Abstract
Human sialic acid binding immunoglobulin-like lectin-8 (Siglec-8) is an inhibitory receptor that triggers eosinophil apoptosis and can inhibit mast cell degranulation when engaged by specific monoclonal antibodies (mAbs) or sialylated ligands. Thus, Siglec-8 has emerged as a critical negative regulator of inflammatory responses in diverse diseases, such as allergic airway inflammation. Herein, we have deciphered the molecular recognition features of the interaction of Siglec-8 with the mAb lirentelimab (2C4, under clinical development) and with a sialoside mimetic with the potential to suppress mast cell degranulation. The three-dimensional structure of Siglec-8 and the fragment antigen binding (Fab) portion of the anti-Siglec-8 mAb 2C4, solved by X-ray crystallography, reveal that 2C4 binds close to the carbohydrate recognition domain (V-type Ig domain) on Siglec-8. We have also deduced the binding mode of a high-affinity analogue of its sialic acid ligand (9-N-napthylsufonimide-Neu5Ac, NSANeuAc) using a combination of NMR spectroscopy and X-ray crystallography. Our results show that the sialoside ring of NSANeuAc binds to the canonical sialyl binding pocket of the Siglec receptor family and that the high affinity arises from the accommodation of the NSA aromatic group in a nearby hydrophobic patch formed by the N-terminal tail and the unique G–G′ loop. The results reveal the basis for the observed high affinity of this ligand and provide clues for the rational design of the next generation of Siglec-8 inhibitors. Additionally, the specific interactions between Siglec-8 and the N-linked glycans present on the high-affinity receptor FcεRIα have also been explored by NMR.
Keywords: Siglec, sialic acid, antibody, X-ray crystallography, NMR
Introduction
Human sialic acid binding immunoglobulin-like lectin-8 (Siglec-8) is an inhibitory receptor located at the surface of mast cells, eosinophils, and, to a lesser extent, basophils.1,2 As with other Siglec receptors, the extracellular domain (ECD) of Siglec-8 contains a unique N-terminal variable (V)-type immunoglobulin (Ig) domain that binds sialylated glycans, followed by two constant 2 (C2)-type Ig-domains3,4 (Figure 1A). Like other inhibitory Siglecs, the cytoplasmic domain of Siglec-8 contains an immunoreceptor tyrosine-based inhibitory motif (ITIM) and an immunoreceptor tyrosine-based switch motif (ITSM) (Figure 1A). In IL-5-primed eosinophils, Siglec-8 mediates cell death via a non-canonical pathway, which involves CD11b/CD18 integrin-mediated adhesion and NADPH oxidase activity,5−8 and Src family kinases, Syk, and SHIP1.9 On mast cells, recruitment of Siglec-8 to the high-affinity receptor for IgE (FcεRI) has been shown to partially suppress immunoglobulin E (IgE)-FcεRI-dependent histamine and prostaglandin D2 release and Ca2+ flux in vitro, without affecting cell survival.10 Binding to anti-Siglec-8 monoclonal antibody (mAb) on FcεRI-activated mast cells globally inhibits kinases involved in initiating the FcεRI signaling cascade via the ITIM present on Siglec-8.11
Figure 1.
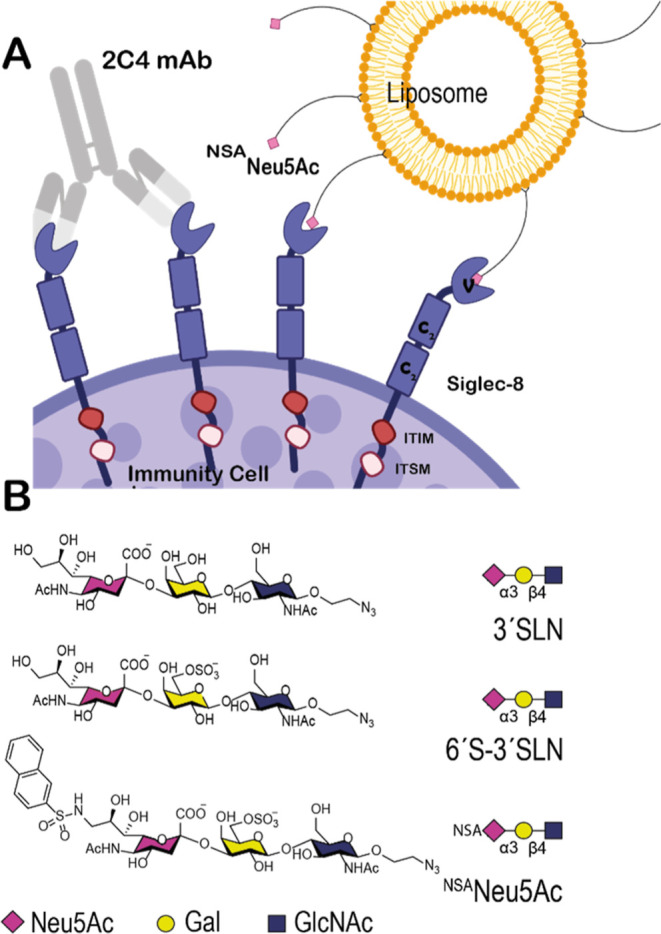
Therapeutically targeting Siglec-8. (A) Schematic representation of the interaction mode of Siglec-8 with 2C4 mAb and liposomes coated with NSANeu5Ac glycomimetic on the surface of immune cells. Siglec-8 is a single-pass transmembrane protein with the ECD composed of one V-type followed by two C2-type Ig domains. The intracellular tail contains ITIM and ITSM for signaling. (B) Chemical structures of three naturally occurring trisaccharides 3′SLN and 6′S–3′SLN and the synthetized glycomimetic NSANeu5Ac. Monosaccharide symbols are indicated.
The selectivity of the binding of Siglec-8 toward the 6′-sulfo sialyl Lewisx (6′S sLex) (Neu5Ac(α2-3)-6-O-sulfo-Gal(β1-4)[Fuc(α1–3]GlcNAc) glycan epitope was first identified by glycan array screening12 and then dissected by structural analysis.13 The NMR-derived structure of the V-Ig domain of Siglec-8 revealed that the unique structural features at G–G′ and C–C′ loops in the canonical ligand binding pocket confer specificity for 6′S sLex. A primary recognition motif for Neu5Ac binding is common to all Siglecs, but the secondary motif for the recognition of Gal6S is exclusive for Siglec-8, while GlcNAc makes few direct and variable interactions which imply only a minor role in specificity. Although Fuc was not found to directly interact with Siglec-8, this sugar moiety might contribute to the affinity by decreasing the entropic penalty associated with the binding.14
Modulation of IgE–FcεRI responses has the potential as a therapeutic strategy in allergy treatment.15 For example, the anti-IgE mAb omalizumab (Xolair), which is currently approved for the treatment of severe asthma and chronic spontaneous urticaria, removes IgE from the blood.16−18 However, immune complexes formed between omalizumab and IgE can induce the side effects of skin inflammation and anaphylaxis through engagement of IgG receptors (FcγRs) in FcγR-humanized mice. An alternative therapeutic approach is to target the inhibitory receptor Siglec-8 by mAbs. In vitro studies showed that the engagement of Siglec-8 with a mAb can induce eosinophil death and inhibit mast cell degranulation.19 Moreover, anti-Siglec-8 mAb administration to humanized mice with human eosinophils and mast cells expressing Siglec-8 confirmed the in vitro findings and identified additional anti-inflammatory effects.19 Thus, Siglec-8 is viewed as a therapeutic target for mAb treatment of eosinophil- and mast cell-associated pathologies.20,21 Currently, the humanized and non-fucosylated anti-Siglec-8 IgG1 antibody clone 2C4 (Allakos Inc) is undergoing clinical investigation for the treatment of allergic, inflammatory, and proliferative diseases and has demonstrated positive activity in eosinophilic gastrointestinal diseases, chronic urticaria, severe allergic conjunctivitis, and indolent systemic mastocytosis.21,22 Herein, we have delineated the epitope on Siglec-8 recognized by 2C4 by solving the crystal structure of the complex. The 3D structure showed that the epitope recognized on Siglec-8 by 2C4 is located at the N-terminal V-set Ig domain that contains the sialic acid binding site.
