Abstract
DNA topoisomerase IIα (TOP2α/170; 170 kDa) and topoisomerase IIβ (TOP2β/180; 180 kDa) are targets for a number of anticancer drugs, whose clinical efficacy is attenuated by chemoresistance. Our laboratory selected for an etoposide-resistant K562 clonal subline designated K/VP.5. These cells exhibited decreased TOP2α/170 and TOP2β/180 expression. We previously demonstrated that a microRNA-9 (miR-9)-mediated posttranscriptional mechanism plays a role in drug resistance via reduced TOP2α/170 protein in K/VP.5 cells. Here, it is hypothesized that a similar miR-9 mechanism is responsible for decreased TOP2β/180 levels in K/VP.5 cells. Both miR-9-3p and miR-9-5p are overexpressed in K/VP.5 compared with K562 cells, demonstrated by microRNA (miRNA) sequencing and quantitative polymerase chain reaction. The 3′-untranslated region (3′-UTR) of TOP2β/180 contains miRNA recognition elements (MRE) for both miRNAs. Cotransfection of K562 cells with a luciferase reporter plasmid harboring TOP2β/180 3′-UTR plus miR-9-3p or miR-9-5p mimics resulted in statistically significant decreased luciferase expression. miR-9-3p and miR-9-5p MRE mutations prevented this decrease, validating direct interaction between these miRNAs and TOP2β/180 mRNA. Transfection of K562 cells with miR-9-3p/5p mimics led to decreased TOP2β protein levels without a change in TOP2β/180 mRNA and resulted in reduced TOP2β-specific XK469-induced DNA damage. Conversely, K/VP.5 cells transfected with miR-9-3p/5p inhibitors led to increased TOP2β/180 protein without a change in TOP2β/180 mRNA and resulted in enhancement of XK469-induced DNA damage. Taken together, these results strongly suggest that TOP2β/180 mRNA is translationally repressed by miR-9-3p/5p, that these miRNAs play a role in acquired resistance to etoposide, and that they are potential targets for circumvention of resistance to TOP2-targeted agents.
SIGNIFICANCE STATEMENT
Results presented here indicate that miR-9-3p and miR-9-5p play a role in acquired resistance to etoposide via decreased DNA topoisomerase IIβ 180 kDa protein levels. These findings contribute further information about and potential strategies for circumvention of drug resistance by modulation of microRNA levels. In addition, miR-9-3p and miR-9-5p overexpression in cancer chemoresistance may lead to future validation as biomarkers of responsiveness to DNA topoisomerase II–targeted therapy.
Introduction
Type II DNA topoisomerases are enzymes that regulate DNA topology by generating transient double-stranded breaks during replication and transcription (Deweese and Osheroff, 2009; Nitiss, 2009; Chen et al., 2013; Pommier et al., 2016; Austin et al., 2018; Austin et al., 2021; Pommier et al., 2022). Vertebrates express two isoforms: 1) topoisomerase IIα (TOP2α/170, 170kDa), which is expressed in a cell cycle–dependent manner (i.e., highly expressed in late S and G2/M phases) (Woessner et al., 1991) and is required for chromosomal segregation during mitosis (Deweese and Osheroff, 2009; Nitiss, 2009), and 2) topoisomerase IIβ (TOP2β/180, 180kDa), which is expressed at a constant level regardless of the cellular proliferation status (Woessner et al., 1991) and is implicated mainly in transcription regulation and tissue differentiation (Austin et al., 2018; Austin et al., 2021).
TOP2α and TOP2β are the molecular targets for a number of drugs used to treat leukemias, lymphomas, and solid tumors (Edwardson et al., 2015; Shanbhag and Ambinder, 2018; Economides et al., 2019). These drugs, referred to as TOP2 poisons/interfacial inhibitors (Pommier and Marchand, 2011), such as etoposide, doxorubicin, epirubicin, daunorubicin, mitoxantrone, and mAMSA, inhibit the religation step of TOP2 enzymatic activity and thus stabilize the normally transient enzyme-DNA cleavage complex. The accumulation of these TOP2-DNA covalent complexes results in loss of cell viability (Austin et al., 2018).
Acquired resistance to TOP2 inhibitors is often associated with a reduction of TOP2α/170 and/or TOP2β/180 expression levels (Harker et al., 1991; Harker et al., 1995; Ritke and Yalowich, 1993; Herzog et al., 1998; Errington et al., 1999; Mirski et al., 2000; Burgess et al., 2008; Pilati et al., 2012; Ganapathi and Ganapathi, 2013; Hermanson et al., 2013; Capelôa et al., 2020). Acquired resistance to etoposide in a human K562 leukemia cell line, K/VP.5, is associated with decreased TOP2α/170 and TOP2β/180 mRNA/protein expression (Kanagasabai et al., 2017).
MicroRNAs (miRNAs) are small, endogenous, noncoding, single-stranded eukaryotic RNAs (∼22 nucleotides) that dynamically regulate gene networks and steady-state gene regulation through posttranscriptional mechanisms (reviewed in Bartel, 2009; O’Brien et al., 2018; Treiber et al., 2019). miRNAs are processed from precursor molecules (pri-miRNAs), and following their processing, miRNAs are assembled into the RNA-induced silencing complex (miRISC) (Treiber et al., 2019). The miRNA then directs the miRISC to the 3′-untranslational region (3′-UTR) of a mRNA target via sequence complementarity to miRNA seed regions, usually comprised of 7 to 8 nucleotides (Treiber et al., 2019). miRNA-mediated posttranscriptional gene silencing subsequently results from target mRNA sequestration or degradation, stimulation of deadenylation, and altering cap protein binding or decapping (Krol et al., 2010; Fabian and Sonenberg, 2012; Jonas and Izaurralde 2015; Bahrami et al., 2022), which impact translation.
Importantly, aberrant/pathologic expression of miRNAs are associated with many forms of cancers, including the four major subtypes of leukemia: chronic lymphocytic leukemia, acute lymphocytic leukemia, acute myeloid leukemia, and chronic myeloid leukemia (reviewed in Gabra and Salmena, 2017; Pekarsky and Croce, 2019; Balatti and Croce, 2022). Additionally, miRNA dysregulation also plays a role in cancer drug resistance in various forms of leukemias (reviewed in Gabra and Salmena, 2017; Corrà et al., 2018; Marima et al., 2021). For example, the oncogenic miR-486 was shown to promote imatinib resistance in chronic myeloid leukemia cells by targeting phosphatase and tensin homolog and forkhead box O1 tumor suppressors (Wang et al., 2015). Additionally, miRNA-125b was demonstrated to be involved in doxorubicin drug resistance in pediatric acute promyelocytic leukemia cells (Zhang et al., 2011). Finally, miRNA-21, which targets BCL2 apoptosis regulator mRNA, was reported to be upregulated in daunorubicin-resistant leukemia cells, whereas miRNA-21 knockdown in these cells was shown to increase daunorubicin cytotoxicity (Löwenberg et al., 2009).
We previously demonstrated that endogenously overexpressed miR-9-3p and miR-9-5p in drug-resistant K/VP.5 cells were responsible for decreased TOP2α/170 protein levels in etoposide-resistant K/VP.5 cells (Kania et al., 2020). In complementary experiments, transfection of miR-9 inhibitors increased TOP2α/170 expression and enhanced etoposide-induced DNA damage in resistant K/VP.5 cells, strongly suggesting that expression of miR-9-3p and miR-9-5p in K/VP.5 cells are determinants of acquired resistance (Kania et al., 2020). In the present study, we hypothesized that miRNA-mediated mechanisms also play a role in drug resistance via decreased expression of TOP2β/180 in etoposide-resistant K/VP.5 cells. To test this hypothesis, we examined whether and to what extent miR-9-3p/-5p 1) directly bind to the 3′-UTR of TOP2β/180, 2) regulated mRNA and protein expression levels of TOP2β/180, and 3) modulated activity of (2-{4-[(7-chloro-2-quinoxalinyl)oxy]phenoxy}propionic acid) (XK469) (Alousi et al., 2007; Gao et al., 1999; Mensah-Osman et al., 2003), a TOP2β selective agent. Together, results indicated that miR-9-3p and miR-9-5p are determinants of acquired drug resistance to TOP2β-targeted drugs.
Materials and Methods
Chemicals, Reagents, and Cell Lines
Etoposide (cat. no. 33419-42-0) and XK469 (cat. no.157435-10-4) were purchased from Sigma-Aldrich (St. Louis, MO) and solubilized in 100% DMSO as concentrated stocks. Human K562 leukemia cells were maintained in Dulbecco’s modified Eagle’s medium (Corning, Manassas, VA) supplemented with 10% FBS. Etoposide-resistant K/VP.5 cells were selected and cloned subsequent to intermittent and eventually continuous exposure of K562 cells to 0.5 μM etoposide as previously described (Ritke and Yalowich, 1993). K/VP.5 cells were maintained in Dulbecco’s modified Eagle’s medium/10% FBS with etoposide (0.5 μM) added every other week. All experiments described below were performed utilizing cells growing in log phase.
Inhibition of Cell Growth
To assess resistance to etoposide, log-phase parental K562 and cloned K/VP.5 cells were adjusted to 1 × 105 cells per ml, and the cells were incubated for 48 hours with 0.01–15 μM etoposide in K562 cells and 0.1–150 μM etoposide in K/VP.5 cells, after which cells were counted on a model Z1 Coulter counter (Beckman Coulter, Danvers, MA). Percentage of growth inhibition for each concentration of drug was determined based on comparison with DMSO control growth.
Quantitative Real-Time Polymerase Chain Reaction Assays
Total RNA was isolated from K562 and K/VP.5 cells using the RNA Easy Plus Mini Kit (cat. no. 74134; Qiagen, Germantown, MD). To ensure complete removal of contaminating DNA, an on-column digestion of DNA with RNase-free DNase (cat. no. 79254; Qiagen) was included during RNA purification. RNA (1 μg) was reverse transcribed using random hexamers and MultiScribe Reverse Transcriptase (High-Capacity cDNA Reverse Transcription Kit, cat. no. 4368814; Thermo-Fisher Scientific, Waltham, MA) as previously described by our laboratory (Kanagasabai et al., 2017). Quantitative real-time polymerase chain reaction (qPCR) evaluations (total reaction volume now modified to 10 µL) were performed using TaqMan Gene Expression hydrolysis probes (ThermoFisher Scientific) as previously described (Kanagasabai et al., 2017, 2018). TOP2β/180 mRNA expression levels were measured using a hydrolysis probe spanning the TOP2β Exon 19/Exon 20 boundary (Assay ID Hs01060678_g1; ThermoFisher Scientific). The relative mRNA expression levels of TOP2β/180 in each cell line were normalized to TATA-binding protein (TaqMan assay Hs99999910_m1; ThermoFisher Scientific) expression using the 2-ΔΔCt method (Schmittgen and Livak, 2008).
