Summary
Hypersensitive response (HR)‐conferred resistance is associated with induction of programmed cell death and pathogen spread restriction in its proximity. The exact role of chloroplastic reactive oxygen species and its link with salicylic acid (SA) signaling in HR remain unexplained.
To unravel this, we performed a detailed spatiotemporal analysis of chloroplast redox response in palisade mesophyll and upper epidermis to potato virus Y (PVY) infection in a resistant potato genotype and its transgenic counterpart with impaired SA accumulation and compromised resistance.
Besides the cells close to the cell death zone, we detected individual cells with oxidized chloroplasts further from the cell death zone. These are rare in SA‐deficient plants, suggesting their role in signaling for resistance. We confirmed that chloroplast redox changes play important roles in signaling for resistance, as blocking chloroplast redox changes affected spatial responses at the transcriptional level.
Through spatiotemporal study of stromule induction after PVY infection, we show that stromules are induced by cell death and also as a response to PVY multiplication at the front of infection. Overall induction of stromules is attenuated in SA‐deficient plants.
Keywords: chloroplast redox state, hypersensitive response (HR)‐conferred resistance, immune signaling, live cell imaging, Solanum tuberosum (potato), spatiotemporal analysis, stromules, virus resistance
Introduction
Plants have evolved a range of constitutive and inducible resistance mechanisms to respond to pathogen attack. The effector‐triggered immunity (ETI) is mediated by intracellular resistance (R) proteins, which recognize pathogen‐derived effectors (Jones & Dangl, 2006). Successful ETI often results in hypersensitive response (HR)‐conferred resistance and is associated with the formation of localized programmed cell death (PCD) (Künstler et al., 2016). This response involves salicylic acid (SA) biosynthesis in cytosol and production of reactive oxygen species (ROS) in chloroplasts and the apoplast (Lu & Yao, 2018; Balint‐Kurti, 2019; Littlejohn et al., 2021). Besides hosting the biosynthesis of SA and ROS, chloroplasts play a central role in plant immunity as integrators of environmental signals and transmitters of pro‐defense signals (Serrano et al., 2016; Kachroo et al., 2021; Li & Kim, 2022). Stromules are stroma‐filled tubules that extend from chloroplasts and are induced in several different processes, including HR (Caplan et al., 2015). Stromules are putatively involved in retrograde signaling after pathogen invasion or light stress (Brunkard et al., 2015; Caplan et al., 2015) and in movement of chloroplasts within the cell (Kumar et al., 2018).
It is established that SA is required for the restriction of pathogens during HR in various interactions, including plant–virus pathosystems (Mur et al., 2008; Baebler et al., 2014; Künstler et al., 2016; Calil & Fontes, 2017). Several studies have also pointed to the essential role of apoplastic ROS, leading to redox state changes in the cytoplasm in HR‐conferred virus resistance (Hernández et al., 2016; Lukan et al., 2020). It has been suggested that chloroplastic ROS are also involved in the signaling for and/or execution of HR cell death in incompatible interactions (Liu et al., 2007; Zurbriggen et al., 2009; Straus et al., 2010; Ishiga et al., 2012; Kim et al., 2012; Xu et al., 2019; Lukan et al., 2020). Zurbriggen et al. (2009) suggested that ROS generated in chloroplasts during nonhost disease resistance are essential for the progress of PCD, but do not contribute to the induction of pathogenesis‐related genes or other signaling components of the response, including SA signaling. On the other hand, Ochsenbein et al. (2006) found that chloroplastic singlet oxygen (1O2) can activate SA‐mediated signaling, although SA is not required for 1O2‐mediated cell death (Ochsenbein et al., 2006). Similarly, Straus et al. (2010) suggested that chloroplastic ROS acts as a flexible spatiotemporal integration point, leading to opposite SA signaling reactions in infected and surrounding tissue (Straus et al., 2010). Moreover, recent evidence suggests that chloroplastic ROS might also be involved in controlling plant immune responses by reprogramming transcription of genes involved in response to pathogen attack as one of the retrograde signals (Ochsenbein et al., 2006; Lee et al., 2007; Maruta et al., 2012; Nomura et al., 2012; Sewelam et al., 2014; Pierella Karlusich et al., 2017). For example, inducible silencing of chloroplastic THYLAKOIDAL ASCORBATE PEROXIDASE increased H2O2 production in chloroplasts, which activated SA biosynthesis and SA‐inducible gene expression (Maruta et al., 2012).
To decipher the consequence of events involving chloroplastic ROS in HR‐conferred resistance, a detailed spatiotemporal analysis of the chloroplast redox state in response to pathogen infection is needed. Nondestructive real‐time measurement of the redox state has been feasible since genetically encoded sensors became available, for example redox state‐sensitive green fluorescent proteins (roGFP1 and roGFP2) in plants (Jiang et al., 2006). Measurement of roGFP fluorescence intensity following excitation at two different excitation maxima permits an evaluation of the relative proportion of roGFP in a reduced or oxidized state. By adding the coding sequence for the RuBisCO small subunit transit peptide to the roGFP coding sequence, pt‐roGFP was constructed for measuring changes of the redox state in chloroplasts (Stonebloom et al., 2012). pt‐roGFP also allows visualization of stromule formation and observation of redox state changes in stromules.
Potato virus Y (PVY), a member of the genus Potyvirus, is the most harmful virus of cultivated potatoes (Karasev & Gray, 2013) and is among the top 10 most economically important plant viruses overall (Quenouille et al., 2013). In response to PVY, HR in potato cv. Rywal is manifested as the formation of necrotic lesions on inoculated leaves at 3 d post‐inoculation (dpi) and the virus is restricted to the site of inoculation (Szajko et al., 2008). We have shown previously that SA regulates HR‐conferred resistance in a spatiotemporal manner, as for some genes involved in immune response the spatiotemporal regulation is completely lost in the SA‐deficient line, whereas other genes show diverse spatiotemporal responses (Lukan et al., 2020). We have also proposed the role of chloroplastic ROS as a signal orchestrating PCD (Lukan et al., 2020). However, the potential role of chloroplastic ROS in viral arrest has not yet been deciphered.
Here, we developed a protocol for confocal imaging of pt‐roGFP potato plants complemented by custom image analysis scripts to simultaneously interrogate the chloroplast redox state and stromule formation in proximity to the cell death zone in palisade mesophyll cells and upper epidermis with spatiotemporal resolution. To decipher the link between cell death, SA signaling, chloroplast redox state and stromule formation, we also analyzed the responses to PVY infection in the SA‐depleted transgenic counterpart of pt‐roGFP (pt‐roGFP‐NahG). We show that chloroplasts are strongly oxidized in the cells adjacent to the cell death zone. More intriguingly, we detected individual cells with chloroplasts in moderately oxidized redox state close to the cell death zone as well as further away from it. The presence of such cells was sparse in SA‐deficient plants, supporting the hypothesis that these cells are involved in the signaling of HR‐conferred resistance.
Materials and Methods
Generation of redox state sensor plants
Potato (Solanum tuberosum L.) transgenic lines pt‐roGFP and pt‐roGFP‐NahG were prepared by introducing pt‐roGFP2 encoding construct (Stonebloom et al., 2012) into cv. Rywal, which is resistant to PVY infection following HR response, and NahG‐Rywal with the SA accumulation reduced to < 10% of that in nontransgenic plants, thus rendering HR ineffective in blocking the viral spread (the depletion of SA renders NahG plants susceptible; Baebler et al., 2014). pCAMBIA1304_pt‐roGFP2 was electroporated into Agrobacterium tumefaciens LBA4404 (Eppendorf Electroporator 2510, Hamburg, Germany) at 2000 V following the manufacturer's protocol. The transformed bacteria were used for the transformation of cv. Rywal and NahG‐Rywal stem internodes from in vitro plantlets as described elsewhere (Baebler et al., 2014), with some modifications (Supporting Information Methods S1). Well‐rooted hygromycin‐resistant plants were subcultured to produce plantlets of the independently transformed lines. The selection of transgenic lines with strong and stable GFP fluorescence in chloroplasts was performed by confocal microscopy (see later). To confirm that the detected signal is specific for GFP emission, we used Rywal and NahG‐Rywal plants as controls.
Transgenic lines were grown in stem node tissue culture. At 2 wk after node segmentation, they were transferred to soil in a growth chamber and kept under controlled environmental conditions as described previously (Baebler et al., 2009).
Construction of PVY‐N605(123)‐GFP infectious clone
PVY‐N605(123)‐GFP was constructed by inserting GFP coding sequence between the coding sequences for viral proteins NIb and CP in the PVY‐N605(123) infectious clone (Bukovinszki et al., 2007) using a similar design as Rupar et al. (2015), allowing the GFP reporter to be excised from the polyprotein following translation.
The GFP coding sequence was amplified from plasmid p2GWF7 (Karimi et al., 2002) using GFPF40 and GFPR40 primers (Methods S2) with Phusion polymerase (New England BioLabs, Ipswich, MA, USA) according to the provider's instructions. The primers were designed with overhangs enabling addition of the PVY‐N605(123) annealing sequence and protease recognition site to the GFP sequence, making the amplicon serve as a megaprimer for restriction‐free insertion with mutagenesis. The reaction was designed using the RF‐Cloning online tool (Bond & Naus, 2012) and carried out using Phusion polymerase with a 1 : 20 molar ratio of PVY‐N605(123) plasmid to GFP amplicon (megaprimer). After DpnI digestion, the mutagenesis mixture was transformed into E. coli XL10‐Gold Ultracompetent Cells following the manufacturer's instructions (Agilent Technologies, Santa Clara, CA, USA). The transformants were plated on LB‐agar with ampicillin selection and grown overnight at 37°C. Grown colonies were screened with colony PCR using CP‐F and UnivR primers and KAPA2G Robust HotStart Kit (Agilent Technologies; Methods S2). Sanger sequencing of the selected clone confirmed the correct sequence of the PVY‐coding part and correct in‐frame insertion of the GFP coding sequence. Constructed PVY‐N605(123)‐GFP was coated onto gold microcarriers and used for Nicotiana clevelandii bombardment with a Helios® gene gun (Bio‐Rad) according to Stare et al. (2020). Detailed PCR reaction conditions and cloning steps are given in Methods S2.
Virus inoculation
Two‐ to four‐week‐old potato plants in soil were inoculated with PVYN‐Wilga (PVYN‐Wi; EF558545), PVY N605‐GFP (Rupar et al., 2015), PVY‐N605(123)‐GFP or mock.
