblA::lacZ fusion.
The plasmid pEVP3, an integrative vector that carries a promoterless E. coli lacZ gene preceded by the B. subtilis spoVG ribosome binding site (4), was used to generate a pblA::lacZ transcriptional fusion. A 0.9-kb BglII fragment internal to pblA was cloned in the BamHI site of pEVP3 and then used to transform E. coli strain DH5α by electroporation. A plasmid with the pblA coding region in the same orientation as lacZ (pSF100AZ) and one with pblA in the reverse orientation (pSF100ArevZ) were each introduced separately to S. mitis strain SF100 by natural transformation, using a competence-stimulating peptide specific for SF100 as described (2). Integration at the expected site was confirmed by Southern blot analysis of chromosomal DNA (not shown).
Bacteriophage inductions.
S. mitis strains were grown for 18 h in THB (SF100 and PS344) or THB containing chloramphenicol (PS291 and P294). Cultures were diluted 1:10 in fresh THB (without antibiotics), incubated for 1 h (SF100 and PS344) or 1.5 h (PS291 and PS294), and then treated either by adding mitomycin C to a final concentration of 0.2 μg per ml or by exposing culture tubes to a UV light source (312 nm wavelength) for 3 min. Bacterial cultures were then incubated at 37°C for an additional 1.5 or 3 h prior to harvesting, as indicated below.
Assay for β-galactosidase activity.
Production of β-galactosidase by PS291 and PS294 was assessed 1.5 h after induction (before phage-mediated cell lysis was likely to occur). Enzyme activity was determined by the method of Miller (17), using the chromogenic substrate o-nitrophenyl β-d-galactoside. Differences in β-galactosidase activity were compared by the Student t test.
Western blot analysis.
Cell surface proteins were extracted from S. mitis strains 3 h after induction (by which time phage is typically assembled and released), using sodium lauroylsarcosinate as described (15). For comparison of uninduced and induced cultures, wells were loaded with proteins extracted from equivalent numbers of bacteria, as determined by optical density at 600 nm (corresponding to approximately 15 μg of total protein per well). For analysis of purified phage, wells were loaded with 400 ng of phage SM1 proteins or 300 ng of phage SM1ΔAB proteins. The cell wall proteins or phage particles were boiled for 10 min in sample buffer prior to loading. The proteins were separated by electrophoresis through sodium dodecyl sulfate–6% polyacrylamide gels under reducing conditions, transferred to Biotrace NT nitrocellulose membranes (Pall Corporation), and incubated with anti-PblA or anti-PblB goat polyclonal antiserum (2). Antibody binding was detected by incubation of the membranes with horseradish peroxidase-conjugated anti-goat immunoglobulin G (Sigma), followed by development with the Super Signal (Pierce) chemiluminescent substrate.
Purification of phage particles.
Phage was isolated from 500-ml cultures of S. mitis strains 3 h after induction with mitomycin C. Cells and debris were removed by centrifugation (3,500 × g at 4°C for 10 min). The supernatant was combined with NaCl (30 g per liter, final concentration) and stirred for 15 min. Phage particles were precipitated by the slow addition of polyethylene glycol 8000 (Fisher) to a final concentration of 10% (wt/vol). The treated supernatant was stirred gently for 1 h, transferred to centrifuge bottles, and incubated at 4°C for 18 h. The precipitated material was recovered by centrifugation (3,500 × g, 4°C, 20 min) and then gently resuspended in 20 ml of SM buffer (0.01% gelatin, 10 mM NaCl, 8 mM MgSO4, 50 mM Tris-HCl [pH 7.5]) and stirred at 4°C for 18 h. The resuspended material was layered onto a step gradient (prepared by layering 2 ml of a 20% sucrose solution [wt/vol in SM buffer] onto 2 ml each of 1.4, 1.5, and 1.6 g of CsCl per ml of SM buffer) and centrifuged at 80,000 × g at 4°C for 2 h. The phage particles, which formed a band within the second CsCl step, were collected by puncturing the side of the centrifuge tube with an 18-gauge needle. The isolated particles were dialyzed against phosphate-buffered saline supplemented with 10 mM MgSO4 and stored at 4°C.