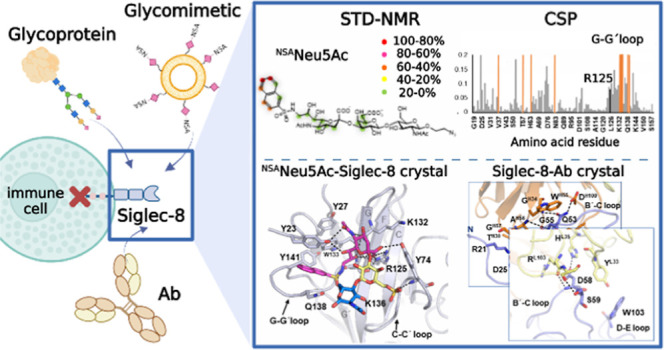
Modulating the activity of the Siglec-8 receptor using high-affinity glycan ligands has been pursued as an alternative to mAbs.12,23,24 Recently, a high-affinity mimetic based on 6′-sulfo-sialyl LewisX was developed using the classical structure–activity relationship (SAR) process.23 In this case, substitution of Gal by a cyclohexane and introduction of naphthyl sulfonamide in the 9 position of the Neu5Ac moiety increased the affinity 20-fold.23 Some of us have previously identified 9-N-[(2-naphtyl)sulfonyl]-Neu5Ac(α2-3)-6-O-sulfo-Gal(β1-4)GlcNAc (NSANeu5Ac) from a library of synthetic C-9 sulfonamide analogues as a strong binder for Siglec-824 (Figure 1B). Antigenic liposomes displaying both an allergen/antigen and NSANeu5Ac were able to suppress the activation of mast cells sensitized with an anti-allergen IgE by a mechanism involving recruitment of Siglec-8 to the IgE–FcεRI complex.1 However, the structural details of the interaction between Siglec-8 and the NSANeu5Ac high-affinity binder have not been solved yet. Herein, we have determined the binding mode of NSANeu5Ac to Siglec-8 using a combined NMR spectroscopy and X-ray crystallography analysis. Based on the results obtained, we propose that the reported high affinity (KD = 2.37 ± 0.67 μM, estimated by NMR) of NSANeu5Ac to Siglec-8 is partially due to the favorable π–π and CH−π interactions generated between the naphthyl substituent at C9 of Neu5Ac and the hydrophobic G–G′ loop. Additionally, in order to investigate how glycan presentation influences their interactions with Siglec-8, we have directly monitored by NMR its binding to the complex N-linked glycans naturally present on the FcεRIα receptor, expressed in HEK293 cells.
Overall, our findings provide key structural knowledge on the molecular recognition features and binding mechanism of an anti-Siglec-8 mAb and high-affinity glycan ligands, both with specific therapeutic potential for the treatment of allergic diseases.
Results
Siglec-8 Epitope Recognized by 2C4 mAb
In order to determine the molecular basis of the recognition mode of 2C4 anti-Siglec-8 mAb, the three-dimensional (3D) structure of the Siglec-8 ECD containing the three Ig domains (Siglec-8d1-d3, 17–352 amino acids according to the UniProt sequence numbering) in complex with 2C4 Fab has been solved by X-ray crystallography (Table 1). To facilitate crystallization of Siglec-8, the high-mannose-type N-linked glycans (N172 in d2 and N249 and N267 in d3) were removed with endoH glycosidase. The complete deglycosylation of the ECD of Siglec-8 does not affect the correct folding of the protein, as demonstrated by circular dichroism (CD) (Figure S1A). The crystal structure was solved at a resolution of 2.5 Å (C121 space group) by molecular replacement, using epratuzumab Fab (PDB ID 5VKK) as the initial search model (26), and the Siglec-8d1 crystal structure in the unbound form was also solved herein (Table 1).
Table 1. Crystallographic Data Collection and Refinement Statistics.
| Siglec-8 | Siglec-8-NSANeu5Ac | Siglec-8-2C4 | |
|---|---|---|---|
| PDB ID | 7QU6 | 7QUI | 7QUH |
| Data Collection Statistics | |||
| wavelength (Å) | 0.97918 | 0.97927 | 1.00003 |
| resolution range (Å) | 83.76–2.34(2.42–2.34) | 45.84–3.35(3.47–3.35) | 42.32–2.87(2.97–2.87) |
| space group | P1211 | P3221 | C121 |
| unit cell a, b, c (Å) | 70.08, 36.37, 167.52 | 91.68, 91.68, 150.13 | 145.34, 44.55, 111.39 |
| α, β, γ (deg) | 90, 90.05, 90 | 90, 90, 120 | 90, 111.41, 90 |
| multiplicity | 6.7 (6.5) | 2.0 (2.0) | 6.7 (6.4) |
| completeness (%) | 99.8 (99.6) | 99.9 (99.8) | 95.4 (100.0) |
| mean I/σI | 10.8 (2.6) | 20.4 (1.5) | 9.0 (2.1) |
| Wilson B-factor (Å2) | 34.8 | 126.2 | 41.5 |
| Rmerge | 0.179 (1.249) | 0.163 (2.123) | 0.233 (1.042) |
| Rpim | 0.075 (0.526) | 0.037 (0.476) | 0.097 (0.446) |
| CC1/2 | 99.2 (63.4) | 99.9 (68.6) | 98.5 (67.8) |
| Refinement Statistics | |||
| resolution (Å) | 83.76–2.34 | 42.30–3.35 | 42.32–2.87 |
| no. reflections | 36,474 (3,624) | 10,954 (1,082) | 14,935 (1575) |
| Rwork/Rfree | 0.206/0.257 | 0.265/0.292 | 0.225/0.284 |
| no. atoms: | |||
| protein | 6727 | 2163 | 4329 |
| ligand | 126 | ||
| water | 305 | - | |
| RMS (bonds) | 0.004 | 0.004 | 0.002 |
| RMS (angles) | 0.688 | 0.635 | 0.605 |
| Ramachandran Statistics | |||
| favored (%) | 97.79 | 96.18 | 96.58 |
| allowed (%) | 2.21 | 3.82 | 3.06 |
| outliers (%) | 0 | 0 | 0.36 |
| rotamer outliers (%) | 0 | 0.86 | 0 |
| average B (Å2): | |||
| protein | 39.36 | 126.71 | 44.87 |
| ligand | - | 120.38 | - |
| water | 42.48 | - | - |
One molecule of Siglec-8 and one molecule of 2C4 Fab were built in the asymmetric unit (Figure 2A). The solved crystal structure showed that 2C4 Fab binds to the V-Ig domain of Siglec-8. The C-type domains 2 and 3 were not visible on the electron density map (Figure S1B), probably because of their intrinsic high flexibility. The crystal structure of the V-Ig like domain of Siglec-8 is composed of one β-sandwich of two antiparallel β-sheets (β-strands ABED and C′CFG) connected by one intra-sheet disulfide linkage. Superposition of the V domains from the crystal structures of Siglec-8 alone and in complex with 2C4 showed only minor differences in the G–G′ loop (Figure S2). Thus, binding to 2C4 Fab does not induce significant conformational changes in the Siglec-8 secondary structural elements. Moreover, these structures are very similar to the solution NMR structure of Siglec-8d1 in the apo form (PDB ID 2N7A) reported by some of us (8) since only slight differences in the orientation of the long G–G′ loop were observed (Figure S2).
Figure 2.
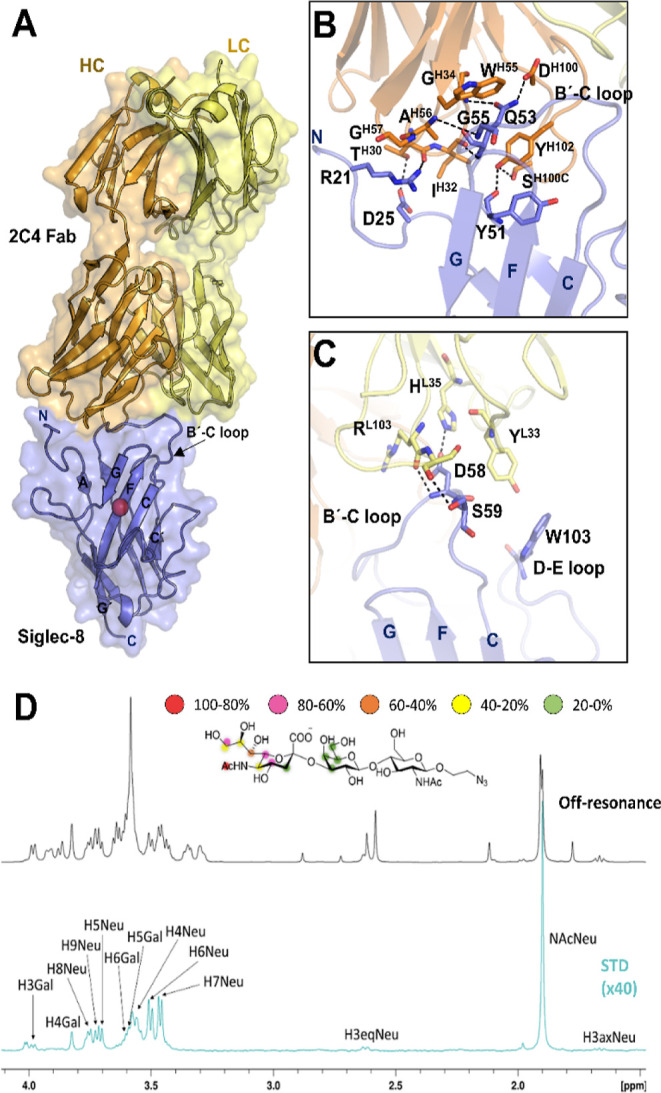
3D structure of Siglec-8 in complex with 2C4 Fab. (A) Crystal structure of Siglec-8 in complex with 2C4 Fab. 2C4 Fab (HC in orange and LC in yellow) binds to the V-domain of Siglec-8 (in purple). The conserved R residue on Siglec-8 that forms the salt bridge with the carboxylate C1 of the sialic acid moiety is represented with a red sphere. 2C4 interacts with the N-terminal and the B′-C loop at the V domain. (B) Interactions of the HC from 2C4 (orange) with the Siglec-8 V domain are mediated by the three HCDRs. (C) Interactions of the LC from 2C4 (yellow) with the Siglec-8 V domain are mediated by LCDRs 1 and 3. (D) 1H STD-NMR experiment for the complex formed by 3′SLN and Siglec-8d1d3 pre-complexed with 2C4 Fab (1:40 molar ratio). Top: the reference spectrum (black, off-resonance). Bottom: the STD-NMR spectrum (blue, on-resonance at the aliphatic region). The 1H NMR signals showing the STD effect are annotated. The epitope mapping (relative STD) is shown in the ligand structure.
