Abstract
Objective
WNT/β-catenin signaling is initiated by binding of a WNT protein to a Frizzled (FZD) receptor and a co-receptor, low-density lipoprotein (LDL) receptor-related protein 5 or 6 (LRP5/6). The objective of this study was to find the genetic variants responsible for dental anomalies found in 4 families.
Methods
Clinical and radiographic examination and whole exome sequencing were performed on 5 patients affected with dental anomalies and the mutant proteins modeled.
Results
Five patients were heterozygous for the WNT10A variants, including c.877C>T; p.Arg293Cys, c.874A>G; p.Ser292Gly, c.1042C>T; p.Arg348Cys, and c.1039G>T; p.347GluX. The p.Arg293Cys and p.Ser292Gly mutations are located in the WNT10A N-terminal domain region with binding sites for FZD receptor, porcupine, WNTLESS, and extracellular binding proteins, so they are likely to have adverse effects on binding these proteins. The p.Arg348Cys mutation, which is located in the binding site of LRP5/6 co-receptors, is postulated to result in impaired binding to these co-receptors. The nonsense mutation p.347GluX is predicted to result in the truncation of most of the C-terminal domain, which is likely to disrupt the binding of WNT10A to WNTLESS, the membrane protein that binds lipid-acylated WNT proteins to carry them from the endoplasmic reticulum to the cell surface and FZD.
Conclusions
Four novel mutations in WNT10A were identified in patients with isolated tooth agenesis. The mutations in the N-terminal domain and the interface between the N- and C-terminal domains of WNT10A in our patients are likely to disrupt its binding with FZD, LRP5/6, and various other proteins involved in WNT10A processing and transport, impair WNT and SHH signaling, and subsequently result in tooth agenesis, microdontia, and root maldevelopment.
Key words: WNT10A mutation, Frizzled binding site, Dental anomaly, Hypodontia, Dental malformation, Dental defect
Introduction
WNT signaling is crucial for a number of developmental, physiologic, and disease processes. It is involved in almost every aspect of embryonic development and controls homeostatic self-renewal in many adult tissues.1 WNT signaling has an important role in development of ectodermally derived organs including teeth, skin, nails, hair, and sweat glands. Genetic variants in WNT10A have been implicated in developmental disorders of ectodermal structures in forms of autosomal dominant or autosomal recessive ectodermal dysplasias or isolated tooth agenesis.2 WNT10A has been reported as a major gene in the etiology of isolated tooth agenesis.3,4
WNT proteins comprise a highly conserved N-terminal domain (NTD) that contains a lipid-modified thumb and a C-terminal domain (CTD).5 Between the NTD and CTD is a peptide region of variable length and sequence. This linking region functions as an NTD–CTD interface, containing the least conserved amino acids that contribute to binding specificity.6 WNTs are hydrophobic as the result of the posttranslational addition of palmitate and/or palmitoleic acid (acylation) by porcupine protein.7 Fatty acylation and glycosylation in the endoplasmic reticulum (ER)-Golgi system are required for WNT intracellular trafficking, FZD binding, secretion, and its full activity when secreted.5,8,9 A carrier protein needed for WNT protein secretion, "?>WNTLESS, also binds near the acylation site.9,10 Glycosylation of Wnts also takes place near the lipid in the NTD,8 an important region for binding to a FZD cysteine-rich receptor domain.11 Once in the extracellular milieu, WNT proteins bind to other extracellular carrier proteins of the lipocalin family, such as secreted WINGLESS-interacting molecule (SWIM) or afamin (AFM), allowing their transport between cells.12,13 The removal of the acyl group by NOTUM and binding to inhibitor proteins in the extracellular matrix may also act to regulate Wnt activity.7 The interaction of WNT proteins with the various processing enzymes, carrier proteins, and inhibitors make WNTsignaling particularly susceptible to perturbation by small changes in the structure or surface properties of WNT proteins.
Here we report on 4 novel WNT10A mutations located in the binding sites of WNT10A in 4 Thai families affected with tooth agenesis, microdontia, single-rooted permanent molars, and taurodontism, a condition defined by enlargement of the pulp chamber of multirooted teeth with apical displacement of the pulpal floor.
Methods
Ethical approval
All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study or, in cases of children, from their parents as their legal guardians.
