Highlights
-
•
Upregulation of circ_0099630 was obtained in iPDLCs.
-
•
Forced circ_0099630 expression restrained iPDLC proliferation and osteogenic differentiation.
-
•
Circ_0099630 repressed iPDLC proliferation and osteogenic differentiation through the miR-212-5p/SPRY1 axis.
Key words: Periodontitis, PDLCs, circ_0099630, miR-212-5p, SPRY1
Abstract
Background
Periodontitis is chronic inflammation that causes damage to periodontal tissues and cementum. It has been reported that circular RNA hsa_circ_0099630 (circ_0099630) was overexpressed in gingival samples from patients with periodontitis. However, the function of circ_0099630 on the osteogenic differentiation of periodontal ligament cells (PDLCs) in periodontitis remains unclear.
Methods
Periodontal ligaments from patients with periodontitis and third molars (termed wisdom teeth) were utilised to isolate inflamed PDLCs (iPDLCs) and healthy PDLCs (hPDLCs). Expression levels of circ_0099630 in isolated PDLCs were assessed using reverse transcription quantitative polymerase chain reaction (RT-qPCR). Effects of circ_0099630 overexpression and silencing on iPDLC viability, proliferation, and cycle progression were determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 5-ethynyl-2’-deoxyuridine (EdU), and flow cytometry assays. The osteogenic differentiation was detected by analysing the alkaline phosphatase (ALP) activity, mineralisation amount, and osteogenic markers osterix (OSX), ALP, and RUNX2 in iPDLCs. The regulatory mechanism of circ_0099630 was predicted by bioinformatics analysis and validated by dual-luciferase reporter, RNA immunoprecipitation, and RNA pull-down assays.
Results
Circ_0099630 was underexpressed in iPDLCs compared to hPDLCs. Overexpression of circ_0099630 repressed iPDLC proliferation and osteogenic differentiation, but circ_0099630 silencing exerted an opposing effect. Mechanically, circ_0099630 sponged miR-212-5p to block the inhibiting effect of miR-212-5p on SPRY1. Elevated expression of SPRY1 partly reversed the promoting effect of circ_0099630 knockdown on iPDLC proliferation and osteogenic differentiation.
Conclusions
Circ_0099630 curbed PDLC proliferation and osteogenic differentiation through elevating SPRY1 expression via sponging miR-212-5p in periodontitis.
Introduction
Periodontitis is a chronic and destructive disease that invades periodontal tissues (gingiva, periodontal ligament, and alveolar bone) and cementum.1 Currently, the diagnosis of the disease is based on clinical rather than etiologic criteria, and it involves a complex dynamic interaction amongst active herpes viruses, specific bacterial pathogens, and destructive immune responses.2 Except for severe types of periodontitis that require surgical intervention, traditional treatment is limited to tooth cleaning, disinfection and irrigation, and occasionally systemic antibiotics.3
It is reported that the grafting of mesenchymal stem cells (MSCs) with multidifferentiation ability is a very promising strategy for periodontal regeneration.4, 5 Periodontal ligament containing MSCs is the connective tissue between tooth root and alveolar bone. Researchers have revealed that periodontal ligament cells (PDLCs), which can differentiate into osteoblasts, cementoblasts, and fibroblasts, are considered to be a very promising stem cell population for periodontal tissue regeneration treatment.6 Therefore, understanding the molecular mechanism of osteogenic differentiation of PDLCs is indispensable for the development of regenerative treatment of periodontal diseases.
Empirical evidence has suggested that circular RNAs (circRNAs) can function as competitive endogenous RNAs (ceRNAs) by sponging specific miRNAs, which in turn mediates the posttranscriptional regulation of gene expression.7,8 CircRNAs are another type of single-stranded molecule.9 They usually have cell type– and tissue type–specific expression levels, which has prompted increasing interest in understanding their potential functions.10 Recent reports showed the involvement of circRNAs in periodontal diseases.11 For instance, circ-CDR1as activates the p38 MAPK signaling through mediating the miR-7/GDF5/SMAD axis, resulting in promotion of PDLC osteoblastic differentiation.12 And circ-Lrp6 contributes to cementoblast differentiation via Zeb2 through repressing miR-145a-5p activity.13 Certain types of circRNAs have been revealed to be dysregulated in the gingival samples of patients with periodontitis based on the results of RNA-seq, but their roles have not yet been explained.14
Hsa_circ_0099630 (circ_0099630), derived from the RMST gene with genomic location chr12: 97885421-97924637, is overexpressed in gingival samples derived from patients with periodontitis.14 The function of circ_0099630 in periodontitis and the mechanism it mediates remain unknown. Online bioinformatics predicted that circ_0099630 might be the upstream target of miR-212-5p. It was exposed that miR-212-5p targeted RUNX2 to decrease osteogenic differentiation of bone marrow–derived mesenchymal stem cells.15 Also, miR-212-5p targeted Myd88 to decrease Porphyromonas gingivalis lipopolysaccharide (LPS)-induced inflammation in PDLCs.16 However, the effect of miR-212-5p on osteogenic differentiation of PDLCs in periodontitis is unclear. In addition, online bioinformatics predicted that SPRY1 might be a downstream target of miR-212-5p. SPRY1 was also reported to be involved in TNF-α–mediated inhibition of osteogenesis in PDLCs.17 At present, the function of SPRY1 in periodontitis needs further verification.
