Abstract
A central response to insufficient cerebral oxygen delivery is a profound reprograming of metabolism, which is mainly regulated by the Hypoxia Inducible Factor (HIF). Among other responses, HIF induces the expression of the atypical mitochondrial subunit NDUFA4L2. Surprisingly, NDUFA4L2 is constitutively expressed in the brain in non-hypoxic conditions. Analysis of publicly available single cell transcriptomic (scRNA-seq) data sets coupled with high-resolution multiplexed fluorescence RNA in situ hybridization (RNA F.I.S.H.) revealed that in the murine and human brain NDUFA4L2 is exclusively expressed in mural cells with the highest levels found in pericytes and declining along the arteriole-arterial smooth muscle cell axis. This pattern was mirrored by COX4I2, another atypical mitochondrial subunit. High NDUFA4L2 expression was also observed in human brain pericytes in vitro, decreasing when pericytes are muscularized and further induced by HIF stabilization in a PHD2/PHD3 dependent manner. In vivo, Vhl conditional inactivation in pericyte targeting Ng2-cre transgenic mice dramatically induced NDUFA4L2 expression. Finally NDUFA4L2 inactivation in pericytes increased oxygen consumption and therefore the degree of HIF pathway induction in hypoxia. In conclusion our work reveals that NDUFA4L2 together with COX4I2 is a key hypoxic-induced metabolic marker constitutively expressed in pericytes coupling mitochondrial oxygen consumption and cellular hypoxia response.
Keywords: Brain, HIF, NDUFA4L2, oxygen, pericyte
Introduction
Mammalian brain’s oxygen requirements are extremely high compared to other organs. In humans, the brain consumes approximately 20% of the body's oxygen derived energy although it only accounts for 2% of body’s weight. 1 Because oxygen cannot be stored, a drop in oxygen levels below normal range immediately triggers a hyperemic response coupled with redistribution of blood flow, thereby increasing cerebral oxygen availability.2–4 When this response fails to restore cerebral oxygen homeostasis, an evolutionary conserved transcriptional and metabolic program to adapt to hypoxia comes into play. This adaptive response is mainly mediated by the hypoxia-inducible factor (HIF) pathway.5–7 HIFs are heterodimeric basic helix-loop-helix (bHLH) transcription factors which consist of an oxygen-regulated α subunit (either HIF1α, HIF2α or HIF3α) and one stable β subunit (HIF1β), also known as the aryl hydrocarbon receptor nuclear translocator (ARNT). 8 The stability of the alpha subunits is controlled by the prolyl-4-hydroxylase domain dioxygenases PHD1, PHD2 and PHD3 through the hydroxylation of specific proline residues of the HIFα subunits in an oxygen and iron-dependent manner.9–12 Under normoxia, the hydroxylated HIFα subunit is targeted by the von Hippel-Lindau (VHL)-E3 ubiquitin ligase complex and degraded via proteasome.9,13 Under hypoxia, this hydroxylation is impaired, the HIFα subunits are not degraded and dimerize with the β subunit in the nucleus, binding to DNA at hypoxia response elements (HREs) and promoting the transcription of genes involved in the regulation of processes that will help the cell to adapt to inadequate levels of oxygenation.5,6 Among the transcriptional program elicited by HIF, the repression and optimization of mitochondrial oxygen consumption which results in a restoration of intracellular oxygen levels has been shown to be mainly mediated by HIF1.14–16 Along this line, the NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 4-like 2 (NDUFA4L2) gene is dramatically induced by HIF1 to repress mitochondrial oxygen consumption. 17 NDUFA4L2 is an atypical paralogue of NDUFA4 that affects mitochondrial oxygen consumption by repressing the mitochondrial complex 1.17 While NDUFA4 is the main isoform present in most cells and NDUFA4L2 is undetectable in many cell lines unless exposed to hypoxia, NDUFA4L2 has been reported to be expressed at low levels in the brain under normoxia. 17 The identity of the NDUFA4L2 expressing cells and the physiological function of such basal levels still remain to be elucidated.
Because understanding cerebral oxygenation is essential to comprehend the brain physiology we sought to identify the NDUFA4L2 expressing cells and investigate the significance of NDUFA4L2 expression in those cells. To this end we first applied single cell transcriptomic (scRNA-seq) analysis from public data sets coupled with high-resolution multiplexed fluorescence RNA in situ hybridization (RNA F.I.S.H.) in mouse brain and found that Ndufa4l2 together with another atypical mitochondrial subunit, Cox4i2, is specifically expressed in pericytes and vascular smooth muscle cells and inversely related to the degree of mural cell muscularization. We then applied single cell transcriptomics analysis from public data sets to human brain samples confirming that NDUFA4L2 is specifically expressed in cerebral pericytes similar to the results observed in mice. Moreover, we found that the high expression of NDUFA4L2 observed in vivo is retained in human brain pericytes in vitro and that it can be further increased by exposure to hypoxia or concomitant genetic inactivation of PHD2 and PHD3 in a HIF1 dependent manner. We expanded these results to mice by conditional inactivating Vhl in Ng2-cre transgenic mice, which stabilize HIF levels in cerebral pericytes, dramatically increasing NDUFA4L2. Finally, NDUFA4L2 deletion increased oxygen consumption which in turn potentiated the effects of hypoxia on human brain pericytes.
In summary, our studies demonstrate that NDUFA4L2 is a specific metabolic marker of pericytes in murine and human brain both in vivo and in vitro and suggest that regulates the response of pericytes to hypoxia. These results together with our previous work point out to pericytes as main players in the oxygen sensing in the brain.
Materials and methods
Generation and genotyping of mice
The generation and genotyping of mice expressing Ng2-cre and floxed alleles for Vhl is described elsewhere.18,19 Briefly, mice carrying floxed alleles were bred with Ng2-cre female mice. Ng2-cre Vhlflox/flox are referred to as NG2-Vhl−/−. Male and female mice aged 9–10 weeks were used for the analysis. For mouse genotyping, ear biopsies were used. The following primers were used for DNA PCR: Ng2-Cre (forward: 5′-AAATCTAAGCGCGGGTCTG-3′; reverse: 5′-GCAAACGGACAGAAGCATTT-3′), Vhl (forward: 5′-GCCCGGCCCCTACCAGTG-3′′; reverse: 5′-TCTGTCTTGGCCTCCTGAGT-3′). All experimental procedures involving mice were first approved by the research ethics committee at the Autonomous University of Madrid (UAM), and they were carried out under the supervision of animal welfare responsible at the UAM in accordance with Spanish RD 53/2013, European (EU Directive 2010/63/EU) and with the ARRIVE guidelines.
RNA extraction and analysis
RNA was isolated using TRItidi G™ (A4051, PanReac AppliChem, Chicago, IL, USA) according to the manufacturer’s instructions. 0,5–1 µg RNA was then reverse – transcribed for cDNA synthesis using GoScript™ Reverse Transcriptase Kit (A5004, Promega, Madison, WI, USA). PCR amplifications were then performed using the SYBR™ Power SYBR™ Green PCR Master Mix (4368708, Applied Biosystems™, Waltham, MA, USA) in a QuantStudio 5 Real-Time PCR system (Applied Biosystems™, Waltham, MA, USA). A detailed list of primer sequences used for qPCR can be found in Supplemental Materials and Methods (Supplemental Table 1). Transcripts of interest were normalized by 28S.
Cell culture
Human brain pericytes (P10363), human astrocytes (P10251), human cortical neurons (P10152) and human brain microvascular endothelial cells (P10361) were obtained from Innoprot (Bizkaia, Spain) and maintained in complete pericyte medium (P60121), complete astrocyte medium (P60101), complete neuronal medium (P60157) and endothelial cell medium (P60104), all from Innoprot (Bizkaia, Spain). Human brain pericytes and human brain microvascular endothelial cells were seeded in plates covered with 0.1% Gelatin Type B from Bovine Skin (G9382, Sigma-Aldrich, San Luis, MO, USA). Human astrocytes and human cortical neurons were seeded in Poly-L-lysine hydrobromide covered plates (P6282, Sigma-Aldrich, San Luis, MO, USA). Cells were maintained at 37°C in an atmosphere with 5% CO2 and 95% air. For hypoxic conditions cells were maintained in 1% O2 for 16 hours. DMOG (dimethyloxaloylglycine) (BML-EI347, Enzo Life Sciences, Inc. Farmingdale, NY, USA), a pan prolyl-4-hydroxylase inhibitor, was used at 0.1 mM for 16 hours.
