Abstract
Fructooligosaccharides (FOS), Ad-fructooligosaccharides (Ad-FOS), resistant maltodextrin (RMD), and maltooligosaccharides (MOS) are commercially available prebiotic oligosaccharides. In this study, the effects of prebiotics on the human gut microbial ecosystem were evaluated using an in vitro gut model. FOS and Ad-FOS showed tolerance to digestion, whereas RMD and MOS showed moderate digestion by digestive enzymes. In in vitro fecal fermentation, Bifidobacterium spp. increased in the following order: FOS, Ad-FOS, MOS, and RMD, whereas Bacteroides spp. increased in RMD medium. Bacteroides xylanisolvens exhibited cross-feeding by enabling the growth of other beneficial bacteria during co-culture in RMD medium. In metabolome analysis, total short-chain fatty acids (SCFAs) were highly produced in the following order: RMD, FOS, MOS, and Ad-FOS; acetate in the order of FOS, MOS/RMD, and Ad-FOS; butyrate in the order of RMD, MOS, FOS, and Ad-FOS; and propionate only in RMD. In addition, the conversion of betaine to trimethylamine was rarely affected in the following order: MOS, RMD, FOS, and Ad-FOS. Lastly, the four oligosaccharides inhibited the adhesion of pathogenic Escherichia coli to human epithelial cells to a similar extent. The comparative analysis results obtained in this study will provide comprehensive information of these substances to manufacturers and customers.
Keywords: fructooligosaccharide, maltooligosaccharide, metabolome, microbiome, prebiotic, resistant maltodextrin
The oligosaccharides, FOS, Ad-FOS, RMD, and MOS, exhibited prebiotic effects, but their action patterns and efficacy vary owing to their different digestibility, fermentability, and cross-feeding interactions in complex microbiome ecosystems.
Introduction
Prebiotics are substrates that are selectively used by host microorganisms to beneficially influence health (Gibson et al. 2017). The important role of prebiotics in host health is well established and has attracted considerable attention over time as an outstanding functional food ingredient (Wang et al. 2019). Representative prebiotics include fructooligosaccharides (FOS), galactooligosaccharides (GOS), xylooligosaccharides (XOS), resistant maltodextrin (RMD), and maltooligosaccharides (MOS) (Hamaker and Tuncil 2014, Moreno et al. 2017, Myhrstad et al. 2020). Among them, FOS, RMD, and MOS have been well commercialized, while GOS and XOS have recently formed new markets. These substances can promote an abundance of beneficial bacteria, such as Lactobacillus and Bifidobacterium spp. (Kruse et al. 1999, Mao et al. 2015), accompanied by the production of several metabolites, such as short-chain fatty acids (SCFAs). In addition, prebiotics have been reported to be involved in mineral absorption (Whisner et al. 2013), control of pathogenic bacterial populations (Carlson et al. 2018), immunomodulation (Frei et al. 2015), and improvement of the gut barrier function (Cani et al. 2009).
A prebiotic was first defined as a ‘nondigestible food ingredient that beneficially affects the host by selectively stimulating the growth and/or activity of one or a limited number of bacteria already resident in the colon’ (Gibson and Roberfroid 1995). However, there have been many variations in the concept of prebiotics between global regulatory agencies because of the many emerging prebiotics that do not fit the definition (Carlson et al. 2018). In 2017, the panel of International Scientific Association of Probiotics and Prebiotics (ISAPP) proposed a broad definition of a prebiotic as ‘a substrate that is selectively utilized by host microorganisms conferring a health benefit’ (Gibson et al. 2017). This has expanded the concept of prebiotics to diverse categories in terms of substances and working sites. Based on the recent definition of prebiotics, previous studies on prebiotics were based on information on intestinal microorganisms and metabolites; these analyses narrowly focused on Bifidobacterium spp. and SCFAs. Therefore, it is necessary to analyze the prebiotic effects of substrates on the gut microbial ecosystem, including changes in the microbiome and metabolome during colonic fermentation. Microbiome and metabolome analyses can elucidate the ecological relationships of beneficial bacteria with commensal and harmful bacteria and provide information on various compounds synthesized by the human gut microbiome, respectively.
In vitro models are useful tools for evaluating food safety and validating health claims, especially in the food industry, to predict the impact of novel and functional foods on the human microbiome (Roupar et al. 2021). Previously, the effects of FOS on human gut bacteria have been evaluated using in vitro models (Mao et al. 2015). RMD has also been investigated for microbial enzyme activity and microbial group composition generated during the in vitro fermentation process (Rösch et al. 2015). In addition, the effect of MOS as a prebiotic was evaluated by analyzing its digestibility, proliferation of Bifidobacterium spp., and changes in SCFAs content (Jang et al. 2020). Although previous studies have reported the prebiotic effects of several oligosaccharides using in vitro systems, these substances have been studied individually, and comparative studies on the prebiotic effects have not yet been performed (Nogacka et al. 2020). Thus, a comparative analysis of the effects of prebiotic oligosaccharides on gut microbiome ecosystems should be conducted to provide comprehensive information on these substances to manufacturers and customers.
Therefore, the aim of this study was to evaluate and compare the prebiotic effects of four commercialized oligosaccharides, FOS, Ad-FOS, RMD, and MOS, focusing on changes in the microbiome and metabolome. For this purpose, their digestibility by digestive enzymes, fermentability through individual bacterial cultivation and co-cultivation, in vitro fecal fermentation, and inhibition of pathogenic E. coli adhesion to intestinal epithelial cells were analyzed. In particular, for in vitro fecal fermentation, we employed the simulated batch fermentation system under pH-controlled anaerobic conditions.
Materials and Methods
Microorganisms, media, and culture conditions
The strains used in this study are listed in Table 1 and were obtained from the Korean Agricultural Culture Collection (KACC, Jeonju, Korea), the Korean Collection for Type Culture (KCTC, Jeongeup, Korea), and the American Type Culture Collection (ATCC, Manassas, USA). They were cultured in De Man, Rogosa, and Sharp medium (MRS; Difco Co., Detroit, MI, USA), brain heart infusion broth medium (BHI; Difco Co.), and supplemented BHI (BHIS) medium (Bacic and Smith 2008) at their optimal temperatures.
Table 1.
Strains used in this study.