Electron microscopy.
Phage particles were negatively stained with a 2% uranyl acetate solution and examined using a Phillips Tecnai 10 transmission electron microscope at an accelerating voltage of 80 kV.
DNA purification.
Chromosomal DNA was extracted from S. mitis strains as described (2). Phage DNA was extracted from purified phage particles using Phase Lock Gel tubes (Eppendorf) as detailed by the manufacturer.
Southern blot analysis.
Following electrophoresis through 0.7% agarose gels, DNA was transferred to nylon membranes, using a Trans-Blot semidry transfer apparatus (Bio-Rad). Membranes were hybridized with digoxigenin-labeled probes, and developed with the CDP-Star chemiluminescent substrate as recommended by the supplier (Roche). Probes were generated by PCR amplification of cloned fragments, using primers corresponding to pblA (5′-AAGGATCCAATAGGAGGTGAGGATTAATGGCTACAG-3′ and 5′-AAGGATCCATTAGATTCCCTCCCTTGC-3′), pblB (5′-AAGGATCCTTGGAGGTATAAAATATGATTTACTT-3′ and 5′-AAGGATCCTTTGTTTGTCCTGTTCGTTCATGC-3′), or pblT (5′-GTCCAATGAAGGCTAAA-3′ and 5′-TTTGATGATTGCCTCTC-3′).
Nucleotide sequence accession number.
The sequence of the SF100 chromosome downstream of pblB was used to supplement the sequence previously deposited under accession number AY007505.
RESULTS
Sequence analysis of a chromosomal segment adjacent to pblB.
Previous analysis of PblA and PblB indicated that these proteins were similar to structural proteins of the lysogenic streptococcal phages r1t and 01205. In these related phages, additional late genes (such as those encoding holin and lysin) are located downstream of genes encoding structural proteins. To determine whether additional phage-like genes were adjacent to pblB, a segment of the SF100 chromosome downstream of pblB was amplified by inverse PCR and sequenced. Five ORFs were identified within this segment (Fig. 1). ORF5, ORF6, and ORF8 encode proteins with no similarity to reported sequences. The protein encoded by ORF7 is 52% similar (31% identical) to a component of unspecified function (ORF5) of the S. pneumoniae lytic phage Dp-1 (21). The most 3′ ORF (lys) encodes a protein that is 85% similar (73% identical) to the first 281 amino acids of the 296-amino-acid residue lysin encoded by Dp-1. The similarity of ORF7 and lys to components of a third streptococcal phage further indicates that the pblAB locus is a mosaic of genes from multiple phages.
Expression of the pblAB locus is induced with mitomycin C and UV light.
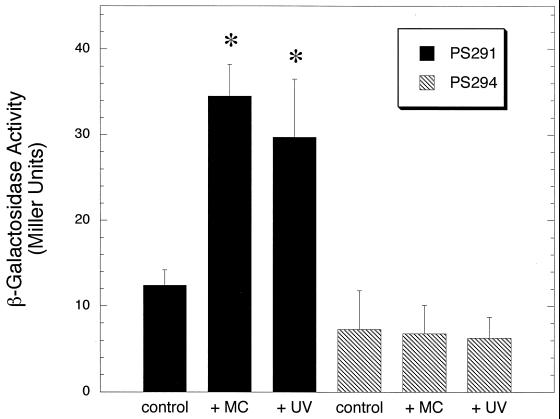
The high similarity of proteins encoded upstream and downstream of pblA and pblB to components of temperate and lytic bacteriophages suggested that the pblAB locus might constitute a portion of a prophage. If so, expression of pblA and pblB was likely to be induced by mitomycin C or UV light, DNA-damaging agents that are typically used to induce the lytic cycle of temperate phages. To monitor transcription of the pblAB locus, a promoterless lacZ reporter was incorporated in pblA, generating strain PS291. Cultures of PS291 were exposed to mitomycin C or UV light, and production of β-galactosidase was measured 1.5 h later. Compared with untreated cultures, those exposed to mitomycin C or UV light showed a significant increase in β-galactosidase activity (Fig. 2). Treatment with mitomycin C led to a 2.8-fold increase (P < 0.0001), and treatment with UV light led to a 2.5-fold increase (P < 0.0001) in β-galactosidase production. Strain PS294, which has lacZ incorporated in pblA in the opposite orientation, showed no increase in β-galactosidase production after exposure to mitomycin C or UV light (P > 0.05 for cultures treated with either agent, compared with untreated cultures of PS294).