Gene silencing
Temporary gene silencing was carried out using the following siRNAs: PHD2 siRNA (sc-45537), PHD3 siRNA (sc-45799), HIF1a siRNA (sc-35561) and NDUFA4L2 siRNA (sc-95677), all of them from Santa Cruz Biotechnology (Dallas, TX, USA). As control, cells were transfected with siRNA consisting of a scrambled sequence (sc-37007, Santa Cruz Biotechnology, Dallas, TX, USA). Human brain pericytes were seeded in 0.1% gelatin coated p60 plates and then transfected using Lipofectamine 2000 (10696153, Invitrogen, Waltham, MA, USA) with 80 nM to 120 nM of the corresponding siRNA. Transfected cells were cultured for 72 hours, and then collected and stored until processing time.
Cellular oxygen consumption analysis
2 × 104/well scramble siRNA and NDUFA4L2 siRNA transfected human brain pericytes were seeded one day before the experiment was performed. The day the experiment was carried out, cells were washed and preincubated for 1 hour at 37°C with Pericyte Medium without FBS and CO2. Oxygen consumption rate (OCR) was measured using an XF24 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA) according to the manufacturer’s instructions. Spare respiratory capacity was calculated as the difference between maximal respiration obtained after the addition of 1 µM Carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) and the basal respiration (OCRmax–OCRbasal). After cellular oxygen consumption experiment, cells were homogenized in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl pH 8.0) supplemented with 5% protease (P8340, Sigma-Aldrich, San Luis, MO, USA) and 1% phosphatase inhibitors (P5726, Sigma-Aldrich, San Luis, MO, USA). Protein levels were quantified after the homogenization of the samples with DC protein Assay kit (5000112, Bio-Rad, Hercules, CA, USA).
Cell survival assay
scramble siRNA or NDUFA4L2 siRNA transfected human brain pericytes were exposed to normoxia or hypoxia (1% O2), trypsinized, collected in medium, centrifuged, stained with 0.4% Trypan Blue and counted with an EVE™ Plus automated cell counter (NanoEnTek, Guro-gu, South Korea). Alternatively, cells were washed with PBS1X and stained with 1:200 Ghost Dye Red 780 viability probe (TONBO biosciences, San Diego, CA, USA) in PBS on ice for 15 minutes and fixed with 4% paraformaldehyde (4% PFA). Samples were analyzed by flow cytometry using a FACSCanto™ II flow cytometer (BD Biosciences, Franklin Lakes, NJ, EEUU) flow cytometer and the data were analyzed with the BD FACSDiva™ software and BD FlowJoTM v10.7 software (Tree Star, Ashland, OR, USA).
Cell proliferation analysis
scramble siRNA and NDUFA4L2 siRNA transfected human brain pericytes were grown in normoxic or hypoxic conditions (1% O2) for 72 hours, washed and fixed in 4% paraformaldehyde (4% PFA) for at least 24 hours, stained with 0.025% Crystal Violet solution (11435027, Fisher Scientific, Hampton, NH, USA) for 20 minutes and washed several times with distilled H2O. Plates were then dried at room temperature and after methanol was added to the stained wells, the optical density was measured at 540 nm.
Western blotting
Mouse cerebral cortex was homogenized and sonicated in RIPA buffer supplemented with 5% protease and 1% phosphatase inhibitors, centrifuged at 1000 g for 10 minutes to remove debris and insoluble material and the supernatant was collected. Protein levels were measured using a DC protein Assay kit (5000112, Bio-Rad, Hercules, CA, USA) and denatured in Laemmli buffer. In vitro cultured cells were directly homogenized and denatured in Laemmli buffer. Samples were loaded in an SDS-polyacrylamide gel and transferred to a 0.45 µm (10600007) or 0.2 µm (10600094) pore size nitrocellulose membranes (Cytiva, Marlborough, MA, USA). Membranes were blocked and probed with antibodies detailed in Supplemental Materials and Methods (Supplemental Table 2). Immunoreactivity was detected by enhanced chemiluminescence using Clarity Western ECL Substrate (1705061, Bio-Rad, Hercules, CA, USA) or Super-Signal West Femto Maximum Sensitivity Substrate (34096, Thermo Scientific, Waltham, MA, USA) and visualized on a digital luminescent image analyzer, Image Quant LAS4000 Mini (GE Healthcare, Chicago, IL, USA). Images were densitometrically analyzed with Image J (NIH image analysis software ImageJ; version 1.47). Protein levels of mitochondrial proteins NDUFA4L2, NDUFA4 and PDK1 were normalized to PORIN and HIF1α and PGK1 levels were normalized to β-ACTIN.
RNA FISH studies
The RNA scope multiplex fluorescent kit was used for the detection of Ndufa4l2, Cox4i2, Pdgfrb, Vtn and Acta2 transcripts in frozen 15 μm thick brain sections as per the manufacturer’s protocol (Advanced Cell Diagnostics, Hayward, CA, USA). Slides were scanned in a Leica TCS-SP5 (Leica Microsystems, Wetzlar, Germany) confocal microscope. A total of 2226 cells from 3 to 7 region of interests (ROI) were analyzed. The proportion of Pdgfrb+ cells that co-expressed Ndufa4l2 or Cox4i2 was determined by dividing the number of Ndufa4l2+ or Cox4i2+Pdgfrb+ cells by the total number of Pdgfrb+ cells. The proportion of Ndufa4l2+ or Cox4i2+ cells that co-expressed Pdgfrb was determined by dividing the number of Ndufa4l2+ or Cox4i2+Pdgfrb+ cells by the total number of Ndufa4l2+ or Cox4i2+ cells. The proportion of Vtn+ cells that co-expressed Ndufa4l2 and the proportion of Ndufa4l2+ that co-expressed Vtn+ was analyzed in a similar way.
scRNAseq analysis
Raw counts from Zeisel et al. 20 were processed using scater 21 (version 1.18.6) and scran 21 (version 1.18.5) packages for R (version 4.0.3). Raw counts were normalized by deconvoluting library size factor and then log-transforming the reads to finally obtain log-normalized counts for downstream analysis. We performed pairwise Welch t-tests between pericytes and every other cell type to get genes that are specifically upregulated in pericytes compared to any other cluster of the data set. In order to further analyze pericytes and vascular smooth muscle cells we subset these cells based on the Taxonomy Group previously annotated by the original authors. Then, we selected high variable genes based on Poisson variance to perform Principal Component Analysis (PCA) and dimensionality reduction by t-distributed stochastic neighbor embedding (tSNE). For Pearson’s correlation analysis we used the log counts of the selected genes in every cell marked as pericytes or vascular smooth muscle cells. To analyze the additional data sets (Vanlandewijck et al. 22 and Allen Institute: https://portal.brain-map.org/atlases-and-data/rnaseq/human-multiple-cortical-areas-smart-seq) we used the normalized values provided by the authors and directly represented them using the corresponding graph. URLs to access the data sets can be found in supplemental materials. Different cell types from Allen Institute data set were named following the Common Cell Type Nomenclature, or CCN. First, name denotes cell type with the exception of neurons that are subdivided in excitatory or inhibitory neurons (exc/inh), followed by the cortical layer the cell body is localized in and a canonical gene. For clarity, we have included the whole name of non-neuronal cell types.
In vivo transient MCAO ischemia model and Infarct volume and edema quantification
This model has previously been established in our group as previously described. 23 Briefly, mice were anesthetized with isoflurane (0.8% oxygen), placed on a heating-pad and maintained at 36.7°C using a rectal probe (Cibertec, Spain). Transient cerebral ischemia was performed under a surgical microscope (Tecnoscopio OPMI pico, Carl Zeiss, Meditec Iberia SA, Spain) by the isolation and permanent ligation of the right common and external carotid arteries. A silicon rubber-coated monofilament (6023910PK10, Doccol Corporation, Sharon, MA, USA) was inserted through a small incision in the common carotid artery and advanced into the internal carotid artery until the tip of the filament was precisely located at the origin of the right middle cerebral artery, blocking the blood flow. The filament was held in place for one hour of ischemia and then carefully removed to initiate the reperfusion. Animals were kept under anaesthesia throughout the procedure and carefully sutured after surgery. Recovery occurred in a temperature-controlled cage in which mice were given postoperative analgesia (0.5% lidocaine, subcutaneously, s.c.). Surgery time per animal did not exceed 15 minutes. Following euthanasia, brains were quickly removed and cut into four 2-mm-thick coronal sections using a mouse brain slice matrix (Harvard Apparatus, Spain). Sections were stained for 15 min at room temperature with 2% 2,3,5-triphenyltetrazolium chloride (TTC) in PBS to visualize infarctions. Indirect infarct volumes were calculated by volumetry (ImageJ software) according to the following equation: Vindirect (mm3) = Vinfarct × (1−[Vih–Vch]/Vch), where the term (Vih-Vch]) represents the volume difference between the ischemic hemisphere and the control hemisphere and (Vih-Vch]/Vch expresses this difference as a percentage of the control hemisphere.