For inoculation with PVYN‐Wilga, 6‐ to 8‐wk‐old PVYN‐Wilga‐infected cv. Pentland plants from tissue cultures were used. For inoculation with PVY N605‐GFP and PVY‐N605(123)‐GFP, upper noninoculated leaves of PVY N605‐GFP‐ and PVY‐N605(123)‐GFP‐inoculated Nicotiana clevelandii at 3–4 dpi were used. Before inoculation, GFP fluorescence in the leaves used for inoculum was confirmed under confocal microscope. The tissue was ground in phosphate buffer supplemented with DIECA in a plant material : buffer ratio of 1 : 4. For mock inoculation (MOCK), noninfected N. clevelandii and cv. Pentland plants were used.
Inoculation was performed as described in Baebler et al. (2009). Briefly, the first three fully developed bottom leaves of potato plants were dusted with carborundum powder. Leaves were rubbed with the inoculum (c. 60 μl per leaf) which was removed after 10 min by rinsing with tap water.
Treatments with oxidant and reductant
The second and third leaves of 2‐ to 4‐wk‐old pt‐roGFP and pt‐roGFP‐NahG potato transgenic lines were treated with an oxidant (200 mM solution of hydrogen peroxide, H2O2, adapted for potato from Jiang et al., 2006), a reductant (200 mM solution of dithiothreitol (DTT) adapted for potato from Jiang et al., 2006) or a control (bidistilled water, ddH2O) by vacuum infiltration using a syringe. We followed the relative redox state in the chloroplasts 30 min after the treatment, to confirm that the redox state sensor roGFP2 is functional in the selected transgenic lines (Fig. S1; Tables S1, S2).
Treatments with ROS inhibitor
Uracil, a ROS inhibitor (Montaña et al., 2009; Abdollahi & Ghahremani, 2011), was infiltrated into PVY‐N605(123)‐GFP‐inoculated leaves of pt‐roGFP L2 plants with 100 μM uracil (Sigma; U0750‐25G in 224 μM NaOH diluted in ddH2O; left side of the leaf) at 4 dpi. As a control, the right side of the leaf was inoculated with 224 μM NaOH diluted in ddH2O. At 48 h post‐treatment, the relative chloroplast redox state close to the lesion was determined to confirm functionality of chloroplastic ROS inhibitor in the selected transgenic line.
Confocal microscopy
A confocal microscope (Leica TCS LSI macroscope with Plan APO ×5 and ×20 objective; Leica Microsystems, Wetzlar, Germany) was used to detect emission of Chl or redox state‐sensitive GFP (pt‐roGFP2) for the selection of transgenic plants, stromule counting and redox state detection.
To select potato transgenic lines with strong and stable roGFP2 fluorescence (emission window 505–520 nm) in the chloroplasts, plants from tissue cultures were analyzed with a ×20 objective, zoom factor set to 3.00, frame average to 2 and z‐stack size adjusted to 10 steps to cover at least 30 μm of the mesophyll after excitation with 488 nm laser.
For redox state detection experiments, confocal imaging was adapted from Jiang et al. (2006). The emission of roGFP2 was followed after excitation with 405 nm (detection of oxidized roGFP2) and 488 nm (detection of reduced roGFP2) laser in the window between 505 and 520 nm. The Chl fluorescence was excited with the 488 nm laser and the emission was measured in the window between 690 and 750 nm. The fluorescence was followed on the upper side of the leaf which was detached from the plant on the particular day post‐inoculation or 30 min after the treatment with an oxidant or a reductant and immediately analyzed. Plants were imaged with a ×20 objective, zoom factor set to 3.00, and frame average to 2. Regions of interest (ROI) were scanned unidirectionally with a scan speed of 400 Hz. Fluorescence emissions were collected sequentially through three channels (roGFP2 fluorescence after excitation with 405 nm laser, roGFP2 fluorescence after excitation with 488 nm laser and Chl fluorescence). z‐stack size was adjusted to 10 steps to cover at least 30 μm of the mesophyll. As the line between epidermal and mesophyll cells in potato leaf is not flat and epidermal cells often reach into the plane of mesophyll cells, epidermal chloroplasts were also analyzed in several images. ROI sizes and magnifications for individual experiments are specified in Tables S1 and S3. The images were processed using Leica las X software (Leica Microsystems) to obtain maximum projections from z‐stacks for each of two or three channels. On mock‐inoculated and oxidant−/reductant‐treated plants, ROIs were selected randomly on the leaf. On PVYN‐Wilga‐inoculated plants, two consecutive ROIs were imaged adjacent to the lesion (ROI1, ROI2) and one ROI was imaged distant from the lesion (CTR) (Fig. 1a). See Table S1 for the number of analyzed plants, leaves, lesions and ROIs for each experiment for each treatment and time point. On PVY‐N605(123)‐GFP‐inoculated plants, ROI1 was imaged adjacent to the lesion (ROI1) or on the front of the virus spread (ROIV) (Fig. 5; Table S1).
Fig. 1.
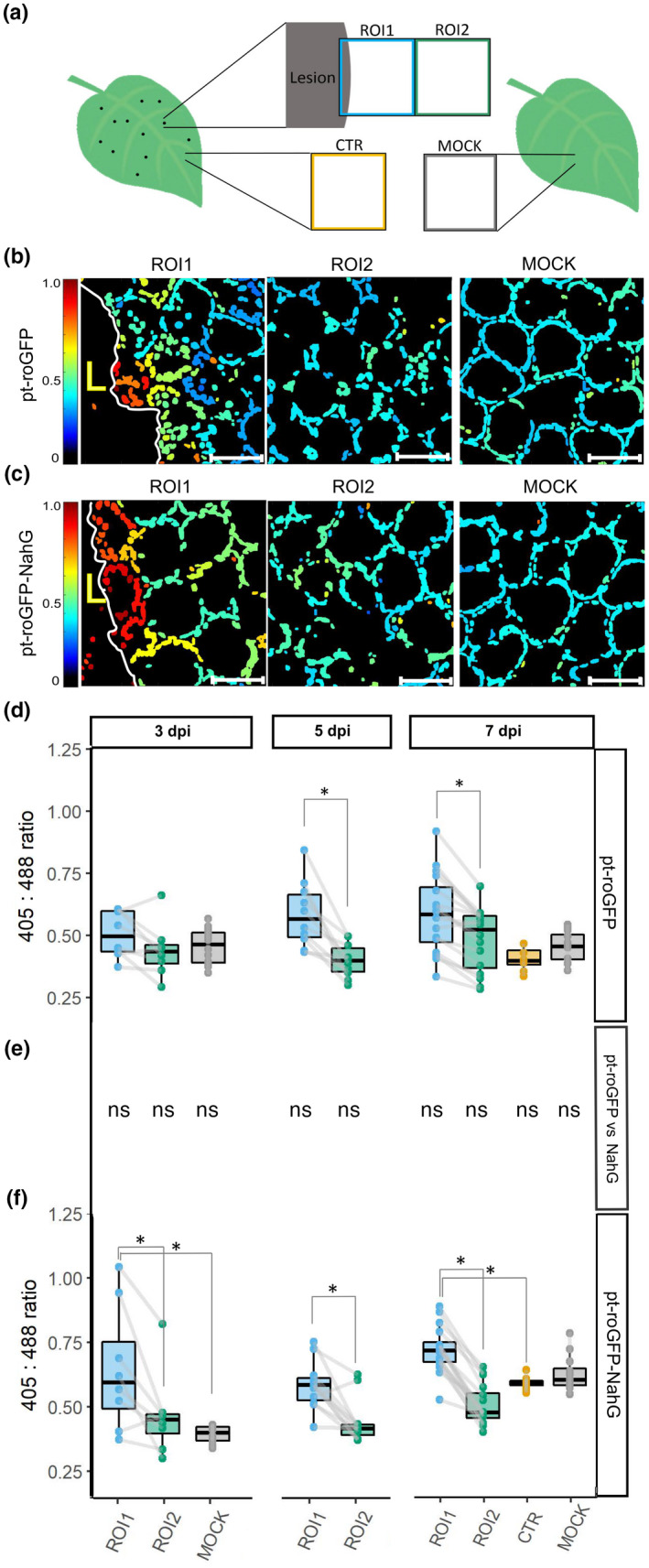
Chloroplast redox state is spatially regulated around the cell death zone during potato virus Y (PVY)‐induced hypersensitive response (HR). (a) After inoculating leaves with PVY, the chloroplast redox state was measured in transgenic sensor plants (pt‐roGFP and salicylic acid (SA)‐deficient pt‐roGFP‐NahG) with redox state‐sensitive green fluorescent protein (roGFP) targeted to chloroplasts. The chloroplast redox state was measured in three regions: the area adjacent to the lesion (region of interest 1, ROI1), the area adjacent to ROI1 (ROI2), and an area distant from the lesion (control, CTR) on the same leaf. As a negative control, the chloroplast redox state was also measured in mock‐inoculated plants (MOCK). To estimate the location of the lesion and determine the border between the cell death zone and normal tissue, the tissue was scanned in the channel for the background chloroplast fluorescence detection (the border of the lesion is marked with mesophyll cells with disorganized chloroplasts that moved away from the cell periphery) and brightfield (to detect dead tissue). In normal tissue, chloroplasts are well arranged between the vacuole and the cell wall, forming a ring in a circular shape of a mesophyll cell. (b, c) Visual presentation of the ratios of fluorescence intensities of roGFP after excitation with 405 and 488 nm laser (405 : 488 ratio, chloroplast redox state) presented on the rainbow scale for pt‐roGFP L2 (b) and pt‐roGFP‐NahG L2 (c) in ROI1, ROI2 and MOCK (from left to right). Images are maximum projections from z‐stacks processed with in‐house matlab script. Higher ratios denote chloroplasts in a more oxidized state (red). Chloroplasts in the cells surrounding the cell death zone are highly oxidized, in contrast with more distant cells. Bar, 50 μm. (d, f) Chloroplast redox state in the earlier‐mentioned leaf areas in pt‐roGFP L2 (d) and pt‐roGFP‐NahG L2 (f) plants at 3, 5 or 7 d post‐inoculation. Results are presented as boxplots with 405 : 488 ratios of each measured ROI shown as dots (Exp3NahG and Exp5NT in Supporting Information Table S1). Gray lines connect ROI1 and ROI2 pairs for each lesion. Asterisks denote statistically significant differences (P < 0.05) between the marked regions (ROI2, CTR or MOCK) and ROI1, determined by the mixed‐effects model (ANOVA). Boundaries of the box, 25th and 75th percentiles; horizontal line, median; vertical line, all points except outliers. The experiment was performed three times independently on three transgenic lines (Redox Exp6NT in Fig. S1; Tables S1, S2; doi: 10.5281/zenodo.6417635). (e) Comparison of 405 : 488 ratios for marked regions (ROI1, ROI2, CTR and MOCK) between genotypes determined by the mixed‐effects model (ANOVA). ns, not statistically significant. See Lukan et al. (2018, 2020) for the figures showing visual and microscopic lesion formation and reactive oxygen species (ROS) staining at different days post‐inoculation (dpi).