| Strains | Medium | Temperature (°C) | |
|---|---|---|---|
| 1 | Limosilactobacillus fermentum KACC 11 441 | MRS | 30 |
| 2 | Limosilactobacillus reuteri KACC 11 452 | MRS | 37 |
| 3 | Lacticaseibacillus rhamnosus KACC 11 953 | MRS | 37 |
| 4 | Lactiplantibacillus plantarum KACC 11 451 | MRS | 37 |
| 5 | Lactobacillus acidophilus KACC 12 419 | MRS | 30 |
| 6 | Lacticaseibacillus casei KACC 12 413 | MRS | 37 |
| 7 | Lactobacillus bulgaricus KACC 12 420 | MRS | 37 |
| 8 | Lactobacillus gasseri KACC 12 424 | MRS | 37 |
| 9 | Lactobacillus helveticus KACC 12 418 | MRS | 37 |
| 10 | Lacticaseibacillus paracasei KACC 12 361 | MRS | 37 |
| 11 | Ligilactobacillus salivarius KACC 10 006 | MRS | 37 |
| 12 | Lactococcus lactis KACC 13 877 | MRS | 37 |
| 13 | Streptococcus thermophilus KACC 11 857 | MRS | 37 |
| 14 | Bifidobacterium lactis KACC 16 638 | MRS + L-cysteine | 37 |
| 15 | Bifidobacterium breve KACC 16 639 | MRS + L-cysteine | 37 |
| 16 | Bifidobacterium bifidum KACC 20 601 | MRS + L-cysteine | 37 |
| 17 | Bifidobacterium longum KCTC 3128 | MRS + L-cysteine | 37 |
| 18 | Anaerostipes hadrus KCTC 15 606 | BHIS | 37 |
| 19 | Bacteroides fragilis KCTC 5013 | BHIS | 37 |
| 20 | Bacteroides ovatus KCTC 5827 | BHIS | 37 |
| 21 | Bacteroides uniformis KCTC 5204 | BHIS | 37 |
| 22 | Bacteroides vulgatus KCTC 25 021 | BHIS | 37 |
| 23 | Bacteroides xylanisolvens KCTC 15 192 | BHIS | 37 |
| 24 | Blautia hansenii KCTC 5951 | BHIS | 37 |
| 25 | Collinsella aerofaciens KCTC 15 038 | BHIS | 37 |
| 26 | Dorea formicigenerans KCTC 15 690 | BHIS | 37 |
| 27 | Enterococcus faecium KCTC 13 225 | BHIS | 37 |
| 28 | Escherichia coli KCTC 2441 | BHIS | 37 |
| 29 | Akkermansia muciniphila ATCC BAA-835 | BHIS | 37 |
| 30 | Escherichia coli O157:H7 ATCC 43 895 | BHIS | 37 |
| 31 | Listeria monocytogenes ATCC 19 115 | BHIS | 37 |
In vitro digestion
In vitro digestion of oligosaccharides was performed using a previously reported method (Minekus et al. 2014). The prebiotic oligosaccharides used in this study were FOS (β-D-fructofuranose-(2→1)-β-D-fructofuranose-(2→1)-α-D-glucopyranose, GF2, 38.69%; β-D-fructofuranose-(2→1)-β-D-fructofuranose-(2→1)-β-D-fructofuranose-(2→1)-α-D-glucopyranose, GF3, 48.44%; β-D-fructofuranose-(2→1)-β-D-fructofuranose-(2→1)-β-D-fructofuranose-(2→1)-β-D-fructofuranose-(2↔1)-α-D-glucopyranose, GF4, 8.25%, 95.38% purity), Ad-FOS (87.96% GF2 and 92.96% purity), RMD (89.5% purity), and MOS (maltotetraose, G4, 59.9% purity in hydrolyzed corn starch). These were provided by an industrial producer (Samyang, Seoul, South Korea). Briefly, for oral phase digestion, 2 ml simulated saliva fluid (SSF) electrolyte stock solution (15.1 mmol/L KCl, 3.7 mmol/L KH2PO4, 13.6 mmol/L NaHCO3, 0.15 mmol/L MgCl2(H2O)6, 0.06 mmol/L (NH4)2CO3, 1.5 mmol/L CaCl2(H2O)2, pH 7.0) was mixed with 80 mg oligosaccharides, 75 U/mL oral enzyme (α-amylase from human saliva Type IX-A, 1000–3000 U/mg protein, Sigma), and 0.75 mM CaCl2 (Junsei, Tokyo, Japan). The reactions were performed at 37°C for 2 min and stopped by boiling for 5 min. For gastric phase digestion, 2 ml simulated gastric fluid (SGF) electrolyte stock solution (6.9 mmol/L KCl, 0.9 mmol/L KH2PO4, 25 mmol/L NaHCO3, 47.2 mmol/L NaCl, 0.1 mmol/L MgCl2(H2O)6, 0.5 mmol/L (NH4)2CO3, 0.15 mmol/L CaCl2(H2O)2, pH 7.0) was mixed with 80 mg oligosaccharides, 2000 U/mL gastric enzymes (pepsin from porcine gastric mucosa 3200–4500 U/mg protein, Sigma), and 0.075 mM CaCl2. The reactions were performed at 37°C for 2 h and stopped by boiling for 5 min. For intestinal phase digestion, 2 ml simulated intestinal fluid (SIF) electrolyte stock solution (6.8 mmol/L KCl, 0.8 mmol/L KH2PO4, 85 mmol/L NaHCO3, 38.4 mmol/L NaCl, 0.33 mmol/L MgCl2(H2O)6, 0.6 mmol/L CaCl2(H2O)2, pH 7.0) was mixed with 80 mg oligosaccharides and 100 U/mL pancreatin (pancreatin from porcine pancreas, Sigma). The reaction was then performed at 37°C for 4 h and stopped by boiling for 5 min. For digestion of the intestinal brush border membrane, 2 ml fresh SIF was mixed with 80 mg oligosaccharides and 2.89 U/mL (maltase activity) of the brush border membrane vesicle (BBMV) enzyme isolated from pig small intestine. The reaction was then carried out at 37°C for 4 h and stopped by boiling for 5 min. The digestion ratios of oligosaccharides in the oral, gastric, intestinal, and BBMV phases were determined based on the free sugar content released during digestion. The reducing sugar content was analyzed using the DNS assay described by Miller (1959). After mixing the DNS solution (300 μL) with 100 μL of each sample, the mixture was heated in boiling water for 5 min and cooled on ice for 5 min. Subsequently, 300 μl of the solution was transferred into each well of a 96-well plate and the absorbance was measured at 550 nm. The reducing sugar content was calculated using a standard curve. In addition, glucose concentrations were analyzed using high-performance liquid chromatography (HPLC) (Young Lin, Yongin, Korea) with a Shodex Asahipak NH2P-50 4E column (Shodex, Showa Denco, Tokyo, Japan).
Individual cultivation of bacterial species
To investigate the capability of oligosaccharides, as carbon sources for intestinal bacteria, the strains listed in Table 1 were individually cultivated in glucose-free MRS or BHI medium containing 1% oligosaccharides (w/v) at their optimal temperatures for 24 h. Thereafter, their growth (OD600 nm) and pH changes were measured.
Co-cultivation using transwell system
To investigate the interactions of different microbial species with each oligosaccharide, a co-cultivation method was employed using 12-well transwell insert plates (Costar, Washington, DC, USA). First, Bacteroides (Ba.) xylanisolvens was inoculated into the lower chamber of the plates, and precultured Lactiplantibacillus (L.) plantarum, Lactobacillus (L.) gasseri, Lactobacillus (L.) helveticus, Bifidobacterium (Bi.) longum, and Akkermansia (Ak.) muciniphila were individually inoculated into the upper chamber. The plates were incubated at 37°C for 24 h under anaerobic conditions (Vinyl Anaerobic Chambers; Coy Lab, Grass Lake Charter Township, MI, USA). Microbial growth in each chamber was analyzed by measuring the optical density at 600 nm after 24 h of cultivation.