FIG. 2.
Induction of pblA transcription by mitomycin C and UV light. S. mitis strains carrying a lacZ transcriptional fusion in pblA in the forward orientation (strain PS291) or in the reverse orientation (strain PS294) were treated with mitomycin C (+MC) or UV light (+UV). Transcription was monitored by measuring the production of β-galactosidase. An asterisk denotes a value that was significantly different (P < 0.0001) from that of untreated cultures (control).
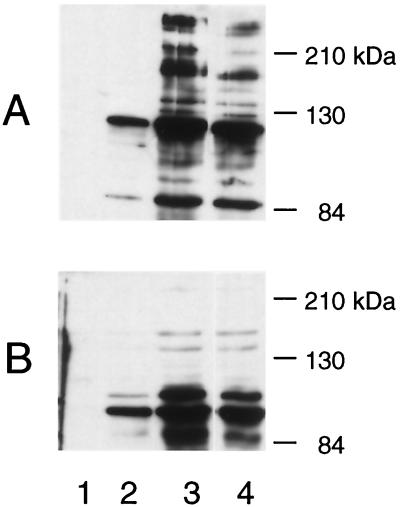
To determine whether increased transcription of the pblAB locus is correlated with increased surface expression of PblA and PblB, the effect of inducing agents on the expression of these cell wall-associated proteins was examined. In Western blots of proteins extracted from uninduced cultures of SF100, the anti-PblA serum reacts predominantly with a protein that migrated at 120 kDa (Fig. 3A, lane 2). Minor proteins were also seen at 110 and 85 kDa (the predicted size of PblA is 107 kDa). The anti-PblB serum reacted with a 110-kDa protein (Fig. 3B, lane 2) and with a minor 120-kDa protein (the predicted size of PblB). Neither antiserum reacted with proteins extracted from uninduced cultures of the PblA− PblB− deletion mutant PS344 (lane 1), confirming the specificity of these reagents.
FIG. 3.
Western blot analysis of PblA and PblB production in uninduced and induced cultures. Proteins were extracted from untreated cultures of PS344 (lane 1), SF100 (lane 2), and cultures of SF100 treated with mitomycin C (lane 3) or UV light (lane 4). Blots were probed with either anti-PblA serum (A) or anti-PblB serum (B).
Following induction with mitomycin C or UV light, an increase in the amount of both PblA and PblB was evident in the cell wall extracts of SF100 (lanes 3 and 4). Numerous higher and lower molecular mass forms of these proteins (ranging from 85 kDa to more than 216 kDa) were also apparent. The combined results indicate that transcription of the pblAB locus is induced with mitomycin C and UV light and that the increased transcription is correlated with an increased expression of PblA and PblB in the bacterial cell wall.
Phage particles are evident in the spent media of induced cultures.
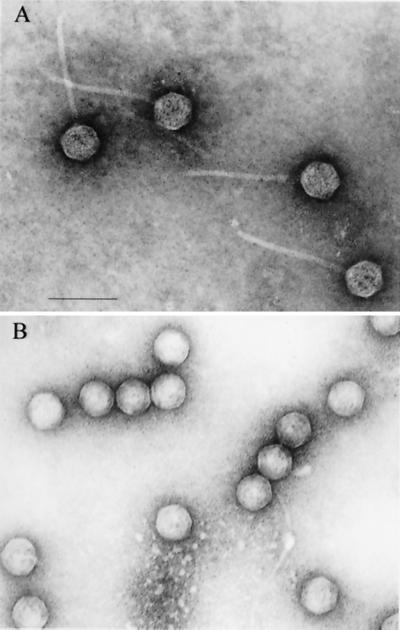
Although the ability to induce expression of the pblAB locus with mitomycin C and UV light indicated that the genes lie within a prophage, it was possible that the phage might be cryptic (i.e., unable to undergo full replication and assembly). To address this issue, the spent medium of an induced culture of SF100 was examined for the presence of phage particles. Using electron microscopy, a small isometric-headed phage, designated SM1, was detected in the material purified from the induced culture supernatant (Fig. 4A). The phage particles had heads approximately 60 nm in diameter and noncontractile tails that were approximately 150 by 8 nm. These features indicate that phage SM1 is a member of the Siphoviridae class of bacteriophages.