Thus, we aimed to explore the effect of circ_0099630 on PDLC osteogenic differentiation in periodontitis. Furthermore, the network mechanism of the circ_0099630/miR-212-5p/SPRY1 was investigated. The analysed data sets generated during the present study are available from the corresponding author on reasonable request.
Materials and methods
Preparation of human tooth
Teeth samples were obtained during tooth extraction from 10 patients with periodontitis who lost more than two-thirds of the alveolar bone and more than 1 pocket (greater than 5 mm in depth). Healthy teeth were collected during the extraction of third molars (wisdom teeth) (n = 10). All participants involved in the study gave their written informed consent and were diagnosed and treated in the First Medical Center of Chinese PLA General Hospital. The protocol executed for this study was approved by the Ethics Committee of the First Medical Center of Chinese PLA General Hospital. The present study was approved by the ethical review committee of First Medical Center of Chinese PLA General Hospital.
Isolation of human PDLCs
Human PDLCs isolated from patients with periodontitis and teeth samples derived from healthy participants were performed as previously described18 and named as inflamed PDLCs (iPDLCs) and healthy PDLCs (hPDLCs). The periodontal ligaments in the middle one-third of the roots of healthy and sick teeth were obtained using a surgical scalpel after cleaning with sterile phosphate-buffered saline. The cut periodontal ligaments were digested with a solution containing 3 mg/mL collagenase I (Sigma-Aldrich) and 4 mg/mL dispase (Sigma-Aldrich ) for 1 hour at 37 °C, followed by filtration with 40-µm cell strainers to obtain single-cell suspensions. Subsequently, these suspensions were seeded into six-well plates (Thermo) with a complete α-Modified Essential Medium (Thermo) containing 10% fetal bovine serum (Sigma-Aldrich) and 1% penicillin/streptomycin (Sigma-Aldrich) and incubated under suitable conditions (37 ˚C and 5% CO2 in air). The culture medium was changed every 3 days, and the cells from 3 to 5 passages were used in subsequent experiments.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
PDLCs were lysed in TRIzol reagent (Thermo) and subjected to Direct-zolTM kit (Zymo Research) to isolate total RNA as per the manufacturer's instructions. RNA quantity and integrity were determined using a UV spectrophotometer (Beckman Coulter) and 1% agarose gel. Complementary DNA was produced through reverse transcription with the RT2 first strand kit (Qiagen) and miScript II RT Kit (Qiagen) following the procedures described by the manufacturer. Amplification reactions were analysed using the polymerase chain reaction Maxima SYBR Green kit (Thermo) or miScript SYBR Green PCR kit (Qiagen) with specific primers (Supplementary Table 1). Data were normalised to GAPDH or U6 and expressed as relative quantity using the 2−ΔΔCt method.19
Western blotting (WB)
Whole-cell lysates of PDLCs were prepared with a RIPA buffer (Thermo) containing a protease inhibitor cocktail (Sigma-Aldrich). Protein concentration was quantised using the BCA protein assay (Beyotime). Total protein was evaluated by WB with primary antibodies against RUNX2 (#sc-390715, 200 µg/mL, SANTA), alkaline phosphatase (ALP; #sc-365765, 1:1000, SANTA), OSX (#sc-393325, 1:1000, SANTA), SPRY1 (#sc-365520, 200 µg/mL, SANTA), and β-actin (#sc-81178, 1:200, SANTA) as previously described.20 Protein bands were visualised with Pierce ECL detection system (Thermo), followed by analysing the intensity using Image Lab (Bio-Rad).
Lentivirus production and infection
To gain and loss of circ_0099630, iPDLCs were infected with lentiviral particles carrying PLKO.1-sh-circ_0099630 (sh-circ_0099630) or pLO5-ciR-circ_0099630 (circ_0099630), which came from the supernatants of 293T cells co-transected with psPAX2, pMD2.G, and sh-circ_0099630/circ_0099630. iPDLCs infected with lentiviral particles carrying PLKO.1-sh-NC (sh-NC) or empty pLO5-ciR vector (vector) were used as controls.
Transient transfection
To overexpress and silence miR-212-5p, iPDLCs were transfected with miR-212-5p mimic (miR-212-5p: 5’-ACCUUGGCUCUAGACUGCUUACU-3’) and miR-212-5p inhibitor (in-miR-212-5p: 5’-AGUAAGCAGUCUAGAGCCAAGGU-3’) using Lipofectamine RNAi-MAX (Thermo), with miR-NC (5’-GGUUCGUACGUACACUGUUCA-3’) and in-miR-NC (5’-CGGUACGAUCGCGGCGGGAUAUC-3’) as controls, respectively. To overexpress SPRY1, iPDLCs were transfected with the pcDNA-SPRY1 plasmid (SPRY1) using the Lipofectamine 3000 (Thermo), with the empty pcDNA vector as a control.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
In short, transfected iPDLCs were incubated in a complete α-modified essential medium (5 × 103 cells/well). Following incubation with the MTT solution (10 μL) (Roche), the purple crystals were dissolved with the solubilisation solution (100 μL) (Roche). The absorbance was measured at 490 nm using a microplate reader (Molecular Devices).