In vitro fecal fermentation
The in vitro human fecal fermentation of oligosaccharides was conducted according to an established protocol (Moon et al. 2016). In detail, 300 ml capacity of water-jacketed fermenter vessels and basal growth medium (2 g/L peptone water, 1 g/L yeast extract, 0.1 g/L NaCl, 0.04 g/L K2HPO4, 0.04 g/L KH2PO4, 0.01 g/L MgSO4·7H2O, 0.01 g/L CaCl2·2H2O, 2 g/L NaHCO3, 0.5 g/L bile salts, 0.5 g/L L-cysteine hydrochloride, 50 mg/L hemin, 10 μL/L vitamin K1, and 2 ml/L Tween 80) were used for the fermentation. Bile salts, L-cysteine hydrochloride, hemin, MgSO4∙7H2O, and NaHCO3 were purchased from Sigma-Aldrich (St. Louis, MO, USA). Tween 80 was purchased from VWR (Radnor, PA, USA). Vitamin K1 was purchased from Wako Pure Chemical Industries (Osaka, Japan). Peptone water and yeast extract were purchased from BD Biosciences (Franklin Lakes, NJ, USA). CaCl2∙2H2O, K2HPO4, KH2PO4, and NaCl were purchased from Junsei (Tokyo, Japan). A total of 135 ml medium was inoculated with 15 ml 10% (w/v) fecal slurry, prepared by homogenizing freshly voided adult feces in 0.1 M phosphate-buffered saline (PBS) (pH 7.0). Fresh fecal samples were collected from 13 healthy adults who had not received antibiotics or pre/probiotics and had no recent history of gastrointestinal disorders. The study protocol and consent forms were approved by the Institutional Review Board of Chungbuk National University (CBNU-201905-BR-839–01). All collected feces were mixed and used as a sample. Oligosaccharides were added at a final concentration of 1% (w/v). The slurry in each vessel was magnetically stirred, and the pH and temperature were maintained at pH 6.8 and 37°C, respectively. The anaerobic conditions were maintained by sparging the vessels with oxygen-free nitrogen gas at a flow rate of 15 ml/min. Samples (5 ml) were taken at 12 h and 24 h for the analysis of bacterial composition and metabolites.
Microbial change analysis using 16S rRNA gene amplicon sequencing
The fecal bacterial communities in the in vitro fecal fermentation samples were determined using tag-encoded 16S rRNA gene MiSeq-based sequencing (Illumina, San Diego, CA, USA). The 16S rRNA gene was amplified using a primer set of 341-F (5′-CCTACGGGNGGCWGCAG-3′) and 785-R (5′-GACTACHVGGGTATCTAATCC -3′) (Klindworth et al. 2013) compatible with the Nextera Index Kit (Illumina). Sequencing analysis was conducted by Macrogen, Inc. (Seoul, Korea) using the Illumina MiSeq platform. Raw sequences were trimmed using a Seqpurge adapter trimmer (Sturm et al. 2016), and the resulting data were analyzed using QIIME2 (Bolyen et al. 2019) and the DADA2 pipeline (Callahan et al. 2016). Taxonomic assignment was performed based on the SILVA 132 reference database (https://www.arb-silva.de/documentation/release-132/) for bacteria (Quast et al. 2012). Sequence alignment was performed using MAFFT (Katoh and Standley 2013).
Metabolite analysis
The production of various metabolites during in vitro fecal fermentation was analyzed using proton nuclear magnetic resonance (1H-NMR), following the methods of Lee et al. (2011). In brief, the extracts were recovered using centrifugation after agitating the dissolved samples in a water bath at 60°C for 30 min. The supernatant was then mixed with an equal volume of deionized water containing 10% deuterium oxide (D2O) and 1 mM sodium 2,2-dimethyl-4-silapentane-1-sulfonic acid (DSS); the pH of the mixture was adjusted to 6 ± 0.01. The mixtures (700 µL) were transferred into 0.5-mm NMR tubes, and 1H-NMR spectra were acquired on a Varian INOVA 400 MHz NMR spectrometer (Varian Inc., Palo Alto, CA, USA). Individual spectra were identified and quantified using the Processor and Profiler module of the Chenomx NMR suite, V.6.1 (Chenomx, Inc., Edmonton, Alberta, Canada).
Inhibitory activity of oligosaccharides against adhesion of E. Coli onto epithelial cells
Caco-2 cells were added to a 24-well tissue culture plate containing 1 ml of Dulbecco's modified Eagle's medium (DMEM; Cytiva, Marlborough, MA, USA) at a concentration of 4.7 × 105 cells/well and incubated at 37°C in an atmosphere of 5% CO2 for 2 weeks. Oligosaccharides suspended in DMEM at a concentration of 10 mg/mL were dispensed into the wells, incubated for 1 h, and washed with PBS to remove unbound oligosaccharides. Then, E. coli O157: H7 precultured at 37°C for 12 h was suspended in DMEM at a concentration of 108 colony-forming units (CFU)/mL, and 1 ml was added to each well. After incubation for 1 h, non-adherent E. coli were removed by washing twice with PBS, and the attached E. coli were treated with a separation solution containing 0.1% Triton X-100 and 0.1% trypsin-EDTA (Sigma) for 15 min. The number of adherent E. coli was counted using BHI agar plates after appropriate dilution and incubation at 37°C for 48 h.
Statistical analysis
Statistical analyses were performed using the SPSS software (version 22.0; SPSS Inc., Chicago, IL, USA). Data analysis was performed using the independent t-test to determine the differences (P ≤ 0.05) between the two groups. Descriptive (mean and standard deviation) were conducted to determine the differences between multiple groups. The Shapiro Wilk test was used to determine the normality of the relative abundance of each group microbiome prior to one-way ANOVA. One-way analysis of variance (ANOVA followed by Tukey's test; for normally distributed data) was used to compare significant differences in relative abundance between the initial and each group microbiome, and all p-values were corrected via FDR correction. Statistical significance was set at P ≤ 0.05. All analyses were performed in duplicate or triplicate, as indicated.
Results
Digestibility of oligosaccharides
The digestibility of each oligosaccharide was evaluated using an in vitro digestion system. Reducing sugar was measured using a DNS assay to determine the degree of digestion by both endo- and exo-hydrolases (Park et al. 2015), and released glucose was analyzed using HPLC to determine the degree of complete digestion. As shown in Table 2, after hydrolysis of the oligosaccharides using an in vitro digestion system, the reducing sugar contents in FOS, Ad-FOS, RMD and MOS were 1.49%, 1.69%, 7.71%, and 17.55%, respectively. In addition, the released glucose contents were 0.37%, 0.18%, 5.74%, and 10.3%, respectively. Comparing the results, FOS and Ad-FOS showed the lowest digestibility, which means that most of them could reach colon, and RMD showed moderate digestibility, and MOS showed relatively high digestibility, indicating that only a partial amount could reach the colon.
Table 2.
Hydrolysis of oligosaccharides in each digestion phase.
| Sample | FOS | Ad-FOS | RMD | MOS | |
|---|---|---|---|---|---|
| Reducing sugar contents (△%*) | Oral | 0.11 | 0.08 | 0.42 | 2.33 |
| Gastric | 0.64 | 0.25 | 0.34 | 0.95 | |
| Intestinal | 0.01 | 0.4 | 0.51 | 2.78 | |
| BBMV | 0.72 | 0.96 | 6.45 | 7.66 | |
| Total** | 1.49 | 1.69 | 7.71 | 17.55 | |
| Glucose (or fructose) contents (△%) | Oral | -*** | - | 0.31 | 0.2 |
| Gastric | 0.37 | 0.18 | 0.22 | 0.17 | |
| Intestinal | - | - | 0.42 | 2.4 | |
| BBMV | - | - | 2.12 | 7.62 | |
| Total | 0.37 | 0.18 | 5.74 | 10.3 | |
Changes in reducing sugar and glucose (or fructose) contents were calculated as (each content after digestion)—(each content before digestion).
Total values were calculated as (total increment of reducing sugar and glucose (or fructose) content/initial sample weight) ×100
‘-’ means that there was no detection using HPLC analysis.