FIG. 4.
Electron micrographs of purified phage SM1 (A) and SM1ΔAB (B). Note the absence of tails in particles of phage SM1ΔAB. Bar, 100 nm.
Because PblA and PblB resemble structural components of phage, we then assessed whether the pblAB deletion mutant PS344 was defective in phage production. When examined by electron microscopy, the supernatant fluid of a mitomycin C-induced culture of PS344 was also found to contain phage particles. The phage, designated SM1ΔAB, had heads similar to those of the wild-type phage but lacked tails (Fig. 4B). This finding indicated that the deleted region (Fig. 1) includes genes required for tail assembly. However, the mutation did not appear to affect other aspects of the lytic cycle, such as head assembly, DNA packaging, and particle release.
Analysis of phage DNA.
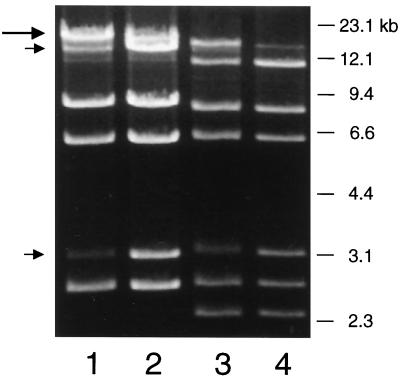
To characterize further the wild-type and mutant phages, the genomic DNA of each phage was characterized by agarose gel electrophoresis. Digestion of DNA from phage SM1 with EcoRI produced seven distinct fragments (Fig. 5, lane 1). To determine whether the phage genome has cohesive ends, DNA samples were heated to 70°C prior to loading (lane 2). A high-molecular-weight fragment resolved to two smaller fragments, indicating the presence of cohesive ends at the genome extremities. Based on the combined sizes of the ethidium bromide-stained restriction fragments (lane 2), the size of the phage SM1 genome is estimated to be 35 kb. DNA from the mutant phage SM1ΔAB had an altered EcoRI digest pattern (Fig. 5, lanes 3 and 4), as would be predicted from the restriction map of the pblAB locus (Fig. 1). The presence of an additional 2.4-kb EcoRI fragment in the SM1ΔAB DNA indicates that the genome was likely to encompass the pblAB region. Furthermore, the presence of distinct fragments in both preparations of phage DNA indicated that each sample contained a homogeneous population of phages.
FIG. 5.
Ethidium bromide stain of phage genomic DNA. DNA from phage SM1 (lanes 1 and 2) or from phage SM1ΔAB (lanes 3 and 4) was digested with EcoRI and then separated by electrophoresis though a 0.7% agarose gel. The samples in lanes 2 and 4 were heated for 10 min at 70°C prior to loading, which led to the shift of a large submolar fragment (large arrow) to two smaller fragments (small arrows).
DNA in purified phage corresponds to the pblA-pblB region.