Fig. 5.
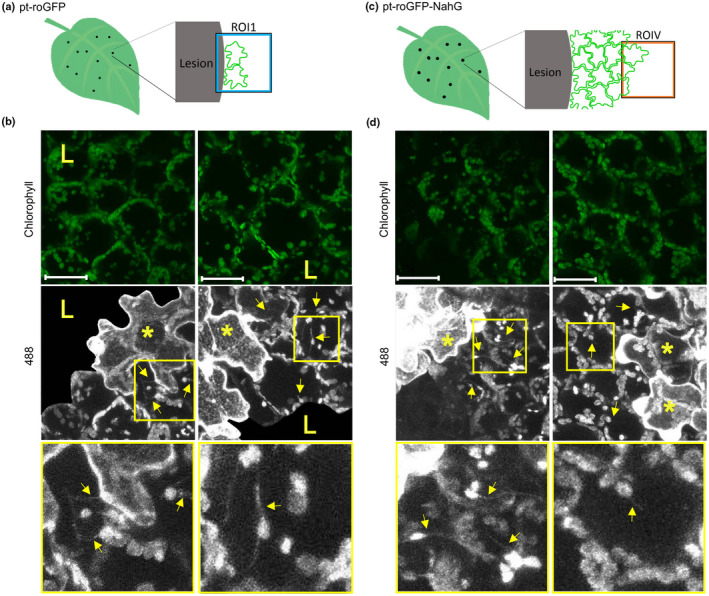
Stromules are induced in immediate proximity to the virus multiplication zone. (a, c) Stromule formation was followed in pt‐roGFP on the border of the cell death zone (ROI1) in the cells adjacent to the cell in which we observed potato virus Y‐green fluorescent protein (PVY‐GFP) accumulation at 4 d post‐inoculation (dpi) (a) or in pt‐roGFP‐NahG on the border of the virus multiplication zone (ROIV) in the cells adjacent to the cell in which we observed PVY‐GFP accumulation at 7 dpi (c). (b, d) 488, stromules (arrows) are induced adjacent to the cell in which we observed PVY‐GFP accumulation (asterisks). Boxed areas show higher magnification of regions with stromules. Owing to high background signal in the cell death zone, as a result of virus‐derived GFP fluorescence, GFP signal is not shown in the cell death zone in 488 images. Owing to a strong background in the 405 nm channel as a result of strong virus‐derived GFP fluorescence, an evaluation of the chloroplast relative redox state as the relative proportion of roGFP in an oxidized or reduced state was not possible, and therefore 405 : 488 ratios are not presented. Left and right panels present different ROIs. Stromules are marked with arrows. Cells in which we observed PVY‐GFP accumulation are marked with asterisks. The cell death zone is marked with an ‘L’. ROI1, region of interest adjacent to cell death zone; ROIV, region of interest adjacent to the virus multiplication zone. Chlorophyll, Chl fluorescence; 488, roGFP fluorescence after excitation with 488 nm laser line showing reduced roGFP (brighter chloroplasts are more reduced). z‐stack size was adjusted to 10 steps to cover the whole epidermis (c. 30 μm) and at least 30 μm of the mesophyll. Bar, 50 μm.
For stromule detection in relation to virus localization, two z‐stacks were taken at the same position. One z‐stack size was adjusted to 10 steps to cover at least 30 μm of the mesophyll (palisade tissue) and the second to cover the whole epidermis (c. 30 μm) and at least 30 μm of the mesophyll (palisade tissue) (see comments in Table S1).
For stromule counting experiments, the emission of roGFP2 was measured after excitation with a 488 nm laser in the window between 505 and 530 nm. Fluorescence emissions were collected sequentially through two channels only (roGFP2 fluorescence after excitation with 488 nm laser and Chl fluorescence).
In stromule counting experiments, PVY N605‐GFP‐inoculated plants were imaged. In Exp3NT and Exp5NahG (Table S3), plants were imaged with a ×5 objective, the frame average was set to 3 and the z‐stack was adjusted to 12–15 steps (1.50–2.00 μm per step). Four consecutive ROIs were imaged adjacent to the lesion (ROI1–ROI4; Fig. S2). In Exp4NT, plants were imaged with ×5 and ×20 objectives, the frame average was set to 3 and the z‐stack size was adjusted to 15 steps (0.8–1.0 μm per step). Two consecutive ROIs were imaged adjacent to the lesion (ROI1 and ROI2; Fig. 4a). In Exp6NTNahG, plants were imaged with a ×20 objective, the frame average was set to 3 and the z‐stack size was adjusted to 15 steps to cover the whole epidermis and at least 50 μm of the mesophyll. Two consecutive ROIs were imaged adjacent to the lesion (ROI1 and ROI2; Fig. 4a). See Table S3 for zoom factor, digital zoom, optical zoom and the numbers of analyzed plants, leaves, lesions and ROIs for each experiment for each treatment and time point.
Fig. 4.
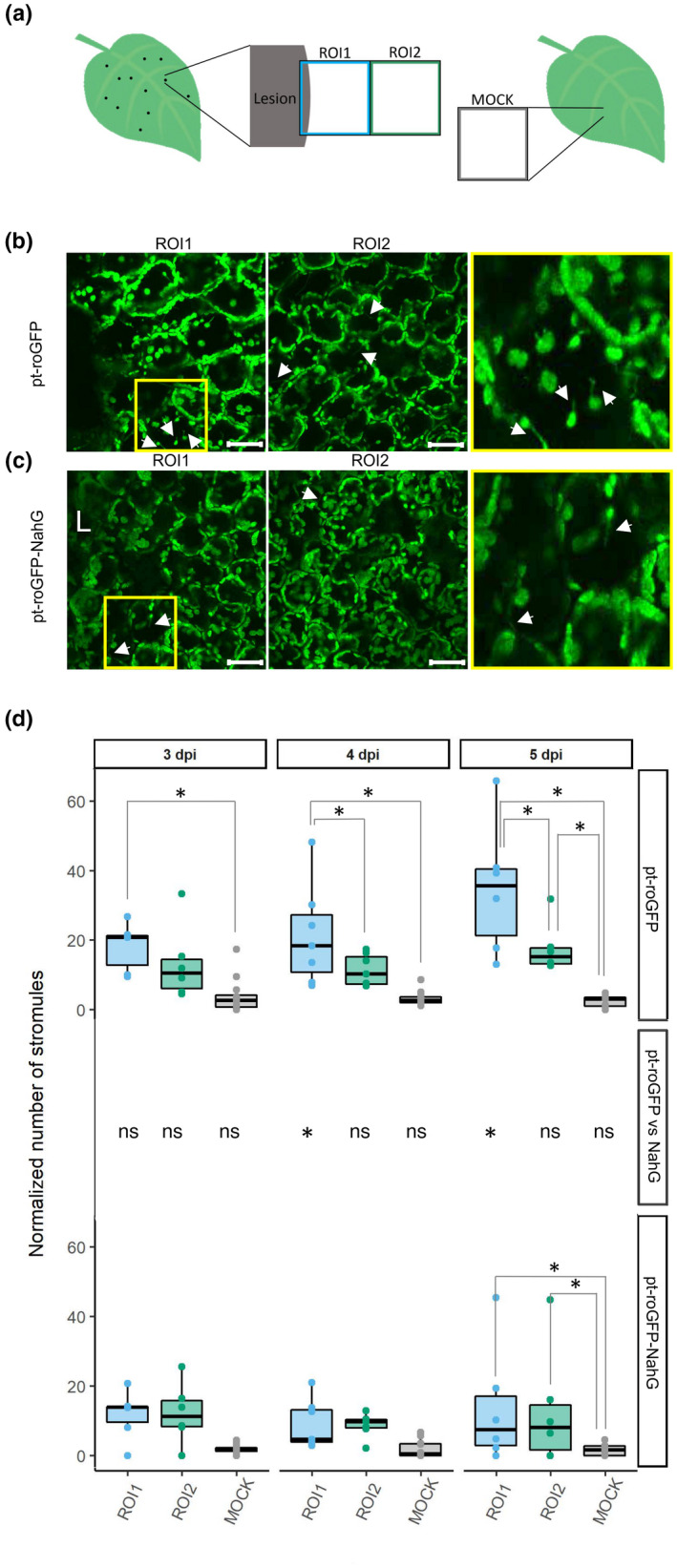
Spatiotemporal regulation of stromule formation around the cell death zone is salicylic acid (SA) signaling‐dependent. (a) Stromule formation was followed in redox state sensor plants after potato virus Y (PVY) inoculation. See Fig. 1 legend for the details. (b, c) Confocal image showing the difference in the number of stromules between region of interest 1 (ROI1, left) and ROI2 (right) in pt‐roGFP (b) and pt‐roGFP‐NahG (c). After inoculation, we observed stromule formation, which was even more pronounced in the proximity of the cell death zone in pt‐roGFP plants. Higher magnification of an area with stromules (boxed in yellow) is shown in the rightmost panel. Arrows show stromules. Bar, 50 μm. (d) Normalized number of stromules, calculated by dividing the number of stromules by the number of chloroplasts counted in the earlier‐mentioned leaf areas in pt‐roGFP L2 and pt‐roGFP‐NahG L2 transgenic lines at 3, 4 and 5 d post‐inoculation (dpi) (Exp6NTNahG in Supporting Information Table S3). Results are presented as boxplots with normalized numbers of stromules for each ROI shown as dots (Stromules Exp6NTNahG in Table S3). Asterisks denote statistically significant differences (P < 0.05) between regions (shown on the boxplots connecting lines; see Stromules Exp6NTNahG in Table S7 for P‐values) or between genotypes (pt‐roGFP vs NahG, shown for each region for each day post‐inoculation in the middle panel; see Stromules Exp6NTNahG in Table S7 for P‐values), determined by the mixed‐effects model (ANOVA). z‐stack size was adjusted to 15 steps to cover the whole epidermis and at least 50 μm of the mesophyll. ns, not statistically significant. Boundaries of the box, 25th and 75th percentiles; horizontal line, median; vertical line, all points except outliers.
In the experiment with chloroplastic ROS inhibitor, two consecutive ROIs were imaged adjacent to the lesion (ROI1, ROI2) in PVY‐N605(123)‐GFP‐inoculated plants on the left (inhibitor‐infiltrated) and right (control‐infiltrated) sides of the leaf with the same settings as in the redox state detection experiments (Fig. 6a). See Table S4 for details.