5-ethynyl-2’-deoxyuridine (EdU) assay
The Yefluor 594 EdU Imaging Kit (YESEN) was utilised for detection of cell proliferation following the manufacturer's instructions. Nuclei were labeled with blue fluorescence (DAPI), and a fluorescence microscope (Olympus) was utilised to photograph.
Flow cytometry for cell cycle analysis
Analysis of cell cycle was conducted using the Cell Cycle and Apoptosis Analysis Kit (YESEN) as per the manufacturer's instructions. The samples were subjected to a BD FACS flow cytometer (BD Biosciences).
Osteogenic induction
PDLCs were grown in a complete α-modified essential medium supplemented with 10 mM β-glycerophosphate (Sigma-Aldrich), 200 μM L-ascorbic acid (Sigma-Aldrich), and 100 nM dexamethasone (Sigma-Aldrich) for the induction of osteogenic differentiation. Osteogenic induction was performed after transfection, and the differentiation medium was supplemented every 3 days for a 14-day differentiation period.
ALP staining and ALP activity
The ALP staining was performed according to the procedure provided by the NBT/BCIP staining kit (CoWin Biotech). Simply put, following osteogenic induction (after 7 days), iPDLCs were fixed in 4% paraformaldehyde for 10 minutes, followed by incubation in an alkaline solution for 20 minutes.
Measurement of the ALP activity was conducted with an Alkaline Phosphatase Activity Detection Kit (YESEN). The results normalised to the total protein concentration analysed by a Pierce protein assay kit (Thermo).
Alizarin red S (ARS) staining
ARS staining was carried out for mineralisation analysis, as described previously.21 Following osteogenic induction (after 14 days), iPDLCs were fixed in 95% ethanol for 30 minutes and then stained with 0.1% ARS (Sigma-Aldrich) for 20 minutes. For mineralisation analysis, the staining was dissolved in 10% cetylpyridinium chloride (Sigma-Aldrich) for 1 hour, and the absorbance at 570 nm was detected by a microplate reader (Molecular Devices); the results were normalised to the total protein concentration.
Dual-luciferase reporter assay
iPDLCs were transfected with a luciferase reporter (200 ng) together with pRL-TK (2 ng) and miR-212-5p mimic or miR-NC (40 nM) using the Lipofectamine 3000 (Thermo). The luciferase reporters circ_0099630 WT, circ_0099630 MUT, SPRY1 3’UTR WT, and SPRY1 3’UTR MUT were constructed using the luciferase vector psiCHECK-2 (Promega), respectively. Forty-eight hours posttransfection, the lysates of iPDLCs were detected for luciferase analysis with the dual-luciferase reporter assay system (Promega).
RNA immunoprecipitation (RIP) assay
The Magna RIP kit (Millipore) was used for this experiment. In brief, iPDLC-derived lysates were incubated with magnetic beads conjugated to IgG/Ago2 antibody (Abcam). The immunoprecipitated RNA eluted from magnetic beads was subjected to RT-qPCR analysis after purification with Direct-zol RNA MiniPrep Kit (Zymo Research).
Biotin-coupled miRNA capture
Briefly, the lysates of iPDLCs transfected with Bio-miR-NC, Bio-miR-212-5p-MUT, or Bio-miR-212-5p-WT (RiboBio) were incubated with activated M-280 streptavidin beads (Sigma-Aldrich) at 4 ˚C overnight. RNA complexes eluted from streptavidin beads were purified using Direct-zol RNA MiniPrep Kit (Zymo Research), followed by analysis with RT-qPCR.
Statistical analysis
Consequences obtained from 3 independent experiments were expressed as mean ± standard deviation. The GraphPad Prism 8 (GraphPad Software) was utilised for calculation, and the SPSS v.20.0 (SPSS) was utilised for data recording. Statistical analysis was performed using standard Student t test for direct comparisons amongst experimental groups and analysis of variance for the determination of significance in multivariable studies. Significance was defined as P < .05.