Individual cultivation
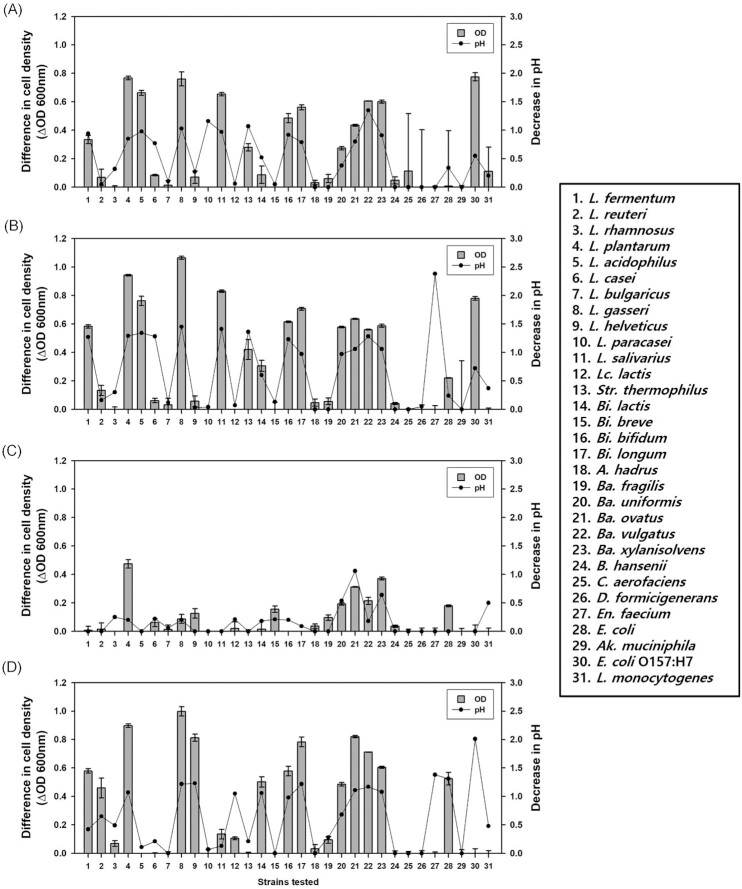
To investigate the utilization of prebiotics by microorganisms, each oligosaccharide was added to the culture media as the sole carbon source, and the growth of 31 microbial strains was analyzed after 24 h (Fig. 1). When FOS was tested, L. plantarum showed significant growth (OD600 = 0.77), followed by L. gasseri, Lactobacillus (L.) acidophilus, Ligilactobacillus (L.) salivarius, Ba. vulgatus, Ba. xylanisolvens, Bi. breve, Bi. longum, Ba. ovatus, and Limosilactobacillus (L.) fermentum (Fig. 1A). Ad-FOS showed higher maximum cell growth than FOS, although the growth patterns were identical for both (Fig. 1B). For RMD, L. plantarum showed significant cell growth (OD600 = 0.47), followed by Ba. xylanisolvens and Ba. ovatus (Fig. 1C). When MOS was tested, L. gasseri showed significant cell growth (OD600 = 1.00), followed by L. plantarum, Ba. ovatus, L. helveticus, Bi. longum, Ba. vulgatus, Ba. xylanisolvens, L. fermentum, Bi. breve, Bi. lactis, Ba. Uniformis, and Limosilactobacillus reuteri (Fig. 1D). Whereas, among the harmful and pathogenic bacteria, E. coli O157: H7 showed significant cell growth (OD600 = 0.77) in FOS and Ad-FOS. In all cultures, pH values decreased along with the growth of the cells. In summary, FOS, Ad-FOS, and MOS showed broad availability by the strains used, but RMD showed narrow availability only by L. plantarum and some Bacteroides spp.
Figure 1.
Changes in cell density and pH after cultivation of individual gut microbiota in optimal medium containing oligosaccharides for 24 h. (A) fructooligosaccharides (FOS), (B) Ad-fructooligosaccharides (Ad-FOS), (C) resistant maltodextrin (RMD), and (D) maltooligosaccharides (MOS).
Co-cultivation of dual bacteria in a transwell plate
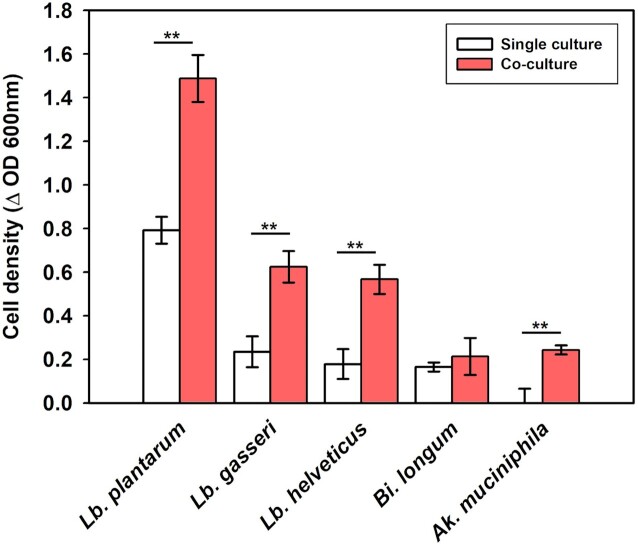
Recent research by Smith et al. (2019) defined cross-feeding as ‘an interaction between bacterial strains in which molecules resulting from the metabolism of one strain are further metabolized by another strain’. Cross-feeding interactions between Bacteroides and other microorganisms have been well established (Luis et al. 2018, Murakami et al. 2021, Kim et al. 2022). In the present study (Fig. 1), Bacteroides grew in all individual cultures containing each oligosaccharide, indicating the possibility of cross-feeding. To investigate the cross-feeding hypothesis, co-cultivation was performed using a transwell system, where Ba. xylanisolvens was cultured with the probiotic bacteria L. plantarum, L. gaseri, L. helveticus, Bi. longum, and Ak. muciniphila which were not grown in the previous individual cultivation test. As shown in Fig. 2, when they were cultured in the medium containing RMD, three probiotic species, L. plantarum, L. gasseri, and L. helveticus, showed significantly higher cell growth compared with the cases of single cultivations. The three bacterial species metabolized RMD during co-cultivation with Ba. xylanisolvens but did not utilize it well in individual cultivation (Fig. 1). These results indicate that Ba. xylanisolvens may confer a cross-feeding effect to facilitate the growth of other beneficial bacteria during co-culture in RMD medium.
Figure 2.
Differences in cell growth during single culture and co-culture of Bacteroides xylanisolvens KCTC 15192 with Lactiplantibacillus plantarum, Lactobacillus gasseri, Lactobacillus helveticus, Bifidobacterium longum, and Akkermansia muciniphila in optimal media containing maltodextrin (RMD) as carbon source after 24 h cultivation.