Patient report
Patient 1 (Family 1)
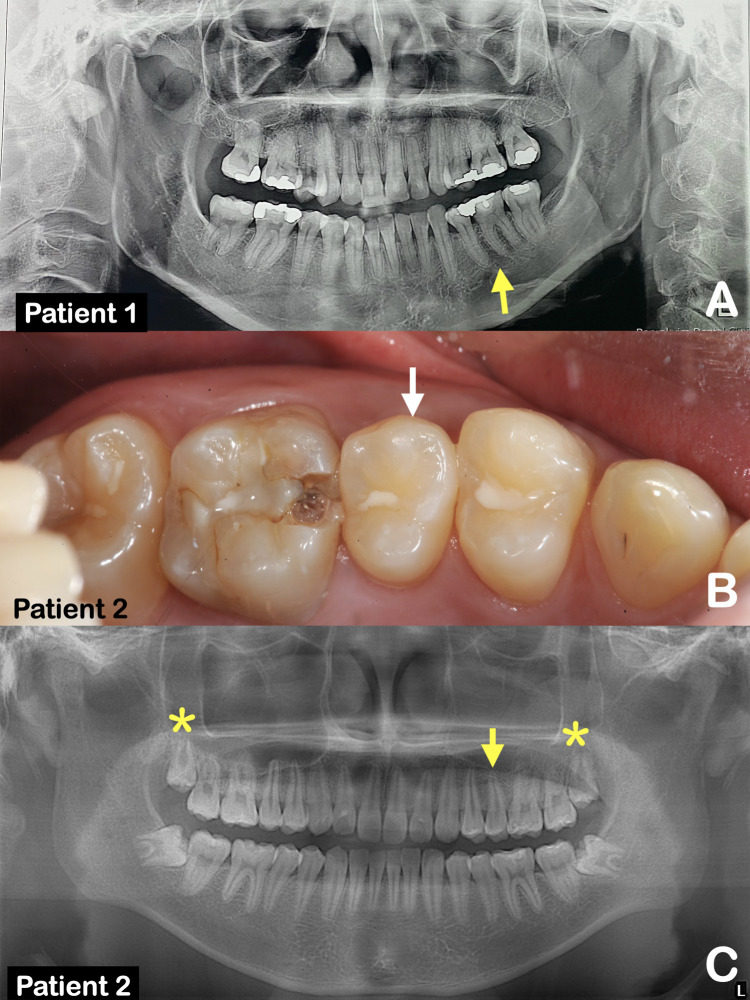
Patient 1 was a 27-year-old Thai woman who had agenesis of the mandibular left permanent first molar. She was otherwise healthy. The second permanent left molar was orthodontically shifted into the place of the first permanent left molar (Figure 1A). Family history was not available.
Fig. 1.
A, Panoramic radiograph of patient 1 at age 30 showing tooth agenesis of the mandibular left permanent first molar (arrow). B, Patient 2. Microdontia of the maxillary left second premolar (arrow). C, Panoramic radiograph of patient 2 at age 17. Microdontia of the maxillary left second premolar (arrow). Single-rooted maxillary third permanent molars are noted (asterisks).
Patient 2 (Family 2)
Patient 2 was a 17-year-old Thai boy who had microdontia of the maxillary left second premolar (Figure 1B). Panoramic radiograph showed single-rooted maxillary second permanent molars (Figure 1C). He was otherwise healthy. Family history was not available.
Patient 3 (Family 3)
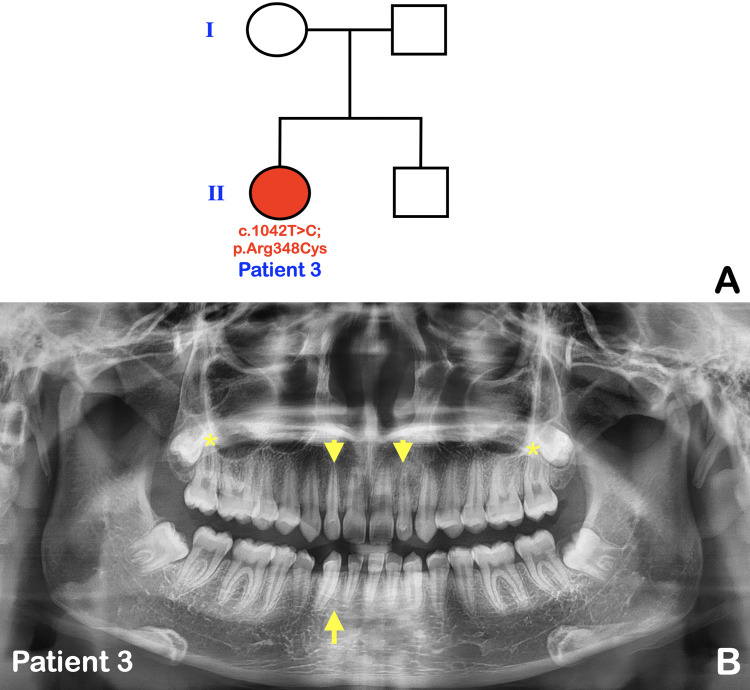
Patient 3 was a 17-year-old Thai girl who presented with isolated tooth agenesis of the mandibular right permanent lateral incisor, microdontia of the maxillary permanent lateral incisors, and taurodontism of the maxillary permanent second molars (Figure 2A, B). She was otherwise healthy. Her family history was unremarkable (Figure 2A).