In addition, K562 and K/VP.5 total RNA samples, isolated from miR-9-3p and miR-9-5p mimic/inhibitor-transfected and control mimic/inhibitor-transfected cells (1 µg), were reverse transcribed using human miR-9-3p, miR-9-5p, and RNU48 antisense primers and MultiScribe Reverse Transcriptase in a 30-µL reaction according to manufacturer instructions. qPCR for miRNA quantification (20 µL per reaction) was performed using 1.33 µL of cDNA with primer/probe sets specific for miR-9-3p (Assay ID 00231), miR-9-5p (Assay ID 000583), and RNU48 (Assay ID 001006). The relative gene expression level of miR-9-3p and/or miR-9-5p in each experimental condition was normalized to RNU48 expression using the 2−ΔΔCt method (Schmittgen and Livak, 2008).
Immunoassays
K562 and K/VP.5 cellular extracts isolated from miR-9-3p and miR-9-5p mimic/inhibitor-transfected and control mimic/inhibitor-transfected cells (with or without XK469 treatment) were subjected to western blot analysis as previously described (Kanagasabai et al., 2017, 2018). Based on protein content in cellular extracts, 16 µg were loaded into each well, and an equal volume of Precision Plus Protein Color Standards was also run for molecular mass reference (cat. no. 1610374; Bio-Rad Laboratories, Hercules, CA). Membranes were incubated overnight at 4°C with one of the following primary antibodies: a mouse monoclonal antibody raised against amino acids 1341–1626 of TOP2β of human origin (H-8) (cat. no. sc-25330 Santa Cruz Biotechnology, Santa Cruz, CA; used at 1:350 dilution), a rabbit polyclonal antibody raised against the human TOP2α raised against amino acids 14–27 (cat. no. C10345 Assay Biotechnology, Sunnyvale, CA; used at 1:1000 dilution), or a mouse monoclonal glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (cat. no. sc-47724; Santa Cruz Biotechnology; used at 1:5000 dilution). The membranes were subsequently incubated at room temperature for ∼3 hours with a donkey anti-rabbit or an anti-mouse secondary antibody (Jackson Immuno Research, West Grove, PA; used at 1:5000 dilution). Finally, antibody-labeled TOP2α/170, TOP2β/180, and GAPDH were detected using the Clarity Max chemiluminescence kit (Bio-Rad Laboratories Hercules, CA). All immunoassay images were acquired with the ChemiDoc XRS+ imaging system and analyzed with ImageLab software (Bio-Rad Laboratories).
TOP2β/180 mRNA/miRNA Bioinformatic Analyses
To predict putative MREs harbored in the 3′-UTR of TOP2β/180 mRNAs, the DIANA-microT-CDS computational algorithm was used (DIANA-microT-CDS, https://dianalab.e-ce.uth.gr/html/dianauniverse/index.php?r=microT_CDS, fifth version of the microT algorithm; Paraskevopoulou et al., 2013). Additionally, the experimentally supported TOP2β/180 mRNA/miRNA targets were also surveyed (DIANA-TarBase v8, https://dianalab.e-ce.uth.gr/html/diana/web/index.php?r=tarbasev8; Karagkouni et al., 2018).
Luciferase Reporter Constructs
The pEZX-MT06/TOP2β/3′-UTR dual reporter plasmid was purchased from GeneCopoeia (cat. # HmiT110765-MT06; Rockville, MD). This plasmid harbors a 419-bp fragment encompassing the entire TOP2β/180 3′-UTR (NCBI Reference Sequence: NM_001330700.1) that was subcloned downstream of a firefly luciferase reporter gene open reading frame, which is expressed under control of the SV40 promoter. The authenticity and orientation of the TOP2β/180 3′-UTR relative to the firefly luciferase gene were confirmed by Sanger sequencing.
The mutant reporter construct designated pEZX/TOP2β/3′-UTR/mut5p was generated utilizing the pEZX-MT06/TOP2β/3′-UTR vector as a template and mutating the predicted miR-9-5p MRE (located from nucleotides 5–11 of the TOP2β 3′-UTR) using a Q5 Site-Directed Mutagenesis kit from New England Biolabs (cat. no. E0554S) following the manufacturer’s instructions. The primers used for mutagenesis were sense (5′-AAGCGTGGCACAAACATTTTTCAACAA-3′) and antisense (5′-GCACGGTACGGAATTCGCGATC-3′). The mutant reporter construct designated pEZX/TOP2β/3′-UTR/mut3p was generated in the same manner as pEZX/TOP2β/3′-UTR/mut5p, but the predicted miR-9-3p MRE (located from nucleotides 165–169 of the TOP2β 3′-UTR) was modified with the following sense (5′-AAATAACTCTTTTTTACTCATTGAAATGTCACG-3′) and antisense (5′-ACTGTATGTGTAAGAACAAAATGTTAAAAG-3′) primers. Importantly, these primers were designed with their 5′ ends annealing back-to-back (inverse polymerase chain reaction). The nucleotides that were mutated are shown in bold print. The mutations of the putative miR-9-3p and miR-9-5p seed sequences harbored in the TOP2β 3′-UTR were confirmed by Sanger sequencing. Finally, transformed bacterial cultures were grown, and each reporter construct was isolated with a ZymoPureTM II Plasmid Maxiprep Kit (cat. no. D4203-A; Zymo Research).
Transfection Experiments
K562 and K/VP.5 cells (2 × 106 in 1 ml per condition) were transfected with the pEZX-MT06/TOP2β/3′-UTR dual luciferase construct utilizing 1 µg of the plasmid, 3.0 µL Lipofectamine 3000, 2 µL P3000 (cat. no. L3000008; ThermoFisher), and 50 µL Opti-Mem Medium (cat. no. 31985062; ThermoFisher). Cotransfection experiments using K562 cells were performed as described above with the respective dual luciferase construct pEZX-MT06/TOP2β/3′-UTR, pEZX/TOP2β/3′-UTR/mut5p, or pEZX/TOP2β/3′-UTR/mut3p and either a miRCURY LNA miRNA Mimic Negative Control 5 (cat. no.339173, GeneGlobe ID-YM00479902-ADA; Qiagen, Germantown, MD), miRCURY LNA miR-9-3p Mimic (cat. no. 339173, GeneGlobe ID-YM00471370-ADA; Qiagen), or miRCURY LNA miR-9-5p Mimic (cat. no. 339173, GeneGlobe ID-YM00471434-ADA; Qiagen). In addition, K562 cells were transfected with only the mimics described above for subsequent TOP2β immunoassays and for DNA damage studies. Finally, K/VP.5 cells were transfected with either a miRCURY LNA miRNA Power Inhibitor (5) Negative Control (cat. no. 339136, GeneGlobe ID-YI00199006-DDA), miRCURY LNA miR-9-3p Power Inhibitor (cat. no. 339131, GeneGlobe ID-YI04100288-DDA), or miRCURY LNA miR-9-5p Power Inhibitor (cat. no. 339131, GeneGlobe ID-YI04100536-DDA). Unless otherwise noted, in transfection experiments employing miRNA mimics/inhibitors, the final concentrations were 25 nM/50 nM, respectively. At 24–48 hours after transfection, total RNA and cells/cellular extracts were prepared for qPCR, luciferase assays, immunoblotting, and DNA damage assays.
Luciferase Assay
Posttransfection (24–48 hours), K562 and K/VP.5 cells were washed and lysed with Passive Lysis Buffer (cat. no. E194A; Promega, Madison, WI), and firefly and Renilla luciferase activities were determined using the Dual-Luciferase Reporter Assay System (cat. no. E1910; Promega) with a Synergy H1 Hybrid Multi-Mode Reader (BioTek, Winooski, VT). Firefly luciferase expression from the pEZX-MT06/TOP2β/3′-UTR and mutant plasmids are produced via an SV40 promoter, whereas Renilla luciferase in this vector is generated via a cytomegalovirus promoter and has been specifically designed to be an intraplasmid transfection normalization reporter. The firefly luciferase signal was therefore normalized to the Renilla luciferase signal.
DNA Damage (Comet) Assays
Alkaline (pH 13; detects primarily single-stranded breaks) single-cell gel electrophoresis (Comet) assays were performed according to the manufacturer’s protocol (CometAssay Kit, cat. no. 4250-050-K; Trevigen, Gaithersburg, MD) and as previously described by our laboratory (Vlasova et al., 2011; Kanagasabai et al., 2017, 2018). Briefly, K562 or K/VP.5 cells were transfected with control miRNA mimics/inhibitors or miR-9 mimics/inhibitors, respectively, as described above. Forty-eight hours after transfection, cells were washed and resuspended in buffer [25 mM HEPES, 10 mM glucose, 1 mM MgCl2, 5 mM KCl, 130 mM NaCl, 5 mM monosodium phosphate (pH 7.4)]. Transfected cells were subsequently incubated with 250 µM XK469 or DMSO (solvent control) for 1 hour at 37°C. The treated cells were washed with ice-cold buffer and resuspended to 0.28 × 106 cells per ml and then further diluted in low-melt agarose. After alkaline electrophoresis (of ∼2000 cells) and subsequent staining with a fluorescence DNA intercalating dye, SYBR Gold, the migrating fragments (comet tail) from the nucleoid (comet head) were visualized, and the images were captured by fluorescence microscopy. The Olive tail moment (Olive, 2002) was quantified by the ImageJ processing program with the open-source software tool Open Comet (Gyori et al., 2014). The Olive tail moment is defined as the product of the percentage of DNA in the tail of visualized “comets” and the distance between the intensity centroids (center of gravity) of the comet head and tail along the x-axis of the comet. Olive tail moments from more than 100 cells per sample condition were determined.
Data Analysis
Statistical analysis was performed using Sigma-Plot 14.5. All data are expressed as the mean ± S.D. qPCR gene expression data (2 -ΔCt values) were subjected to log transformation to assure distribution normality prior to paired Student’s t test analysis (Ganger et al., 2017). Groupwise differences were analyzed using a two-tailed paired Student’s t test with no adjustment for multiple comparisons. A P value less than 0.05 was considered statistically significant.