Changes in gut microbial community during fecal fermentation
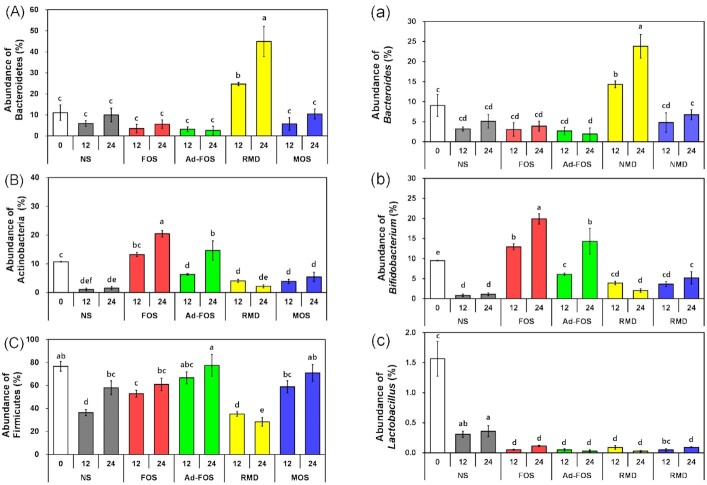
To investigate the effects of oligosaccharides on the intestinal microbial community, in vitro fecal fermentation was conducted using a fresh human fecal mixture and 1% oligosaccharides. The bacterial communities in the fecal samples were determined using tag-encoded 16S rRNA gene MiSeq-based sequencing. As shown in Table S2, the raw sequences were trimmed, and the resulting data were analyzed; the average values of total reads, GC content, and Q30 were 319449, 54%, and 80%, respectively. The microbial changes at the phylum and genus levels are shown in Fig. S1 (Supporting Information) and summarized in Fig. 3. At the phylum level, Bacteroidetes increased mainly in RMD (44.96%); Actinobacteria increased in FOS (20.46%) and Ad-FOS (14.62%); and Firmicutes increased in FOS (60.96%), Ad-FOS (77.51%), and MOS (71.00%) after 24 h. In addition, at the genus level, Bifidobacterium spp. increased mainly in FOS (19.92%) and Ad-FOS (14.33%), followed by MOS and RMD, after 24 h. Bacteroides increased mainly in RMD (23.81%), whereas Lactobacillus did not increase in any tested oligosaccharides. Therefore, the oligosaccharides used in this study increased the abundance of beneficial and commensal microorganisms, and FOS, a well-known prebiotic, was the most effective for increasing Bifidobacterium spp.
Figure 3.
Changes of relative abundance (%) of intestinal bacteria at phylum level, (A) Bacteroidetes, (B) Actinobacteria, and (C) Firmicutes on left side and at genus level, (a) Bacteroides, (b) Bifidobacterium, and (c) Lactobacillus on right side after 12 h and 24 h of in vitro fecal fermentation in the presence of oligosaccharides. Significant differences are compared between samples at the same time (P ≤ 0.05). NS, no substrate addition; FOS, fructooligosaccharide addition; Ad-FOS, Ad-fructooligosaccharide addition; RMD, resistant maltodextrin addition; MOS, maltooligosaccharide addition. NS0 represents a relative abundance of intestinal bacteria at 0 h as a baseline for comparison.
Analysis of fermentation metabolites
Synthesis of short-chain fatty acids
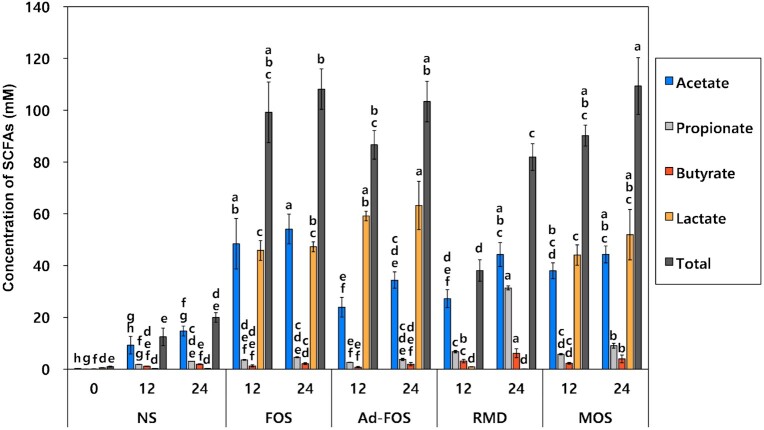
Within addition to in vitro fecal fermentation, the amounts of SCFAs (acetate, propionate, butyrate, and lactate) were analyzed (Fig. 4). Acetate and lactate were highly produced by FOS (54.13 ± 5.77 mM), RMD (31.38 ± 1.70 mM), Ad-FOS (63.20 ± 9.31 mM) after 24 h. In addition, propionate and butyrate were highly produced by RMD (31.38 ± 1.70 and 6.17 ± 0.78 mM) after 24 h. Fermentation of FOS resulted in high production of acetate and lactate showing concentrations of 54.13 ± 5.77 mM and 47.27 ± 1.94 mM after 24 h, respectively, while fermentation of Ad-FOS showed higher production of (63.20 ± 9.31 mM) than acetate (34.41 ± 3.16 mM). This result is consistent with the data presented in Fig. 3, showing a higher increase in the growth of Bifidobacterium spp. in FOS than in Ad-FOS. Fermentation of MOS showed a similar pattern of SCFA production to that of Ad-FOS. However, fermentation of RMD resulted in high production of acetate and propionate at concentrations of 44.26 ± 4.60 mM and 31.38 ± 1.70 mM, respectively. This result is also consistent with the result presented in Fig. 3, which shows a significant increase in the growth of Bacteroides. In summary, acetate was highly produced in oligosaccharides in the order FOS, MOS, RMD, and Ad-FOS, whereas propionate was highly produced only in RMD.
Figure 4.
Changes in short-chain fatty acids (SCFAs) and lactic acid concentrations after 12 h and 24 h during in vitro fecal fermentation in the presence of oligosaccharides. NS, no substrate addition; FOS, fructooligosaccharide addition; Ad-FOS, Ad-fructooligosaccharide addition; RMD, resistant maltodextrin addition; MOS, maltooligosaccharide addition. NS0 represents an initial SCFA concentration at 0 h as a baseline for comparison.
Metabolite changes
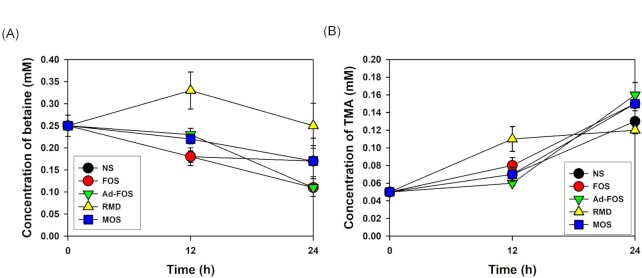
The metabolites produced during gut fermentation significantly affect human and animal health. Betaine, a choline compound involved in cardiovascular diseases, is converted into trimethylamine (TMA) by the gut microbiome and oxidized to trimethylamine N-oxide (TMAO) in the liver (Wang et al. 2011, Tang et al. 2019). During the in vitro fecal fermentation of the oligosaccharides tested, changes in indole derivatives, choline metabolites, phenolic derivatives, vitamins, polyamines, and amino acids were analyzed (Fig. 5; Table S1 and Fig. S2, Supporting Information). Among the choline compounds, the concentration of betaine was maintained for up to 24 h in RMD, and it was less degraded in the order MOS, FOS, and Ad-FOS. Consistent with this result, TMA was less produced in the order RMD, MOS, FOS, and Ad-FOS. Therefore, RMD is the promising oligosaccharide among the tested samples in terms of the low conversion of betaine to TMA, which is related to cardiovascular diseases.
Figure 5.
Changes in (A) betain and (B) trimethylamine concentrations at 0, 12, and 24 h during in vitro fecal fermentation in the presence of oligosaccharides. NS, no substrate addition; FOS, fructooligosaccharide addition; Ad-FOS, Ad-fructooligosaccharide addition; RMD, resistant maltodextrin addition; MOS, maltooligosaccharide addition.
