Abstract
Our laboratory is studying an extraintestinal pathogenic isolate of Escherichia coli (CP9) as a model pathogen. We have been using human urine, ascites, and blood ex vivo to identify genes with increased expression in these media relative to expression in Luria-Bertani (LB) broth. Such genes may represent new or unrecognized virulence traits. In this study, we report the identification of a new gene, ireA (iron-responsive element). This gene has an open reading frame of 2,049 nucleotides, and its peptide has a molecular mass of 75.3 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Its expression is increased a mean of 3.6-fold in human urine, 16.2-fold in human ascites, and 6.6-fold in human blood relative to expression in LB medium, and it is Fe repressible. IreA also exhibits peptide similarities (48 to 56%) to previously identified proteins that function as siderophore receptors, suggesting that IreA is involved in iron acquisition. PCR-based analysis of ireA's phylogenetic distribution detected ireA in none (0%) of 14 fecal isolates that represented probable commensal strains, but in 13 (26%) of 50 random urine and blood clinical isolates (P = 0.05) and in 5 (100%) of 5 representatives of the J96-like, clonal group of which CP9 is a member (P < 0.001). In a mouse urinary tract infection model, the presence of ireA contributed significantly to CP9's ability to colonize the bladder (P < 0.02), evidence that IreA is a urovirulence factor. Taken together, these findings demonstrate that ireA encodes a new virulence factor, which is likely involved in Fe acquisition.
Extraintestinal pathogenic isolates of Escherichia coli (ExPEC) cause infections of nearly every organ and anatomic site and occur frequently in all age groups (23). Common infections include urinary tract infection (UTI), diverse intra-abdominal infection, pneumonia (particularly in hospitalized and institutionalized patients), meningitis (mainly in neonates and following neurosurgery), intravascular device infection, osteomyelitis, and soft tissue infection, which usually occurs in the setting of tissue compromise. Bacteremia can accompany infection at any of these sites (7). The scope and magnitude of infection caused by ExPEC is as great as any invasive bacterial pathogen. Although these bacteria do not make the headlines, billions of health care dollars, millions of workdays, and thousands of lives are lost to this group of pathogens each year.
Identification of new or unrecognized bacterial factors important in the pathogenesis of these infections may enable the development of an effective vaccine or new treatment modalities. One approach to accomplish this goal is to identify genes with increased expression in vivo. Such genes are likely to play a role in pathogenesis (16). Further, those bacterial traits which are surface exposed, regardless of their role in pathogenesis, are potential vaccine candidates. A variety of methods have been successfully developed for this purpose (14, 38, 41). One approach has been to identify genes with increased expression ex vivo after exposure to body fluids (e.g., urine) or eucaryotic tissue culture cells (17, 19, 22, 42). Subsequent evaluation of genes identified by in this manner in animal models has validated its utility for identifying virulence traits (22, 42). Our laboratory and others have successfully used this method to identify several differentially expressed genes from ExPEC strains. To date, genes for arginine biosynthesis (argC) or transport (artJ), genes for iron acquisition (iroN) or regulation (airR), and a gene of unknown function (ure1) have been reported (19, 22, 42).
Our laboratory has continued to use human body fluids ex vivo as a means to identify new or unrecognized virulence traits from a well-described ExPEC strain (CP9). A previously constructed TnphoA mutant library generated from CP9 (21) was screened for mutant derivatives with increased phoA activity in the presence of human urine, ascites, and blood relative to Luria-Bertani (LB) medium. In this report, we describe the identification and initial characterization of the gene ireA, which has increased expression in these body fluids.
MATERIALS AND METHODS
Bacterial strains and media.
The model pathogen CP9 is an E. coli blood isolate cultured from a patient with sepsis hospitalized at the National Institutes of Health and is being used as a model pathogen. It has been previously described in detail (13, 19, 21). CP9 possesses all of the characteristics of ExPEC strains (23) and is highly virulent in a urinary tract infection (UTI) model (18), an intraperitoneal (IP) infection model (25), and a pneumonitis model (20). All strains were maintained at −80°C in 50% Luria-Bertani (LB) medium and 50% glycerol. LB broth consisted of 5 g of yeast extract, 10 g of tryptone, and 10 g of NaCl per liter. Incubations were performed at 37°C unless otherwise described. For plates, 15 g of Bacto-Agar (Difco Laboratories, Detroit, Mich.) was added per liter and kanamycin (kan) (40 μg/ml) or ampicillin (200 μg/ml) (Amresco, Solon, Ohio) was added where appropriate. For gene expression studies, urine was used that was (i) fresh and unfiltered, (ii) fresh and filtered with a 0.22-μm-pore-size filter, or (iii) filtered and stored at 4°C. Urine was obtained and used from individuals who have had or never had a UTI. Filter-sterilized ascites (peritoneal fluid) was obtained from an asymptomatic patient hospitalized at Erie County Medical Center, divided into multiple aliquots, and frozen at −80°C. Blood was used fresh and was obtained from a single, healthy donor. It was collected in sterile, 8.3-ml Vacutainer tubes (Becton Dickinson, Rutherford, N.J.) which contained 1.7 ml of sodium polyanetholesulfonate (0.35%) and NaCl (0.85%) (nonbactericidal) as the anticoagulant.
Transposon mutagenesis and mutant library construction.
A mutant library consisting of 527 CP9 derivatives that contained random active TnphoA fusions had been previously constructed and was used in this study (21).
Identification of genes with increased expression in urine, ascites, and blood.
The TnphoA mutant library was screened to identify genes that coded for extracytoplasmically located gene products, which had increased expression in human urine, ascites, and blood ex vivo. A colorimetric assay was used with growth in human urine, whereas a fluorescent assay was used with growth in ascites and blood (19). Mutants that appeared to have increased expression in blood or ascites relative to that in LB broth via these qualitative screens were confirmed with quantitative assays.
Quantitative alkaline phosphatase assays in human urine, ascites, and blood.
CP197 contained an active ireA::phoA fusion. Therefore, quantitative expression of ireA was determined by measuring alkaline phosphatase activity in the CP197 background. Assays in human urine and LB medium were performed as previously described (19, 22). Preliminary experiments established that ireA expression was the same for log phase and stationary phase grown cells. In brief, CP9 and CP197 (ireA) were grown overnight in LB broth and human urine. Cells were then washed and permeabilized, and p-nitrophenyl phosphate was added for the detection of alkaline phosphatase activity. To control for both endogenous bacterial alkaline phosphatase and any activity from the growth media that may have persisted despite washing, alkaline phosphatase activity from CP9 was subtracted from the measured activity of CP197. The ratio of the specific activity of CP197 grown in urine versus LB medium defined the fold induction.