Fig. 2.
A, Pedigree of patient 3. B, Panoramic radiograph of patient 3 at age 17 shows agenesis of the mandibular right permanent lateral incisor (arrow), microdontia of the maxillary permanent lateral incisors (arrowheads), and taurodontism of the maxillary permanent second molars (asterisks).
Patients 4 and 5 (Family 4)
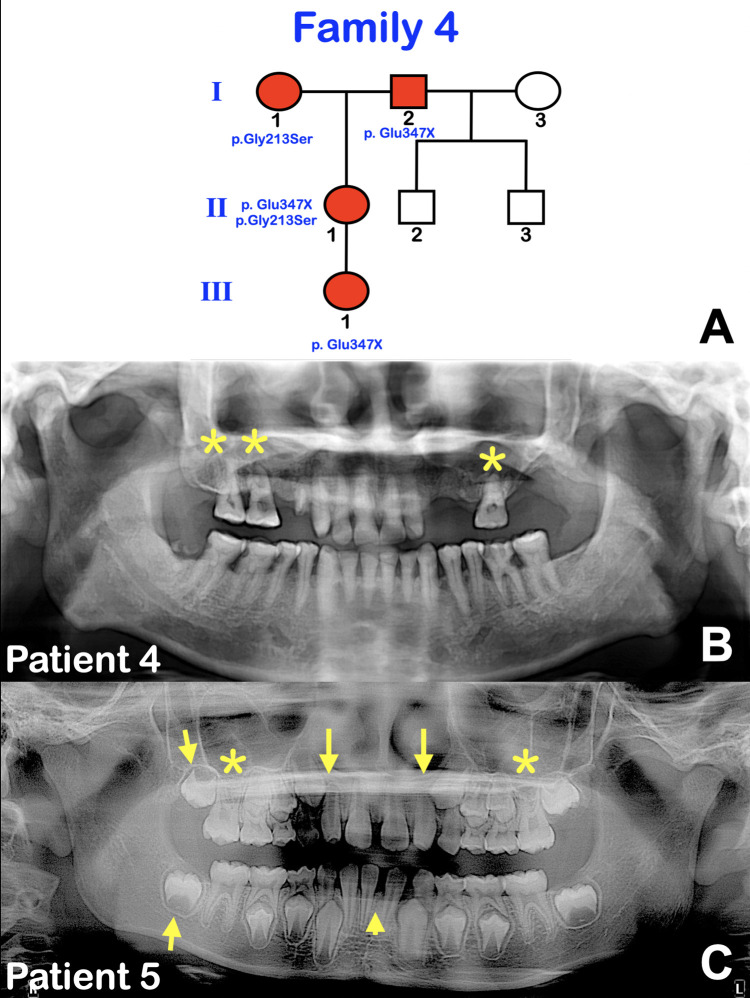
Patient 4 (I-2) is the father of the previously reported female patient (II-1) with tricho-odonto-onycho-dermal dysplasia (OODD; Figure 3A) (Patient 314). At 58 years, he had lost several teeth as a result of dental decay. Panoramic radiograph showed taurodontism of the maxillary right first permanent molar and maxillary second permanent molars. Pulp stones were observed in those teeth. Severe pulp obliteration was evident in the remaining teeth (Figure 3B). His health was normal. Patient 5 (III-1), a granddaughter of patient 4 (I-2) (Figure 3A), had dental anomalies including microdontia of the maxillary permanent lateral incisors and agenesis of the mandibular left first central incisor. Panoramic radiograph showed agenesis of the mandibular first left central incisor and taurodontism of the maxillary first permanent molars. Microdontia of the maxillary permanent lateral incisors and the developing maxillary and mandibular second permanent molars was noted (Figure 3C).
Fig. 3.
Patients 4 and 5. A, Pedigree. Patient 4 (I-2) is the grandfather of patient 5 (III-1), whose mother has a compound heterozygous mutation c.1039G>T; p.Glu347X and c.673G>A; p.Gly213Ser in WNT10A. Patients 4 and 5 are heterozygous for the c.1039G>T; p.Glu347X variant in WNT10A. B, Panoramic radiograph of patient 4 at age 58 shows taurodontism and pulp stones in the maxillary right first permanent molar and maxillary second permanent molars (asterisks). Severe pulp obliteration is evident in most teeth. C, Panoramic radiograph of patient 5 at age 8 shows mixed dentition, agenesis of the mandibular first left central incisor (arrowhead), and taurodontism of the maxillary first permanent molars (asterisks). Microdontia of the maxillary permanent lateral incisors and the developing maxillary and mandibular right second permanent molars was noted (arrows).
Whole exome sequencing and bioinformatic analysis
Details of whole exome sequencing and bioinformatic analysis can be found in supplemental information 1.