Inhibition of E. Coli adhesion on human epithelial cells
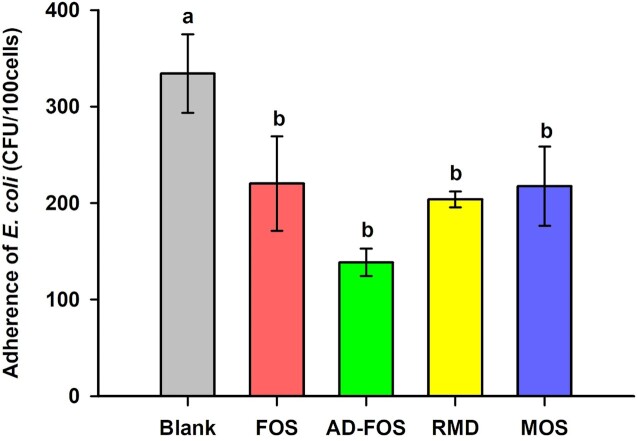
To investigate the ability of oligosaccharides to inhibit the adhesion of E. coli O157: H7 to the intestine, Caco-2 cells were treated with oligosaccharides, and the number of E. coli attached to the cells was measured (Fig. 6). As a result, the colony-forming unit (CFU) of E. coli attached to 100 cells without oligosaccharides were 334.24 CFU/100 cells, and the adhesion rate was 6.35%. All four prebiotic candidates inhibited the adhesion of pathogenic E. coli and their inhibition efficiencies were similar (p<0.05), with adhered bacterial counts ranging 138.59–220.11 CFU/100 cells and adhesion rates ranging 2.64–4.19%. These results indicate that the four oligosaccharides inhibited the adhesion of pathogenic Escherichia coli to human epithelial cells to a similar extent.
Figure 6.
Adherence inhibition of E. coli O157: H7 to Caco-2 cells in the presence of oligosaccharides. Blank, (E. coli O157: H7 only); FOS, fructooligosaccharide addition; Ad-FOS, Ad-fructooligosaccharide addition; RMD, resistant maltodextrin addition; MOS, maltooligosaccharide addition.
Discussion
In the in vitro digestion analyses, the prebiotic oligosaccharides tested showed low digestibility at all digestion stages. FOS and Ad-FOS were weakly degraded at each digestion step because of the β-(1→2) bonds in their structures (Krupa-Kozak et al. 2016), as has been reported in other studies (Nobre et al. 2018). However, RMD and MOS showed relatively higher degradation rates during the BBMV digestion step due to its enzyme activity to degrade disaccharides and oligosaccharides. BBMV contains various enzymatic complex that are mainly involved in α(1→4) bond cleavage, and may also participate in α(1→2) and α(1→6) bond cleavage (Hooton et al. 2015). Representative enzymes in BBMV are the sucrase-isomaltase complex associated with the cleavage of α(1→2), α(1→4), and α(1→6) bonds and maltase, which degrades α(1→4) and α(1→6) bonds. In addition, the maltase-glucoamylase complex is present in BBMV (McConnell et al. 2011). Moreover, Kondo et al. (2017) reported that the digestion rates of various RMDs by BBMV extracted from rat small intestine were approximately 25%, similar to that of the present study. Lee and Hamaker (2018) also reported that human mucosal maltase and glucoamylase mainly contribute to the hydrolysis of MOS. Therefore, FOS and Ad-FOS are slightly digested by digestive enzymes, and most can reach the large intestine, whereas RMD and MOS show relatively high digestibility and only a fraction can reach the large intestine.
For investigating the effects of foods or ingredients on the gut microbiome, in vivo experiments in humans or animals are the best models. However, these models are difficult to analyze by factors such as age, sex, diet, geography, genetic background, and antibiotic use (Venema and van den Abbeele 2013). Additionally, they are expensive and time-consuming and have limitations that are difficult to control. Therefore, effective alternatives such as in vitro gut models are needed to study prebiotic effects by controlling these factors (Roupar et al. 2021). In vitro models are invaluable tools for scientists to investigate the effects of foods and functional materials on the gut microbiome, providing easy, fast, and inexpensive means of using one or more gut regions (Roupar et al. 2021). Furthermore, the in vitro gut model can utilize pure, mixed cultures or human feces by precisely controlling pH and temperature from batch to continuous culture which provides similar results to in vivo experiments (Song et al. 2004). However, in vitro models have limitations such as difficulties in the absorption of metabolites and water and incapable adhesion of microbiome on the intestinal epithelial cells, even though models using dialysis membrane have been developed (Le Blay et al. 2010; Van den Abbeele et al. 2010).
The fermentability of the prebiotic oligosaccharide was analyzed in individual cultures. The 31 microorganisms used in this study were selected among the culturable species that play an important role in health and the intestinal ecosystem. Lactobacillus spp. (now reclassified as Limosilactobacillus, Lactiplantibacillus, Lacticaseibacillus, and Lactobacillus spp.) and Bifidobacterium spp. are the most well-known beneficial bacteria and provide health benefits by producing SCFAs. Accordingly, the Korean Ministry of Food and Drug Safety (MFDS) approved 17 species (No. 1–17 in this study) of Lactobacillus spp. and Bifidobacterium spp. as probiotics. In addition, Bacteroides spp. are symbiotic bacteria that can break down various polysaccharides and form a symbiotic relationship between intestinal microbes (Wexler and Goodman 2017). Furthermore, the harmful bacteria used in this study were microorganisms associated with various diseases and food poisoning. The total 31 strains including beneficial, commensal, and harmful bacteria, were used in this study (Table 1). Among the 17 probiotics, in FOS containing medium, Bi. longum, Bi. breve, L. salivarius, L. gasseri, L. acidophilus, L. plantarum, and L. fermentum were cultured individually. Ad-FOS showed identical patterns of cell growth but higher optical densities than FOS. In in vitro fecal fermentation with FOS and Ad-FOS, Bifidobacterium spp. increased the most, and Bacteroides spp. decreased relatively. Inulin, oligofructose, and fructooligosaccharide, possessing β(2→1) bonds, are the most studied prebiotics. According to Meyer and Stasse-Wolthuis (2009), the administration of inulin and FOS extracted from chicory to adult participants had an important bifidogenic effect on the composition of the colonic microbiota. Interestingly, the microbial diversity patterns of FOS and Ad-FOS were different, and the bifidogenic effect of FOS was greater than that of Ad-FOS. The content of 1-kestose (GF2) in FOS and Ad-FOS was 38.69% and 87.96%, respectively. Tochio et al. (2018) reported that Bifidobacterium spp. grew faster and more abundantly in medium with 1-kestose than nystose (GF3), which is consistent with the results of the present study. In contrast, Sannohe et al. (2008) reported a species-dependent preference of Bifidobacterium spp. for GF2 or nystose (GF3), the major components of FOS. Based on these results, the different ratios of components in Ad-FOS and FOS, and the type of strain in the cultures may change their bifidogenic effects. Similarly, in the individual culture, AD-FOS showed a higher bifidogenic effect than FOS; however, in the mixed culture, the bifidogenic effect of FOS was greater than that of Ad-FOS. Whereas pathogenic E. coli O157: H7 was grown in single culture supplemented with FOS, but its adverse effects on the actual intestine will be prevented by Bifidobacterium spp. in the gut, which are known to inhibit E. coli O157: H7 (Fukuda et al. 2011). Meanwhile, in our result of MOS-containing media, many strains were grown in the order of L. gasseri, L. plantarum, Ba. ovatus, L. helveticus, Bi. longum, Ba. vulgatus, Ba. xylanisolvens, L. fermentum, Bi. breve, E. coli, Bi. lactis, Ba. uniformis, and L. reuteri. According to Crittenden and Playne (1996), only a small portion of MOS reaches the colon due to its hydrolysis and absorption in the intestine; thus, its prebiotic effects on the gut were insignificant despite the fermentability of many commensal and beneficial microorganisms. Taken together, FOS, Ad-FOS and RMD were regarded as relatively effective prebiotics for the growth of beneficial microbes among the tested, although all oligosaccharides showed growth stimulation activities for commensal and beneficial microorganisms.