Statistical analysis
Data are reported as mean ± SD. Data collection and analyses were not performed blindly to the conditions of the experiments. To asses data distribution, Shapiro-Wilk normality test was performed. Statistical analyses were performed with Prism 9 software (GraphPad Software, Inc., La Jolla, CA, USA) using the unpaired two-tailed Student's t-test with Welch’s correction as needed, or a one-way or two way analysis of variance (ANOVA) followed by Tukey’s post hoc analysis for multi-group comparisons. P values < 0.05 were considered statistically significant.
Results
NDUFA4L2 expression is confined to pericytes and vascular smooth mucle cells in the murine brain
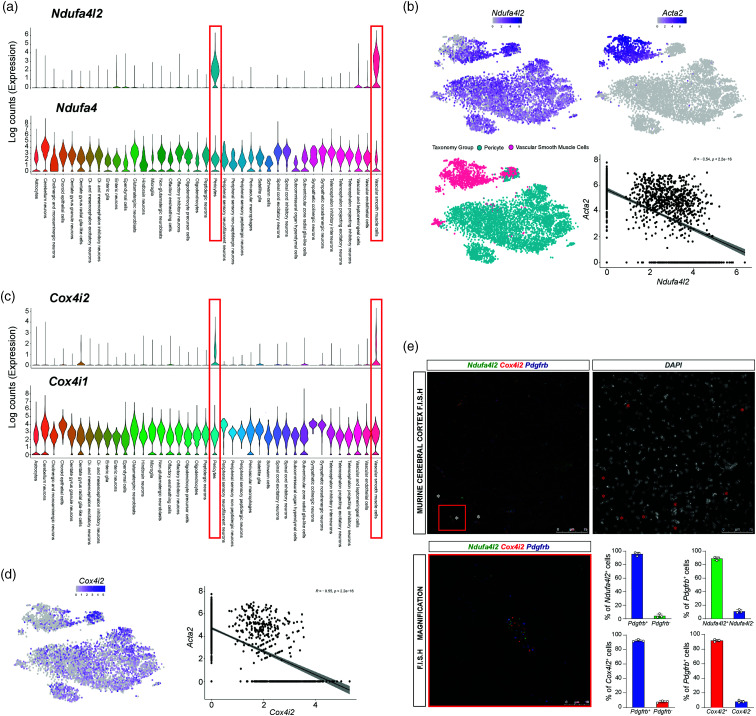
In order to identify which cells express NDUFA4L2 in the normal adult mouse brain, we began by exploring one of the most comprehensive publicly available scRNA-seq data set published to date, 20 with more than 160000 single cell transcriptomes including all main cell types of the brain, i.e neurons, glia, oligodendrocytes, inmune and vascular cells. After normalizing and log-transforming the raw counts based on library size factors we segmented the data in 39 different clusters according to the original annotation provided by the authors. To our surprise, Ndufa4l2 transcript was enriched in two closely related cell types, pericytes and vascular smooth muscle cells (Figure 1 (a)). Pericytes and vascular smooth muscle cells surround capilaries and arterioles and arteries respectively and altogether comprise the so called mural cells 24 which modulate endothelial cell behaviour.25,26 Interestingly, endothelium related clusters showed negligible expression of Ndufa4l2 (Figure 1 (a)). On the other hand, its paralog Ndufa4 was consistently expressed across all populations in the data set (Figure 1(a)). While pericytes and vascular smooth muscle cells share molecular markers such as platelet-derived growth factor receptor β (PDGFRB) and neural/glial antigen 2 (NG2) they differ in their morphology, location and functions and are usually distinguised by the expression of the protein α smooth muscle actin or α-SMA (encoded by the gene Acta2), since pericytes do not express it.24,27,28 To ascertain whether there is a relationship between the degree of cell muscularization (reflected by Acta2 expression) and Ndufa4l2 levels we co-related both parameters. As shown in the Figure 1(b), we found an inverse correlation between Acta2 and Ndufa4l2 levels (Pearson R = −0.54), indicating that the less muscularized the cell, the more Ndufa4l2 expresses.
Figure 1.
Ndufa4l2 and Cox4i2 expression is confined to vascular smooth muscle cells and pericytes in mouse brain. (a) Violin plots showing normalized log counts of Ndufa4l2 and Ndufa4 expression within 39 clusters identified in the mouse nervous system. (b) TSNE plots showing Ndufa4l2 and Acta2 expression (top panels), color-coded Taxonomy group (bottom left panel) and scatter plots showing Pearson’s correlation between Ndufa4l2 and Acta2 expression (bottom right panel). (c) Violin plots showing normalized log counts of Cox4i2 and Cox4i1 expression within the 39 clusters previously identified. (d) TSNE plots showing Cox4i2 expression and scatter plots showing Pearson’s. Data were extracted and reanalyzed from scRNA-seq data set (Zeisel et al. 20 ) and (e) (Left) Representative images of multiplex RNA F.I.S.H. of mouse cerebral cortex detecting Ndufa4l2 (green fluorescent signal), Cox4i2 (red fluorescent signal) and Pdgfrb-expressing cells (blue fluorescent signal) (n = 3). White asterisks depict examples of cells positive for the 3 transcripts. (Right) DAPI nuclear staining corresponding to the left image. Red asterisks depict the same cells positive for the 3 transcripts from the left image. Lower panels show magnified an example of a Pdgfrb-expressing cell positive for Ndufa4l2 and Cox4i2 and the percentage of Ndufa4l2 or Cox4i2+ and Ndufa4l2 or Cox4i2− expressing Pdgfrb mRNA, and the percentage of Pdgfrb+ and Pdgfrb− expressing Ndufa4l2 or Cox4i2 mRNA (n = 3). Data are represented as mean ± SD.
To validate these results we explored another publicly available scRNA-seq data set with special focus in the vasculature of the adult mouse brain, 22 which in agreement with our data showed that Ndufa4l2 transcript was specifically found in mural cells, with pericytes and venular smooth muscle cells (which express significantly less Acta2 than their arteriolar counterparts) showing the strongest expression (Figure S1A). This expression gradually diminished along the arteriole-arterial smooth muscle cell axis supporting the inverse correlation between Acta2 and Ndufa4l2 levels (Figure 1(b)) and identifying the pericytes as the main contributors to NDUFA4L2 expression in the brain.