Results
Circ_0099630 was overexpressed whilst osteogenic markers OSX, ALP, and RUNX2 were downregulated in iPDLCs
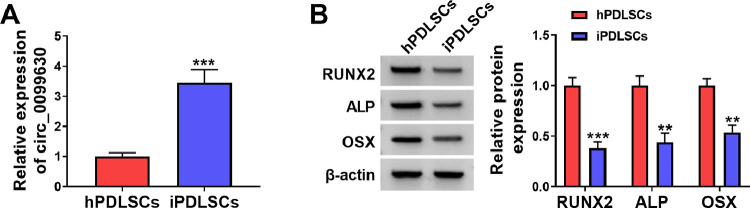
The circ_0099630 has been uncovered to be overexpressed in gingival samples derived from patients with periodontitis.14 In this study, we isolated iPDLCs and hPDLCs from teeth with periodontitis and healthy teeth, respectively, to investigate the function of circ_0099630. RT-qPCR exhibited an about 3-fold higher expression of circ_0099630 in iPDLCs with respect to hPDLCs (Figure 1A). Osteogenic differentiation markers OSX, ALP, and RUNX2 were overtly decreased in iPDLCs relative to hPDLCs (Figure 1B). These findings manifested that circ_0099630 might be related to the osteogenic differentiation of PDLCs.
Fig. 1.
Circ_0099630 and osteogenic markers were overexpressed in iPDLCs. A, RT-qPCR evaluation of circ_0099630 expression in iPDLCs and hPDLCs. B, WB detection of OSX, ALP, and RUNX2 protein levels in iPDLCs and hPDLCs.
**P < .01 and ***P < .001.
ALP, alkaline phosphatase; hPDLC, healthy periodontal ligament cell; iPDLC, inflamed periodontal ligament cell; OSX, osterix; RT-qPCR, reverse transcription quantitative polymerase chain reaction; WB, Western blotting.
Circ_0099630 restrained iPDLC proliferation and osteogenic differentiation
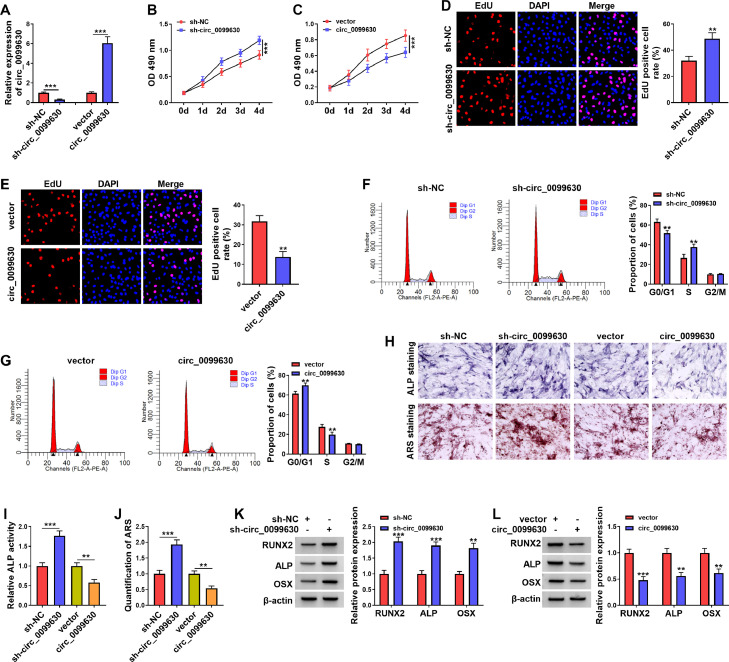
To explain the effect of circ_0099630 on the osteogenic differentiation of PDLCs, we overexpressed and silenced circ_0099630 in iPDLCs. As exhibited in Figure 2A, the transduction of sh-circ_0099630 into iPDLCs caused a distinct decrease in circ_0099630 expression, whilst the transfection of the circ_0099630 overexpression plasmid resulted in an overt increase in circ_0099630 expression. MTT and EdU assays exhibited that circ_0099630 silencing elevated iPDLC viability and facilitated iPDLC proliferation, whereas the exogenetic expression of circ_0099630 reduced iPDLC viability and curbed iPDLC proliferation (Figure 2B–E). Cell cycle analysis exhibited that circ_0099630 silencing caused a decrease in the number of cells in the G0/G1 phase and an increase in the number of cells in the S phase, indicating that circ_0099630 knockdown induced cell cycle progression (Figure 2F and G). Conversely, the forced expression of circ_0099630 increased the number of cells in G0/G1 phase but decreased in the S phase, suggesting that circ_0099630 overexpression retarded cell cycle progression (Figure 2F and G). Circ_0099630 silencing elevated the ALP activity at 7 days after osteogenic induction and the amounts of mineralisation at 14 days after osteogenic induction, but circ_0099630 overexpression exerted the opposing function (Figure 2H–J). In addition, OSX, ALP, and RUNX2 protein levels were upregulated in circ_0099630-inhibited iPDLCs and downregulated in circ_0099630-overexpressed iPDLCs at 14 days after osteogenic induction (Figure 2K and L). Together, circ_0099630 exerted an inhibiting effect on iPDLC proliferation and osteogenic differentiation.
Fig. 2.