Fig. 6.
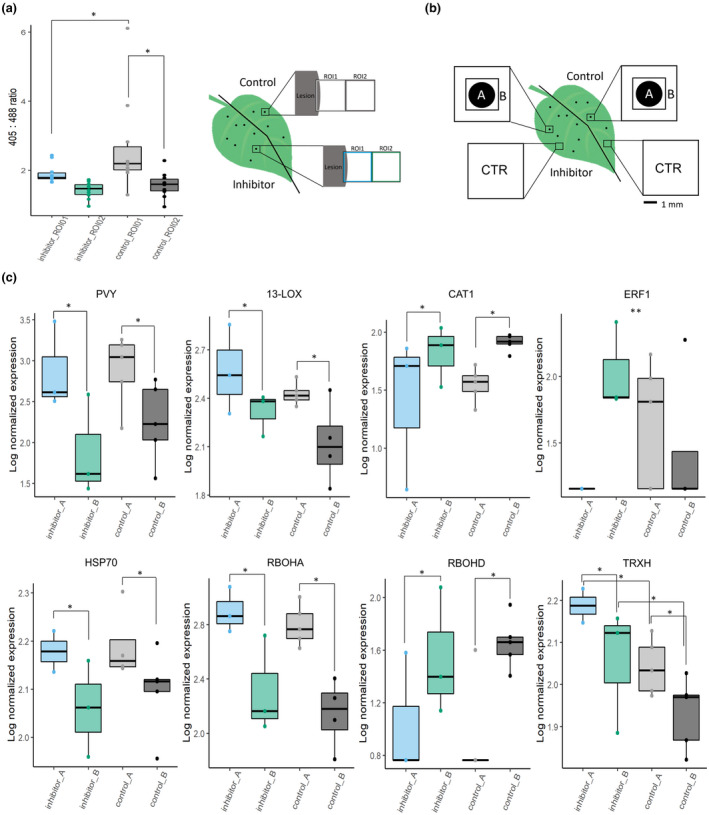
Chloroplastic redox changes regulate spatial expression of selected genes. (a) Uracil is efficiently blocking chloroplastic reactive oxygen species (ROS) production near potato virus Y (PVY) infection lesions. Left and right sides of the PVY‐green fluorescent protein (PVY‐GFP)‐inoculated leaves of pt‐roGFP plants were infiltrated with chloroplastic ROS inhibitor and control at 4 d post‐inoculation (dpi). At 48 h post‐treatment, two consecutive regions of interest (ROIs) were imaged adjacent to the lesion (ROI1, ROI2) on the left (inhibitor‐infiltrated) and right (control‐infiltrated) sides of the leaf with the same settings as in redox state detection experiments using confocal microscopy. The relative chloroplast redox state was determined to confirm functionality of the chloroplastic ROS inhibitor in the selected transgenic line. (b) Schematic overview of sampling for transcriptomics. PVY‐GFP‐inoculated leaves of pt‐roGFP L2 plants were infiltrated as described earlier. At 5 dpi, two tissue sections were sampled: lesion (section A) and a 1 mm section adjacent to section A (section B). As a control (CTR), tissue sections of the same size as the A and B sections together were sampled further away from the lesions on both the inhibitor‐ and control‐treated sides of the leaf. (c) Viral RNA abundance (PVY) and expression profiles of seven genes (HSP70, heat shock protein 70; RBOHD, potato respiratory burst oxidase homolog D; RBOHA, potato respiratory burst oxidase homolog A; CAT1, catalase 1; 13‐LOX, 13‐lipoxygenase; ERF1, potato ethylene responsive transcription factor 1a; TRXH, thioredoxin H) are presented as boxplots with logarithmic normalized relative expression in sections A and B for chloroplastic ROS inhibitor‐treated and control‐treated lesions shown as dots. Abundance/expression is presented as a log2 ratio between relative expression in each section and averaged relative expression in CTR sections. Significance for effects of treatment, tissue section and interaction between the two was tested using a linear model (ANOVA), and significant relationships are marked by an asterisk (*) in the figure for treatment and tissue section and two asterisks (**) for the interaction (Supporting Information Table S9). See Tables S4 and S6 for the details, number of analyzed lesions and P‐values for all analyzed genes. Asterisks (* and **) denote statistically significant differences (P < 0.05). Boundaries of the box, 25th and 75th percentiles; horizontal line, median; vertical line, all points except outliers.
Raw and analyzed imaging data were deposited at Zenodo (doi: 10.5281/zenodo.6417635).
Image analysis
For the detection of chloroplast redox state, maximum projections from z‐stacks for each of three channels – Chl fluorescence, roGFP2 fluorescence after excitation with a 405 nm laser line, and GFP fluorescence after excitation with a 488 nm laser line – were, for each ROI, exported from leica las X software as a tif file.
Image analysis was performed using an in‐house Matlab script (MATLAB and Image Processing Toolbox Release 2019b). The images were analyzed with the following steps: import of tif files, conversion to grayscale, filtering out pixels of low intensity, conversion to binary format using spatial adaptive thresholding, a round or erosion and dilation to remove single pixel noise around the chloroplasts, followed by size‐based segmentation of individual chloroplasts. The ratios of fluorescence intensities 405 : 488 were then calculated for each pixel belonging to the chloroplast masks obtained in the previous step. Results were calculated per image (normalized to the fraction of pixels belonging to chloroplasts) and per individual chloroplast in each image (see 405_488 in Table S1 for 405 : 488 ratios). The ratios present the relative redox state and were not normalized to ratios obtained by treatment with H2O2 and DTT, as oxidation of potato chloroplasts in close proximity to HR‐PCD was stronger than oxidation with H2O2, which is most probably a result of the ineffective uptake of the solution into the potato cells.
In the next step, ROI1 and ROI2 were further divided into five regions (bins; Fig. S3a), depending on the distance from the lesion (bin 1 being closest and bin 5 furthest from the lesion). Again, the 405 : 488 ratios were determined for each pixel inside chloroplast masks, and then the first four bins were normalized to the 405 : 488 ratio in bin 5, which was set to 1 (Table S5).
For counting stromules, maximum projections from z‐stacks for each of two channels (Chl and 488) were, for each ROI, exported from Leica las X software as a tif file. The stromules were counted manually on the 488 tif images, and the number of the chloroplasts was determined as described earlier. The number of stromules was normalized to the number of chloroplasts for the corresponding ROI (Table S3). Details of all the parameters used in the image analysis can be found in the analysis scripts, which are available on Github (https://github.com/NIB‐SI/SensorPlantAnalysis).
Statistics
To determine which factors contributed most to the variability in measured 405 : 488 ratio values, we used a linear mixed effects model (LME):
where F is the fluorescence (405 : 488 ratio) measured on day j after treatment/ROI i, on leaf o of plant n belonging to plant line k; T i is the mean (fixed) effect of treatment/ROI i; D j is the mean (fixed) effect of time after treatment/viral infection j; L k is the mean (fixed) effect of the specific plant line k; P n is the (random) effect of plant n, assumed to be normally distributed with mean 0; LE no is the (random) effect of the chosen leaf n on plant o, assumed to be normally distributed with mean 0; and e ijkno is the residual error, assumed to be normally distributed with mean 0. If factors were not relevant for individual experiments (e.g. experiments run on a single line, or measured on a single day after treatment), they were removed from the mixed‐effects model. The final LME model was fitted with maximum likelihood to model the 405 : 488 ratio as the dependent variable, treatment/ROI, dpi, transgenic line/genotype as fixed effects, and plant and leaf as random effects.
Statistical analysis was implemented as an R‐based script, with significance calculation of the fixed factors performed using the ‘anova’ function (Satterthwaite's method) from the package lmertest (Kuznetsova et al., 2017) and post hoc pairwise comparisons within levels of significant fixed factors performed using the ‘emmeans’ function (Kendall–Roger method) available from the package emmeans (Lenth et al., 2021). The post hoc pairwise comparisons were performed between levels of individual fixed factors (e.g. between ROI1 and ROI2 after viral infection), with and without stratification by the other fixed factors. For instance, comparison between ROI1 and ROI2 after viral infection was performed for the whole experiment, as well as for the data available only for a single line, on a single day post‐infection, and for all combinations between line and day post‐infection (Table S2).
The results of the experiments with the ROS inhibitor were analyzed using a linear model with factors treatment, tissue sections and interaction between the two. Significance was calculated using the ‘anova’ and ‘emmeans’ function, as for the mixed model.
Frequency of ROI2 with individual cells with moderately oxidized chloroplasts was calculated with Fisher's exact test.
All scripts were deposited on Github (https://github.com/NIB‐SI/SensorPlantAnalysis).
Spatial RT‐qPCR expression analysis
Left and right sides of the first fully developed leaf of PVY‐N605(123)‐GFP‐inoculated pt‐roGFP L2 transgenic plants were infiltrated with ROS inhibitor/control as described earlier. At 5 dpi, for each of the 12 lesions from four plants, two tissue sections were sampled: the lesion and its close proximity (A) and a 1 mm section adjacent to section A (B; Fig. 6b). As a control, tissue sections of the same size as the A and B sections together were sampled further from the lesions on both the inhibitor‐ and control‐treated sides of the leaf (Fig. 6b). Lesions in which we did not detect viral RNA or with inaccurate sampling were excluded from the analysis. See Table S4 for the details.
Total RNA was extracted from the sampled A and B sections using 50 μl of TRizol reagent (Thermo Fisher Scientific, Waltham, MA, USA) combined with RNA purification on Zymo‐Spin columns (Direct‐zol RNA MicroPrep Kit; Zymo Research, Irvine, CA, USA) according to the manufacturer's protocols. To elute RNA, 15 μl of prewarmed (80°C) DNase/RNase‐free water was added to the Zymo‐Spin columns and incubated at room temperature for 10 min before centrifugation at 16 000 g for 1 min. DNase‐treated (0.5 μl DNase per μg RNA; Qiagen) total RNA was quality‐controlled using 2100 Bioanalyzer and RNA 6000 Pico LabChip Kit (Agilent Technologies) and then reverse‐transcribed using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific).
Samples were analyzed in the setup for quantitative PCR (qPCR) as previously described (Lukan et al., 2020). The expression of 13 genes involved in different steps of immune signaling (Table S6 for a full list of genes) was analyzed and normalized to the expression of two validated reference genes, COX and elongation factor 1 (EF‐1), as described previously (Petek et al., 2014; see Lukan et al., 2020 for primer and probe information). Additionally, CALS (Glucan synthase like 1/GSL1, Sotub07g019600.1.1.) gene expression was analyzed (forward primer: 5′‐GAACACGAACTGGAGGATATTTTACC‐3′, probe: FAM/TTGTCCTGGTGGTTCCAGAGTCGG‐3′ Zen Iowa BlackTM FQ, reverse primer: 5′‐GATTCCACGACCCACAAACG‐3′). In parallel with the gene expression of those genes, the relative quantity of PVY RNA was measured (Lukan et al., 2020). The standard curve method was used for relative gene expression quantification using quantGenius (http://quantgenius.nib.si; Baebler et al., 2017).