COX4I2 is another atipical mitochondrial subunit specifically expressed in brain pericytes and vascular smooth muscle cells
We next applied an unbiased differentially expressed analysis of upregulated genes in pericytes compared to the rest of the clusters represented, which revealed Ndufa4l2 as the 16th top upregulated gene in pericytes (based on log-FoldChange (logFC) and significant False Discovery Rate (FDR) values). As shown in figure S1B, Ndufa4l2 shows an average logFC ∼ 2 in pericytes compared to any other population in the data set. To our surprise, we found that Cox4i2, another atipical mitochondrial subunit induced by HIF, 29 also scored among the most differentially expressed genes between pericytes and vascular smooth muscle cells and the rest of the cells in the murine brain while its paralog Cox4i1 was consistently expressed across all clusters (Figure 1 (c) and S1C). Interestingly, Cox4i2 expression shared a similar pattern of capillary/arteriole/artery zonation with Ndufa4l2 ( Figure 1 (d)), suggesting that the expression of atipical subunits of the mitochondria is a common feature of capillary pericytes in the brain. To unequivocally confirm the mural cell specificity of these transcripts, we performed high-resolution multiplexed fluorescence RNA in situ hybridization (RNA F.I.S.H.) in cortex from 3 mice with specific probes for Cox4i2, Ndufa4l2 and Pdgfrb, which is a well-recognized molecular marker of pericytes and vascular smooth muscle cells in the brain.30,31 As shown in the Figure 1(e), 95.77 ± 2.8% of Ndufa4l2 and 92.3 ± 1.49% of Cox4i2 transcripts were confined to Pdgfrb-positive cells, which were positive for Ndufa4l2 and Cox4i2 transcripts in the 89.1 ± 4.3% and 91.83 ± 1.8% of the cases, respectively. Moreover, we found that Ndufa4l2 and Cox4i2 positive signals were intimately associated as most of the cells expressed both transcripts at the same time. To further confirm these results, we performed multiplex RNA F.I.S.H of Ndufa4l2 and 2 different markers of mural cells, the mouse specific pericytes marker vitronectin (Vtn) and the smooth muscle cell marker Acta2 (Figure S2A and B). In line with the single cell analysis and RNA F.I.S.H data, Ndufa4l2 positive signal was predominantly associated with Vtn expressing cells while a less clear signal was associated with Acta2 expressing cells (Figure S2B). Altogether, these results demonstrate that the atypical mitochondrial subunits NDUFA4L2 and COX4I2 are specific molecular markers of murine mural cells in the brain, most robustly expressed by pericytes and gradually declining along the precapillary/arteriolar/arterial axis.
NDUFA4L2 and COX4I2 expression in pericytes is conserved in the human brain
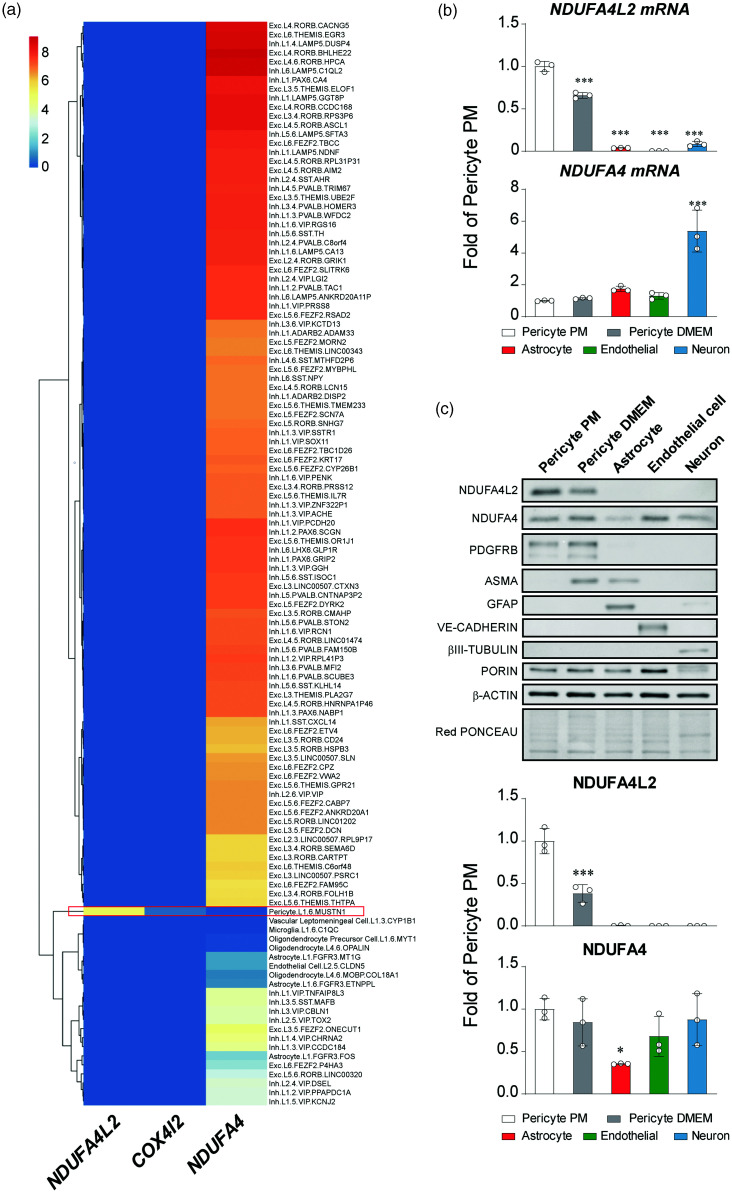
Because a molecular marker is as good as it is conserved between species we next explored scRNA-seq data sets obtained from multiple human cortical areas from the Allen Institute which include single-nucleus transcriptomes from 49,495 nuclei. After proccessing the trimmed means provided by the authors (i.e. gene expression aggregated per cell type, we found that NDUFA4L2 expression was restricted to the cluster identified as pericyte (Figure 2(a) and S3A). We confirmed these results by assesing the expression of NDUFA4L2 in pericytes in 4 other different studies with scRNA-seq data available that encompass brain samples from development into the adulthood (data not shown).32–35 We also investigated the expression of COX4I2, which we have identified in mice as specifically expressed in mural cells. Interestingly we found that the expresssion of COX4I2 in the human brain was weak, which is a plausible explanation for why we could only detect it in 3 of the 5 scRNA-seq data sets interrogated. Nevertheless, when detectable, it was confined to the pericyte/mural cell cluster (Figure 2(a) and references33,34).
Figure 2.
NDUFA4L2 expression is confined to vascular smooth muscle cells and pericytes in human brain in vivo an in vitro. (a) Heatmap showing the normalized trimmed means (gene expression aggregated per cell type) of NDUFA4L2, COX4I2 and NDUFA4 within 120 clusters identified in the Allen Multiple Cortical Areas – SMART-seq data set. Red rectangle marks pericytes cluster. (b) Relative mRNA levels of NDUFA4L2 and NDUFA4 in primary cultures of different cell types of human brain (n = 3) and (c) Representative western blot of NDUFA4L2 and NDUFA4 in primary cultures of different cell types of human brain and molecular markers of each cell type: PDGFRB for pericytes and Smooth muscle cells, αSMA for Smooth muscle cells, GFAP for astrocytes, VE-CADHERIN for endothelial cells and β-III TUBULIN for neurons. PORIN was used as mitochondrial loading control; β-ACTIN and Red PONCEAU were used as loading control. Below is shown quantification of NDUFA4L2 and NDUFA4 signals normalized by PORIN (n = 3). Data are represented as mean ± SD; Statistical analysis was performed using one-way ANOVA followed by Tukey’s post hoc analysis; *P < 0.05, ***P < 0.001, compared with Pericyte PM.
NDUFA4L2 expression but not COX4I2 is retained in human brain pericytes in vitro
In order to test if the expression pattern observed in the scRNA-seq data set was retained in vitro, we analized NDUFA4L2 expression in several human brain primary cells; pericytes, astrocytes, microvascular endothelial cells and cortical neurons. To explore the effect of muscularization in mural cells on the expression levels of NDUFA4L2 we also cultured human brain pericytes in DMEM + 10% FBS because pericytes in vitro acquire a phenotype resembling to vascular smooth muscle cells when cultured in these conditions36,37 (Figure S3B). In line with the previously interrogated scRNA-seq data, human brain pericytes showed a robust NDUFA4L2 expression by mRNA and protein (Figure 2(b) and (c)). Moreover, its expression decreases as the pericyte muscularizes, somehow mimicking in vitro the gradation observed in vivo along the precapillary/arteriolar/arterial axis (Figure 2(b) and (c)). Of note, we did not detect NDUFA4L2 in astrocytes, microvascular endothelial cells or cortical neurons although we did detect a clear positive signal corresponding to NDUFA4 in every cell type tested (Figure 2(b) and (c)). Surprisingly we could not detect mRNA or protein of COX4I2 in our in vitro settings (data not shown), posibly reflecting the low quantity observed in the scRNA-seq data sets. We cannot exclude however the possibility that human brain pericytes lose COX4I2 when cultured.