Circ_0099630 repressed iPDLC proliferation and osteogenic differentiation. A, RT-qPCR demonstrated the transfection efficiency of sh-circ_0099630 and circ_0099630 overexpression plasmid. B–G, Effects of circ_0099630 overexpression and knockdown on iPDLC viability, proliferation, and cell cycle progression were determined using MTT, EdU, and flow cytometry assays. H–J, ALP staining, ARS staining, ALP activity, and ARS quantification were carried out to verify the effects of circ_0099630 overexpression and knockdown on iPDLC osteogenic differentiation. K and L, WB detection of the effects of circ_0099630 overexpression and knockdown on OSX, ALP, and RUNX2 protein levels in iPDLCs.
**P < .01 and ***P < .001.
ALP, alkaline phosphatase; ARS, alizarin red S; EdU, 5-ethynyl-2’-deoxyuridine; iPDLC, inflamed periodontal ligament cell; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; OSX, osterix; RT-qPCR, reverse transcription quantitative polymerase chain reaction; WB, Western blotting.
Circ_0099630 was a miR-212-5p sponge
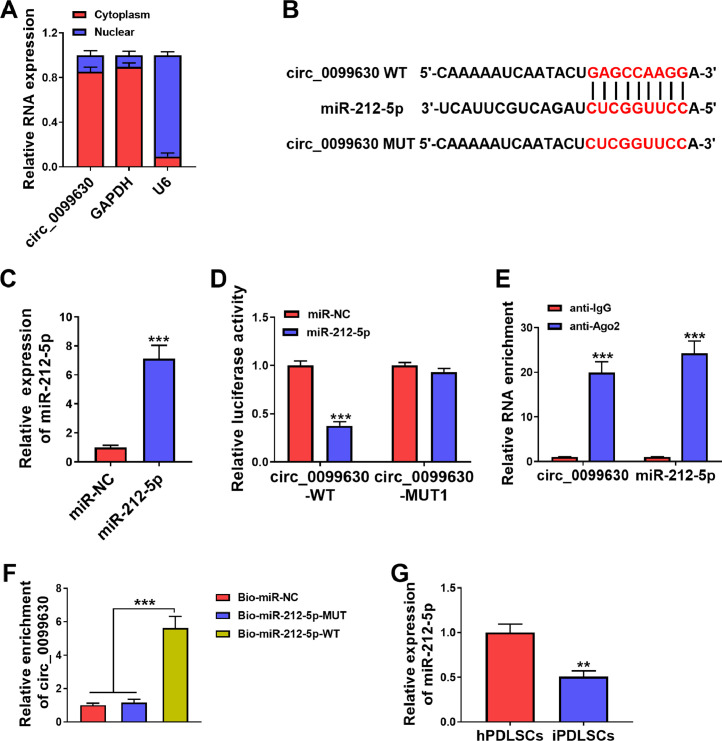
We determined the subcellular location of circ_0099630 in iPDLCs. Nuclear and cytoplasmic RNA fractionation with RT-qPCR showed that circ_0099630 was mainly localised in the cytoplasm, implying that circ_0099630 might function as a miRNA sponge (Figure 3A). Through circBank prediction, we discovered that circ_0099630 might be a target of miR-212-5p (Figure 3B). To verify the effect of miR-212-5p on the luciferase activity of luciferase plasmids, miR-212-5p mimic was transfected into iPDLCs to overexpress miR-212-5p (Figure 3C). Moreover, miR-212-5p could suppress the reporter activity in the circ_0099630-WT group but not the circ_0099630-MUT group (Fig 3D). RIP assay showed that circ_0099630 and miR-212-5p were significantly enriched by the anti-Ago2 antibody (Figure 3E). RNA pull-down assay exhibited a significant enrichment of circ_0099630 in the Bio-miR-212-5p-WT group (Figure 3F). As expected, miR-212-5p was underexpressed in iPDLCs compared to hPDLCs (Figure 3G). Collectively, circ_0099630 acted as a miR-212-5p sponge.
Fig. 3.
Circ_0099630 functioned as a miR-212-5p sponge. A, RT-qPCR showed the subcellular location of circ_0099630 in iPDLCs. B, A schematic drawing showing the putative binding sites between miR-212-5p and circ_0099630. C, The transfection efficiency of miR-212-5p mimic. D–F, The interaction between miR-212-5p and circ_0099630 was verified by dual-luciferase reporter, RIP, and RNA pull-down assays. G, RT-qPCR analysed the expression of miR-212-5p in iPDLCs and hPDLCs.
**P < .01 and ***P < .001.
RT-qPCR, reverse transcription quantitative polymerase chain reaction; hPDLC, healthy periodontal ligament cell; iPDLC, inflamed periodontal ligament cell; RIP, RNA immunoprecipitation.
