Abstract
Objective: Ovarian cancer is a kind of common gynecological malignancy in women. PARP inhibitors (PARPi) have been approved for ovarian cancer treatment. However, the primary and acquired resistance have limited the application of PARPi. The mechanisms remain to be elucidated.
Methods: In this study, we characterized the expression profiles of mRNA and nonconding RNAs (ncRNAs) and constructed the regulatory networks based on RNA sequencing in PARPi Olaparib-induced ovarian cancer cells.
Results: We found that the functions of the differentially expressed genes were enriched in “PI3K/AKT signaling pathway,” “MAPK signaling pathway” and “metabolic process”. The functions of DELs (cis) were enriched in “Human papillomavirus infection”“tight junction” “MAPK signaling pathway”. As the central regulator of ceRNAs, the differentially expressed miRNAs were enriched in “Human papillomavirus infection” “MAPK signaling pathway” “Ras signaling pathway”. According to the degree of interaction, we identified 3 lncRNAs, 2 circRNAs, 7 miRNAs, and 12 mRNA as the key regulatory ceRNA axis, in which miR-320b was the important mediator.
Conclusion: Here, we revealed the key regulatory lncRNA (circRNA)-miRNA-mRNA axis and their involved pathways in the PARPi resistant ovarian cancer cells. These findings provide new insights into exploring the ceRNA regulatory networks and developing new targets for PARPi resistance.
Keywords: Ovarian cancer, PARPi resistance, ceRNA network, mRNA, DNA, SSB
1. INTRODUCTION
Ovarian cancer (OC) is a common gynecological malignancy in women. The standard therapies include cisplatin-based chemotherapy and cytoreductive surgery [1]. PARP (poly-ADP ribose polymerase) is an enzyme that helps DNA single-strand breaks (SSBs) to repair [2]. PARP inhibitors catalyze the addition of poly-ADP-ribose polymers to target molecules and prevent the SSB DNA repair. PARPi leads to DNA repair defects and the generation of DNA double-strand breaks (DSBs) when the tumor cells have defects in repair by homologous recombination (HR) [3]. The unrepaired SSBs eventually progress to double-strand breaks (DSBs), which are highly cytotoxic to cells [4]. BRCA1/2 deficient tumor cells are more sensitive to PARPi through synthetic lethality [5]. PARP inhibitors have been approved for the treatment of ovarian cancer, fallopian tube cancer and peritoneal cancer. Despite the efficacy of PARP on ovarian cancer patients, challenges remain due to primary and secondary resistance. More than 40% of BRCA1/2-deficient patients failed to respond to PARPi [6]. HR repair restoration, DNA replication fork protection and other mechanisms have been reported to be involved in PARPi resistance [6]. On the other hand, studies have been conducted to test the effect of PARPi in BRCA wild-type patients [4]. A single-agent Olaparib showed favorable responses in patients with platinum-recurrent ovarian cancer, including those with BRCA wild-type [7, 8]. In the phase II CLEO study, patients with platinum resistance were randomly assigned to Olaparib or other chemotherapy. The patients with BRCA wild-type had a 13% response rate compared to 6% with chemotherapy [8]. Therefore, the primary and acquired PARPi resistance need to be further elucidated.
Noncoding RNAs (ncRNAs), including microRNA (miRNA), long ncRNA (lncRNA) and circular RNA (circRNA) are important regulators, regulating the function of mRNAs directly or indirectly. Whole-transcriptome analysis with total RNA sequencing detects both coding and noncoding RNA, identifying known and novel features of the transcriptome. miRNAs bind to the 3’ end of mRNA and mediate mRNA degradation. LncRNA and circRNAs are considered to be competitive endogenous RNAs of miRNAs and interfere with the degradation of mRNA [9].
In this study, we aim to elucidate the molecular alteration in PARPi acquired resistant ovarian cancer cells. We identified RNA transcript profiles in PARPi resistant ovarian cancer cells and further analyzed the crosstalk and regulatory networks among ncRNA and mRNA. Our findings identified the key ceRNA networks that regulate the PARPi acquired resistance, which provide the insight for further exploring the molecular mechanisms and therapeutic targets for PARPi resistance.
2. MATERIALS AND METHODS
2.1. Cell Culture
The human BRCA1 wild-type OC cell lines SKOV3 were cultured in Dulbecco's modified Eagle medium containing 10% FBS and maintained at 37 °C in a humidified incubator with 5% CO2. Olaparib (AZD-2281) was purchased from Selleckshem (Suffolk, UK). Olaparib was dissolved in DMSO and diluted to its final concentration with culture media. Resistant SKOV3 cells were generated by being exposed to gradually increasing doses of Olaparib from 10μM to 75μM for 6 months.
2.2. Whole-transcriptome Analysis with Total RNA Sequencing (RNA-Seq)
Total RNA from AKOV3 cells was isolated using Trizol (ThermoFisher) as recommended by the manufacturer’s manual. Purified total RNA was reverse-transcribed into cDNA and was followed by constructing cDNA library. cDNAs were subjected to illumina sequencing. High-quality clean reads were aligned to the human reference genome using STAR. The reads were assembled with StringTie. The raw data for this study can be found in the (SRA, PRJNA780335, NCBI).
2.3. Screening for Differentially Expressed mRNA and ncRNAs
The expression levels for each of the genes were normalized as fragments per kilobase of exon model per million mapped reads (FPKM) by Expectation-Maximization (RSEM). FPKM values were calculated by StringTie. Raw data were normalized using the quantile algorithm and were subsequently analysed using an unpaired t-test. Differentially expressed RNAs were identified by a log2 fold-change of more than 1 and p values less than 0.05.