Results
TOP2β/180 Expression is Decreased in Etoposide-Resistant K562 Cells
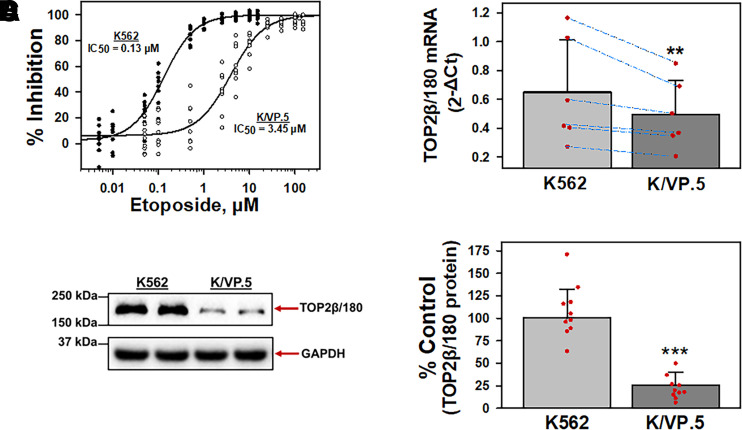
Our laboratory previously established an etoposide-resistant K562 cell line that was derived by first periodic and then continuous exposure of K562 cells to 0.5 µM etoposide, followed by limiting dilution cloning to isolate and then characterize a single clone, designated K/VP.5 cells (Ritke and Yalowich, 1993; Ritke et al., 1994). Cell growth inhibition of K562 and K/VP.5 cells was assessed by 48-hour incubation with increasing etoposide concentrations. Results indicated that K/VP.5 cells were ∼27-fold resistant to etoposide (Fig. 1A), similar to previously published results with these cell lines (Ritke and Yalowich, 1993; Ritke et al., 1994; Kanagasabai et al., 2017; Kania et al., 2020). To investigate the relative TOP2β/180 mRNA expression levels, K562 and K/VP.5 cDNAs were subjected to qPCR utilizing TaqMan hydrolysis primers/probes. Results indicated that TOP2β/180 mRNA expression levels were statistically significantly reduced in K/VP.5 cells to 76.4% of that found in parental K562 cells (P = 0.003; Fig. 1B). Accordingly, TOP2β/180 protein was also decreased in K/VP.5 cells (Fig. 1C) to 25.5% the level expressed in parental K562 cells (P = 3.8 × 10−7; Fig. 1D), recapitulating the reduction of TOP2β/180 mRNA and protein expression in K/VP.5 cells found previously (Kanagasabai et al., 2017).
Fig. 1.
K/VP.5 cell resistance to etoposide is related to decreased TOP2β/180 mRNA and protein levels. (A) Parental K562 and etoposide-resistant K/VP.5 cells were incubated with increasing concentrations of etoposide for 48 hours; following which, cells were counted. The extent of growth (beyond initial concentration) in drug-treated versus controls was expressed as % Inhibition growth. Results are shown as a scattergram from eight independent experiments performed on separate days. (B) qPCR experiments were performed utilizing K562 and K/VP.5 cDNAs and a TaqMan hydrolysis assay specific for TOP2β/180. Results shown are the mean ± S.D. from six RNA/cDNA isolations/determinations performed on separate days. Calculated 2-ΔCt values were log transformed to assure distribution normality prior to analysis using a two-tailed paired Student’s t test comparing the differences in mean calculated values for K/VP.5 versus K562 TOP2β/180 mRNA; P = 0.003. Blue lines document daily paired evaluations for K562 and K/VP.5 mRNA. Paired evaluations shown are biologic replicates from separate experiments. (C) Representative immunoassay (from 10 experiments performed on separate days) using K562 and K/VP.5 cellular lysates. Blots were probed with antibodies specific for TOP2β/180 (i.e., amino acids 1341–1626) or for GAPDH. Results/data points shown in (D) are biologic replicates from the separate experiments performed. (D) Expression of TOP2β/180 protein levels in K562 and K/VP.5 cells. Averaging results from 10 separate paired collections of K562 and K/VP.5 cells on different days, there was a statistically significant reduction of TOP2β/180 in K/VP.5 cells to 25.5% the level compared with parental K562 cells; P = 3.8 × 10−7, taking into account the GAPDH loading control. **P < 0.01; ***P < 0.001. Statistical analysis was performed using a two-tailed paired Student’s t test, as documented in Materials and Methods.
miR-9-3p and miR-9-5p Overexpression in K/VP.5 Cells
Given that miRNAs can play important roles in regulating gene expression (O’Brien et al., 2018), we hypothesized that TOP2β/180 expression was decreased in K/VP.5 cells, in part, by a miRNA-mediated mechanism. miRNA sequencing (miRNA-seq) data from our previous study [Gene Expression Omnibus (GEO) accession number: GSE141687; Kania et al., 2020] demonstrated that 87 mature miRNAs were differentially expressed (twofold or greater) in these cells, with 73 miRNAs overexpressed in K/VP.5 compared with K562 cells. The top 15 overexpressed miRNAs in K/VP.5 cells are listed in Table 1 with the seven miRNAs predicted by the DIANA-micro-T-CDS algorithm (Paraskevopoulou et al., 2013) to interact with the human TOP2β/180 3′-UTR bolded (Table 1).
TABLE 1.
The top 15 miRNAs overexpressed (fold change > 2; adjusted P < 0.05) in K/VP.5 compared with K562 cells. Fold change determined from miRNA-seq data (GEO accession number: GSE141687; Kania et al., 2020) are listed below. The DIANA-microT-CDS algorithm (Paraskevopoulou et al., 2013) revealed that the TOP2β/180 3′-UTR harbored MREs for seven (bolded) of the 15 overexpressed miRNAs.
| miRNA | Fold Expression (K/VP.5)/(K562) | Adjusted P Value |
|---|---|---|
| hsa-miR-9-3p | ∼75 | 6.8 × 10−5 |
| hsa-miR-372-3p | ∼45 | 3.5 × 10−4 |
| hsa-miR-7706 | ∼32 | 2.3 × 10−3 |
| hsa-miR-504-3p | ∼28 | 8.5 × 10−8 |
| hsa-miR-383-5p | ∼27 | 6.0 × 10−4 |
| hsa-miR-9-5p | ∼26 | 9.2 × 10−19 |
| hsa-miR-451a | ∼16 | 4.6 × 10−5 |
| hsa-miR-493-5p | ∼16 | 3.6 × 10−10 |
| hsa-miR-675-5p | ∼16 | 1.9 × 10−10 |
| hsa-miR-127-3p | ∼15 | 4.4 × 10−8 |
| hsa-miR-196a-5p | ∼15 | 1.0 × 10−15 |
| hsa-miR-543-3p | ∼14 | 6.6 × 10−4 |
| hsa-miR-3681-5p | ∼13 | 5.0 × 10−10 |
| hsa-miR-493-3p | ∼13 | 3.4 × 10−7 |
| hsa-miR-1299-3p | ∼13 | 2.1 × 10−3 |
To prioritize the subsequent evaluation of the seven miRNAs predicted to interact with the TOP2β/180 3′-UTR, all of the experimentally supported TOP2β/180 mRNA/miRNA targets were surveyed utilizing the DIANA-TarBase version 8 database (Karagkouni et al., 2018). Importantly, of the seven algorithm-predicted miRNAs, only miR-9-5p (Kameswaran et al., 2014) and miR-451a (Boudreau et al., 2014) were shown to directly interact with the TOP2β/180 mRNA utilizing the high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation methodology(Haecker et al., 2012; Balakrishnan et al., 2014). Since miR-9-5p overexpression was greater than miR-451a (Table 1), we chose to focus subsequent evaluations on the effects of miR-9-5p.
miR-9-3p was also evaluated for its biologic effects since 1) miR-9-3p and miR-9-5p are matured simultaneously from the same pre-miRNA, hsa-mir-9 (Packer et al., 2008; Schraivogel et al., 2011, Nowek et al., 2016, 2018), 2) miR-9-3p levels were the most overexpressed in K/VP.5 compared with K562 cells (Table 1), and 3) manual sequence inspection (Elton and Yalowich, 2015) of the TOP2β/180 3′-UTR identified a putative miR-9-3p MRE.
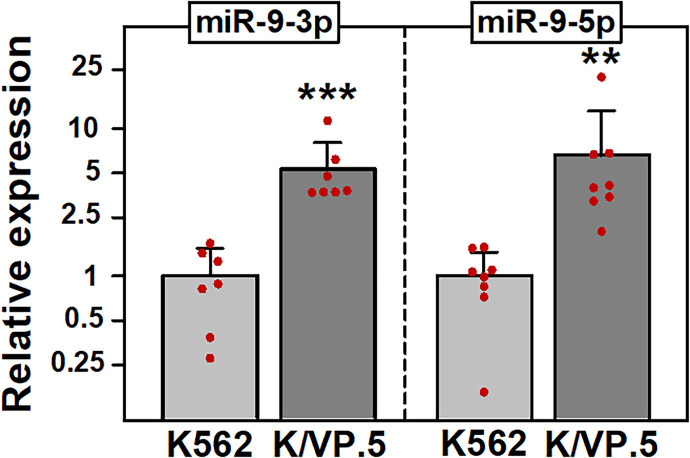
To independently validate the miR-9 miRNA-seq data (Table1; Kania et al., 2020), qPCR experiments were performed using miR-9-3p and miR-9-5p TaqMan primer/probe sets. As expected, miR-9-3p and miR-9-5p were statistically significantly overexpressed in K/VP.5 cells compared with K562 cells: 6.6-fold (P = 0.008) and 8.9-fold (P = 0.01), respectively (Fig. 2).
Fig. 2.
miR-9-3p and miR-9-5p are overexpressed in K/VP.5 cells. (A) qPCR utilizing K562 and K/VP.5 cDNAs and TaqMan hydrolysis assays specific for miR-9-3p and miR-9-5p. Results shown are the mean ± S.D. from multiple determinations made from separate RNA/cDNA preparations made on separate days, comparing K/VP.5 to K562 cell levels of miR-9-3p (N = 7; P = 0.0003) and miR-9-5p (N = 8; P = 0.005), respectively. **P < 0.01; ***P < 0.001. All data points represent biologic replicates from the separate experiments performed (depicted N values). Statistical analysis was performed using a two-tailed paired Student’s t test, as documented in Materials and Methods.
miR-9-3p and miR-9-5p Bind Directly to the TOP2β/180 3′-UTR
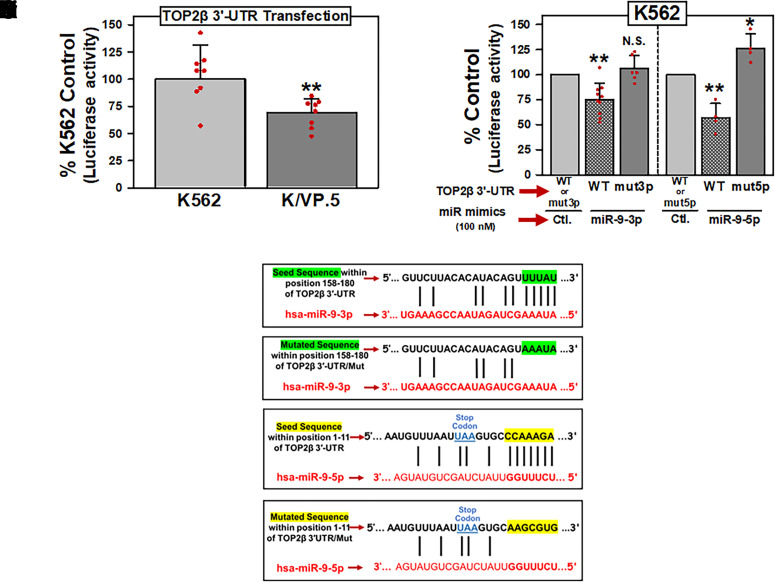
miRNAs can exert their function by association with MREs located in the 3′-UTR of target mRNAs (Treiber et al., 2019). Therefore, a dual luciferase reporter plasmid harboring the TOP2β/180 3′-UTR (419-bp, NCBI Reference Sequence: NM_001330700.1), subcloned downstream of a firefly luciferase reporter gene open reading frame, was purchased from GeneCopoeia and was designated pEZX-MT06/TOP2β/180/3′-UTR. This plasmid was subsequently transfected into K562 and K/VP.5 cells, and luciferase activity was measured. There was a statistically significant decrease in luciferase activity in K/VP.5 cells compared with K562 cells (P = 0.006), suggesting a miRNA-mediated mechanism regulating TOP2β/180 protein expression levels (Fig. 3A).