Protein modelling
The hWNT10A protein sequence (NP_079492.2) was submitted to the SWISS-MODEL server (https://swissmodel.expasy.org) for homology modelling15 with the hWNT3structure (PDB code: 6AHY) as a template.16 Models were visualised in Pymol (Schrödinger LLC).
Results
Whole exome sequencing showed heterozygous mutations c.877 C>T; p.Arg293Cys (rs563548971), c.874A>G; p.Ser292Gly (rs767665930), and c.1042C>T; p.Arg348Cys (rs978088338) in patients 1 through 3, respectively. The c.1039G>T; p.347GluX variant was identified in patients 4 and 5. All 4 mutations were confirmed by Sanger direct sequencing (Supplemental Figure 1).
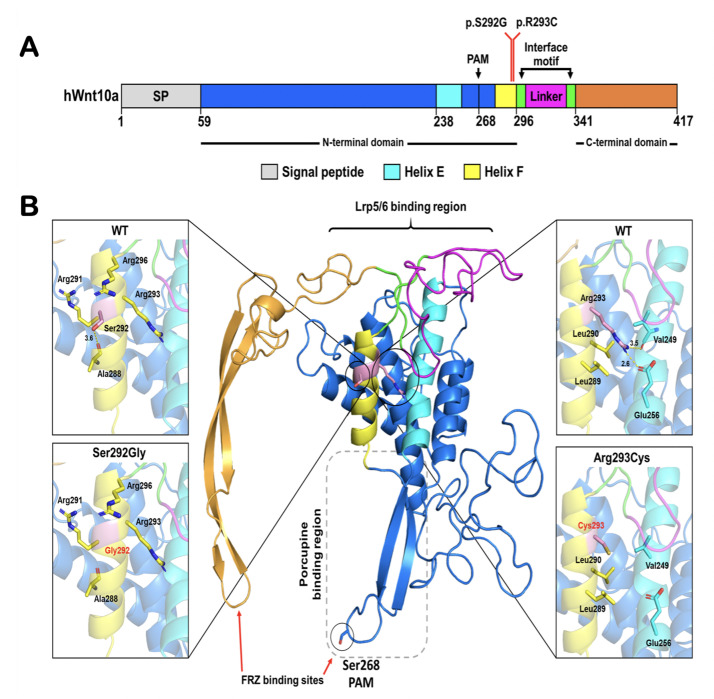
Ser292Gly
Ser292 is found on the solvent side of alpha-helix F in the N-terminal palm domain of the WNT10A structural model. The mutation of Ser292 to Gly may increase the flexibility and decrease the stability of the helix (Figure 4A, B). This residue is highly conserved across species but not conserved in other WNT proteins (Supplemental Figure 2A). As such, it may be partially responsible for protein binding specificity.
Fig. 4.
Predicted structure of WNT10A showing the position of the p.Ser292Gly and p.Arg293Cys mutation. A, Schematic diagram of the primary structure of human WNT10A (signal peptide [SP], N-terminal domain, interface motif, linker region, and C-terminal domain) based on the structure of WNT3. B, The structural model of WNT10A based on the crystal structure of WNT3 in complex with mouse FZD 8 Cys-Rich domain (CRD) (PDB: 6AHY).16 The position of the Ser292Gly mutation and the surrounding residues are shown in the expanded views of alpha-helix F on the left, whilst Arg293 in the wild-type and Cys293 created by the Arg293Cys mutation are emphasised in the expanded views of alpha-helix F on the right. Helix E is coloured cyan to emphasise that changing Arg293 to Cys affects interaction between helix E and helix F.
Arg293Cys
Arg293Cys, a highly conserved amino acid (Supplemental Figure 2A), is also located on the solvent side of alpha-helix F in the N-terminal palm domain of the WNT10A structural model. It is part of a positively charged patch that includes Arg291 and Arg296 (Figure 4A, B). The mutation to Cys would decrease the charge in this region, possibly affecting its interaction with other proteins. The introduction of an additional unpaired Cys may also cause it to compete with other Cys side chains for disulfide bond formation during protein folding, thereby trapping the protein in unproductive conformations and reducing the efficiency of folding.
Arg348Cys
The highly conserved amino acid residues Glu347 and Arg348 (Supplemental Figure 2B) are located at the top surface of the CTD, near the interface between the NTD and CTD. Arg348Cys is located near the region that has been proposed to be a binding site of the LRP5/6 receptor (Figures 5A, B).5 The disruption of the surface charge by removing the arginine may decrease such interactions, whilst the introduction of an additional cysteine may interfere with formation of neighboring disulfide bonds, such as that between Cys346 and Cys377 by competing for disulfide bond formation (Figure 5A, B).
Fig. 5.