miR-212-5p directly targeted SPRY1
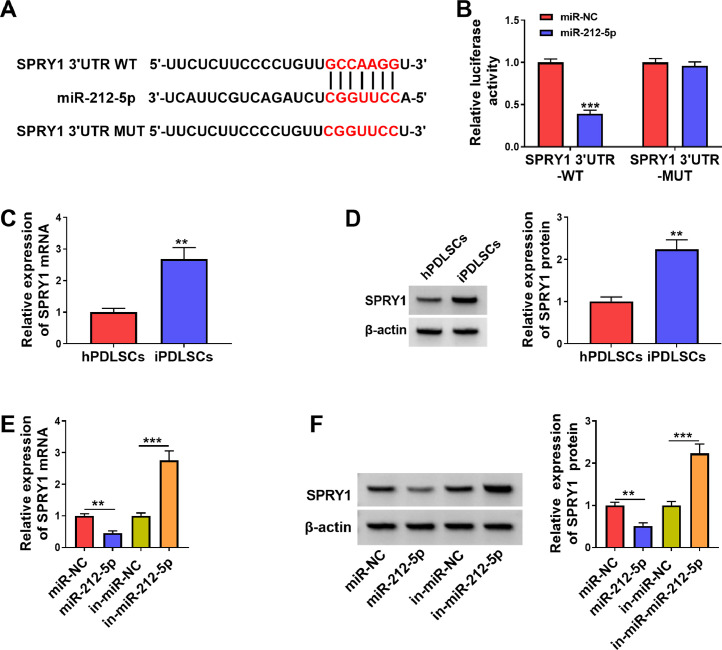
Through pre-analysis, SPRY1 was selected as a candidate gene for miR-212-5p, and their potential binding sites were exhibited in Figure 4A. Luciferase assays demonstrated that miR-212-5p repressed the luciferase activity of the SPRY1 3’UTR WT reporter instead of the SPRY1 3’UTR MUT reporter (Figure 4B). We discovered that SPRY1 mRNA and protein levels were upregulated in iPDLCs compared to hPDLCs (Figure 4C and D). As expected, SPRY1 mRNA and protein levels were repressed in miR-212-5p-overexpressed iPDLCs and elevated in miR-212-5p-silenced iPDLCs (Figure 4E and F). Also, circ_0099630 silencing decreased SPRY1 mRNA and protein levels, but this reduction driven by circ_0099630 inhibition was weakened after miR-212-5p knockdown (Supplementary Figure 1A and B). In sum, circ_0099630 mediated SPRY1 expression through binding to miR-212-5p.
Fig. 4.
SPRY1 was a miR-212-5p target. A, A schematic drawing showing the putative binding sites between the 3’UTR of SPRY1 and miR-212-5p. B, The interaction between SPRY1 and miR-212-5p was verified by dual-luciferase reporter assay. C and D, Expression levels of SPRY1 mRNA and protein in iPDLCs and hPDLCs. E and F, Effects of miR-212-5p upregulation and knockdown on the expression levels of SPRY1 mRNA and protein in iPDLCs were estimated.
**P < .01 and ***P < .001.
hPDLC, healthy periodontal ligament cell; iPDLC, inflamed periodontal ligament cell.
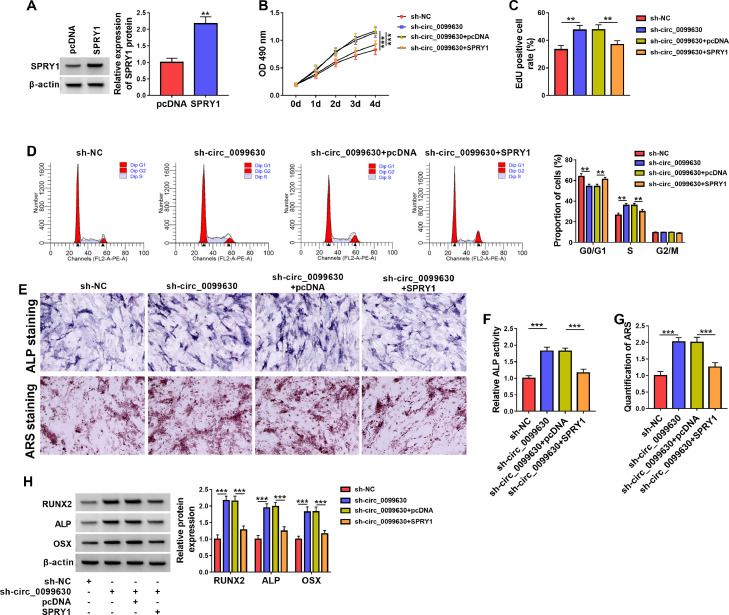
Circ_0099630 suppressed iPDLC proliferation and osteogenic differentiation via SPRY1
The transfection efficiency of the SPRY1 overexpression plasmid is shown in Figure 5A. Moreover, the promoting effects of circ_0099630 silencing on iPDLC viability, proliferation, and cycle progression were impaired after SPRY1 overexpression (Figure 5B–D). Also, the increased ALP activity and the amounts of mineralisation in sh-circ_0099630-transfected iPDLCs were crippled after SPRY1 upregulation (Figure 5E and F). Congruously, the elevated protein levels of OSX, ALP, and RUNX2 in circ_0099630-inhibiting iPDLCs were mitigated by exogenous expression of SPRY1 (Figure 5G). Together, circ_0099630 restrained iPDLC proliferation and osteogenic differentiation via SPRY1.
Fig. 5.