Fig. 3.
miR-9-3p and miR-9-5p directly interact with the TOP2β/180 3′-UTR. (A) K562 and K/VP.5 cells were transfected with the pEZX-MT06/TOP2β/180/3′-UTR luciferase reporter construct (1 µg). Twenty-four hours later, luciferase activities were measured. Firefly luciferase activity was normalized to Renilla luciferase activity. Results shown are the % K562 Control luciferase activities, depicted as the mean ± S.D. from eight experiments performed on separate days; P = 0.006; K/VP.5 luciferase versus K562 luciferase activity. Statistical analysis was performed using a two-tailed paired Student’s t test, as documented in Materials and Methods. All data points represent biologic replicates from the separate experiments performed. (B) K562 cells were transfected with 1 µg pEZX-MT06/TOP2β/3′-UTR [wild type (WT)], pEZX/TOP2β/3′-UTR/mut3p (mut3p), or pEZX/TOP2β/3′-UTR/mut5p (mut5p) luciferase reporter constructs together (cotransfected) with either Control, miR-9-3p, or miR-9-5p mimics (100 nM). Twenty-four hours later, luciferase activities were measured. Results shown are the respective % Control luciferase activities, depicted as the mean ± S.D. from multiple experiments performed on separate days, comparing the differences in mean values for control mimic versus miR-9-3p mimic (N = 10; P = 0.001) and control mimic versus miR-9-5p mimic (N = 4; P = 0.008) after pEZX-MT06/TOP2β/3′-UTR (wild type) cotransfection. In addition, results shown are the respective % Control luciferase activities, depicted as the mean ± S.D. from experiments performed on separate days, comparing the differences in mean values for control mimic versus miR-9-3p mimic (N = 6; P = 0.241) and control mimic versus miR-9-5p mimic (N = 4; P = 0.042) after pEZX/TOP2β/3′-UTR/mut3p (mut3p) and pEZX/TOP2β/3′-UTR/mut5p (mut5p) cotransfections, respectively. Statistical analysis was performed using a two-tailed paired Student’s t test, as documented in Materials and Methods. All data points represent biologic replicates from the separate experiments performed (depicted N values). (C) Schematic representation of the “Seed Sequence” complementarity is shown between miR-9-3p and the putative TOP2β/180 3′-UTR MRE (Ci). Schematic representation of the mutated miR-9-3p “Seed Sequence” to eliminate the complementarity with the putative TOP2β/180 3′-UTR MRE (Cii). Schematic representation of the “Seed Sequence” complementarity is shown between miR-9-5p and the putative TOP2β/180 3′-UTR MRE (Ciii). Schematic representation of the mutated miR-9-5p “Seed Sequence” to eliminate the complementarity with the putative TOP2β/180 3′-UTR MRE (Civ). *P < 0.05; **P < 0.01. Ctl., control; N.S, not significant.
To assess the putative role of miR-9-3p and miR-9-5p in regulating the expression of TOP2β/180, gain-of-inhibitory-function experiments were performed utilizing K562 cells since these cells express low levels of these miRNAs compared with K/VP.5 cells (Fig. 2; Table 1; Kania et al., 2020). K562 cells were transfected with 1 µg pEZX-MT06/TOP2β/3′-UTR (wild type), pEZX/TOP2β/3′-UTR/mut3p (mut3p), or pEZX/TOP2β/3′-UTR/mut5p (mut5p) luciferase reporter constructs together (cotransfected) with miRNA mimics (i.e., chemically modified double-stranded RNAs that mimic endogenous miRNAs): either control, miR-9-3p, or miR-9-5p mimics (100 nM). Twenty-four hours later, cellular lysates were isolated from the reporter/mimic cotransfected K562 cells and subjected to a dual luciferase assay. Results shown are the respective percent control luciferase activities, depicted as the mean ± S.D. from multiple experiments performed on separate days (Fig. 3B). For cotransfections performed with the pEZX-MT06/TOP2β/180/3′-UTR (wild type) plasmid, there were statistically significant reductions in luciferase activity with the miR-9-3p mimic to 75% of the nontargeted miRNA mimic control (P = 0.012) and with miR-9-5p mimic to 57% of the nontargeted miRNA mimic control (P = 0.008) (Fig. 3B). Together, these results strongly suggested that both miR-9-3p and miR-9-5p can interact with MREs harbored in the TOP2β/180 3′-UTR.
The DIANA-micro-T-CDS algorithm (Paraskevopoulou et al., 2013) predicted a miR-9-5p MRE in the 3′-UTR of TOP2β/180 mRNA with the “7 mer” complementary seed sequence shown in Fig. 3Ciii. Although the computational algorithm did not predict the presence of a miR-9-3p binding site in the TOP2β/180 3′-UTR, the luciferase data (Fig. 3B) suggested that there would be a target sequence in this region. Therefore, the TOP2β/180 3′-UTR sequence was manually inspected (Elton and Yalowich, 2015), and a putative miR-9-3p MRE with a “5 mer” complementary seed sequence was identified (Fig. 3, C–I).
To verify that miR-9-3p and miR-9-5p directly interact with the MREs shown in Fig. 3, C–I and Fig. 3Ciii, the complementary seed sequences were individually mutated (AAATA for miR-9-3p and AAGCGTG for miR-9-5p) in the parental pEZX-MT06/TOP2β/3′-UTR plasmid, and the resulting mutant plasmids were designated pEZX/TOP2β/3′-UTR/mut3p (Fig. 3Cii) and pEZX/TOP2β/3′-UTR/mut5p (Fig. 3C-IV), respectively. For cotransfections with the mutated expression plasmids pEZX/TOP2β/3′-UTR/mut3p plus miR-9-3p mimic or pEZX/TOP2β/3′-UTR/mut5p plus miR-9-5p mimic, these miRNA mimics no longer reduced luciferase activity compared with controls (Fig. 3B). Taken together, results from Fig. 3B demonstrated that miR-9-3p and miR-9-5p can interfere with luciferase expression by directly interacting with the predicted miR-9-3p and miR-9-5p MREs harbored within TOP2β/180/3′-UTR.
miR-9-3p and miR-9-5p Overexpression Results in Decreased TOP2β/180 Protein and Reduced XK469-Induced DNA Damage in K562 Cells
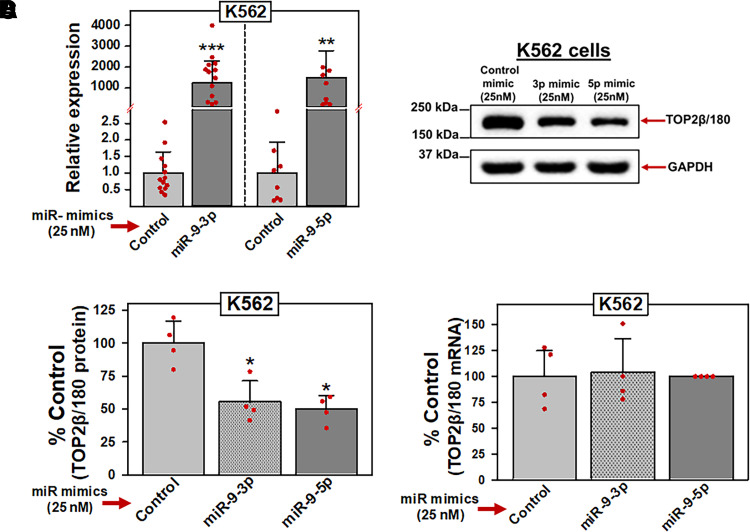
If TOP2β/180 mRNA is a target of miR-9-3p and miR-9-5p, then experimental manipulation of the endogenous levels of these two miRNAs should correlate with predictable changes in protein expression levels and drug-induced effects on TOP2β/180. Therefore, K562 cells were transfected with 25 nM miR-9-3p, miR-9-5p, or control (nontargeting) miRNA mimics. Forty-eight hours after transfection, qPCR experiments demonstrated that both miR-9-3p and miR-9-5p levels were increased greater than 1200-fold compared with miR mimic controls (Fig. 4A). In separate transfection experiments under the same conditions, TOP2β/180 protein levels were evaluated by immunoblotting experiments. Consistent with our hypothesis, miR-9-3p and miR-9-5p mimic overexpression resulted in decreased TOP2β/180 protein levels (i.e., gain of inhibitory function) (Fig. 4, B and C). Averaging results from four independent transfection and immunoblotting experiments run on separate days, there was a reduction of TOP2β/180 levels to 55.3% (P = 0.015) and 49.6% (P = 0.012) of miR mimic control in cells transfected with miR-9-3p and miR-9-5p mimic, respectively (Fig. 4C).
