Abstract
A putative LysR-type transcriptional activator, Hre20, was identified previously in an in vivo expression technology screen designed to identify factors which are expressed early during infection by Yersinia enterocolitica (G. M. Young and V. L. Miller, Mol. Microbiol. 25:319–328, 1997). An insertion in hre20, now designated rscR, resulted in increased splenic dissemination of bacteria during infection in a BALB/c mouse model. A nonpolar mutation was generated in rscR, and examination of this strain in the BALB/c mouse model demonstrated that the mutation in rscR was responsible for the increased dissemination to the spleen that was seen in the original experiments. RscR is homologous to the LysR family of transcriptional regulators; thus, a screen was undertaken to identify genes regulated by RscR. A strain containing an insertion in the chromosomal rscR gene and carrying rscR on a plasmid under the control of the inducible araBAD promoter was mutagenized with an mTn5Km-2 transposon containing a promoterless lacZY. Eighteen insertions were identified which appeared to respond to levels of RscR, and these were classified into four allelic groups based on Southern blot hybridization analysis. Representative members were sequenced from three allelic groups. Sequencing revealed insertions in an ORF with no known homologues, a homologue of OmpF of Serratia marcescens, and a locus (designated rscBAC) with similarity to the hmwABC locus of Haemophilus influenzae. The hmwABC locus promotes adherence of H. influenzae to host cells (S. J. Barenkamp and J. W. St. Geme III, Infect. Immun. 62:3320–3328, 1994; J. W. St. Geme III, S. Falkow, and S. J. Barenkamp, Proc. Natl. Acad. Sci. USA 90:2875–2879, 1993). A strain containing a deletion mutant of rscA, the hmwA homologue, exhibits increased splenic dissemination of bacteria during infection in a BALB/c mouse model, similar to the rscR mutant. This suggests that the phenotype of an rscR mutant is due to the loss of RscA.
Yersinia enterocolitica infects a wide range of animal hosts, including humans, and is transmitted through ingestion of contaminated food and water (4). In humans, the infection generally leads to an acute gastroenteritis which is self-limiting and manifests as fever, diarrhea, and abdominal pain. However, in patients who are iron overloaded or immunocompromised, a systemic infection, which frequently is fatal, may ensue. Y. enterocolitica is a lymphotropic organism. After ingestion, the bacteria are able to cross the intestinal epithelium by invasion of specialized M cells and colonize the underlying lymphoid follicles, called Peyer's patches (PP), that line the small intestine (1). From the PP, the bacteria can progress to the mesenteric lymph nodes (MLN) and eventually establish a systemic infection (5).
A large number of virulence factors reside on a well-characterized 70-kb virulence plasmid (7). Several of these genes encode a type III secretion system that exports factors, also encoded on the virulence plasmid, into host cells which disrupt cellular function. Other factors are chromosomally encoded, such as invasin, which has been shown to bind to β1 integrins on the surface of M cells and in Y. enterocolitica is important in the initial steps of invasion (13, 21, 38). Previously, an in vivo expression technology (IVET) screen was performed in order to identify additional chromosomally encoded factors which are expressed early during infection (36). In that study, random fragments of Y. enterocolitica DNA were transcriptionally fused to a promoterless cat gene and integrated onto the chromosome of wild-type Y. enterocolitica. Fusion of cat to an active promoter confers to the bacterium resistance to chloramphenicol (CHL). Strains containing cat fusions were used to orally infect BALB/c mice. Y. enterocolitica strains in which the promoter upstream of cat was transcriptionally active during infection (i.e., in vivo) were enriched for by administration of CHL to the mice. Fusions that were enriched in the animal were then screened for the absence of expression under standard laboratory conditions. One of the genes discovered by that screen was designated hre20. Hre20 was found by BLAST analysis to be 67% identical to YeiE from Escherichia coli, which is a hypothetical protein with homology to the LysR family of transcriptional regulators. An insertion in hre20 resulted in increased splenic dissemination of bacteria during infection in a BALB/c mouse model, indicating a role for the hre20 locus during infection. Based on the observed in vivo phenotype, hre20 was designated rscR for reduced splenic colonization regulator.
Due to the apparent role in the progression of Y. enterocolitica infection, rscR was further characterized in the present study. Since RscR is proposed to be a transcriptional regulator, the genes which it regulates are likely to encode the effectors of the observed in vivo phenotype. A screen was undertaken to identify genes regulated by RscR in order to further understand the effect that these factors have on the course of infection.
MATERIALS AND METHODS
Growth conditions.
All cultures were grown in Luria-Bertani (LB) broth unless otherwise noted. E. coli was grown at 37°C and Y. enterocolitica was grown at 26°C, with aeration on a roller drum. The following antibiotics were used at the indicated concentrations: ampicillin, 100 μg/ml; CHL, 12.5 μg/ml; kanamycin (KAN), 100 μg/ml; nalidixic acid (NAL), 20 μg/ml; streptomycin, 50 μg/ml; and spectinomycin, 50 μg/ml. As a chromogenic substrate for β-galactosidase, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used at 40 μg/ml. For regulation of the PBAD promoter, arabinose and glucose were used at 0.2% in LB.