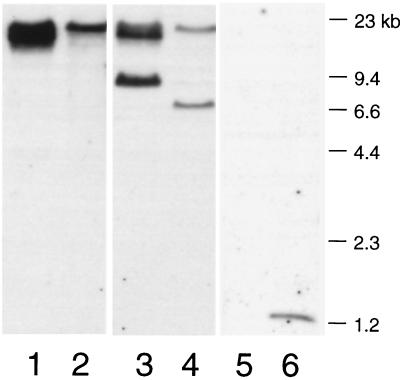
The production of defective phage particles by the pblAB deletion mutant strongly suggested that the phage SM1 genome includes the pblAB locus. However, many bacterial strains have been reported to carry more than one prophage (10, 11). To confirm that the pblAB locus was part of the phage SM1 genome, the purified phage DNA was examined by Southern blot analysis. A pblA probe hybridized with a 15-kb EcoRI fragment of the phage DNA (Fig. 6, lane 1) and with the same-sized fragment of the SF100 chromosomal DNA (lane 2). A pblB probe hybridized with two fragments of the phage and chromosomal DNAs (lanes 3 and 4, respectively). One fragment, which includes the 5′ portion of pblB, was the 15-kb EcoRI fragment that also hybridized with the pblA probe. The other, which includes the 3′ portion of pblB, was a 9-kb fragment of phage DNA or a 7-kb fragment of chromosomal DNA. This difference in fragment sizes indicates that SM1 has an attachment site within the region downstream of pblB. Upon integration of the phage into the host chromosome, this region becomes altered, resulting in the observed change in size of the restriction fragment. A probe from a chromosomal region not linked to the pblAB locus (pblT [2]) hybridized with a 1.2-kb fragment of the chromosomal DNA (lane 6), but did not hybridize with the phage DNA (lane 5). The results clearly demonstrate that pblA and pblB are carried by phage SM1.
FIG. 6.
Southern blot of phage SM1 genomic DNA and SF100 chromosomal DNA. Lanes 1, 3, and 5 contain phage DNA. Lanes 2, 4, and 6 contain chromosomal DNA. DNA was digested with EcoRI, and membranes were hybridized with probes corresponding to pblA (lanes 1 and 2), pblB (lanes 3 and 4), or pblT (lanes 5 and 6). One of two pblB fragments is a different size in the phage DNA (lane 3) than in the SF100 chromosomal DNA (lane 4), which indicates the presence of an attachment site within these fragments.
PblA and PblB are incorporated into phage particles.
Although the expression of cell wall-associated forms of PblA and PblB was greatly increased with phage induction, it was unknown whether the proteins were also structural components of the phage. As is true with phage-encoded toxins, it was possible that the platelet-binding proteins were merely encoded within the phage genome but were not an integral part of the phage structure. To determine whether PblA and PblB were incorporated into phage particles, the purified phages were examined by Western blotting.
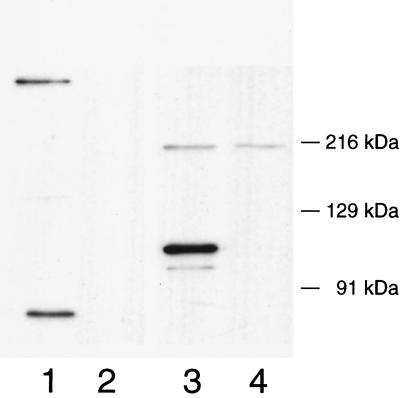
Using anti-PblA serum, predominant proteins of 85 kDa and of a very high molecular mass (in excess of 216 kDa) were detected in phage SM1 (Fig. 7, lane 1). A 130-kDa protein was also present in trace amounts. None of these proteins was detected in particles of phage SM1ΔAB (lane 2). Although each of these forms of PblA was seen among proteins extracted from the cell wall of SF100 following induction (Fig. 3A, lanes 3 and 4), the most predominant form of PblA in cell walls was the 120-kDa form.
FIG. 7.
Western blot analysis of PblA and PblB in purified phage. Lanes 1 and 3 contain phage SM1, and lanes 2 and 4 contain phage SM1ΔAB. Lanes 1 and 2 were probed with anti-PblA serum. Lanes 3 and 4 were probed with anti-PblB serum.
Using anti-PblB serum, a predominant protein of 120 kDa (the predicted size of PblB) and a minor protein of 110 kDa were detected in the purified wild-type phage (Fig. 7, lane 3) but not in particles produced by PS344 (lane 4). This is opposite to the relative proportion of the 120- and 110-kDa forms of PblB extracted from the SF100 cell wall (Fig. 3B, lane 2). Particles of SM1ΔAB, which lack tails, were noted to contain a 215-kDa protein that cross-reacts with the anti-PblB serum (Fig. 7, lane 4).