Results
Chloroplast redox state is highly oxidized in the cells surrounding the cell death zone
Our previous studies of HR in potato–PVY interactions suggested the role of chloroplastic ROS as a signal orchestrating PCD (Lukan et al., 2020). To further explore the role of chloroplastic ROS in PCD induction in a spatiotemporal manner, we constructed transgenic plants of cv. Rywal with redox state‐sensitive GFP2 (roGFP2) targeted to chloroplasts (hereafter pt‐roGFP plants). Measurement of roGFP2 emission following excitation at 405 and 488 nm in constructed transgenic plants permits an evaluation of the chloroplast relative redox state as the relative proportion of roGFP2 in an oxidized or reduced state (405 : 488 ratio) (Jiang et al., 2006). Transgenic lines (pt‐roGFP L2, L4 and L15) with strong and stable roGFP fluorescence in the chloroplasts were selected for further work (Table S1).
To determine if the chloroplast redox state is spatially and/or temporally regulated around the cell death zone, we measured the relative redox state in the palisade mesophyll chloroplasts of pt‐roGFP plants in the areas adjacent to the lesion (ROI1) and adjacent to ROI1 (ROI2) at 3, 5, and 7 dpi with PVY, as well as in the area distant from the lesion (control, CTR) at 7 dpi (Fig. 1a). Within the ROI1, chloroplasts in the cells surrounding the cell death zone were highly oxidized, in contrast to more distant cells (more distant from cell death zone within ROI1, in ROI2, and in CTR) (Fig. 1b,d; doi: 10.5281/zenodo.6417635). As a control of the normal physiological redox state in potato chloroplasts, we also measured the chloroplast redox state in mock‐inoculated pt‐roGFP plants at 3 and 7 dpi (Fig. 1a). The chloroplast redox state was highly uniform across all analyzed mock samples and comparable to the redox state of chloroplasts in cells distant to the cell death zone in PVY‐infected plants (Fig. 1b,d; doi: 10.5281/zenodo.6417635).
We also imaged the epidermal cell layer in the majority of the analyzed areas but not in all, as a result of unevenness of the leaf cell layer distribution. We analyzed the redox state of their chloroplasts. In most cases, the chloroplasts in epidermal cells were in a similar redox state to chloroplasts in the underlying palisade mesophyll cells. Interestingly, in ROI1, chloroplasts in some of the epidermal cells were more reduced than those in mesophyll cells which we inspected in more detail in further experiments.
The measured variability of chloroplast redox state in ROI1 of the mesophyll cells was higher than in mock samples (Fig. 1d), as a result of the highly oxidized chloroplasts in cells of the PCD transition zone (Fig. 1b,d). To perform a more detailed spatiotemporal analysis of the redox state, we separated each ROI into five regions (bins), according to the distance from the cell death zone (Fig. S3a). The results show that the chloroplast redox state is indeed the most oxidized close to the cell death zone (bin 1) and is getting less oxidized with increasing distance from the cell death zone at different time points (Figs S3b, S4). In the most distant bin from the cell death zone (bin 5 of ROI1), the redox state stabilizes and remains comparable in all five bins of ROI2 (Fig. S3b).
Individual cells with moderately oxidized chloroplasts spread further away from the cell death zone
In cells at the border of the cell death zone, within ROI1, strongly oxidized chloroplasts often became more disorderly in their arrangement as they moved from the cell periphery towards the interior (Fig. 1b). These cells probably represent mesophyll cells that are in transition between normal cells and cells undergoing PCD, as supported by electron microscopy in our previous work (Lukan et al., 2020). However, we also observed chloroplasts in a moderately oxidized redox state that were well arranged, following the shape of a normal mesophyll cell further from the cell death zone in ROI2 (Figs 2a, S5). Such cells, which we named signaling cells, were rarely observed in mock‐inoculated plants (Fig. 1b).
Fig. 2.
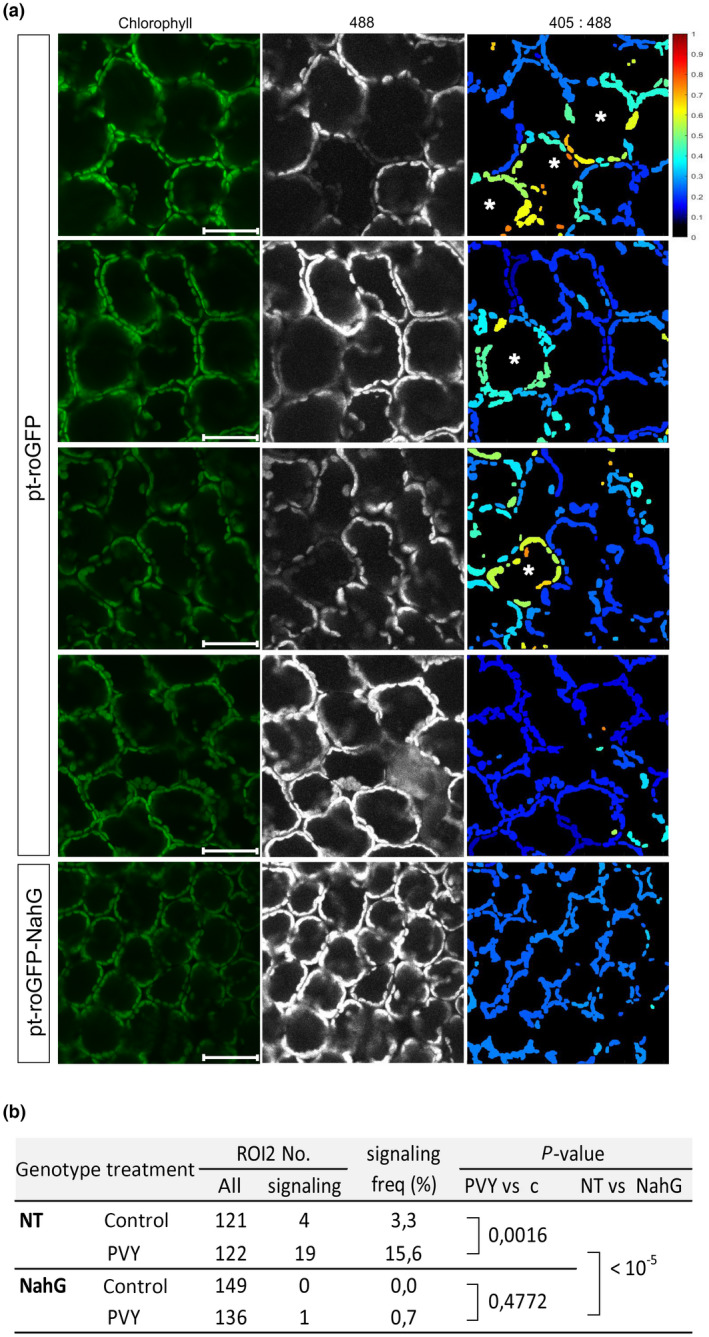
Individual cells with moderately oxidized chloroplasts (asterisks) can be observed further from the cell death zone (region of interest 2, ROI2). (a) The first four panels present ROI2 in pt‐roGFP2 plants with individual cells with moderately oxidized chloroplasts (signaling cells, first three panels) or with no signaling cells (fourth panel). The bottom panel presents ROI2 with no signaling cells in a salicylic acid (SA)‐deficient pt‐roGFP‐NahG plant for comparison. Cells with oxidized chloroplasts further away from the cell death zone are marked with asterisks. Chlorophyll, Chl fluorescence; 488, roGFP fluorescence after excitation with 488 nm laser line showing reduced roGFP (brighter chloroplasts are more reduced); 405 : 488, relative redox state (405 : 488 ratio) presented on a rainbow scale. Higher ratios denote more oxidized chloroplasts (red). Bar, 50 μm. (b) Frequency of ROI2 with signaling cells for pt‐roGFP (NT) and pt‐roGFP‐NahG (NahG). P‐value comparing frequency of ROI2 with signaling cells between potato virus Y (PVY) and control treatments (PVY vs c) and between PVY treatments of each genotype (NT vs NahG) was calculated with Fisher's exact test. PVY, ROI2 of PVY‐inoculated plants; control, combined MOCK and CTR inspected ROIs (Fig. 1a).
SA‐dependent presence of cells with oxidized chloroplasts further away from the cell death zone suggests their role in signal transmission in HR‐conferred resistance
To study the link between SA signaling and chloroplast redox state in PCD and resistance, we also introduced roGFP2 in the SA‐depleted transgenic counterpart NahG‐Rywal (hereafter pt‐roGFP‐NahG plants). Potato genotype NahG‐Rywal is a transgenic counterpart of cv. Rywal, depleted in the accumulation of SA (reduced to < 10% of native values). The depletion of SA renders NahG plants susceptible to PVY (catechol‐independent; Baebler et al., 2014), allowing the virus to spread throughout the plant (Baebler et al., 2014). Transgenic lines (pt‐roGFP‐NahG L2 and L7) with strong and stable GFP fluorescence in the chloroplasts were selected for further work. We next followed the chloroplast redox state after viral infection in different regions, close to or further away from the cell death zone at three time points. Similarly as in pt‐roGFP plants (Fig. 1b), within the ROI1, chloroplasts in the cells surrounding the cell death zone were highly oxidized (Fig. 1c). This was further confirmed by image analysis, as the chloroplast redox state was more oxidized in ROI1 and was statistically significantly different from the chloroplast redox state in ROI2 at all analyzed time points (Figs 1f, S1; Tables S1, S2). We conclude that the chloroplast redox state is precisely spatially regulated around the cell death zone in both pt‐roGFP and SA‐deficient pt‐roGFP‐NahG plants. Interestingly, however, the cells with moderately oxidized chloroplasts, further away from the cell death zone, were rarely observed in SA‐deficient plants (Fig. 2; doi: 10.5281/zenodo.6417635). This result suggests that chloroplast oxidation distant from the cell death zone requires SA signaling, whereas chloroplast oxidation near the cell death zone occurs independently of SA signaling. As SA is required to prevent the viral spread, but not for the development of PCD (Lukan et al., 2018, 2020), we hypothesize that chloroplast oxidation in distant cells participates in signaling for resistance, whereas chloroplast oxidation near the cell death zone is related to PCD.