2.4. Gene Function Annotation of Differentially Expressed mRNA and ncRNAs
Differentially expressed mRNA and ncRNAs were subjected to analysis of ontology classification based on gene annotation through DAVID (Database for Annotation, Visualization and Integrated Discovery). Predicted target genes were assigned to functional groups based on molecular function, biological processes and specific pathways.
2.5. Construction of the lncRNA-miRNA-mRNA Network
The lncRNA-miRNA-mRNA and circRNA-miRNA-mRNA networks were constructed by differentially expressed genes (DEGs), differentially expressed lncRNAs (DELs), and differentially expressed miRNAs (DEM) and differentially expressed circRNA (DEC). miRnada was used to predict the miRNA responsive elements (MRE) of target genes and binding sites of miRNAs with lncRNAs and mRNAs. Expression correlations between lncRNAs (circRNAs), miRNA and mRNAs were calculated using the Pearson correlation coefficient. The identified miRNA-targets were filtered with Pearson correlation coefficient absolute values bigger than 0.7 and p values less than 0.05. We predicted ceRNA by ceRNA scores calculated from the ratio of the number of MREs for shared miRNA and the number of MREs for lncRNA(circRNA)-miRNA pairs. The lncRNA-miRNA-mRNA and circRNA-miRNA-mRNA networks were visualized by using Cytoscape software.
3. RESULTS
3.1. Differentially Expressed mRNA, miRNA, circRNA and lncRNA
BRCA1 wild-type SKOV3 were treated with a gradually increasing dose of Olaparib for about 6 months. To explore the differential expressed genes and regulatory roles of noncoding RNA in the PARPi resistant OC cells, we performed high-throughput RNA sequencing in untreated SKOV3 and Olaprib-induced resistant SKOV3. Cells were collected after treatment with long-term low dose of PARP inhibitor. After constructing cDNA library, cDNAs were subjected to illumina sequencing. Clean reads were obtained and mapped to the reference genome sequence. We performed machine learning-based methods, including Coding Potential Calculator (CPC), Coding Potential Assessment Tool (CPAT), Coding-Non-Coding Indel (CNCI) predictor of long noncoding RNAs and messenger RNAs based on an improved k-mer scheme (PLEK) to identify lncRNA. Among transcripts detected, 1813 lncRNA were identified by all these four methods (Fig. 1A). These lncRNA can be divided into 4 classes: intronic lncRNA (54.5%), antisense lncRNA (5.4%), intergenic lncRNA (38.1%) and birectional lncRNA (36.2%). Of these lncRNA, the transcript types are known lncRNA, predicted lncRNA and known protein-coding mRNA (Fig. 1B).
Fig. (1).
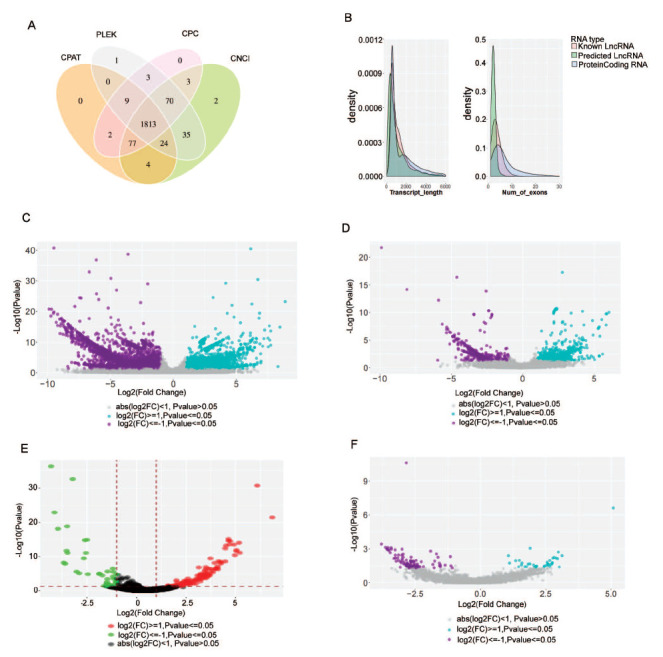
Differentially expressed mRNA, miRNA, circRNA and lncRNA in PARPi-resistant ovarian cancer cells and untreated ovarian cancer cells. (A) The venn diagram of the noncoding transcripts identified by. The number in the circle represents the number of lncRNA predicted by different methods. The intersection of the four methods was shown as the final lncRNA identified. (B) The density plots of distribution of protein-coding RNA, known lncRNA and predicted lncRNA. The number of The volcano plot of differentially expressed mRNAs(C), lncRNAs (D), circRNAs (E) and miRNAs (F). Green and red dots represent downregulated and upregulated mRNAs and ncRNAs, respectively.
For mRNA and lncRNA transcript, we evaluated the transcript expression using FPKM (Fragments Per Kilobase of transcript per Million fragments mapper). There were dramatic alterations in the expression profiles of lncRNAs and mRNA. Differentially expressed RNAs were identified by a log2 fold-change of more than 1 and p values less than 0.05. A total of 11545 differentially expressed genes (DEGs) were identified between control cells and resistant cancer cells, including 4580 upregulated and 6965 downregulated (Fig. 1C). Also, 1289 differentially expressed lncRNA(DEL) included 795 upregulated and 494 downregulated lncRNA (Fig. 1D). In addition to lncRNA, we also evaluate the expression profiles of other noncoding RNA, including microRNA and circRNA. Forty upregulated and 113 downregulated circRNAs (DECs) (Fig. 1E) and 115 upregulated miRNA, 42 downregulated miRNA were identified (Fig. 1F). The top 20 DECs, DELs, DEMs and DEGs are presented in Table 1.
Table 1.