A fluorescent assay was used to measure ireA::phoA fusion expression in human ascites and blood with 4-methylumbelliferonephosphate as the substrate. This assay was previously described in detail (19). The net sample rate in blood or ascites relative to that in LB broth established the fold induction. The net sample rate/milliliter was (SR) calculated as follows: {[(fluorescence cycle B − fluorescence cycle A over the linear portion of the curve)/(elapsed time)] × 20} − (CP9 SR). Specific activities were determined by dividing net sample rates by the CFU per milliliter. The sensitivities of the colorimetric and fluorescent assays were similar. When grown in pooled human urine, the specific activity of CP197 was determined to be 8.9 and 7.3 by the colorimetric and fluorescent assays, respectively.
Genetic and DNA manipulations and analyses.
Transduction of a transposon insertion back into the wild-type strain CP9 was accomplished using the bacteriophage T4 (21). Whole-cell DNA preparation, restriction enzyme (New England Biolabs, Beverly, Mass.)-mediated DNA digestion, and Southern hybridization using PCR-generated radioactive probes was performed as previously described (21, 25). Primers 63 (5′ GATCAAGAGACAGGATGA 3′) and 64 (5′ TGATCCTCGCCGTACTGC 3′) were used to amplify a 4.0-kb internal fragment of TnphoA (contained in pRT291 [21]), which were used to probe for the TnphoA insertion. Southern analysis of BglII-digested whole-cell DNA containing TnphoA produced a 2.8-kb internal fragment and two variable junction fragments per copy. Lipopolysaccharide, capsular polysaccharide, and outer membrane protein profiles were determined as previously described (25).
Construction of ireA subclone.
A subclone of the gene loci 5′ to the TnphoA insertion in CP9.197(ireA) was obtained by restricting whole-cell DNA with BamHI, which recognizes a site located 3′ to the kanamycin resistance gene in TnphoA. Ligation of this restriction into pBSII SK(−), electroporation into XL1 Blue cells (Stratagene, La Jolla, Calif.) and selection of ampicillin (100 μg/ml)- and kanamycin (40 μg/ml)-resistant transformants resulted in the identification of the subclone p197.1. A subclone of the IreA gene locus 3′ to the TnphoA insertion in CP9.197 was obtained by restricting whole-cell DNA with ClaI which recognizes a site 5′ to the kanamycin resistance gene in TnphoA. Ligation of this restriction into pBSII SK(−), electroporation into XL1 Blue cells (Stratagene), and selection of ampicillin- and kanamycin-resistant transformants resulted in the identification of the subclone p197.2.
DNA sequencing and analysis.
DNA sequence was determined by the dideoxy chain termination method of Sanger (28) using the subclones p197.1 and p197.2 as the DNA templates. DNA sequencing of p197.1 initially utilized a TnphoA fusion joint primer (5′ AATATCGCCCTGAGC 3′), which established the location for the TnphoA insertion. Sequencing of the gene subclone p197.2 initially utilized the TnphoA primer (5′ CATGTTAGGAGGTCACAT 3′). Subsequent DNA sequence was determined using primers derived from the deduced sequences of the gene subclones. p197.2 did not contain the last 66 bases of ireA. Sequence for these bases was determined by using whole chromosome as the template. A consensus sequence for ireA was generated by assembling and editing the DNA sequence obtained from 25 overlapping but independent sequencing reactions using AssemblyLIGN 1.0.2 (Oxford Molecular Group, Beaverton, Oreg.). Both strands of the gene sequences submitted in this report were sequenced. Sequence analysis, comparisons, and CLUSTAL alignments were performed, in part, using MacVector (version 6.0; Oxford Molecular Group). Comparisons were also performed via BLAST analysis of the nonredundant GenBank, EMBL, DDBJ, and PDB sequences. TargetP V1.0 was used for identification of signal sequences (4). Motif analysis utilized Profile Scan from the ISREC Bioinformatics Group (www.expasy.ch).
Cloning and expression of ireA.
Primers (forward, 5′ CGCGCGGGATCCTCTGATAAAAAAGAAGAT 3′; reverse, 5′ ATATATAAGCTTGAAGGATACTCTTACATT 3′) based on the ireA sequence were designed for PCR-mediated amplification of the entire ireA gene, excepting its signal sequence (1,971 bp). A single band of the expected size was PCR amplified from CP9 chromosomal DNA. The DNA was gel purified and ligated into the kanamycin resistance pET28a T7/His tag expression vector. The pET28a::ireA construct was electroporated into XL1 Blue cells and selected for on LB plates containing kanamycin. The ireA gene in the selected clone was confirmed to be correct by DNA sequencing. This excluded the possibility that an error was introduced into the cloned ireA during PCR amplification. The clone was subsequently electroporated into the expression cell line AD494(DE3)pLysS for expression of IreA. AD494(DE3)pLysS pET28a::ireA was grown overnight in LB media plus kanamycin. The next morning, 1 ml of the overnight culture was transferred into 11 ml of LB media plus kanamycin and grown at 37°C for 2.5 h, with shaking. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM to induce the expression of IreA. One-milliliter aliquots of the induced culture as well as an uninduced control culture were taken in 30-min intervals. The samples were prepared for gel electrophoresis and run on an SDS–7.5% PAGE gel.
Ex vivo growth in human urine, ascites, and blood.
The urine, ascites, and blood used for these studies was collected as described above. CP9 and CP197(ireA) were grown overnight in 2 ml of the body fluid to be evaluated. The next day, the bacterial cells were diluted into fresh body fluid to achieve a starting concentration of approximately 1.0 × 102 to 1.0 × 103 CFU/ml, since this titer is at the lower end of the spectrum for what is considered significant for UTI in symptomatic young women (30). During incubation at 37°C, aliquots were removed at intervals and bacterial titers were established by plating 10-fold serial dilutions in 1× phosphate-buffered saline in duplicate on appropriate media. For competition experiments, approximately equal titers of both CP9 and CP197 were added to the appropriate body fluid. The titer of CP197 was determined from the enumeration of LB medium plus kanamycin (40 μg/ml), and the titer of CP9 was established by subtracting the titer of CP197 from the total bacterial titer enumerated from LB medium.
Gene regulation studies.
M9 minimal medium was utilized for some gene regulation studies. Fe was either chelated from M9 medium by mixing 200 ml of medium with 21.2 g of washed (two times with 1 liter of dH2O) iminodiacetic acid (Chelex 100; Sigma, St. Louis, Mo.) for 90 min followed by filter sterilization or adding exogenous Fe (0.1 mM FeNO3) to generate a defined medium depleted or replete with Fe. Gene regulation studies also utilized urine to which exogenous Fe (0.1 mM FeNO3) or glucose (0.5%) was added. For additional studies, an individual, filter-sterilized urine sample had its pH adjusted to 5.0, 6.0, and 7.0 with either HCl or NaOH. For these studies, quantitative alkaline phosphatase assays were performed as described above and the ratio of the specific activities of CP197 grown in the two growth media being compared defined the fold induction.
PCR-mediated detection of ireA from various strains of E. coli.