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are described in Table 1. A designation of “v” indicates the presence of the 70-kb virulence plasmid. Electroporations were performed as described previously (25). Restriction enzymes and DNA ligase were purchased from New England Biolabs. PCR was carried out using recombinant PFU (Stratagene). Sequencing was performed with BigDye Terminator (Amersham) and reactions were processed by the Protein and Nucleic Acid Chemistry Lab at Washington University.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Y. enterocolitica strains | ||
| JB580v | r−m+ Nalr | 15 |
| YVM557 | rscR::lacZYA Cmr | This study |
| YVM617 | rscR::Ω(Sm/Sp) | This study |
| YVM690 | ΔrscR | This study |
| YVM776 | ΔrscA | This study |
| YVM777 | rscA::Ω(Sm/Sp) | This study |
| YVM796 | rscA::lacZYA Cmr | This study |
| YVM797 | rscA::lacZYA ΔrscR Cmr | This study |
| YVM641 | rovA::erm | 8a |
| YVM885 | rscA::lacZYA rovA::erm Cmr | This study |
| Plasmids | ||
| pBAD18 | Inducible promoter vector; Kanr | 10 |
| pBAD33 | Inducible promoter vector; Cmr | 10 |
| pEP185.2 | Suicide vector | 15 |
| pFUSE | Suicide vector; lacZYA | 3 |
| pGY31 | 1.1-kb ClaI-EcoRI rscR in pWKS30; Ampr | This study |
| pHG329 | Cloning vector, medium copy number; Ampr | 29 |
| pIVET8 | Suicide vector; lacZY | 18 |
| pKN7 | rscR in pFUSE; Cmr | This study |
| pKN10.2 | ΔrscR in pEP185.2; Cmr | This study |
| pKN11 | rscR::Ω(Sm/Sp) in pEP185.2; Cmr Smr Spr | This study |
| pKN14 | PBAD−rscR Cmr | This study |
| pKN15 | PBAD−rscR Kanr | This study |
| pKN16 | ΔrscA in pEP185.2 Cmr | This study |
| pKN17 | rscA::Ω(Sm/Sp) in pEP185.2; Cmr Smr Spr | This study |
| pKN21 | rscA in pFUSE; Cmr | This study |
| pRev10 | mTn5Km2-lacZY; Ampr Kanr | This study |
| pTnMax2 | TnMax22 Ermr | 11 |
| pUTmTn5Km2 | mTn5Km2, Ampr Kanr | 8 |
| pWKS30 | Cloning vector, low copy number; Ampr | 34 |
Chromosomal lacZYA fusions to rscR and rscA were generated as follows. Cloned fragments or PCR products were subcloned into the vector pFUSE upstream of lacZYA. In the case of the rscR fusion, a 564-bp PCR fragment was amplified from the chromosome utilizing primers hre20-F1 (5′ GCTCTAGAGCGGTAACAGTCAGGATGTGAT 3′) and hre20-R1 (5′ CAATACGCTTTGCCGAAAACCACC 3′). hre20-F1 creates a cleavage site at the end for XbaI, and hre20-R1 leaves a blunt end. The product was digested with XbaI and cloned into the XbaI-SmaI site of pFUSE, and this construct was designated pKN7. For the rscA fusion, the 21B2 mutant clone (see “Cloning and sequencing” below) was digested with XbaI/StuI and cloned into the XbaI-SmaI site of pFUSE to generate plasmid pKN21. Replication of the pFUSE vector is pir dependent, and thus it cannot replicate in Y. enterocolitica which does not contain pir. The pFUSE suicide vector carrying these fragments was conjugated into the desired strain by mixing 500 μl of 16- to 18-h cultures of the donor and recipient strains and plating onto LB agar. After growth for 16 to 20 h at 26°C, the bacteria were collected from the plate and dilutions were plated onto LB containing NAL, to select for Y. enterocolitica, and CHL, to select for plasmid integration onto the chromosome. Integration was confirmed by Southern blot hybridization (data not shown).
The rscR::Ω(Sm/Sp) strain, YVM617, used in the screen was constructed as follows. A plasmid, pGY31 (pWKS30 plus ClaI-EcoRI hre20), was utilized which carried a partial sequence of rscR, from the upstream ClaI site to the EcoRI site within the gene. The Ω(Sm/Sp) cassette from pSmuC was cloned into the HindIII site of the rscR gene in pGY31. The pSmuC plasmid was provided by Hao Shen and was constructed by cloning the Ω(Sm/Sp) cassette from pRU875 into pMTL24 and then cloning the cassette into pUC1129 (32). The rscR-Ω(Sm/Sp) fragment was isolated by digestion with ClaI/SacII and was ligated into these sites of the suicide vector pEP185.2 to generate the construct pKN11. This vector was conjugated as described above into strain JB580v. A double-crossover event to completely replace the wild-type copy of rscR was identified by selecting for the Ω(Sm/Sp) cassette and not CHL resistance, which would have selected for integration of the whole plasmid. Clones were then tested for sensitivity to CHL. The crossover was confirmed by Southern blot hybridization (data not shown). The plasmid pKN14 (see below) was electroporated into this strain.
The strains YVM690 (ΔrscR) and YVM776 (ΔrscA)were generated by the same strategy. Flanking regions to the deletion were amplified. The primers used to construct ΔrscR were as follows: IFD1 (5′ GGGAACAAAAGCTGGGTACCG 3′) and IFD2 (5′ GGAAGATCTGCTGGCAGCAATGCGTAGTG 3′) to amplify the upstream region and IFD3 (5′GGAAGATCTGAGCTTGGGAATTCTGAGGC 3′) and IFD4 (5′ GCTCTAGAGGTCAACGGCGATAGTCACC 3′) to amplify the downstream region. The primers used to construct ΔrscA were as follows: del1 (5′ CCATCGATGGCTTACGGTATTGGCGAAG 3′) and del2 (5′ GGAAGATCTGCCATTCACCGGCAATGAAG 3′) to amplify the upstream region and del3 (5′ GGAAGATCTGTAGCGGGGTGGATATTGGC 3′) and del4 (5′ GCTCTAGACCTGAGGCTGCGTTATCTGC 3′) to amplify the downstream region. These fragments were cloned sequentially into pEP185.2, with a BglII linker joining the fragments, to create pKN10.2 and pKN16, and the deletions were confirmed by sequence analysis. These plasmids were conjugated into JB580v as described above. Each plasmid was integrated by a single-crossover event at the locus of interest, and this was confirmed by Southern blot hybridization (data not shown). Cycloserine enrichment was utilized to resolve the merodiploid (19). Southern blot hybridization then allowed identification of clones retaining the deleted copy of the gene. Additionally, the Ω(Sm/Sp) cassette was cloned into the BglII site of pKN16 (ΔrscA/pEP185.2), and this new construct, designated pKN17, was crossed onto the chromosome of JB580v by selection for the Ω(Sm/Sp) cassette. This strain was confirmed by Southern blot hybridization and was designated YVM777 [ΔrscA::Ω(Sm/Sp)] (data not shown).