The crystal structure of the complex shows that the complementary-determining regions of the heavy chains (HCs) 2 and 3 (CDRH2 and 3) of 2C4 Fab highly contribute to the interaction with Siglec-8 [with a total buried surface area (BSA) of 525 Å2 (Table S1)]. CDRH2 and CDRH3 establish polar interactions with the N-terminal and the B′-C loop on the Siglec-8 V domain (Figure 2B). On the other hand, the CDRs of the light chains (LCs) 1 and 2 (CDRL1 and 2) interact with loops B′–C and D–E [BSA of 229 Å2 (Table S1)] (Figure 2C). The detailed inspection of the structure shows that the epitope recognized by 2C4 on Siglec-8 does not include any amino acid of the sialic acid binding site. This fact was further corroborated using competition saturation-transfer difference NMR experiments (STD-NMR) with a ligand of Siglec-8, the α2,3-linked sialyl-N-acetyl-lactosamine (3′SLN), and Siglec-8d1-d3 pre-complexed with 2C4 Fab. Even in the presence of Fab, intense STD-NMR signals were observed for several protons of the sialic acid and Gal moieties of 3′SLN, thus demonstrating that 2C4 Fab does not preclude the interaction of Siglec-8 with this glycan ligand (Figure 2D). Nevertheless, it is still possible that the 2C4 mAb may sterically block ligand binding on Siglec-8 on the surface of cells. As previously reported,1 liposomes decorated with this glycomimetic and an allergen, in the presence of anti-Siglec-8 2C4 Ab, can relieve inhibition of degranulation caused by its interaction with Siglec-8, preventing its recruitment to IgE-FcεRI in the mast cell surface.
Binding of the Sulfonamide Sialoside Glycomimetic NSANeu5Ac to Siglec-8
Receptor-Based NMR Analysis
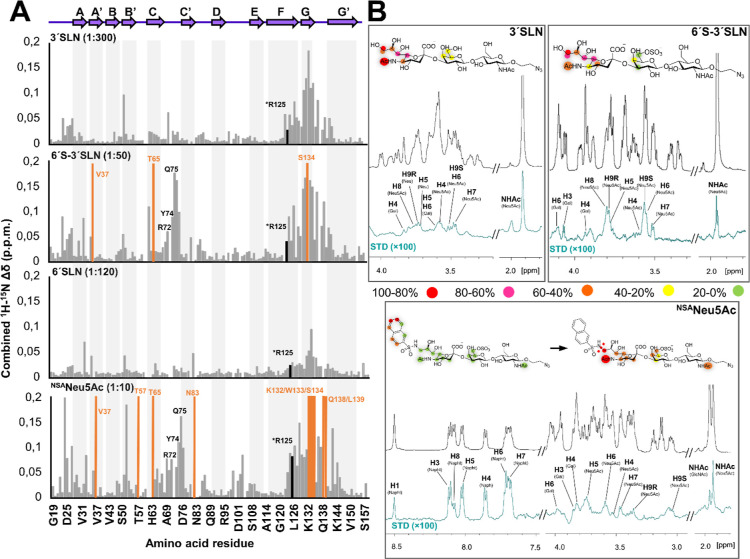
The interaction of Siglec-8d1 with NSANeu5Ac was studied by NMR using receptor-based 1H–15N TROSY NMR experiments25,26 (Figure S3). Information of ligand recognition by Siglec-8 was obtained from the chemical shift perturbations (CSPs) measured upon subsequent additions of the ligand to the 15N uniformly labeled Siglec-8 V-Ig-like domain (Siglec-8d1).27 As control experiments, the binding to two naturally occurring trisaccharides (Neu5Ac(α2–3)Gal(β1–4)GlcNAc (3′SLN) and 6′-O-sulfo Neu5Ac(α2–3)Gal(β1–4)GlcNAc (6′S-3′SLN) was also assessed using the same protocol (Figure 1B).
Specific CSPs in the F and G β-strands, at the sialic acid binding site, were clearly observed in the presence of 3′SLN (Figure 3). The analysis of the CSPs induced by this ligand indicated that the exchange rate falls into the fast exchange regime in the chemical shift timescale, with an estimated low binding affinity of ∼4 mM (Figure S4). In contrast, for the sulfated analogue 6′S-3′SLN, a fast/intermediate exchange was determined for most cross-peaks along the titration. In this case, the CSP analysis showed that saturation was reached using relatively low molar equivalents of the ligand versus the protein (Figure S3). More perturbed residues are located at the (expected) canonical binding site, which include the F and G β-sheets, but also in the C–C′ loop (Figure 3). In fact, residues R72, Y74, and Q75 in the loop C–C′ displayed significant CSPs, not present for the titrations with 3′SLN, strongly suggesting that the sulfate group of the Gal moiety is directly involved in the interaction with Siglec-8 at this region. The CSP data obtained from the titration were used to estimate the dissociation constant (KD) of 6′S-3′SLN, which was in the micromolar range (140 ± 20 μM). Thus, the presence of the sulfate group increases the affinity by more than 1 order of magnitude compared to that of 3′SLN, in agreement with the observation of Pröpster et al. for the 6′sulfo sialyl LeX analogue.13 Interestingly, and in contrast to the observations for the natural ligands 3′SLN and 6′S-3′SLN, the modified ligand NSANeu5Ac showed a slow-intermediate exchange in the chemical shift timescale, although a similar CSP profile to that of 6′S-3′SLN was obtained (Figure 3). Indeed, although some signals were lost when 0.5 molar equiv of the ligand was added, those peaks were recovered when the protein was saturated using ∼10 equiv. Furthermore, additional CSPs in the N-terminal region and at the G–G′ loop were observed. This fact strongly suggests that the naphthyl group at C9 of the Neu5Ac residue is located close to the N-terminus. Moreover, at 0.5 equiv of NSANeu5Ac, some residues, such as D24 and P123, located near the ligand binding site, showed simultaneous cross-peaks for the apo and bound forms. The dissociation constant estimated by integration of the free and bound peak volumes (Figure S4) yielded an average value of 2.4 ± 0.7 μM. Thus, the addition of the NSA group boosts the binding strength by 60-fold compared to the sulfated 6′S-3′SLN.
Figure 3.
Binding of 3′SLN, 6′-3′SLN, and NSANeu5Ac to Siglec-8 measured by NMR spectroscopy. (A) Combined 1H–15N CSPs (Δδ) of the 15N-labeled Siglec-8d1 residues observed upon titration with the different ligands and at different molar ratios, plotted vs amino acid residues. Secondary structure elements derived from the herein determined crystal structure of Siglec-8d1 are represented at the top. The residues that experience dramatic signal line broadening during the titration are represented with the orange bars. Residues involved in the interaction with the ligands are also depicted. (B) 1H STD-NMR experiments for the complex formed by 3′SLN, 6′-3′SLN, and NSANeu5Ac and Siglec-8 (1:40 molar ratio with 3′SLN and 6′-3′SLN and 1:50 for NSANeu5Ac). Top: the reference spectrum (black, off-resonance). Bottom: the STD-NMR spectrum (blue) on-resonance was set at the aliphatic region). The 1H NMR signals showing the STD effect are annotated. The epitope mapping (relative STD) is shown in the ligand structure. In the case of NSANeu5Ac, the epitope mapping was normalized in percentage of the largest signal H6 Napht (left), while the NSA group was not considered, and the epitope was normalized in percentage of the H9S Neu5Ac signal (right).
Ligand-Based NMR Analysis
The ligand binding epitopes for 3′SLN, 6′S-3′SLN, and NSANeu5Ac in complex with Siglec-8d1 were elucidated (Tables S2-5) using STD-NMR experiments.28,29 Not surprisingly, the terminal Neu5Ac in all the sialylated ligands displayed the strongest STD-NMR signals, especially the protons at the glycerol chain and the N-acetyl moiety (Figure 3). Only modest STDs were observed at the Gal moieties. For NSANeu5Ac, besides the clear STD-NMR signals observed for the Neu5Ac and Gal residues, the strongest STD signals arose from the aromatic NSA substituent. Within the NSA protons, H6 and H7 display the largest intensities, followed by H3, H4, and H5. H1 and H8, at the opposite face of the ring, showed weaker intensities.
The conformational behavior of sialyl oligosaccharides in their free state and when bound to different lectins has been extensively analyzed using NMR.30 It has been demonstrated that the glycosidic Neu5Acα2-3Gal linkages may display three major basic conformers (I–III) in solution (Figures S6 and S7), which differ in conformation around Φ/Ψ of the glycosidic linkages.30 In particular, conformer I is defined by Φ/Ψ −170°/–15°, while the corresponding torsions for II and III are in the same area of the energy map (−80°/–60° and −65°/–5°, respectively). For 6′S-3′SLN and NSANeu5Ac, a similar strategy was employed for their analyses in the free and bound states (NOESY and trNOESY, respectively), assisted by molecular dynamics (MD) simulations (Figure S6). As expected, for both molecules, the analysis of the NOEs demonstrated the existence of conformational equilibrium around the Neu5Acα2-3Gal linkages, given the presence of three exclusive NOEs (H3eq Neu5Ac–H3Gal and H3ax Neu5Ac–H4Gal exclusive for conformer I, H6Neu5Ac–H4Gal exclusive for conformer II, and H3eq Neu5Ac–H4Gal exclusive for conformer III). In addition, H8 Neu5Ac–H3Gal NOE cross-peaks were observed, which are compatible with both conformers II and III.30 Interestingly, in the presence of Siglec-8, the conformer I exclusive H3ax Neu5Ac–H3Gal and H3ax Neu5Ac–H4Gal NOEs disappeared in the trNOESY spectra, suggesting the existence of a conformational selection process. In fact, the inter-residue NOE cross-peaks exclusive for conformers II and III were experimentally detected for the complexes of 6′S-3′SLN and NSANeu5Ac with Siglec-8, pointing out the recognition of a geometry compatible with conformers II and III by Siglec-8, but not with conformer I. Such finding highlights the existence of certain flexibility on 6′S-3′SLN and NSANeu5Ac ligands even in the bound state.