Chromosomal DNA was prepared from the strains to be tested as described, and 1 μl was used as DNA template. Three independent primer pairs were used for PCR-mediated amplification of ireA. Primer set 1 (forward, 5′ TGGTCTTCAGCTATATGG 3′; reverse, 5′ ATCTATGATTGTGTTGGT 3′) amplified a 415-bp fragment, primer set 2 (forward, 5′ ATTTCCCCGCATCCAGG 3′; reverse, 5′ CCCTGTATGGTTCTGATGC 3′) amplified a 315-bp fragment, and primer set 3 (forward, 5′ TCTGATAAAAAAGAAGATACG 3′; reverse, 5′ GAAGGATACTCTTACATT 3′) amplified a 1,971-bp fragment. There was excellent agreement between primer sets with >95% concordance. Concentrations per PCR were as follows: primers, 50 pmol; deoxynucleoside triphosphates, 10 mM; MgCl2, 2 mM; and Amplitaq Gold (Perkin-Elmer), 1.25 U. The PCR conditions were one cycle of 10 min at 95°C, followed by 25 cycles of 15 s at 95°C, 1 min at 55°C, and 30 s at 72°C, and a terminal extension of 7 min at 72°C. CP9 and XL1 Blue were the positive and negative control strains, respectively. Experimental strains consisted of seven groups: group 1, 14 unique fecal isolates that had been previously established not to contain pap, hly, or cnf-1 (presumably commensal strains) (10); group 2, five unique fecal isolates that possessed some combination of pap, hly, or cnf-1 (presumably ExPEC strains) (10); group 3, 19 unique first-time UTI isolates (26); group 4, five unique recurrent UTI isolates (26); group 5, 21 blood isolates (19); group 6, five J96-like strains (12); group 7, the 50 clinical isolates from groups 2 to 5. Since group 1 was the most representative of nonpathogenic strains, it was used in statistical comparisons against the random clinical isolates from groups 2 to 7.
Mouse UTI model.
Mouse UTI experiments were done using an ascending, atraumatic mouse model of UTI as previously described (18). However, to minimize the impact of mouse-to-mouse variation and to maximize the sensitivity for identifying differences between strains, dual-infection (competition) experiments were done. Proportional differences between CP9 and CP197 were analyzed on a per-mouse basis, thereby enabling each mouse to serve as its own control. Briefly, 6- to 10-week-old female BALB/c mice were anesthetized and inoculated transurethrally, via the use of a Harvard infusion pump, with 1.0 μl/g of body weight of a suspension of the two bacterial strains in approximately equal concentrations (as confirmed by quantitative culture). A challenge inoculum of approximately 4.0 × 109 CFU of each strain was delivered to each mouse. Inoculation conditions were utilized that have been shown to avoid inoculation-induced vesicoureteral reflux (9), as reconfirmed for the present study in pilot experiments. Mice were euthanatized 2 days after inoculation and underwent sterile harvesting of urine, bladder, and kidneys. Urine and organ homogenates were cultured quantitatively via serial 10-fold dilutions as well as inoculated undiluted into broth to enable detection of low-level titers. Colonies from LB medium without antibiotics were replica plated onto selective medium (LB kanamycin, 40 μg/ml) to determine the relative proportions of CP9 and CP197. Quantitative culture results were adjusted for the relative proportion of the two test strains in the inoculum suspension. If in the postmortem cultures both test strains yielded isolated colonies on the direct plates or were detected only in broth cultures but could be assessed for relative prevalence by replica plating of colonies from the broth cultures, actual colony counts were used to derive the estimated ratio of CP9 to CP197. If instead only one of the test strains was detected in the direct platings and yielded at least five colonies but both test strains were detected in broth culture, the first strain was presumed to be 1,000-fold more prevalent than (i.e., to exhibit a log ratio of 3.0 in comparison with) the other strain. Similarly, if one strain was detected in broth (with or without colonies on the direct plates) and the other strain was not detected at all, i.e., even in broth cultures, the first strain was presumed to be 10,000-fold more prevalent than (i.e., to exhibit a log ratio of 4.0 in comparison with) the other strain. Selected colonies obtained after harvest underwent genetic, PCR, and random amplified polymorphic DNA analysis (11, 39) to exclude contamination. In separate pilot experiments, it was established that the genotype and phenotype of CP197 was stable in the presence of in vivo selection pressure.
Mouse systemic infection model.
Mouse systemic infections were performed as previously described (24). In brief, outbred Swiss mice (20 ± 2 g) (mean ± standard error [SE]) (Sprague-Dawley, Indianapolis, Ind.) were challenged with CP9 (wt) in parallel with CP197 (ireA) by IP challenge. The strains evaluated were grown overnight in LB medium for experiments 1 and 2. For experiments 3 and 4, the strains were grown overnight in M9 minimal medium (27) containing 0.5% lactose instead of glucose and treated with Chelex to remove any trace Fe present (M9-lactose-Chelex treated). This medium simulates the Fe, nutrient-limiting environment within the host. Subsequently, cells were diluted in 1× PBS such that an IP injection of 0.5 ml resulted in the delivery of 105 to 107 organisms. Bacterial titers were performed on dilutions to precisely determine the number of organisms inoculated. The measured endpoint was death within the initial 24 h postinoculation. No deaths occurred after 24 h.
Statistical analysis.
Fisher's exact test was used for the comparisons of proportions. McNemar's test was used with the mouse UTI model to assess the proportion of mice in which one bacterial strain outcompeted the other (5). For paired comparisons of outcomes in the mouse UTI model competition experiments, the log ratio of CP9 to CP197 from each urine and bladder culture was used to derive a relative CFU number for each strain, based on an arbitrary 10,000 CFU scale in which a log ratio of 0.0 would convert to 5,000 CFU for each strain and a log ratio of 4.0 would convert to 9,999 CFU of one strain and 1 CFU of the other (and vice versa, for a log ratio of −4.0). The Wilcoxon test, a nonparametric paired t test equivalent, was used to compare these semiderived culture results for the two strains based on rank order. Logistic regression was used to derive the 50% lethal doses (LD50s) for the IP model for each experiment separately and for the pooled data from the four experiments combined. A P value of ≤0.05 was the criterion for statistical significance throughout.
Nucleotide sequence accession number.
The accession number of the complete nucleotide sequence for ireA is AF320691.
RESULTS
Identification of CP9.197 (ireA).
In order to identify genes from an extraintestinal pathogenic isolate of E. coli with increased expression in human body fluids ex vivo, we screened a library of 527 CP9 derivatives that contained active phoA (TnphoA) fusions. Initially, a qualitative microtiter fluorescent assay was used to measure PhoA activity for each of these strains in ascites, blood, and LB medium. Mutants with an apparent increase in activity when exposed to ascites and/or blood relative to LB medium underwent quantitative evaluation for confirmation. This screen resulted in the identification of CP9.197.