The top 20 DECs, DELs, DEMs and DEGs.
| miRNA | Fold Change | P Value |
|---|---|---|
| hsa-miR-3182 | 5.171447956 | 8.96E-15 |
| hsa-miR-4532 | 5.039138394 | 2.52E-12 |
| hsa-miR-4516 | 4.699657981 | 2.91E-14 |
| hsa-miR-4492 | 4.474779583 | 2.56E-09 |
| hsa-miR-518b | 4.209453366 | 3.57E-09 |
| hsa-miR-6800-3p | 4.061111514 | 2.09E-07 |
| hsa-miR-3656 | 3.989946335 | 3.52E-07 |
| hsa-miR-146a-5p | 3.730705216 | 3.48E-10 |
| hsa-miR-3141 | 3.6127161 | 2.86E-07 |
| hsa-miR-4508 | 3.544320516 | 4.25E-06 |
| hsa-miR-4488 | 3.499032322 | 1.16E-05 |
| hsa-miR-103a-3p | -4.308189328 | 4.11E-37 |
| hsa-miR-107 | -4.133692684 | 1.26E-23 |
| hsa-miR-891a-5p | -3.959661071 | 7.38E-19 |
| hsa-miR-888-5p | -3.666756592 | 5.97E-09 |
| hsa-miR-891b | -3.596935142 | 1.20E-08 |
| hsa-miR-424-5p | -3.52007297 | 1.97E-12 |
| hsa-miR-196b-5p | -3.495147674 | 1.38E-19 |
| hsa-miR-551b-3p | -3.473415848 | 8.66E-12 |
| hsa-miR-16-5p | -3.215792525 | 2.23E-33 |
| hsa-miR-4798-5p | -3.017921908 | 2.46E-06 |
| circRNA | ||
| 14:61970132|61996542 | 5.085897294 | 2.39E-07 |
| 3:13500713|13502980 | 3.12706581 | 0.003910453 |
| 1:172555869|172557794 | 3.019610665 | 0.038970624 |
| 7:64068891|64069609 | 2.984748856 | 0.001897136 |
| 5:141580744|141583615 | 2.950093053 | 0.007299037 |
| 12:20875235|20883602 | 2.897978363 | 0.006418302 |
| 3:136980331|136995556 | 2.820973096 | 0.005566833 |
| 13:113510238|113520733 | 2.772614508 | 0.006455488 |
| 13:75326197|75349302 | 2.755261187 | 0.016778459 |
| 7:75199651|75202613 | 2.748753713 | 0.016768676 |
| 6:85557976|85574378 | -3.792147616 | 0.000363825 |
| 17:50667554|50669528 | -3.639036524 | 0.000718271 |
| 7:103025063|103029418 | -3.586144156 | 0.001020785 |
| 18:36195257|36203189 | -3.577038905 | 0.000991828 |
| 2:219565242|219568324 | -3.544409257 | 0.000756582 |
| 20:34072066|34078553 | -3.532936856 | 0.001238831 |
| 11:61437625|61438113 | -3.498893925 | 0.001290298 |
| 16:72957427|72960194 | -3.458066112 | 0.002156597 |
| 10:110964125|110985765 | -3.424938218 | 0.000795012 |
| 1:30992390|31007102 | -3.414339996 | 0.001024219 |
| lncRNA ID (Gene) | ||
| ENST00000624034 C9orf163 | 6.040245281 | 9.16E-11 |
| ENST00000661335 FTX | 5.891854737 | 1.63E-10 |
| ENST00000442383 OSER1-AS1 | 5.83547037 | 1.79E-08 |
| ENST00000649047 SNHG20-216 | 5.830133751 | 1.74E-10 |
| MSTRG.61850.5 | 5.59018646 | 0.003259312 |
| ENST00000461864 NCK1-AS1 | 5.530478529 | 1.92E-09 |
| MSTRG.16384.1 | 5.395867717 | 1.88E-06 |
| ENST00000663395 | 5.351647602 | 3.68E-08 |
| MSTRG.31221.1 | 5.35118555 | 0.007769284 |
| MSTRG.38592.1 | 5.34916988 | 0.007042967 |
| ENST00000541196 HCP5 | -9.894406324 | 1.74E-22 |
| ENST00000354376 C9orf163 | -8.112122365 | 6.58E-15 |
| ENST00000559458 XXbac-BPG300A18.13 | -5.95318985 | 0.042087404 |
| ENST00000660956 SNHG14-270 | -5.894849706 | 5.70E-13 |
| ENST00000666458 LINC00665-220 | -5.641815381 | 1.23E-08 |
| ENST00000665759 | -5.341851416 | 4.57E-07 |
| ENST00000669788 LINC00355-204 | -5.332818908 | 1.11E-07 |
| ENST00000661899 LINC01572-212 | -5.326204909 | 4.53E-08 |
| ENST00000436612 TMEM99-202 | -5.233511706 | 1.01E-07 |
| ENST00000380604 B3GALT5-AS1 | -5.136792674 | 3.10E-07 |
| mRNA | ||
| PLEKHA6 | 10.16650299 | 2.71E-26 |
| ITSN1 | 9.489488838 | 5.97E-24 |
| VARS2 | 8.478490774 | 1.31E-15 |
| IQGAP1 | 10.91035318 | 0.009915323 |
| WNK3 | 8.029618474 | 2.42E-14 |
| PTPRS | 11.21318751 | 1.04E-09 |
| SDCBP | 7.957421777 | 2.28E-11 |
| HERC6 | 7.636711801 | 5.20E-16 |
| ANLN | 9.909792805 | 3.43E-09 |
| ALDOA | 7.525818281 | 5.09E-12 |
| CCT3 | -12.35879265 | 3.13E-33 |
| CAD | -11.96096413 | 8.60E-31 |
| ERGIC3 | -11.86601695 | 7.99E-30 |
| TRRAP | -11.53161308 | 9.59E-28 |
| COL12A1 | -10.90682949 | 1.90E-25 |
| EI24 | -10.33839825 | 2.56E-23 |
| PHLDB1 | -10.24484246 | 5.93E-24 |
| EP400 | -10.23146705 | 4.98E-24 |
| DGKQ | -10.18671302 | 1.22E-22 |
| CLCN7 | -10.1278134 | 2.73E-19 |
3.2. GO and KEGG Analysis of DECs, DELs, DEMs and DEGs
The functions of these differentially expressed molecules were analyzed by GO and KEGG. The GO analysis showed that the functions of DEGs were enriched in “cellular process” “metabolic process” et al. (Fig. 2A). KEGG analysis revealed the functions of DEGs were enriched in “PI3K/AKT signaling pathway”“MAPK signaling pathway” et al. (Fig. 2B).