Fig. 4.
miR-9-3p and miR-9-5p overexpression results in decreased TOP2β/180 protein levels. (A) K562 cells were transfected with either control, miR-9-3p, or miR-9-5p mimics (25 nM). Forty-eight hours later, qPCR experiments were performed utilizing K562 cDNAs and TaqMan hydrolysis assays specific for miR-9-3p and miR-9-5p after isolation of total RNA and reverse transcription. Results shown are the mean ± S.D. from multiple experiments performed on separate days, comparing control mimic versus miR-9-3p mimic (N = 13; P = 8 × 10−12) and miR-9-5p mimic (N = 8; P = 3.1 × 10−7), respectively. qPCR values (2 -ΔCt) were log transformed to assure normal distribution prior to data analysis by use of a two-tailed paired Student’s t test. All data points represent biologic replicates from the separate experiments performed (depicted N values). (B) Representative immunoassay (from four separate experiments performed on separate days) using cellular lysates from K562 cells transfected (48 hours) with control, miR-9-3p, or miR-9-5p mimics (25 nM). Blots were probed with antibodies specific for TOP2β or for GAPDH. (C) Expression of TOP2β/180 protein levels in K562 cells transfected (48 hours) with either control, miR-9-3p, or miR-9-5p mimics (25 nM). Results shown are the mean ± S.D. from multiple experiments performed on separate days, comparing control mimic versus miR-9-3p mimic (N = 4; P = 0.015) and miR-9-5p mimic (N = 4; P = 0.012), respectively. All data points represent biologic replicates from the separate experiments performed (depicted N values). (D) TOP2β/180 mRNA levels in K562 cells measured by qPCR 48 hours after transfection with either control, miR-9-3p, or miR-9-5p mimics (25 nM). Results shown are the mean ± S.D. from four experiments performed on separate days, comparing control mimic versus miR-9-3p mimic (P = 0.882) and miR-9-5p mimic (P = 0.653), respectively. *P < 0.05; ***P < 0.001. All data points represent biologic replicates from the separate experiments performed. For panels (C) and (D), statistical analyses were performed using a two-tailed paired Student’s t test, as documented in Materials and Methods. Data points represent biologic replicates from the separate experiments performed.
Since miRISC-bound target mRNAs are subjected to mRNA degradation and/or translational repression (i.e., inhibition of translation initiation) (Krol et al., 2010; Fabian and Sonenberg, 2012; Jonas and Izaurralde 2015; Bahrami et al., 2022), we next investigated whether TOP2β/180 protein levels were reduced by a miR-9-3p- and/or miR-9-5p–mediated TOP2β/180 mRNA degradation mechanism. To investigate this potential mechanism, total RNA was isolated from K562 cells transfected (48 hours) with miR-9-3p, miR-9-5p, or control (nontargeting) miRNA mimics, and qPCR was used to quantify TOP2β/180 mRNA levels. In four separate experiments performed on separate days, transfection with miR-9-3p or miR-9-5p mimics did not affect TOP2β/180 steady-state mRNA levels (Fig. 4D), suggesting that the observed decrease in TOP2β/180 protein expression levels in transfected K562 cells (Fig. 4B) was not due to TOP2β/180 mRNA degradation but rather TOP2β/180 mRNA translational repression.
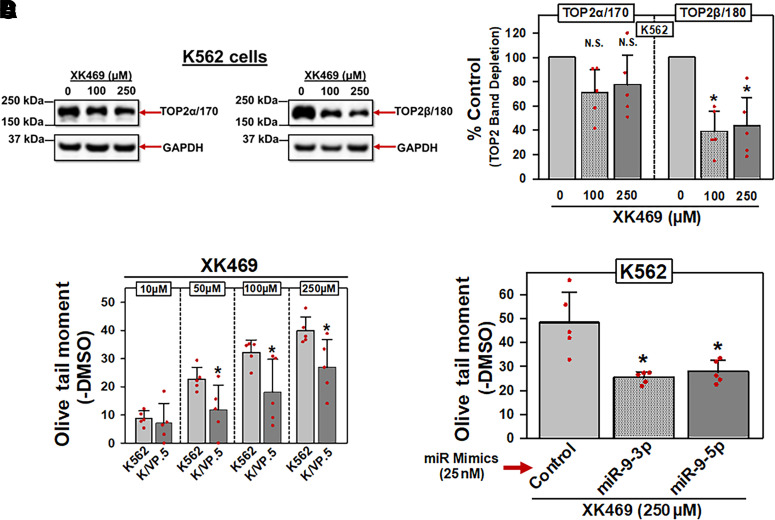
Next, a TOP2β/180-specific inhibitor, XK469, was used to evaluate miR-9-3p and miR-9-5p effects on drug-induced activity. Specificity of XK469 was validated by incubating K562 cells with either DMSO vehicle, 100 µM XK469 or 250 µM XK469 for 1 hour, followed by immunoblot/band depletion assays (Kaufmann and Svingen, 1999) (Fig. 5A). XK469-induced “band depletion” of TOP2β/180 (Fig. 5A, right), consistent with XK469-induced formation of high-molecular-mass TOP2/β-DNA covalent complexes that were unable to enter the PAGE gel (Kaufmann and Svingen, 1999). Averaging results from five experiments run on separate days, there was a statistically significant depletion of TOP2β/180 protein at 100 µM XK469 (P = 0.016) and at 250 µM XK469 (P = 0.035) compared with control levels (Fig. 5B). In contrast, there was no statistically significant decrease in TOP2α/170 at 100 µM or 250 µM XK469 (Fig. 5B). Results are consistent with XK469 specificity for TOP2β/180 at these concentrations.
Fig. 5.
miR-9-3p and miR-9-5p overexpression in K562 cells results in decreased XK469-induced DNA damage. (A) Representative immunoassay (from five experiments performed on separate days) using cellular lysates from K562 cells treated with DMSO or XK469 (100 and 250 µM) for 1 hour. Blots were probed with antibodies specific for the TOP2α/170 and the TOP2β/180 or for GAPDH. (B) TOP2α/170 and TOP2β/180 protein levels in K562 cells treated with DMSO or XK469 (100 and 250 µM) for 1 hour. For TOP2α/170 protein, results shown are the mean ± S.D. from five experiments performed on separate days, comparing DMSO control versus 100 µM XK469 (P = 0.089) and 250 µM XK469 (P = 0.129), respectively. For TOP2β/180 protein, results shown are the mean ± S.D. from five experiments performed on separate days, comparing DMSO control versus 100 µM XK469 (P = 0.016) and 250 µM XK469 (P = 0.035), respectively. Statistical analysis was performed using a two-tailed paired Student’s t test, as documented in Materials and Methods. All data points represent biologic replicates from the separate experiments performed. (C) K562 and KVP.5 cells were incubated with DMSO vehicle or XK469 (10–250 µM) for 1 hour, followed by alkaline Comet assay evaluation. Results shown are the mean ± S.D. from five experiments performed on separate days, comparing effects of XK469 in K562 versus K/VP.5 cells at 10 µM (P = 0.624), 50 µM (P = 0.038), 100 µM (P = 0.036), and 250 µM (P = 0.029). Statistical analysis was performed using a two-tailed paired Student’s t test, as documented in Materials and Methods. All data points represent biologic replicates from the separate experiments performed on separate days. (D) K562 cells transfected (48 hours) with either control, miR-9-3p, or miR-9-5p mimics (25 nM) were incubated with DMSO vehicle or 250 µM XK469 for 1 hour, followed by alkaline Comet assays. Results shown are the mean ± S.D. from five experiments performed on separate days, comparing effects of XK469 in miR mimic controls versus miR-9-3p (P = 0.018) and mir-9-5p mimic (P = 0.016), respectively. *P < 0.05. Statistical analysis was performed using a two-tailed paired Student’s t test, as documented in Materials and Methods. All data points represent biologic replicates from the separate experiments performed. N.S, not significant.
Compared with parental K562 cells, K/VP.5 cells contain reduced TOP2β/180 protein, as shown in Fig. 1, C and D. Since XK469 exhibited specificity for targeting TOP2β/180 (Fig. 5, A and B), it was expected that XK469 would induce less DNA damage in K/VP.5 compared with K562 cells as a result of decreased TOP2β/180 protein in these cells. Consistent with this expectation, Fig. 5C results demonstrated concentration-dependent XK469-induced DNA damage as measured by alkaline single-cell gel electrophoresis (Comet) assays, which was attenuated in K/VP.5 cells, further validating XK469 specificity at the level of TOP2β/180.
We next investigated whether forced expression of miR-9-3p and miR-9-5p would diminish XK469 activity secondary to a reduction in TOP2β/180 levels (as demonstrated in Fig. 4, B and C). K562 cells were transfected (48 hours) with 25 nM miR-9-3p, mir-9-5p, or a nontargeting control miRNA mimic; following which, cells were incubated for 1 hour with XK469 (250 μM) or DMSO vehicle. The cells were subsequently evaluated for DNA damage by alkaline Comet assays (Kanagasabai et al., 2017, 2018) (Fig. 5D). XK469-induced DNA strand breaks were attenuated in miR-9-3p and miR-9-5p mimic-transfected K562 cells compared with control miR mimic (Fig. 5D). Averaging results from five experiments run on separate days, there was a reduction in XK469-induced DNA damage to 52.6% (P = 0.018) and 57.6% (P = 0.016) of miRNA mimic control in K562 cells transfected with miR-9-3p and miR-9-5p mimics, respectively (Fig. 5D). Together, these results demonstrated that forced overexpression of miR-9-3p or miR-9-5p reduced TOP2β/180 protein expression and, consequently, decreased XK469-mediated DNA damage.
miR-9-3p and miR-9-5p Inhibitors Increased TOP2β/180 Protein and Enhanced XK469-Induced DNA Damage in K/VP.5 Cells
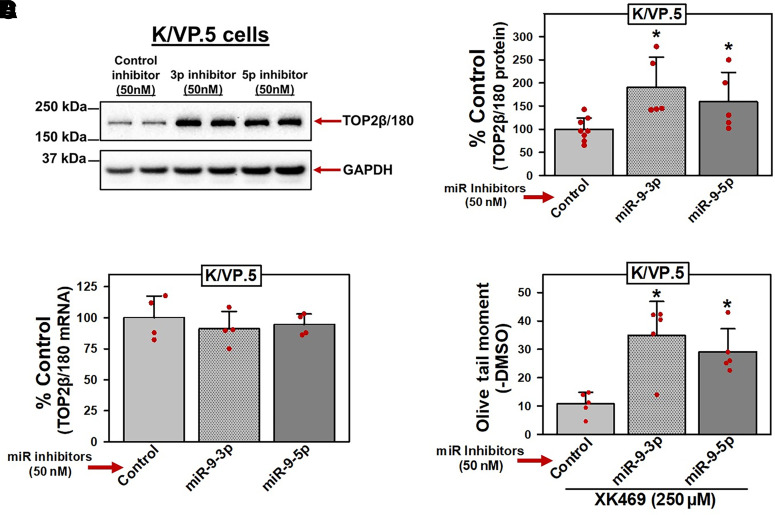
Since transfection of miRNA mimics generally results in supra-physiologic concentrations of miRNAs (Fig. 4A) (Bracken et al., 2008; Khan et al., 2009), miRNA mimic forced expression (gain-of-inhibitory-function) experiments documented above (Figs. 4C and 5D) may lead to off-target effects. Therefore, reciprocal loss-of-miR-9-inhibitory-function experiments were performed utilizing miRNA inhibitors targeting miR-9-3p/5p in K/VP.5 cells since these cells express high levels of miR-9-3p or miR-9-5p compared with K562 cells (Table 1; Fig. 2; Kania et al., 2020). miRNA inhibitors are chemically modified single-stranded antisense oligonucleotides with perfect sequence complementary to endogenous miRNAs designed to bind to and inhibit their function. Using miRNA inhibitors specific to miR-9-3p or miR-9-5p, effects on TOP2β/180 protein expression and subsequent XK469-induced DNA damage were investigated. K/VP.5 cells were transfected with 50 nM miR-9-3p, miR-9-5p, or control miRNA inhibitors (48 hours). Cellular lysates were isolated, and immunoblotting was employed to evaluate TOP2β/180 protein levels. Immunoassays revealed that TOP2β/180 protein levels were increased after transfection of miR-9-3p and miR-9-5p inhibitors (Fig. 6A). Averaging results from five separate experiments run on separate days, there was an increase in TOP2β/180 levels to 190% (P = 0.043) and 159% (P = 0.014) of miRNA inhibitor control in K/VP.5 cells treated with miR-9-3p and miR-9-5p inhibitors, respectively (Fig. 6B).