Studies were performed to confirm that the gene into which TnphoA inserted in CP9.197, and not a cryptic mutation(s) acquired during the mutagenesis procedure, was responsible for the increased PhoA activity of CP9.197 in blood and ascites. Southern analysis demonstrated that CP9.197 had a single TnphoA insertion. T4 transduction of this insertion back into the wild-type strain (CP9) resulted in a derivative (CP197) that possessed the same degree of PhoA activity in human body fluids ex vivo relative to LB medium as the original mutant (CP9.197). Southern analysis demonstrated that the TnphoA insertion in CP197 was physically in the same location as in the original mutant, CP9.197. In addition, no differences were observed in capsule, lipopolysaccharide, and outer membrane profiles between CP9, CP9.197, and CP197 (data not shown). However, because CP197 contained a TnphoA insertion, a polar effect needed to be ruled out. This possibility was excluded when subsequent sequence analysis established that 3′ to ireA was a truncated transposase (see below). These results support CP9.197 and its T4-generated transductant CP197 as being isogenic derivatives of their wild-type parent CP9. CP197 was chosen for use in future studies.
Genomic analysis of ireA.
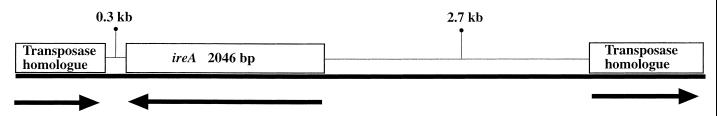
Analysis of DNA sequences from the subclones p197.1 and p197.2 and from CP9 chromosomal DNA determined that the TnphoA insertion in CP197 was located within a 2,049-bp open reading frame which was designated ireA (iron responsive element). Comparison of this sequence with entries in GenBank did not reveal any significant DNA matches, establishing ireA as a new gene. Regions of DNA that are not present in E. coli K-12 and which appear to have been acquired by horizontal transmission have been termed pathogenicity-associated islands (PAI). ireA appeared to be part of such a locus based on several observations. First, the guanosine-plus-cytosine content was 43% for ireA compared to the 51% observed for E. coli K-12. Secondly, the translated DNA sequence beginning 317 bases from the 3′ end of ireA possesses 34 to 44% identity and 50 to 61% similarity to defined or probable transposases from a wide variety of bacteria (size range, 390 to 444 amino acids [aa]). From highest to lowest homology, these transposases were identified from Yersinia pestis pCD1 (8), an intestinal pathogenic E. coli (IS1414) (15), Burkholderia cepacia (37), Rhizobium meliloti (ISRm3) (40), Mycobacterium ulcerans (32), and Bacillus halodurans (33). Interestingly, the DNA sequence encoding the C-terminal portion of these transposases was absent (range, 41 to 61 aa) in CP9. Additionally, located 2.7 kb 5′ to the start of ireA is a DNA sequence coding for a putative protein product that possesses 44% identity and 48% similarity to the first 337 aa to a transposase that was recently described for the E. coli Nissle 1917 fimB gene (31). However, CP9 does not possess sequence homologous to the last 49 aa of this transposase, similar to what was observed with the transposases 3′ to ireA. At present it is unclear whether the DNA sequences flanking ireA encode functional full-length transposases with C termini that are divergent from their nearest homologues or whether the gene products are truncated and nonfunctional. Nonetheless, these data further suggest that ireA was obtained by horizontal transfer. The 2.7 kb of DNA sequence 5′ to ireA does not possess any DNA or protein homology to known sequences. The genomic organization of this region is depicted in Fig. 1.
FIG. 1.
Schematic diagram of the ireA gene and the flanking sequence described in this report. (Left to right) (i) The DNA sequence beginning 317 bases from the 3′ end of ireA possesses 34 to 44% identity and 50 to 61% similarity to defined or probable transposases from a wide variety of bacteria, (ii) the ireA gene, a putative Fur DNA binding site, was present 5′ to ireA (bases −27 to minus −45), (iii) 2.7 kb of DNA that does not possess any DNA or protein homology to known sequences, (iv) 2.7 kb 5′ to the start of ireA is a DNA sequence whose putative protein product possesses 44% identity and 48% similarity to the first 337 aa of a transposase that was recently described for the E. coli Nissle 1917 fimB gene. The arrows below the solid line indicate the direction of transcription for ireA and the transposase homologues.
The predicted IreA protein consists of 682 aa and has a putative molecular mass of 75,351 kDa and an estimated pI of 6.0. A putative signal sequence was identified (aa 1 to 25) (4). Comparisons of the deduced IreA sequence identified a variety of protein homologues, the majority of which are siderophore receptors. The identities and similarities of these comparisons as derived via CLUSTAL alignments are summarized in Table 1. Further, motif analysis of the deduced IreA sequence revealed a TonB-dependent receptor protein signature 1 (aa 31 to 38) and a TonB-dependent receptor C-terminal region (aa 575 to 682). In addition, a putative Fur DNA binding site was present 5′ to ireA (bases −27 to minus −45), with 15 out of 19 bases conserved when compared to the Fur consensus sequence (3). A possible Shine-Delgarno sequence (three of six bases) was identified at bases −8 to −10. However, no promoter −10 and −35 binding sites were identified. Taken together, this proteogenomic analysis supports the contention that IreA is a new TonB-dependent siderophore receptor.
TABLE 1.
Deduced protein sequence identity and similarity of IreA to various homologues
| Organism | Characteristics of IreA homologues
|
||||
|---|---|---|---|---|---|
| Name | % Identity | % Similarity | Function | Regulation | |
| Vibrio cholerae | IrgA | 37 | 56 | ? Siderophore receptor | Fe |
| Escherichia coli | CirA | 38 | 55 | ? Fe transport, colicin receptor (I) | Fe, cAMP/CRP |
| Escherichia coli | R4/Iha | 36 | 56 | ? Siderophore receptor | ? |
| Bordetella bronchiseptica | BfrA | 36 | 54 | Siderophore receptor | ? |
| Campylobacter jejuni | CfrA | 33 | 54 | ? Fe uptake | ? |
| Escherichia coli | FepA | 31 | 53 | Catecol siderophore receptor colicin receptor (B,D) | Fe |
| Escherichia coli | IroN | 30 | 49 | ? Siderophore receptor | Fe |
| Salmonella enterica | IroN | 29 | 48 | ? Siderophore receptor, benzoic acid receptor | ? |
Cloning and expression of ireA.
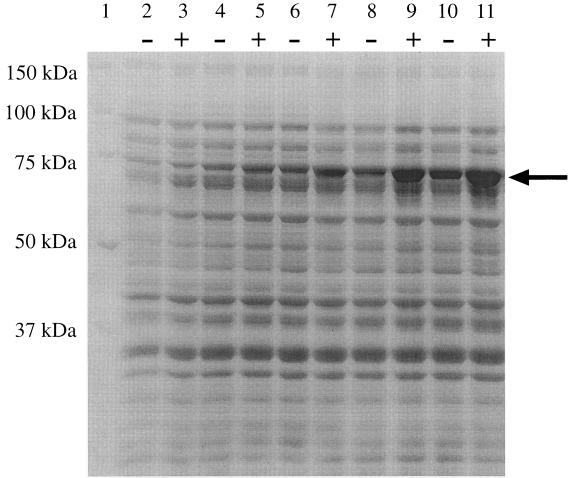
Both for future studies and to confirm that ireA encoded a full-length product, ireA was cloned, minus its signal sequence, into the expression vector pET28a::ireA. IreA was successfully expressed with its molecular mass of 72.7 kDa being the same as its predicted molecular mass without a signal sequence (Fig. 2).