Fig. (2).
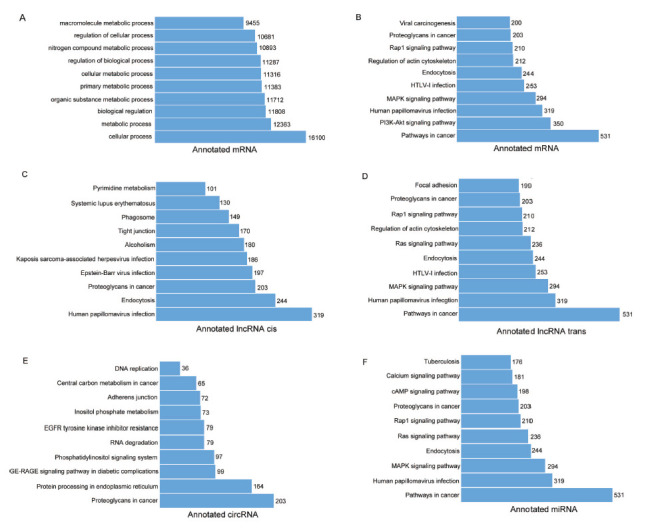
GO and KEGG analysis of DECs, DELs, DEMs, and DEGs. (A) GO functional analysis for DEGs (top 10 GO terms). KEGG pathway analysis (top 10) for DEGs (B), DEL cis (C), DEL trans (D), DECs (E), DEMs (F).
Noncoding RNAs are considered to be the crucial regulator of gene expression. We further investigated the functions of the differentially expressed noncoding RNA. The functions of DELs (cis) were enriched in “Human papillomavirus infection”“tight junction” “Pyrimidine metabolism” et al.(Fig. 2C). The functions of DELs (trans) were enriched in “Human papillomavirus infection”“MAPK signaling pathway” “Focal adhesion” et al. (Fig. 2D). The functions of DECs were enriched in “Proteoglycans in cancer” protein processing in endoplasmic reticulum” “Adherens junction” et al. (Fig. 2E). The functions of DEMs were enriched in “Human papillomavirus infection” “MAPK signaling pathway” “Ras signaling pathway” et al. (Fig. 2F).
4. CONSTRUCTION OF THE ceRNA NETWORK
To further examine the regulatory networks involved in the drug-resistant cancer cells, we constructed the ceRNA networks, including lncRNA-miRNA-mRNA and circRNA-miRNA-mRNA networks. The DEGs, DELs, DEM and DEC were used to construct the initial ceRNA network. We firstly used miRnada to predict the miRNA responsive elements (MRE) of target genes and binding sites of miRNAs with lncRNAs and mRNAs. The expressions of miRNA and related mRNAs or lncRNAs were analyzed by Pearson’s correlation coefficient. The identified miRNA-targets were filtered with Pearson correlation coefficient absolute values bigger than 0.7 and p value less than 0.05. We found 6338 filtered lncRNA-miRNA pairs and 4776 filtered miRNA-mRNA pairs (Fig. 3A, 3B). ceRNAs are natural decoys that compete for the miRNAs with shared MREs. We firstly conducted Pearson’s correlation analysis to identify lncRNA-mRNA and cirRNA-mRNA co-expression networks. Positive-correlation analysis identified the lncRNA-mRNA pairs and cirRNA-mRNA pairs. We predicted ceRNA by ceRNA scores which are calculated from the ratio of the number of MREs for shared miRNA and the number of MREs for lncRNA-miRNA pairs. A total of 13290 lncRNA-mRNA pairs were filtered according to ceRNA score and expression level (Fig. 3C). Similarly, a total of 795 filtered circRNA-miRNA pairs and 1910 circRNA-mRNA pairs were identified (Fig. 3D, 3E). Finally, a ceRNA network was drawn by cytoscape. As presented in Figs. (3F and 3G), the ceRNA network contained the top 500 lncRNA-mRNA pairs and circRNA-mRNA pairs. Based on the relationships between lncRNA and mRNA and the role of miRNA as a middle regulatory molecule, we constructed a lncRNA-miRNA-mRNA ceRNA network to predict the functions of DEL and DEM.
Fig. (3).