Spatiotemporal regulation of stromule formation around the cell death zone is SA signaling‐dependent
Although our experimental setup was focused on measuring the redox state in the mesophyll cells, because of the unevenness of the leaf cell layer distribution, we also imaged epidermal cells layer in the majority of investigated areas. Chloroplasts in epidermal cells were in a similar redox state as chloroplasts in underlying palisade mesophyll cells. Interestingly, we observed individual epidermal cells with highly reduced chloroplasts, often in the area between the cell death zone and signaling mesophyll cells (Fig. 3a,b). In those cells, we were also able to observe clustering of chloroplasts and intensive connectivity via stromules (Fig. 3a,b). We confirmed that the clustering of chloroplasts is the result of their movement towards the nucleus (Video S1). Our observation that chloroplasts with stromules are highly reduced is in disagreement with the results of some other studies (Brunkard et al., 2015; Caplan et al., 2015; Barton et al., 2018; Ding et al., 2019). It is possible, however, that our findings are specific for cell‐to‐cell signaling in this particular plant–virus interaction, which was not yet studied in this aspect. Moreover, we observed the majority of chloroplasts with stromules in epidermal cells, which is in agreement with the literature showing that stromules are a characteristic cell‐specific feature and are more frequently present in epidermal cells (reviewed in Natesan et al., 2005).
Fig. 3.
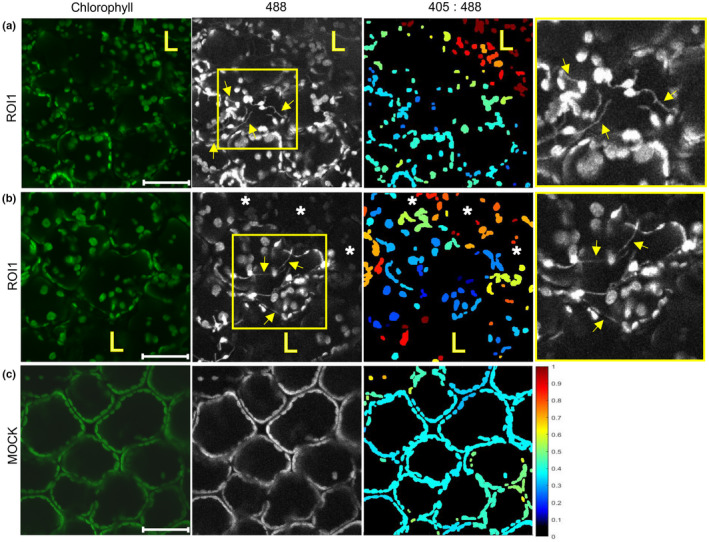
Stromules are induced in cells adjacent to the hypersensitive response (HR) cell death zone. (a, b) Chloroplasts with stromules (arrows) in the cells on the border of the cell death zone in ROI1 (a) or in the cells between the cell death zone and the cells with oxidized chloroplasts in ROI1 (b) in leaf epidermis of potato virus Y (PVY)‐inoculated pt‐roGFP plants. A strongly reduced redox state is shown as a high signal in 488 channel, black or dark blue on 405 : 488 ratio images. Higher magnification of an area with stromules (boxed with yellow) is shown in the rightmost panel. (c) Epidermal chloroplasts in the mock‐inoculated pt‐roGFP plant for the comparison of redox state. Images were taken in PVY‐ and mock‐inoculated pt‐roGFP plants at 5 and 3 d post‐inoculation, respectively. Stromules are marked with arrows. Cells with oxidized chloroplasts further away from the cell death zone are marked with asterisks. The cell death zone is marked with an ‘L’. ROI1, region of interest adjacent to cell death zone (Fig. 1a); chlorophyll, Chl fluorescence; 488, roGFP fluorescence after excitation with 488 nm laser line showing reduced roGFP (brighter chloroplasts are more reduced); 405 : 488, relative redox state (405 : 488 ratio) presented on a rainbow scale. Higher ratios denote more oxidized chloroplasts (red). z‐stack size was adjusted to 10–15 steps to cover at least 30 μm of the mesophyll. As the line between epidermal and mesophyll cells in potato leaf is not flat and epidermal cells often reach into the plane of mesophyll cells, in several images we were also able to observe epidermal chloroplasts. Bar, 50 μm.
We performed additional experiments to investigate these results further. In these experiments, we adjusted z‐stack size to cover the whole epidermis and at least 50 μm of the mesophyll. We observed increased stromule formation frequency in epidermal chloroplasts after inoculation at all time points, which was even more pronounced in the cell death zone proximity in pt‐roGFP plants (Fig. 4). When comparing the frequency of stromules between ROI1 and ROI2 in pt‐roGFP plants, the difference was statistically significant at 4 and 5 dpi. By contrast, in pt‐roGFP‐NahG plants, the number of stromules was similar between ROI1 and ROI2 at all time points (Fig. 4d; Tables S3, S7). The frequency of stromules of PVY‐inoculated plants was higher if compared with random regions on the leaf of mock‐inoculated plants at all time points in pt‐roGFP plants (Fig. 4d; Tables S3, S7). The induction of stromules was, however, less pronounced in pt‐roGFP‐NahG plants. We also compared the frequency of stromules between genotypes, and the difference was statistically significant at 4 and 5 dpi for ROI1, but not for ROI2 (Fig. 4d). We thereby conclude that in PVY‐induced HR in potato, stromule formation is spatiotemporally regulated around the cell death zone and is SA signaling‐dependent.
Stromules are induced in immediate proximity to virus multiplication zone
To investigate why the stromule frequency is 5 dpi in pt‐roGFP‐NahG plants induced in both ROI1 and ROI2, whereas in pt‐roGFP plants the induction was visible only in ROI1, we decided to expand our characterization of the spatiotemporal responses to PVY infection and check stromule frequency in ROI3 and ROI4 as well. We followed stromule formation in four consecutive regions adjacent to the cell death zone at different time points following inoculation in both genotypes (ROI1–ROI4; Fig. S2a,b,d). This experiment confirmed the previous results showing that stromule formation is differentially spatiotemporally regulated between genotypes. In pt‐roGFP‐NahG plants, induction of stromule frequency was, besides ROI2, higher also in ROI3, whereas in pt‐roGFP plants, the induction was visible only in ROI1 (Figs 4, S2a,b,d; Tables S3, S7).
In our previous study, we reported that PVY accumulated in the cells on the border of the cell death zone in nontransgenic plants at all analyzed time points (most frequently at 4 dpi), but the virus did not spread beyond this zone. In NahG plants, however, we observed concentric virus spread to adjacent cells, such that PVY reached the outer borders of ROI2 within 6–7 dpi (Lukan et al., 2018). The viral spread correlates with stromule frequency, which in pt‐roGFP is statistically higher in ROI1 than in ROI2, ROI3, ROI4 or MOCK, while in pt‐roGFP‐NahG, both ROI1 and ROI2 have higher stromule frequency as compared with ROI3, ROI4 or MOCK (Figs 4d, S2a,b,d).
To confirm the relationship between stromule frequency and virus multiplication, we followed the cells on the border of the virus multiplication zone in pt‐roGFP plants at 4 dpi and in pt‐roGFP‐NahG plants at 7 dpi using GFP‐tagged PVY (Fig. 5a,c; Table S1). We collected two z‐stacks in the same ROI, one covering the whole epidermis with at least 50 μm of the mesophyll and one only 50 μm of the mesophyll. In both genotypes, we observed epidermal chloroplasts with stromules in the cell adjacent to the cell with PVY‐GFP accumulation (Fig. 5b,d). We therefore conclude that stromule formation was more pronounced in the cell adjacent to the infected cell, which could be a result of plant‐mediated response to the presence of the virus or virus multiplication. Owing to a strong background in the 405 nm channel as a result of strong PVY‐GFP fluorescence, an evaluation of the chloroplast relative redox state as the relative proportion of roGFP in an oxidized or reduced state (405 : 488 ratio) was not possible.
Blocking the chloroplastic redox changes affects the spatial response at the transcriptional level in HR
To further study the mechanism of this chloroplast redox state‐dependent signaling and its downstream targets, we treated plants with ROS scavenger uracil (Abdollahi & Ghahremani, 2011) and performed spatial transcriptomics in cell death zone proximity at 5 dpi (Fig. 6). The target genes were selected according to their involvement in redox potential homeostasis (CAT1, PRX28, RBOHD, RBOHA, TRXH), JA signaling (13‐LOX, 9‐LOX, ACX3), ET signaling (ERF1), stability of R proteins (HSP70) and actuation of defense (BGLU, PR1B, CALS) and all showed spatial regulation following PVY inoculation (Lukan et al., 2020). We show that induction of chloroplast oxidation around the site of viral foci was successfully blocked when treated with uracil (Fig. 6a). Genes show different spatial transcriptional responses (Figs 6c, S6). ERF1 was upregulated further from the cell death zone (section B) when ROS signaling was hampered compared with induction in the cell death zone (section A) in the control (Fig. 6; Tables S4, S6, S8, S9). RBOHD expression is, by contrast, higher in section A of inhibitor‐treated plants, and RBOHA, HSP70 and CAT1 showed similar spatial regulation in the control and inhibitor‐treated tissue. Interestingly, TRXH expression is lower if ROS is generated as its expression was higher in inhibitor‐treated tissue in both the A and B zones. Therefore, ERF1, RBOHD and TRXH are downstream chloroplast redox state‐dependent signaling for resistance.
Discussion
Programmed cell death has been associated with increased chloroplastic ROS production (Wagner et al., 2004; Liu et al., 2007; Straus et al., 2010). Generation of chloroplastic ROS was proposed as a signal orchestrating PCD, rather than only being a consequence of PCD (Zurbriggen et al., 2010; Pierella Karlusich et al., 2017; Su et al., 2018). Our results showing that the disordered chloroplasts in the cells adjacent to the cell death zone were strongly oxidized (Figs S3b,c (top panel), S4) are in agreement with and extend these previous conclusions. In addition, close to the border of the cell death zone (ROI1; Fig. S3b,c), as well as further away from the cell death zone (ROI2; Figs 2a, S5; Table S1), we observed the so‐called signaling cells, individual mesophyll cells with moderately oxidized chloroplasts. These cells were sparse in SA‐deficient plants with compromised resistance, which indicates their role in signal transmission in HR‐conferred resistance. This is supported by the results of several studies showing that decreased chloroplastic ROS production leads to compromised resistance (Su et al., 2018; Xu et al., 2019; Schmidt et al., 2020). Here we also show that chloroplastic redox changes are modulating spatial transcriptional response of immunity‐related genes (Fig. 6). Indeed, the regulation of genes involved in ethylene signaling (ERF1) and redox state sensing (TRXH) was changed further away from the viral foci when oxidation of chloroplast was blocked, confirming the signaling role of chloroplastic redox changes in PVY‐induced HR response (Fig. 6c). The chloroplast redox state has previously been studied during HR‐PCD, triggered by N‐mediated recognition of a fragment of the helicase domain of the TMV replicase (p50) with the HyPer2 H2O2 sensor (Caplan et al., 2015). The authors detected higher concentrations of ROS in chloroplasts at 28 h post‐agroinfiltration to express p50. As their approach causes even expression of p50 across all cells and does not track an actual virus infection, the spatial dynamics of redox state changes during HR and the effect on signaling could not be directly interrogated. Our study thus brings a new perspective to redox signaling during HR resistance.