The inducible rscR construct, pKN14, was generated by amplifying the rscR open reading frame (ORF), including the ribosome binding site but excluding the putative promoter region. Primers hreR F2 (5′ CGGGATCCGATGATAGCGTCCTCCATTCT 3′) and hreR R2 (5′ GCTCTAGACACCAATTCAGGGAAGAAGG 3′) amplified a 934-bp product with an XbaI cleavage site at the 3′ end of the ORF. This was digested with XbaI and cloned into the SmaI-XbaI sites of pBAD33 to generate pKN14. The correct construct was confirmed by sequencing. The fragment was moved from pKN14 into the SacI-PstI sites of pBAD18 to generate pKN15. This was used for some experiments in order to have an inducible construct with Kanr rather than Cmr.
The transposon delivery plasmid, pRev10, was constructed as follows: the SphI fragment of pIVET8, carrying lacZY, was cloned into the SphI site of pUTmTn5-Km2. This was the delivery plasmid utilized in the mutagenesis to identify rscR regulated genes.
Animal experiments.
Six- to seven-week-old BALB/c mice were used. Oral infections were performed using a 1-ml syringe with 2 in. of intramedic tubing encompassing a 21-guage needle. Bacteria were grown 16 to 18 h at 26°C and then resuspended in sterile phosphate-buffered saline to the desired concentration. Bacterial suspension (200 μl) was administered. On the indicated day postinfection, the tissues of interest were harvested and homogenized in sterile phosphate-buffered saline. The homogenates were diluted and plated to determine viable cell counts. Y. enterocolitica was selected for using NAL. Bacterial load was reported as CFU per gram of tissue. The significance of the results was assessed using the Mann-Whitney test.
Mutagenesis.
The strain YVM618 was mutagenized by delivery of mTn5Km2-lacZY from pRev10. Conjugation was carried out by filter mating of YVM617 [rscR::Ω(Sm/Sp)] carrying pKN14 and E. coli S17-1λpir containing pRev10. An aliquot (100 μl) of a 16- to 18-h culture of each strain was added to 3 ml of sterile 10 mM MgSO4 and was pushed through a 0.45-μm-pore-size filter. The filter was placed on minimal medium without glucose to prevent outgrowth and was incubated overnight at 26°C. Filters were vortexed in 1 ml of MgSO4, and 100 μl of a 1:4 dilution was plated on LB containing NAL (to select against the E. coli donor strain), KAN (to select for transposition), and CHL (to select for pKN14). Plates were incubated at 26°C for 30 h, and then each was replica plated to two plates—one containing NAL, KAN, CHL, X-Gal, and 0.2% arabinose and one containing NAL, KAN, CHL, X-Gal, and 0.2% glucose. Colonies were compared visually, with inspection for differences in color after growth on arabinose compared to glucose. Colonies displaying potential differential β-galactosidase expression were purified, and enzyme assays were performed.
Enzyme assays.
β-Galactosidase assays were performed as previously described (19). Cultures (1 ml) were grown for 16 to 18 h at 26°C in LB containing appropriate antibiotics and either 0.2% arabinose or 0.2% glucose as described above for the plate assays. In initial screening, single assays were performed with each mutant, and those displaying differential expression were then repeated and assays were performed in duplicate. Fusions selected for further characterization displayed fivefold or more induction in arabinose or glucose in the repeated assays.
Screen controls.
To identify fusions to genes encoding products involved in arabinose utilization, bacteria containing fusions were patched onto minimal medium containing arabinose as the only carbon source. To identify fusions responding to glucose or arabinose rather than the presence or absence of RscR, the rscR expression plasmid, pKN14, was cured by culturing without selection for 3 days. These cultures were diluted and plated on LB agar containing no antibiotics. After 2 days of growth, colonies were patched to two plates, one containing KAN and one containing CHL. Clones which retained Kanr and lost Cmr were selected. If both resistances were lost, this indicated a transposon insertion on the plasmid. Selected plasmid-cured fusions were analyzed as above for differential β-galactosidase levels in arabinose and glucose. Fusions displaying induction after the loss of the plasmid were removed from further consideration as it was likely that the differential expression observed was due to the carbon source rather than the presence or absence of RscR.
Cloning and sequencing.
Chromosome-transposon junctions were cloned as follows. Total genomic DNA preparations of the mutant strains were digested with restriction enzymes known to have a unique restriction site between the KAN resistance gene and the lacZY genes of the transposon. Using the KAN resistance gene as a probe, Southern hybridization analysis was used to determine which fragment contained the fusion. After identification of potential fragments for cloning, the genomic DNA was digested with the selected enzyme and separated on an agarose gel. The approximately sized fragments, as determined from the Southern blot, were excised from the gel and the DNA was purified by Gene-Clean. These fragments were then ligated into the cloning vector pHG329 which had been linearized with the same enzyme. After the clones were obtained, these were sequenced using a primer, P6, designed from the transposon end which allows sequencing through the junction and into the disrupted gene (12).
To obtain additional sequence for the rsc locus, a clone was obtained containing the wild-type locus from an existing cosmid library described previously (37), and the clone containing the junction of the 21B2 fusion contained 12 kb of the locus, beginning within the rscB gene, was utilized. Subcloning and TnMax mutagenesis, as described previously (11), allowed compilation of the entire sequence of the region. The sequence of both strands of the DNA was determined.
Nucleotide sequence accession numbers. The sequences for rscR and rscBAC may be found under accession numbers AF394928 and AF394927, respectively.
RESULTS
The rscR mutation is responsible for increased dissemination to the spleen.
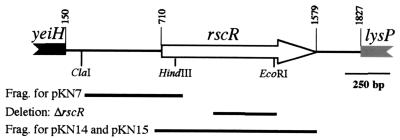
The nucleotide sequence of rscR, formerly hre20, and the surrounding DNA was determined (Fig. 1). The predicted amino acid sequence of RscR is 68% identical to the hypothetical protein YeiE and 24% identical to LysR of E. coli. Upstream of rscR is a divergently transcribed ORF with 55% identity to YeiH, the hypothetical protein encoded upstream of yeiE in E. coli. YeiH has no homology to any other known proteins in the database. The potential translational start site of the yeiH homologue is 560 bp from the rscR initiation codon. Downstream of rscR is an ORF with 80% identity to LysP of E. coli, which is also located downstream of yeiE in E. coli. The translational start site of the LysP homologue is 244 bp from the stop codon of rscR.
FIG. 1.