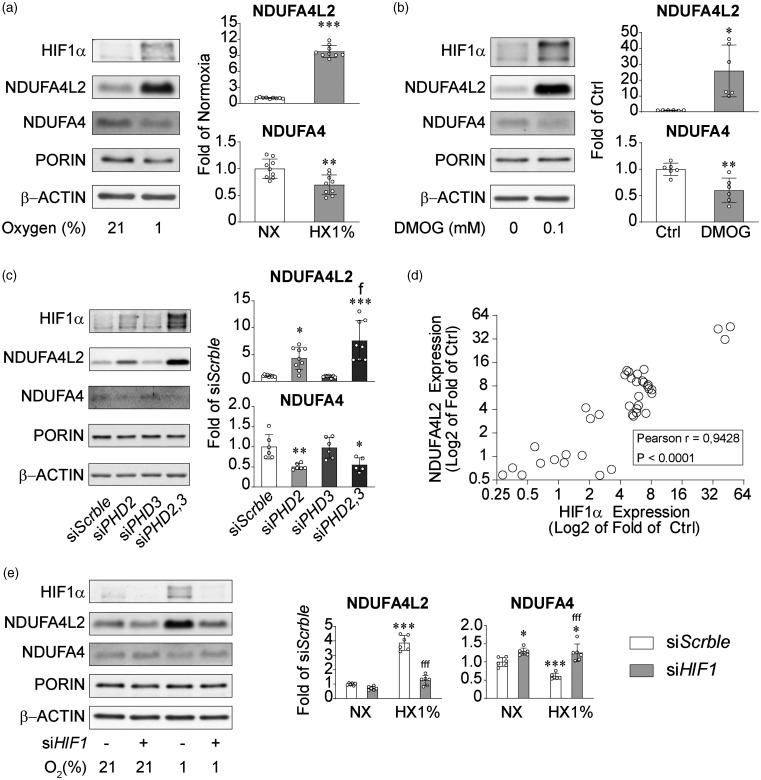
Hypoxic induction of NDUFA4L2 in pericytes in vitro is controlled by HIF1 and PHD2/3
We next evaluated the potential of human brain pericytes to increase NDUFA4L2 levels under hypoxia. Although their basal levels are notably high, exposure to environmental (1% O2) or chemical hypoxia (DMOG, 100 µM) for 16 hours robustly increased NDUFA4L2 protein (Figure 3(a) and (b) upper graph bars). In parallel, levels of its paralog NDUFA4, which were already low, decreased even more (Figure 3(a) and (b) lower bar graphs). HIF stability is controlled by HIF-prolyl hydroxilases 7 and PHD2 inactivation alone can stabilize HIFα in multiple cell types.38,39 Silencing of PHD2 in human brain pericytes (Figure S4A) significantly increased NDUFA4L2 levels while simultaneously decreasing NDUFA4 levels (Figure 3(c)). Although PHD2 is considered the main HIF-prolyl hydroxilase,39,40 we have previously shown that brain pericytes as well as a subset of renal EPO producing cells in the kidney are insensitive to PHD2 inactivation.18,19,41 Because silencing of PHD2 significantly increased the transcript of PHD3 (Figure S4B), we hypothesized that PHD3 acts as a negative feedback compensating the loss of PHD2 function and blunting the HIF mediated response. To test this hypothesis we silenced PHD3 alone or in combination with PHD2 (Figure S4A and S4B). Although inactivation of PHD3 had no effect on NDUFA4L2 or NDUFA4 levels, co-inactivation of PHD2 and PHD3 significantly increased NDUFA4L2 compared with PHD2 silenced cells (Figure 3(c)). Interestingly, NDUFA4 levels were reduced to the same extent by both treatments (Figure 3(c)) suggesting that while NDUFA4 inhibition is mediated by PHD2 activity, NDUFA4L2 activation has multiple levels of induction and is controlled by both PHD2 and PHD3 activity. Along this line, we observed that HIF1 and NDUFA4L2 levels, showed a very robust correlation (Pearson’s r = 0.95) between both variables (Figure 3(d)), indicating that NDUFA4L2 is exquisitely regulated by HIF1 and reflects the degree of HIF1 activity. To probe the causality suggested by the correlational study, we silenced HIF1A in normoxia and hypoxia (1% O2). As shown in Figure 3(e), silencing of HIF1A had little effect on NDUFA4L2 levels in normoxia but completely prevented its induction by hypoxia. On the other hand silencing of HIF1A significantly increased NDUFA4 levels under normoxia and averted hypoxia mediated decrease on its levels (Figure 3(e)). These results demonstrate that in human brain pericytes in vitro, NDUFA4L2 induction by hypoxia is mediated by HIF1α although its basal levels are not. Moreover, our results also indicate that NDUFA4 levels decrease under hypoxia through a PHD2 mediated mechanism and that this suppression is controlled by HIF1α.
Figure 3.
NDUFA4L2 is induced by HIF1 in a PHD2/3 dependent manner. (a and b) Representative western blot and quantification of NDUFA4L2 and NDUFA4 expression in human brain pericytes exposed to (a) 1% O2 (n = 9) or (b) DMOG 0.1 mM (n = 6) for 16 hours. Control cells were exposed to normoxia (21% O2) or vehicle PBS respectively. (c) Representative western blot and quantification of NDUFA4L2 and NDUFA4 expression in transient PHD2-, PHD3- and PHD2/3-silenced human brain pericytes and their corresponding siScrble cells (n = 7–9). (d) Correlation between NDUFA4L2 and HIF1α expression in human brain pericytes exposed to different hypoxic stimulus and (e) Representative western blot and quantification of NDUFA4L2 and NDUFA4 expression in transient HIF1A-silenced human brain pericytes and their corresponding siScrble cells exposed to normoxia (21% O2) or hypoxia (1% O2) (n = 6). Data are represented as mean ± SD; Statistical analysis was performed using two-tailed Student’s t test for (a), (b) or one-way ANOVA followed by Tukey’s post hoc analysis for (c) and (e); *P < 0.05, **P < 0.01, ***P < 0.001, compared with controls; fP < 0.05, fffP < 0.001 compared with siPHD2 cells (c) or hypoxic siScrble (e).
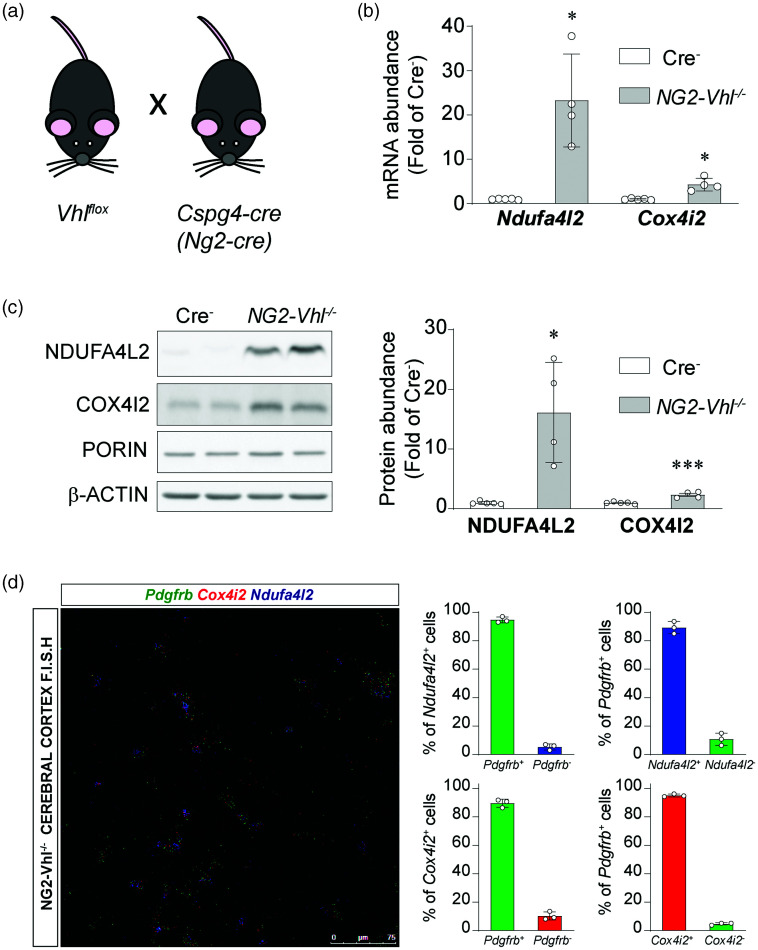
HIF stabilization induces NDUFA4L2 and COX4I2 in pericytes in vivo
We also assesed whether pericytes induce NDUFA4L2 when HIF is stabilized in vivo. To that end we took advantage of transgenic mice that express Cre-recombinase under control of the Ng2 promoter to target pericytes.18,42 To stabilize HIF in these animals, we targeted the VHL tumour suppressor, as its inactivation abrogates HIFα degradation. 5 Thus, we generated Ng2-cre Vhlflox/flox mice (Figure 4(a)), from hereon refered to as NG2-Vhl−/−. NG2-Vhl−/− are characterized by elevated hematocrit and red blood cell counts as a consequence of increased EPO production (data not shown). 18 mRNA analysis of cerebral cortices from adult NG2-Vhl−/− revealed a dramatic increase of 20 fold in Ndufa4l2 transcript, which translated into 15 fold increase in NDUFA4L2 protein, compared with control animals (Figure 4(b) and (c)). In addition, we evaluated the expression of COX4I2, another atypical mitochondrial subunit previously described to be exclusively expressed in cerebral mural cells, under conditions of HIF stabilization. In line with the results obtained for NDUFA4L2, Cox4i2 mRNA showed a 4.3 fold induction, which translated into a 2.3 fold increase in COX4I2 protein, compared with control animals (Figure 4(b) and (c)).