Reactive oxygen species production in plant organelles triggers intracellular communication pathways that alter intercellular communication via plasmodesmata (PD) (Stonebloom et al., 2012). They showed that oxidized plastids resulted in decreased PD transport, whereas reduced plastids can facilitate PD transport. Thus, we could assume that the highly oxidized chloroplasts at the border of the cell death zone have PD closed to prevent the spread of the pathogen. Interestingly, the cells with high frequency of stromules that we observed in proximity to the cell death zone or to the cell in which we observed PVY‐GFP accumulation had reduced chloroplasts (Fig. 5), meaning they could have increased PD transport. This is in accordance with Caplan et al. (2015), who suggested that stromule function is HR‐PCD signaling. However, their experimental approach with transient expression of HR‐triggering proteins only focused on signaling for PCD while signaling for viral arrest was not studied. We additionally showed that the increase in stromule formation observed in SA‐deficient plants in ROI2 (Fig. S2) was linked to the border of the virus multiplication zone (Fig. 5).
Maruta et al. (2012) suggested that the chloroplastic H2O2 and SA activate each other and that this positive feedback loop is involved in the plant immune response. By contrast, we found no major differences between the chloroplast redox state of SA‐deficient pt‐roGFP‐NahG compared with pt‐roGFP plants close to the lesion (Figs 1, S3). However, the frequency of previously mentioned signaling cells with moderately oxidized chloroplasts was lower in SA‐depleted plants (Fig. 2). Additionally, we studied SA involvement in stromule formation. Previous studies showed that an oxidative environment induces stromule formation (Brunkard et al., 2015; Caplan et al., 2015; Barton et al., 2018; Ding et al., 2019). Indeed, in our study, the number of stromules was induced in lesion proximity. Interestingly, however, the frequency of stromules was statistically significantly higher in pt‐roGFP than in SA‐deficient pt‐roGFP‐NahG plants (Fig. 4), although the oxidative state of chloroplasts adjacent to the cell death zone was the same (Fig. 1). This is in agreement with results from Caplan et al. (2015), who showed that application of an SA analog induces stromules when HR is triggered by pathogen recognition. Therefore, we conclude that stromule induction does not depend solely on the redox state of chloroplasts in adjacent cells.
Our results collectively show that the increase in mesophyll chloroplast redox state is linked to PCD signaling, which is SA‐independent, and also to signaling for HR‐conferred resistance to the virus, which is SA‐dependent. The later phenomenon is supported by the presence of mesophyll cells with moderately oxidized chloroplasts further away from the cell death zone which are linked to epidermal cells with reduced chloroplasts and high stromule connectivity and changes in transcriptional regulation, indicating intensive intracellular and cell‐to‐cell communication activity.
Competing interests
None declared.
Author contributions
KG and TL designed the research; TMP, MJ, MK and TL performed the research; AŽ, TMP and TL contributed new analytic/computational/imaging tools; TL, KG, AŽ, TMP, MK, JB and ŠB analyzed the data. TL, KG, AŽ, TMP, MJ, ŠB and JB contributed to the writing or revision of the article.
Supporting information
Fig. S1 Relative chloroplast redox state after PVY and MOCK inoculation and H2O2 and DTT treatments.
Fig. S2 Stromule formation around the cell death zone is differentially spatiotemporally regulated between potato genotypes.
Fig. S3 Chloroplast redox state is highly oxidized around the cell death zone.
Fig. S4 Detailed spatial analysis of relative chloroplast redox state around the cell death zone.
Fig. S5 Individual cells with chloroplasts in oxidized redox state in ROI2 in PVY‐inoculated redox state sensor plants.
Fig. S6 Impact of chloroplast inhibitor on transcriptional response.
Methods S1 Stable transformation.
Methods S2 Construction of PVY‐N605(123)‐GFP infectious clone.
Table S1 Sample information and 405 : 488 ratios for chloroplast redox detection experiments.
Table S2 Statistical evaluation of observed relative redox changes in different conditions in individual experiments.
Table S3 Sample information and normalized number of stromules for stromule counting experiments.
Table S4 Sample information and 405 : 488 ratios for chloroplast redox detection experiments with the inhibitor.
Table S5 405 : 488 ratios in five bins of ROI1 and ROI2.
Table S6 Chloroplast redox regulated spatial expression of selected genes.
Table S7 Statistical evaluation of stromule counts in different conditions in individual experiments.
Table S8 Statistical evaluation of observed relative redox changes after chlROS inhibitor and control treatment.
Table S9 Statistical analysis of the effects of chloroplastic ROS inhibitor (chlROS) and control treatment on expression of selected genes.
Video S1 Epidermal chloroplasts cluster around the nucleus in potato plants in response to PVY infection.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank Dr Fabrizio Cillo (Institute for Sustainable Plant Protection, Italy) for kindly providing the PVY‐N605(123) plasmid, Prof. Andrej Blejec for help with statistical analysis, and Lidija Matičič, Katja Stare, Barbara Dušak, Matej Rebek, Maja Križnik, Karmen Pogačar and Barbara Jaklič for technical assistance. This research was financially supported by the Slovenian Research Agency (research core funding no. P4‐0165 and projects J4‐7636, J4‐1777, 1000‐21‐0105, N4‐0199, Z4‐3217), the European Community's H2020 Program ADAPT (grant agreement 862858), and the US National Institutes of Health (DP5‐OD023072).
Data availability
Raw and analyzed imaging data were deposited at Zenodo and are openly available at doi: 10.5281/zenodo.6417635. Scripts were deposited at Github and are openly available at https://github.com/NIB‐SI/SensorPlantAnalysis.
References
- Abdollahi H, Ghahremani Z. 2011. The role of chloroplasts in the interaction between Erwinia amylovora and host plants. Acta Horticulturae 896: 215–222. [Google Scholar]
- Baebler Š, Krečič‐Stres H, Rotter A, Kogovšek P, Cankar K, Kok EJ, Gruden K, Kovač M, Žel J, Pompe‐Novak M et al. 2009. PVYNTN elicits a diverse gene expression response in different potato genotypes in the first 12 h after inoculation. Molecular Plant Pathology 10: 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baebler Š, Svalina M, Petek M, Stare K, Rotter A, Pompe‐Novak M, Gruden K. 2017. QuantGenius: implementation of a decision support system for qPCR‐based gene quantification. BMC Bioinformatics 18: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baebler Š, Witek K, Petek M, Stare K, Tušek‐Žnidarič M, Pompe‐Novak M, Renaut J, Szajko K, Strzelczyk‐Zyta D, Marczewski W et al. 2014. Salicylic acid is an indispensable component of the Ny‐1 resistance‐gene‐mediated response against Potato virus Y infection in potato. Journal of Experimental Botany 65: 1095–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balint‐Kurti P. 2019. The plant hypersensitive response: concepts, control and consequences. Molecular Plant Pathology 20: 1163–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton KA, Wozny MR, Mathur N, Jaipargas EA, Mathur J. 2018. Chloroplast behaviour and interactions with other organelles in Arabidopsis thaliana pavement cells. Journal of Cell Science 131: jcs202275. [DOI] [PubMed] [Google Scholar]
- Bond SR, Naus CC. 2012. RF‐Cloning.org: an online tool for the design of restriction‐free cloning projects. Nucleic Acids Research 40: 209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkard JO, Runkel AM, Zambryski PC. 2015. Chloroplasts extend stromules independently and in response to internal redox signals. Proceedings of the National Academy of Sciences, USA 112: 10044–10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukovinszki Á, Götz R, Johansen E, Maiss E, Balázs E. 2007. The role of the coat protein region in symptom formation on Physalis floridana varies between PVY strains. Virus Research 127: 122–125. [DOI] [PubMed] [Google Scholar]
- Calil IP, Fontes EPB. 2017. Plant immunity against viruses: antiviral immune receptors in focus. Annals of Botany 119: 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan JL, Kumar AS, Park E, Padmanabhan MS, Hoban K, Modla S, Czymmek K, Dinesh‐Kumar SP. 2015. Chloroplast stromules function during innate immunity. Developmental Cell 34: 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Jimenez‐Gongora T, Krenz B, Lozano‐Duran R. 2019. Chloroplast clustering around the nucleus is a general response to pathogen perception in Nicotiana benthamiana . Molecular Plant Pathology 20: 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández JA, Gullner G, Clemente‐Moreno MJ, Künstler A, Juhász C, Díaz‐Vivancos P, Király L. 2016. Oxidative stress and antioxidative responses in plant–virus interactions. Physiological and Molecular Plant Pathology 94: 134–148. [Google Scholar]
- Ishiga Y, Ishiga T, Wangdi T, Mysore KS, Uppalapati SR. 2012. NTRC and chloroplast‐generated reactive oxygen species regulate Pseudomonas syringae pv. tomato disease development in tomato and Arabidopsis . Molecular Plant–Microbe Interactions 25: 294–306. [DOI] [PubMed] [Google Scholar]
- Jiang K, Schwarzer C, Lally E, Zhang S, Ruzin S, Machen T, Remington SJ, Feldman L. 2006. Expression and characterization of a redox‐sensing green fluorescent protein (reduction‐oxidation‐sensitive green fluorescent protein) in Arabidopsis. Plant Physiology 141: 397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. 2006. The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kachroo P, Burch‐Smith TM, Grant M. 2021. An emerging role for chloroplasts in disease and defense. Annual Review of Phytopathology 59: 423–445. [DOI] [PubMed] [Google Scholar]
- Karasev AV, Gray SM. 2013. Continuous and emerging challenges of Potato virus Y in potato. Annual Review of Phytopathology 51: 571–586. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY™ vectors for Agrobacterium‐mediated plant transformation. Trends in Plant Science 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kim C, Meskauskiene R, Zhang S, Lee KP, Ashok ML, Blajecka K, Herrfurth C, Feussner I, Apela K. 2012. Chloroplasts of Arabidopsis are the source and a primary target of a plant‐specific programmed cell death signaling pathway. Plant Cell 24: 3026–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar AS, Park E, Nedo A, Alqarni A, Ren L, Hoban K, Modla S, McDonald JH, Kambhamettu C, Dinesh‐Kumar SP et al. 2018. Stromule extension along microtubules coordinated with actin‐mediated anchoring guides perinuclear chloroplast movement during innate immunity. eLife 7: e23625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künstler A, Bacsó R, Gullner G, Hafez YM, Király L. 2016. Staying alive – is cell death dispensable for plant disease resistance during the hypersensitive response? Physiological and Molecular Plant Pathology 93: 75–84. [Google Scholar]
- Kuznetsova A, Brockhoff PB, Christensen RHB. 2017. lmertest package: tests in linear mixed effects models. Journal of Statistical Software 82: 1–26. [Google Scholar]
- Lee KP, Kim C, Landgraf F, Apel K. 2007. EXECUTER1‐ and EXECUTER2‐dependent transfer of stress‐related signals from the plastid to the nucleus of Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 104: 10270–10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenth RV, Buerkner P, Herve M, Love J, Riebl H, Singmann H. 2021. Package ‘emmeans’ estimated marginal means, aka least‐squares means . [WWW document] URL http://cran.r‐project.org/package=emmeans [accessed 09 February 2021].