Schematic of the rscR locus showing the gene arrangement. DNA fragments and restriction sites are indicated for relevant constructs. Only partial gene length is shown for the yeiH and lysP homologues. Numbering begins at left from position 0. The fragments (Frag.) subcloned to generate pKN7, pKN14, and pKN15 are shown. In addition the region deleted to generate the in-frame ΔrscR mutation is indicated.
The original rscR mutant contained an insertion which could have exerted polar effects on the lysP homologue. To test if an insertion in rscR affects transcription of the downstream lysP homologue, a Φ(lysP-lacZYA) fusion was constructed. This fusion was integrated into the parental strain (JB580v), a strain containing an in-frame deletion of rscR [ΔrscR (YVM690)] and an rscR::Ω(Sm/Sp) [YVM617] strain background (mutant construction described below). Comparisons of β-galactosidase activity from Φ(lysP-lacZYA) in the three strain backgrounds demonstrated a modest polar effect of an insertional mutation on the transcriptional levels of the lysP homologue. The rscR::Ω(Sm/Sp) strain had 50% less β-galactosidase activity than the wild type and the ΔrscR mutant (data not shown). These assays demonstrated that an insertion in rscR, rscR::Ω(Sm/Sp), did affect lysP transcription, while the in-frame deletion of rscR, ΔrscR, did not exert any polar effects.
To determine if the increased dissemination to the spleen of the originally reported rscR mutant was a result of the mutation in rscR rather than a result of polar effects on downstream genes, the animal infection studies were repeated using the rscR in-frame deletion strain, ΔrscR. To create ΔrscR, an rscR mutant containing a 360-bp deletion was exchanged for the wild-type rscR by homologous recombination (see Materials and Methods). Mice were inoculated orally with 5 × 107 CFU of either JB580v or the ΔrscR mutant. Mice were sacrificed on days 3, 5, and 7 postinfection. Viable cell counts of bacteria recovered from the PP, MLN, and spleen were determined. This experiment was performed twice and demonstrated increased dissemination to the spleen by the strain containing the ΔrscR mutation on day 5 postinfection. This result is similar to what was previously observed for the original insertional mutant (data not shown). Therefore, the rscR mutation, not the polar effect on downstream transcription, was responsible for the increased dissemination to the spleen.
Screening for rscR-regulated genes.
rscR has been demonstrated to play a role in the course of infection by Y. enterocolitica. Since RscR has homology to the LysR family of transcriptional regulators, it was presumed that RscR does not have a direct effect on the course of infection but instead may regulate one or more genes involved in pathogenesis. Therefore, it was of interest to determine what genes are regulated by RscR, because these would be expected to be the effectors of the virulence phenotype. The screen for RscR-regulated genes utilized transposon mutagenesis (Fig. 2). The transposon used was an mTn5 derivative containing a KAN resistance gene and a promoterless lacZY on the plasmid designated pRev10. Transposition into a given gene in the correct orientation will generate a transcriptional fusion to lacZY.
FIG. 2.
Flowchart depicting the screen for RscR-regulated genes.
Fusions to genes which are regulated by RscR would be expected to display differential β-galactosidase activity in response to different levels of RscR. Conditions permitting control of the levels of RscR needed to be identified. Additionally, it was of interest to characterize the expression patterns of rscR since under standard in vitro growth conditions (LB, 26°C) rscR was not expressed. Various in vitro conditions were examined using β-galactosidase assays in order to identify those which induced expression of Φ(rscR::lacZYA) and thus could be utilized in the screen for RscR-regulated genes. The conditions tested were the following: temperature (26 and 37°C), O2 levels (high aeration in a flask, low aeration in a screw-cap tube), pH {20 mM morpholineethanesulfonic acid (MES), pH 5.5; 20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 6.5; 20 mM N-tris[hydroxymethyl]methyl-3-amino-propanesulfonic acid (TAPS), pH 8.0}, RPMI, minimal medium, iron-depleted LB (100 μM dipyridyl), calcium-depleted LB (20 mM MgCl2, 20 mM Na oxalate), and growth phase (2-h time points for 10 h, beginning at an optical density at 600 nm of 0.05). None of the conditions tested affected activity from the rscR reporter more than twofold, and therefore none were suitable to screen for RscR-regulated genes (data not shown). To control levels of RscR, plasmid pKN14, which carried the rscR gene under control of the araBAD promoter (PBAD), was constructed (see Materials and Methods). This construct allowed for the identification of genes whose transcription was affected by RscR.
The strain that was mutagenized was rscR::Ω(Sm/Sp) (YVM617), which contains an Ω(Sm/Sp) cassette disruption in the HindIII site of the rscR gene. rscR::Ω(Sm/Sp) was created by allelic exchange utilizing selection for the resistance marker (see Materials and Methods). pKN14 was electroporated into rscR::Ω(Sm/Sp) and allows differential expression of rscR; growth on arabinose induces expression, and growth on glucose represses expression. pRev10 was mated into the strain rscR::Ω(Sm/Sp) containing the plasmid pKN14, and transposon insertions were selected using KAN. Kanr colonies were then replica plated to plates containing X-Gal and either arabinose or glucose. Approximately 40,000 colonies from 38 independent matings were examined for a difference in color on arabinose or glucose, to identify fusions to genes whose expression responds to the presence or absence of RscR. Approximately 250 mutants displaying differential expression of lacZY on plates containing arabinose or glucose were then analyzed by β-galactosidase assays. Thirty-two mutants demonstrated an induction of fivefold or more on either arabinose or glucose and were examined further.
Several control assays were performed on these 32 mutants to identify fusions that were differentially expressed due to factors other than RscR levels. Mutants were tested for ability to grow on minimal medium supplemented with arabinose in order to identify genes whose products are involved in arabinose utilization. Two mutants were unable to grow on minimal medium supplemented with arabinose. Additionally, mutants were cultured without selection in order to cure the pKN14 plasmid and then tested for induction by arabinose or glucose. This allowed identification of genes responding to the presence of arabinose or glucose and not to RscR; three mutants fell into this group. Curing the plasmid also allowed identification of transposition events in which the transposon inserted into the PBAD region of pKN14, which resulted in lacZY expression that was controlled by the PBAD promoter. Nine mutants were identified which contained plasmid insertions. After these control assays, 18 mutants remained which appeared to respond to levels of RscR.
Identification and sequencing of RscR-regulated genes.