Circ_0099630 repressed iPDLC proliferation and osteogenic differentiation through SPRY1. A, WB assessed the transfection efficiency of the SPRY1 overexpression plasmid. B–D, Impacts of SPRY1 upregulation on cell viability, proliferation, and cycle progression in circ_0099630-inhibiting iPDLCs were determined. E–G, Effects of SPRY1 on cell osteogenic differentiation were evaluated by analysing the ALP activity, mineralisation amount, and osteogenic markers OSX, ALP, and RUNX2 in circ_0099630-inhibiting iPDLCs.
**P < .01 and ***P < .001.
ALP, alkaline phosphatase; iPDLC, inflamed periodontal ligament cell; OSX, osterix; WB, Western blotting.
Discussion
Here, our findings uncovered that circ_0099630 could adsorb miR-212-5p to restrain PDLC proliferation and osteogenic differentiation through SPRY1, offering a novel circRNA-related pathway that mediates PDLC osteogenic differentiation in periodontitis.
A recent report showed an elevation in circ_0099630 expression in gingival samples from patients with periodontitis.14 In this study, we isolated iPDLCs and hPDLCs from the periodontal ligaments of patients with periodontitis and third molars, respectively, to investigate the function of circ_0099630 in periodontitis. A conspicuous increase in circ_0099630 expression was obtained in iPDLCs with respect to hPDLCs. Moreover, circ_0099630 overexpression repressed iPDLC proliferation, decreased ALP activity and the amount of mineralisation, and downregulated osteogenic marker proteins OSX, ALP, and RUNX2 in iPDLCs, whilst circ_0099630 silencing played an opposite function. These results illustrated that circ_0099630 exerted an inhibiting effect on iPDLC proliferation and osteogenic differentiation.
Cytoplasmic RNA transcripts containing miRNA response elements can communicate and regulate each other by binding to miRNAs.22 Here, the nuclear and cytoplasmic RNA fractionation showed the main localisation of circ_0099630 in the cytoplasm of iPDLC, implying that circ_0099630 might exert function through serving as a miRNA sponge. Hua et al reported that miR-212-5p targeted Myd88 in LPS-stimulated PDLCs, leading to a reduced inflammatory response.16 A series of experiments such as dual-luciferase reporter, RIP, and RNA pull-down assays confirmed that circ_0099630 served as a miR-212-5p sponge. SPRY1 is a member of the Sprouty family that antagonises the Ras/Erk signaling.23 Furthermore, the activation of Ras/Erk signaling exerts a vital role in osteogenic differentiation.24, 25, 26 A previous report showed that miR-21 facilitated MSC osteogenic differentiation by targeting SPRY1 in inflammatory conditions.27 In this study, SPRY1 was overexpressed in iPDLCs. Moreover, SPRY1 was identified as a miR-212-5p target, and circ_0099630 mediated SPRY1 expression through functioning as a miR-212-5p sponge. And the upregulation of SPRY1 weakened the promoting effects of circ_0099630 silencing on iPDLC proliferation and osteogenic differentiation. These results urged us to conclude that circ_0099630 repressed PDLC proliferation and osteogenic differentiation by mediating the miR-212-5p/SPRY1 in periodontitis.
In summary, our findings revealed that circ_0099630 mediated iPDLC proliferation and osteogenic differentiation via sponging and sequestering miR-212-5p and subsequently liberalising the repressive effect of miR-212-5p on SPRY1 in periodontitis. This study provides insights into the osteogenic differentiation of PDLCs in periodontitis. Targeting circ_0099630 may be a possible strategy for clinical periodontal reconstruction, but more studies are needed to further validate this result.
Conflict of interest
None disclosed.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.identj.2022.06.025.