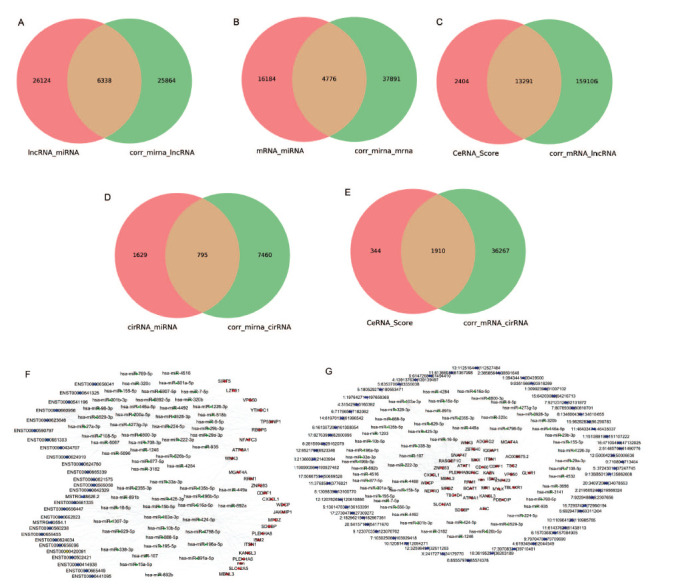
PARPi resistance-related ceRNA network. A. Venn diagram of lncRNA-miRNA shows the filtered lncRNA-target. The filtered lncRNA-miRNA were identified by miRnada and Pearson correlation coefficient analysis. B. Venn diagram of miRNA-mRNA shows the filtered miRNA-target. MiRnada and Pearson correlation coefficient analysis identified the filtered miRNA-mRNA. C. Venn diagram of ceRNA and filtered lncRNA-mRNA. The ceRNAs were filtered through Pearson correlation coefficient analysis. D. Venn diagram of circRNA-miRNA show the filtered circRNA-target. The filtered circRNA-miRNA were identified by miRnada and Pearson correlation coefficient analysis. E. Venn diagram of ceRNA and filtered circRNA-mRNA. The ceRNAs were filtered through Pearson correlation coefficient analysis. F. lncRNA-miRNA-mRNA ceRNA network. G. circRNA-miRNA-mRNA ceRNA network.
5. NETWORK HUB ANALYSIS
Hubs with high degrees are crucial for regulatory processes. We then analyzed the hubs with the top 3 highest degrees of nodes in both ceRNA networks (Table 2, Table 3). LncRNA ENST00000660956, ENST00000624919, and ENST00000623646 are the top 3 lncRNA with the highest degree (Fig. 4A). 5:83537007|83555038, 14:61970132| 61996542, 17:27304780|27309272 are the top 3 circRNAs with the highest degree (Fig. 4B). Among the differentially expressed miRNA, miR-7-5p, miR-1273g-3p, miR-320c, miR-320b, miR-310-3p, miR-526b-5p, miR-155-5p, miR-29a-3p were regulated by these 3 lncRNAs (Fig. 4C). Meanwhile, miR-320b was regulated simultaneously by 5:83537007iv|83555038 and 17:27304780|27309272 (Fig. 4D). The expression of these miRNAs was negatively correlated with its target genes in resistant cancer cells (Fig. 4E). The downregulation of these target genes, including BCAT1, WNK3, SIRT5 YTHDC1, ATP8A1, TSC2 and TP53INP1 was also confirmed in the Olaparib resistant A2780 cells (GSE 153867). We then suggested that miR-320b may be the key mediator in the ceRNA networks.
Table 2.
The degrees of nodes in lncRNA-miRNAs ceRNA network.
| Target | Degree |
|---|---|
| ENST00000660956 | 152 |
| ENST00000624919 | 143 |
| ENST00000623646 | 140 |
| ENST00000670085 | 128 |
| ENST00000590797 | 127 |
| ENST00000665339 | 125 |
| ENST00000656041 | 121 |
| ENST00000434707 | 114 |
| ENST00000661889 | 107 |
| MSTRG.1192.2 | 107 |
| ENST00000655923 | 106 |
| ENST00000671323 | 105 |
| ENST00000424333 | 98 |
| ENST00000661303 | 98 |
| ENST00000595310 | 97 |
| ENST00000650393 | 97 |
| ENST00000666458 | 95 |
| ENST00000664033 | 93 |
| ENST00000466953 | 92 |
| ENST00000660040 | 91 |
| ENST00000380604 | 91 |
| ENST00000647102 | 87 |
| ENST00000655613 | 86 |
| ENST00000609207 | 85 |
| ENST00000669283 | 84 |
| ENST00000655379 | 83 |
| ENST00000606285 | 83 |
| ENST00000661181 | 82 |
| ENST00000590838 | 81 |
| ENST00000666884 | 80 |
| ENST00000366153 | 80 |
| ENST00000428289 | 79 |
| ENST00000603468 | 78 |
| ENST00000669788 | 77 |
| ENST00000615168 | 76 |
| ENST00000661193 | 76 |
| ENST00000624760 | 74 |
| ENST00000670637 | 74 |
| ENST00000595394 | 73 |
| ENST00000518732 | 72 |
| ENST00000666013 | 70 |
| ENST00000424435 | 69 |
| ENST00000504349 | 69 |
| ENST00000660381 | 69 |
| ENST00000541196 | 69 |
Table 3.