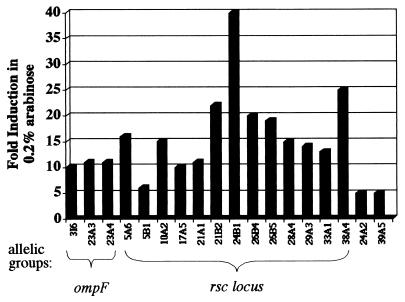
Southern blot analysis indicated that the eighteen mutants belong to four allelic groups (Fig. 3). Southern blotting was performed using the KAN resistance gene as a probe. Allelic groups were determined on the basis of the same size fragments containing the KAN resistance gene for at least two different restriction enzyme digests (data not shown). To identify the potential RscR-regulated genes, transposon-chromosome junctions were cloned. Sequence data were obtained by using a primer that annealed to sequences at the transposon end. This allowed the DNA sequence of the cloned junction to be determined. The sequence was then analyzed for potential ORFs and used to search available databases for sequence homology. Other than the mutant 39A5, representative sequence data were obtained for each allelic group.
FIG. 3.
Chart displaying levels of induction of putative RscR-regulated fusions. Fold induction represents β-galactosidase activity in 0.2% arabinose divided by β-galactosidase activity in 0.2% glucose. The allelic groups are represented by braces, and identified loci are indicated. Representative assays are shown. Assays were performed three times on at least two different days.
The chromosome-transposon junction of mutant 24A2 was cloned and sequenced. An ORF was present but displayed no significant homology to any proteins in the database. No apparent motifs were present to indicate function. Sequence collected from the 3I6, 23A3, and 23A4 allelic group indicates insertions in an ompF homologue most closely related to ompF from Serratia marcescens. The partial DNA sequence obtained from the clones was 71% identical and 84% similar to the S. marcescens OmpF. This allelic group retained threefold induction after curing of the plasmid but was still included because the original induction in the presence of RscR was 10-fold or greater. β-Galactosidase assays were performed comparing activity without NaCl and with 0.3 M NaCl. Expression was 10-fold higher without NaCl, as would be expected from a fusion to an ompF homologue (data not shown) (23). These insertions have not been studied further.
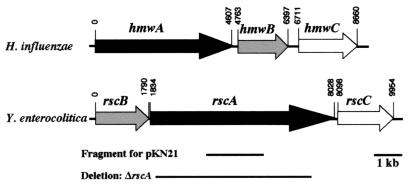
The remaining 13 mutants contained insertions in a homologue of hxuB from Haemophilus influenzae. The chromosome-transposon junctions were cloned from mutants 5A6, 21B2, and 38A4. Utilizing an internal fragment of the hxuB homologue as a probe, the remaining insertions were confirmed by Southern blot analysis to be disrupted at the same locus. Additionally, localization of each fusion to an approximately 3-kb XbaI fragment indicated that all insertions were in the hxuB homologue within this locus. Sequence data were compiled which encompassed 1.5 kb upstream and 8.5 kb downstream of the 1.8-kb hxuB homologue. This revealed two additional ORFs downstream of the hxuB homologue which appeared to be in an operon with this gene. The products of these ORFs were homologous to HmwA and HmwC of H. influenzae. These have been designated as follows: the HxuB homologue, RscB; the HmwA homologue, RscA; and the HmwC homologue, RscC (Fig. 4). In H. influenzae, the locus contains hmwABC in that order. The first ORF, rscB, in the Y. enterocolitica locus is 31% identical to hxuB, which is a homologue of hmwB of H. influenzae. HmwB and HxuB of H. influenzae are 23% identical and belong to a family of transporters which function to secrete an effector across the outer membrane (30). ShlB of S. marcescens is a well-characterized member of the family that includes HmwB and HxuB and functions to activate and secrete the ShlA hemolysin across the outer membrane (27). HxuB is proposed to secrete the heme-hemopexin binding protein HxuA (6), and HmwB has been demonstrated to secrete the adhesin HmwA across the outer membrane of nontypeable H. influenzae (30). RscA is 24% identical to HmwA, and although rscA is 1,587 bp greater in length than hmwA, the homology extends across the entire rscA ORF. The role of HmwC is not clear, but it is essential for proper processing and secretion of HmwA (30). RscC is 41% identical to HmwC and is the first described homologue of HmwC.
FIG. 4.
Comparison of the H. influenzae hmwABC locus and the Y. enterocolitica rscBAC locus. Nucleotides are numbered starting from the putative translational start sites of hmwA and rscB. The fragment utilized for creation of Φ(rscA::lacZYA) and the fragment deleted in ΔrscA are indicated.
Expression of the rsc locus.
The 13 insertions in the rscBAC locus all map to the hxuB homologue, rscB, although the gene arrangement suggests an operon structure in which insertions could have been isolated from each of the three ORFs. To begin to address the question of why transposon insertions were not isolated from rscA and rscC, a chromosomal lacZYA fusion to rscA, the putative effector, was generated by integration of a suicide plasmid, pFUSE, carrying a fragment of rscA. Φ(rscA::lacZYA) was generated in JB580v, ΔrscR, and rovA::erm (YVM641), to create strains YVM796, YVM797, and YVM885, respectively. RovA regulates the expression of inv and is expected to regulate other genes in Y. enterocolitica (24). Levels of expression from the rscA promoter as determined by monitoring β-galactosidase activity from Φ(rscA::lacZYA) from these three strains were equivalent (141 ± 2, 140 ± 3, and 141 ± 2 Miller units, respectively [results are given as means ± standard deviations]). To recreate the conditions of the screen, plasmid pKN15 containing an arabinose inducible rscR was moved into the ΔrscR mutant containing Φ(rscA::lacZYA). When rscR was induced with 0.2% arabinose the β-galactosidase activity from Φ(rscA::lacZYA) was 318 ± 31 Miller units. When rscR expression was repressed by 0.2% glucose, the β-galactosidase activity from Φ(rscA::lacZYA) was 84 ± 2 Miller units. In the presence of RscR, rscA was induced fourfold, which is an induction level that would have been excluded in the original screen. As a control, plasmid pBAD18 was also moved into the ΔrscR mutant containing Φ(rscA::lacZYA). No induction was observed with the control plasmid; β-galactosidase activity from Φ(rscA::lacZYA) was 84 ± 3 Miller units in arabinose and 100 ± 3 Miller units in glucose. This suggests that transposon insertions in rscA or rscC may not have displayed sufficient differential expression to be selected in the screen, and this could explain why none were identified.