Predicted structure of WNT10A showing the position of the Glu347X and Arg348Cys mutation. A, Schematic map of the primary structure of human WNT10A based on the structure of WNT3. The signal sequence (SP) is followed by the N-terminal domain (NTD), thumb hairpin, interface motif, linker region, C-terminal domain (CTD), helix G, and index finger hairpin. B, Structural model of WNT10A based on the crystal structure of WWNT3 in complex with mouse FZD 8 Cys-Rich domain (CRD) (PDB: 6AHY).16 Arg348Cys is shown in an expanded view of the region of the mutation in wild-type (Arg348) and mutant (Cys348), with the mutated and neighboring amino acid side chains shown in stick representation. Since Glu347X results in the loss of the C-terminal part including helix G and index finger hairpin, this region is shown in faded colours.
Glu347X
The Glu347X results in a truncated protein losing 70 amino acid residues from the C-terminus, including the 2-stranded β-sheet hairpin known as the index finger, which contributes to binding to the FZD receptor (Figure 5A, B).5,16 This index finger is also involved in binding to WNTLESS, which escorts WNT proteins from the ER to secretion out of the cell,10 so loss of this region will likely block secretion and subsequent WNT/β-catenin signaling.
Discussion
We report 5 Thai patients from 4 families with dental anomalies and novel heterozygous WNT10A mutations. Patient 1, who carried a c.877 C>T; p.Arg293Cys mutation in WNT10A, was affected with tooth agenesis of the mandibular left first permanent molar. Patient 2, who carried a c.874A>G; p.Ser292Gly mutation in WNT10A, was affected with microdontia and single-rooted maxillary permanent third molars. A de novo WNT10A mutation NM_025216.2: c.1042C>T; NP_079492.2: p.Arg348Cys (rs978088338) was identified in patient 3, who was affected with agenesis of the mandibular permanent right lateral incisor, microdontia of the maxillary permanent lateral incisors, and taurodontism of the maxillary permanent second molars. Her parents had normal teeth and did not have the mutation. The c.1039G>T; p.Glu347X mutation in WNT10A was identified in patients 4 and 5. Patient 4 lost a number of teeth as a result of dental decay; therefore, it was not possible to evaluate tooth agenesis. He had taurodontism with dental pulp stones and generalised pulp obliteration. Patient 5, a granddaughter of patient 4, had tooth agenesis, microdontia, and taurodontism.
We are convinced that these 4 variants were associated with the dental anomalies found in the patients and are not coincidental findings. First, according to gnomAD (https://gnomad.broadinstitute.org), the c.877 C>T; p.Arg293Cys and c.874A>G; p.Ser292Gly variants are very rare in the general population. The c.877 C>T; p.Arg293Cys variant is seen in only one of 146,326 alleles, with an allele frequency of 0.000006834 and without any homozygous variation. This variant was not found in our in-house exome database of 775 individuals. The c.874A>G; p.Ser292Gly variant is seen in 38 of 183,346 alleles with allele frequency of 0.0002073 and without any homozygous variation. This variant is reported only in the East Asian population. Even though these variants have been seen in the general population, common anomalies such as microdontia might have escaped clinical assessment and without panoramic radiography the dental anomalies such as taurodontism and root anomalies might have been undetected.
Second, the amino acid residues Arg293 and Ser292 are highly conserved across species (Supplemental Figure 2). Third, the p.Arg293Cys mutation is predicted as deleterious, possibly damaging, and damaging by MutationTaster, PolyPhen-2, and SIFT prediction, respectively. The p.Ser292Gly mutation is predicted as damaging by SIFT.
Both p.Ser292Gly and p.Arg293Cys have not been reported to be associated with dental anomalies. It is interesting to note that the c.874A>G; p.Ser292Gly variant has been reported in a patient with severe scoliosis, hypotrichosis, hypohidrosis, hearing loss, seborrheic keratoses, multiple cherry angioma, basal cell carcinoma, clinodactyly, osteoporosis, and mild stenosis of both carotid arteries.17 We are convinced that dental anomalies might have been detected if panoramic radiography had been performed.18
The WNT/β-catenin signaling pathway is initiated by binding of a WNT protein to a FZD receptor and a co-receptor, low-density lipoprotein receptor-related protein 5 or 6 (LRP5/6).6 FZD, the 7-pass transmembrane receptor, is known to be critical for nearly all WNT signaling pathways. The linker between the NTD and CTD of the WNT protein is required for LRP6 binding (Figure 4B).6 WNT posttranslational modification includes O-acylation at the Ser site (Ser268, WNT10A) that is performed in the ER by porcupine to generate palmitileic acid or palmitate modified WNT (PAM WNT). Porcupine acyltransferase makes extensive contacts with the thumb hairpin loop that is extended from the palm region where the identified mutations are located (Figure 4B). This modification is necessary for the PAM WNT-WNTLESS interaction responsible for transport through the Golgi to the cell surface, which also makes extensive interactions over the thumb hairpin stabilised by the palm region where these mutations are located.10 In addition, PAM WNT is critical to form the PAM WNT–FZD complex on the surface of the cell for WNT signal transduction.9 The p.Arg293Cys and p.Ser292Gly mutations in WNT10A found in our patients are located in the NTD, a binding site for FZD-cysteine-rich domain (Figure 4A, B),5 and they are likely to have effects on binding of porcupine, WNTLESS, extracellular binding proteins, and/or the FZD receptor.