Appendix. Supplementary materials
REFERENCES
- 1.Gasner NS, Schure RS. StatPearls. StatPearls Publishing; Treasure Island, FL: 2021. Periodontal disease. [Google Scholar]
- 2.Slots J. Periodontitis: facts, fallacies and the future. Periodontol 2000. 2017;75(1):7–23. doi: 10.1111/prd.12221. [DOI] [PubMed] [Google Scholar]
- 3.Fischer RG, Lira Junior R. Periodontal disease and its impact on general health in Latin America. Section V: Treatment of periodontitis. 2020;34(Supp1 1):e026. [DOI] [PubMed]
- 4.Hernández-Monjaraz B, Santiago-Osorio E, Monroy-García A, Ledesma-Martínez E, Mendoza-Núñez VM. Mesenchymal stem cells of dental origin for inducing tissue regeneration in periodontitis: a mini-review. Int J Mol Sci. 2018;19(4) doi: 10.3390/ijms19040944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ledesma-Martínez E, Mendoza-Núñez VM, Santiago-Osorio E. Mesenchymal stem cells for periodontal tissue regeneration in elderly patients. J Gerontol A Biol Sci Med Sci. 2019;74(9):1351–1358. doi: 10.1093/gerona/gly227. [DOI] [PubMed] [Google Scholar]
- 6.Hosoya A, Shalehin N, Takebe H, et al. Stem cell properties of Gli1-positive cells in the periodontal ligament. J Oral Biosci. 2020;62(4):299–305. doi: 10.1016/j.job.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 8.Kristensen LS, Ebbesen KK, Sokol M. Spatial expression analyses of the putative oncogene ciRS-7 in cancer reshape the microRNA sponge theory. 2020;11(1):4551. [DOI] [PMC free article] [PubMed]
- 9.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB. The biogenesis, biology and characterization of circular RNAs. 2019;20(11):675–91. [DOI] [PubMed]
- 10.Gokool A, Loy CT, Halliday GM, Voineagu I. Circular RNAs: the brain transcriptome comes full circle. Trends Neurosci. 2020;43(10):752–766. doi: 10.1016/j.tins.2020.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Jiao K, Walsh LJ. The emerging regulatory role of circular RNAs in periodontal tissues and cells. 2021;22(9). [DOI] [PMC free article] [PubMed]
- 12.Li X, Zheng Y, Zheng Y, et al. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res Ther. 2018;9(1):232. doi: 10.1186/s13287-018-0976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Du M, Wang Y, Zhu J, Pan J, Cao Z. CircRNA Lrp6 promotes cementoblast differentiation via miR-145a-5p/Zeb2 axis. 2021;56(6):1200–1212. [DOI] [PubMed]
- 14.Li J, Xie R. Circular RNA expression profile in gingival tissues identifies circ_0062491 and circ_0095812 as potential treatment targets. 2019, 120(9):14867–74. [DOI] [PubMed]
- 15.Zhang Y, Jiang Y, Luo Y, Zeng Y. Interference of miR-212 and miR-384 promotes osteogenic differentiation via targeting RUNX2 in osteoporosis. Exp Mol Pathol. 2020;113 doi: 10.1016/j.yexmp.2019.104366. [DOI] [PubMed] [Google Scholar]
- 16.Hua B, Xiang J, Guo L, Lu D. MicroRNA-212-5p regulates the inflammatory response of periodontal ligament cells by targeting myeloid differentiation factor 88. Arch Oral Biol. 2020;118 doi: 10.1016/j.archoralbio.2020.104831. [DOI] [PubMed] [Google Scholar]
- 17.Yang N, Li Y, Wang G, Ding Y, Jin Y, Xu Y. Tumor necrosis factor-α suppresses adipogenic and osteogenic differentiation of human periodontal ligament stem cell by inhibiting miR-21/Spry1 functional axis. Differentiation. 2017;97:33–43. doi: 10.1016/j.diff.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Wang P, Li Y, Meng T, et al. KDM6A promotes chondrogenic differentiation of periodontal ligament stem cells by demethylation of SOX9. 2018;51(3):e12413. [DOI] [PMC free article] [PubMed]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) Method. Methods (San Diego, Calif) 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Xia P, Gu R, Zhang W, et al. MicroRNA-200c promotes osteogenic differentiation of human bone mesenchymal stem cells through activating the AKT/β-Catenin signaling pathway via downregulating Myd88. 2019;234(12):22675–86. [DOI] [PubMed]
- 21.Huang Y, Zheng Y, Jia L, Li W. Long noncoding RNA H19 promotes osteoblast differentiation via TGF-β1/Smad3/HDAC signaling pathway by deriving miR-675. Stem Cells. 2015;33(12):3481–3492. doi: 10.1002/stem.2225. [DOI] [PubMed] [Google Scholar]
- 22.Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guy GR, Jackson RA, Yusoff P, Chow SY. Sprouty proteins: modified modulators, matchmakers or missing links? J Endocrinol. 2009;203(2):191–202. doi: 10.1677/JOE-09-0110. [DOI] [PubMed] [Google Scholar]
- 24.Peng S, Zhou G, Luk KD, et al. Strontium promotes osteogenic differentiation of mesenchymal stem cells through the Ras/MAPK signaling pathway. Cell Physiol Biochem. 2009;23(1-3):165–174. doi: 10.1159/000204105. [DOI] [PubMed] [Google Scholar]
- 25.Feng L, Xue D, Chen E, et al. HMGB1 promotes the secretion of multiple cytokines and potentiates the osteogenic differentiation of mesenchymal stem cells through the Ras/MAPK signaling pathway. Exp Ther Med. 2016;12(6):3941–3947. doi: 10.3892/etm.2016.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Zhou K, Wu L, Gu H, Huang Z, Xu J. Downregulation of microRNA-143 promotes osteogenic differentiation of human adipose-derived mesenchymal stem cells through the k-Ras/MEK/ERK signaling pathway. Stem Cells. 2020;46(3):965–976. doi: 10.3892/ijmm.2020.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang N, Wang G, Hu C, et al. Tumor necrosis factor α suppresses the mesenchymal stem cell osteogenesis promoter miR-21 in estrogen deficiency-induced osteoporosis. J Bone Miner Res. 2013;28(3):559–573. doi: 10.1002/jbmr.1798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.