3D Structure of the Complex between Siglec-8 and NSANeu5Ac Solved by X-ray Crystallography
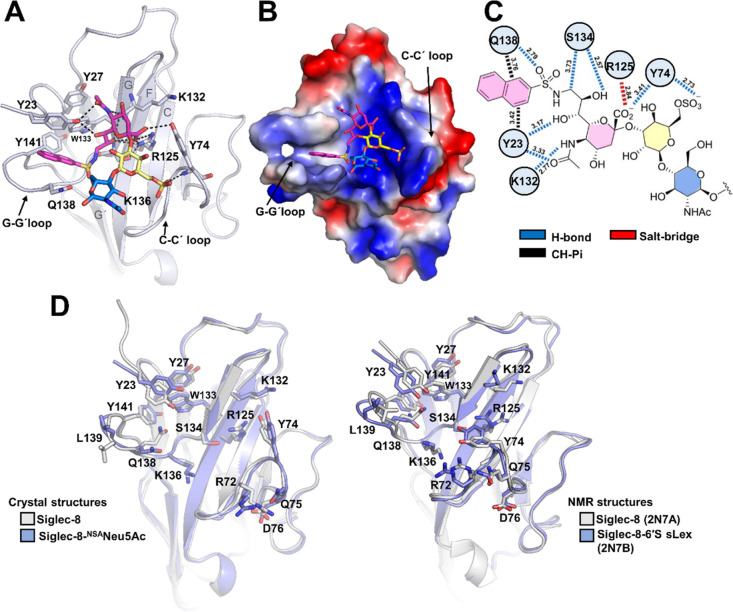
To further define and understand the structural basis of the recognition of NSANeu5Ac by Siglec-8, its V-set sialic acid binding domain was co-crystallized with this ligand (Figure 4A). The crystal structure was solved at a resolution of 3.3 Å, showing two Siglec-8 molecules (A and B), each one bound to one molecule of NSANeu5Ac in the asymmetric unit (Table 1 and Figure S8). The superposition of A and B monomers of the asymmetric unit (Figure S9) showed that there are no major conformational differences between them (the Cα rmsd is only 0.37 Å). Accordingly, we focus below on the structural features of the complex of NSANeu5Ac with Siglec-8 monomer A. The analysis of the Neu5Acα2-3Gal glycosidic torsions allowed deducing that the conformations of the two ligands bound to the two Siglec-8 monomers in the asymmetric unit correspond to the geometries (Φ/Ψ −78/–1° and −86°/10°) in between the region defined by conformers II and III in perfect agreement with the NMR-based conclusions described above (Figure S7). Overall, the structure of Siglec-8 in complex with NSANeu5Ac only showed minor differences (the Cα rmsd is ca. 0.4–0.5 Å) in the key G–G′ loop, compared to the unliganded Siglec-8 crystal structure (Figures 4D and S9), indicating that the Siglec-8 binding site is largely preorganized to host this ligand. Interestingly, side chain rearrangements were observed involving residues Y23 at the N-terminus, Q138 in the G–G′ loop, and R72, Y74, Q75, and D76 in the C–C′ loop (Figure 4D).
Figure 4.
Structural details of the interaction of Siglec-8 and NSANeu5Ac. (A) Crystal structure of Siglec-8d1 (in cartoon with blue color) in complex with NSANeu5Ac. Neu5Ac (pink), Gal (yellow), and GlcNAc (blue) are depicted as sticks. Intermolecular H-bond interactions are indicated by black dashed lines. (B) Electrostatic surface representation of the V-Ig like domain of Siglec-8 in complex with NSANeu5Ac (in sticks). The electrostatic surface was calculated with the APBS software (51), prepared using PyMol (41), and displayed on a scale of −5 kT/e (red) to 5 kT/e (blue). (C) Schematic representation of the interactions between Siglec-8 amino acid residues from the sialic acid binding site of monomer A from the crystal structure and NSANeu5Ac. (D) Structural comparison of unliganded (gray) and liganded (blue) crystal (left) and NMR (right) structures. Amino acid residues involved in the interaction with sialylated ligands (in sticks) are depicted as sticks.
As in the Siglec family, and as previously described by NMR of Siglec-8 (13), the carboxylate group (COO–) of the Neu5Ac moiety interacts via an ion–pair interaction with the guanidium group of the highly conserved R125 (Figure 4A,C). Nevertheless, the COO– of Neu5Ac also interacts via a H-bond with the hydroxyl group of the side chain of Y74 (Table S6) (Figure 4A,C). In addition, the N-acetyl amide group is hydrogen-bonded to the side-chain hydroxyl of Y23 and the backbone carbonyl group of K132. Other polar interactions involve the glycerol lateral chain, which establishes hydrogen bonds through Neu5Ac O7 with Y23 and Neu5Ac O8 and O9 with S134. The orientation of the aromatic fragment is also well defined. Significant interactions occur through the NSA substituent at C9 of the sialic acid portion of the ligand (440 Å2 of BSA for the NSA substituent, out of a total of 570 Å2 for the entire trisaccharide ligand). The non-polar naphthyl group is accommodated in the hydrophobic aromatic pocket composed by Y23, Y27, W133, and Y141 residues (Figure 4B). The side chain of Y23 at the N-terminus and Q138 at the G–G′ loop suffered a drastic shift with respect to its orientation in the free state, likely to accommodate the large NSA group (Figure 4D). In this context, this naphthyl ring establishes a T-shaped antiparallel π–π interaction with Y23 and a CH−π interaction with the methylene group of Q138 (Figure 4C), leading most probably to the affinity enhancement of the mimetic.
The 6-O-sulfo Gal moiety interacts through its sulfate group with the main chain of Y74 in loop C–C′ (Figure 4A,C). As mentioned above, the NMR structure (13) of Siglec-8 bound to 6′S sLex (6′S-3′SLN with one additional Fuc moiety) has been reported (PDB ID 2N7B). Interestingly, the crystal and NMR structure complexes show different orientations of R72, Y74, Q75, and D76 residues at the C–C′ loop (Figures 4D and S9). In addition, the sulfate group of NSANeu5Ac is not engaged with R72 and Q75 residues, as in the 6′S sLex NMR complex, but with the backbone amide of Y74. This interaction also contrasts with the NMR-based complex, for which Y74 is involved in a CH/π stacking interaction with the Gal moiety.
For this reason, the conformational stability of the complex formed between Siglec-8 and NSANeu5Ac was evaluated by MD simulations (simulation time 500 ns). The crystal structure was shown to be perfectly stable during the simulation time, keeping the key polar contacts between the carboxylate group of Neu5Ac and R125 for over 80% of the simulation time (Figure S10). The sulfate group also maintained polar interactions with the backbone amide of the Y74 main chain, which was not involved in stacking interactions with the Gal moiety. In addition, the sulfate group also established salt bridge interactions with the side chains of R72 and Q75 amino acids throughout the MD simulation, which nicely fit with the CSPs observed by NMR exclusively for the sulfated ligands. The NSA substituent was preserved at the aromatic pocket, while the glycerol side chain retained the intermolecular hydrogen bonds observed in the crystal structure. Fittingly, when the sulfate group was removed and new MD simulations were carried out, the key interactions of Y74 with the sugar moieties were absent, and the motion of the Neu5Ac residue was largely increased. Such finding highlights the relevance of the sulfate group in stabilizing the interactions with the canonical sialic acid binding site in Siglec-8.
Overall, the interactions observed in the crystal structure perfectly fits with the receptor-based and ligand-based NMR data described above. The observed CSP for NSANeu5Ac (Figure 3) suggests the major involvement of amino acid residues at the N-terminal region (Y23 and Y27), strands F (R125) and G (K132, W133, and S134), and loops C–C′ (R72, Y74, Q75, and D76) and G–G′ (K136, Q138, and Y141), as observed in the crystal structure (Figure 4A). The bound pose of the NSANeu5Ac ligand is also in full agreement with the STD-NMR experiments (Figure 3), which showed major intensities for the aromatic protons, followed by those at the glycerol and acetyl side chains of the Neu5Ac moiety, with minor contacts for the Gal ring and none for the GlcNAc residues.