- Li M, Kim C. 2022. Chloroplast ROS and stress signaling. Plant Communications 3: 100264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlejohn GR, Breen S, Smirnoff N, Grant M. 2021. Chloroplast immunity illuminated. New Phytologist 229: 3088–3107. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ren D, Pike S, Pallardy S, Gassmann W, Zhang S, Life B. 2007. Chloroplast‐generated reactive oxygen species are involved in hypersensitive response‐like cell death mediated by a mitogen‐activated protein kinase cascade. The Plant Journal 51: 941–954. [DOI] [PubMed] [Google Scholar]
- Lu Y, Yao J. 2018. Chloroplasts at the crossroad of photosynthesis, pathogen infection and plant defense. International Journal of Molecular Sciences 19: 3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukan T, Baebler Š, Pompe‐Novak M, Guček K, Zagorščak M, Coll A, Gruden K. 2018. Cell death is not sufficient for the restriction of potato virus Y spread in hypersensitive response‐conferred resistance in potato. Frontiers in Plant Science 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukan T, Pompe‐Novak M, Baebler Š, Tušek‐Žnidarič M, Kladnik A, Križnik M, Blejec A, Zagorščak M, Stare K, Dušak B et al. 2020. Precision transcriptomics of viral foci reveals the spatial regulation of immune‐signaling genes and identifies RBOHD as an important player in the incompatible interaction between potato virus Y and potato. The Plant Journal 104: 645–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta T, Noshi M, Tanouchi A, Tamoi M, Yabuta Y, Yoshimura K, Ishikawa T, Shigeoka S. 2012. H2O2‐triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress. Journal of Biological Chemistry 287: 11717–11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaña MP, Blasich N, Haggi E, García NA. 2009. Oxygen uptake in the vitamin B2‐sensitized photo‐oxidation of tyrosine and tryptophan in the presence of uracil: kinetics and mechanism. Photochemistry and Photobiology 85: 1097–1102. [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Kenton P, Lloyd AJ, Ougham H, Prats E. 2008. The hypersensitive response; the centenary is upon us but how much do we know? Journal of Experimental Botany 59: 501–520. [DOI] [PubMed] [Google Scholar]
- Natesan SKA, Sullivan JA, Gray JC. 2005. Stromules: a characteristic cell‐specific feature of plastid morphology. Journal of Experimental Botany 56: 787–797. [DOI] [PubMed] [Google Scholar]
- Nomura H, Komori T, Uemura S, Kanda Y, Shimotani K, Nakai K, Furuichi T, Takebayashi K, Sugimoto T, Sano S et al. 2012. Chloroplast‐mediated activation of plant immune signalling in Arabidopsis . Nature Communications 3: 1–10. [DOI] [PubMed] [Google Scholar]
- Ochsenbein C, Przybyla D, Danon A, Landgraf F, Göbel C, Imboden A, Feussner I, Apel K. 2006. The role of EDS1 (enhanced disease susceptibility) during singlet oxygen‐mediated stress responses of Arabidopsis. The Plant Journal 47: 445–456. [DOI] [PubMed] [Google Scholar]
- Petek M, Rotter A, Kogovšek P, Baebler Š, Mithöfer A, Gruden K. 2014. Potato virus Y infection hinders potato defence response and renders plants more vulnerable to Colorado potato beetle attack. Molecular Ecology 23: 5378–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierella Karlusich JJ, Zurbriggen MD, Shahinnia F, Sonnewald S, Sonnewald U, Hosseini SA, Hajirezaei MR, Carrillo N. 2017. Chloroplast redox status modulates genome‐wide plant responses during the non‐host interaction of Tobacco with the hemibiotrophic bacterium Xanthomonas campestris pv. vesicatoria . Frontiers in Plant Science 8: 1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenouille J, Vassilakos N, Moury B. 2013. Potato virus Y: a major crop pathogen that has provided major insights into the evolution of viral pathogenicity. Molecular Plant Pathology 14: 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupar M, Faurez F, Tribodet M, Gutierrez‐Aguirre I, Delaunay A, Glais L, Kriznik M, Dobnik D, Gruden K, Jacquot E et al. 2015. Fluorescently tagged Potato virus Y: a versatile tool for functional analysis of plant–virus interactions. Molecular Plant–Microbe Interactions 28: 739–750. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Mächtel R, Ammon A, Engelsdorf T, Schmitz J, Maurino VG, Voll LM. 2020. Reactive oxygen species dosage in Arabidopsis chloroplasts can improve resistance towards Colletotrichum higginsianum by the induction of WRKY33. New Phytologist 226: 189–204. [DOI] [PubMed] [Google Scholar]
- Serrano I, Audran C, Rivas S. 2016. Chloroplasts at work during plant innate immunity. Journal of Experimental Botany 67: 3845–3854. [DOI] [PubMed] [Google Scholar]
- Sewelam N, Jaspert N, Van Der Kelen K, Tognetti VB, Schmitz J, Frerigmann H, Stahl E, Zeier J, Van Breusegem F, Maurino VG. 2014. Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Molecular Plant 7: 1191–1210. [DOI] [PubMed] [Google Scholar]
- Stare K, Coll A, Gutiérrez‐Aguirre I, Tušek Žnidarič M, Ravnikar M, Kežar A, Kavčič L, Podobnik M, Gruden K. 2020. Generation and in planta functional analysis of potato virus Y mutants. Bio‐Protocol 10: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonebloom S, Brunkard JO, Cheung AC, Jiang K, Feldman L, Zambryski P. 2012. Redox states of plastids and mitochondria differentially regulate intercellular transport via plasmodesmata. Plant Physiology 158: 190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus MR, Rietz S, Ver Loren Van Themaat E, Bartsch M, Parker JE. 2010. Salicylic acid antagonism of EDS1‐driven cell death is important for immune and oxidative stress responses in Arabidopsis. The Plant Journal 62: 628–640. [DOI] [PubMed] [Google Scholar]
- Su J, Yang L, Zhu Q, Wu H, He Y, Liu Y, Xu J, Jiang D, Zhang S. 2018. Active photosynthetic inhibition mediated by MPK3/MPK6 is critical to effector‐triggered immunity. PLoS Biology 16: e2004122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajko K, Chrzanowska M, Witek K, Strzelczyk‐Zyta D, Zagórska H, Gebhardt C, Hennig J, Marczewski W. 2008. The novel gene Ny‐1 on potato chromosome IX confers hypersensitive resistance to Potato virus Y and is an alternative to Ry genes in potato breeding for PVY resistance. Theoretical and Applied Genetics 116: 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Przybyla D, Op Den Camp R, Kim C, Landgraf F, Keun PL, Würsch M, Laloi C, Nater M, Hideg E et al. 2004. The genetic basis of singlet oxygen‐induced stress response of Arabidopsis thaliana . Science 306: 1183–1185. [DOI] [PubMed] [Google Scholar]
- Xu Q, Tang C, Wang X, Sun S, Zhao J, Kang Z, Wang X. 2019. An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nature Communications 10: 5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurbriggen MD, Carrillo N, Hajirezaei MR. 2010. ROS signaling in the hypersensitive response: when, where and what for? Plant Signaling and Behavior 5: 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurbriggen MD, Carrillo N, Tognetti VB, Melzer M, Peisker M, Hause B, Hajirezaei MR. 2009. Chloroplast‐generated reactive oxygen species play a major role in localized cell death during the non‐host interaction between tobacco and Xanthomonas campestris pv. vesicatoria . The Plant Journal 60: 962–973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Relative chloroplast redox state after PVY and MOCK inoculation and H2O2 and DTT treatments.
Fig. S2 Stromule formation around the cell death zone is differentially spatiotemporally regulated between potato genotypes.
Fig. S3 Chloroplast redox state is highly oxidized around the cell death zone.
Fig. S4 Detailed spatial analysis of relative chloroplast redox state around the cell death zone.
Fig. S5 Individual cells with chloroplasts in oxidized redox state in ROI2 in PVY‐inoculated redox state sensor plants.
Fig. S6 Impact of chloroplast inhibitor on transcriptional response.
Methods S1 Stable transformation.
Methods S2 Construction of PVY‐N605(123)‐GFP infectious clone.
Table S1 Sample information and 405 : 488 ratios for chloroplast redox detection experiments.
Table S2 Statistical evaluation of observed relative redox changes in different conditions in individual experiments.
Table S3 Sample information and normalized number of stromules for stromule counting experiments.
Table S4 Sample information and 405 : 488 ratios for chloroplast redox detection experiments with the inhibitor.
Table S5 405 : 488 ratios in five bins of ROI1 and ROI2.
Table S6 Chloroplast redox regulated spatial expression of selected genes.
Table S7 Statistical evaluation of stromule counts in different conditions in individual experiments.
Table S8 Statistical evaluation of observed relative redox changes after chlROS inhibitor and control treatment.
Table S9 Statistical analysis of the effects of chloroplastic ROS inhibitor (chlROS) and control treatment on expression of selected genes.
Video S1 Epidermal chloroplasts cluster around the nucleus in potato plants in response to PVY infection.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
Raw and analyzed imaging data were deposited at Zenodo and are openly available at doi: 10.5281/zenodo.6417635. Scripts were deposited at Github and are openly available at https://github.com/NIB‐SI/SensorPlantAnalysis.