ISAPP proposed a broad definition of a prebiotic as ‘a substrate that is selectively utilized by host microorganisms conferring a health benefit’ (Gibson et al. 2017). When analyzing selective utilization of a substrate, it is necessary to consider the ecological relationships of beneficial bacteria with commensal and harmful bacteria owing to cross-feeding phenomenon in microbiome. Cross-feeding is defined as a symbiotic microbial interaction in which specific microorganisms break down complex compounds into low-molecular-weight substances that are used for the growth of other microorganisms (Sung et al. 2017). Therefore, it was predicted that there would be a cross-feeding interaction between microorganisms during the co-cultivation of dual bacteria. As shown in Fig. 1, Bacteroides spp. showed broad fermentability for all oligosaccharides used in this study. They have polysaccharide utilization loci, which encodes various glycolytic enzymes; therefore, they can degrade various poly- and oligosaccharides, such as RMD (Grondin et al. 2017). As shown in Fig. 2, when Ba. xylanisolvens was cultured in the medium containing RMD, three probiotic species, L. plantarum, L. gasseri, and L. helveticus, showed significantly higher cell growth compared with the single cultivations. These results indicate that Ba. xylanisolvens could confer a cross-feeding effect to facilitate the growth of other beneficial bacteria during co-culture in RMD medium. However, the same distinct change was not observed in in vitro fecal fermentation with RMD (Fig. 3). While the abundance of Bacteroides spp. significantly increased after consumption of RMD, the abundance of Bifidobacterium spp. slightly increased (p<0.05), but Lactobacillus spp. decreased. Rösch et al. (2015) reported a similar result; RMD enriched the abundance of Bacteroides spp., whereas the abundance of Bifidobacterium spp. and Lactobacillus spp. decreased after in vitro fecal fermentation. These results imply that the cross-feeding effect demonstrated in the co-cultivation experiment focusing on dual interactions may not be generalized in fecal fermentation, where a complex interaction exists in the microbial ecosystem.
In the analyses of SCFA produced from in vitro fecal fermentation, acetate and lactate were mainly produced in FOS, Ad-FOS, and MOS, while acetate and propionate were produced in RMD. Propionate plays a role in weight and blood sugar control by inhibiting lipogenesis in the liver, and together with acetate, it stimulates FFAR2 (free fatty acid receptor) to suppress the secretion of ghrelin (an appetite-increasing hormone). Therefore, these results indicate that oligosaccharides are fermented by human fecal microbiota to produce a large amount of SCFAs, which is beneficial to human health. In the metabolome analysis, the amount of betaine decreased as fermentation progressed along with an increase in TMA because betaine was converted to TMA by the intestinal microflora (Tang et al. 2019). However, TMA is absorbed into the body, moves to the liver, and is subsequently converted to TMAO in the liver, causing various cardiovascular diseases, such as arteriosclerosis and myocardial infarction. Gut microbiome that converts betaine into TMA are mainly Proteobacteria, Firmicutes, and Actinobacteria, but Bacteroidetes cannot (Craciun and Balskus 2012). Therefore, the health benefits of the remaining betaine and the reduced risk of cardiovascular disease are expected, as RMD supplementation will lower the conversion of betaine to TMA, with an increased abundance of Bacteroides spp. In comparison, in RMD-containing media, the TMA concentration was maintained until 24 h, whereas in the medium containing FOS, Ad-FOS, or MOS, a high amount of TMA was produced.
In conclusion, the oligosaccharides tested in this study showed typical prebiotic effects to promote the growth of beneficial and commensal bacteria and produce SCFAs. However, their action patterns and efficacy in the colon vary owing to their different digestibility, fermentability, and cross-feeding interactions in complex microbiome ecosystems. The analysis results obtained in this study will provide comprehensive information of these substances to manufacturers and customers.
Supplementary Material
Contributor Information
Seongwon Cheon, Brain Korea 21 Center for Bio-Health Industry, Department of Food Science and Biotechnology, Chungbuk National University, Cheongju 28644, Republic of Korea.
Geonhee Kim, Brain Korea 21 Center for Bio-Health Industry, Department of Food Science and Biotechnology, Chungbuk National University, Cheongju 28644, Republic of Korea.
Jae-Han Bae, Brain Korea 21 Center for Bio-Health Industry, Department of Food Science and Biotechnology, Chungbuk National University, Cheongju 28644, Republic of Korea.
Dong Hyeon Lee, Brain Korea 21 Center for Bio-Health Industry, Department of Food Science and Biotechnology, Chungbuk National University, Cheongju 28644, Republic of Korea.
Hyunbin Seong, Brain Korea 21 Center for Bio-Health Industry, Department of Food Science and Biotechnology, Chungbuk National University, Cheongju 28644, Republic of Korea.
Da Hye Kim, Brain Korea 21 Center for Bio-Health Industry, Department of Food Science and Biotechnology, Chungbuk National University, Cheongju 28644, Republic of Korea.
Jung-Sook Han, Samyang Corp. 295 Pangyo-ro, Samyang Corporation Food Biotech R&D Center, Bundang-gu, Seongnam-si Gyeonggi-do 13488, Republic of Korea.
Su-Youn Lim, Samyang Corp. 295 Pangyo-ro, Samyang Corporation Food Biotech R&D Center, Bundang-gu, Seongnam-si Gyeonggi-do 13488, Republic of Korea.
Nam Soo Han, Brain Korea 21 Center for Bio-Health Industry, Department of Food Science and Biotechnology, Chungbuk National University, Cheongju 28644, Republic of Korea.
Conflicts of interest statement
Jung-Sook Han and Su-Youn Lim are employed by the Samyang Corp. The authors declare no other conflicts of interest.
Funding
This study was supported by a research fund from the Samyang Corp.