Figure 4.
NDUFA4L2 and COX4I2 are induced in cerebral mural cells in NG2-Vhl−/− mice. (a) Graphical representation of breeding scheme. (b) Relative cortical mRNA transcript levels of Ndufa4l2 and Cox4i2 in homogenates from cortex of Cre− control and NG2-Vhl−/− mice (n = 4-5). (c) Representative western blot and quantification of NDUFA4L2 and COX4I2 expression in homogenates from cerebral cortex of Cre− control and NG2-Vhl−/− mice (n = 4-5) and (d) (Left) Representative image of multiplex RNA F.I.S.H. of NG2-Vhl−/− mouse cerebral cortex detecting Pdgfrb (green fluorescent signal), Cox4i2 (red fluorescent signal) and Ndufa4l2-expressing cells (blue fluorescent signal). (Right) Percentage of Pdgfrb+ and Pdgfrb− expressing Ndufa4l2 or Cox4i2 mRNA, and percentage of Ndufa4l2 or Cox4i2+ and Ndufa4l2 or Cox4i2− expressing Pdgfrb mRNA (n = 3) Data are represented as mean ± SD; Statistical analysis was performed using two-tailed Student’s t test with Welch’s correction when needed. *P < 0.05, ***P < 0.001, compared with Cre− control mice.
To ascertain the cellular localization of these induced transcripts, we performed Multiplex RNA F.I.S.H. with probes for Ndufa4l2, Cox4i2 and the mural cell marker Pdgfrb (Figure 4(d)). Quantification revealed that 94.69 ± 2.01% of Ndufa4l2 positive cells were also Pdgfrb positive cells and that 89.3 ± 4.3% of all Pdgfrb positive cells were Ndufa4l2 positive cells, which do not differ from control mice (95.77 ± 2.8% and 89.1 ± 4.3%; p = 0.6218 and 0.9477 respectively, (n = 3) (Figure 4(d)). With regard to Cox4i2 expression, quantification revealed that 89.7 ± 2.86% of Cox4i2 positive signal was expressed by Pdgfrb positive cells, which do not differ from control animals (92.3 ± 1.49%; p = 0.24 n = 3) and that 95.09 ± 0.98% of Pdgfrb positive cells expressed the Cox4i2 transcript, which was a slightly higher than the controls (91.8 ± 1.8; p = 0.05 n = 3) (Figure 4(d)). Interestingly and in line with the specific induction of NDUFA4L2 and COX4I2 expression upon pericyte HIF activation in NG2-Vhl−/− mice, we have observed that these mice show a smaller infarct volume than Cre− mice in a model of cerebral ischemic stroke (MCAO), where oxygen delivery is drastically reduced (Figure S5).
These results demonstrate that NDUF4L2 induction observed in human brain pericytes in vitro is operative in brain mural cells in vivo. Moreover, we show that COX4I2 is also induced pon HIF activation in brain in mural cells in the mouse brain, although to a lesser extent that NDUFA4L2. Finally, our data support a protective role of these two proteins in pathological conditions of oxygen deprivation such as ischemic stroke.
NDUFA4L2 represses oxygen consumption and controls the degree of hypoxia response in pericytes
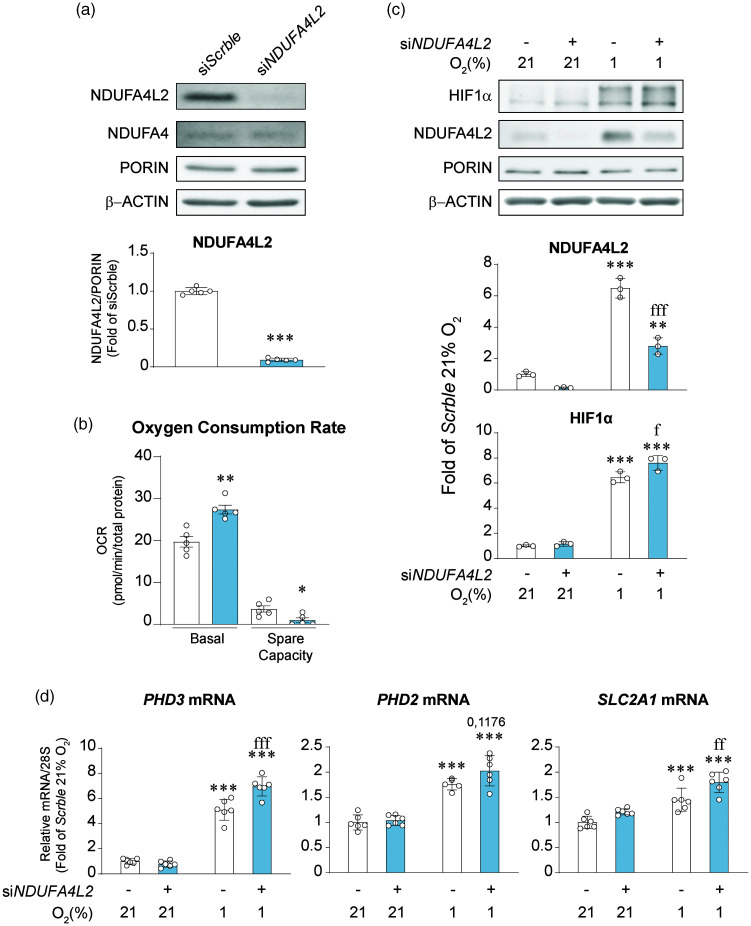
To investigate the possible function of the highly expressed NDUFA4L2 in brain pericytes, we silenced NDUFA4L2 with siRNAs resulting in a ∼90% reduction of protein levels (Figure 5(a)), without affecting NDUFA4 expression (Data not shown). It did however affect cell proliferation in normoxia (Figure S6A). Moreover, NDUFA4L2 silencing abolished the increased proliferation of human brain pericytes induced by hypoxic conditions (Figure S6A), although this effect was not attributable to a decrease in cell viability or survival (Figure S6 B-D). NDUFA4L2 induction under hypoxia has been previously reported to inhibit oxygen consumption.17 We hypothesized that basal levels of NDUFA4L2 in human brain pericytes are so high that can already repress mitochondrial respiration. Inactivation of NDUFA4L2 increased basal oxygen consumption rate (OCR) in human brain pericytes by 40% in normoxia (Figure 5(b)) confirming our hypothesis. In addition, we evaluated the spare respiratory capacity, which represents the mitochondrial plasticity to an increased demand of energy. Surprisingly, while siScrble treated human brain pericytes showed an increase in OCR after the addition of FCCP and a spare respiratory capacity of about a 20% of basal OCR, NDUFA4L2 silenced human brain pericytes could not further elevate its OCR. Since hypoxia is a balance between oxygen availability and oxygen consumption,43,44 we assesed whether NDUFA4L2 silencing affected the HIF-dependent response to hypoxia of human brain pericytes. Exposure to hypoxia (1% O2) for 16 hours increased NDUFA4L2 levels in control pericytes (Figure 5(c)) as expected. Silencing of NDUFA4L2 significanly blunted this response although did not completely avert it. Nevertheless, the inhibition was enough to facilitate a small but significant stronger stabilization of HIF1α (Figure 5(c)) that was associated with a more robust induction of HIF targets such as PHD3, PHD2 and SLC2A1 mRNAs when compared with control group (Figure 5(d)). These results not only support the role of NDUFA4L2 as a repressor of mitochondrial respiration in human brain pericytes but also suggest that participates as a regulator of the HIF transcriptional response.