Interaction of Siglec-8 with Complex N-linked Glycans Present on the Human Glycoprotein Assessed by NMR Spectroscopy
The interaction between naturally occurring N-linked glycans and Siglec-8 was also investigated in order to unveil how a different presentation of the glycan epitope could influence its recognition. Given the biological relevance of the high-affinity receptor FcεRI for IgE in relation to Siglec-8, the interaction between the α-subunit of FcεRI, which contains seven N-linked glycans (31), and the ECD of Siglec-8 was assessed by NMR spectroscopy. Using the NMR technique, it has previously been shown that the α-subunit contains pauci-mannose, high-mannose, hybrid, and bi-, tri-, and tetra-antennary complex-type N-glycans with different degrees of fucosylation and sialylation (31). O-linked glycans have not been found (31). FcεRIα was expressed and purified using HEK293F cells, employing 13C-labeled glucose as the carbon source, as described before.31 Under these conditions, all the glycans decorating FcεRIα are 13C-labeled and may be used as probes to monitor intermolecular interactions by NMR. As previously deduced in our lab (31), 6′-sulfated sialyl Lewis X glycans, which based on glycan arrays are the preferred ligands for Siglec-8 32, are absent in FcεRIα.33
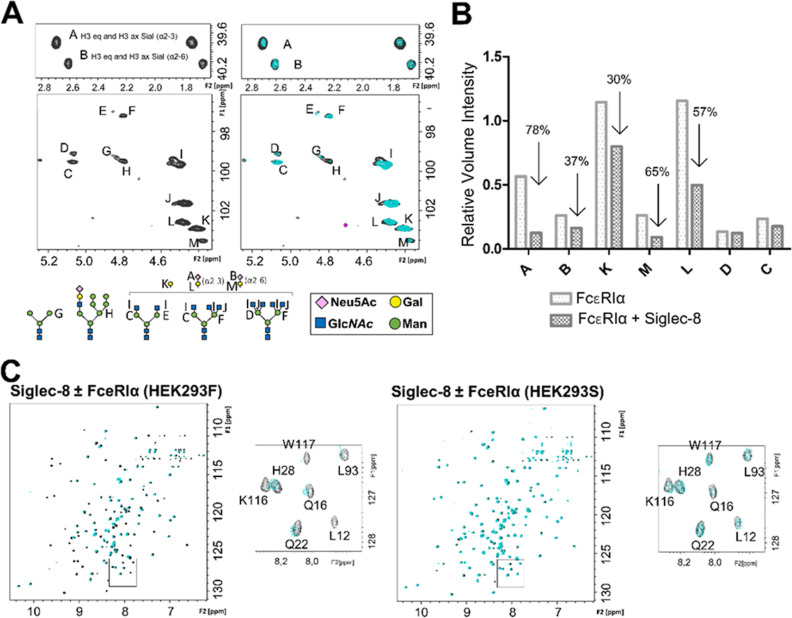
The 1H–13C HSQC NMR spectrum of the FcεRIα glycans in the absence and presence of the Siglec-8 V domain showed significant changes in the cross-peak pattern, especially evident in the spectral region containing the axial and equatorial H3 protons of the Neu5Ac moieties on FcεRIα (Figure 5).
Figure 5.
Interaction of the ECD of Siglec-8 with the sialylated N-linked glycans of FcεRI as measured by NMR. (A) Expansion of the anomeric region and the region containing the axial and equatorial H3 protons of the Neu5Ac residues of FcεRIα expressed in HEK293F cells. On the right-hand side, the glycan composition of FcεRIα under physiological conditions is represented (black), and on the left-hand side, the superimposition of glycan spectra in the absence (black) and presence of 5 equiv of Siglec-8d1 (cyan). Note the selective loss of intensity for the NMR signals of sialic acid. The glycan composition of FcεRIα was deduced by direct analysis of the [1H,13C]-HSQC NMR spectra. Each carbohydrate is identified by a letter. The different N-glycans present on the glycoprotein expressed in HEK293F cells are represented with Symbol Nomenclature for Glycans [SNFG] symbols. (B) Bar graph representing intensities corresponding to the signals of sialic acid, with and without Siglec-8. All signals were normalized with respect to trimethylsilylpropanoic acid (TSP). (C) Superimposition of 1H,15N-TROSY experiment of Siglec-8d1 in the apo form (black) and after the addition of 1 equiv of FcεRIα (green) expressed in HEK293F cells (left) and in HEK293S cells (green) (right).
The 1H–15N TROSY of 15N-labeled Siglec-8d1 (the V domain) in the presence of glycosylated FcεRIα showed a dramatic decrease in the intensities of many cross-peaks of the Siglec-8 V domain (Figure 5). The careful analysis of the induced CSPs and signal line broadening of the 1H–15N TROSY cross-peaks upon addition of glycosylated FcεRIα confirmed that the interaction occurs through the canonical ligand binding site of Siglec-8 (Figure S11). Fittingly, the largest intensity losses were observed for the signals corresponding to amino acids located in the binding site, especially in the F and G β-strands (Figure S11).
Additionally, a competition assay between FcεRIα and the NSA-containing glycomimetic was performed by NMR (Figure S11). When 10 equiv of NSANeu5Ac was added to this NMR tube, many cross-peaks of Siglec-8d1 were recovered, further demonstrating that the N-glycans on FcεRIα and the glycomimetic compete for the same binding site on Siglec-8 (Figure S11). To confirm the sialic acid binding specificity, FcεRIα was expressed in HEK293S cells, which lack N-acetylglucosaminyltransferase I GnTI activity, and thus, the glycoproteins exclusively display high-mannose-type N-glycans and no sialic acid residues. Indeed, no CSP or intensity changes were observed in the 1H–15N TROSY cross-peaks of Siglec-8 in the presence of FcεRIα expressed in HEK293S (Figure 5), indicating that Siglec-8 does not interact with FcεRIα when decorated with high-Man N-glycans.
Discussion
Siglec-8 exhibits a highly restricted expression on human eosinophils and mast cells and to a lesser extent on basophils, making it an attractive target for cell-type-specific treatment of inflammatory responses that contribute to allergic and non-allergic diseases mediated by these cells. Already, promising strategies have been developed using anti-Siglec-8 mAbs or multivalent sialoglycan ligands to induce apoptosis/cell death of eosinophils and suppress and/or desensitize mast cells from degranulation upon allergen challenge.34,35 These results have opened the potential for the treatment of a wide range of allergic and inflammatory diseases associated with eosinophils and mast cells. The humanized 2C4 mAb was generated for mast cell- and eosinophil-mediated diseases.19 It was demonstrated that the divalent interaction of 2C4 IgG mAb to the ECD of Siglec-8 occurs with high affinity (Kd < 1 pM), while it does not cross-react with other recombinant Siglecs.19 Moreover, 2C4 binds to mast cells and eosinophils and, at a lower level, to basophils in human blood and tissues via selective engagement of Siglec-8. The analysis of the obtained crystal structure described herein for the Siglec-8/2C4 complex shows that Siglec-8 binds at the tip of the V domain in the ECD. Moreover, NMR-based competition assays with sialic acid ligands and the Siglec-8/2C4 Fab complex shows that 2C4 does not block the binding site of Siglec-8 for sialylated ligands. Nevertheless, it is still possible that the 2C4 mAb may sterically block ligand binding on Siglec-8 on the surface of cells,1 even though there are no direct interactions between the Fab region and the ligand binding site of Siglec-8.
As an alternative to targeting Siglec-8 with antibodies, the use of specific glycomimetics also has the potential for regulating mast cell immune responses.23 Co-displaying an allergen and the high-affinity NSANeu5Ac ligand for Siglec-8 on liposomal nanoparticles can suppress IgE-mediated degranulation on mouse bone marrow-derived mast cells (BMMCs) or rat basophilic leukemia cells expressing Siglec-8 by recruitment of Siglec-8 to the IgE/FcεRI receptor complex.1,24 Herein, we have also analyzed in detail the binding of Siglec-8 to NSANeu5Ac using a variety of structural biology and biophysics methodologies. The observed high binding affinity (Kd = 7.1 ± 4 μM) of NSANeu5Ac can be now rationalized in the context of the structural data reported here. Comparison of the binding of NSANeu5Ac versus the natural trisaccharide 6′S-3′SLN was particularly relevant for the contribution of the NSA group in the dramatic increase of affinity (60-fold) and selectivity toward Siglec-8. Indeed, the primary recognition motif for Neu5Ac is maintained, while the NSA group is accommodated in the hydrophobic pocket formed by residues Y23, Y27, Y142, and W133, which results in a conformational rearrangement of the side chains of these amino acids with respect to the unbound form of Siglec-8. The additional CH−π interaction established by the NSA group with Y23 and Q138 residues significantly contributes to the increment in the binding affinity of NSANeu5Ac.
Moreover, we have assessed the binding preference of Siglec-8 for naturally occurring complex N-linked glycans present in FcεRIα. Siglec-8 indeed recognizes the Neu5Ac moiety on the N-linked glycans attached to the glycoprotein.12,32 However, the biological implications of glycan-dependent interactions between the ECDs of Siglec-8 and FceRI should be further studied. Indeed, direct interactions between Siglec-8 and FcεRI have already been detected in BMMCs.11
Overall, this work establishes a solid structural basis for the rational design of improved molecules with anti-inflammatory capacity targeting Siglec-8, either based on antibodies or on glycomimetics.
Materials and Methods
Cloning, Expression, and Purification of Siglec-8, 2C4, and FcERIα Proteins
Following the construction of the Siglec-8 V domain for its expression in Escherichia coli cells,36 we designed the Siglec-8d1 (UniprotKB Q9NYZ4, amino acid residues 17–155) construct containing C42S mutation and a removable 6× His tag. The DNA for Siglec-8d1 was synthesized and cloned into pET-43.1(a) plasmid by GenScript. The expression in Rosetta-gami B (DE3) competent cells (Novagen) was conducted as described elsewhere.36 Siglec-8d1 was purified from the soluble fraction of lysed cells. After ultracentrifugation, the supernatant was passed through a nickel affinity column, followed by size exclusion chromatography (Superdex 75; GE Healthcare). The purity of the protein was analyzed by SDS–PAGE and mass spectrometry, and the protein folding was determined by a 1H–15N-HSQC NMR experiment.