DISCUSSION
We have previously shown that platelet binding by S. mitis strain SF100 is mediated in part by streptococcal surface proteins PblA and PblB (2). The results presented here now demonstrate that PblA and PblB are encoded by a lysogenic phage of SF100, designated SM1. In addition to being cell wall-associated proteins of S. mitis, PblA and PblB were found to be integral components of phage SM1. These proteins are likely to be involved in tail assembly, since deletion of a chromosomal region spanning pblA and pblB resulted in the production of phage particles that lacked tails. The combined results indicate that PblA and PblB are multifunctional proteins, in that they are important both for platelet binding by SF100 and for the normal morphogenesis of phage tails.
The finding that PblA and PblB are structural proteins of SM1 is unusual, because most phage-encoded virulence factors are not integral components of the phages that encode them. For example, the outer membrane proteins Lom and Bor encoded by phage lambda are not required for phage assembly or propagation (1). In addition, most phage-encoded toxins are not incorporated into phage particles. A possible exception may be the accessory enterotoxin of Vibrio cholera, which has been proposed to be a minor coat protein of the lysogenic phage CTX (28, 29). PblA and PblB, on the other hand, not only show similarity to phage structural proteins, but also were found by Western blot analysis to be present in purified SM1 particles. These findings, in conjunction with the morphology of SM1ΔAB seen in electron micrographs, indicate that PblA and PblB are structural components of SM1.
The mechanism by which PblA and PblB are selectively partitioned to the S. mitis cell wall or to phage particles is unknown. The predominant forms of PblA and PblB in the S. mitis cell wall differ in size from the counterparts in phage SM1. In SM1 particles, PblA appears to be present in both processed (85-kDa) and multimeric (in excess of 216-kDa) forms. Protein processing and the formation of multimeric complexes have been noted in the assembly of two phages of gram-negative bacteria (6, 13). The multiple forms of PblA and PblB in the cell wall may similarly correspond to proteins that have undergone posttranslational modification, or that have formed heteromeric complexes with other cell wall-associated components.
Several lines of evidence indicate that the expression of PblA and PblB by SF100, and hence the ability of this organism to bind platelets, is linked to the life cycle of SM1. First, pblA and pblB are apparently not transcribed independently of the other phage genes. In a previous report, the size of pblB transcripts in uninduced cultures was found to be much greater than 6.9 kb (2). In addition, sequence analysis of the pblAB locus did not reveal any likely promoters immediately upstream of pblA or pblB. In studies presented here, transcription of pblA was increased two- to threefold by agents that induced the production of phage SM1. Thus, pblA and pblB appear to be transcribed only in concert with other phage genes.
The subsequent export of PblA and PblB to the SF100 cell wall may also be linked to the biology of SM1. PblB, in particular, has no apparent signal sequence for transport that would be mediated by components of the general secretory pathway, indicating that another mechanism for export may exist. One possibility is that the surface presentation of PblA and PblB requires phage-mediated lysis. We have detected small quantities of phage SM1 DNA in the culture supernatants of SF100, even when this organism is grown in the absence of inducing agents (data not shown), indicating that low levels of phage-mediated lysis occur constitutively. Thus, it is conceivable that PblA and PblB are released via lysis of a small number of cells, and that the proteins then interact exogenously with the cell walls of surviving bacteria. An alternative possibility is that SM1 products facilitate export of PblA and PblB by a means that does not involve lysis of the host. For example, the SM1 holin might facilitate translocation of PblA and PblB across the SF100 cytoplasmic membrane, without subsequent bacterial lysis.
The encoding of PblA and PblB by a phage has implications for the transmission of these possible virulence determinants to other strains of S. mitis and to closely related species, including Streptococcus oralis and S. pneumoniae. Although the host range of SM1 has not yet been determined, there is precedent for phage transfer between the latter two species (18). In addition, the genome of Streptococcus pyogenes contains a pblA homolog within phage r1t-like sequences, and the Enterococcus faecalis genome includes both a pblA homolog within r1t-like sequences and a pblB homolog within phage 01205-like sequences (2). These findings suggest that SM1 and related phages may be capable of disseminating pblA and pblB to other organisms.