The change from Arg293 to Cys293 in patient 1 may cause this Cys293 to compete with other Cys for disulfide bond formation during protein folding, thereby placing the protein in unproductive conformations and impairing the efficiency of folding. The Ser292Gly mutation in patient 2 may increase the flexibility and decrease the stability of the helix. Arg293Cys and Ser292Gly may have an effect on the WNT secretory pathway by affecting the stability of helix E and helix F, which are near the porcupine binding region9 and may contribute to the O-acylation that is necessary for PAM WNT to function.
We are also convinced that the c.1042C>T; p.Arg348Cys mutation in patient 3 was also associated with dental anomalies, because this mutation was de novo and the amino acid residue Arg348 is conserved across vertebrate species except for a few animals such as African elephants (Loxodonta africana), in which the amino acid residue at this position is histidine instead of arginine (Supplemental Figure 2B), a relatively conservative substitution. In addition, the c.1042C>T; p.Arg348Cys variant is extremely rare in the global population. This variant is not reported in Lovd (https://www.lovd.nl) and HGMD (http://www.hgmd.cf.ac.uk) databases. According to gnomAD (https://gnomad.broadinstitute.org), this variant has been seen in one of 220,610 individuals with allele frequency of 0.000004533. It has not been seen in Asian populations or been reported to be pathogenic. This mutation is predicted to be probably damaging by PolyPhen-2.
The protein model showed that the amino acid residue Arg348 is located near the interface between the NTD and CTD. The NTD comprises a 6-helix core and 2 protruding β-hairpins, one of which is acylated with a fatty acid. The CTD is a cytokine-like hairpin loop. The fatty acyl group of the NTD protruding β-hairpin interacts with the FRZ co-receptor, as do residues on the tip of CTD-hairpin (Figure 5A, B).5,11,16 The amino acid residue Arg348 near the interface between the NTD and CTD is on a surface postulated to bind to LRP5/6-type co-receptors,5,6 so its change as a result of mutation is likely to affect binding to the co-receptor. This region mostly contains charged amino acids, located at loop F linked to helix G (Figure 5A, B). Because of loop flexibility, Arg348 might form a charge–charge interaction with Glu347 and Glu349 and hydrogen bonds with Ser354 and Thr357. The change from Arg to Cys at amino acid residue 348 might affect loop flexibility by reducing ionic interactions in loop F. Cys348 might also form an alternative disulfide bond to disrupt that between Cys346 to Cys377 near helix G, thereby disrupting the structure of this region of the protein (Figure 5A, B). Indeed, it was recently reported that a similar mutation on a nearby loop, Arg360Cys, causes lower protein expression and reduced binding to the FZD5 receptor.19 The change in surface charge and possible disruption of the local protein folding could cause aberrant interaction between WNT10A protein and LRP5/6-type co-receptors and result in aberrant WNT signaling. All lines of evidence suggest that the p.Arg348Cys variant found in the family with isolated tooth agenesis and microdontia is de novo and novel and might disrupt protein folding and stability and the surface charge required for proper interactions with other proteins, especially LRP5/6 co-receptors. p.Glu347X has been reported as a variant of a compound heterozygous mutation in WNT10A in a Thai woman who was a daughter of patient 4 (family 4: II-2). She was affected with OODD (MIM 257980).14 This is the first time the heterozygous state of this variant is reported in patients with isolated dental anomalies. This p.Glu347X variant is a nonsense mutation located in the last exon of WNT10A, so it is not expected to go through nonmediated mRNA decay and result in haploinsufficiency. Instead, the translated protein would miss the CTD index finger structure, which is a disulfide-bond-stabilised cytokine-like hairpin loop that is critical for binding to the FZD co-receptor and also for binding to WNTLESS for secretion (Figure 5A, B).9,10,20 Pulp stones and severe pulp obliteration in patient 4 suggesting that the p.Glu347X variant predisposed the patient to dysregulation of calcification in the dental pulp.