FIG. 2.
Expression of IreA. pET28a::ireA was constructed as described in Materials and Methods. AD494 (DE3) pLysS pET28a::ireA was grown overnight in LB medium plus kanamycin (40 μg/ml) and the next morning was diluted 1 to 12 in fresh medium and grown at 37°C for 2.5 h. Cultures were either induced with IPTG (final concentration, 1 mM) (+) or uninduced (−) for detection of IreA expression. One-milliliter aliquots of the culture as well as an uninduced control culture were taken in 30-min intervals. The samples were prepared for gel electrophoresis and run on an SDS–7.5% PAGE gel and stained with Coomassie blue. Lane 1, molecular mass markers; lanes 2 and 3, 30-min samples; lanes 4 and 5, 60-min samples; lanes 6 and 7, 90-min samples; lanes 8 and 9, 120-min samples; lanes 10 and 11, 150-min samples. The arrow demonstrates the increased expression of IreA over time. Although IPTG induction resulted in increased expression of IreA (+), a lesser degree of expression was seen without induction (−).
Expression of ireA in human ascites, blood, and urine.
ExPEC cells commonly infect the peritoneal cavity, urinary tract, and bloodstream. ireA was initially identified via a screen for genes with increased expression in human urine, ascites, and blood. Subsequent quantitative assessments were done in these body fluids ex vivo. ireA had increased expression in all of these fluids relative to that in LB medium (Table 2). Since the composition of human urine has the potential to be variable both within and between individuals, assays were performed using 22 independent urine samples collected from 10 different volunteers. Five of these individuals were women with a prior history of UTIs. The 22 independent urine samples used were filter sterilized and stored at 4°C prior to use. The results of these quantitative assays are summarized in Table 2. Although there was variability in the degree of increased ireA expression from urine sample to urine sample, increased expression was seen in all urine samples evaluated. The degree of expression of ireA was similar in urine samples from individuals with and without a prior history of UTI. To determine if the processing of urine affected gene expression, assays were performed in parallel using four independent urine samples that were either (i) fresh and unfiltered, (ii) fresh and filtered with a 0.22-μm-pore-size filter, or (iii) filtered and stored at 4°C. The expression of ireA was similar, regardless of how the urine was processed (data not shown).
TABLE 2.
Ex vivo expression of ireA in human blood, ascites, and urine relative to that in LB medium
| Body fluid | Fold increase in expression of each isolate | Mean fold increase in expression ± SEa (range) |
|---|---|---|
| Ascitesb(n = 5) | 24.6, 24.6, 17.4, 7.7, 6.7 | 16.2 ± 3.9 (6.7–24.6) |
| Bloodb(n = 8) | 4.3, 5.6, 4.0, 8.5, 3.7, 11.8, 10.3, 4.9 | 6.6 ± 1.1 (3.7–11.8) |
| Urinec(n = 22) | 1.5, 3.1, 6.4, 2.1, 4.9, 2.8, 4.5, 1.6, 5.4, 5.3, 3.1, 4.1, 1.2, 3.6, 5.6, 2.2, 3.3, 2.2, 5.8, 2.4, 5.9, 1.9 | 3.6 ± 0.35 (1.2–6.4) |
CP197 (ireA::phoA) was utilized.
The net sample rate/milliliter (SR) = {[(fluorescence cycle B − fluorescence cycle A over the linear portion of the curve)/(elapsed time)] × 20} − (CP9 SR). Specific activities were determined by dividing net sample rates by CFU/milliliter.
PhoA specific activity is expressed as micromoles of para-nitrophenyl phosphate hydrolyzed per minute per A600 unit.
Modulation of ireA expression by Fe.
As described above, DNA sequence analysis strongly suggested that ireA encoded a siderophore receptor. Therefore, in vitro studies were performed to determine if the ireA::phoA fusion was Fe regulated. CP197 was grown in M9 minimal media that was either Fe depleted via chelation or Fe replete via exogenous administration of FeNO3 (final concentration, 0.1 mM). The expression of ireA relative to LB medium was 6.11 ± 0.68 (mean ± SE) in M9 Fe-depleted medium and 0.89 ± 0.30 in M9 Fe-repleted medium. CP197 was also grown in two independent urine samples and a pooled urine sample (five donors) to which Fe was added exogenously. The addition of Fe significantly repressed the expression of ireA. The addition of Fe to a urine sample decreased expression of ireA 10- to 12-fold relative to expression in LB medium (from 2.2- to 0.28-fold in the two independent urine samples and from 8.7- to 0.73-fold in the pooled urine sample without and with exogenous Fe, respectively). Therefore, expression of ireA is Fe repressible. These findings were consistent with the genomic analyses, which suggested that ireA encoded a siderophore receptor.
Modulation of ireA expression by other environmental signals.
The effect of glucose and pH were evaluated as potential modulators of ireA expression. The addition of exogenous glucose did not repress the increased expression of ireA in human urine relative to that in LB medium (3.1- versus 3.0-fold in urine without and with exogenous glucose, respectively). Therefore, ireA is not regulated by catabolite repression. The effect of urinary pH on ireA expression was also evaluated. At a urinary pH of 5.0, 6.0, and 7.0, the mean (± SE) fold increase in expression of ireA relative to that in LB medium was 0.9 ± 0.26, 2.8 ± 0.5, and 4.4 ± 0.44, respectively. These results demonstrate the ireA expression is modulated by pH, increasing as the pH increases over a physiologic range observed in urine. This may reflect a diminished solubility of Fe with increasing pH.
Phylogenetic distribution of ireA.
The prevalence of ireA among various isolates of E. coli was evaluated by PCR-mediated gene fragment amplification (Table 3). In summary, ireA was detected in none (0%) of 14 fecal isolates that presumptively represented commensal strains (negative for pap, hly, or cnf-1). In contrast, ireA was detected in 13 (26%) of 50 random clinical isolates (P = 0.05) versus presumed fecal commensals. In a selected subset of strains that had been previously established as being part of a widely disseminated group of J96-like ExPEC strains that are characterized in part by possessing group 3 capsules, the O4-specific antigen, and classes 1 and 3 Pap adhesins, ireA was present in five (100%) of five strains (P < 0.001 versus presumed fecal commensals) (12, 13). These results establish the association of ireA with clinical but not commensal isolates of E. coli.
TABLE 3.