Fig. 6.
miR-9-3p and miR-9-5p inhibitors increase TOP2β/180 protein and enhance XK469-induced DNA damage in K/VP.5 cells. (A) Representative immunoassay (from five experiments performed on separate days) using cellular lysates from K/VP.5 cells transfected (48 hours) with control, miR-9-3p, or miR-9-5p inhibitors (50 nM). Blots were probed with antibodies specific for TOP2β or for GAPDH. (B) Expression of TOP2β/180 protein levels in K/VP.5 cells transfected (48 hours) with either control, miR-9-3p, or miR-9-5p inhibitors (50 nM). Results shown are the mean ± S.D. from five experiments performed on separate days, comparing control inhibitor versus miR-9-3p inhibitor (P = 0.043) and miR-9-5p mimic (P = 0.014), respectively. Statistical analysis was performed using a two-tailed paired Student’s t test, as documented in Materials and Methods. All data points represent biologic replicates from the separate experiments performed on separate days. (C) TOP2β/180 mRNA levels in K/VP.5 cells transfected (48 hours) with either control, miR-9-3p, or miR-9-5p inhibitors (50 nM). Results shown are the mean ± S.D. from four experiments performed on separate days, comparing control mimic versus miR-9-3p mimic (P = 0.427) and miR-9-5p mimic (P = 0.605), respectively. Statistical analysis was performed using a two-tailed paired Student’s t test, as documented in Materials and Methods. Data points represent biologic replicates from the separate experiments performed on separate days. (D) K/VP.5 cells transfected (48 hours) with either control, miR-9-3p, or miR-9-5p inhibitors (50 nM) were incubated with XK469 (250 µM) or DMSO for 1 hour, followed by performance of alkaline Comet assays. Results shown are the mean ± S.D. from five experiments performed on separate days, comparing effects of XK469 in miR-inhibitor controls versus miR-9-3p inhibitor (P = 0.016) and mir-9-5p inhibitor (P = 0.011), respectively. *P < 0.05. Statistical analysis was performed using a two-tailed paired Student’s t test, as documented in Materials and Methods. All data points represent biologic replicates from the separate experiments performed on separate days.
miR-9-3p and miR-9-5p inhibitors did not alter TOP2β/180 mRNA expression in K/VP.5 cells (Fig. 6C). These results are similar to those obtained with miR-9-3p/5p mimic transfection of K562 cells (Fig. 4D), again strongly suggesting that miR-9 impacts translation of TOP2β/180 mRNA without altering mRNA stability.
Finally, we investigated whether transfection of K/VP.5 cells with miR-9-3p and miR-9-5p inhibitors would increase XK469 activity secondary to augmented TOP2β/180 protein levels (Fig. 6, A and B). K/VP.5 cells were transfected with 50 nM miR-9-3p, miR-9-5p, or control miRNA inhibitors. Forty-eight hours later, cells were incubated for 1 hour with XK469 (250 µM) or DMSO vehicle and evaluated for DNA damage by alkaline single-cell gel electrophoresis (Comet) assays (Kanagasabai et al., 2017, 2018). Averaging results from five separate experiments performed on separate days, there was an increase in XK469-induced DNA damage to 349% (P = 0.016) and 291% (P = 0.011) of DMSO control in K/VP.5 cells transfected with miR-9-3p and miR-9-5p, respectively (Fig. 6D). Together these results demonstrated that increased TOP2β/180 protein expression by transfection of miR-9-3p and miR-9-5p inhibitors in K/VP.5 cells resulted in enhanced XK469-induced DNA damage.
Discussion
Intrinsic and acquired chemoresistance continues to be a major therapeutic challenge in the treatment of cancers. Cancer drug resistance can result from a variety of molecular mechanisms: 1) alteration of drug targets by modulation of expression and/or mutation, 2) modification of cellular pharmacokinetics/drug metabolism (e.g., uptake, efflux, and detoxification), 3) abnormal cell cycling, 4) cell DNA damage/repair dysregulation, and 5) reduced susceptibility to apoptosis and cell death (Zahreddine and Borden, 2013; Cree and Charlton, 2017). Clinical chemoresistance to TOP2-targeted agents used to treat leukemias (Edwardson et al., 2015; Economides et al., 2019) are frequently associated with alteration of drug targets by reduced expression (i.e., the reduced expression of TOP2α and TOP2β isoforms), suggesting that resistance can be mediated, in part, through a reduction in the formation of TOP2α/2β-DNA covalent cleavage complexes, which, in turn, decreases drug-induced DNA damage and cytotoxicity (Harker et al., 1991; Ritke and Yalowich, 1993; Harker et al., 1995 Herzog et al., 1998; Errington et al., 1999; Mirski et al., 2000; Burgess et al., 2008; Pilati et al., 2012; Ganapathi and Ganapathi, 2013; Hermanson et al., 2013; Capelôa et al., 2020).
Numerous studies in leukemias have established that aberrant expression of miRNAs is involved in cancer drug resistance by directly interacting with specific mRNAs and targeting these mRNAs for degradation and/or inhibiting their translation, thus altering the biologic pathways described above (reviewed in Gabra and Salmena, 2017; Corrà et al., 2018; Marima et al., 2021).
Several studies have been published investigating the miRNA-mediated expression of TOP2α/170 and chemoresistance. Chen et al. (2011) established that TOP2α/170 expression was reduced in a teniposide-resistant human lymphoblastic leukemia CEM cell line (CEM/VM-1-5) compared with parental CEM cells. These investigators further showed that when miR-485-3p expression was reduced, nuclear transcription factor Y subunit β was increased and negatively regulated TOP2α/170 expression in teniposide-resistant CEM/VM-1-5 cells (Chen et al., 2011). Importantly, they were able to replicate their results utilizing drug-sensitive and -resistant human rhabdomyosarcoma Rh30 cells (Chen et al., 2011). Additionally, Srikantan et al. (2011) demonstrated that ELAV-like RNA binding protein 1 enhanced TOP2α/170 translation by competing with miR-548c-3p at a common MRE harbored in the TOP2α/170 3′-UTR. Their combined actions determined the effectiveness of doxorubicin by controlling the TOP2α/170 expression levels. Finally, our laboratory demonstrated that miR-9-3p and miR-9-5p play a role in etoposide resistance by reducing TOP2α/170 protein levels in K/VP.5 cells through direct binding to specific MREs localized to the TOP2α/170 3′-UTR and inhibiting the translation of TOP2α/170 mRNAs (Kania et al., 2020).
Two reports of miRNA-mediated chemoresistance associated with TOP2β/180 have been published (Yu et al., 2010; Hatzl et al., 2020). Yu et al. (2010) established that miR-23a-3p was overexpressed in a cisplatin-resistant human tongue squamous cell carcinoma line (Tca/cisplatin) compared with cisplatin-sensitive parental Tca8113 cells. These investigators subsequently demonstrated that transfection of resistant Tca/cisplatin with miR-23a-3p inhibitors increased TOP2β/180 protein expression and sensitivity to cisplatin, suggesting that this miRNA contributes to cisplatin resistance by targeting TOP2β/180 mRNA (Yu et al., 2010). Recently, Hatzl et al. (2020) also determined that increased expression of miR-23a-3p mediated chemoresistance to cytarabine in acute myeloid leukemia cell lines associated with and determined by downregulation of TOP2β/180 protein expression.
Our present study focused on whether the observed decrease in TOP2β/180 protein levels (Fig. 1C) and resistance to etoposide in K/VP.5 cells (Fig. 1A) resulted, in part, from miR-9-3p and miR-9-5p, which are highly overexpressed in this etoposide-resistant cell line (Fig. 2) (Kania et al., 2020). Importantly, cotransfection of luciferase constructs harboring the TOP2β/180 3′-UTR and miR-9-3p and miR-9-5p mimics established that TOP2β/180 mRNA is a direct target of both miRNAs (Fig. 3, B and C). Additionally, when parental K562 cells were transfected with either miR-9-3p or miR-9-5p mimics (i.e., forced overexpression), TOP2β protein levels decreased without a change in TOP2β/180 mRNA expression and resulted in reduced TOP2β-specific XK469-induced DNA damage (i.e., gain of inhibitory function) (Figs. 4 and 5). In contrast, when etoposide-resistant K/VP.5 cells were transfected with either miR-9-3p or miR-9-5p inhibitors, TOP2β protein levels were increased (i.e., loss of inhibitory function), which resulted in augmented TOP2β-specific XK469-induced DNA damage (Fig. 6). Together, these results strongly support miR-9-3p and miR-9-5p as determinants of acquired drug resistance to a TOP2β-targeted drug. The functional importance of both miR-9-3p or miR-9-5p mediating chemoresistance establishes their potential as drug targets for circumvention of and/or biomarkers for resistance to TOP2 interfacial inhibitors.
In support of miR-9-3p and/or miR-9-5p playing an important role in mediating chemoresistance, a number of recent (i.e., 2020–2022) studies have been published (greater than 25 as per PubMed). For example, Wang et al., (2020) established a relationship between the resistance of breast cancer cells to doxorubicin and interactions of long noncoding RNA taurine-upregulated 1 (TUG1) and miR-9-5p regulation of eukaryotic translation initiation factor 5A-2 (EIF5A2). Additionally, Wang et al., (2021) demonstrated that miR-9-3p regulated resistance of breast cancer cells to gemcitabine by targeting a metastatic adhesion factor/protein, metadherin (MTDH), which is highly expressed in tumors and promotes angiogenesis, tumor cell proliferation, invasion, and metastasis. Finally, Chen et al., (2021) established that miR-9-5p directly targets ATP binding cassette subfamily C member 1 (ABCC1, also known as MRP) and that forced expression of this miRNA enhanced temozolomide sensitivity in glioma cells by decreasing ABCC1 expression.