Full-length ECD of Siglec-8 (UniprotKB Q9NYZ4, residues 17–352) (Siglec-8d1-d3) was codon-optimized for expression in human cells and synthesized by GenScript. The construct was subcloned into the pHLsec vector37 using restriction enzymes AgeI and KpnI such that a 6× His-tag was at the C terminus of the construct to facilitate affinity purification. Siglec-8d1-d3 was transiently transfected into HEK293F (Thermo Fisher Scientific) suspension cells. The cells were split into 200 mL cultures at 0.8 × 106 cells/mL. DNA (50 μg) was added to the cells in a 1:1 ratio with the transfection reagent FectoPRO (Polyplus Transfections). The cells were incubated at 37 °C, 130 rpm, 8% CO2, and 70% humidity for 6–7 days. The cells were harvested by centrifugation at 5000g for 20 min, and supernatants were retained and filtered using a 0.45 μm Steritop filter (EMD Millipore). The supernatants were passed through a HisTrap Ni-NTA column (GE Healthcare) and eluted in 20 mM Tris pH 8.0 and 300 mM NaCl buffer with an increasing gradient of imidazole (up to 500 mM). Fractions containing Siglec-8d1-d3 were pooled and separated on a Superdex 200 Increase size exclusion column (GE Healthcare) in 20 mM Tris pH 8.0 and 300 mM NaCl buffer to achieve size homogeneity.
The sequences of the CDRs of the HC and LC of 2C4 mAb were copied into HC and LC Fab sequences. The Fab HC and LC were synthesized and cloned into a pHLsec vector37 between the AgeI and KpnI restriction sites by GenScript. The Fab expression in HEK293F cells was performed by transfecting the HC/LC at 2:1 ratio and following the same procedure as with Siglec-8d1–d3. The supernatant containing the 2C4 Fab protein was passed through a KappaSelect affinity column (GE Healthcare) and eluted with 100 mM glycine pH 3.5. The eluted fractions were immediately neutralized with 1 M Tris–HCl pH 9.0. Fractions containing the protein were pooled and run on a Superdex 200 Increase gel filtration column (GE Healthcare) to obtain purified samples.
The Siglec-8d1–d3-2C4 Fab complex was obtained by transiently co-transfecting Siglec-8d1–d3 with the HC and LC of 2C4 Fab into HEK293S (Thermo Fisher Scientific) suspension cells. The expression was achieved following the same procedure described for Siglec-8d1–d3 alone. Supernatants were first purified by affinity on a HisTrap Ni-NTA column (GE Healthcare) and then separated on a Superdex 200 Increase size exclusion column (GE Healthcare) in 20 mM Tris pH 8.0 and 300 mM NaCl buffer. Prior to crystallization, the Siglec-8d1d3-2C4 complex was treated with the enzyme EndoH (New England Biolabs) for 1 h at 37 °C. Deglycosylated protein was purified further via a second size exclusion chromatography.
The FcεRIα protein33 was transiently expressed in HEK293F or HEK293S GnTI– (ATCC) cells following similar conditions to those of Siglec-8d1d3. For purification, we followed the protocol described elsewhere.33 The expression and purification of 13C-labeled proteins was performed according to the protocol described elsewhere.31
Crystallization, X-ray Data Collection, and Structure Solution of Siglec-8d1 and Siglec-8d1d3-2C4
Purified Siglec-8d1 protein was concentrated to 10 mg mL–1 in a buffer containing 20 mM Tris pH 8.0 and 300 mM NaCl. Crystals were obtained by hanging drop vapor diffusion at 291 K in 0.1 M Bis–Tris HCl pH 6.5 and 27% PEG 3350 in 24-well plates after mixing 0.5 and 0.5 μL of protein and solution, respectively. The crystals were cryo-protected by soaking them in the mother liquor solution containing 25% glycerol and flash-cooled in liquid nitrogen. X-ray diffraction data was collected at the XALOC synchrotron beamline at ALBA (Spain). The data was processed using XDS38 in the P1211 space group at a resolution of 2.3 Å. The structure was solved by molecular replacement using Siglec-8 (PDB ID 2N7A) NMR structure as a model in Phaser.39
Crystals of Siglec-8d1 in complex with NSANeu5Ac were obtained by co-crystallization, after mixing 10 mg mL–1 protein with 0.3 equiv of NSANeu5Ac. The complex was crystallized by sitting drop vapor diffusion at 277 K in 0.075 M HEPES pH 7.5, 15% PEG 10 000, and 25% glycerol in 96-well plates using a Mosquito Crystal (SPT Labtech) crystallization robot. X-ray diffraction data was collected at the SLS synchrotron beamline at PXIII in Swiss Light Source (Switzerland). Data was processed using XDS38 in the P3221 space group at a resolution of 3.35 Å. The structure was solved by molecular replacement using Siglec-8d1 crystal structure as a search model in Phaser.39
Crystals of the Siglec-8d1–d3-2C4 Fab complex were obtained by sitting drop vapor diffusion at 291 K in 0.2 M ammonium chloride and 20% PEG 3350 pH 6.3 in 96-well plates after mixing 0.1 μL of protein (10 mg mL–1) with 0.1 μL of solution. X-ray diffraction data was collected at the SLS synchrotron beamline at PXIII in Swiss Light Source (Switzerland). Data was processed using XDS38 in the C121 space group at a resolution of 2.87 Å. The structure was solved by molecular replacement using the LC and HC of epratuzumab Fab (PDB ID 5VKK) and Siglec-8d1 crystal structures as search models in Phaser.39
All structures were refined by manual building in Coot and using phenix.refine.40 PyMOL41 was utilized for structure analysis and figure rendering. All BSA values reported were calculated using EMBL-EBI PDBePISA.42 The crystal structures, reported in this manuscript, have been deposited in the Protein Data Bank, www. rcsb.org (PDB ID: 7QU6, 7QUI, and 7QUH).
Circular Dichroism
The secondary structure of Siglec-8d1d3 expressed in HEK293F before and after PNGase F (NEB P0704S) at 40 μM concentration was analyzed by CD spectroscopy on a Jasco-815 spectropolarimeter (Jasco, Easton, MD) in 20 mM Tris buffer (pH 8) and 150 mM NaCl. The CD spectra were recorded using a quartz cell of 2 mm optical path length over a wavelength range of 200–250 nm with a scanning speed of 50 nm/min and a response time of 4 s at 20 °C.
Ligands
The compounds were prepared as described before.24
NMR Studies
NMR spectra were acquired either on a Bruker AVANCE 2 600 MHz spectrometer equipped with a standard triple-channel probe or in a Bruker 800 MHz Bruker spectrometer with a cryoprobe (Bruker, Billerica, MA, USA). Samples were measured in phosphate-buffered saline (sodium phosphate 20 mM, NaCl 40 mM, pH 7.4), containing either 10% (1H–15N TROSY HSQC) or 100% (STD and NOESY) (vol/vol) D2O. 1H Saturation-transfer difference (1H STD-NMR) experiments were performed at 298 K in a 600 MHz spectrometer using the stddiff.3 standard sequence from the Bruker library. The ligand/protein (Siglec-8) ratio was 50/1 for molecules 3′SLN and 6′S-3′SLN and 40:1 for NSANeuAc. The protein concentration was of 50 μM in all cases. For 3′SLN and 6′S-3′SLN, the saturation time was set to 2 s, while for NSANeuAc, different saturation times (0.5, 1, 2, and 4 s) were employed in order to generate a build-up curve, which was used to normalize the saturation of protons at the different residues. The following monoexponential function was applied:
where 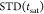 stands for the observed STD intensity at
a given saturation time (t), STDmax is
the maximal obtainable STD signal when the longest saturation time
is applied, and ksat indicates the observable
saturation rate constant. After fitting, multiplication of STDmax and ksat gives the slope of
the curve when the saturation times goes to zero.
stands for the observed STD intensity at
a given saturation time (t), STDmax is
the maximal obtainable STD signal when the longest saturation time
is applied, and ksat indicates the observable
saturation rate constant. After fitting, multiplication of STDmax and ksat gives the slope of
the curve when the saturation times goes to zero.
STD-NMR experiments were acquired with a train of 50 ms Gaussian-shaped pulses and 4 s relaxation delay. The off-resonance frequency was set at 100 ppm, and the on-resonance frequency was set at 0.7 ppm. When the Siglec-8d1-d3 construct was used, no spin-lock filter was necessary. Instead, for the smaller Siglec-8d1 construct, a spin-lock filter at 20 ms was used to clean the STD spectrum from the protein signals. Further, blank 1H STD-NMR experiments were also performed for the Siglec-8d1 apo protein. Thus, the resulting 1H STD-NMR spectra are presented as the difference between the STD spectra of the protein in the presence and absence of ligands (STDD).
NOESY experiments were acquired at 298 K at an 800 MHz spectrometer with mixing times of 400 ms. tr-NOESY experiments were performed at a protein concentration of 50 μM and a protein/ligand ratio of 1:10 for ligands 6′3′SLN and NSANeuAc. The mixing time was set to 200 ms.