Phylogenetic distribution of ireA among various isolates of E. colia
| Group | % (n) of isolates with ireA | P valuec |
|---|---|---|
| 1 (presumed fecal commensals) | 0 (14) | |
| 2 (presumed fecal ExPEC) | 40 (5) | 0.059 |
| 3 (first-time UTI) | 16 (19) | 0.24 |
| 4 (recurrent UTI) | 40 (5) | 0.059 |
| 5 (blood) | 29 (21) | 0.06 |
| 6 (J96-like) | 100 (5) | <0.001 |
| 7 (groups 2–5) | 26 (50) | 0.05 |
Group 1, 14 unique fecal isolates that had been previously established not to contain pap, hly, or cnf-1 (presumably commensal strains) (10); group 2, five unique fecal isolates that possessed some combination of pap, hly, or cnf-1 (presumably ExPEC strains) (10); group 3, 19 unique first-time UTI isolates (26); group 4, five unique recurrent UTI isolates (26); group 5, 21 blood isolates (19); group 6, five J96-like strains (13); group 7, the 50 clinical or fecal ExPEC isolates from groups 2 to 5.
Fisher's exact test was used for proportions.
All comparisons are versus group 1. A P value of ≤0.05 was considered significant.
Growth of CP197 in human ascites, blood, and urine.
The growth of CP9 (wt) and CP197 (ireA) cells in urine, ascites, and blood, was evaluated via enumeration of bacterial titers. The growth of CP197 was equivalent to its wild-type parent CP9 in four independent urine, blood, and ascites samples when grown alone (data not shown). Further, the growth of CP197 and CP9 were also equivalent in competition experiments in urine, ascites, and blood where the starting inocula of each strain ranged from 103 to 105 CFU (data not shown). Therefore, despite the known limiting concentration of Fe in each of these body fluids, no difference in the growth of CP197 compared to its wild-type parent in these body fluids ex vivo could be demonstrated.
Virulence of CP9 (wt) and CP197 (ireA) in mouse UTI model.
To determine the relative urovirulence of CP9 and CP197, 20 mice were challenged simultaneously with approximately 109 CFU of both CP9 and CP197 as a mixed intravesicular challenge inoculum via urethral catheterization. Overall, CP9 equaled or outcompeted CP197 according to every parameter of bladder and urine colonization ability (Table 4). In urine cultures, the median log ratio of CP9 to CP197 was 0.75, reflecting a median 5.6-fold excess of CP9 over CP197 (P > 0.05 [Wilcoxon test]). CP9 exceeded CP197 with respect to all four categorical culture endpoints (Table 4), although none of these differences was statistically significant. In bladder cultures, the median log ratio of CP9 to CP197 was 0.08, reflecting a median 1.2-fold excess of CP9 over CP197 (P > 0.05 [Wilcoxon test]). Although both strains were detectable in all bladder specimens by broth culture, CP9 was somewhat more likely than CP197 to yield colonies in the direct platings and was significantly more likely to exhibit a >15-fold excess over the comparison strain (P < 0.02 [McNemar's test]). Taken together, these findings support the contention that ireA contributes to CP9's ability to colonize both the bladder and the urine; hence, it appears to be a urovirulence factor.
TABLE 4.
Virulence of CP9 (wt) and CP197 (ireA) in a mouse UTI model
| Result | No. positive in bladder (n = 20)
|
No. positive in urine (n = 14a)
|
||
|---|---|---|---|---|
| CP9 | CP197 | CP9 | CP197 | |
| Any CFU in broth | 20 | 20 | 7b | 4b |
| Any CFU on plates | 20 | 17 | 13 | 12 |
| >1 × other strain | 10 | 10 | 9 | 5 |
| >15 × other strain | 7c | 0c | 6 | 3 |
Postmortem urine was available from 14 of 20 mice.
Broth urine cultures were done for 8 of 14 urine samples.
For CP9 versus CP197, the P value was 0.02 (McNemar's test).
Virulence of CP9 (wt) and CP197 (ireA) in mouse systemic infection model.
To determine the impact of IreA on systemic virulence, mice were challenged in parallel with CP9(wt) and CP197(ireA) via IP challenge in four separate experiments (Table 5). In the first two experiments, CP9 and CP197 were grown overnight in LB medium prior to IP challenge. In the next two experiments, CP9 and CP197 were grown prior to challenge in Fe-depleted minimal medium to minimize bacterial intracellular Fe stores, which likely occurs within their colonic reservoir. Prior growth in rich medium (LB broth) resulted in lower LD50s than when CP9 and CP197 were grown in Fe-depleted minimal medium. In all four experiments, the LD50 was unexpectedly lower for CP197 than for CP9. The combined LD50 titer for CP197 (2.55 × 106) was only 45% of that for CP9 (5.37 × 106) (range, 32 to 91%). However, this difference did not reach statistical significance (P > 0.05). Whether this small, but reproducible, difference is biologically significant is unclear.
TABLE 5.
Virulence of CP9 (wt) and CP197 (ireA) in a mouse systemic infection model
| Expt | Strain | LD50 (CFU) |
|---|---|---|
| 1 | CP9 | 3.82 × 106 |
| CP197 | 1.22 × 106 | |
| 2 | CP9 | 3.95 × 106 |
| CP197 | 3.56 × 106 | |
| 3 | CP9 | 5.06 × 106 |
| CP197 | 2.93 × 106 | |
| 4 | CP9 | 1.2 × 107 |
| CP197 | 4.17 × 106 |
DISCUSSION
Through the use of human body fluids ex vivo we have identified a new gene, ireA, which has increased expression in urine, blood, and ascites. Sequence analysis of its putative gene product disclosed significant identities (29 to 38%) and similarities (48 to 56%) with a variety of siderophore receptors. Further, 5′ to the coding region for ireA is a Fur box and the expression of ireA is Fe repressed. Thus, although we have not experimentally established a function for IreA, it is likely involved with Fe acquisition.
IreA is not the only putative siderophore receptor present in CP9. We have previously identified IroN, an additional putative siderophore receptor in CP9 (19) and a homologue of FyuA (29), a siderophore receptor identified in Yersinia (J. Johnson, personal communication). Further, other siderophore receptors or other Fe acquisition systems likely exist using E. coli K-12 as a paradigm (2). These findings raise the obvious question as to why such a functional redundancy exists. Clearly, Fe acquisition is a critical need for any microorganism and particularly for a pathogen that must grow within a host that actively attempts to limit Fe availability. Perhaps the maintenance of multiple active siderophore systems maximizes the chances for successful Fe procurement and as a result confers a selective advantage. Further, possession of multiple systems may afford the organism “insurance” in the event that one system becomes dysfunctional due to mutation, genomic rearrangements, or other mechanisms. Alternatively, certain receptors, and their cognate siderophores, may be more effective in certain environmental milieu outside of the host, within the gastrointestinal tract, or at specific sites of infection within the host (1, 36). Clearly, the nearly ubiquitous presence of Fe acquisition systems among the human and animal pathogens evaluated to date and the increased expression of such systems in vivo or ex vivo (e.g., iroN and ireA) strongly supports a functional requirement for them within the host. It remains unclear, however, whether certain siderophore systems are site specific. The testing of this hypothesis awaits the discovery of all of the Fe acquisition systems for a given pathogen, the generation of single and multiple isogenic mutants for each of these, and subsequent testing in various in vitro and in vivo model systems.