Numerous temperate phages are known to carry determinants that confer increased virulence to the bacterial host. These factors have been predominantly secreted toxins, such as the streptococcal erythrogenic toxin, staphylococcal enterotoxin A, diphtheria toxin, cholera toxin, and the E. coli Shiga toxins (reviewed in reference 28). Other phage-encoded virulence determinants include extracellular enzymes (staphylokinase [5] and streptococcal hyaluronidase [14]), enzymes that alter the antigenic properties of the host strain (30), and outer membrane proteins that confer an increased serum resistance (1). However, phage-encoded adhesins for human tissues have not been reported previously. Although the precise mechanism by which PblA and PblB mediate platelet binding by S. mitis has not been delineated, it is likely that one of these proteins binds platelets directly. Thus, the encoding of PblA and PblB by lysogenic SM1 may represent a novel class of phage-mediated virulence determinants.
ACKNOWLEDGMENTS
This work was supported in part by grant AI41513 from the National Institutes of Health and by the Department of Veterans Affairs.
We thank the staff of the Cell Imaging Laboratory at the San Francisco VA Medical Center for assistance with electron microscopy.
REFERENCES
- 1.Barondess J J, Beckwith J. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature. 1990;346:871–874. doi: 10.1038/346871a0. [DOI] [PubMed] [Google Scholar]
- 2.Bensing B A, Rubens C, Sullam P M. Genetic loci of Streptococcus mitis that mediate binding to human platelets. Infect Immun. 2001;69:1373–1380. doi: 10.1128/IAI.69.3.1373-1380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun L, Dramsi S, Dehoux P, Bierne H, Lindahl G, Cossart P. InlB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol Microbiol. 1997;25:285–294. doi: 10.1046/j.1365-2958.1997.4621825.x. [DOI] [PubMed] [Google Scholar]
- 4.Claverys J P, Dintilhac A, Pestova E V, Martin B, Morrison D A. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene. 1995;164:123–128. doi: 10.1016/0378-1119(95)00485-o. [DOI] [PubMed] [Google Scholar]
- 5.Coleman D C, Sullivan D J, Russell R J, Arbuthnott J P, Carey B F, Pomeroy H M. Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of beta-lysin, staphylokinase and enterotoxin A: molecular mechanism of triple conversion. J Gen Microbiol. 1989;135:1679–1697. doi: 10.1099/00221287-135-6-1679. [DOI] [PubMed] [Google Scholar]
- 6.Duda R L. Protein chainmail: catenated protein in viral capsids. Cell. 1998;94:55–60. doi: 10.1016/s0092-8674(00)81221-0. [DOI] [PubMed] [Google Scholar]
- 7.Durack D T. Experimental bacterial endocarditis. IV. Structure and evolution of very early lesions. J Pathol. 1975;115:81–89. doi: 10.1002/path.1711150204. [DOI] [PubMed] [Google Scholar]
- 8.Durack D T, Beeson P B. Experimental bacterial endocarditis. I. Colonization of a sterile vegetation. Br J Exp Pathol. 1972;53:44–49. [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson D J, McColm A A, Savage T J, Ryan D M, Acred P. A morphological study of experimental rabbit staphylococcal endocarditis and aortitis. I. Formation and effect of infected and uninfected vegetations on the aorta. Br J Exp Pathol. 1986;67:667–678. [PMC free article] [PubMed] [Google Scholar]
- 10.Ferretti J J, McShan W M, Ajdic D, Savic D J, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov A N, Kenton S, Lai H S, Lin S P, Qian Y, Jia H G, Najar F Z, Ren Q, Zhu H, Song L, White J, Yuan X, Clifton S W, Roe B A, McLaughlin R. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figueroa-Bossi N, Bossi L. Inducible prophages contribute to Salmonella virulence in mice. Mol Microbiol. 1999;33:167–176. doi: 10.1046/j.1365-2958.1999.01461.x. [DOI] [PubMed] [Google Scholar]
- 12.Fischetti V A, Pancholi V, Schneewind O. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol Microbiol. 1990;4:1603–1605. doi: 10.1111/j.1365-2958.1990.tb02072.x. [DOI] [PubMed] [Google Scholar]