When the WNT protein binds to the FZD receptor, the 2-domain WNT structure resembles a right hand where the NTD β-hairpin is like the thumb and CTD β-hairpin is like the index finger of a hand grasping the FZD receptor at 2 distinct binding sites (Figure 5A, B).5,21 The lipid group at the tip of the NTD (thumb) is inserted into the deep groove in the WNT-binding domain of the FZD receptor, whereas the CTD (index finger) interacts with the residues on the opposite side of the receptor.5 The truncation of most of the CTD is also likely to disrupt the binding of WNT10A to WNTLESS, the membrane protein that binds lipid-acylated WNT proteins to carry them from the ER to the cell surface, since WNTLESS makes extensive interactions with the WNT index finger hairpin.10 As such, little or none of the truncated WNT10A is likely to make it to the outside of the cell, and that which does will bind poorly to FZD, to result in aberrant WNT signaling, disrupted SHH signaling and subsequent dental anomalies. In addition, the mutant protein may result in a misfolded protein stress response in the ER as well.9,20
The heterozygous mutations p.Arg293Cys, p.Ser292Gly, p.Arg348Cys, and p.Glu347X are considered novel because they have not previously been reported to be pathogenic. Disruption of the binding between WNT10A and LRP5/6 co-receptors in our patients might lead to disruptive protein folding and/or stabilisation, decreased WNT signaling, concomitant aberrant expression of relevant tooth development genes including PAX9, RUNX2, MSX1, and AXIN2,19 and subsequent tooth agenesis and microdontia.
Taurodontism and single-rooted maxillary permanent molars in our patients emphasise the roles of WNT10A in root development. WNT10A expression has been demonstrated in the cervical loop of the enamel organ epithelial cells.22 The apical extension of the outer and inner dental epithelium along the cervical loop subsequently forms the Hertwig epithelial root sheath, a signaling centre for tooth root formation and guides for root development.23 Root development requires WNT and SHH signaling.24,25 It is hypothesised that WNT10A mutations in our patients resulted in aberrant WNT and SHH signaling, maldevelopment of tooth root, and subsequent taurodontism. Of note, the permanent molars do not have deciduous predecessors; they develop from the extensions of the dental epithelium behind the second deciduous molar.26 The presence of the second and third molars and agenesis of the first permanent molar in patient 1 raises the question of how the 2 posterior molars were formed.
Conclusions
Dysregulation of the highly conserved WNT–FZD interactions has been reported to be associated with a number of human hereditary diseases.5 The mutations in the NTD and the interface between the NTD and CTD of WNT10A protein in our patients may disrupt the binding of the protein with FZD and various other proteins involved in WNT10A processing and transport, impair WNT and SHH signaling, and subsequently result in tooth agenesis, microdontia, and taurodontism.
Conflict of interest
None disclosed.
Acknowledgments
Acknowledgements
We thank our patients and their families for their kind cooperation and for allowing us to use their medical and dental information for the benefit of other patients.
Funding
This work was supported by Genomics Thailand Research Grant of Health System Research Institute (HSRI) of Thailand.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.identj.2022.04.006.
Appendix. Supplementary materials
Supplemental Fig. 1 Sequencing chromatograms. A, Patient 1. Heterozygous single base substitution c.877 C>T; p.Arg293Cys in WNT10A. B, Patient 2. Heterozygous single base substitution c.874A>G; p.Ser292Gly in WNT10A. C, Patient 3. Heterozygous single base substitution c.1042C>T; p.Arg348Cys in WNT10A. D, Patients 4 and 5. Heterozygous single base substitution c.1039G>T; p.Glu347X in WNT10A.
Supplemental Fig. 2 Amino acid sequence alignment of Wnt10A proteins. A, The amino acid residues Ser292 and Arg293 in Wnt10A proteins are conserved across species. B, The amino acid residue Arg 348 is conserved across the vertebrate species shown, except for elephant Loxodonta africana (White square).