The degrees of nodes in circRNA-miRNAs ceRNA network.
| Target | Degree |
|---|---|
| 5:83537007|83555038 | 131 |
| 14:61970132|61996542 | 68 |
| 17:27304780|27309272 | 62 |
| 11:61366045|61367998 | 59 |
| 8:134600436|134610655 | 58 |
| 9:710804|713464 | 57 |
| 16:72957427|72960194 | 49 |
| 18:21366027|21403994 | 44 |
| 15:64203082|64216713 | 42 |
| 7:92123120|92131872 | 41 |
| 5:136147832|136163391 | 41 |
| 5:180529275|180553471 | 39 |
| 1:151088190|151107222 | 39 |
| 5:69294793|69311204 | 38 |
| 12:120782655|120810886 | 37 |
| 12:50004235|50006636 | 36 |
| 8:13098399|13100770 | 36 |
| 5:61472681|61494410 | 35 |
| 20:54157169|54171670 | 34 |
| 7:80789306|80810701 | 32 |
| 2:61485768|61490776 | 31 |
| 11:3768581|3776021 | 31 |
| 11:61437625|61438113 | 30 |
| 8:28155942|28162078 | 30 |
| 9:123070350|123076762 | 30 |
| 11:46433474|46435037 | 29 |
| 15:80978766|80982182 | 27 |
| 4:61934840|62044549 | 26 |
| 2:38585644|38591648 | 25 |
| 15:96282832|96290783 | 25 |
| 10:110964125|110985765 | 25 |
| 2:219565242|219568324 | 25 |
| 17:50667554|50669528 | 24 |
| 1:30992390|31007102 | 24 |
| 16:74660932|74661451 | 23 |
| 20:34072066|34078553 | 23 |
| 13:112516440|112527484 | 23 |
| 7:92294889|92307656 | 22 |
| 1:39434414|39439500 | 22 |
| 6:7176655|7182082 | 22 |
| 6:85557976|85574378 | 21 |
| 9:93515667|93516269 | 19 |
| 10:12081472|12094271 | 18 |
| 9:36246031|36246482 | 18 |
| 12:32598497|32611283 | 17 |
| 9:79704783|79709699 | 17 |
| 7:103025063|103029418 | 16 |
| 2:182962159|182967361 | 16 |
| 17:82763965|82800996 | 15 |
| 12:6521798|6523346 | 15 |
| 2:241089188|241094432 | 14 |
| 6:136694140|136698682 | 12 |
| 1:151427823|151442205 | 12 |
| 18:36195257|36203189 | 12 |
| 16:47109483|47132025 | 12 |
| 6:157036835|157084905 | 12 |
| 17:42322282|42323625 | 11 |
| 9:135850137|135862608 | 11 |
| 4:3154293|3160392 | 11 |
| 6:125045211|125058106 | 11 |
| X:13666317|13668383 | 10 |
| 4:139137630|139139497 | 10 |
| 2:190900564|190927482 | 9 |
| 5:170878097|170896115 | 9 |
| 21:37420299|37430400 | 8 |
| 18:23507985|23519174 | 8 |
| 17:45117994|45121372 | 6 |
| 17:55072900|55081183 | 6 |
| 5:37243013|37247745 | 6 |
| 6:149676260|149680457 | 5 |
| 12:48939655|48941188 | 5 |
| 17:28122603|28172618 | 5 |
| 1:197642711|197658369 | 5 |
| 17:39708321|39710481 | 4 |
| 1:233135013|233208689 | 4 |
| 6:57379901|57537625 | 3 |
| X:24172715|24179770 | 3 |
| 11:130260856|130261929 | 3 |
| 17:39490557|39492890 | 2 |
| 6:163455279|163478896 | 2 |
| 6:161307209|161308054 | 2 |
| 11:120475340|120477526 | 2 |
| 19:11513056|11514221 | 2 |
| 4:74174506|74225394 | 1 |
| 7:116695750|116700284 | 1 |
| 5:151790323|151791062 | 1 |
| 15:29761139|29773357 | 1 |
| 17:39724726|39725853 | 1 |
| 5:112985835|113004077 | 1 |
Fig. (4).
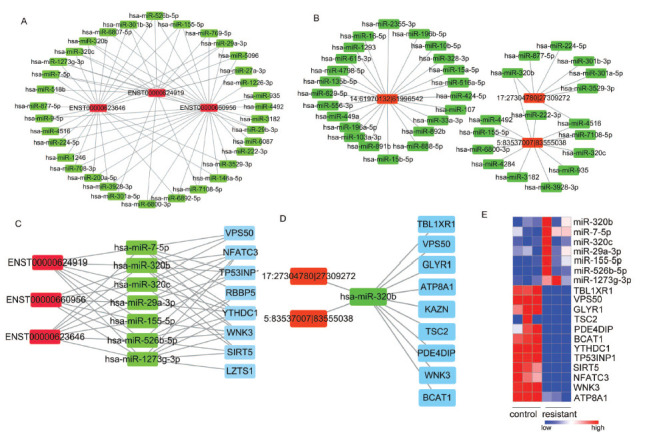
The hub gene associated with ceRNA. The hubs with the top 3 highest degree of nodes in lncRNA-miRNA (A) and circRNA-miRNAs (B). (C) The ceRNA network with the top 3 highest degrees of lncRNA-miRNAs. (D) The top highest degree of circRNA-mRNA ceRNA network with shared target miRNAs. (E) Heatmap of FPKM value of miRNA-targets.
6. DISCUSSION
The ceRNA networks, including lncRNA-miRNA-mRNA and circRNA-miRNA-mRNA identified in our study, provide the possible regulatory mechanisms for Olaparib resistance. According to the hub analysis, we identified the key hub nodes, including 3 lncRNAs and 2 circRNAs. miR-320b was predicted to be regulated these lncRNAs and circRNAs.