A multifunctional role for siderophore receptors is another potential explanation for their redundancy. Recently, the new adhesin Iha was identified from an O157:H7 strain of E. coli (34). This adhesin possesses significant homology with a variety of established or putative siderophore receptors, including IreA and IroN. Further IreA also posseses significant homology with IrgA, an Fe-regulated virulence factor in Vibrio cholera. IrgA contributes to growth in vivo in the rabbit ileal loop model and enhances virulence in an infant mouse model, suggesting a possible role in colonization (6, 35). Perhaps IreA and IroN also possess the ability to serve as adhesins. If so, particularly if each siderophore receptor has a specific cognate ligand, an evolutionary advantage for such multifunctional proteins can be more easily envisioned. In this regard, our findings demonstrating a contribution of IreA to bladder colonization is consistent with such a role. Studies designed to specifically assess both IreA and IroN as adhesins are presently in progress.
If the functions of different siderophore receptors are equivalent or overlapping, the demonstration of the importance of a single receptor may be problematic. This may account for our inability, despite urine, ascites, and blood being Fe-limiting growth environments, to demonstrate decreases in ex vivo growth in these body fluids with CP197 (ireA) compared to its wild-type parent CP9. However, the combination of the presence of ireA on a putative PAI, the increased expression of ireA in human urine, ascites, and blood, and the contribution of IreA to urine and bladder colonization in vivo support a role for IreA in at least urovirulence. Although our demonstration that IreA does not contribute to virulence in an IP systemic infection model suggests that it may possess site-specific virulence properties, this by no means excludes the possibility that this factor may be important for virulence at other sites of infection. An increasing body of epidemiological and experimental evidence supports the contention that many virulence factors identified in ExPEC strains are important for virulence at multiple sites of extraintestinal infection (18, 20, 23, 24).
The frequency, severity, and cost of infections due to ExPEC strains defines the need for an efficacious vaccine. IreA possesses a number of characteristics that make it a potential vaccine candidate. An ideal vaccine candidate needs to be surface exposed, to be broadly prevalent among clinical extraintestinal isolates of E. coli, to possess epitopes which are conserved and elicit a protective immune response. Other desirable characteristics include increased expression at the site of infection and a role in the pathogenesis of disease. IreA fulfills or has the potential to fulfill all of these characteristics. However, it is unlikely that an efficacious vaccine will consist of a single protein (or portion thereof). ExPEC strains are genomically and phenotypically diverse. The probability that all or nearly all of the strains in this group will express the same protein antigen against which protective antibodies can be developed is small. However, it is possible that multiple antibodies directed against multiple protein targets will result in broad, maximal bactericidal activity and protection. Therefore, development of a polyvalent vaccine will probably be needed to achieve optimal success. IreA is a candidate protein for such a vaccine.
ACKNOWLEDGMENTS
We thank Connie Clabots and Adam Stell (Medical Service, Minneapolis VA Medical Center) for assistance with the mouse UTI model and assessments of isolates from mice, Michelle Detwiler and the Roswell-Park Cancer Institute Biopolymer Facility (RPCI) for their assistance with DNA sequencing and Michael Kuskowski (Geriatric Research, Education, and Clinical Center, Minneapolis VA Medical Center) for assistance with statistical analyses. This material is based upon work supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (T.A.R., J.R.J.), National Institutes of Health grants AI 42059 (T.A.R.), and DK 47504 (J.R.J.), and a Cancer Center Support Grant CA16056 (RPCI).
REFERENCES
- 1.Bearden S, Perry R. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol Microbiol. 1999;32:403–414. doi: 10.1046/j.1365-2958.1999.01360.x. [DOI] [PubMed] [Google Scholar]
- 2.Crosa J. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol Rev. 1989;53:517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Lorenzo V, Giovannini F, Herrero M, Neilands J. Metal iron regulation of gene expression. Fur repressor-operator interaction at the promoter region of the aerobactin system of pColV-K30. J Mol Biol. 1988;203:875–884. doi: 10.1016/0022-2836(88)90113-1. [DOI] [PubMed] [Google Scholar]
- 4.Emanuelsson O, Nielson H, vov Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 5.Fleiss J. Statistical methods for rates and proportions. New York, N.Y: John Wiley & Sons; 1981. pp. 112–137. [Google Scholar]
- 6.Goldberg M, DiRita V, Calderwood S. Identification of an iron-regulated virulence determinant in Vibrio cholerae, using TnphoA mutagenesis. Infect Immun. 1990;58:55–60. doi: 10.1128/iai.58.1.55-60.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gransden W, Eykyn S, Phillips I, Rowe B. Bacteremia due to Escherichia coli: a study of 861 episodes. Rev Infect Dis. 1990;12:1008–1018. doi: 10.1093/clinids/12.6.1008. [DOI] [PubMed] [Google Scholar]
- 8.Hu P, Elliott J, McCready P, Skowronski E, Garnes J, Kobayashi A, Brubaker R, Garcia E. Structural organization of virulence-associated plasmids of Yersinia pestis. J Bacteriol. 1998;180:5192–5202. doi: 10.1128/jb.180.19.5192-5202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson J, Brown J. Defining inoculation conditions for the mouse model of ascending urinary tract infection that avoid immediate vesicoureteral reflux yet produce renal and bladder infection. J Infect Dis. 1996;173:746–749. doi: 10.1093/infdis/173.3.746. [DOI] [PubMed] [Google Scholar]
- 10.Johnson J, Brown J, Carlino U, Russo T. Colonization with and acquisition of uropathogenic Escherichia coli as revealed by polymerase chain reaction-based detection. J Infect Dis. 1998;177:1120–1124. doi: 10.1086/517409. [DOI] [PubMed] [Google Scholar]
- 11.Johnson J, O'Bryan T, Low D, Ling G, Delavari P, Fasching C, Russo T, Carlino U, Stell A. Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli that express PapG allele III. Infect Immun. 2000;68:3327–3336. doi: 10.1128/iai.68.6.3327-3336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson J, Russo T, Scheutz F, Brown J, Zhang L, Palin K, Rode C, Bloch C, Marrs C, Foxman B. Discovery of a disseminated J96-like clone of uropathogenic Escherichia coli O4:H5 containing both genes for both PapGJ96 (“Class I”) and PrsGJ96 (“Class III”) Gal(alpha1–4)Gal-binding adhesins. J Infect Dis. 1997;175:983–988. doi: 10.1086/514006. [DOI] [PubMed] [Google Scholar]