REFERENCES
- 1.Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 2.Doolan BJ, Onoufriadis A, Kantaputra P, McGrath JA. WNT10A, dermatology and dentistry. Br J Dermatol. 2021;185(6):1105–1111. doi: 10.1111/bjd.20601. [DOI] [PubMed] [Google Scholar]
- 3.van den Boogaard MJ, Créton M, Bronkhorst Y, et al. Mutations in WNT10A are present in more than half of isolated hypodontia cases. J Med Genet. 2012;49(5):327–331. doi: 10.1136/jmedgenet-2012-100750. [DOI] [PubMed] [Google Scholar]
- 4.Williams MA, Letra A. The changing landscape in the genetic etiology of human tooth agenesis. Genes (Basel) 2018;9(5):255. doi: 10.3390/genes9050255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science. 2012;337(6090):59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu ML, Ahn VE, Choi HJ, Daniels DL, Nusse R, Weis WI. Structural studies of Wnts and identification of an LRP6 binding site. Structure. 2013;21(7):1235–1242. doi: 10.1016/j.str.2013.05.006.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kakugawa S, Langton PF, Zebisch M, et al. Notum deacylates Wnt proteins to suppress signalling activity. Nature. 2015;519(7542):187–192. doi: 10.1038/nature14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takada R, Satomi Y, Kurata T, et al. Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev Cell. 2006;11(6):791–801. doi: 10.1016/j.devcel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Nile AH, Hannoush RN. Fatty acylation of Wnt proteins. Nat Chem Biol. 2016;12(2):60–69. doi: 10.1038/nchembio.2005. [DOI] [PubMed] [Google Scholar]
- 10.Zhong Q, Zhao Y, Ye F, et al. Cryo-EM structure of human Wntless in complex with Wnt3a. Nat Commun. 2021;12(1):4541. doi: 10.1038/s41467-021-24731-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bazan JF, de Sauvage FJ. Structural ties between cholesterol transport and morphogen signaling. Cell. 2009;138(6):1055–1056. doi: 10.1016/j.cell.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Mulligan KA, Fuerer C, Ching W, Fish M, Willert K, Nusse R. Secreted Wingless-interacting molecule (Swim) promotes long-range signaling by maintaining Wingless solubility. Proc Natl Acad Sci U S A. 2012;109(2):300–307. doi: 10.1073/pnas.1119197109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mihara E, Hirai H, Yamamoto H, et al. Active and water-soluble form of lipidated Wnt protein is maintained by a serum glycoprotein afamin/α-albumin. eLife. 2016;5:e11621. doi: 10.7554/eLife.11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kantaputra P, Kaewgahya M, Jotikasthira D, Kantaputra W. Tricho-odonto-onycho-dermal dysplasia and WNT10A mutations. Am J Med Genet Part A. 2014;164A(4):1041–1048. doi: 10.1002/ajmg.a.36388. [DOI] [PubMed] [Google Scholar]
- 15.Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirai H, Matoba K, Mihara E, Arimori T, Takagi J. Crystal structure of a mammalian Wnt–frizzled complex. Nat Struct Mol Biol. 2019;26(5):372–379. doi: 10.1038/s41594-019-0216-z. [DOI] [PubMed] [Google Scholar]
- 17.Koguchi-Yoshioka H, Wataya-Kaneda M, Nakano H, et al . Severe scoliosis associated with the WNT10A mutation. J Dermatol. 2015;42(3):322–323. doi: 10.1111/1346-8138.12762. [DOI] [PubMed] [Google Scholar]
- 18.Kantaputra P, Olsen B, McGrath JA. WNT10A variant and severe scoliosis? J Dermatol. 2022 doi: 10.1111/1346-8138.16313. [DOI] [PubMed] [Google Scholar]
- 19.Zeng Y, Baugh E, Akyalcin S, Letra A. Functional effects of WNT10A rare variants associated with tooth agenesis. J Dent Res. 2021;100(3):302–309. doi: 10.1177/0022034520962728.20. [DOI] [PubMed] [Google Scholar]
- 20.Nygaard R, Yu J, Kim J, et al. Structural basis of WLS/Evi-mediated Wnt transport and secretion. Cell. 2021;184(1):194–206. doi: 10.1016/j.cell.2020.11.038. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerekes K, Bányai L, Patthy L. Wnts grasp the WIF domain of Wnt Inhibitory Factor 1 at two distinct binding sites. FEBS Lett. 2015;589(20 pt B):3044–3051. doi: 10.1016/j.febslet.2015.08.031. [DOI] [PubMed] [Google Scholar]
- 22.Liu F, Chu EY, Watt B, et al. Wnt/beta-catenin signaling directs multiple stages of tooth morphogenesis. Dev Biol. 2018;313(1):210–224. doi: 10.1016/j.ydbio.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Parada C, Chai Y. Cellular and molecular mechanisms of tooth root development. Development. 2017;144(3):374–384. doi: 10.1242/dev.137216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Fen JQ. Signaling pathways critical for tooth root formation. J Dent Res. 2017;96(11):1221–1228. doi: 10.1177/0022034517717478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosoya A, Shalehin N, Takebe H, Shimo T, Irie K. Sonic hedgehog signaling and tooth development. Int J Mol Sci. 2020;21(5):1587. doi: 10.3390/ijms21051587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hovorakova M, Lesot H, Peterka M, Peterkova R. Early development of the human dentition revisited. J Anat. 2018;233(2):135–145. doi: 10.1111/joa.12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1 Sequencing chromatograms. A, Patient 1. Heterozygous single base substitution c.877 C>T; p.Arg293Cys in WNT10A. B, Patient 2. Heterozygous single base substitution c.874A>G; p.Ser292Gly in WNT10A. C, Patient 3. Heterozygous single base substitution c.1042C>T; p.Arg348Cys in WNT10A. D, Patients 4 and 5. Heterozygous single base substitution c.1039G>T; p.Glu347X in WNT10A.
Supplemental Fig. 2 Amino acid sequence alignment of Wnt10A proteins. A, The amino acid residues Ser292 and Arg293 in Wnt10A proteins are conserved across species. B, The amino acid residue Arg 348 is conserved across the vertebrate species shown, except for elephant Loxodonta africana (White square).