Pri-miRNAs (mir-9s) are the source of miR-9-3p and miR-9-5p via transcription from three genes (MIR9-1, MIR9-2, and MIR9-3) located on three separate chromosomes (Yuva-Aydemir et al., 2011). Interestingly, all three human MIR9 genes transcribe distinct primary transcripts, which are processed into identical mature miR-9-3p and miR-9-5p miRNAs (Yuva-Aydemir et al., 2011). Given that the three pri-miRNA transcripts are divergent from each other, our laboratory previously used qPCR with TaqMan primer/probe sets specific for each MIR9 gene to establish that only the MIR9-1 gene is significantly upregulated in K/VP.5 cells (Kania et al., 2020), likely leading to the observed increase in mature miR-9-3p and miR-9-5p levels. The activating mechanism(s) for MIR9-1 gene expression in K/VP.5 cells is unknown. However, it has been determined that the MIR9-1 promoter can be silenced by binding of the runt-related family transcription factor 1 (RUNX1) which is associated with leukemogenesis (Fu et al., 2017). Since it has been demonstrated that RUNX1 is a direct target of miR-9-5p (Tian et al., 2015; Raghuwanshi et al., 2018), and our RNA-seq data established that RUNX1 expression is decreased 3.3-fold in K/VP.5 cells compared with K562 cells (GEO accession number: GSE163013; Hernandez et al., 2021), we speculate that the MIR9-1 gene (i.e., miR-9-3p and miR-9-5p expression) and RUNX1 expression are coregulated by a feedback loop mechanism. Our laboratory is currently testing this hypothesis.
Because cisplatin and cytarabine resistance were mediated, in part, by miR-23a-3p overexpression and subsequent decrease in TOP2β/180 protein expression (i.e., gain of inhibitory function) (Yu et al., 2010; Hatzl et al., 2020), miRNA-seq data from our previous study (GEO accession number: GSE141687; Kania et al., 2020) was reexamined to determine miR-23a-3p expression in K562 and etoposide-resistant K/VP.5 cells. Although miR-23a-3p was not expressed in these cell lines, a related miRNA, miR-23c-3p, a miRNA with an identical seed sequence as miR-23a-3p (https://www.mirbase.org/), was overexpressed 3.7-fold in K/VP.5 compared with K562 cells (GSE141687). miR-23c-3p was also shown to directly interact with TOP2β/180 mRNA utilizing high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (DIANA-TarBase v8; Kameswaran et al., 2014). Therefore, future studies will investigate whether miR-23c-3p also plays a role in etoposide-mediated resistance and may interact with miR-9-3p and miR-9-5p. Finally, since miR-451a is also overexpressed in K/VP.5 cells (GEO accession number: GSE141687; Kania et al., 2020; Table 1) and is experimentally supported to interact with TOP2β/180 mRNA (DIANA-TarBase v8; Kameswaran et al., 2014), this miRNA will also be included in future analyses.
In conclusion, TOP2β/180 is a target for clinically effective anticancer agents and a determinant of sensitivity/resistance. Results demonstrated that overexpressed miR-9-3p and miR-9-5p in drug-resistant K/VP.5 cells negatively regulated the expression of TOP2β/180, most likely by inhibition of translation: the subject of future investigations. In addition, miR-9-3p and miR-9-5p modulated the DNA damage induced by the TOP2β/180-specific agent XK469 secondary to the effects on TOP2β/180 protein levels. Hence, miR-9-3p and miR-9-5p are functional determinants of acquired drug resistance and potential targets for drugs that target TOP2β/180 and/or biomarkers predictive for response to these agents.
Abbreviations
- GEO
Gene Expression Omnibus
- miRNA-seq
miRNA sequencing
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- K/VP.5
etoposide (VP-16)-resistant human K562 leukemia cell line
- miRNA
microRNA
- miRISC
miRNA-induced silencing complex
- MRE
miRNA recognition elements
- pri-miRNA
primary miRNA
- qPCR
quantitative polymerase chain reaction
- RUNX1
runt-related family transcription factor 1
- Tca/cisplatin
cisplatin-resistant human tongue squamous cell carcinoma line
- TOP2α
DNA topoisomerase IIα
- TOP2α/170
TOP2α 170 kDa
- TOP2β
DNA topoisomerase IIβ
- TOP2β/180
TOP2β 180 kDa
- XK469
(2-{4-[(7-chloro-2-quinoxalinyl)oxy]phenoxy}propionic acid)
- 3′-UTR
3′-untranslated region
Authorship Contributions
Participated in research design: Carvajal-Moreno, Yalowich, Elton.
Conducted experiments: Carvajal-Moreno, Hernandez, Wang.
Performed data analysis: Carvajal-Moreno, Li, Yalowich, Elton.
Wrote or contributed to the writing of the manuscript: Carvajal-Moreno, Yalowich, Elton.
Footnotes
This work was supported by National Institutes of Health National Cancer Institute [Grant R01-CA226906-01A1] (to J.C.Y. and T.S.E.).
No author has an actual or perceived conflict of interest with the contents of this article.
References
- Alousi AM, Boinpally R, Wiegand R, Parchment R, Gadgeel S, Heilbrun LK, Wozniak AJ, DeLuca P, and LoRusso PM (2007) A phase 1 trial of XK469: toxicity profile of a selective topoisomerase IIbeta inhibitor. Invest New Drugs 25:147–154. [DOI] [PubMed] [Google Scholar]
- Austin CA, Lee KC, Swan RL, Khazeem MM, Manville CM, Cridland P, Treumann A, Porter A, Morris NJ, Cowell IG (2018) TOP2B: The First Thirty Years. Int J Mol Sci 19:2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CA, Cowell IG, Khazeem MM, Lok D, Ng HT (2021) TOP2B’s contributions to transcription. Biochem Soc Trans 49:2483–2493. [DOI] [PubMed] [Google Scholar]
- Bahrami A, Jafari A, Ferns GA (2022) The dual role of microRNA-9 in gastrointestinal cancers: oncomiR or tumor suppressor? Biomed Pharmacother 145:112394. [DOI] [PubMed] [Google Scholar]
- Balakrishnan I, Yang X, Brown J, Ramakrishnan A, Torok-Storb B, Kabos P, Hesselberth JR, Pillai MM (2014) Genome-wide analysis of miRNA-mRNA interactions in marrow stromal cells. Stem Cells 32:662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balatti V, Croce CM (2022) Small Non-Coding RNAs in Leukemia. Cancers (Basel) 14:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau RL, Jiang P, Gilmore BL, Spengler RM, Tirabassi R, Nelson JA, Ross CA, Xing Y, Davidson BL (2014) Transcriptome-wide discovery of microRNA binding sites in human brain. Neuron 81:294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ (2008) A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res 68:7846–7854. [DOI] [PubMed] [Google Scholar]
- Burgess DJ, Doles J, Zender L, Xue W, Ma B, McCombie WR, Hannon GJ, Lowe SW, Hemann MT (2008) Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc Natl Acad Sci USA 105:9053–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelôa T, Benyahia Z, Zampieri LX, Blackman MCNM, Sonveaux P (2020) Metabolic and non-metabolic pathways that control cancer resistance to anthracyclines. Semin Cell Dev Biol 98:181–191. [DOI] [PubMed] [Google Scholar]
- Chen CF, He X, Arslan AD, Mo YY, Reinhold WC, Pommier Y, Beck WT (2011) Novel regulation of nuclear factor-YB by miR-485-3p affects the expression of DNA topoisomerase IIα and drug responsiveness. Mol Pharmacol 79:735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Chan NL, Hsieh TS (2013) New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem 82:139–170. [DOI] [PubMed] [Google Scholar]
- Chen XR, Zhang YG, Wang Q (2021) miR-9-5p Mediates ABCC1 to Elevate the Sensitivity of Glioma Cells to Temozolomide. Front Oncol 11:661653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrà F, Agnoletto C, Minotti L, Baldassari F, Volinia S (2018) The network of non-coding RNAs in cancer drug resistance. Front Oncol 8:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cree IA, Charlton P (2017) Molecular chess? Hallmarks of anti-cancer drug resistance. BMC Cancer 17:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deweese JE, Osheroff N (2009) The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res 37:738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economides MP, McCue D, Borthakur G, Pemmaraju N (2019) Topoisomerase II inhibitors in AML: past, present, and future. Expert Opin Pharmacother 20:1637–1644. [DOI] [PubMed] [Google Scholar]
- Edwardson DW, Narendrula R, Chewchuk S, Mispel-Beyer K, Mapletoft JP, Parissenti AM (2015) Role of drug metabolism in the cytotoxicity and clinical efficacy of anthracyclines. Curr Drug Metab 16:412–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton TS, Yalowich JC (2015) Experimental procedures to identify and validate specific mRNA targets of miRNAs. EXCLI J 14:758–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington F, Willmore E, Tilby MJ, Li L, Li G, Li W, Baguley BC, Austin CA (1999) Murine transgenic cells lacking DNA topoisomerase IIbeta are resistant to acridines and mitoxantrone: analysis of cytotoxicity and cleavable complex formation. Mol Pharmacol 56:1309–1316. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N (2012) The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 19:586–593. [DOI] [PubMed] [Google Scholar]
- Fu LShi JLiu AZhou LJiang MFu HXu KLi DDeng AZhang Q, et al. (2017) A minicircuitry of microRNA-9-1 and RUNX1-RUNX1T1 contributes to leukemogenesis in t(8;21) acute myeloid leukemia. Int J Cancer 140:653–661. [DOI] [PubMed] [Google Scholar]
- Gabra MM, Salmena L (2017) microRNAs and acute myeloid leukemia chemoresistance: a mechanistic overview. Front Oncol 7:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi RN, Ganapathi MK (2013) Mechanisms regulating resistance to inhibitors of topoisomerase II. Front Pharmacol 4:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganger MT, Dietz GD, Ewing SJ (2017) A common base method for analysis of qPCR data and the application of simple blocking in qPCR experiments. BMC Bioinformatics 18:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Huang KC, Yamasaki EF, Chan KK, Chohan L, Snapka RM (1999) XK469, a selective topoisomerase IIbeta poison. Proc Natl Acad Sci USA 96:12168–12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyori BM, Venkatachalam G, Thiagarajan PS, Hsu D, Clement MV (2014) OpenComet: an automated tool for comet assay image analysis. Redox Biol 2:457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker I, Gay LA, Yang Y, Hu J, Morse AM, McIntyre LM, Renne R (2012) Ago HITS-CLIP expands understanding of Kaposi’s sarcoma-associated herpesvirus miRNA function in primary effusion lymphomas. PLoS Pathog 8:e1002884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker WG, Slade DL, Drake FH, Parr RL (1991) Mitoxantrone resistance in HL-60 leukemia cells: reduced nuclear topoisomerase II catalytic activity and drug-induced DNA cleavage in association with reduced expression of the topoisomerase II beta isoform. Biochemistry 30:9953–9961. [DOI] [PubMed] [Google Scholar]