Figure 5.
NDUFA4L2 represses oxygen consumption and HIF1 signaling in human brain pericytes in vitro. (a) Representative western blot and quantification of NDUFA4L2 and NDUFA4 expression in transient NDUFA4L2-silenced human brain pericytes and their corresponding siScrble cells (n = 5). (b) Oxygen consumption rate and spare respiratory capacity of transient NDUFA4L2-silenced human brain pericytes and their corresponding control cells (n = 5). (c) Representative western blot and quantification of HIF1α and NDUFA4L2 expression in transient NDUFA4L2-silenced human brain pericytes and their corresponding siScrble cells exposed to normoxia (21% O2) and hypoxia (1% O2) for 16 hours (n = 3) and (d) Relative mRNA transcript levels of hypoxia induced genes PHD3, PHD2 and SLC2A1 (n = 5–6). Data are represented as mean ± SD; Statistical analysis was performed using two-tailed Student’s t test for (a) and (b) and two-way ANOVA followed by Tukey’s post hoc analysis for (c) and (d); *P < 0.05, **P < 0.01, ***P < 0.001, compared with normoxic siScrble cells; fP < 0.05, ffP <0.01, fffP < 0.001 compared with hypoxic siScrble.
Discussion
Here we have used a combination of scRNA-seq data analysis coupled with high-resolution multiplexed fluorescence RNA in situ hybridization (RNA F.I.S.H.) and primary cell culture to identify the NDUFA4L2 expressing cells in mouse and human brain. Our results establish that NDUFA4L2 is a distinctive metabolic feature of mural cells of murine and human brain not only in vivo but also in vitro. Mural cell is an umbrella term covering both vascular smooth muscle cells, pericytes and cells with mixed characteristics of both smooth muscle cells and pericytes, all of which line and modulate the cerebral vasculature.4,24,25,28 Although these populations share classical molecular markers like PDGFRB and NG2, they can be differentiated by their morphology, location and α-SMA expression.25,28 Pericytes are found in capillary networks with a characteristic bump on a log form, thin processes that run along the capillaries, and negative expression of α-SMA.20,22,28 Smooth muscle cells on the other hand are found in higher caliber vessels, have a ring-shaped form and express a variable amount of α-SMA depending on whether they are located in arteries, arterioles or pre-capilary arterioles.20,22,28
We show that among mural cells, NDUFA4L2 expression is most robustlty expressed in capillary pericytes and that this expression declines along the precapillary/arteriolar/arterial axis in parallel with increasing cell muscularization. Moreover, when we increase pericyte muscularization in vitro we obtain a similar response to the observed in the scRNA-seq data sets; lower NDUFA4L2 levels; suggesting that both variables are inextricably ligated and that NDUFA4L2 expression defines the identity of the mural cell.
Now, why is this atipical subunit of the mitochondria exclusively expressed in mural cells and more robustly in pericytes? Cerebral oxygenation decreases along the arterial/arteriolar/capillary axis. 45 Intriguingly NDUFA4L2 expression and PO2 are inversely associated. Pericytes around capillaries, where the PO2 is lowest, show the highest expression of NDUFA4L2, with this expression decreasing with increasing PO2 along the arteriolar/arterial axis. Our data show that NDUFA4L2 represses oxygen consumption in normoxic conditions, in line with its role previously described under hypoxia. 19 The exact physiological relevance of this is yet unknown, but because NDUFA4L2 represses oxygen consumption, it is tempting to speculate that pericytes reduce their use of oxygen to favor parenchymal oxygenation. Interestingly, endothelial cells display a highly glycolytic metabolism, obtaining 85% of their ATP via glucose conversion to lactate46,47 therefore consuming less oxygen than other cells on the parenchyma. Since most of the energy in the brain is used by neurons, which are more oxidative than glycolytic and rely on oxygen to generate energy, it would be highly efficient to avoid excessive oxygen consumption between red blood cells and neurons so the latter are provided with the oxygen required to carry out their function in the most efficient way. Along this line, we show here that pericytes express at least 2 atipical mitochondrial subunits involved in repressing (NDUFA4L2) 17 and optimizing (COX4I2) 29 oxygen consumption which in addition to the glycolytic metabolism of endothelial cells, would result in a maximization of oxygen delivery into the parenchyma. To our surprise, although NDUFA4L2 is conserved in mouse and human mural cells, both in vivo and in vitro, COX4I2 expression in mural cells seems to be lower in humans than in mice. In fact, it was not detected in some scRNA-seq data sets (which detected NDUFA4L2) and we could not detect it by mRNA or protein in human brain pericytes in vitro, possibly as a result of its low expression which is likely lost during the process of cell isolation. Nevertheless, brain pericytes show features of cells that are exposed to hypoxia, which are charaterized by reduced oxygen consumption. We have previously showed that pericytes have the ability to function as oxygen sensors in the brain through the combined action of PHD2 and PHD3.18,19 Here we expand our previous research by showing that the already high basal levels of NDUFA4L2 in cerebral pericytes can be further induced under hypoxia in a mechanism dependent on HIF1 and the coordinate function of PHD2 and PHD3. In the presence of molecular oxygen and iron these prolyl hydroxylases catalize the hydroxylation of critical residues of HIF that will lead to its degradation.9–13 When oxygen levels fall below a threshold, this reaction does not occur, HIF is stabilized and promotes the cellular adaptation to hypoxia. Because oxygen levels are a balance between availability and consumption, processes that affect the use of oxygen, such as mitochondrial respiration are of critical importance. We show that under hypoxia NDUFA4L2, which represses the oxygen consumption rate, controls the degree of HIF stabilization and therefore the response to hypoxia. Taken together our results suggest that capillary pericytes have the ability to act as cellular oxygen sensors in the brain through a NDUFA4L2/PHD2,3/HIF axis.
In summary, our data provide new insights into the oxygen metabolism and the molecular characteristics specifics of brain mural cells. Our data may help to understand not only the normal microvascular physiology of the brain but also pathological scenarios, in which hypoxia is a common feature.
Supplemental Material
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X221118236 for Unique expression of the atypical mitochondrial subunit NDUFA4L2 in cerebral pericytes fine tunes HIF activity in response to hypoxia by Claudia Mesa-Ciller, Guillermo Turiel, Andrea Guajardo-Grence, Ana Belen Lopez-Rodriguez, Javier Egea, Katrien De Bock, Julián Aragonés and Andrés A Urrutia in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-pdf-2-jcb-10.1177_0271678X221118236 for Unique expression of the atypical mitochondrial subunit NDUFA4L2 in cerebral pericytes fine tunes HIF activity in response to hypoxia by Claudia Mesa-Ciller, Guillermo Turiel, Andrea Guajardo-Grence, Ana Belen Lopez-Rodriguez, Javier Egea, Katrien De Bock, Julián Aragonés and Andrés A Urrutia in Journal of Cerebral Blood Flow & Metabolism
Acknowledgements
The data that support the findings of this study are available from the corresponding author upon reasonable request. The authors would like to thank Francisca Molina Jiménez (Fundación de Investigación Biomédica del Hospital la Princesa) for assistance with confocal microscopy experiments and Enrique Martín-Gayo for advice and expertise with FACS analysis.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grant SI1/PJI/2019-00399 from Universidad Autónoma de Madrid and Comunidad Autónoma de Madrid. CMC is supported by the FPI fellowship from Ministerio de Ciencia e Innovación. GT is funded through an ERC Starting grant by KDB (716140). KDB is endowed by the Schulthess foundation. AGG is supported by the FPI UAM fellowship from Universidad Autónoma de Madrid. AU is supported by the Comunidad de Madrid “Atracción de Talento Program”.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions: AAU conceived and designed the research studies, analyzed and interpreted data, wrote the manuscript and made the figures. CMC, GT, AGG, ABLR and AAU performed experiments and acquired data. JA, JE and KDB analyzed and interpreted data.
Supplemental material: Supplemental material for this article is available online.