1H STD-NMR and NOESY Spectra Were Processed and Analyzed in TopSpin 3.5pl6 (Bruker)
All the 1H–15N TROSY HSQC experiments for apo and bound Siglec-8 were acquired at 293 K at an 800 MHz field. Uniformly 15N-labeled Siglec-8d1 (50 μM) was titrated with increasing equivalents of ligands until complete or almost complete saturation was achieved. An in-house BEST-TROSY experiment was used for which 72 scans were acquired with 256 (t1) × 2048 (t2) complex data points in 15N and 1H, respectively.
CccpNmr Analysis software was employed for data analysis. Average chemical shift changes were calculated by using the following equation
For determining the KD of the ligands, CcpNmr Analysis was used using the following non-linear least-squares fit
Since the ligand NSANeuAc was in the slow exchange regime, KD was calculated by integrating the NMR peaks corresponding to the protein in its apo [P] and bound [PL] states at 0.5 equiv of the ligand [L]. The following equation can be solved to obtain the corresponding dissociation constant
The 1H–13C HSQC experiments for FcεRIα in the absence and presence of Siglec-8d1 were performed at 298 K with a Bruker AVANCE 2 800 MHz spectrometer equipped with a cryoprobe. FcεRIα with 13C-labeled glycans was concentrated to 40 μM in buffered (PBS1× pD = 7.4) D2O, and the experiments were performed at 298 K. 1H–13C HSQC spectra of FcεRIα with 13C-labeled glycans were recorded for 0, 1, 3, and 6 equiv of Siglec-8d1. Trimethylsilylpropanoic acid was used as the internal reference.
MD Simulations
MD simulations were carried out with the AMBER 20 package43 implemented with ff14SB(44) and the general Amber force field (GAFF2)45 for the protein and ligands, respectively. Parameters for the ligand were generated following an in-house-developed protocol for combining GLYCAM06j-146 and GAFF245 force fields in a semiautomatic manner. The initial structures were neutralized with either Na+ or Cl– ions and set at the center of a cubic TIP3P47 water box with a buffering distance between the solute and box of 10 Å. A two-stage geometry optimization approach was performed. The first stage minimizes only the positions of solvent molecules and ions, and the second stage is an unrestrained minimization of all the atoms in the simulation cell. The systems were then heated by incrementing the temperature from 0 to 300 K under a constant pressure of 1 atm and periodic boundary conditions. Harmonic restraints of 10 kcal mol–1 were applied to the solute under the Andersen temperature coupling scheme.48 The time step was kept at 1 fs during the heating stages, allowing potential inhomogeneities to self-adjust. Water molecules were treated with the SHAKE algorithm49 such that the angle between the hydrogen atoms is kept fixed through the simulations. Long-range electrostatic effects were modeled using the particle mesh Ewald method.50 An 8 Å cut-off was applied to non-bonded interactions. Each system was equilibrated for 2 ns with a 2 fs time step at a constant volume and temperature of 300 K. Five independent production trajectories were then run for additional 100 ns under the same simulation conditions, leading to accumulated simulation times of 500 ns for each system.51
Acknowledgments
This work was supported by operating grant PID2019-107770RA-I00 (J.E.-O.) from the Agencia Estatal Investigación of Spain and by the European Research Council (ERC-2017-AdG, 788143-RECGLYCANMR to J.J.-B.). We also thank the Marie-Skłodowska-Curie actions (ITN Glytunes grant agreement no. 956758 to J.E.-O and ITN BactiVax under grant agreement no. 860325 to U.A.). Additional funding was provided by CIBER, an initiative of Instituto de Salud Carlos III (ISCIII), Madrid, Spain. We also thank the Ikerbasque Basque Foundation of Science and the Spanish Ministry of Economy, Industry and Competitiveness (for the postdoctoral contract Juan de la Cierva Incorporación to J.E-O). X-ray diffraction experiments described in this paper were performed using the XALOC synchrotron beamline at ALBA (Spain) and PXIII in Swiss Light Source (Switzerland).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.2c00592.
Additional NMR experimental data, including 1H–15N TROSY titrations for all ligands and tr-NOESY for conformational analysis in free and bound states, and information on the STD epitopes and the residues involved in binding in the crystal structures (PDF)
Author Contributions
M.P.L., U.A., J.E.-O., and J.J-B. contributed to experimental conception and design; M.P.L., U.A., C.N., I.O., S.D., R.N.-F., C.T.G.-M., and J.E.-O. contributed to data acquisition; M.P.L., U.A., A.F., H.C., G.J.-O., L.U., M.S., J.E.-O., and J.J.-B. contributed to analysis of data; and M.P.L., U.A., A.F., L.U., M.S., J.C.P., J.E.-O., and J.J.-B. contributed to drafting the article or revising it critically for important intellectual content. CRediT: Maria Pia Lenza investigation, writing-review & editing.
The authors declare no competing financial interest.
Supplementary Material
References
- Duan S.; Arlian B. M.; Nycholat C. M.; Wei Y.; Tateno H.; Smith S. A.; Macauley M. S.; Zhu Z.; Bochner B. S.; Paulson J. C. Nanoparticles Displaying Allergen and Siglec-8 Ligands Suppress IgE-FcεRI-Mediated Anaphylaxis and Desensitize Mast Cells to Subsequent Antigen Challenge. J. Immunol. 2021, 206, 2290–2300. 10.4049/jimmunol.1901212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. W.; Kelly E. A.; Nguyen C. L.; Jarjour N. N.; Bochner B. S. Characterization of Siglec-8 Expression on Lavage Cells after Segmental Lung Allergen Challenge. Int. Arch. Allergy Immunol. 2018, 177, 16–28. 10.1159/000488951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenza M. P.; Atxabal U.; Oyenarte I.; Jiménez-Barbero J.; Ereño-Orbea J. Current Status on Therapeutic Molecules Targeting Siglec Receptors. Cells 2020, 9, 2691. 10.3390/cells9122691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S.; Paulson J. C. Siglecs as Immune Cell Checkpoints in Disease. Annu. Rev. Immunol. 2020, 38, 365–395. 10.1146/annurev-immunol-102419-035900. [DOI] [PubMed] [Google Scholar]
- Nutku E.; Hudson S. A.; Bochner B. S. Mechanism of Siglec-8-Induced Human Eosinophil Apoptosis: Role of Caspases and Mitochondrial Injury. Biochem. Biophys. Res. Commun. 2005, 336, 918–924. 10.1016/j.bbrc.2005.08.202. [DOI] [PubMed] [Google Scholar]
- Nutku-Bilir E.; Hudson S. A.; Bochner B. S. Interleukin-5 Priming of Human Eosinophils Alters Siglec-8-Mediated Apoptosis Pathways. Am. J. Respir. Cell Mol. Biol. 2008, 38, 121–124. 10.1165/rcmb.2007-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na H. J.; Hudson S. A.; Bochner B. S. IL-33 Enhances Siglec-8 Mediated Apoptosis of Human Eosinophils. Cytokine 2012, 57, 169–174. 10.1016/j.cyto.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. J.; O’Sullivan J. A.; Nix D. B.; Cao Y.; Tiemeyer M.; Bochner B. S. Sialic acid-binding immunoglobulin-like lectin 8 (Siglec-8) is an activating receptor mediating β2-integrin-dependent function in human eosinophils. J. Allergy Clin. Immunol. 2018, 141, 2196–2207. 10.1016/j.jaci.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. J.; Cao Y.; Bochner B. S.; O’Sullivan J. A. Siglec-8 Signals Through a Non-Canonical Pathway to Cause Human Eosinophil Death In Vitro. Front. Immunol. 2021, 12, 737988. 10.3389/fimmu.2021.737988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi H.; Choi O. H.; Hubbard W.; Lee H. S.; Canning B. J.; Lee H. H.; Ryu S. D.; von Gunten S.; Bickel C. A.; Hudson S. A.; et al. Inhibition of FcεRI-dependent mediator release and calcium flux from human mast cells by sialic acid-binding immunoglobulin-like lectin 8 engagement. J. Allergy Clin. Immunol. 2008, 121, 499–505. 10.1016/j.jaci.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Korver W.; Wong A.; Gebremeskel S.; Negri G. L.; Schanin J.; Chang K.; Leung J.; Benet Z.; Luu T.; Brock E. C.; et al. The Inhibitory Receptor Siglec-8 Interacts With FcεRI and Globally Inhibits Intracellular Signaling in Primary Mast Cells Upon Activation. Front. Immunol. 2022, 13, 833728. 10.3389/fimmu.2022.833728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. S.; Alvarez R. A.; Mehta P.; Bovin N. V.; Blixt O.; White J. R.; Schnaar R. L. Glycan Array Screening Reveals a Candidate Ligand for Siglec-8*. J. Biol. Chem. 2005, 280, 4307–4312. 10.1074/jbc.M412378200. [DOI] [PubMed] [Google Scholar]