- 13.Gilakjan Z A, Kropinski A M. Cloning and analysis of the capsid morphogenesis genes of Pseudomonas aeruginosa bacteriophage D3: another example of protein chain mail? J Bacteriol. 1999;181:7221–7227. doi: 10.1128/jb.181.23.7221-7227.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hynes W L, Hancock L, Ferretti J J. Analysis of a second bacteriophage hyaluronidase gene from Streptococcus pyogenes: evidence for a third hyaluronidase involved in extracellular enzymatic activity. Infect Immun. 1995;63:3015–3020. doi: 10.1128/iai.63.8.3015-3020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkinson H F. Cell-surface proteins of Streptococcus sanguis associated with cell hydrophobicity and coaggregation properties. J Gen Microbiol. 1986;132:1575–1589. doi: 10.1099/00221287-132-6-1575. [DOI] [PubMed] [Google Scholar]
- 16.McGowan D A, Gillett R. Scanning electron microscopic observations of the surface of the initial lesion in experimental streptococcal endocarditis in the rabbit. Br J Exp Pathol. 1980;61:164–171. [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 18.Ronda C, Garcia J L, Lopez R. Infection of Streptococcus oralis NCTC 11427 by pneumococcal phages. FEMS Microbiol Lett. 1989;53:187–192. doi: 10.1016/0378-1097(89)90389-3. [DOI] [PubMed] [Google Scholar]
- 19.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneewind O, Model P, Fischetti V A. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 21.Sheehan M M, Garcia J L, Lopez R, Garcia P. The lytic enzyme of the pneumococcal phage Dp-1: a chimeric lysin of intergeneric origin. Mol Microbiol. 1997;25:717–725. doi: 10.1046/j.1365-2958.1997.5101880.x. [DOI] [PubMed] [Google Scholar]
- 22.Stanley E, Fitzgerald G F, Le Marrec C B, Fayard B, van Sinderen D. Sequence analysis and characterization of phi O1205, a temperate bacteriophage infecting Streptococcus thermophilus CNRZ1205. Microbiology. 1997;143:3417–3429. doi: 10.1099/00221287-143-11-3417. [DOI] [PubMed] [Google Scholar]
- 23.Sullam P M. Host-pathogen interactions in the development of bacterial endocarditis. Curr Opin Infect Dis. 1994;7:304–309. [Google Scholar]
- 24.Sullam P M, Bayer A S, Foss W M, Cheung A L. Diminished platelet binding in vitro by Staphylococcus aureus is associated with reduced virulence in a rabbit model of infective endocarditis. Infect Immun. 1996;64:4915–4921. doi: 10.1128/iai.64.12.4915-4921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullam P M, Frank U, Yeaman M R, Tauber M G, Bayer A S, Chambers H F. Effect of thrombocytopenia on the early course of streptococcal endocarditis. J Infect Dis. 1993;168:910–914. doi: 10.1093/infdis/168.4.910. [DOI] [PubMed] [Google Scholar]
- 26.Tjalsma H, Bolhuis A, Jongbloed J D, Bron S, van Dijl J M. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol Mol Biol Rev. 2000;64:515–547. doi: 10.1128/mmbr.64.3.515-547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Sinderen D, Karsens H, Kok J, Terpstra P, Ruiters M H, Venema G, Nauta A. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol Microbiol. 1996;19:1343–1355. doi: 10.1111/j.1365-2958.1996.tb02478.x. [DOI] [PubMed] [Google Scholar]
- 28.Waldor M K. Bacteriophage biology and bacterial virulence. Trends Microbiol. 1998;6:295–297. doi: 10.1016/s0966-842x(98)01320-1. [DOI] [PubMed] [Google Scholar]
- 29.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 30.Wright A. Mechanism of conversion of the salmonella O antigen by bacteriophage epsilon 34. J Bacteriol. 1971;105:927–936. doi: 10.1128/jb.105.3.927-936.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yother J, White J M. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J Bacteriol. 1994;176:2976–2985. doi: 10.1128/jb.176.10.2976-2985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]