ORCID iDs
Andrea Guajardo-Grence https://orcid.org/0000-0002-4437-8529
Andrés A Urrutia https://orcid.org/0000-0001-7945-5515
References
- 1.Raichle ME, Gusnard DA. Appraising the brain's energy budget. Proc Natl Acad Sci U S A 2002; 99: 10237–10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Girouard H, Iadecola C. Neurovascular coupling in the normal brain and in hypertension, stroke, and Alzheimer disease. J Appl Physiol (1985) 2006; 100: 328–335. [DOI] [PubMed] [Google Scholar]
- 3.Mistry N, Mazer CD, Sled JG, et al. Red blood cell antibody-induced anemia causes differential degrees of tissue hypoxia in kidney and brain. Am J Physiol Regul Integr Comp Physiol 2018; 314: R611–R622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iadecola C. The neurovascular unit coming of age: a journey through neurovascular coupling in health and disease. Neuron 2017; 96: 17–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaelin WG, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 2008; 30: 393–402. [DOI] [PubMed] [Google Scholar]
- 6.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell 2012; 148: 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivan M, Kaelin WG. The EGLN-HIF O2-sensing system: multiple inputs and feedbacks. Mol Cell 2017; 66: 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang GL, Jiang BH, Rue EA, et al. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A 1995; 92: 5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 2001; 292: 464–468. [DOI] [PubMed] [Google Scholar]
- 10.Epstein AC, Gleadle JM, McNeill LA, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 2001; 107: 43–54. [DOI] [PubMed] [Google Scholar]
- 11.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 2001; 294: 1337–1340. [DOI] [PubMed] [Google Scholar]
- 12.Hon WC, Wilson MI, Harlos K, et al. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature 2002; 417: 975–978. [DOI] [PubMed] [Google Scholar]
- 13.Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001; 292: 468–472. [DOI] [PubMed] [Google Scholar]
- 14.Papandreou I, Cairns RA, Fontana L, et al. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 2006; 3: 187–197. [DOI] [PubMed] [Google Scholar]
- 15.Semenza GL. Hypoxia-inducible factor 1: regulator of mitochondrial metabolism and mediator of ischemic preconditioning. Biochim Biophys Acta 2011; 1813: 1263–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas LW, Ashcroft M. Exploring the molecular interface between hypoxia-inducible factor signalling and mitochondria. Cell Mol Life Sci 2019; 76: 1759–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tello D, Balsa E, Acosta-Iborra B, et al. Induction of the mitochondrial NDUFA4L2 protein by HIF-1α decreases oxygen consumption by inhibiting complex I activity. Cell Metab 2011; 14: 768–779. [DOI] [PubMed] [Google Scholar]
- 18.Urrutia AA, Afzal A, Nelson J, Davidoff O, et al. Prolyl-4-hydroxylase 2 and 3 coregulate murine erythropoietin in brain pericytes. Blood 2016; 128: 2550–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urrutia AA, Guan N, Mesa-Ciller C, et al. Inactivation of HIF-prolyl 4-hydroxylases 1, 2 and 3 in NG2-expressing cells induces HIF2-mediated neurovascular expansion independent of erythropoietin. Acta Physiol (Oxf) 2021; 231: e13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeisel A, Hochgerner H, Lönnerberg P, et al. Molecular architecture of the mouse nervous system. Cell 2018; 174: 999–1014.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy DJ, Campbell KR, Lun AT, et al. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 2017; 33: 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanlandewijck M, He L, Mäe MA, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 2018; 554: 475–480. [DOI] [PubMed] [Google Scholar]
- 23.Palomino-Antolin A, Narros-Fernández P, Farré-Alins V, et al. Time-dependent dual effect of NLRP3 inflammasome in brain ischaemia. Br J Pharmacol 2022; 179: 1395–1410. [DOI] [PubMed] [Google Scholar]
- 24.Grutzendler J, Nedergaard M. Cellular control of brain capillary blood flow: in vivo imaging veritas. Trends Neurosci 2019; 42: 528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armulik A, Genové G, Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell 2011; 21: 193–215. [DOI] [PubMed] [Google Scholar]
- 26.Sweeney MD, Zhao Z, Montagne A, et al. Blood-brain barrier: from physiology to disease and back. Physiol Rev 2019; 99: 21–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Attwell D, Mishra A, Hall CN, et al. What is a pericyte? J Cereb Blood Flow Metab 2016; 36: 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grant RI, Hartmann DA, Underly RG, et al. Organizational hierarchy and structural diversity of microvascular pericytes in adult mouse cortex. J Cereb Blood Flow Metab 2019; 39: 411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukuda R, Zhang H, Kim JW, et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 2007; 129: 111–122. [DOI] [PubMed] [Google Scholar]
- 30.Winkler EA, Bell RD, Zlokovic BV. Pericyte-specific expression of PDGF beta receptor in mouse models with normal and deficient PDGF beta receptor signaling. Mol Neurodegener 2010; 5: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He L, Vanlandewijck M, Raschperger E, et al. Analysis of the brain mural cell transcriptome. Sci Rep 2016; 6: 35108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polioudakis D, de la Torre-Ubieta L, Langerman J, et al. A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron 2019; 103: 785–801.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han X, Zhou Z, Fei L, et al. Construction of a human cell landscape at single-cell level. Nature 2020; 581: 303–309. [DOI] [PubMed] [Google Scholar]
- 34.Garcia FJ, Sun N, Lee H, et al. Single-cell dissection of the human brain vasculature. Nature 2022; 603: 893–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang AC, Vest RT, Kern F, et al. A human brain vascular atlas reveals diverse mediators of Alzheimer's risk. Nature 2022; 603: 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dubrac A, Künzel SE, Künzel SH, et al. NCK-dependent pericyte migration promotes pathological neovascularization in ischemic retinopathy. Nat Commun 2018; 9: 3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tigges U, Welser-Alves JV, Boroujerdi A, et al. A novel and simple method for culturing pericytes from mouse brain. Microvasc Res 2012; 84: 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minamishima YA, Kaelin WG. Reactivation of hepatic EPO synthesis in mice after PHD loss. Science 2010; 329: 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berra E, Benizri E, Ginouvès A, et al. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J 2003; 22: 4082–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Appelhoff RJ, Tian YM, Raval RR, et al. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem 2004; 279: 38458–38465. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi H, Liu Q, Binns TC, et al. Distinct subpopulations of FOXD1 stroma-derived cells regulate renal erythropoietin. J Clin Invest 2016; 126: 1926–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu X, Bergles DE, Nishiyama A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development 2008; 135: 145–157. [DOI] [PubMed] [Google Scholar]
- 43.Hagen T, Taylor CT, Lam F, et al. Redistribution of intracellular oxygen in hypoxia by nitric oxide: effect on HIF1alpha. Science 2003; 302: 1975–1978. [DOI] [PubMed] [Google Scholar]
- 44.Pan Y, Mansfield KD, Bertozzi CC, et al. Multiple factors affecting cellular redox status and energy metabolism modulate hypoxia-inducible factor prolyl hydroxylase activity in vivo and in vitro. Mol Cell Biol 2007; 27: 912–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakadžić S, Mandeville ET, Gagnon L, et al. Large arteriolar component of oxygen delivery implies a safe margin of oxygen supply to cerebral tissue. Nat Commun 2014; 5: 5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Bock K, Georgiadou M, Schoors S, et al. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell 2013; 154: 651–663. [DOI] [PubMed] [Google Scholar]
- 47.Rohlenova K, Veys K, Miranda-Santos I, et al. Endothelial cell metabolism in health and disease. Trends Cell Biol 2018; 28: 224–236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X221118236 for Unique expression of the atypical mitochondrial subunit NDUFA4L2 in cerebral pericytes fine tunes HIF activity in response to hypoxia by Claudia Mesa-Ciller, Guillermo Turiel, Andrea Guajardo-Grence, Ana Belen Lopez-Rodriguez, Javier Egea, Katrien De Bock, Julián Aragonés and Andrés A Urrutia in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-pdf-2-jcb-10.1177_0271678X221118236 for Unique expression of the atypical mitochondrial subunit NDUFA4L2 in cerebral pericytes fine tunes HIF activity in response to hypoxia by Claudia Mesa-Ciller, Guillermo Turiel, Andrea Guajardo-Grence, Ana Belen Lopez-Rodriguez, Javier Egea, Katrien De Bock, Julián Aragonés and Andrés A Urrutia in Journal of Cerebral Blood Flow & Metabolism