Analysis of an rscA mutant.
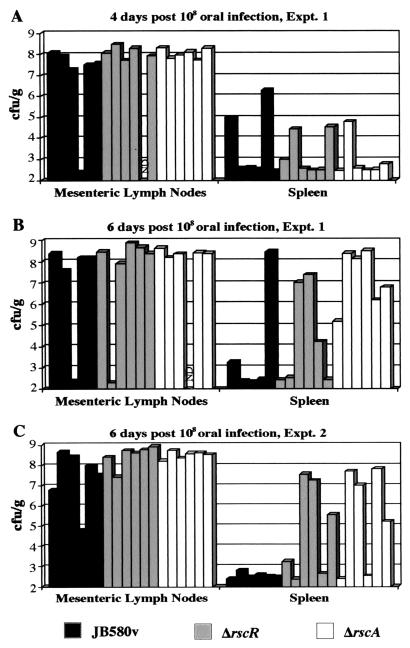
In order to determine if the rscBAC locus encodes the factor responsible for the increased splenic colonization observed for an rscR mutant, a deletion was made in the rscA gene, designated ΔrscA (YVM776). This gene was chosen because by analogy to H. influenzae RscA would be the predicted effector and RscB and RscC would be accessory proteins. The ORF of rscA is 6,195 bp, and the deletion removes 5,131 bp to create the ΔrscA mutant (see Materials and Methods). BALB/c mice were infected orally with 108 CFU of the ΔrscA mutant, the ΔrscR mutant, or JB580v; the ΔrscR strain and JB580v were tested in parallel for comparison. For each strain, 12 mice were infected. On days 4 and 6 postinfection, six mice per strain were sacrificed, and the MLN and spleen were harvested and viable cell counts were assessed for each tissue. On day 4, mice infected with the parental strain looked similar to mice infected with the ΔrscA mutant or the ΔrscR mutant (Fig. 5A). By day 6, more mice infected with either the ΔrscA mutant or the ΔrscR mutant had a high bacterial load in the spleen compared to mice infected with the parental strain (Fig. 5B). A second experiment again demonstrated increased bacterial dissemination to the spleen of mice infected with the ΔrscA mutant or the ΔrscR mutant compared to JB580v at day 6 postinfection (Fig. 5C). These results were significantly different compared to those from wild-type-infected mice, with P values of 0.008 and 0.03 for ΔrscA and ΔrscR, respectively. In both experiments, it appeared that the rscA mutation caused a more severe phenotype than the rscR mutation (P = 0.06). These results demonstrate that a mutation in rscA mimicked the phenotype observed when mice were infected with the rscR mutant and potentially a mutation in rscA causes an even more severe phenotype than a mutation in rscR.
FIG. 5.
Kinetics of infection of wild type, ΔrscR, and ΔrscA. Viable cell counts recovered from the MLN and spleen are indicated (log scale). Each bar represents data from one animal. ND, no data. (A) Viable cell counts recovered from mice at day 4 postinfection. (B) Viable cell counts recovered from mice at day 6 postinfection. Results are from the same experiment as shown in panel A. (C) Viable cell counts recovered from mice at day 6 postinfection. These results are from a separate experiment from that shown in panels A and B.
DISCUSSION
A previous study by Young and Miller examined the effect on infection of an insertion in rscR (formerly called hre20) and found that the mutant spread more rapidly to the liver and spleen of infected mice than did the parental strain (36). Sequencing of the region surrounding rscR showed a gene arrangement that is similar to the organization of yeiE and lysP in E. coli, with a homologue of lysP downstream of rscR. In this study, we demonstrated that an in-frame deletion in the rscR gene also results in increased dissemination of bacteria to the spleen after oral infection of BALB/c mice. This suggests that the phenotype of the previously described rscR mutant was not a result of polar effects on the downstream gene. This was confirmed by comparing the kinetics of infection for mice infected with wild-type bacteria to mice infected with a mutant containing the nonpolar ΔrscR allele.
After establishing that RscR plays a role in the course of infection, we wanted to identify genes that were regulated by RscR, because RscR is predicted to be a LysR-type transcriptional regulator. Members of the LysR family of transcriptional regulators typically utilize a coinducer for optimal function (26). Hence, it was of interest to determine in vitro conditions which could produce optimal levels of RscR and would more likely be permissive for a coinducer to be present if one is required. In vitro conditions which allowed expression of rscR from its own promoter were not identified, and the presence or absence of a necessary coinducer can only be speculated. This made it necessary to artificially induce expression of rscR from a PBAD promoter for the purpose of screening for genes regulated by RscR. A strain containing an inducible rscR was mutagenized with the mTn5Km2-lacZY transposon. However, because RscR levels may not be physiological or a necessary coinducer may not be present, some genes regulated by RscR may have been missed in the screen and the level of regulation observed is not likely to be representative of true in vivo values.
Several RscR-regulated fusions were identified in the screen and placed into four allelic groups based on Southern blot analysis. The 39A5 mutant was not cloned or sequenced. The second group also contained only one fusion, 24A2, and although this junction was cloned and sequenced, the sequence did not display significant similarity to any known proteins in the databases. The third group contained three fusions to a gene exhibiting homology to ompF. These fusions demonstrated differential expression in response to RscR levels (≥10-fold induction in arabinose) but also displayed differential expression to arabinose and glucose in the absence of RscR (threefold induction in arabinose). Studies in other systems have shown ompF to be regulated by many factors, including the carbon source (23). The fourth group of 13 insertions was located in a 12-kb region of the chromosome. This region was sequenced, and three ORFs were identified which were designated rscBAC (for reduced splenic colonization).