- 13.Johnson J, Stapleton A, Russo T, Scheutz F, Brown J, Maslow J. Characteristics and prevalence within serogroup O4 of a J96-like clonal group of uropathogenic Escherichia coli O4:H5 containing the Class I and Class III alleles of papG. Infect Immun. 1997;65:2153–2159. doi: 10.1128/iai.65.6.2153-2159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahan M, Slauch J, Mekalanos J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 15.McVeigh A, Fasano A, Scott D, Jelacic S, Moseley S, Robertson D, Savarino S. IS1414, an Escherichia coli insertion sequence with a heat-stable enterotoxin gene embedded in a transposase-like gene. Infect Immun. 2000;68:5710–5715. doi: 10.1128/iai.68.10.5710-5715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mekalanos J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson K, Wolf-Watz H. Modulation of virulence factor expression by pathogen target cell contact. Science. 1996;273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- 18.Russo T, Brown J, Jodush S, Johnson J. The O4 specific antigen moiety of lipopolysaccharide but not the K54 group 2 capsule is important for urovirulence in an extraintestinal isolate of Escherichia coli. Infect Immun. 1996;64:2343–2348. doi: 10.1128/iai.64.6.2343-2348.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russo T, Carlino U, Mong A, Jodush S. Identification of genes in an extraintestinal isolate of Escherichia coli with increased expression after exposure to human urine. Infect Immun. 1999;67:5306–5314. doi: 10.1128/iai.67.10.5306-5314.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russo T, Davidson B, Prior R, Carlino U, Helinski J, Knight P I. Capsular polysaccharide and O-specific antigen divergently modulate pulmonary neutrophil influx in a rat Escherichia coli model of gram-negative pneumonitis. Infect Immun. 2000;68:2854–2862. doi: 10.1128/iai.68.5.2854-2862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russo T, Guenther J, Wenderoth S, Frank M. Generation of isogenic K54 capsule-deficient Escherichia coli strains through TnphoA-mediated gene disruption. Mol Microbiol. 1993;9:357–364. doi: 10.1111/j.1365-2958.1993.tb01696.x. [DOI] [PubMed] [Google Scholar]
- 22.Russo T, Jodush S, Brown J, Johnson J. Identification of two previously unrecognized genes (guaA, argC) important for uropathogenesis. Mol Microbiol. 1996;22:217–229. doi: 10.1046/j.1365-2958.1996.00096.x. [DOI] [PubMed] [Google Scholar]
- 23.Russo T, Johnson J. A proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis. 2000;181:1753–1754. doi: 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- 24.Russo T, Liang Y, Cross A. The presence of K54 capsular polysaccharide increases the pathogenicity of Escherichia coli in vivo. J Infect Dis. 1994;169:112–118. doi: 10.1093/infdis/169.1.112. [DOI] [PubMed] [Google Scholar]
- 25.Russo T, Sharma G, Brown C, Campagnari A. The loss of the O4 antigen moiety from the lipopolysaccharide of an extraintestinal isolate of Escherichia coli has only minor effects on serum sensitivity and virulence in vivo. Infect Immun. 1995;63:1263–1269. doi: 10.1128/iai.63.4.1263-1269.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russo T, Stapleton A, Wenderoth S, Hooton T, Stamm W. Chromosomal restriction fragment length polymorphism analysis of Escherichia coli strains causing recurrent urinary tract infections in young women. J Infect Dis. 1995;172:440–445. doi: 10.1093/infdis/172.2.440. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;85:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schubert S, Rakin A, Karch H, Carniel E, Heeseman J. Prevalence of the “high pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun. 1998;66:480–485. doi: 10.1128/iai.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stamm W, Counts G, Running K, Fihn S, Turck M, Holmes K. Diagnosis of coliform infection in acutely dysuric women. N Engl J Med. 1982;307:463–468. doi: 10.1056/NEJM198208193070802. [DOI] [PubMed] [Google Scholar]
- 31.Stentebjerg-Olesen B, Chakraborty T, Klemm P. Type I fimbriation and phase switching in a natural Escherichia coli fimB null strain, Nissle 1917. J Bacteriol. 1999;181:7470–7478. doi: 10.1128/jb.181.24.7470-7478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stinear T, Ross B, Davies J, Marino L, Robins-Browne R, Oppedisano F, Sievers A, Johnson P. Identification and characterization of IS2404 and IS2606: two distinct repeated sequences for detection of Mycobacterium ulcerans by PCR. J Clin Microbiol. 1999;37:1018–1023. doi: 10.1128/jcm.37.4.1018-1023.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takami H, Takaki Y, Nakasone K, Sakiyama T, Maeno G, Sasaki R, Hirama C, Fuji F, Masui N. Genetic analysis of the chromosome of alkaliphilic Bacillus halodurans C-125. Extremophiles. 1999;3:227–233. doi: 10.1007/s007920050120. [DOI] [PubMed] [Google Scholar]
- 34.Tarr P, Bilge S, Vary J J, Jelacic S, Habeeb R, Ward T, Baylor M, Besses T. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect Immun. 2000;68:1400–1407. doi: 10.1128/iai.68.3.1400-1407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tashima K, Carroll P, Rogers M, Calderwood S. Relative importance of three iron-regulated outer membrane proteins for in vivo growth of Vibrio cholerae. Infect Immun. 1996;64:1756–1761. doi: 10.1128/iai.64.5.1756-1761.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsolis R, Baumler A, Heffron F, Stojiljkovic I. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect Immun. 1996;64:4549–4556. doi: 10.1128/iai.64.11.4549-4556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyler S, Rozee D, Johnson W. Identification of IS1356, a new insertion sequence, and its association with IS402 in epidemic strains of Burkholderia cepacia infecting cystic fibrosis patients. J Clin Microbiol. 1996;34:1610–1616. doi: 10.1128/jcm.34.7.1610-1616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valdivia R, Hromockyj A, Monack D, Ramakrishnan L, Falkow S. Applications for green fluorescent protein (GFP) in the study of host-pathogen interactions. Gene. 1996;173:47–52. doi: 10.1016/0378-1119(95)00706-7. [DOI] [PubMed] [Google Scholar]
- 39.Wang G, Whittam T, Berg C, Berg D. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993;21:5930–5933. doi: 10.1093/nar/21.25.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wheatcroft R, Laberge S. Identification and nucleotide sequence of Rhizobium meliloti insertion sequence ISRm3: similarity between the putative transposase encoded by ISRm3 and those encoded by Staphylococcus aureus IS256 and Thiobacillus ferrooxidans IST2. J Bacteriol. 1991;173:2530–2538. doi: 10.1128/jb.173.8.2530-2538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young G, Miller V. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol Microbiol. 1997;25:319–328. doi: 10.1046/j.1365-2958.1997.4661829.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, Normark S. Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science. 1996;273:1234–1236. doi: 10.1126/science.273.5279.1234. [DOI] [PubMed] [Google Scholar]