References
- Bacic MK, Smith CJ.. Laboratory maintenance and cultivation of Bacteroides species. Curr Protoc Microbiol. 2008;9:13C.11.11-13C.11.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MRet al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJet al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Possemiers S, Van de Wiele Tet al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JL, Erickson JM, Lloyd BBet al. Health effects and sources of prebiotic dietary fiber. Curr Develop Nutrit. 2018;2:nzy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craciun S, Balskus EP.. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci. 2012;109:21307–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden Ra, Playne MJ.. Production, properties and applications of food-grade oligosaccharides. Trends Food Sci Technol. 1996;7:353–61. [Google Scholar]
- Frei R, Akdis M, O'Mahony L. Prebiotics, probiotics, synbiotics, and the immune system: experimental data and clinical evidence. Curr Opin Gastroenterol. 2015;31:153–8. [DOI] [PubMed] [Google Scholar]
- Fukuda S, Toh H, Hase Ket al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–7. [DOI] [PubMed] [Google Scholar]
- Gibson GR, Hutkins R, Sanders MEet al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14:491–502. [DOI] [PubMed] [Google Scholar]
- Gibson GR, Roberfroid MB.. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995;125:1401–12. [DOI] [PubMed] [Google Scholar]
- Grondin JM, Tamura K, Déjean Get al. Polysaccharide utilization loci: fueling microbial communities. J Bacteriol. 2017;199:e00860–00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaker BR, Tuncil YE.. A perspective on the complexity of dietary fiber structures and their potential effect on the gut microbiota. J Mol Biol. 2014;426:3838–50. [DOI] [PubMed] [Google Scholar]
- Hooton D, Lentle R, Monro Jet al. (eds). Reviews of Physiology, Biochemistry and Pharmacology, vol. 168, Springer: Cham, 2015, 59–118. [DOI] [PubMed] [Google Scholar]
- Jang EY, Ahn Y, Suh HJet al. Amylase-producing maltooligosaccharide provides potential relief in rats with loperamide-induced constipation. Evid-Based Complement Altern Med. 2020;2020:5470268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G, Bae J-H, Cheon Set al. Prebiotic activities of dextran from Leuconostoc mesenteroides SPCL742 analyzed in the aspect of the human gut microbial ecosystem. Food Funct. 2022;13:1256–67. [DOI] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer Tet al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Handa K, Genda Tet al. Digestion-resistant dextrin derivatives are moderately digested in the small intestine and contribute more to energy production than predicted from large-bowel fermentation in rats. J Nutr. 2017;147:330–6. [DOI] [PubMed] [Google Scholar]
- Krupa-Kozak U, Świątecka D, Bączek Net al. Inulin and fructooligosaccharide affect in vitro calcium uptake and absorption from calcium-enriched gluten-free bread. Food Funct. 2016;7:1950–8. [DOI] [PubMed] [Google Scholar]
- Kruse H-P, Kleessen B, Blaut M.. Effects of inulin on faecal bifidobacteria in human subjects. Br J Nutr. 1999;82:375–82. [DOI] [PubMed] [Google Scholar]
- Le Blay G, Chassard C, Baltzer Set al. Set up of a new in vitro model to study dietary fructans fermentation in formula-fed babies. Br J Nutr. 2010;103:403–11. [DOI] [PubMed] [Google Scholar]
- Lee B-H, Hamaker BR.. Maltase has most versatile α-hydrolytic activity among the mucosal α-glucosidases of the small intestine. J Pediat Gastroenterol Nutri. 2018;66:S7–S10. [DOI] [PubMed] [Google Scholar]
- Lee J-E, Lee B-J, Chung J-Oet al. (2011) 1H NMR-based metabolomic characterization during green tea (Camellia sinensis) fermentation. Food Res Int. 2011;44:597–604. [Google Scholar]
- Luis AS, Briggs J, Zhang Xet al. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nature Microbiology. 2018;3:210–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B, Li D, Zhao Jet al. In vitro fermentation of fructooligosaccharides with human gut bacteria. Food & Function. 2015;6:947–54. [DOI] [PubMed] [Google Scholar]
- McConnell RE, Benesh AE, Mao Set al. Proteomic analysis of the enterocyte brush border. Am J Physiol-Gastrointest Liver Physiol. 2011;300:G914–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Stasse-Wolthuis M.. The bifidogenic effect of inulin and oligofructose and its consequences for gut health. Eur J Clin Nutr. 2009;63:1277–89. [DOI] [PubMed] [Google Scholar]
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–8. [Google Scholar]
- Minekus M, Alminger M, Alvito Pet al. A standardised static in vitro digestion method suitable for food–an international consensus. Food Funct. 2014;5:1113–24. [DOI] [PubMed] [Google Scholar]
- Moon JS, Joo W, Ling Let al. In vitro digestion and fermentation of sialyllactoses by infant gut microflora. J Funct Foods. 2016;21:497–506. [Google Scholar]
- Moreno FJ, Corzo N, Montilla Aet al. Current state and latest advances in the concept, production and functionality of prebiotic oligosaccharides. Curr Opin Food Sci. 2017;13:50–5. [Google Scholar]
- Murakami R, Hashikura N, Yoshida Ket al. Growth-promoting effect of alginate on Faecalibacterium prausnitzii through cross-feeding with Bacteroides. Food Res Int. 2021;144:110326. [DOI] [PubMed] [Google Scholar]
- Myhrstad MC, Tunsjø H, Charnock Cet al. Dietary fiber, gut microbiota, and metabolic regulation—Current status in human randomized trials. Nutrients. 2020;12:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobre C, Sousa S, Silva Set al. In vitro digestibility and fermentability of fructo-oligosaccharides produced by Aspergillus ibericus. J Funct Foods. 2018;46:278–87. [Google Scholar]
- Nogacka AM, Salazar N, Arboleya Set al. In vitro evaluation of different prebiotics on the modulation of gut microbiota composition and function in morbid obese and normal-weight subjects. Int J Mol Sci. 2020;21:906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-M, Jang M-U, Oh GWet al. (2015) Synergistic action modes of arabinan degradation by exo-and endo-arabinosyl hydrolases. J Microbiol Biotechnol. 2015;25:227–33. [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz Pet al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösch C, Venema K, Gruppen Het al. Characterisation and in vitro fermentation of resistant maltodextrins using human faecal inoculum and analysis of bacterial enzymes present. Bioactive Carbohydrates and Dietary Fibre. 2015;6:46–53. [Google Scholar]
- Roupar D, Berni P, Martins JTet al. Bioengineering approaches to simulate human colon microbiome ecosystem. Trends Food Sci Technol. 2021;112:808–22. [Google Scholar]
- Sannohe Y, Fukasawa T, Koga Jet al. Comparison of the growth of bifidobacteria in two culture media containing either 1-kestose (GF2) or nystose (GF3). Biosci Microflora. 2008;27:13–7. [Google Scholar]
- Smith NW, Shorten PR, Altermann Eet al. The classification and evolution of bacterial cross-feeding. Front Ecol Evolut. 2019;7:153. [Google Scholar]
- Song Y, Liu C, Finegold SM.. Real-time PCR quantitation of Clostridia in feces of autistic children. Appl Environ Microbiol. 2004;70:6459–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm M, Schroeder C, Bauer P.. SeqPurge: highly-sensitive adapter trimming for paired-end NGS data. BMC Bioinf. 2016;17:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung J, Kim S, Cabatbat JJTet al. Global metabolic interaction network of the human gut microbiota for context-specific community-scale analysis. Nat Commun. 2017;8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Li DY, Hazen SL.. Dietary metabolism, the gut microbiome, and heart failure. Nat Rev Cardiol. 2019;16:137–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z-Z, Chen G, Hong Qet al. Multi-omic analysis of the microbiome and metabolome in healthy subjects reveals microbiome-dependent relationships between diet and metabolites. Frontiers in Genetics. 2019;10:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochio T, Kadota Y, Tanaka Tet al. 1-Kestose, the smallest fructooligosaccharide component, which efficiently stimulates faecalibacterium prausnitzii as well as bifidobacteria in humans. Foods. 2018;7:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Abbeele P, Grootaert C, Marzorati Met al. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for bacteroidetes and Clostridium cluster IX. Appl Environ Microbiol. 2010;76:5237–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema K, Van den Abbeele P.. Experimental models of the gut microbiome. Best Pract Res Clin Gastroenterol. 2013;27:115–26. [DOI] [PubMed] [Google Scholar]
- Wang L, Li C, Huang Qet al. In vitro digestibility and prebiotic potential of a novel polysaccharide from Rosa roxburghii Tratt fruit. J Funct Foods. 2019;52:408–17. [Google Scholar]
- Wang Z, Klipfell E, Bennett BJet al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler AG, Goodman AL. An insider's perspective: B acteroides as a window into the microbiome. Nat Microbiol. 2017;2:17026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisner CM, Martin BR, Schoterman MHCet al. Galacto-oligosaccharides increase calcium absorption and gut Bifidobacteria in young girls: a double-blind cross-over trial. Br J Nutr. 2013;110:1292–303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.