Although indirect, the observed in vivo phenotype of the ΔrscA mutant indicates that RscR regulates expression of rscBAC in the host environment. If the presence of RscR is necessary for proper expression of the rscBAC locus, then one would predict that a mutation in the regulator would have an effect on infection similar to that of a deletion of one of the genes in the locus. A comparison of the kinetics of infection of the parental strain, the ΔrscR mutant, and the ΔrscA mutant demonstrated that for both mutants there were increased numbers of mice with bacterial colonization of the spleen, with the ΔrscA mutant colonizing the largest number of spleens by day 6 postinfection. The phenotype of a ΔrscA mutant appears slightly more severe than that of a ΔrscR mutant, which could be explained if there is a basal level of transcription of the rscBAC locus in the absence of RscR. If this were the case, then a ΔrscR mutant would be producing a low level of RscA during infection and this could prevent the phenotype from being as severe as a ΔrscA mutant which expresses no functional RscA. These data along with the reporter fusion studies suggest that RscR, directly or indirectly, activates rscA expression.
The rscBAC locus is most similar to the hmwABC locus of H. influenzae. HmwA is a surface-expressed protein which promotes adherence to various epithelial cell lines in vitro and has been demonstrated to be expressed in clinical isolates from patients with acute otitis media (17). HmwA is expressed as a 150-kDa protein which contains a nontraditional signal sequence of 68 amino acids (9). These 68 amino acids are cleaved after secretion across the inner membrane by the general secretory pathway. A second cleavage event takes place after amino acid 441, concurrent with secretion across the outer membrane, to produce the mature 125-kDa protein, though this processing has been shown not to be essential for adherence. HmwB is essential for surface expression and the second cleavage of HmwA (30). Mutations in hmwB lead to the presence of unprocessed HmwA and probable degradation in the periplasm. Mutations in hmwC result in reduced levels of HmwA, and the protein is present in the unprocessed form. Null mutations in both hmwB or hmwC result in loss of adherence.
RscA has a putative nontraditional signal sequence which contains a number of charged residues in the first 47 amino acids followed by a typical signal sequence with a predicted cleavage site after residue 69. This is the same distribution of residues present in the HmwA 68-amino-acid signal sequence (9). Additionally, the second cleavage event of HmwA at amino acid 441 contains WLLDP, and this motif is present in RscA at residues 460 to 464, with predicted cleavage after residue 463. Another motif, NPNGI, is present in the amino terminus of HmwA (residues 150 to 154) as well as three related proteins: ShlA of S. marcescens, HpmA of Proteus mirabilis, and HhdA of Haemophilus ducreyi (20, 27, 30, 33). This family of proteins is grouped together based upon the mechanism of processing and secretion. Each protein has an associated outer membrane protein that functions in processing the protein to its mature form and in transport across the outer membrane. The NPNGI sequence is suggested to be a point of contact with the outer membrane transporter, which is identified as HmwB for HmwA (14, 28). The NPNGI motif is also present in RscA at residues 154 to 158, suggesting that RscA is a new member of this family of proteins. The hmwABC locus is unique in that it encodes a third functional protein, HmwC, which does not have homologues in the other related loci (2). RscC is the first identified homologue of HmwC, and this could suggest that HmwABC and RscBAC are functionally similar. RscA has the highest similarity to the adhesin HmwA, and the similarity spans the entire protein. However, RscA does have significant homology to other outer membrane and some secreted proteins with diverse functions and activities. For example, RscA is similar to a filamentous hemagglutinin-like protein of H. ducreyi which is suspected to be involved in forming lesions in the rabbit model of infection (35). RscA is also similar to the ShdA protein of Salmonella enterica, which is involved in fecal shedding of the bacteria and believed to be important in the spread of the bacteria among livestock and domestic fowl populations (16).
Adhesins have been shown to be important in the pathogenesis of numerous bacteria. Initial colonization of a pathogen at a particular site often requires a surface molecule which promotes adherence to the host cells. From these experiments it does not appear that RscA would function in initial colonization. Kinetic analysis of the ΔrscR and ΔrscA did not demonstrate a defect in the colonization of the PP, the initial site of a Y. enterocolitica infection (data not shown). In contrast, a mutation in inv, the gene encoding the surface protein invasin, results in greatly decreased colonization of the PP at early time points postinfection (21). Adhesins can function in later steps of infection as well. The YadA adhesin of Y. enterocolitica has been demonstrated to be important in the persistence of bacterial colonization in the PP (22). It is possible that RscA could function as an adhesin for a particular cell type during infection, and loss of adherence in an rscA mutant results in the altered kinetics seen in this study. Future experiments will examine the ability of the rscBAC locus to promote adherence to different cell types in vitro.
This study demonstrated that the RscR protein affects the normal progression of disease in the BALB/c mouse model of Y. enterocolitica infection. The rscBAC locus was identified in a transposon mutagenesis screen to be regulated by RscR, and a deletion in the rscA gene results in an alteration of the normal infection kinetics of Y. enterocolitica similar to that of an rscR mutant. The rapid dissemination to the spleen of the rscR and rscA mutants is an unusual phenotype. This alteration of infection kinetics could impact the virulence of Y. enterocolitica. Further study should elucidate more specifically the role that the rscBAC locus and related loci in other pathogenic bacteria play during infection.
ACKNOWLEDGMENTS
We thank A. S. Gort for excellent assistance in inoculations of all animals used in this study and critical review of the manuscript. We also thank P. A. Revell for construction of the pRev10 plasmid and the members of the Miller laboratory for valuable discussions.
This work was supported by National Institute of Health (NIH) grant AI 42736 awarded to V. L. Miller. K. M. Nelson was also supported by the NIH Cellular and Molecular Biology training grant 5 T32 GM07067 between 1 September 1997 and 1 August 2000.