- Pröpster J. M.; Yang F.; Rabbani S.; Ernst B.; Allain F. H.; Schubert M. Structural Basis for Sulfation-Dependent Self-Glycan Recognition by the Human Immune-Inhibitory Receptor Siglec-8. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, E4170–E4179. 10.1073/pnas.1602214113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno A.; Delgado S.; Valverde P.; Bertuzzi S.; Berbís M. A.; Echavarren J.; Lacetera A.; Martín-Santamaría S.; Surolia A.; Cañada F. J.; et al. Minimizing the Entropy Penalty for Ligand Binding: Lessons from the Molecular Recognition of the Histo Blood-Group Antigens by Human Galectin-3. Angew. Chem., Int. Ed. Engl. 2019, 58, 7268–7272. 10.1002/anie.201900723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli S. J.; Tsai M. IgE and Mast Cells in Allergic Disease. Nat. Med. 2012, 18, 693–704. 10.1038/nm.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbino B.; Herviou P.; Godon O.; Stackowicz J.; Goff O. R.; Iannascoli B.; Sterlin D.; Brûlé S.; Millot G. A.; Harris F. M.; et al. The anti-IgE mAb omalizumab induces adverse reactions by engaging Fcγ receptors. J. Clin. Invest. 2020, 130, 1330–1335. 10.1172/jci129697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama Y.; Matsumoto H.; Odajima H.; Takahagi S.; Hide M.; Okubo K. Roles of Omalizumab in Various Allergic Diseases. Allergol. Int. 2020, 69, 167–177. 10.1016/j.alit.2020.01.004. [DOI] [PubMed] [Google Scholar]
- Tiotiu A.; Oster J. P.; Roux P. R.; Nguyen Thi P. L.; Peiffer G.; Bonniaud P.; Dalphin J. C.; de Blay F. Effectiveness of Omalizumab in Severe Allergic Asthma and Nasal Polyposis: A Real-Life Study. J Investig. Allergol. Clin. Immunol. 2020, 30, 49–57. 10.18176/jiaci.0391. [DOI] [PubMed] [Google Scholar]
- Youngblood B. A.; Brock E. C.; Leung J.; Falahati R.; Bryce P. J.; Bright J.; Williams J.; Shultz L. D.; Greiner D. L.; Brehm M. A.; et al. AK002, a Humanized Sialic Acid-Binding Immunoglobulin-Like Lectin-8 Antibody that Induces Antibody-Dependent Cell-Mediated Cytotoxicity against Human Eosinophils and Inhibits Mast Cell-Mediated Anaphylaxis in Mice. Int. Arch. Allergy Immunol. 2019, 180, 91–102. 10.1159/000501637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiwamoto T.; Kawasaki N.; Paulson J. C.; Bochner B. S. Siglec-8 as a Drugable Target to Treat Eosinophil and Mast Cell-Associated Conditions. Pharmacol. Ther. 2012, 135, 327–336. 10.1016/j.pharmthera.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B. A.; Leung J.; Falahati R.; Williams J.; Schanin J.; Brock E. C.; Singh B.; Chang A. T.; O’Sullivan J. A.; Schleimer R. P.; et al. Discovery, Function, and Therapeutic Targeting of Siglec-8. Cells 2020, 10, 19. 10.3390/cells10010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellon E. S.; Peterson K. A.; Murray J. A.; Falk G. W.; Gonsalves N.; Chehade M.; Genta R. M.; Leung J.; Khoury P.; Klion A. D.; et al. Anti-Siglec-8 Antibody for Eosinophilic Gastritis and Duodenitis. N. Engl. J. Med. 2020, 383, 1624–1634. 10.1056/NEJMoa2012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroezen B. S.; Conti G.; Girardi B.; Cramer J.; Jiang X.; Rabbani S.; Müller J.; Kokot M.; Luisoni E.; Ricklin D.; et al. A Potent Mimetic of the Siglec-8 Ligand 6’-Sulfo-Sialyl Lewis x. ChemMedChem 2020, 15, 1706–1719. 10.1002/cmdc.202000417. [DOI] [PubMed] [Google Scholar]
- Nycholat C. M.; Duan S.; Knuplez E.; Worth C.; Elich M.; Yao A.; O’Sullivan J.; McBride R.; Wei Y.; Fernandes S. M.; et al. A Sulfonamide Sialoside Analogue for Targeting Siglec-8 and -F on Immune Cells. J. Am. Chem. Soc. 2019, 141, 14032–14037. 10.1021/jacs.9b05769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K.; Riek R.; Wider G.; Wüthrich K. Attenuated T 2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 12366–12371. 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solyom Z.; Schwarten M.; Geist L.; Konrat R.; Willbold D.; Brutscher B. BEST-TROSY Experiments for Time-Efficient Sequential Resonance Assignment of Large Disordered Proteins. J. Biomol. NMR 2013, 55, 311–321. 10.1007/s10858-013-9715-0. [DOI] [PubMed] [Google Scholar]
- Williamson M. P. Using Chemical Shift Perturbation to Characterise Ligand Binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 73, 1–16. 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Mayer M.; Meyer B. Characterization of Ligand Binding by Saturation Transfer Difference NMR Spectroscopy. Angew. Chem., Int. Ed. Engl. 1999, 38, 1784–1788. . [DOI] [PubMed] [Google Scholar]
- Meyer B.; Peters T. NMR Spectroscopy Techniques for Screening and Identifying Ligand Binding to Protein Receptors. Angew. Chem., Int. Ed. Engl. 2003, 42, 864–890. 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- Siebert H.-C.; André S.; Lu S.-Y.; Frank M.; Kaltner H.; van Kuik J. A.; Korchagina E. Y.; Bovin N.; Tajkhorshid E.; Kaptein R.; et al. Unique Conformer Selection of Human Growth-Regulatory Lectin Galectin-1 for Ganglioside GM1 versus Bacterial Toxins,. Biochemistry 2003, 42, 14762–14773. 10.1021/bi035477c. [DOI] [PubMed] [Google Scholar]
- Lenza M. P.; Oyenarte I.; Diercks T.; Quintana J. I.; Gimeno A.; Coelho H.; Diniz A.; Peccati F.; Delgado S.; Bosch A.; et al. Structural Characterization of N-Linked Glycans in the Receptor Binding Domain of the SARS-CoV-2 Spike Protein and their Interactions with Human Lectins. Angew. Chem., Int. Ed. Engl. 2020, 59, 23763–23771. 10.1002/anie.202011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno H.; Crocker P. R.; Paulson J. C. Mouse Siglec-F and human Siglec-8 are functionally convergent paralogs that are selectively expressed on eosinophils and recognize 6′-sulfo-sialyl Lewis X as a preferred glycan ligand. Glycobiology 2005, 15, 1125–1135. 10.1093/glycob/cwi097. [DOI] [PubMed] [Google Scholar]
- Unione L.; Lenza M. P.; Ardá A.; Urquiza P.; Laín A.; Falcón-Pérez J. M.; Jiménez-Barbero J.; Millet O. Glycoprofile Analysis of an Intact Glycoprotein As Inferred by NMR Spectroscopy. ACS Cent. Sci. 2019, 5, 1554–1561. 10.1021/acscentsci.9b00540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr S. C.; Gonzalez J. R.; Schanin J.; Peters M. C.; Lambrecht B. N.; Brock E. C.; Charbit A.; Ansel K. M.; Youngblood B. A.; Fahy J. V. An anti-siglec-8 antibody depletes sputum eosinophils from asthmatic subjects and inhibits lung mast cells. Clin. Exp. Allergy 2020, 50, 904–914. 10.1111/cea.13681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanin J.; Gebremeskel S.; Korver W.; Falahati R.; Butuci M.; Haw T. J.; Nair P. M.; Liu G.; Hansbro N. G.; Hansbro P. M.; et al. A Monoclonal Antibody to Siglec-8 Suppresses non-Allergic Airway Inflammation and Inhibits IgE-independent Mast Cell Activation. Mucosal Immunol. 2021, 14, 366–376. 10.1038/s41385-020-00336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröpster J. M.; Yang F.; Ernst B.; Allain F. H.; Schubert M. Functional Siglec Lectin Domains from Soluble Expression in the Cytoplasm of Escherichia coli. Protein Expr. Purif. 2015, 109, 14–22. 10.1016/j.pep.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Aricescu A. R.; Lu W.; Jones E. Y. A Time- and Cost-Efficient System for High-Level Protein Production in Mammalian Cells. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 1243–1250. 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. 10.1107/s0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. Phasercrystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkóczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; et al. PHENIX: a Comprehensive Python-based System for Macromolecular Structure Solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. 10.1107/s0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger L.The PyMol Molecular Graphics System, Versión 1.8. Thomas Hold, 2015. .
- Krissinel E.; Henrick K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Case D. A.; Belfon K.; Ben-Shalom I. Y.; Brozell S. R.; Cerutti D. S.; Cheatham T. E. III; Cruzeiro V. W. D.; Darden T. A.; Duke R. E.; Giambasu G.; et al. AMBER2020; University of California: San Francisco, CA, USA, 2020.
- Maier J. A.; Martinez C.; Kasavajhala K.; Wickstrom L.; Hauser K. E.; Simmerling C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Wolf R. M.; Caldwell J. W.; Kollman P. A.; Case D. A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Kirschner K. N.; Yongye A. B.; Tschampel S. M.; González-Outeiriño J.; Daniels C. R.; Foley B. L.; Woods R. J. GLYCAM06: a Generalizable Biomolecular Force Field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Andersen H. C. Molecular Dynamics Simulations at Constant Pressure and/or Temperature. J. Chem. Phys. 1980, 72, 2384–2393. 10.1063/1.439486. [DOI] [Google Scholar]
- Miyamoto S.; Kollman P. A. Settle: An Analytical Version of the SHAKE and RATTLE Algorithm for Rigid Water Models. J. Comput. Chem. 1992, 13, 952–962. 10.1002/jcc.540130805. [DOI] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle mesh Ewald: AnN·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Unni S.; Huang Y.; Hanson R. M.; Tobias M.; Krishnan S.; Li W. W.; Nielsen J. E.; Baker N. A. Web Servers and Services for Electrostatics Calculations with APBS and PDB2PQR. J. Comput. Chem. 2011, 32, 1488–1491. 10.1002/jcc.21720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.