The identification of the ceRNAs network in PARPi resistance may represent promising new therapeutic targets. The complementary principle is widely used to predict the binding of DNA-RNA, RNA-RNA. RNA molecules are involved in computational, algorithmic processes, including RNA editing and RNA-based regulatory networks [11]. miRanda was used to predict the sequences that miRNAs bind with. miRanda algorithm is based on the free energy of association of A-T and G-C base pairs. The interactions between two molecules are based on the overlap of electron clouds [12]. Similar to DNA, RNA molecules are also regarded as electronic devices, transferring charges. Many limitations to biological computation still remain. The RNA-based logic operations are accompanied by an increase in noise. Computational approaches such as molecular dynamics and quantum mechanical techniques are applied to RNA, particularly in relation to complementary experimental studies. Density functional theory (DFT) calculation is usually used to optimize the hybridization of RNA-RNA [13]. Although the usage of PARPi has improved the therapeutic effects of ovarian cancer, some patients progressed after PARPi. The mechanisms of resistance to PARPi include PARP1 mutation, restoration of homologous recombination, BRCA1 promoter alterations, generation of hypomorphic BRCA proteins, decreased proteasomal degradation et al. [14] In addition, other mechanisms were involved in PARPi-related resistance, including PI3K/ATK pathway activation, epigenetic modification, and restoration of PARylation [6, 10]. Noncoding RNAs act as post-transcriptional regulators affecting mRNA expression. LncRNAs and circRNAs are considered to be the important regulators in cancer progression and therapeutic resistance, acting as “sponges” for miRNAs and, in turn affecting the expression of target genes [15]. In this study, ENST00000660956 (SNHG14), ENST 00000624919(RP4-671O14.6), ENST00000623646 (NKAIN3-IT1:8) were obviously downregulated in resistant cancer cells. LncRNAs and circRNAs are new classes of epigenetic regulators and their functions are much less well understood. SNHG14 was reported to be involved in the proliferation, invasion and DDP-resistance in cancer progression through regulating miRNA [16-19]. The expression of SNHG14 was higher in ovarian cancer tissues and cell lines than in normal controls [20]. In this study, the expression of SNHG14 was obviously reduced in PARPi resistant ovarian cancer cells, suggesting the alternative roles of SNHG14 during the process of PARPi therapy. miR-320 family members play important roles in various cancers. The downregulations of miR-320a, miR-320b and miR-320c suggest that miR-320 family act as an antioncogenes in certain cancer types [21, 22]. The differential effects of noncoding RNA are mostly attributed to downstream target genes. The target genes of miR-320b were involved in lipid transporting ATPase (ATP8A1), transcellular calcium ion transportation (WNK3), regulator of TP53 (TP53INP1), Glucose metabolism and NAD metabolism (SIRT5, BCAT1), m6A RNAs alternative splicing (YTHDC1), CoA biosynthesis (BCAT1), negative regulator mTORC1 signaling (TSC2), regulator of p38 MAP kinase (GLYR1), endocytic recycling (VPS50) and nuclear receptor transcriptional activation (TBL1XR1). The reduced expression of ATP8A1, which play roles in transmembrane lipid homeostasis, may abolish the modulatory roles of PARPs in the lipid homeostasis including lipoprotein homeostasis and fatty acid oxidation [23, 24]. The WNK lysine deficient protein kinase 3 gene (WNK3) lacks the catalytic lysine in subdomain II and has a conserved lysine in subdomain I, which was reported to play a role in the cell survival in a caspase-3-dependent pathway. WNK3 and TAOK3 were reported to be involved in paclitaxel resistance [25].
CONCLUSION
The downregulation of WNK3 by miR-320b identified in this study suggested the involvement of WNK3 in the PARPi resistance. However, the clues for the ceRNA regulatory networks in PARPi resistance, more functional and mechanistic studies are needed to be explored.
ACKNOWLEDGEMENTS
Declared none.
AUTHORS’ CONTRIBUTIONS
The work was carried out as a collaboration between all authors. Kong L and Xu J carried out the experiments, including the construction of resistant cancer cells and RNA extraction. Yu L, Liu S, Liu Z, and Xiang J designed and planned the research. Kong L and Xiang J analyzed the data and drafted the manuscript. All authors contributed to the article and approved the submitted version.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available within the article.
FUNDING
This work was supported by the Science and Technology Development Fund of Beijing Rehabilitation Hospital, Capital Medical University (2020-057).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Vescarelli E., Gerini G., Megiorni F., Anastasiadou E., Pontecorvi P., Solito L., De Vitis C., Camero S., Marchetti C., Mancini R., Benedetti Panici P., Dominici C., Romano F., Angeloni A., Marchese C., Ceccarelli S. MiR-200c sensitizes Olaparib-resistant ovarian cancer cells by targeting Neuropilin 1. J. Exp. Clin. Cancer Res. 2020;39(1):3. doi: 10.1186/s13046-019-1490-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen A. PARP inhibitors: Its role in treatment of cancer. Chin. J. Cancer. 2011;30(7):463–471. doi: 10.5732/cjc.011.10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu H., Wei M., Xu J., Hua J., Liang C., Meng Q., Zhang Y., Liu J., Zhang B., Yu X., Shi S. PARP inhibitors in pancreatic cancer: Molecular mechanisms and clinical applications. Mol. Cancer. 