- Harker WG, Slade DL, Parr RL, Feldhoff PW, Sullivan DM, Holguin MH (1995) Alterations in the topoisomerase II alpha gene, messenger RNA, and subcellular protein distribution as well as reduced expression of the DNA topoisomerase II beta enzyme in a mitoxantrone-resistant HL-60 human leukemia cell line. Cancer Res 55:1707–1716. [PubMed] [Google Scholar]
- Hatzl SPerfler BWurm SUhl BQuehenberger FEbner STroppmair JReinisch AWölfler ASill H, et al. (2020) Increased Expression of Micro-RNA-23a Mediates Chemoresistance to Cytarabine in Acute Myeloid Leukemia. Cancers (Basel) 12:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson DL, Das SG, Li Y, Xing C (2013) Overexpression of Mcl-1 confers multidrug resistance, whereas topoisomerase IIβ downregulation introduces mitoxantrone-specific drug resistance in acute myeloid leukemia. Mol Pharmacol 84:236–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez VA, Carvajal-Moreno J, Papa JL, Shkolnikov N, Li J, Ozer HG, Yalowich JC, Elton TS(2021) CRISPR/Cas9 Genome Editing of the Human Topoisomerase IIα Intron 19 5' Splice Site Circumvents Etoposide Resistance in Human Leukemia K562 Cells. Mol Pharmacol 99:226–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog CE, Holmes KA, Tuschong LM, Ganapathi R, Zwelling LA (1998) Absence of topoisomerase IIbeta in an amsacrine-resistant human leukemia cell line with mutant topoisomerase IIalpha. Cancer Res 58:5298–5300. [PubMed] [Google Scholar]
- Jonas S, Izaurralde E (2015) Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16:421–433. [DOI] [PubMed] [Google Scholar]
- Kameswaran VBramswig NCMcKenna LBPenn MSchug JHand NJChen YChoi IVourekas AWon KJ, et al. (2014) Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab 19:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagasabai R, Serdar L, Karmahapatra S, Kientz CA, Ellis J, Ritke MK, Elton TS, Yalowich JC (2017) Alternative RNA processing of topoisomerase IIα in etoposide-resistant human leukemia K562 cells: intron retention results in a novel C-terminal truncated 90-kDa isoform. J Pharmacol Exp Ther 360:152–163. [DOI] [PubMed] [Google Scholar]
- Kanagasabai R, Karmahapatra S, Kientz CA, Yu Y, Hernandez VA, Kania EE, Yalowich JC, Elton TS (2018) The novel C-terminal truncated 90-kDa isoform of topoisomerase IIα (TOP2α/90) is a determinant of etoposide resistance in K562 leukemia cells via heterodimerization with the TOP2α/170 isoform. Mol Pharmacol 93:515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania EE, Carvajal-Moreno J, Hernandez VA, English A, Papa JL, Shkolnikov N, Ozer HG, Yilmaz AS, Yalowich JC, Elton TS (2020) hsa-miR-9-3p and hsa-miR-9-5p as Post-Transcriptional Modulators of DNA Topoisomerase IIα in Human Leukemia K562 Cells with Acquired Resistance to Etoposide. Mol Pharmacol 97:159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagkouni DParaskevopoulou MDChatzopoulos SVlachos ISTastsoglou SKanellos IPapadimitriou DKavakiotis IManiou SSkoufos G, et al. (2018) DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 46 (D1):D239–D245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann SH, Svingen PA (1999) Immunoblot analysis and band depletion assays. Methods Mol Biol 94:253–268. [DOI] [PubMed] [Google Scholar]
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS (2009) Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol 27:549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11:597–610. [DOI] [PubMed] [Google Scholar]
- Löwenberg BOssenkoppele GJvan Putten WSchouten HCGraux CFerrant ASonneveld PMaertens JJongen-Lavrencic Mvon Lilienfeld-Toal M, et al. ; Dutch-Belgian Cooperative Trial Group for Hemato-Oncology (HOVON); German AML Study Group (AMLSG); Swiss Group for Clinical Cancer Research (SAKK) Collaborative Group (2009) High-dose daunorubicin in older patients with acute myeloid leukemia. N Engl J Med 361:1235–1248. [DOI] [PubMed] [Google Scholar]
- Marima R, Francies FZ, Hull R, Molefi T, Oyomno M, Khanyile R, Mbatha S, Mabongo M, Owen Bates D, Dlamini Z (2021) MicroRNA and Alternative mRNA Splicing Events in Cancer Drug Response/Resistance: Potent Therapeutic Targets [published correction appears in N Engl J Med (2010) 362:1155]. Biomedicines 9:1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensah-Osman E, Al-Katib A, Dandashi M, Mohammad R (2003) XK469, a topo IIbeta inhibitor, induces apoptosis in Waldenstrom’s macroglobulinemia through multiple pathways. Int J Oncol 23:1637–1644. [PubMed] [Google Scholar]
- Mirski SE, Sparks KE, Yu Q, Lang AJ, Jain N, Campling BG, Cole SP (2000) A truncated cytoplasmic topoisomerase IIalpha in a drug-resistant lung cancer cell line is encoded by a TOP2A allele with a partial deletion of exon 34. Int J Cancer 85:534–539. [DOI] [PubMed] [Google Scholar]
- Nitiss JL (2009) Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer 9:338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowek KSun SMBullinger LBindels EMExalto CDijkstra MKvan Lom KDöhner HErkeland SJLöwenberg B, et al. (2016) Aberrant expression of miR-9/9* in myeloid progenitors inhibits neutrophil differentiation by post-transcriptional regulation of ERG. Leukemia 30:229–237. [DOI] [PubMed] [Google Scholar]
- Nowek K, Wiemer EAC, Jongen-Lavrencic M (2018) The versatile nature of miR-9/9* in human cancer. Oncotarget 9:20838–20854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J, Hayder H, Zayed Y, Peng C (2018) Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) 9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive PL (2002) The comet assay. An overview of techniques. Methods Mol Biol 203:179–194. [DOI] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL (2008) The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci 28:14341–14346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG (2013) DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 41:W169–W173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y, Croce CM (2019) Noncoding RNA genes in cancer pathogenesis. Adv Biol Regul 71:219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati P, Nitti D, Mocellin S (2012) Cancer resistance to type II topoisomerase inhibitors. Curr Med Chem 19:3900–3906. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Marchand C (2011) Interfacial inhibitors: targeting macromolecular complexes. Nat Rev Drug Discov 11:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Sun Y, Huang SN, Nitiss JL (2016) Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol 17:703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Nussenzweig A, Takeda S, Austin C (2022) Human topoisomerases and their roles in genome stability and organization. Nat Rev Mol Cell Biol 23:407–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuwanshi S, Gutti U, Kandi R, Gutti RK (2018) MicroRNA-9 promotes cell proliferation by regulating RUNX1 expression in human megakaryocyte development. Cell Prolif 51:e12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritke MK, Yalowich JC (1993) Altered gene expression in human leukemia K562 cells selected for resistance to etoposide. Biochem Pharmacol 46:2007–2020. [DOI] [PubMed] [Google Scholar]
- Ritke MK, Roberts D, Allan WP, Raymond J, Bergoltz VV, Yalowich JC (1994) Altered stability of etoposide-induced topoisomerase II-DNA complexes in resistant human leukaemia K562 cells. Br J Cancer 69:687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- Schraivogel DWeinmann LBeier DTabatabai GEichner AZhu JYAnton MSixt MWeller MBeier CP, et al. (2011) CAMTA1 is a novel tumour suppressor regulated by miR-9/9* in glioblastoma stem cells. EMBO J 30:4309–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag S, Ambinder RF (2018) Hodgkin lymphoma: A review and update on recent progress. CA Cancer J Clin 68:116–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan SAbdelmohsen KLee EKTominaga KSubaran SSKuwano YKulshrestha RPanchakshari RKim HHYang X, et al. (2011) Translational control of TOP2A influences doxorubicin efficacy. Mol Cell Biol 31:3790–3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Rui K, Tang X, Ma J, Wang Y, Tian X, Zhang Y, Xu H, Lu L, Wang S (2015) MicroRNA-9 regulates the differentiation and function of myeloid-derived suppressor cells via targeting Runx1. J Immunol 195:1301–1311. [DOI] [PubMed] [Google Scholar]
- Treiber T, Treiber N, Meister G (2019) Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol 20:5–20. [DOI] [PubMed] [Google Scholar]
- Vlasova II, Feng WH, Goff JP, Giorgianni A, Do D, Gollin SM, Lewis DW, Kagan VE, Yalowich JC (2011) Myeloperoxidase-dependent oxidation of etoposide in human myeloid progenitor CD34+ cells. Mol Pharmacol 79:479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LSLi LLi LChu SShiang KDLi MSun HYXu JXiao FJSun G, et al. (2015) MicroRNA-486 regulates normal erythropoiesis and enhances growth and modulates drug response in CML progenitors. Blood 125:1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Cheng M, Zheng X, Zheng L, Liu H, Lu J, Liu Y, Chen W (2020) Interactions Between lncRNA TUG1 and miR-9-5p Modulate the Resistance of Breast Cancer Cells to Doxorubicin by Regulating eIF5A2. OncoTargets Ther 13:13159–13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dong L, Wan F, Chen F, Liu D, Chen D, Long J (2021) MiR-9-3p regulates the biological functions and drug resistance of gemcitabine-treated breast cancer cells and affects tumor growth through targeting MTDH. Cell Death Dis 12:861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woessner RD, Mattern MR, Mirabelli CK, Johnson RK, Drake FH (1991) Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ 2:209–214. [PubMed] [Google Scholar]
- Yuva-Aydemir Y, Simkin A, Gascon E, Gao FB (2011) MicroRNA-9: functional evolution of a conserved small regulatory RNA. RNA Biol 8:557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZW, Zhong LP, Ji T, Zhang P, Chen WT, Zhang CP (2010) MicroRNAs contribute to the chemoresistance of cisplatin in tongue squamous cell carcinoma lines. Oral Oncol 46:317–322. [DOI] [PubMed] [Google Scholar]
- Zahreddine H, Borden KL (2013) Mechanisms and insights into drug resistance in cancer. Front Pharmacol 4:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Luo XQ, Feng DD, Zhang XJ, Wu J, Zheng YS, Chen X, Xu L, Chen YQ (2011) Upregulation of microRNA-125b contributes to leukemogenesis and increases drug resistance in pediatric acute promyelocytic leukemia. Mol Cancer 10:108. [DOI] [PMC free article] [PubMed] [Google Scholar]