REFERENCES
- 1.Autenrieth I B, Firsching R. Penetration of M cells and destruction of Peyer's patches by Yersinia enterocolitica: an ultrastructural and histological study. J Med Microbiol. 1996;44:285–294. doi: 10.1099/00222615-44-4-285. [DOI] [PubMed] [Google Scholar]
- 2.Barenkamp S J, St. Geme J W., III Genes encoding high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are part of gene clusters. Infect Immun. 1994;62:3320–3328. doi: 10.1128/iai.62.8.3320-3328.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumler A J, Tsolis R M, van der Veldon A W M, Stojiljkovic I, Anic S, Heffron F. Identification of a new iron regulated locus of Salmonella typhi. Gene. 1996;183:207–213. doi: 10.1016/s0378-1119(96)00560-4. [DOI] [PubMed] [Google Scholar]
- 4.Bottone E J. Yersinia enterocolitica: the charisma continues. Clin Microbiol Rev. 1997;10:257–276. doi: 10.1128/cmr.10.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter P B. Pathogenicity of Yersinia enterocolitica for mice. Infect Immun. 1975;11:164–170. doi: 10.1128/iai.11.1.164-170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cope L D, Yogev R, Muller-Eberhard U, Hansen E J. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type B. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 8.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Dube, P. H., P. A. Revell, D. D. Chaplin, R. G. Lorenz, and V. L. Miller. A role for IL-1α in inducing pathologic inflammation during bacterial infection. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 9.Grass S, St. Geme J W., III Maturation and secretion of the non-typeable Haemophilus influenzae HMW1 adhesin: roles of the N-terminal and C-terminal domains. Mol Microbiol. 2000;36:55–67. doi: 10.1046/j.1365-2958.2000.01812.x. [DOI] [PubMed] [Google Scholar]
- 10.Guzman L, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas R, Kahrs A F, Facius D, Allmeier H, Schmitt R, Meyer T F. TnMax—a versatile mini-transposon for the analysis of cloned genes and shuttle mutagenesis. Gene. 1993;130:23–31. doi: 10.1016/0378-1119(93)90342-z. [DOI] [PubMed] [Google Scholar]
- 12.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 13.Isberg R R, Leong J M. Multiple β1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell. 1990;60:861–871. doi: 10.1016/0092-8674(90)90099-z. [DOI] [PubMed] [Google Scholar]
- 14.Jacob-Dubuisson F, Buisine C, Willery E, Renauld-Mongeniw G, Locht C. Lack of functional complementation between Bordetella pertussis filamentous hemagglutinin and Proteus mirabilis HpmA hemolysin secretion machineries. J Bacteriol. 1997;179:775–783. doi: 10.1128/jb.179.3.775-783.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R−M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 16.Kingsley R A, van Amsterdam K, Kramer N, Baumler A J. The shdA gene is restricted to serotypes of Salmonella enterica subspecies I and contributes to efficient and prolonged fecal shedding. Infect Immun. 2000;68:2720–2727. doi: 10.1128/iai.68.5.2720-2727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krasan G P, Cutter D, Block S L, St. Geme J W., III Adhesin expression in matched nasopharyngeal and middle ear isolates of nontypeable Haemophilus influenzae from children with acute otitis media. Infect Immun. 1999;67:449–454. doi: 10.1128/iai.67.1.449-454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahan M J, Tobias J W, Slauch J M, Hanna P C, Collier R J, Mekalanos J J. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc Natl Acad Sci USA. 1995;92:669–673. doi: 10.1073/pnas.92.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 20.Palmer K L, Munson R S., Jr Cloning and characterization of the genes encoding the haemolysin of Haemophilus ducreyi. Mol Microbiol. 1995;18:821–830. doi: 10.1111/j.1365-2958.1995.18050821.x. [DOI] [PubMed] [Google Scholar]
- 21.Pepe J C, Miller V L. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc Natl Acad Sci USA. 1993;90:6473–6477. doi: 10.1073/pnas.90.14.6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pepe J C, Wachtel M R, Wagar E, Miller V L. Pathogenesis of defined invasion mutants of Yersinia enterocolitica in a BALB/c mouse model of infection. Infect Immun. 1995;63:4837–4848. doi: 10.1128/iai.63.12.4837-4848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pratt L A, Hsing W, Gibson K E, Silhavy T J. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol Microbiol. 1996;20:911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- 24.Revell P A, Miller V L. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol Microbiol. 2000;35:677–685. doi: 10.1046/j.1365-2958.2000.01740.x. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 27.Schiebel E, Schwarz H, Braun V. Subcellular location and unique secretion of the hemolysin of Serratia marcescens. J Biol Chem. 1989;264:16311–16320. [PubMed] [Google Scholar]
- 28.Schonherr R, Tsolis R, Focareta T, Braun V. Amino acid replacements in the Serratia marcescens hemolysin ShlA define sites involved in activation and secretion. Mol Microbiol. 1993;6:1229–1237. doi: 10.1111/j.1365-2958.1993.tb01252.x. [DOI] [PubMed] [Google Scholar]
- 29.Stewart G S A B, Lubinsky-Mink S, Jackson C G, Kassel A, Kuhn J. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid. 1986;15:172–181. doi: 10.1016/0147-619x(86)90035-1. [DOI] [PubMed] [Google Scholar]
- 30.St. Geme J W, III, Grass S. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol Microbiol. 1998;27:617–630. doi: 10.1046/j.1365-2958.1998.00711.x. [DOI] [PubMed] [Google Scholar]
- 31.St. Geme J W, III, Falkow S, Barenkamp S J. High-molecular-weight proteins of non-typeable Haemophilus influenzae mediate attachment to human epithelial cells. Proc Natl Acad Sci USA. 1993;90:2875–2879. doi: 10.1073/pnas.90.7.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ubben D, Schmitt R. A transposable promoter and transposable promoter probes derived from Tn1721. Gene. 1987;53:127–134. doi: 10.1016/0378-1119(87)90100-4. [DOI] [PubMed] [Google Scholar]
- 33.Uphoff T S, Welch R A. Nucleotide sequencing of the Proteus mirabilis calcium-independent genes (hpmA and hpmB) reveals sequence similarity with the Serratia marcescens hemolysin genes (shlA and shlB) J Bacteriol. 1990;172:1206–1216. doi: 10.1128/jb.172.3.1206-1216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 35.Ward C K, Lumbley S R, Cope L D, Hansen E J. Haemophilus ducreyi secretes a filamentous hemagglutinin-like protein. J Bacteriol. 1998;180:6013–6022. doi: 10.1128/jb.180.22.6013-6022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young G M, Miller V L. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol Microbiol. 1997;25:319–328. doi: 10.1046/j.1365-2958.1997.4661829.x. [DOI] [PubMed] [Google Scholar]
- 37.Young G M, Smith M J, Minnich S A, Miller V L. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J Bacteriol. 1999;181:2823–2833. doi: 10.1128/jb.181.9.2823-2833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young V B, Falkow S, Schoolnik G K. The invasin protein of Yersinia enterocolitica: internalization of invasin-bearing bacteria is associated with reorganization of the cytoskeleton. J Cell Biol. 1992;116:197–207. doi: 10.1083/jcb.116.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]