2020;19(1):49. doi: 10.1186/s12943-020-01167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim D.S., Camacho C.V., Kraus W.L. Alternate therapeutic pathways for PARP inhibitors and potential mechanisms of resistance. Exp. Mol. Med. 2021;53(1):42–51. doi: 10.1038/s12276-021-00557-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dedes K.J., Wilkerson P.M., Wetterskog D., Weigelt B., Ashworth A., Reis-Filho J.S. Synthetic lethality of PARP inhibition in cancers lacking BRCA1 and BRCA2 mutations. Cell Cycle. 2011;10(8):1192–1199. doi: 10.4161/cc.10.8.15273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H., Liu Z.Y., Wu N., Chen Y.C., Cheng Q., Wang J. PARP inhibitor resistance: The underlying mechanisms and clinical implications. Mol. Cancer. 2020;19(1):107. doi: 10.1186/s12943-020-01227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ledermann J.A., Pujade-Lauraine E. Olaparib as maintenance treatment for patients with platinum-sensitive relapsed ovarian cancer. Ther. Adv. Med. Oncol. 2019;11:1758835919849753. doi: 10.1177/1758835919849753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vanderstichele A., Nieuwenhuysen S.H., Els V., Concin T.V.G., Berteloot P., Neven P., Busschaert P., Lambrechts D., Vergote I. Randomized phase II CLIO study on olaparib monotherapy versus chemotherapy in platinum-resistant ovarian cancer. ASCO Meeting 2019. 2019;1 [Google Scholar]
- 9.Kartha R.V., Subramanian S. Competing endogenous RNAs (ceRNAs): New entrants to the intricacies of gene regulation. Front. Genet. 2014;5:8. doi: 10.3389/fgene.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tapodi A., Debreceni B., Hanto K., Bognar Z., Wittmann I., Gallyas F., Jr, Varbiro G., Sumegi B. Pivotal role of Akt activation in mitochondrial protection and cell survival by poly(ADP-ribose)polymerase-1 inhibition in oxidative stress. J. Biol. Chem. 2005;280(42):35767–35775. doi: 10.1074/jbc.M507075200. [DOI] [PubMed] [Google Scholar]
- 11.Akhlaghpour H. An RNA-based theory of natural universal computation. J. Theor. Biol. 2022;537:110984. doi: 10.1016/j.jtbi.2021.110984. [DOI] [PubMed] [Google Scholar]
- 12.Kandagalla S., Rimac H., Potemkin V.A., Grishina M.A. Complementarity principle in terms of electron density for the study of EGFR complexes. Future Med. Chem. 2021;13(10):863–875. doi: 10.4155/fmc-2020-0265. [DOI] [PubMed] [Google Scholar]
- 13.Glossman-Mitnik D. Density functional theory. Intechopen. 2019;2019:76822. doi: 10.5772/intechopen.76822. [DOI] [Google Scholar]
- 14.Lee E.K., Matulonis U.A. PARP Inhibitor Resistance Mechanisms and Implications for Post-Progression Combination Therapies. Cancers (Basel) 2020;12(8):E2054. doi: 10.3390/cancers12082054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Militello G., Weirick T., John D., Döring C., Dimmeler S., Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief. Bioinform. 2017;18(5):780–788. doi: 10.1093/bib/bbw053. [DOI] [PubMed] [Google Scholar]
- 16.Xu L., Xu Y., Yang M., Li J., Xu F., Chen B.L. LncRNA SNHG14 regulates the DDP-resistance of non-small cell lung cancer cell through miR-133a/HOXB13 pathway. BMC Pulm. Med. 2020;20(1):266. doi: 10.1186/s12890-020-01276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Yang P., Zhang D., Lu M., Zhang C., Sun Y. LncRNA SNHG14 promotes cell proliferation and invasion in colorectal cancer through modulating miR-519b-3p/DDX5 axis. J. Cancer. 2021;12(16):4958–4970. doi: 10.7150/jca.55495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao Z., Zhang H., Su C., Liu F., Liu Y., Song J., Zhu H., Fan Y., Zhang X., Dong W., Chen X., Liang H., Zhang B. Long noncoding RNA SNHG14 promotes hepatocellular carcinoma progression by regulating miR-876-5p/SSR2 axis. J. Exp. Clin. Cancer Res. 2021;40(1):36. doi: 10.1186/s13046-021-01838-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun B., Ke K.B., Liu D.F., Wang Q., Li Y.N., Chen J.H., Zhang J.H. Long noncoding RNA SNHG14 acts as an oncogene in prostate cancer via targeting miR-613. Eur. Rev. Med. Pharmacol. Sci. 2020;24(21):10919. doi: 10.26355/eurrev_202011_23556. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y.L., Huang Y.M. LncSNHG14 promotes ovarian cancer by targeting microRNA-125a-5p. Eur. Rev. Med. Pharmacol. Sci. 2019;23(8):3235–3242. doi: 10.26355/eurrev_201904_17683. [DOI] [PubMed] [Google Scholar]
- 21.Wan C., Wen J., Liang X., Xie Q., Wu W., Wu M., Liu Z. Identification of miR-320 family members as potential diagnostic and prognostic biomarkers in myelodysplastic syndromes. Sci. Rep. 2021;11(1):183. doi: 10.1038/s41598-020-80571-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lv Q., Hu J.X., Li Y.J., Xie N., Song D.D., Zhao W., Yan Y.F., Li B.S., Wang P.Y., Xie S.Y. MiR-320a effectively suppresses lung adenocarcinoma cell proliferation and metastasis by regulating STAT3 signals. Cancer Biol. Ther. 2017;18(3):142–151. doi: 10.1080/15384047.2017.1281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaubey P.M., Hofstetter L., Roschitzki B., Stieger B. Proteomic analysis of the rat canalicular membrane reveals expression of a complex system of P4-ATPases in liver. PLoS One. 2016;11(6):e0158033. doi: 10.1371/journal.pone.0158033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szántó M., Gupte R., Kraus W.L., Pacher P., Bai P. PARPs in lipid metabolism and related diseases. Prog. Lipid Res. 2021;84:101117. doi: 10.1016/j.plipres.2021.101117. [DOI] [PubMed] [Google Scholar]
- 25.Lai T.C., Fang C.Y., Jan Y.H., Hsieh H.L., Yang Y.F., Liu C.Y., Chang P.M., Hsiao M. Kinase shRNA screening reveals that TAOK3 enhances microtubule-targeted drug resistance of breast cancer cells via the NF-κB signaling pathway. Cell Commun. Signal. 2020;18(1):164. doi: 10.1186/s12964-020-00600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available within the article.


